Abstract
Ribosomes are the cellular machines responsible for protein synthesis. Ribosome biogenesis, the production of ribosomes, is a complex process involving pre-ribosomal RNA (rRNA) cleavages and modifications as well as ribosomal protein assembly around the rRNAs to create the functional ribosome. The SSU processome is a large ribonucleoprotein (RNP) in eukaryotes required for the assembly of the small subunit (SSU) of the ribosome as well as for the maturation of the 18S rRNA. Despite the fundamental nature of the SSU processome to the survival of any eukaryotic cell, mutations in SSU processome components have been implicated in human diseases. Three SSU processome components and their related human diseases will be explored in this review: hUTP4/Cirhin, implicated in North American Indian Childhood Cirrhosis (NAIC); UTP14, implicated in infertility, ovarian cancer, and scleroderma; and EMG1, implicated in Bowen-Conradi Syndrome (BCS). Diseases with suggestive, though inconclusive, evidence for the involvement of the SSU processome in their pathogenesis are also discussed, including a novel putative ribosomopathy.
Keywords: SSU processome, ribosomopathies, hUTP4/Cirhin, UTP14, EMG1
1. Introduction
Ribosome biogenesis is a fundamental process that is necessary for dividing or metabolically active cells. Interestingly, all three RNA polymerases (RNAPs) and over 200 assembly factors are required to manufacture the ribosome in the eukaryotic nucleolus. In humans, RNAPI transcribes the 47S precursor pre-rRNA that is processed into the 18S, 5.8S, and 28S rRNAs. RNAPII transcribes the mRNAs for the ribosomal proteins and ribosome biogenesis proteins [1]. RNAPII also transcribes the U3, U8, and U13 small nucleolar (sno) RNAs, which are non-coding RNAs required for 18S rRNA maturation [2]. Finally, RNAPIII transcribes the 5S rRNA. This transcriptional complexity is matched by the numerous pre-rRNA processing events required to make the fully mature rRNAs, the functional catalytic (large subunit; LSU) and recognition (small subunit; SSU) components of the ribosome [1].
To make the small subunit of the ribosome, eukaryotic cells use a large ribonucleoprotein (RNP) called the SSU processome [3]. The SSU processome is assembled co-transcriptionally with the 47S pre-rRNA in the nucleolus [4] and contains the U3 snoRNA and over 70 associated proteins including the U3 proteins (UTPs). The assembly of the small ribosomal subunit and the maturation of the 18S rRNA, which is incorporated in the small subunit, require the SSU processome [5]. The SSU processome was first identified and purified from the yeast, Saccharomyces cerevisiae [3], and later identified in humans [6].
Since ribosome biogenesis is a vital cellular process, it is surprising that some defects in ribosome assembly cause human disease in specific cell-types without affecting all tissues. These diseases, termed ribosomopathies, are caused by mutations in various factors involved in ribosome biogenesis including RNAPI transcription in Treacher Collins syndrome [7], large subunit biogenesis in alopecia, neurological defects, and endocrinopathy (ANE) syndrome and Shwachman-Diamond syndrome, snoRNPs in Dyskeratosis congenita and Prader-Willi syndrome [8], and ribosomal proteins in Diamond-Blackfan Anemia (DBA) [9] and isolated congenital asplenia (ICA) [10]. Mutations in components of the SSU processome also cause human diseases.
Three ribosomopathies caused by mutations in components of the SSU processome resulting in nucleolar dysfunction will be described here. These SSU processome components and their respective diseases are: hUTP4/Cirhin-North American Indian Childhood Cirrhosis (NAIC); UTP14-infertility, ovarian cancer, and scleroderma; and EMG1-Bowen-Conradi syndrome (BCS). As with all ribosomopathies, the tissue proclivity of these diseases is a conundrum, and therein lies the need to elucidate the molecular mechanisms of their pathogenesis. In addition, studies suggesting that SSU processome components modify Neurofibromatosis type 1 and primary open angle glaucoma are also examined. Finally, evidence suggestive that Williams-Beuren syndrome is a novel ribosomopathy is proposed.
2. Disease of hUTP4/Cirhin: North American Indian Childhood Cirrhosis (NAIC)
North American Indian childhood cirrhosis (NAIC) (OMIM: 604901), first discovered in 1970, is a rare, autosomal recessive familial cholestasis found exclusively in Ojibway-Cree children from a First Nations Canadian population. The incidence of the disease is estimated to be from 1 in 250 to 1 in 750 Ojibway-Cree children. The most recent clinical report on the disease states that 12 of the total 36 patients diagnosed with NAIC have had liver transplantation, with no recurrence of disease after transplantation. With a survival to adulthood of less than 50%, only 17 patients total were still alive [11]. The disease first presents with neonatal jaundice progressing to biliary cirrhosis and portal hypertension. Unfortunately, the only known treatment is liver transplantation [12]. Histologically, NAIC is identical to the pathology seen in extrahepatic biliary tree obstruction in most cases [11].
Using DNA pooling, the candidate gene for the disease, which was named CIRH1A, was localized to chromosome 16q22 [13]. Subsequently, SNP analysis of this region revealed an R565W mutation in human UTP4/Cirhin (hUTP4/Cirhin) to be present in the patient population [14]. It has an unusually high estimated carrier frequency of 8–12% in the Ojibway-Cree population [12]. Utp4 was first described in yeast as part of the original purification of the SSU processome [3]. Additionally, yeast Utp4 was shown to be a member of the t-Utp/UtpA subcomplex of the SSU processome, required for pre-rRNA processing and transcription as well as for the assembly of the SSU processome [4]. hUTP4/Cirhin is also a member of the human t-UTP/UTPA subcomplex, is required for pre-18S rRNA processing, but surprisingly is not required for pre-rRNA transcription [6]. The identification of this candidate gene in patients with NAIC is unexpected as the requirement for ribosome biogenesis is ubiquitous in a developing organism and would not be just liver-specific. In situ hybridization showed that mouse UTP4/Cirhin (mUTP4/Cirhin) is expressed highly in the fetal liver of day-11.5 mouse embryos [14]. However, it is also highly expressed in other developing tissues.
Since the elucidation of the R565W mutation in NAIC, efforts have been made to study the molecular pathogenesis of NAIC using model organisms. Unfortunately in mice, knockout (−/−) of mUTP4/Cirhin (also known as TEX292) is embryonic lethal [11] and heterozygotes (+/−) are phenotypically normal [15]. No knock-in mouse with the R565W mutation has been documented nor has a liver-specific hUTP4/Cirhin knockout been made.
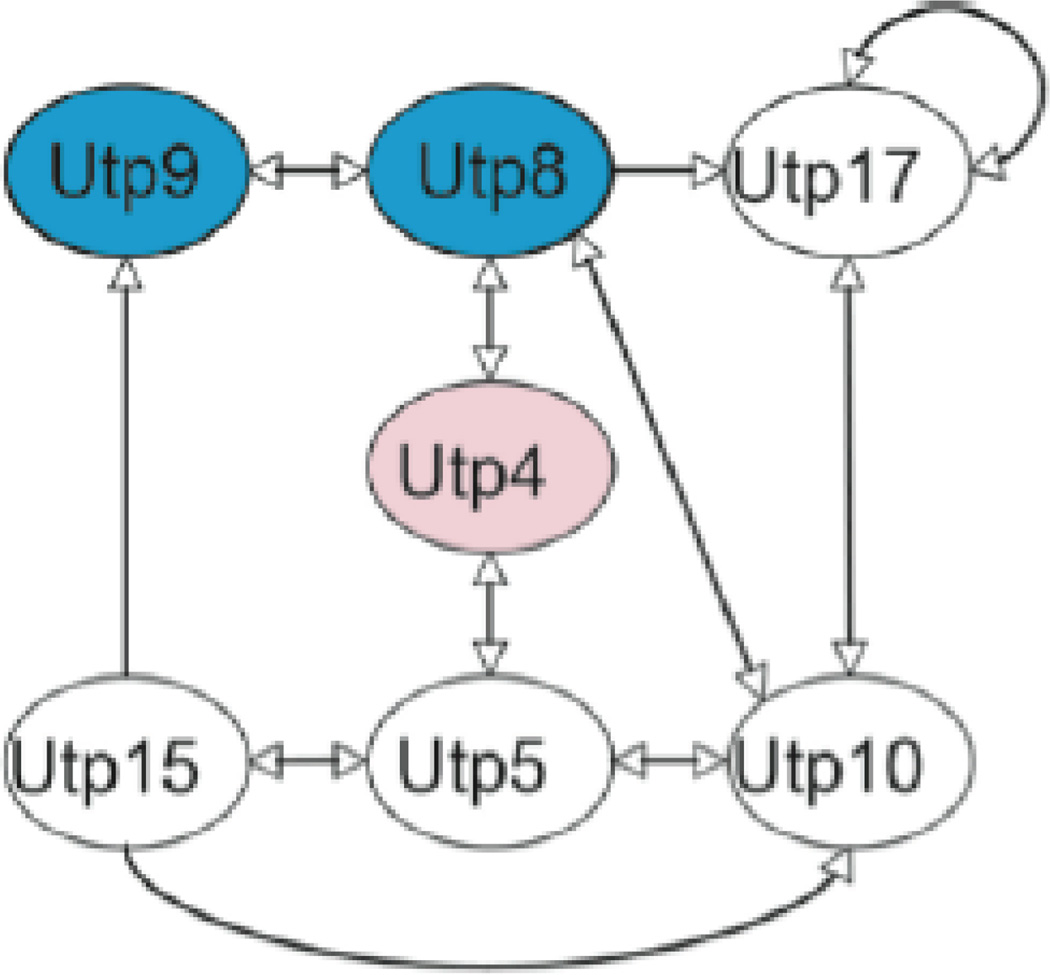
Efforts were also made to use yeast to model NAIC. Yet, introducing the analogous mutation into yeast Utp4 did not affect growth or ribosome biogenesis in yeast [16]. Despite this, yeast proved a valuable model system to study the molecular genetics of Utp4 and its role in NAIC. Yeast two-hybrid (Y2H) analysis revealed a map of the protein-protein interactions (PPIs) among the yeast t-Utp/UtpA subcomplex members. Its members in yeast include Utp4, Utp5, Utp8, Utp9, Utp10, Utp15, and Utp17 [4, 17, 18] (Fig. 1). All are conserved to humans except Utp8 and Utp9 [6]. Interestingly, the C-terminus of yeast Utp4 (yUtp4), where the R565W mutation resides, is required for interaction with Utp8 [16]. This led to the hypothesis that the R565W mutation disrupts the interaction between hUTP4/Cirhin and the then unidentified human analog of Utp8.
Figure 1.
Interactome map of the yeast Utp4-containing protein t-Utp/UtpA subcomplex. Blue shading indicates proteins not conserved to metazoans [16]. Reprinted with permission.
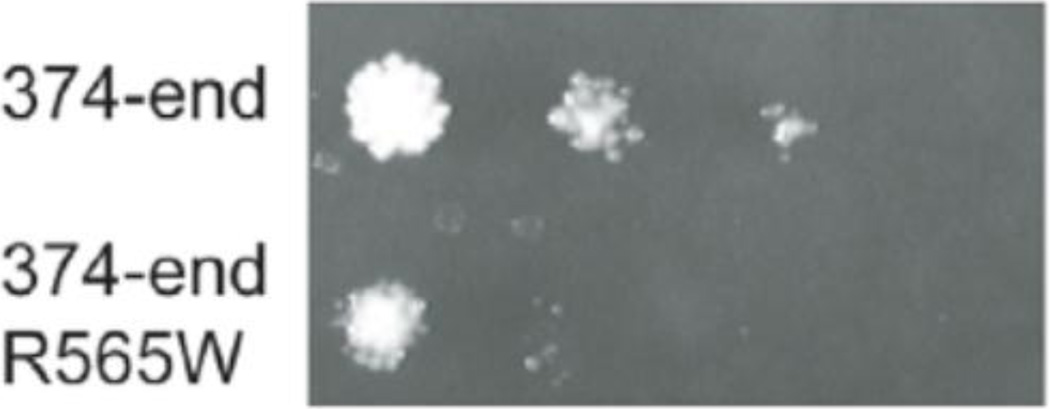
To identify the human analog of Utp8, Freed et al. [19] sought out the interacting partners of hUTP4/Cirhin. Y2H analysis of a human liver cDNA library revealed that hUTP4/Cirhin interacts with the metazoan-specific, nucleolar protein, NOL11. Affinity purification followed by mass spectrometry of a tagged form of hUTP4/Cirhin also identified NOL11 as well as other members of the human t-UTP/UTPA subcomplex including hUTP5, hUTP15, and hUTP17. A subsequent study also found that hUTP4/Cirhin is in a complex with hUTP15 [20], supporting the protein co-purification of t-UTP/UTPA subcomplex members by Freed et al. [19]. Probing its function revealed that NOL11 is required for pre-rRNA processing and transcription as well as for normal nucleolar morphology, establishing that NOL11 is the functional human analog of yeast Utp8. Intriguingly, the R565W mutation in a C-terminal fragment of hUTP4/Cirhin weakened but did not abolish its interaction with NOL11 in the Y2H system [19] (Fig. 2). This suggests a possible molecular etiology for NAIC: the R565W mutation abrogates the PPI between hUTP4/Cirhin and NOL11, which may result in defects in pre-rRNA processing. It is not yet known whether this weakened PPI results in a pre-rRNA processing defect.
Figure 2.
The R565W NAIC mutuation in the context of the C-terminal portion of hUTP4/Cirhin causes reduced interaction with NOL11 by Y2H analysis [19]. Reprinted with permissions.
In an attempt to understand the pathogenesis of NAIC, another group also sought out interacting partners of hUTP4/Cirhin [15]. Using Y2H analysis, hUTP4/Cirhin was found to interact with Cirip, a protein required for transcription of the HIV-1 LTR enhancer element. However, hUTP4/Cirhin is nucleolar [19, 21], while Cirip is nuclear [15] indicating that an interaction between these two proteins is not likely required for the role of hUTP4/Cirhin in ribosome biogenesis.
We are still left to wonder, is nucleolar dysfunction the root cause of NAIC? If so, how is it that a disease of ribosome biogenesis only affects the liver? Answers to these questions will have a significant impact both on the understanding of this congenital liver disease and on human ribosome biogenesis in general.
3. Diseases of UTP14: Infertility, Scleroderma, and Ovarian Cancer
A genetic cause of infertility in some men is due to a mutation in the human ribosome biogenesis factor, hUTP14c. Utp14 was first described in yeast with the discovery of the SSU processome [3] and is therefore involved in SSU biogenesis and required for the maturation of the pre-18S rRNA. hUTP14c is one of three copies of UTP14 in the human genome; these include hUTP14a, hUTP14b, and hUTP14c. hUTP14c is unusual in that it is a retrogene. Retrogenes originate from fully or partially spliced mRNAs that are reverse transcribed into DNA and incorporated into the genome and therefore lack some or all of the introns of their original sequence [22]. Many genes on the X chromosome have retrogene analogs within autosomes so that their expression can occur during X chromosome silencing during meiosis [23].
Although Utp14 is required for the formation and function of the SSU processome, the specific function of Utp14 within the SSU processome has remained elusive. Recently, an A758G mutation in Utp14 rescued the growth defect found in bud23Δ yeast [24]. Bud23 is required for A2 cleavage of pre-rRNA and for SSU biogenesis. Wild-type (wt) Utp14 co-purified with Bud23 and deletion of BUD23 prevented the nucleolar localization of Utp14. Utp14 was found in sucrose gradient fractions containing the SSU processome (early-stage SSU biogenesis), the U3 snoRNA, and the pre-40S particle containing 20S pre-rRNA (late-stage SSU biogenesis). Loss of Bud23 prevented association of Utp14 with the 20S pre-rRNA but not with U3 while the A758G mutation in Utp14 partially restored the association with the 20S prerRNA. These data suggest a role for Utp14 in both early and late stage SSU biogenesis in concert with Bud23. The exact role of Utp14 in these ribosome biogenesis steps remains to be identified [24].
The mouse Utp14b (mUtp14b) is of particular significance in a mouse model of male infertility. Two groups [25, 26] independently identified a truncation in mUtp14b due to a premature stop codon as the causative agent of the infertility phenotype in the juvenile spermatogonial depletion (jsd) mouse model first described by Beamer et al. [27]. Male jsd mice undergo a single wave of spermatogenesis and subsequently fail to differentiate spermatogonia. Interestingly, they showed that mUtp14b is expressed exclusively in the testes of mice while the mUtp14a gene on the X chromosome [25] is expressed ubiquitously [26]. Specifically, mUtp14b is expressed in the germ cells [26]. This is in agreement with evidence that infertility could be rescued in jsd mice by transplanting wild-type spermatogonia [25].
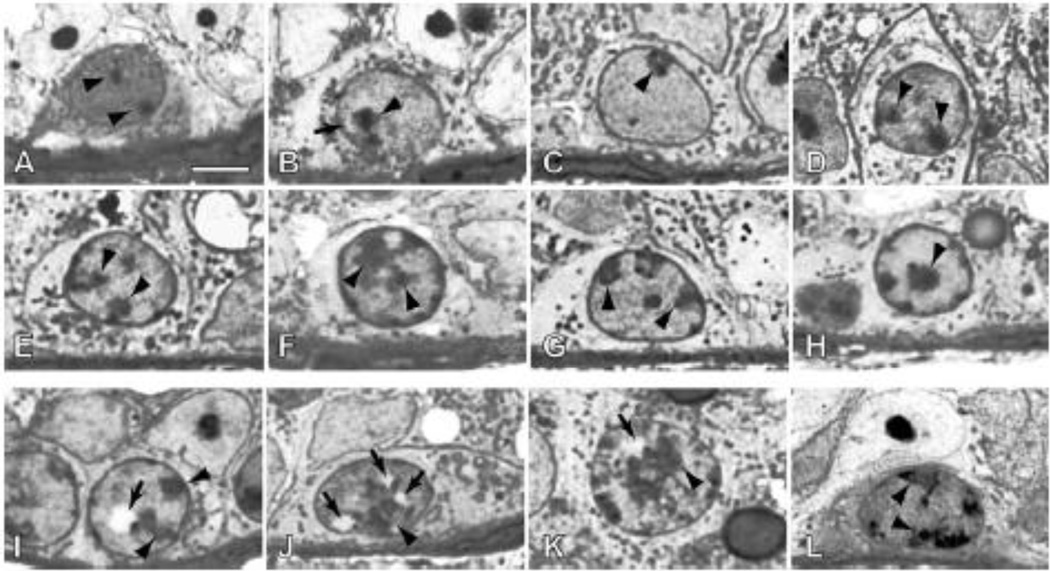
The mutation identified in jsd mice replaces a GG dinucleotide in chromosome 1 in mice with CTTTTC. This introduces a premature stop codon, truncating the mUtp14b protein product [25, 26]. Intriguingly, the genetic background of a mouse affects the jsd phenotype by altering the average percentage of tubules with differentiating germ cells [28]. A similar phenomenon has been demonstrated previously in the case of mouse model of the ribosomopathy, Treacher Collins syndrome [29]. The reason for the effect of genetic background on these phenotypes is as of yet unknown. To explain why mUtp14b is uniquely expressed in spermatogonia, it is hypothesized that mUtp14b is required to produce 18S rRNA during meiosis when mUtp14a on the X chromosome is silenced [26]. As might be expected, abnormal nucleoli and nuclear vacuoles are found in spermatogonia of jsd mice [30] (Fig. 3) supporting the hypothesis that a defect in nucleolar function results in aberrant sperm maturation. Furthermore, mUtp14b is expressed at higher levels when mUtp14a is silenced during sperm maturation [23].
Figure 3.
Spermatogonia with apparently normal (A–H) and abnormal (I–L) morphologies in the testes of Utp 14bjsd mutant mice. Arrowheads indicate nucleoli; arrows indicate vacuoles. Large (I) or numerous (J) nuclear vacuoles are present in nuclei that have diffuse nucleoli and heterochromatin typical differentiated type A sprematogonia. Other abnormalities include large nuclei with increased amounts of hetrochromatin (K) and those with highly reticulated and dispersed nucleoli (L). Bar = 5 µm [30]. Reprinted with permission.
The human genome contains an analogous gene to mUtp14b. Unlike mUtp14b, the human UTP14b (hUTP14b) has degenerated into a non-functional retrogene, or pseudoretrogene [26, 31]. However, another UTP14 retrogene was found in humans in the 3’ UTR of the GT8 gene (a glycosyl transferase) on chromosome 13, called hUTP14c. hUTP14c is an intronless copy of the hUTP14a gene that is found on the human X chromosome [31] and is 66% identical and 77% similar to the amino acid sequence of mUtp14b. Similar to the situation in the mouse, hUTP14a is expressed ubiquitously while hUTP14c is expressed in human testes. It was hypothesized that mutations in hUTP14c cause infertility in men comparable to the mutation in jsd mice. Screening 234 DNA samples from infertile men revealed that 3 unrelated Caucasian patients had a Y738X mutation in hUTP14c resulting in a truncated protein product similar to that found in jsd mice [31]. These patients were all heterozygous for the mutation indicating a dominant negative effect of the truncation of hUTP14c. This specific form of infertility is indeed a ribosomopathy since a defect in ribosome biogenesis results in human infertility without any other associated abnormalities.
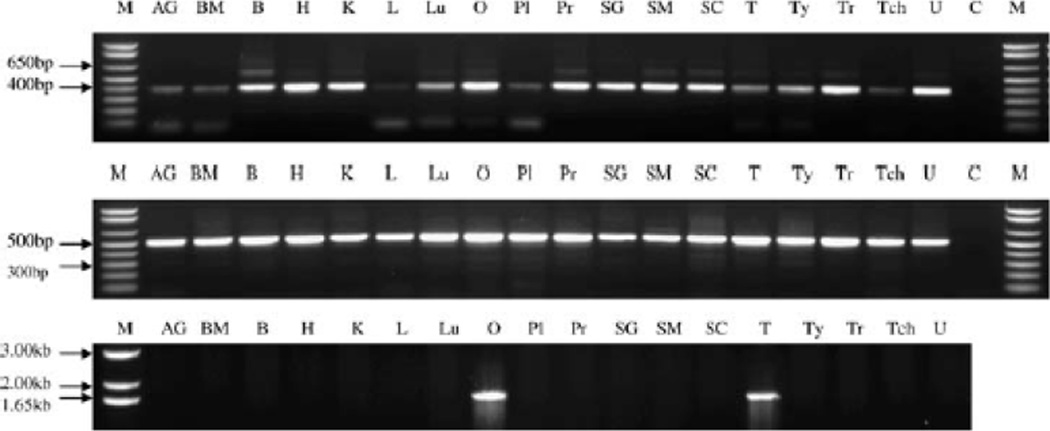
In addition to male infertility, it has been suggested the hUTP14c plays a role in ovarian cancer. Besides its expression in testes, hUTP14c is also expressed in human ovaries (Fig. 4). RT-PCR analysis of normal human ovarian tissue showed that hUTP14c is expressed in some but not all individuals. This is consistent with the finding that other retrogenes, also thought to be exclusively expressed in the testis, are found in some but not all normal ovaries [22]. hUTP14a interacts directly with p53 and knockdown of hUTP14a resulted in increased levels of p53 while over expression of hUTP14a led to decreased p53 levels. Furthermore, the interaction between hUTP14a and p53 targets the latter for degradation [32]. Since hUTP14a and hUTP14c have 90% identical and 94% similar amino acid sequences, Rohozinski et al. [33] propose that expression of hUTP14c in the ovary might also result in decreased levels of p53 similar to over expression of hUTP14a. Lower expression of p53 may then lead to susceptibility to tumor generation. This led to the hypothesis that factors involved in spermatogenesis in the testis may either pre-dispose or result in the generation of tumors in the ovary [33]. This fascinating hypothesis has yet to be tested.
Figure 4.
RT-PCR expression analysis of hUTP14c. Top) Expression of the ancestral X-linked gene, hUp14a, is ubiquitous, which is consistent with its role in 18S ribosomal RNA processing. Center) Similarly, GT8 also is expressed in all 18 tissues. Bottom) expression pattern of hUP14c, which is restricted to the ovary and testis. AG, adrenal gland; B, brain; BM, bone marrow; C, control reaction minus cDNA; H, heart; K, kidney; L, liver; Lu, lung; M, marker; O, ovary; PI, placenta; Pr, prostate; SC, spinal cord; SG, salivary gland; SM, skeletal muscle; T, testis; Tch, trachea; Ty, thymus; Tr, thymus; Tr, thyroid; U, uterus [31]. Reprinted with permission.
Recently, hUTP14a was also implicated in a human disease. Scleroderma is a disorder resulting in small blood vessel disease, autoimmune problems, and fibrosis of connective tissue. In twins affected with scleroderma, the hUTP14a gene was found to be hypermethylated, which decreases the expression of a gene, compared to unaffected twins [34]. The role of hypermethylation of hUTP14a in susceptibility to scleroderma is still unclear.
Unlike the other ribosomopathies, the tissue proclivity of infertility and ovarian cancer caused by mutations in hUTP14c is easily explained because of the unusual tissue-specific expression of this ribosome biogenesis factor. Perhaps tissue-specific splicing variants or retrogenes of other ribosome biogenesis factors are responsible for the tissue proclivity of other ribosomopathies?
4. Disease of EMG1: Bowen-Conradi Syndrome
The Hutterites are a small religious community currently comprising about 40,000 members primarily living in the Canadian Prairies and Great Plains of the United States. These 40,000 Hutterites are all descended from 100 original founders [35]. Consanguineous unions are common making rare genetic diseases more likely in this isolated population. One such apparently genetic and devastating disease affecting Hutterite neonates was first described by Bowen and Conradi in 1976 [36]. The original report of the disease described two brothers, born years apart, whose parents were second cousins. Both had a poor sucking reflex, undescended testes, a prominent nose, micrognathia (underdeveloped lower jaw), dolichocephaly (increased height of the skull in relation to the width), “rocker-bottom” feet, general developmental delays, and failure to thrive. Unusually, their karyotypes and laboratory findings were normal. Both brothers died before their first birthdays [36]. Disease presentation was consistent with either an X-linked or autosomal recessive trait as the parents were unaffected and both affected individuals were males. However, Hunter et al. reported 6 additional cases of Bowen-Conradi syndrome (BCS) (OMIM: 211180) in detail in 1979, three of which were female infants, and it was therefore concluded that the syndrome had an autosomal recessive inheritance pattern [37].
Since these early reports, many cases of BCS have been identified. All patients present with symptoms similar to the original description of the syndrome provided by Bowen and Conradi. However, because the signs and symptoms of BCS are very similar to trisomy 18 and cerebro-oculo-facial-skeletal syndrome (COFS), it is important to identify the genetic factors of each for proper diagnosis and management [38]. As patients with BCS have a normal karyotype, it is distinct from trisomy 18. Furthermore, genome-wide scan and linkage analysis showed that BCS and its related syndrome, COFS, are distinct syndromes as BCS failed to map to loci associated with COFS. This analysis also revealed that the causative gene for BCS was in the chromosome region 12p13.3. [39].
In 2009, Armistead et al. [38] identified a mutation in the gene, essential for mitotic growth 1 (EMG1), as the causative agent of BCS using SNP analysis. EMG1, termed nucleolar essential protein 1 (Nep1) in yeast, is a member of the SSU processome and is thus necessary for small ribosomal subunit assembly and pre-18S rRNA maturation [40]. As expected, the function of EMG1 is necessary for normal mammalian development, as demonstrated by the pre-implantation lethality for EMG1 null mice [41]. EMG1 is highly conserved throughout archaea and eukaryotes. Structural studies in Methanocaldococcus jannaschii revealed that EMG1 is an N1-specific pseudouridine methyltransferase that catalyzes methylation of the pseudouridine at position 914 of M. jannaschii 16S rRNA. This position is analogous to the pseudouridine in position 1248 in human 18S rRNA, which is the only known methylated pseudouridine [42]. A crystal structure of the yeast Nep1 dimer further supports its role as an N1-specific methyltransferase [43].
Patients with BCS have a D86G mutation in EMG1 that results in decreased levels of EMG1 protein without affecting mRNA transcript levels. The aspartate residue at position 86 is the most highly conserved region of the EMG1 protein and is conserved throughout eukaryotes and archaea [38] (Fig. 5). In fact, human EMG1 is able to functionally replace yeast Nep1 in vivo [44]. In order to elucidate the molecular details of the pathogenesis of BCS, studies in yeast with a D90G mutation in Nep1 (the mutation analogous to D86G in human EMG1) revealed that the mutant protein retained its methyltransferase activity. EMG1 dimerizes naturally and, a 10-fold increase for the interaction between monomers of D86G EMG1 as compared to wild-type was observed in a Y2H analysis. It is speculated that the increased dimerization or aggregation of the mutant EMG1 is responsible for preventing its localization to the nucleolus. Thus, because enzymatic activity is not affected, the D86G mutation in EMG1 may cause disease because of a role in ribosome assembly and not rRNA modification [45].
Figure 5.
Protein sequence alignment via Clustal W of the region of the EMG1 protein containing the c.400A/G, p.D86G mutation. The residues that are completely conserved in all orthologs are indicated with an asterisk, and the Asp (D) that is mutated in BCS is shown in red [38]. Reprinted with permission.
Indeed, EMG1 plays a critical role in ribosome biogenesis as a ribosome assembly factor. Studies in yeast show that EMG1 genetically interacts with Utp30, another component of the SSU processome. This interaction supports incorporation of the ribosomal protein Rps19 into the assembling pre-ribosome [46]. In addition, overexpression of Rps19 rescues growth defects in EMG1 deficient yeast [47]. This is rather intriguing as human Rps19 is mutated in 25% of patients with the ribosomopathy, Diamond-Blackfan Anemia (DBA). DBA is a bone marrow failure syndrome characterized by decreased or absent red blood cell progenitors. Patients can also present with various other abnormalities including craniofacial defects, various skeletal abnormalities, cancer predisposition, and heart defects [48]. Despite the associated functions of EMG1 and Rps19, a connection, if one exists, between their respective ribosomopathies, BCS and DBA, has yet to be uncovered.
5. Potential Diseases Modified by the SSU Processome
Two components of the SSU processome have been found to possibly modify human diseases. One of these components is hUTP6/HCA66, which may modify Neurofibromatosis type 1 (NF1), an autosomal dominant disease marked by the development of neural tumors. About 5% of NF1 cases are associated with heterozygous microdeletions in genes near the NF1 allele causing additional symptoms of craniofacial defects, heart malformations, and an increased number of neurofibromas. One such gene that may contain deletions in patients with NF1 is hUTP6/HCA66 [49]. It has been suggested that the interaction between hUTP6/HCA66 and the pro-apoptotic factor, Apaf-1, may play a role in the modification of NF1 [50]. However, how deletions in hUTP6/HCA66 would worsen disease remains to be elucidated. In addition to NF1, primary open angle glaucoma (POAG) has been proposed to be modified by the SSU processome component hUTP21/WDR36 [51]. POAG results in degeneration of the optic nerve and increased intraocular pressure in patients over 35. Indeed, mutations in hUTP21/WDR36 affect axon growth in the retina in mice [52], and loss of hUTP21/WDR36 in zebrafish activates the p53 response [53]. Conversely, some studies claim no association between POAG and hUTP21/WDR36 [54] while others claim the effect of mutations in hUTP21/WDR36 is dependent upon genetic background [55]. More investigation into the molecular contribution of both of these SSU processome components is necessary before we can conclude that they are involved in the pathogenesis of NF1 or POAG.
6. Putative Ribosomopathy: Williams-Beuren Syndrome
Utp14, whose human analog hUTP14C is mutated in some forms of male infertility, and the methyltransferase, Bud23, interact with each other within the yeast SSU processome [24]. Interestingly, Bud23 may be implicated in the human disease, Williams-Beuren syndrome [56]. This disease is marked by congenital heart defects, craniofacial dysmorphology (also found in ribosomopathies such as Treacher Collins syndrome and Diamond-Blackfan Anemia [8]), premature aging, short stature, hypercalcemia, and cognitive defects. A protein BLAST search using the amino acid sequence of yeast Bud23 revealed the putative human methyltransferase, WBSCR22 (NP_059998.2), which is 49% identical and 67% similar to Bud23 in its amino acid sequence. WBSCR22 is deleted in patients with Williams-Beuren syndrome [56]. Furthermore, another gene deleted in some patients with Williams-Beuren syndrome is WBSCR20, which has an amino acid sequence similar to the nucleolar protein, NOL1. Doll and Grzeschik [56] hypothesized that WBSCR20 may have a role in pre-rRNA processing or ribosome assembly. The deletions in WBSCR20 and WBSCR22 result in hemizygosity and thus haploinsufficiency of the gene products similar to many other ribosomopathies [57]. Due to the homology of WBSCR22 to Bud23, an SSU processome component, and the predicted role of WBSCR20 in ribosome biogenesis, we propose that Williams-Beuren syndrome may be a novel ribosomopathy.
7. Conclusions and Perspectives
The complex and vital process of ribosome biogenesis in eukaryotes is carried out by over 200 factors. Several of these factors associate with the U3 snoRNA to form a large RNP known as the SSU processome, which is necessary for the production of fully functional small ribosomal subunits. As all cells need proteins and therefore ribosomes, it is surprising that mutations that cause defects in the SSU processome would be compatible with life. Yet, in the last decade, some genetic diseases have been attributed to mutations in SSU processome components. This begs the question, how can only some cell-types be affected by defects in a process assumed to be necessary for all cells?
Tissue proclivity is of particular interest in NAIC and BCS. The literature reports only liver disease in patients with NAIC [11]. Despite the detrimental congenital defects of patients with BCS, few internal malformations are noted [38]. Whether BCS shows the extent of tissue specificity as other ribosomopathies is less clear. In the case of hUTP14c, a cell-type specific form of the UTP14 SSU processome component easily explains the tissue proclivity. Are there cell-type specific forms of hUTP4/Cirhin and EMG1?
There are other possible mechanisms by which defects in ribosome biogenesis could affect some tissues and not others. Perhaps defects in making the ribosome are sensed differently by different cell-types. All ribosomes may not be the same [1, 58], or all nucleoli may not be the same in each cell of a multicellular organism. Certain tissues may sense aberrant translation resulting from dysfunctional ribosomes differently as well. These hypotheses are not mutually exclusive; they all may contribute to the pathogenesis of ribosomopathies.
Highlights.
The small subunit (SSU) processome is a large ribonucleoprotein responsible for the maturation of the 18S rRNA.
Mutations in components of the SSU processome cause ribosomopathies.
North American Indian childhood cirrhosis is caused by a missense mutation in human UTP4/Cirhin.
Infertility can be caused by truncations of human UTP14c, an SSU processome component specific to developing male germ cells.
A missense mutation in EMG1 causes Bowen-Conradi syndrome.
Acknowledgements
The authors would like to thank the members of the Baserga lab for their helpful comments and suggestions during the preparation of this manuscript. This work was supported by the National Institutes of Health (R01 GM52581 to S.J.B. and MSTP TG T32GM07205).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cellular and molecular life sciences : CMLS. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith CM, Steitz JA. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 3.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, Hunt DF, Baserga SJ. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes & development. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phipps KR, Charette J, Baserga SJ. The small subunit processome in ribosome biogenesis-progress and prospects, Wiley interdisciplinary reviews. RNA. 2011;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prieto JL, McStay B. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes & development. 2007;21:2041–2054. doi: 10.1101/gad.436707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannan KM, Sanij E, Rothblum LI, Hannan RD, Pearson RB. Dysregulation of RNA polymerase I transcription during disease. Biochimica et biophysica acta. 2013;1829:342–360. doi: 10.1016/j.bbagrm.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Molecular bioSystems. 2010;6:481–493. doi: 10.1039/b919670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolze A, Mahlaoui N, Byun M, Turner B, Trede N, Ellis SR, Abhyankar A, Itan Y, Patin E, Brebner S, Sackstein P, Puel A, Picard C, Abel L, Quintana-Murci L, Faust SN, Williams AP, Baretto R, Duddridge M, Kini U, Pollard AJ, Gaud C, Frange P, Orbach D, Emile JF, Stephan JL, Sorensen R, Plebani A, Hammarstrom L, Conley ME, Selleri L, Casanova JL. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science (New York, N.Y.) 2013;340:976–978. doi: 10.1126/science.1234864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richter A, Mitchell GA, Rasquin A. North American Indian childhood cirrhosis (NAIC) Medecine sciences: M/S. 2007;23:1002–1007. doi: 10.1051/medsci/200723111002. [DOI] [PubMed] [Google Scholar]
- 12.Drouin E, Russo P, Tuchweber B, Mitchell G, Rasquin-Weber A. North American Indian cirrhosis in children: a review of 30 cases. Journal of pediatric gastroenterology and nutrition. 2000;31:395–404. doi: 10.1097/00005176-200010000-00013. [DOI] [PubMed] [Google Scholar]
- 13.Betard C, Rasquin-Weber A, Brewer C, Drouin E, Clark S, Verner A, Darmond-Zwaig C, Fortin J, Mercier J, Chagnon P, Fujiwara TM, Morgan K, Richter A, Hudson TJ, Mitchell GA. Localization of a recessive gene for North American Indian childhood cirrhosis to chromosome region 16q22-and identification of a shared haplotype. American journal of human genetics. 2000;67:222–228. doi: 10.1086/302993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chagnon P, Michaud J, Mitchell G, Mercier J, Marion JF, Drouin E, Rasquin-Weber A, Hudson TJ, Richter A. A missense mutation (R565W) in cirhin (FLJ14728) in North American Indian childhood cirrhosis. American journal of human genetics. 2002;71:1443–1449. doi: 10.1086/344580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu B, Mitchell GA, Richter A. Cirhin up-regulates a canonical NF-kappaB element through strong interaction with Cirip/HIVEP1. Experimental cell research. 2009;315:3086–3098. doi: 10.1016/j.yexcr.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Freed EF, Baserga SJ. The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis. Nucleic acids research. 2010;38:4798–4806. doi: 10.1093/nar/gkq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, Beattie BK, Lalev A, Zhang W, Davierwala AP, Mnaimneh S, Starostine A, Tikuisis AP, Grigull J, Datta N, Bray JE, Hughes TR, Emili A, Greenblatt JF. High-definition macromolecular composition of yeast RNA-processing complexes. Molecular cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Molecular and cellular biology. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed EF, Prieto JL, McCann KL, McStay B, Baserga SJ. NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS genetics. 2012;8:e1002892. doi: 10.1371/journal.pgen.1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, Babu M, Craig SA, Hu P, Wan C, Vlasblom J, Dar VU, Bezginov A, Clark GW, Wu GC, Wodak SJ, Tillier ER, Paccanaro A, Marcotte EM, Emili A. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu B, Mitchell GA, Richter A. Nucleolar localization of cirhin, the protein mutated in North American Indian childhood cirrhosis. Experimental cell research. 2005;311:218–228. doi: 10.1016/j.yexcr.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Rohozinski J, Anderson ML, Broaddus RE, Edwards CL, Bishop CE. Spermatogenesis associated retrogenes are expressed in the human ovary and ovarian cancers. PloS one. 2009;4:e5064. doi: 10.1371/journal.pone.0005064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao M, Rohozinski J, Sharma M, Ju J, Braun RE, Bishop CE, Meistrich ML. Utp14b: a unique retrogene within a gene that has acquired multiple promoters and a specific function in spermatogenesis. Developmental biology. 2007;304:848–859. doi: 10.1016/j.ydbio.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sardana R, White JP, Johnson AW. The rRNA methyltransferase Bud23 shows functional interaction with components of the SSU processome and RNase MRP. RNA (New York, N.Y.) 2013;19:828–840. doi: 10.1261/rna.037671.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradley J, Baltus A, Skaletsky H, Royce-Tolland M, Dewar K, Page DC. An X-to-autosome retrogene is required for spermatogenesis in mice. Nature genetics. 2004;36:872–876. doi: 10.1038/ng1390. [DOI] [PubMed] [Google Scholar]
- Rohozinski J, Bishop CE. The mouse juvenile spermatogonial depletion (jsd) phenotype is due to a mutation in the X-derived retrogene, mUtp14b. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11695–11700. doi: 10.1073/pnas.0401130101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beamer WG, Cunliffe-Beamer TL, Shultz KL, Langley SH, Roderick TH. Juvenile spermatogonial depletion (jsd): a genetic defect of germ cell proliferation of male mice. Biology of reproduction. 1988;38:899–908. doi: 10.1095/biolreprod38.4.899. [DOI] [PubMed] [Google Scholar]
- 28.Shetty G, Weng CC, Porter KL, Zhang Z, Pakarinen P, Kumar TR, Meistrich ML. Spermatogonial differentiation in juvenile spermatogonial depletion (jsd) mice with androgen receptor or follicle-stimulating hormone mutations. Endocrinology. 2006;147:3563–3570. doi: 10.1210/en.2006-0159. [DOI] [PubMed] [Google Scholar]
- 29.Dixon J, Dixon MJ. Genetic background has a major effect on the penetrance and severity of craniofacial defects in mice heterozygous for the gene encoding the nucleolar protein Treacle. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;229:907–914. doi: 10.1002/dvdy.20004. [DOI] [PubMed] [Google Scholar]
- 30.Bolden-Tiller OU, Chiarini-Garcia H, Poirier C, Alves-Freitas D, Weng CC, Shetty G, Meistrich ML. Genetic factors contributing to defective spermatogonial differentiation in juvenile spermatogonial depletion (Utp14bjsd) mice. Biology of reproduction. 2007;77:237–246. doi: 10.1095/biolreprod.107.060087. [DOI] [PubMed] [Google Scholar]
- 31.Rohozinski J, Lamb DJ, Bishop CE. UTP14c is a recently acquired retrogene associated with spermatogenesis and fertility in man. Biology of reproduction. 2006;74:644–651. doi: 10.1095/biolreprod.105.046698. [DOI] [PubMed] [Google Scholar]
- 32.Hu L, Wang J, Liu Y, Zhang Y, Zhang L, Kong R, Zheng Z, Du X, Ke Y. A small ribosomal subunit (SSU) processome component, the human U3 protein 14A (hUTP14A) binds p53 and promotes p53 degradation. The Journal of biological chemistry. 2011;286:3119–3128. doi: 10.1074/jbc.M110.157842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohozinski J, Edwards CL, Anderson ML. Does expression of the retrogene UTP14c in the ovary pre-dispose women to ovarian cancer? Medical hypotheses. 2012;78:446–449. doi: 10.1016/j.mehy.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 34.Selmi C, Feghali-Bostwick CA, Lleo A, Lombardi SA, De Santis M, Cavaciocchi F, Zammataro L, Mitchell MM, Lasalle JM, Medsger T, Jr, Gershwin ME. X chromosome gene methylation in peripheral lymphocytes from monozygotic twins discordant for scleroderma. Clinical and experimental immunology. 2012;169:253–262. doi: 10.1111/j.1365-2249.2012.04621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Souza RA. Mystery behind Bowen-Conradi syndrome solved: a novel ribosome biogenesis defect. Clinical genetics. 2010;77:116–118. doi: 10.1111/j.1399-0004.2009.01304.x. [DOI] [PubMed] [Google Scholar]
- 36.Bowen P, Conradi GJ. Syndrome of skeletal and genitourinary anomalies with unusual facies and failure to thrive in Hutterite sibs. Birth defects original article series. 1976;12:101–108. [PubMed] [Google Scholar]
- 37.Hunter AG, Woerner SJ, Montalvo-Hicks LD, Fowlow SB, Haslam RH, Metcalf PJ, Lowry RB. The Bowen-Conradi syndrome -- a highly lethal autosomal recessive syndrome of microcephaly, micrognathia, low birth weight, and joint deformities. American journal of medical genetics. 1979;3:269–279. doi: 10.1002/ajmg.1320030305. [DOI] [PubMed] [Google Scholar]
- 38.Armistead J, Khatkar S, Meyer B, Mark BL, Patel N, Coghlan G, Lamont RE, Liu S, Wiechert J, Cattini PA, Koetter P, Wrogemann K, Greenberg CR, Entian KD, Zelinski T, Triggs-Raine B. Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen-Conradi syndrome. American journal of human genetics. 2009;84:728–739. doi: 10.1016/j.ajhg.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamont RE, Loredo-Osti J, Roslin NM, Mauthe J, Coghlan G, Nylen E, Frappier D, Innes AM, Lemire EG, Lowry RB, Greenberg CR, Triggs-Raine BL, Morgan K, Wrogemann K, Fujiwara TM, Zelinski T. A locus for Bowen-Conradi syndrome maps to chromosome region 12p13.3. American journal of medical genetics. Part A. 2005;132A:136–143. doi: 10.1002/ajmg.a.30420. [DOI] [PubMed] [Google Scholar]
- 40.Bernstein KA, Gallagher JE, Mitchell BM, Granneman S, Baserga SJ. The small-subunit processome is a ribosome assembly intermediate. Eukaryotic cell. 2004;3:1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, Sandhu S, Patel N, Triggs-Raine B, Ding H. EMG1 is essential for mouse pre-implantation embryo development. BMC developmental biology. 2010;10:99. doi: 10.1186/1471-213X-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wurm JP, Meyer B, Bahr U, Held M, Frolow O, Kotter P, Engels JW, Heckel A, Karas M, Entian KD, Wohnert J. The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic acids research. 2010;38:2387–2398. doi: 10.1093/nar/gkp1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas SR, Keller CA, Szyk A, Cannon JR, Laronde-Leblanc NA. Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic acids research. 2011;39:2445–2457. doi: 10.1093/nar/gkq1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eschrich D, Buchhaupt M, Kotter P, Entian KD. Nep1p (Emg1p), a novel protein conserved in eukaryotes and archaea, is involved in ribosome biogenesis. Current genetics. 2002;40:326–338. doi: 10.1007/s00294-001-0269-4. [DOI] [PubMed] [Google Scholar]
- 45.Meyer B, Wurm JP, Kotter P, Leisegang MS, Schilling V, Buchhaupt M, Held M, Bahr U, Karas M, Heckel A, Bohnsack MT, Wohnert J, Entian KD. The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Psi1191 in yeast 18S rRNA. Nucleic acids research. 2011;39:1526–1537. doi: 10.1093/nar/gkq931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schilling V, Peifer C, Buchhaupt M, Lamberth S, Lioutikov A, Rietschel B, Kotter P, Entian KD. Genetic interactions of yeast NEP1 (EMG1), encoding an essential factor in ribosome biogenesis. Yeast. 2012;29:167–183. doi: 10.1002/yea.2898. [DOI] [PubMed] [Google Scholar]
- 47.Buchhaupt M, Meyer B, Kotter P, Entian KD. Genetic evidence for 18S rRNA binding and an Rps19p assembly function of yeast nucleolar protein Nep1p. Molecular genetics and genomics : MGG. 2006;276:273–284. doi: 10.1007/s00438-006-0132-x. [DOI] [PubMed] [Google Scholar]
- 48.Campagnoli MF, Ramenghi U, Armiraglio M, Quarello P, Garelli E, Carando A, Avondo F, Pavesi E, Fribourg S, Gleizes PE, Loreni F, Dianzani I. RPS19 mutations in patients with Diamond-Blackfan anemia. Human mutation. 2008;29:911–920. doi: 10.1002/humu.20752. [DOI] [PubMed] [Google Scholar]
- 49.Bonnart C, Gerus M, Hoareau-Aveilla C, Kiss T, Caizergues-Ferrer M, Henry Y, Henras AK. Mammalian HCA66 protein is required for both ribosome synthesis and centriole duplication. Nucleic acids research. 2012;40:6270–6289. doi: 10.1093/nar/gks234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddubnyak V, Rigou P, Michel L, Rain JC, Geneste O, Wolkenstein P, Vidaud D, Hickman JA, Mauviel A, Poyet JL. Positive regulation of apoptosis by HCA66, a new Apaf-1 interacting protein, and its putative role in the physiopathology of NF1 microdeletion syndrome patients. Cell death and differentiation. 2007;14:1222–1233. doi: 10.1038/sj.cdd.4402122. [DOI] [PubMed] [Google Scholar]
- 51.Blanco-Marchite C, Sanchez-Sanchez F, Lopez-Garrido MP, Inigez-de-Onzono M, Lopez-Martinez F, Lopez-Sanchez E, Alvarez L, Rodriguez-Calvo PP, Mendez-Hernandez C, Fernandez-Vega L, Garcia-Sanchez J, Coca-Prados M, Garcia-Feijoo J, Escribano J. WDR36 and P53 gene variants and susceptibility to primary open-angle glaucoma: analysis of genegene interactions. Investigative ophthalmology & visual science. 2011;52:8467–8478. doi: 10.1167/iovs.11-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chi ZL, Yasumoto F, Sergeev Y, Minami M, Obazawa M, Kimura I, Takada Y, Iwata T. Mutant WDR36 directly affects axon growth of retinal ganglion cells leading to progressive retinal degeneration in mice. Human molecular genetics. 2010;19:3806–3815. doi: 10.1093/hmg/ddq299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skarie JM, Link BA. The primary open-angle glaucoma gene WDR36 functions in ribosomal RNA processing and interacts with the p53 stress-response pathway. Human molecular genetics. 2008;17:2474–2485. doi: 10.1093/hmg/ddn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fingert JH, Alward WL, Kwon YH, Shankar SP, Andorf JL, Mackey DA, Sheffield VC, Stone EM. No association between variations in the WDR36 gene and primary open-angle glaucoma. Archives of ophthalmology. 2007;125:434–436. doi: 10.1001/archopht.125.3.434-b. [DOI] [PubMed] [Google Scholar]
- 55.Footz TK, Johnson JL, Dubois S, Boivin N, Raymond V, Walter MA. Glaucoma-associated WDR36 variants encode functional defects in a yeast model system. Human molecular genetics. 2009;18:1276–1287. doi: 10.1093/hmg/ddp027. [DOI] [PubMed] [Google Scholar]
- 56.Doll A, Grzeschik KH. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenetics and cell genetics. 2001;95:20–27. doi: 10.1159/000057012. [DOI] [PubMed] [Google Scholar]
- 57.McCann KL, Baserga SJ. Genetics. Mysterious ribosomopathies. Science (New York, N.Y.) 2013;341:849–850. doi: 10.1126/science.1244156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nature reviews. Molecular cell biology. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]