Abstract
The risk of developing papillary thyroid carcinoma (PTC), the most frequent form of thyroid malignancy, is elevated up to 8.6-fold in first-degree relatives of PTC patients. The familial risk could be explained by high-penetrance mutations in yet unidentified genes, or polygenic action of low-penetrance alleles. Since the DNA-damaging exposure to ionizing radiation is a known risk factor for thyroid cancer, polymorphisms in DNA repair genes are likely to affect this risk. In a search for low-penetrance susceptibility alleles we employed Sequenom technology to genotype deleterious polymorphisms in ATM, CHEK2, and BRCA1 in 1,781 PTC patients and 2,081 healthy controls. As a result of the study, we identified CHEK2 rs17879961 (OR = 2.2, P = 2.37e-10) and BRCA1 rs16941 (odds ratio [OR] = 1.16, P = 0.005) as risk alleles for PTC. The ATM rs1801516 variant modifies the risk associated with the BRCA1 variant by 0.78 (P = 0.02). Both the ATM and BRCA1 variants modify the impact of male gender on clinical variables: T status (P = 0.007), N status (P = 0.05), and stage (P = 0.035). Our findings implicate an important role of variants in the ATM- CHEK2- BRCA1 axis in modification of the genetic predisposition to PTC and its clinical manifestations.
INTRODUCTION
Thyroid carcinoma with nearly 60,000 new cases per year (in USA only) is the most common malignancy of the endocrine system (Siegel et al., 2012). Inherited predisposition to papillary thyroid carcinoma (PTC), the most frequent form of thyroid malignancy, is well established, as evidenced by epidemiologic studies showing that the risk of developing PTC is elevated up to 8.6-fold (Goldgar et al., 1994; Frich et al., 2001) in first-degree relatives of PTC patients. The observed familial risk could be partially explained by high-penetrance mutations in yet unidentified genes (Canzian et al., 1998; McKay et al., 2001; He et al., 2009), and a polygenic action of low-penetrance alleles is an alternative explanation (Houlston and Peto, 2004; Jazdzewski et al., 2008; Adjadj et al., 2009; Gudmundsson et al., 2009, 2012; Landa and Robledo, 2011; Liyanarachchi et al., 2013). This hypothesis is supported by the paucity of families with more than two affected members. The role of regulatory RNA genes, especially microRNAs, is widely discussed (Jazdzewski and de la Chapelle, 2009; Jazdzewski et al., 2009; de la Chapelle and Jazdzewski, 2011; Swierniak et al., 2013). Weak linkage signals from low penetrance disease alleles make the use of association studies comparing the frequency of polymorphic genotypes in patients and control subjects, more adequate for the identification of such susceptibility alleles than the classic genome-wide linkage study (Botstein and Risch, 2003). Among the etiological culprits, exposure to ionizing radiation during childhood is a known risk factor for thyroid cancer (Sigurdson et al., 2005). The mechanism by which ionizing radiation promotes carcinogenesis consists mainly of its ability to induce DNA double-strand breaks. In mammalian cells, double-strand DNA breaks activate the ataxia-telangiectasia-mutated (ATM) kinase, which phosphorylates and activates checkpoint yeast homolog 2 (CHEK2). Subsequently, CHEK2 phosphorylates the breast cancer 1 gene (BRCA1) and triggers DNA repair or, if failed, leads to cellular apoptosis (Dasika et al., 1999). The functionality of the ATM-BRCA1-CHEK2 pathway is affected by polymorphisms and mutations within the involved genes, underlying inefficient DNA repair and leading to tumorigenic changes within the cells.
A number of studies showed that the rs1801516 (G>A, Asp1853Asn) polymorphism in ATM, rs17879961 (T>C, Ile157Thr) in CHEK2 and rs16941 (A>G, Glu1038Gly) in BRCA1 predispose to a number of human cancers and might affect the functioning of the whole DNA repair pathway. ATM rs1801516 is a missense mutation (Asp1853Asn). Although the 30-AA sequence surrounding the rs1801516 SNP does not contain known functional domains, it is highly conserved among species from Xenopus to humans, with almost 100% conservation among primates, indicating its potentially important role for the proper functioning of the ATM protein. The variant allele of rs1801516 has been associated with increased telomere length in blood cells of breast cancer patients (Kim et al., 2012) that could indicate slower cell division rates in such individuals and potentially protect against immediate loss of genomic stability in cancer cells. CHEK2 is a tumour suppressor gene and the rs17879961 (Ile157Thr) variant has been documented to produce protein defective in its ability to bind p53 and BRCA1, and therefore unable to exert its cellular function (Falck et al., 2001; Li et al., 2002). Rs17879961 in CHEK2 was shown to be associated with increased risk for several cancers, including prostate, breast cancer, and leukaemia and was proposed to be a founder mutation in ethnically diverse populations (Cybulski et al., 2004; Dufault et al., 2004; Kilpivaara et al., 2004, 2006; Rudd et al., 2006; Williams et al., 2006; Brennan et al., 2007; Kaufman et al., 2009). The molecular function of the BRCA1 rs16941 polymorphism has not been thoroughly studied, but the SNP has been documented to be associated with an increased risk of prostate cancer (Leongamornlert et al., 2012).
Thus, variants in ATM, BRCA1, and CHEK2 predispose to several cancers and potentially lead to inefficient repair of ionizing radiation-induced DNA breaks. Simultaneously, since ionizing radiation is a known risk for thyroid cancer, polymorphisms in the ATM-BRCA1-CHEK2 axis are likely to affect this risk. To elucidate the role of the ATM Asp1853Asn, BRCA1 Glu1038Gly, and CHEK2 Ile157Thr polymorphisms in tailoring the genetic predisposition to PTC, we genotyped 1,781 patients and 2,081 healthy controls from a Central European Caucasian population and identified a complex association between the ATM-CHEK2-BRCA1 axis and the predisposition to papillary thyroid carcinoma.
MATERIALS AND METHODS
Patients and Control Subjects
Patients with PTC (1781 total; 1599 women, 182 men; median age at diagnosis 48 years, SD ± 14.1) were recruited at the Medical University of Warsaw, Poland, and Maria Skłodowska-Curie Memorial Cancer Center - Institute of Oncology in Warsaw, Poland. After approval of the Institutional Review Board, blood samples were obtained with informed consent. DNA was extracted using the conventional salting-out method and quantified using Nanodrop 2000. Final diagnosis was confirmed through histopathological examination. Clinicopathological information was obtained from the medical records. TNM staging was defined on the basis of the 2010 American Joint Commission on Cancer staging system (Edge et al., 2010). A positive history of head and neck radiotherapy previous to the diagnosis of thyroid cancer has been documented in eight patients.
Control samples (2,081 total; 1,079 women, 1,002 men; median age 37 years, SD ± 10.6) were provided by the Department of Medical Genetics, Medical University of Warsaw, Poland, and consisted of consenting healthy volunteers. All patients and control subjects were white and Caucasian. Demographic information for all cases and controls can be found in Table 1.
TABLE 1.
Demographic Data of Analyzed Samples
| Cohort | Variable | PTC | Control |
|---|---|---|---|
| Total | Female (%) | 1599 (90%) | 1079 (52%) |
| Cohort | Male (%) | 182 (10%) | 1002 (48%) |
| Age (median ± SD) | 48 ± 14.1 | 37 ± 10.6 | |
| Total number of samples | 1781 | 2081 | |
| Age- and | Female (%) | 712 (80%) | 712 (80%) |
| Gender- | Male (%) | 173 (20%) | 173 (20%) |
| Matched | Age (median ± SD) | 39 ± 11.5 | 38 ± 11.4 |
| Cohort | Total number of samples | 885 | 885 |
PTC, papillary thyroid carcinoma; SD, standard deviation.
Due to a significant difference in age and gender between cases and controls, equal numbers of age- and gender-matched samples from available cases and controls were selected to create matched cohorts of cases and controls. Each matched cohort consisted of 712 women and 173 men (885 total; median age at diagnosis of cases 39 years, SD ± 11.5, median age of controls 38 years, SD ± 11.4; Table 1).
SNP Genotyping and Confirmation
All SNPs were genotyped using the Sequenom genotyping technology (Sequenom Inc., USA). For each sample, 20ng of genomic DNA was genotyped using iPLEX Gold system. Primers and probes for the analysis were designed using MassARRAY® Assay Design v3.1. Data were visualized and analysed in MassARRAY Typer Viewer v4.0.24. DNA samples scoring lower than “moderate” as defined by the software were excluded. Finally, for confirmation, each SNP was directly sequenced in 96 random samples, and no discrepancies were found between the sequencing and Sequenom genotyping results.
Statistical Analysis
Statistical analyses were performed using the R software package (http://www.r-project.org/). For each SNP, Hardy-Weinberg equilibrium was tested in control samples by Fisher’s exact test. All SNP variants satisfied Hardy-Weinberg equilibrium in control samples with a P-value > 0.05. The risks associated with each SNP were estimated by analyzing the difference between allelic frequencies in patients and control subjects using Fisher’s exact test. Two further tests were performed based on 2 × 2 tables combining the heterozygotes with either the common or rare homozygotes to derive the statistics under recessive or dominant models, respectively. The risks associated with each SNP were estimated by allelic, dominant, and recessive odds ratios (ORs) with associated 95% CIs using conditional Maximum Likelihood Estimate (MLE). The impact of the SNPs, gender and age on selected clinical variables was evaluated by fitting the main effects regression model: logistic for binary variable M status, or linear for categorical variables: N status, stage, and T status. The interactive effects of the SNPs and age or gender on the risk for PTC or selected clinical variables were examined by fitting the full regression model for each pair (logistic for case/control studies and M status, linear for other clinical variables).
RESULTS
Demographic Characteristics
Demographic characteristics of the analysed case and control subjects are shown in Table 1. Age is reported as age at diagnosis in the cases, and age at blood draw in controls. Controls were significantly younger than cases (P <2.2e-16), and there were significantly more females among the cases compared with controls (P <2.2e-16). The frequencies of each SNP variant did not show significant difference between males and females (P >0.05). Nevertheless, all the analyses were performed in both unmatched and age- and gender-matched cohorts.
Risk of PTC Associated with SNPs
Rs17879961 of CHEK2 and rs16941 of BRCA1 showed significant association with susceptibility to PTC with allelic OR = 2.2 (95%CI 1.71–2.86; P = 2.37e-10) and OR = 1.16 (95%CI 1.04–1.28; P = 0.005), respectively (Table 2). There was no association between rs1801516 of ATM and PTC (P = 1). Similar results were found in age- and gender-matched cohorts (Table 2). Some of the cases were follicular variant PTC (PTCfv, n = 232). We performed an independent calculation of the risks by excluding the PTCfv cases and obtained similar results (Table 2). Similarly, we performed association analysis after exclusion of microPTC cases (pT1a, n = 740). The results were essentially the same (Table 2). Thus, we propose that inclusion of PTCfv or microPTC cases did not have an undue influence on the results.
TABLE 2.
Association of the Analyzed SNPs with Predisposition to Papillary Thyroid Carcinoma
| Gene | SNP | Variant allele | MAF | No of cases/controls | No of genotypes in cases | No of genotypes in controls | Allelic statistics | Dominant/recessive statisticsa | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| OR (95% CI) | PA | OR (95% CI) | PD/R | ||||||||
| Total Cohort | CHEK2 | rs17879961 | T>C | 0.052 | 1,700/2,056 | 1531/161/8 | 1,958/96/2 | 2.2 (1.71–2.86) | 2.37e–10 | 2.21 (1.69–2.88)D | 9.60e–10D |
| BRCA1 | rs16941 | A>G | 0.302 | 1,635/2,021 | 807/667/161 | 1,074/792/155 | 1.16 (1.04–1.28) | 0.005 | 1.31 (1.04–1.67)R | 0.021R | |
| ATM | rs1801516 | G>A | 0.114 | 1,603/1,844 | 1,261/319/23 | 1,455/357/32 | 1 (0.86–1.16) | 1 | 0.82 (0.46–1.46)R | 0.499R | |
| PTCfv excluded | CHEK2 | rs17879961 | T>C | 0.054 | 1,477/2,056 | 1,324/146/7 | 1,958/96/2 | 2.3 (1.77–2.99) | 8.79e–11 | 2.31 (1.76–3.04)D | 2.82e–10D |
| BRCA1 | rs16941 | A>G | 0.303 | 1,419/2,021 | 704/570/145 | 1,074/792/155 | 1.16 (1.04–1.29) | 0.006 | 1.37 (1.07–1.75)R | 0.01R | |
| ATM | rs1801516 | G>A | 0.112 | 1,391/1,844 | 1,099/272/20 | 1,455/357/32 | 0.98 (0.84–1.15) | 0.812 | 0.83 (0.45–1.5)R | 0.573R | |
| pT1a excluded | CHEK2 | rs17879961 | T>C | 0.050 | 993/2,056 | 897/92/4 | 1,958/96/2 | 2.13 (1.59–2.85) | 2.13e–07 | 2.14 (1.58–2.9)D | 4.8e–07D |
| BRCA1 | rs16941 | A>G | 0.305 | 958/2,021 | 465/402/91 | 1,074/792/155 | 1.17 (1.04–1.32) | 0.011 | 1.2 (1.03–1.41)D | 0.021D | |
| ATM | rs1801516 | G>A | 0.121 | 939/1,844 | 730/191/18 | 1,455/357/32 | 1.07 (0.89–1.27) | 0.479 | 1.11 (0.58–2.04)R | 0.763R | |
| Age- & Gender-Matched | CHEK2 | rs17879961 | T>C | 0.050 | 885/885 | 800/82/3 | 841/44/0 | 2.05 (1.4–3.04) | 1.20e–04 | 2.03 (1.38–3.03)D | 0.0002D |
| BRCA1 | rs16941 | A>G | 0.304 | 848/848 | 417/347/84 | 462/326/60 | 1.22 (1.05–1.42) | 0.010 | 1.44 (1.01–2.08)R | 0.045R | |
| ATM | rs1801516 | G>A | 0.109 | 793/793 | 629/155/9 | 634/149/10 | 1.03 (0.81–1.29) | 0.864 | 0.9 (0.32–2.48)R | 1R | |
SNP, single nucleotide polymorphism; MAF, minor allele frequency in cases; OR, odds ratio; CI, confidence interval; PTCfv, follicular variant of papillary thyroid carcinoma; PA, allelic P-value; PD/R, P-value under dominant or recessive model; Ddominant model; Rrecessive model;
most significant association under a dominant or recessive model.
Covariates and Interactions
Evaluation of pairwise interactions of risk alleles of the 3 SNPs resulted in identification of an interaction between BRCA1 and ATM. The minor allele of ATM seems to alleviate the pathogenic role of the rare BRCA1 variant by the mode of 0.78 (95%CI 0.62–0.97; P = 0.023, Table 3).
TABLE 3.
Gene-Gene and Gene-Covariates Interactions and the Risk of Papillary Thyroid Carcinoma
| Interaction | OR (95% CI) | P value |
|---|---|---|
| CHEK2 x BRCA1 | 0.73 (0.48–1.11) | 0.141 |
| CHEK2 x ATM | 1.30 (0.75–2.26) | 0.352 |
| BRCA1 x ATM | 0.78 (0.62–0.97) | 0.023a |
| CHEK2 x gender | 1.34 (0.74–2.43) | 0.330 |
| BRCA1 x gender | 0.89 (0.67–1.18) | 0.407 |
| ATM x gender | 0.54 (0.33–0.87) | 0.012a |
| CHEK2 x age | 1.01 (0.99–1.03) | 0.365 |
| BRCA1 x age | 0.99 (0.98–1) | 0.081 |
| ATM x age | 1 (0.99–1.01) | 0.898 |
Statistically significant interaction.
OR, odds ratio; CI, confidence interval.
We also evaluated the effect of interaction between each of the SNPs and age or gender on tailoring the risk for PTC, and found a positive interaction between the ATM variant and gender. Male gender modifies the risk associated with the minor allele of ATM by the mode of 0.54 (95%CI 0.33–0.87; P = 0.012, Table 3). We did not observe interaction between the CHEK2 or BRCA1 variants and selected variables (P >0.05, Table 3).
Impact of the SNPs on Clinical Phenotype
We evaluated the impact of particular SNPs, separately or in interaction with age or gender, on selected clinical variables (T, N, M status, stage). The distribution of patients with disease Stages I, II, III, and IV was as follows: 50.3%, 8.4%, 17.0%, 10.5%, respectively. The disease stage has not been specified for 13.9% patients. We observed no effect of any one of the SNPs alone, or in interaction with other SNPs on the selected variables (P >0.05). However, we found that the clinical phenotype is influenced by the interactive impact of the SNPs in combination with gender or age.
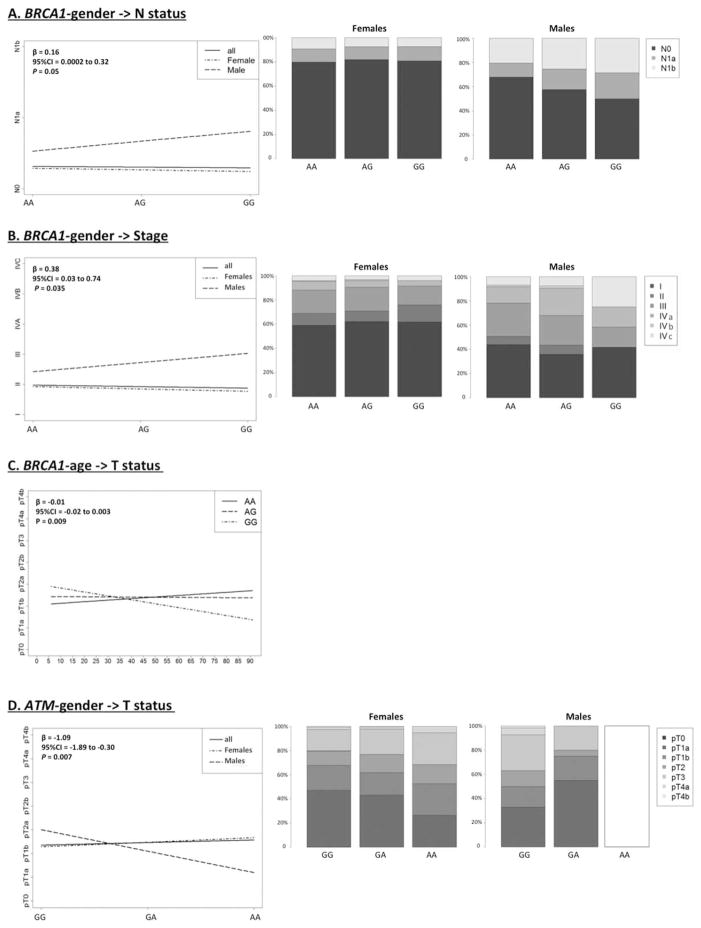
Interaction between the BRCA1 variant and gender affects N status and stage (P = 0.05 and 0.035, respectively) of PTC. Even though gender by itself has a significant impact on metastasis to lymph nodes and on the stage of cancer, the presence of the BRCA1 variant significantly increases the metastatic potential and stage of cancer in males when compared with females (Figs. 1A and 1B). Interestingly, analysis of the interaction between BRCA1 and age revealed an interactive, protective impact on T status; the risk of higher T-status significantly decreases in the presence of the BRCA1 variant allele (P = 0.009, Fig. 1C).
Figure 1.
Interactive impact of the SNPs, gender and age on clinical covariates (T, N, M, stage). The interactive effects of BRCA1 rs16941 and gender on N-status (A) and stage (B), BRCA1 rs16941 and age on T-status (C), and between ATM and gender on T-status were examined by fitting the full regression model for each pair (logistic for M status, linear for other clinical variables). No male cases with the rare AA genotype of ATM were observed.
The ATM variant also showed an interaction with gender, having a protective impact against higher T status (P = 0.007). Gender by itself has a significant impact on T-status, with males having higher risk of presenting more advanced disease compared with women. However, the ATM polymorphism modifies this trend. The risk for worse T-status significantly decreases with each additional variant allele (hetero- or homozygous state) in males compared to females (P = 0.007; Fig. 1D).
DISCUSSION
To comprehensively elucidate the role of three deleterious, multicancer culprit SNPs in ATM (rs1801516), CHEK2 (rs17879961), and BRCA1 (rs16941) with the risk for papillary thyroid carcinoma, we genotyped germinal DNA from 1781 patients and 2081 healthy controls. The implication of these polymorphisms in the pathogenesis of the thyroid gland has previously been investigated only in small-scale studies (173–273 PTC cases). More importantly, the SNPs have never been analyzed interactively as a group (Cybulski et al., 2004; Akulevich et al., 2009; Xu et al., 2012), although the functional linkage between ATM, CHEK2 and BRCA1 suggests a potential cooperative influence of their variations in the pathogenesis of cancer. The significance of the SNPs likely stems from their ability to impair the activity of ATM, BRCA1, and CHEK2, important regulators of the DNA damage response pathway. The main factor responsible for causing DNA breaks is ionizing radiation, which is a known risk factor for thyroid cancer. It was therefore of great interest to analyse whether deleterious variants of genes, whose products are responsible for the protection of genomic stability against ionizing radiation, can contribute to the predisposition to thyroid cancer.
Variants in ATM are associated with a number of cancer types (Lavin, 2008). Interestingly, the variant allele was reported to be protective against cutaneous melanoma (OR = 0.84, P = 3.4e-9) (Barrett et al., 2011), a malignancy that shares the common occurrence of BRAF mutations with PTC. The risk of PTC in melanoma patients is elevated 2.17-fold, suggesting a common genetic background (Goggins et al., 2006). Our study did not show any association of the ATM rs1801516 with the risk of PTC, but the variant proved to have an important role as a modifier of PTC risk associated with BRCA1, alleviating the pathogenic role of the rare BRCA1 variant by the mode of 0.78 (P = 0.023). Further clinicopathological analysis revealed interaction between ATM and gender (see below).
Rs17879961 (Ile157Thr) in CHEK2 was shown to be associated with increased risk for prostate and breast cancer, and leukaemia (Cybulski et al., 2004; Dufault et al., 2004; Kilpivaara et al., 2004; Kilpivaara et al., 2006; Rudd et al., 2006; Williams et al., 2006; Brennan et al., 2007; Kaufman et al., 2009). A multicancer study by Cybulski et al. (focused mainly on breast, colon, and prostate cancers) tested 173 cases of thyroid cancer and showed association of the CHEK2 rs17879961 variant with thyroid cancer (OR = 1.9, P = 0.04) (Cybulski et al., 2004). Of note, pathological subtypes of thyroid cancer were not reported, and the study has never been validated. Our study, performed in a large cohort of 1781 papillary thyroid cancer cases revealed significantly higher risk of PTC associated with CHEK2 rs17879961 variant (OR = 2.2, P = 2.4e-10) than previously reported.
BRCA1 is a tumour suppressor gene with a well-documented role in breast and ovarian cancer susceptibility and the rs16941 polymorphism is associated with an increased risk of prostate cancer (Leongamornlert et al., 2012). Our study revealed evidence of a moderately increased risk for PTC associated with the minor allele of BRCA1 (OR = 1.16, P = 0.005). The same SNP has been previously analysed in 303 thyroid cancer patients, including 273 PTC cases, and no association has been found (Xu et al., 2012). The difference may be explained by the disparity in the number of cases (power) and also by the different ethnicities analyzed, as 30% of cases were reported as other than non-Hispanic white in the study by Xu et al.
Apart from information on the simple associations between the analysed SNPs and PTC, our study brings new insight into the age and gender-related outcome of thyroid cancer. Age and gender are two variables with a known, significant impact on the biology of PTC: the cancer carries a worse prognosis in older age and affects women significantly more often than men, but in males the course of the disease is more aggressive (Mazzaferri and Jhiang, 1994). Concomitantly, the primary tumor diameter, risk of lymph node, and distant metastases is higher in males compared to females (Machens et al., 2006). However, a subgroup analysis showed that although disease-specific survival for females diagnosed under 55 years of age was much more favourable than for males (Hazard Ratio = 0.33, CI 0.13–0.81), this effect was less obvious for women diagnosed at 55–69 years (Jonklaas et al., 2012).
Our study showed an apparent role of age and gender in shaping the clinical phenotype of PTC that was significantly modified by the BRCA1 and ATM variants. The impact of gender on lymph node metastasis (N-status) and cancer stage was significantly strengthened by the BRCA1 variant (P = 0.05 and 0.035, respectively). Interestingly, the same variant alleviated the effect of age on tumor size (P = 0.009) with a significantly favorable outcome for patients older than 55 years, which, notably, is an age by which most women have reached menopause. As already mentioned, also the ATM variant seems to exert a protective role, alleviating the effect of gender on tumor size (P = 0.009), and on developing a more advanced disease (T-status) in males (P = 0.007). This fact can be explained by the observation that the ATM variant allele is associated with increased telomere length, which could indicate slower cell division rates, and thus protect from the loss of genomic stability in cancer cells (Kim et al., 2012). All these observations add to the complexity of the protective ATM-gender interaction in shaping the risk of PTC.
Importantly, the biological relevance of low-penetrance alleles of ATM, CHEK2, and BRCA1/2 has been shown for breast cancer and lymphocytic leukemia (Rudd et al., 2006) and patients with these cancers present an elevated risk of thyroid cancer (Familial Relative Risk, FRR = 1.69 and 2.36, respectively) (Goldgar et al., 1994). This observation supports our findings implicating the variants in the same DNA damage repair axis with the risk of papillary thyroid carcinoma. In conclusion, our study reports the complex association between genetic variants of the ATM-CHEK2-BRCA1 axis and the predisposition to papillary thyroid carcinoma. Furthermore, the study supports previous findings regarding the influence of age and gender on the clinical outcome of the disease and shows that this effect is significantly altered by the minor alleles of the analyzed genes, emphasizing their importance in the pathogenesis of PTC.
Acknowledgments
Supported by: Foundation for Polish Science, Programme FOCUS and Programme TEAM, co-financed by the European Union Regional Development Fund. The Laboratory of Genomic Medicine of the Medical University of Warsaw participates in Bastion a Program funded by the European Commission under the 7th Framework Program and by the Ministry of Science and Higher Education, Grant numbers: FP7-REGPOT-2012-CT2012-316254-BASTION; NCI, Grant numbers: P30 CA16058, P01 CA124570.
References
- Adjadj E, Schlumberger M, de Vathaire F. Germ-line DNA polymorphisms and susceptibility to differentiated thyroid cancer. Lancet Oncol. 2009;10:181–190. doi: 10.1016/S1470-2045(09)70020-8. [DOI] [PubMed] [Google Scholar]
- Akulevich NM, Saenko VA, Rogounovitch TI, Drozd VM, Lushnikov EF, Ivanov VK, Mitsutake N, Kominami R, Yamashita S. Polymorphisms of DNA damage response genes in radiation-related and sporadic papillary thyroid carcinoma. Endocr Relat Cancer. 2009;16:491–503. doi: 10.1677/ERC-08-0336. [DOI] [PubMed] [Google Scholar]
- Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, Akslen LA, Armstrong BK, Avril MF, Azizi E, Bakker B, Bergman W, Bianchi-Scarra G, Bressac-de Paillerets B, Calista D, Cannon-Albright LA, Corda E, Cust AE, Debniak T, Duffy D, Dunning AM, Easton DF, Friedman E, Galan P, Ghiorzo P, Giles GG, Hansson J, Hocevar M, Hoiom V, Hopper JL, Ingvar C, Janssen B, Jenkins MA, Jonsson G, Kefford RF, Landi G, Landi MT, Lang J, Lubinski J, Mackie R, Malvehy J, Martin NG, Molven A, Montgomery GW, van Nieuwpoort FA, Novakovic S, Olsson H, Pastorino L, Puig S, Puig-Butille JA, Randerson-Moor J, Snowden H, Tuominen R, Van Belle P, van der Stoep N, Whiteman DC, Zelenika D, Han J, Fang S, Lee JE, Wei Q, Lathrop GM, Gillanders EM, Brown KM, Goldstein AM, Kanetsky PA, Mann GJ, Macgregor S, Elder DE, Amos CI, Hayward NK, Gruis NA, Demenais F, Bishop JA, Bishop DT. Genome-wide association study identifies three new melanoma susceptibility loci. Nat Genet. 2011;43:1108–1113. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Risch N. Discovering genotypes underlying human phenotypes: Past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33:228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- Brennan P, McKay J, Moore L, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chow WH, Rothman N, Chabrier A, Gaborieau V, Odefrey F, Southey M, Hashibe M, Hall J, Boffetta P, Peto J, Peto R, Hung RJ. Uncommon CHEK2 mis-sense variant and reduced risk of tobacco-related cancers: Case control study. Hum Mol Genet. 2007;16:1794–1801. doi: 10.1093/hmg/ddm127. [DOI] [PubMed] [Google Scholar]
- Canzian F, Amati P, Harach HR, Kraimps JL, Lesueur F, Barbier J, Levillain P, Romeo G, Bonneau D. A gene predisposing to familial thyroid tumors with cell oxyphilia maps to chromosome 19p13.2. Am J Hum Genet. 1998;63:1743–1748. doi: 10.1086/302164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski C, Gorski B, Huzarski T, Masojc B, Mierzejewski M, Debniak T, Teodorczyk U, Byrski T, Gronwald J, Matyjasik J, Zlowocka E, Lenner M, Grabowska E, Nej K, Castaneda J, Medrek K, Szymanska A, Szymanska J, Kurzawski G, Suchy J, Oszurek O, Witek A, Narod SA, Lubinski J. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75:1131–1135. doi: 10.1086/426403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasika GK, Lin SC, Zhao S, Sung P, Tomkinson A, Lee EY. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- de la Chapelle A, Jazdzewski K. MicroRNAs in thyroid cancer. J Clin Endocrinol Metab. 2011;96:3326–3336. doi: 10.1210/jc.2011-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufault MR, Betz B, Wappenschmidt B, Hofmann W, Bandick K, Golla A, Pietschmann A, Nestle-Kramling C, Rhiem K, Huttner C, von Lindern C, Dall P, Kiechle M, Untch M, Jonat W, Meindl A, Scherneck S, Niederacher D, Schmutzler RK, Arnold N. Limited relevance of the CHEK2 gene in hereditary breast cancer. Int J Cancer. 2004;110:320–325. doi: 10.1002/ijc.20073. [DOI] [PubMed] [Google Scholar]
- Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, AT . AJCC Cancer Staging Manual. 7. New York, NY: Springer; 2010. [Google Scholar]
- Falck J, Lukas C, Protopopova M, Lukas J, Selivanova G, Bartek J. Functional impact of concomitant versus alternative defects in the Chk2-p53 tumour suppressor pathway. Oncogene. 2001;20:5503–5510. doi: 10.1038/sj.onc.1204811. [DOI] [PubMed] [Google Scholar]
- Frich L, Glattre E, Akslen LA. Familial occurrence of nonmedullary thyroid cancer: A population-based study of 5673 first-degree relatives of thyroid cancer patients from Norway. Cancer Epidemiol Biomarkers Prev. 2001;10:113–117. [PubMed] [Google Scholar]
- Goggins W, Daniels GH, Tsao H. Elevation of thyroid cancer risk among cutaneous melanoma survivors. Int J Cancer. 2006;118:185–188. doi: 10.1002/ijc.21300. [DOI] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst. 1994;86:1600–1608. doi: 10.1093/jnci/86.21.1600. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Sigurdsson A, Bergthorsson JT, He H, Blondal T, Geller F, Jakobsdottir M, Magnusdottir DN, Matthiasdottir S, Stacey SN, Skarphedinsson OB, Helgadottir H, Li W, Nagy R, Aguillo E, Faure E, Prats E, Saez B, Martinez M, Eyjolfsson GI, Bjornsdottir US, Holm H, Kristjansson K, Frigge ML, Kristvinsson H, Gulcher JR, Jonsson T, Rafnar T, Hjartarsson H, Mayordomo JI, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Kong A, Stefansson K. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet. 2009;41:460–464. doi: 10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Masson G, He H, Jonasdottir A, Sigurdsson A, Stacey SN, Johannsdottir H, Helgadottir HT, Li W, Nagy R, Ringel MD, Kloos RT, de Visser MC, Plantinga TS, den Heijer M, Aguillo E, Panadero A, Prats E, Garcia-Castano A, De Juan A, Rivera F, Walters GB, Bjarnason H, Tryggvadottir L, Eyjolfsson GI, Bjornsdottir US, Holm H, Olafsson I, Kristjansson K, Kristvinsson H, Magnusson OT, Thorleifsson G, Gulcher JR, Kong A, Kiemeney LA, Jonsson T, Hjartarson H, Mayordomo JI, Netea-Maier RT, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Rafnar T, Stefansson K. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat Genet. 2012;44:319–322. doi: 10.1038/ng.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Nagy R, Liyanarachchi S, Jiao H, Li W, Suster S, Kere J, de la Chapelle A. A susceptibility locus for papillary thyroid carcinoma on chromosome 8q24. Cancer Res. 2009;69:625–631. doi: 10.1158/0008-5472.CAN-08-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlston RS, Peto J. The search for low-penetrance cancer susceptibility alleles. Oncogene. 2004;23:6471–6476. doi: 10.1038/sj.onc.1207951. [DOI] [PubMed] [Google Scholar]
- Jazdzewski K, de la Chapelle A. Genomic sequence matters: A SNP in microRNA-146a can turn anti-apoptotic. Cell Cycle. 2009;8:1642–1643. [PMC free article] [PubMed] [Google Scholar]
- Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, de la Chapelle A. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci USA. 2009;106:1502–1505. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonklaas J, Nogueras-Gonzalez G, Munsell M, Litofsky D, Ain KB, Bigos ST, Brierley JD, Cooper DS, Haugen BR, Ladenson PW, Magner J, Robbins J, Ross DS, Skarulis MC, Steward DL, Maxon HR, Sherman SI. The impact of age and gender on papillary thyroid cancer survival. J Clin Endocrinol Metab. 2012;97:E878–E887. doi: 10.1210/jc.2011-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman B, Laitman Y, Gronwald J, Winqvist R, Irmejs A, Lubinski J, Pylkas K, Gardovskis J, Miklasevics E, Friedman E. Haplotypes of the I157T CHEK2 germline mutation in ethnically diverse populations. Fam Cancer. 2009;8:473–478. doi: 10.1007/s10689-009-9269-1. [DOI] [PubMed] [Google Scholar]
- Kilpivaara O, Vahteristo P, Falck J, Syrjakoski K, Eerola H, Easton D, Bartkova J, Lukas J, Heikkila P, Aittomaki K, Holli K, Blomqvist C, Kallioniemi OP, Bartek J, Nevanlinna H. CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer. 2004;111:543–547. doi: 10.1002/ijc.20299. [DOI] [PubMed] [Google Scholar]
- Kilpivaara O, Alhopuro P, Vahteristo P, Aaltonen LA, Nevanlinna H. CHEK2 I157T associates with familial and sporadic colorectal cancer. J Med Genet. 2006;43:e34. doi: 10.1136/jmg.2005.038331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Parks CG, Xu Z, Carswell G, DeRoo LA, Sandler DP, Taylor JA. Association between genetic variants in DNA and histone methylation and telomere length. PLoS ONE. 2012;7:e40504. doi: 10.1371/journal.pone.0040504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa I, Robledo M. Association studies in thyroid cancer susceptibility: Are we on the right track? J Mol Endocrinol. 2011;47:R43–R58. doi: 10.1530/JME-11-0005. [DOI] [PubMed] [Google Scholar]
- Lavin MF. Ataxia-telangiectasia: From a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- Leongamornlert D, Mahmud N, Tymrakiewicz M, Saunders E, Dadaev T, Castro E, Goh C, Govindasami K, Guy M, O’Brien L, Sawyer E, Hall A, Wilkinson R, Easton D, Goldgar D, Eeles R, Kote-Jarai Z. Germline BRCA1 mutations increase prostate cancer risk. Br J Cancer. 2012;106:1697–1701. doi: 10.1038/bjc.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell. 2002;9:1045–1054. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- Liyanarachchi S, Wojcicka A, Li W, Czetwertynska M, Stachlewska E, Nagy R, Hoag K, Wen B, Ploski R, Ringel MD, Kozlowicz-Gudzinska I, Gierlikowski W, Jazdzewski K, He H, de la Chapelle A. Cumulative risk impact of five genetic variants associated with papillary thyroid carcinoma. Thyroid. 2013 doi: 10.1089/thy.2013.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machens A, Hauptmann S, Dralle H. Disparities between male and female patients with thyroid cancers: Sex difference or gender divide? Clin Endocrinol (Oxf) 2006;65:500–505. doi: 10.1111/j.1365-2265.2006.02623.x. [DOI] [PubMed] [Google Scholar]
- Mazzaferri EL, Jhiang SM. Long-term impact of initial surgical and medical therapy on papillary and follicular thyroid cancer. Am J Med. 1994;97:418–428. doi: 10.1016/0002-9343(94)90321-2. [DOI] [PubMed] [Google Scholar]
- McKay JD, Lesueur F, Jonard L, Pastore A, Williamson J, Hoffman L, Burgess J, Duffield A, Papotti M, Stark M, Sobol H, Maes B, Murat A, Kaariainen H, Bertholon-Gregoire M, Zini M, Rossing MA, Toubert ME, Bonichon F, Cavarec M, Bernard AM, Boneu A, Leprat F, Haas O, Lasset C, Schlumberger M, Canzian F, Goldgar DE, Romeo G. Localization of a susceptibility gene for familial nonmedullary thyroid carcinoma to chromosome 2q21. Am J Hum Genet. 2001;69:440–446. doi: 10.1086/321979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MF, Sellick GS, Webb EL, Catovsky D, Houlston RS. Variants in the ATM-BRCA2-CHEK2 axis predispose to chronic lymphocytic leukemia. Blood. 2006;108:638–644. doi: 10.1182/blood-2005-12-5022. [DOI] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Sigurdson AJ, Hauptmann M, Alexander BH, Doody MM, Thomas CB, Struewing JP, Jones IM. DNA damage among thyroid cancer and multiple cancer cases, controls, and long-lived individuals. Mutat Res. 2005;586:173–188. doi: 10.1016/j.mrgentox.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Swierniak M, Wojcicka A, Czetwertynska M, Stachlewska E, Maciag M, Wiechno W, Gornicka B, Bogdanska M, Koperski L, de la Chapelle A, Jazdzewski K. In-depth characterization of the microrna transcriptome in normal thyroid and papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013;98:E1401–E1409. doi: 10.1210/jc.2013-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LH, Choong D, Johnson SA, Campbell IG. Genetic and epigenetic analysis of CHEK2 in sporadic breast, colon, and ovarian cancers. Clin Cancer Res. 2006;12:6967–6972. doi: 10.1158/1078-0432.CCR-06-1770. [DOI] [PubMed] [Google Scholar]
- Xu L, Doan PC, Wei Q, Liu Y, Li G, Sturgis EM. Association of BRCA1 functional single nucleotide polymorphisms with risk of differentiated thyroid carcinoma. Thyroid. 2012;22:35–43. doi: 10.1089/thy.2011.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]