Abstract
Motivation: Large-scale phenotyping projects such as the Sanger Mouse Genetics project are ongoing efforts to help identify the influences of genes and their modification on phenotypes. Gene–phenotype relations are crucial to the improvement of our understanding of human heritable diseases as well as the development of drugs. However, given that there are ∼20 000 genes in higher vertebrate genomes and the experimental verification of gene–phenotype relations requires a lot of resources, methods are needed that determine good candidates for testing.
Results: In this study, we applied an association rule mining approach to the identification of promising secondary phenotype candidates. The predictions rely on a large gene–phenotype annotation set that is used to find occurrence patterns of phenotypes. Applying an association rule mining approach, we could identify 1967 secondary phenotype hypotheses that cover 244 genes and 136 phenotypes. Using two automated and one manual evaluation strategies, we demonstrate that the secondary phenotype candidates possess biological relevance to the genes they are predicted for. From the results we conclude that the predicted secondary phenotypes constitute good candidates to be experimentally tested and confirmed.
Availability: The secondary phenotype candidates can be browsed through at http://www.sanger.ac.uk/resources/databases/phenodigm/gene/secondaryphenotype/list.
Contact: ao5@sanger.ac.uk or ds5@sanger.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
A causative gene has not yet been identified for almost half of the existing human heritable diseases (Schofield et al., 2012). Without the knowledge of the molecular basis of a disease, treatment possibilities are limited to treating symptoms instead of curing the underlying defects. In order to be able to find cures and prevention mechanisms for human genetic disorders, we need to comprehensively understand how each disease originates and progresses over time. A collection of human diseases together with confirmed and speculative causes is available from resources such as the Online Mendelian Inheritance in Man (OMIM) (Amberger et al., 2011) or Orphanet (Aymé, 2003) database.
In the quest for identifying causative genes for human genetic disorders, model organisms have gained increasing importance due to the opportunities arising from targeted gene modifications. For example, the mouse shares 99% of genes with humans, and gene modifications leading to phenotypes characteristic for a disease may offer clues to the origins of this disease (Rosenthal and Brown, 2007). Experimental results of mutagenesis experiments are stored in species-specific Model Organism Database (MOD)s (Leonelli and Ankeny, 2012), e.g. the Sanger Mouse Genetics Project (Sanger-MGP) (White et al., 2013), WormBase (Yook et al., 2012), the Mouse Genome Database (MGD) (Bult et al., 2012) or FlyBase (Drysdale and FlyBase Consortium, 2008).
The Sanger-MGP is part of the International Mouse Phenotyping Consortium (IMPC) project that aims to identify the phenotypic implications of 20 000 genes by 2021 (Brown and Moore, 2012). In the framework of this project genetically modified mouse models are assessed according to 20 pre-defined standard operating procedures (SOPs) that are linked to measurable physical parameters to ascertain the implications of genetic mutations on phenotypes (Mallon et al., 2012). An example of a SOP is the assessment of the grip strength of mice at the age of 9 weeks to assess their neuromuscular function as muscle strength (https://www.mousephenotype.org/impress/protocol/83/7). Mammalian Phenotype Ontology (MP) (Smith and Eppig, 2009) annotations are assigned using a reference range method followed by an expert review. Later studies explore the application of other statistical methods to assign MP phenotype annotations based on the obtained parameter readings (Beck et al., 2009; Karp et al., 2012a), however, these methods only cover a subset of the phenotypes covered by the 20 SOPs.
The process of assessing physical measurements in accordance with the 20 pre-defined SOPs is referred to as primary phenotyping (Justice, 2008). According to the European Mouse Disease Clinic (EUMODIC) web page (http://www.eumodic.org/) “A distributed network of centres with in depth expertise in a number of phenotyping domains will undertake more complex, secondary phenotyping screens and apply them to a subset of the mice which have shown interesting phenotypes in the primary screen.”. However, with the increasing amount of genes being assessed in the primary phenotyping screen, a manual investigation for interesting results from the primary screens becomes impossible. In addition, the manual assessment of experimental results is time consuming, expensive and requires trained biologists. Therefore, automated methods enabling the search for promising secondary phenotypes are needed to complement the results obtained from the primary screens.
Existing automated solutions include the prediction of phenotypes based on orthologous genes (Groth et al., 2007; McGary et al., 2010) as well as functional annotations of genes (King et al., 2003). However, orthologous genes do not necessarily exhibit the same function or expression patterns across different species and therefore, do not always provide reliable answers. A solution relying only on existing phenotype annotations could overcome the problem in differing gene function and resulting phenotypes across different species. To the best of our knowledge, the prediction of secondary phenotypes from primary screen annotations in combination with literature-curated phenotypes in mouse has not been addressed before.
Here, we present an association rule mining approach that enables the identification of potential secondary phenotype screens in mouse using data from MGD, and complementing the primary phenotype screens in Sanger-MGP. Applying association rule mining, we were able to discover 188 rules, covering 242 phenotypes and leading to 1967 predictions for secondary phenotypes for 244 genes contained in the Sanger-MGP database. These 1967 suggested gene–phenotype associations include 136 unique phenotypes for which new assays can be defined for. The predicted associations are neither contained in MGD nor Sanger-MGP. We automatically as well as manually evaluated the secondary phenotype predictions and can demonstrate that our results show viable candidates. In conclusion, we believe that novel biological hypotheses and secondary phenotype screens can be formulated from the predicted secondary phenotypes.
2 METHODS
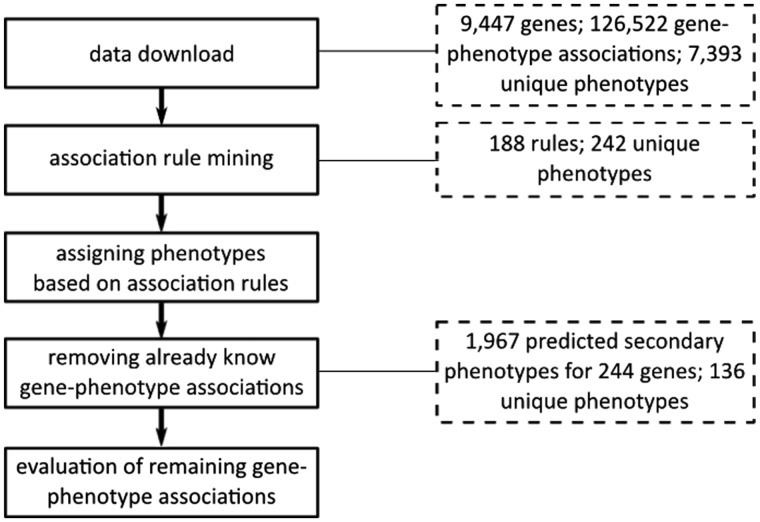
Figure 1 illustrates the overall workflow of this study. The following subsections describe the prediction approach and the utilized datasets in detail.
Fig. 1.
Overall workflow of the study. After determining-related phenotypes, the primary phenotype annotations assigned to genes in Sanger-MGP are enriched with potentially related phenotypes. The additional, predicted secondary phenotypes are evaluated in several steps
2.1 Prediction of secondary phenotype candidates for mouse genes
To determine candidates for secondary phenotyping, we first analysed MGD’s phenotype annotations for mouse mutants. We hypothesized that phenotypes that significantly co-occur with each other more often than expected by chance, given the overall amount of phenotype annotations, constitute good candidates for the secondary phenotype experiments. For example, it is known that body weight correlates with bone density or grip strength and changes in body weight often lead to changes in the correlated phenotypes (Karp et al., 2012a, b; Valdar et al., 2006). Using a large dataset of phenotype annotations, we can determine pairs of phenotypes that may be biologically linked.
Association rule mining was originally used to find patterns of items that are frequently purchased together in one transaction in a supermarket. Each association rule assigns a probability to an implication based on the dataset, e.g. how likely is it that someone who bought bread and milk also purchased butter. In the Bioinformatics domain, association rule mining has been previously successfully applied to large annotation sets with the aim to find relationships between gene functions described with Gene Ontology (GO) (Botstein et al., 2000; Kumar et al., 2004; Manda et al., 2012). As the goal of determining significantly co-occurring concepts to define relationships is the same here, association rule mining can also be applied. We used the apriori (http://www.borgelt.net/doc/apriori/apriori.html) software implementation (Agrawal et al., 1996; Borgelt, 2003) with the following parameter settings:
-tr -s-6 -m2 -n2 -c90 -ep -v ‘‘%e’’
with -tr to enforce the output of association rules instead of item sets, -s-6 to obtain only rules that are supported by at least six item sets, -m2 to include only rules with a minimum of two items, -n2 to include only rules with a maximum of two items, -c90 to only allow rules with a confidence of 90%, -ep to provide P-values for each rule and -v “%e” to add the P-value separated by space to each of the rules. The input to the apriori software was the set of literature-curated phenotype annotations of mouse genes and the output of rules of the type phenotype_1 → phenotype_2. As a starting point, we limited the output to rules including only two phenotypes to avoid complex dependencies between the annotations. However, in future work we aim to extend the approach to address more complex dependencies between phenotypes.
The two parameters that are used to narrow down the associations’ rules to obtain meaningful, biologically related phenotypes, are support and confidence. We set the support for association rules to six which means that a minimum of six genes have to be annotated with both phenotype_1 and phenotype_2. The confidence corresponds to the ratio of genes being annotated with phenotype_1 as well as phenotype_2 over the genes that are only annotated with phenotype_1. In our case, at least 90% of the genes annotated with phenotype_1 must have also phenotype_2 as annotation in order for this rule to be reported. Changing either parameter may lead to the report of different association rules in the output. We considered this to be conservative settings for an initial study, and the determination of the ideal settings for both parameters is subject to future work.
Rules are returned together with their corresponding P-value to enable potential further filtering and user confidence, e.g.
MP:0004725 <- MP:0009448 0
MP:0005606 <- MP:0009448 3.9905e-212
MP:0005606 <- MP:0009557 4.22726e-182
MP:0000245 <- MP:0011171 2.64518e-125
All extracted rules are then sorted according to the phenotypes including them, i.e. one phenotype may potentially be associated with more than one secondary phenotype candidate. As shown by the rules given before as an example, a decreased platelet ATP level phenotype (MP:0009448) would be associated with an increased bleeding time phenotypes (MP:0005606) and a decreased platelet serotonin level (MP:0004725). Following this procedure will lead to a list of mapped phenotypes including the phenotypes from the high-throughput assessment in the primary phenotype screening defined in the SOPs. Therefore, the mapped phenotypes are then filtered to exclude the phenotypes covered by the Sanger-MGP SOPs. Because of this filtering, we obtain only predictions for secondary phenotypes that have not been included in the primary screens.
For all the genes contained in Sanger-MGP, we then generated a list of secondary phenotype annotation predictions by going through all the existing phenotype annotations for a gene and adding those phenotypes that have been mapped based on co-occurrence. Then we removed all gene–phenotype associations that have been identified already and are contained in either MGD or Sanger-MGP to only generate potentially novel links between genes and phenotypes. We refer to the remaining phenotype annotations as predicted secondary phenotypes.
We chose the MGD phenotype annotations for gene knockouts as basis for our predictions and downloaded the report file on July 20, 2013. The downloaded file comprised 126 522 MP annotations for 9447 genes, covering 7393 unique MP concepts. The Sanger-MGP covers 20 SOPs that correspond to 367 MP annotations. Deducting the 367 MP that are covered by the SOPs from the unique number of MP concepts in MGD, provides the target phenotype annotation space. This means that 7027 unique phenotype concepts can be potentially associated with any of the 725 genes that had been assessed by the primary screens in the Sanger-MGP at the time this study was conducted. All the phenotype annotation datasets were applied without conducting a taxonomic closure on the annotations. However, once the secondary phenotypes have been predicted, corresponding assays would have to be determined to test the generated phenotype hypotheses.
2.2 Evaluation of secondary phenotype predictions
We evaluated the secondary phenotype predictions automatically as well as manually. The automated evaluation was realized by applying the secondary phenotype candidates in two use cases for phenotype annotations: the clustering of genes according to phenotypes leading to clusters of gene function, and the prediction of disease gene candidates by comparing disease phenotypes with phenotypes that have been determined to be affected by a gene mutation. In both use cases, we applied first phenotype annotations determined during the primary screens, and after that a combination of the primary screen annotations together with the predictions for secondary phenotypes. We assume that if the performance improves when adding the secondary phenotype predictions, the secondary phenotypes possess biological validity. In addition, we manually investigated five diseases further where the predictability of at least one known causative gene improved. More information about the evaluation of the secondary phenotypes is provided in the following subsections.
2.2.1 Automated evaluation based on gene function
In previous studies, it has been demonstrated that phenotype annotations can be used to determine biologically meaningful clusters with respect to gene function and protein interactions (van Driel et al., 2006). Oti et al. extended the method to validate the content of three human phenotype databases with respect to consistency and completeness. We assume that the secondary phenotype predictions once added to the annotations assigned in the primary screens improve consistency and completeness of the phenotype data. Therefore, we applied the method introduced by Oti et al. relying on the biological coherence of gene clusters built on phenotype similarity. The biological coherence is calculated based on the overlap of GO annotations among all the genes falling into one cluster.
To assess the biological coherence without and with the predicted secondary phenotypes, we generated gene clusters based on the primary phenotypes solely, as well as clusters based on the primary and secondary phenotype data in conjunction. Before the actual clustering step, we performed a taxonomic closure based on MP, which means that all superclasses for each assigned phenotype annotation were added to a gene’s phenotype annotation set. Clusters were formed based on the phenotype similarities, and the similarity between pairs of genes based on their phenotype annotations using a Jaccard coefficient (the ratio of shared phenotypes over the unique set of phenotypes assigned to both the genes). Genes were clustered with respect to their phenotype similarity using average linkage clustering. Clusters were determined by applying the Dynamic Treecut package in R (Langfelder et al., 2008) to the obtained dendrogram. We set the parameters of the Dynamic Treecut package to require a minimum of two genes falling into one cluster. For each of the determined cluster, the biological coherence was calculated with
| (1) |
where C(i,j) is the term overlap between gene i and gene j, and n is the number of genes in this cluster. The overall biological coherence score for all clusters is obtained by averaging the individual scores for the clusters:
| (2) |
where m is the number of clusters formed for a particular dataset.
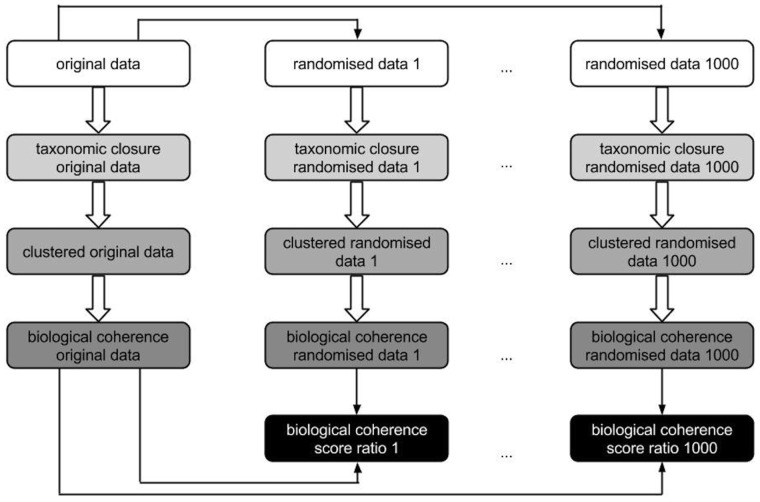
In compliance with the method described in (Oti et al., 2009), the datasets are not directly compared, instead they are compared to randomized datasets to correct for gene annotation biases. For this purpose, we randomized each of the two phenotype annotation sets—primary and secondary—maintaining the number and uniqueness of phenotype annotations per gene. We randomized the original set of annotations 1000 times leading to 2002 phenotype annotation sets in total. We increased the number of randomizations from 30 to 1000 to compensate for the fact that Sanger-MGP contains a number of genes that are poorly described in terms of gene function and may generate a high variation of coherence score ratios otherwise. For each phenotype annotation set, the ratio of the overall biological coherence Ct of the respective original phenotype annotation set (either primary or secondary) over the randomized data is calculated. If this ratio is >1, the biological coherence of the original dataset is greater than the randomization data; vice versa for scores in the range [0,1], the biological coherence for the randomized data exceeds the coherence of the respective original dataset. The ratio scores are then summarized in box plots and the difference between all the ratios of both datasets is calculated with a non-parametric, two-sided Wilcoxon rank-sum test implemented in R. Figure 2 illustrates this evaluation step.
Fig. 2.
Illustration of the calculation of biological coherence scores to evaluate secondary phenotype predictions. Boxes that possess the same background colour are based on the same analysis scripts, only the input data differ (either randomized or original data). Black boxes symbolize the ratio of the biological coherence original versus randomized data which are used as input for the box plots depicted in Figure 3
We assessed gene cluster coherence based on functional annotations of mouse genes. For this purpose, we downloaded the GO annotations of mouse genes from the MGD database on July 12, 2013 (ftp://ftp.informatics.jax.org/pub/reports/gene_association.mgi). The dataset comprised annotations for 25 499 MGD marker accession identifiers with 13 551 unique GO concepts, with an average of 11.64 GO annotations per gene. Using the original Sanger-MGP dataset with primary annotations only, we obtain 33 clusters for the 480 genes investigated that are then assessed for the biological coherence based on their gene function annotations.
2.2.2 Automated evaluation based on disease gene candidate predictions
Among other tools for disease gene candidate prediction, PhenoDigm uses phenotype annotations to predict gene candidates underlying a disease (Smedley et al., 2013). Disease gene candidates are predicted based on the primary phenotype annotations assigned to mouse and zebrafish models and their phenotypic similarity to human genetic disorders described in OMIM (http://www.human-phenotype-ontology.org/contao/index.php/downloads.html). The better the overlap of phenotypes between a model and a disease, the higher the corresponding knockout gene is ranked for this disease.
To assess the performance of a ranking algorithm, commonly Receiver Operating Characteristic (ROC) curves are used that are calculated based on a benchmark dataset. In our case, we used known gene–disease associations to assess the value of the predictions. If the secondary phenotypes add value to the predictions, then a performance increase should be visible from the Area Under Curve (AUC) of the two ROC curves (one for the predictions based on primary phenotype data, and one for the predictions based on primary and secondary phenotype data). To test whether the increase in the AUC is significant, we used a two-sided test for ROC curves available online (http://vassarstats.net/roc_comp.html; Hanley and McNeil, 1982).
Using PhenoDigm as an automated evaluation algorithm of the secondary phenotype predictions required a benchmark set of known gene–disease associations. If the secondary phenotype annotations improve the phenotypic overlap of genes and diseases, the ROC curves used for the evaluation should show an improvement. To generate the ROC curves, we used the gene–disease associations contained in OMIM’s MorbidMap file (http://omim.org/downloads), which was downloaded on July 20, 2013. This dataset comprised 3781 gene–disease associations, including 2530 genes and 3158 diseases.
2.2.3 Manual evaluation
To evaluate some of the secondary phenotype predictions, we manually investigated some of the cases were the predictability of known disease genes improved when adding the predicted secondary phenotypes. We chose five gene–disease associations where the gene improved with respect to its rank for the disease and looked based on which annotations the match between disease and gene could be made. The information concerning the matched phenotype annotations of a disease and a gene is contained in Supplementary Material S1.
2.3 Implementation of PhenoDigm extension to provide secondary phenotype predictions online
To enable access to the predicted secondary phenotypes, we implemented an extension to our online tool PhenoDigm (Smedley et al., 2013) that predicts causative genes for human heritable disorders. The extension is, as well as the original tool, implemented using the Play! Framework (http://www.playframework.com/) (version 1.2.5), jQuery (http://jquery.com/) (version 1.6.4) and jQuery UI (http://jqueryui.com/) (version 1.9.1). The secondary phenotype predictions were imported into PhenoDigm’s underlying MySQL database (http://www.mysql.com/) by extending the database schema. However, secondary phenotype predictions are not incorporated into PhenoDigm’s disease gene candidate predictions available from the web page, unless experimentally confirmed and integrated into one of the phenotype annotation databases.
3 RESULTS
Applying association rule mining, we were able to identify 188 rules (provided in Supplementary Material S1) that lead to secondary phenotype hypotheses for 244 Sanger-MGP genes. In total, we could predict 1967 novel gene–phenotype associations containing 136 unique phenotypes that are not contained in MGD. Out of these 1967 gene–phenotype relationships, 47 were covered by the taxonomy of the ontology, i.e. the predicted phenotypes were ancestor concepts of annotations already used for one particular gene. The 136 unique phenotype concepts span 23 of MGD’s 30 top level phenotypes, such as tumorigenesis (MP:0002006), nervous system phenotype (MP:0003631) or muscle phenotype (MP:0005369), showing the diversity of phenotypes that could be added to the annotation of genes. The 136 phenotypes also cover different hierarchy levels in the ontology spanning from the second to the 11th level, with the highest group falling into level 6 (all measured as shortest distance from the root node of the MP ontology). For example, adenohypophysis hypoplasia (MP:0008365) as well as abnormal cranium size (MP:0010031) are suggested as secondary phenotypes. In general, the deeper an ontology term is the more specific is the concept it is representing. This means that the predictions not only span a variety of different high level phenotypes but also add detailed information to the genes they are associated with which allows for a better characterization of individual genes. Supplementary Material S1 provides all the predicted secondary phenotype annotations together with additional information such as the high level phenotype, term name and frequency of occurrence in the prediction dataset.
3.1 Predicted secondary phenotypes significantly improve the biological coherence of gene clusters
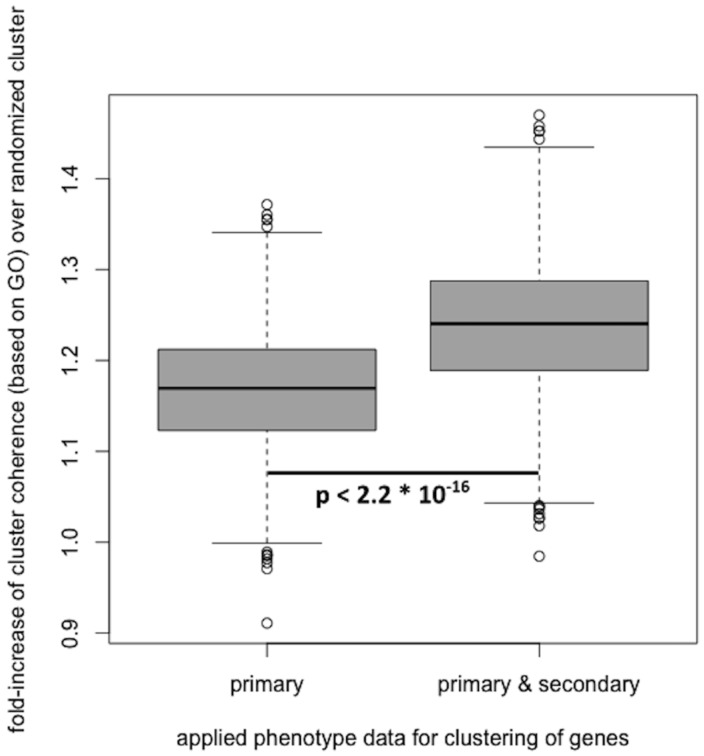
In recent studies, phenotypes have successfully been applied to determine disease gene candidates and gene function (Smedley et al., 2013; van Driel et al., 2006). In order to asses the validity and quality of the predicted secondary phenotype annotations, we assessed the biological coherence of gene clusters, built based on phenotype similarity between genes. Applying the method described by Oti et al. first to the primary phenotype annotations only, and then to both primary and predicted secondary phenotype annotations, shows that the biological coherence of clusters increases when adding the predicted secondary phenotype annotations. The obtained results are depicted in Figure 3.
Fig. 3.
Adding the predicted secondary phenotype annotation to the Sanger-MGP genes with reference range annotations and using these to create gene clusters based on phenotype similarity, improves the biological coherence of the obtained gene clusters
In addition to calculating the biological coherence for both datasets, we determined the significance of the fold-increase of the coherence of the clusters. Using a two-sided Wilcoxon signed-rank test (as implemented in R, α = 0.05), we obtained a P-value of 2.2 × 10−16, indicating a significant improvement when adding secondary phenotype annotations to the previously confirmed in the primary phenotype scans. These results suggest that the predicted secondary phenotypes possess biological validity but will have to be experimentally verified in secondary phenotype screens.
3.2 Secondary phenotypes significantly improve the predictability of disease gene candidates
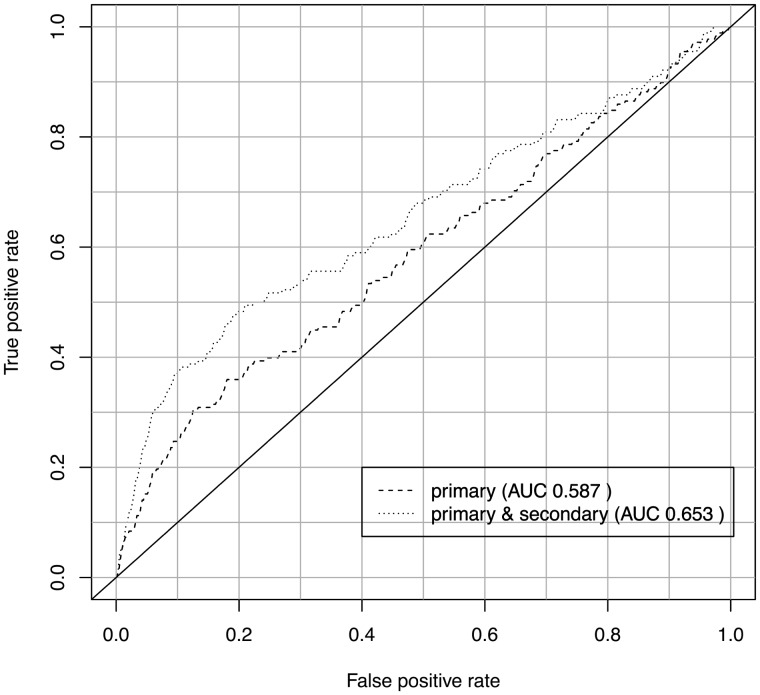
In addition to assessing the biological coherence of gene clusters, we also verified the predicted secondary phenotype annotations by applying them in a second application use case: the prediction of disease gene candidates. One tool that already uses the primary phenotype data of genes to predict disease gene candidates is PhenoDigm (Smedley et al., 2013). To assess whether the secondary phenotypes possess biological validity, we first used only the primary annotations to predict disease gene candidates and then added the secondary phenotype annotations. For both predictions, we calculated the ROC curves, using gene–disease associations contained in MGD as benchmark dataset. Both the obtained ROC curves are depicted in Figure 4. Applying a one-tailed Student’s t-test (α = 0.05) to the ROC curves, we obtain a P-value of P = 0.02.
Fig. 4.
Accumulating the predicted secondary phenotypes together with reference range annotations for Sanger-MGP genes improves the predictability of causative disease genes using PhenoDigm
3.3 The Coq9 mouse improves as a model for primary coenzyme Q10 deficiency 5
To further assess the value added by the predicted secondary phenotypes, we manually assessed diseases that show rank improvements for known causative genes. We determined the number and particular phenotypes that could be matched between models and diseases, where causative genes improved in the ranking as disease candidates. In the best case, the gene Coq9 (MGI:1915164) that has been recognized as a being disrupted in cases of Primary coenzyme Q10 defiency 5 (COQ10D5; MIM:#614654) improves from rank 181 to rank 2 based on the secondary phenotype annotations. Using the annotations assigned in the primary screens, only one pair of matching phenotypes can be determined: Hyperreflexia (HP:0001347) and hyperactivity (MP:0001399). Applying in addition the predicted secondary phenotypes, other signs and symptoms of this disease, such as Postnatal microcephaly (HP:0005484) and Left ventricular hypertrophy (HP:0001712), are detected.
Another example for gene rank improvement is that the Cfh (MGI:88385) gene was ranked in second place after adding the predicted secondary phenotypes (rank 43 when using only primary phenotypes) for Complement factor H deficiency (MIM:#609814). Including the predicted secondary phenotypes allows for a coverage of the following additional phenotypes: Thickening of the glomerular basement membrane (HP:0004722), Progressive renal insufficiency (HP:0000106) and Hematuria (HP:0000790). Interestingly, the Cfh gene is not only associated with Complement factor H deficiency but also with Atypical hemolytic uremic syndrome 1 (MIM:#235400), and adding the secondary phenotype information, the gene also obtained an improved rank for this disease (rank 177 with primary phenotypes only and rank 41 with inserting secondary phenotype information). From the improvement in both cases, we conjecture that the secondary phenotypes cover correct functional aspects of the gene that could not have been identified with the primary phenotype screens.
This information together with additional information for another three diseases and their respective genes is provided in Supplementary Material S1.
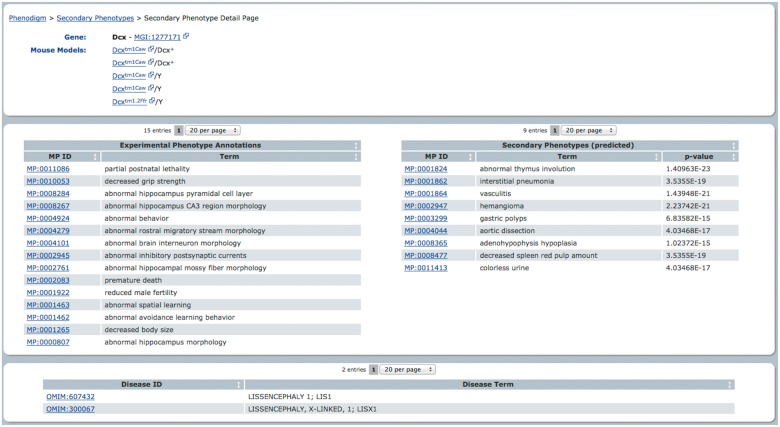
3.4 Browsing the secondary phenotype predictions online
To provide access to the secondary phenotype predictions, we implemented an extension to our disease gene candidate prediction tool PhenoDigm (Smedley et al., 2013). The results can be browsed at http://www.sanger.ac.uk/resources/databases/phenodigm/. The web interface provides all the genes that possess secondary phenotype candidates as a list and the user can select individual genes for further investigation. Upon selecting a gene, the user is provided with all the details available for this gene, i.e. diseases the gene has been confirmed to be a cause for, phenotype annotations from the primary screens, the literature-curated annotations from MGD and the suggested phenotypes for secondary screens. Providing this information together, a biologist or clinician could easily assess whether the secondary phenotype candidates are worthwhile to be tested in a biological experiment. Figure 5 provides a snapshot of the available gene-centric information through PhenoDigm’s web interface.
Fig. 5.
An extension of PhenoDigm’s web interface holds the secondary phenotype predictions
4 DISCUSSION
In this study, we applied an association rule mining approach to mine secondary phenotypes and computationally verify their biological validity using two automated and a limited manual evaluation. We used the manually assigned phenotype annotations contained in MGD for learning phenotype co-occurrence patterns and merged the identified patterns with phenotypes that were experimentally verified in primary screens and are available from the Sanger-MGP (White et al., 2013).
Using data from particular resources creates dependencies towards those data resources. For example, annotation guidelines such as those employed by MGD ensure a consistency of human annotators but can also create artefacts in the predictions generated from the data. In our particular case, we may find phenotype co-occurrence patterns that are intrinsic to annotation guidelines and not purely due to their co-occurrence. This may occur when the annotation guidelines cover rules that enforce a set of phenotypes to be annotated in particular circumstances instead of only one. However, as these guidelines exist to ensure biological correctness of the data, we expect that those cases still constitute biologically interesting, though known connections between individual phenotypes.
The implementation of the secondary phenotype prediction pipeline relies solely on an association rule mining approach. Using the pipeline in conjunction with 7027 unique phenotypes (see Section 2.1), the obtained result of 188 new rules seems comparatively small. As the number of rules is directly related to the settings for the apriori software, the number of potential hypotheses may be increased by changing these parameters. However, and as with any prediction tool, we applied conservative measures that would reduce the likelihood of creating false hypotheses. In addition, starting with a small subset of rules enables better verification possibilities and selection mechanisms for biological experiment design. Potential areas of extension are the incorporation of additional pattern recognition methods that could then be used to form a support system and provide provenance for identified patterns, e.g. only predictions that are made by a number of systems are more likely to be secondary phenotypes.
Using phenotype patterns to generate secondary phenotype predictions implies that phenotypes that co-occur often with each other, are likely to always co-occur. For some biological phenomena this assumption has been validated, e.g. the correlation of body weight with bone density or blood calcium levels (Karp et al., 2012b). Given that the secondary phenotypes perform well in the evaluation, we feel that the assumption can still be used for forming secondary phenotype hypotheses. However, in future work we envisage a more complex filtering strategy for assigning phenotype annotations using not only primary phenotypes, but also gene function annotations and disease involvement to reduce the number of falsely associated genes and phenotypes in addition to voting from different prediction algorithms.
4.1 Predicted secondary phenotypes improve the biological coherence of the clusters
Using automated evaluation procedures that apply predictions in biological use cases may mask-specific problems that can only be spotted by human curators, e.g. if it is known that one particular gene does not cause a particular phenotype in particular circumstances. However, the methods can provide a summarized judgement over all the results instead of providing details for all cases. As demonstrated by van Driel et al., clustering genes according to phenotypes leads to clusters consistent with gene function and protein interaction networks. The same evaluation mechanism that has been applied here, had been successfully applied to assess the quality of existing human phenome databases (Oti et al., 2009). Using gene function annotation to determine biological coherence is directly influenced by the number of annotations available.
4.2 Secondary phenotypes improve the predictability of causative disease genes
In addition to the use case of gene characterization, phenotype annotations are applied to identify the underlying mechanisms of human heritable diseases (Hoehndorf et al., 2011; Robinson et al., 2013; Smedley et al., 2013; Washington et al., 2009). As the primary screens only cover 20 SOPs, the outcome of phenotype annotations is limited by the screens. Using the predicted secondary phenotype annotations may highlight phenotypes of genes that are currently limiting the predictability of certain diseases or groups of diseases. Our results show that adding the secondary phenotype annotations improve the predictability of disease genes and the characterization of genes on a phenotype level can be improved with the suggested phenotypes. However, experimental verification is necessary and assays would have to be incorporated to test for the 137 identified phenotypes.
4.3 Secondary phenotypes further characterize genes assessed with primary phenotyping
As illustrated with a small subset of selected diseases, adding the predicted secondary phenotypes leads to an increase of matched phenotypes between diseases and models. As discussed before, these results indicate that the predictions indeed possess biological validity and constitute good biological hypotheses to guide the design of experimental setups for secondary screens. The selection of diseases, however, was limited to the cases where an improvement for the causative gene happened. In future work, we will also have to extend our analysis to genes where no improvement or rank decrease was experienced. However, we note here that the rank changes of known causative disease genes are only indicators for performance changes and need manual investigation. Some of the OMIM diseases may possess multiple causative genes, some of which have not yet been discovered or listed in OMIM. As a consequence, those genes will be recognized as false positives during the evaluation. If one of the causative genes that have not been listed in OMIM improves tremendously over those genes that are listed, we could obtain a rank decrease for genes that are listed in OMIM.
4.4 Future development of the PhenoDigm extension
The data have been made available through a web interface that provides a gene-centric view on the data. All the predictions can be assessed and are provided in the context of diseases and information about the primary phenotype screens for easy verification and hypothesis derivation. As more data become available, e.g. through additional automated, statistical screens such as suggested by Karp et al., further information can be included such as effect size and additional phenotype annotations.
Furthermore, even though the annotations have been validated using the predicted secondary phenotype annotations in PhenoDigm’s disease prediction algorithm, the disease gene candidate predictions based on the secondary phenotype data are not yet available. A possible extension of the web interface in future work could be the inclusion of these predictions. If the predictions are included, possible new emerging disease groups relevant for a gene could be easily spotted from the list and guide new experiments.
5 CONCLUSION
Here, we presented a method to predict secondary phenotype candidates based on existing large-scale phenotype annotation sources and primary screens for genes. We verified the secondary phenotype candidates by applying it in two use cases and could demonstrate that the predictions add value in either use case and, therefore, seem biologically relevant to the genes they are predicted for. We could show that the phenotype candidates not only increase the biological coherence of gene clusters, but also improve the candidate prediction of genes for human heritable diseases. In conclusion, we provide a set of gene–phenotype associations that can be further assessed in biological experiments and guide the experimental design to further investigate specific genes or gene groups. All the data are freely available online from http://www.sanger.ac.uk/resources/databases/phenodigm/.
In future work, we aim to further improve the method by determining the best parameter settings for the association rule learning, but also investigate other phenotype co-occurrence pattern recognition methods. One possibility is the application of a hypergeometric distribution and find support for patterns that have been identified with the association rule mining approach. We further intend to provide update results through the web interface and improve the integration with other existing resources.
Acknowledgements
A.O.E. designed the experimental setup, implemented the secondary phenotype prediction pipeline, and executed part of the automated and manual evaluation. D.S. conducted the evaluation using PhenoDigm and IP contributed and verified the manual assessment of the prediction results. J.J. implemented the web pages required to provide the data online. All authors contributed and approved the final manuscript.
Funding: This work was supported by the Wellcome Trust grant [098051] and National Institutes of Health grant (NIH) [1 U54 HG006370-01].
Conflict of Interest: none declared.
References
- Agrawal R, et al. Fast discovery of association rules. In: Fayyad U, et al., editors. Advances in Knowledge Discovery and Data Mining. Menlo Park, California: AAAI Press; 1996. pp. 307–328. [Google Scholar]
- Amberger J, et al. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM®) Hum. Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- Aymé S. Orphanet, an information site on rare diseases. Soins. 2003;672:46–47. [PubMed] [Google Scholar]
- Beck T, et al. Practical application of ontologies to annotate and analyse large scale raw mouse phenotype data. BMC Bioinformatics. 2009;10(Suppl. 5):S2. doi: 10.1186/1471-2105-10-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgelt C. Workshop of Frequent Item Set Mining Implementations (FIMI 2003) 2003. Efficient implementations of apriori and eclat. [Google Scholar]
- Botstein D, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SDM, Moore MW. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm. Genome Off. J. Int Mamm. Genome Soc. 2012;23:632–640. doi: 10.1007/s00335-012-9427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, et al. The Mouse Genome Database: genotypes, phenotypes, and models of human disease. Nucleic Acids Res. 2012;41:885–891. doi: 10.1093/nar/gks1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale R, FlyBase Consortium FlyBase: a database for the Drosophila research community. Methods Mol. Biol. (Clifton, N.J.) 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. [DOI] [PubMed] [Google Scholar]
- Groth P, et al. PhenomicDB: a new cross-species genotype/phenotype resource. Nucleic Acids Res. 2007;35:D696–D699. doi: 10.1093/nar/gkl662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- Hoehndorf R, et al. PhenomeNET: a whole-phenome approach to disease gene discovery. Nucleic Acids Res. 2011;39:e119. doi: 10.1093/nar/gkr538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice MJ. Removing the cloak of invisibility: phenotyping the mouse. Dis. Models Mech. 2008;1:109–112. doi: 10.1242/dmm.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp NA, et al. Robust and sensitive analysis of mouse knockout phenotypes. PLoS One. 2012a;7:e52410. doi: 10.1371/journal.pone.0052410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp NA, et al. The fallacy of ratio correction to address confounding factors. Lab. Anim. 2012b;46:245–252. doi: 10.1258/la.2012.012003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, et al. Predicting phenotype from patterns of annotation. Bioinformatics (Oxford, England) 2003;19(Suppl. 1):i183–i189. doi: 10.1093/bioinformatics/btg1024. [DOI] [PubMed] [Google Scholar]
- Kumar A, et al. Proceedings of the 3rd International Workshop on Computational Terminology. 2004. Dependence relationships between Gene Ontology terms based on TIGR gene product annotations. [Google Scholar]
- Langfelder P, et al. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics (Oxford, England) 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- Leonelli S, Ankeny RA. Re-thinking organisms: the impact of databases on model organism biology. Stud. Hist. Philos. Biol. Biomed. Sci. 2012;43:29–36. doi: 10.1016/j.shpsc.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Mallon A-M, et al. Accessing data from the International Mouse Phenotyping Consortium: state of the art and future plans. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2012;23:641–652. doi: 10.1007/s00335-012-9428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manda P, et al. Cross-ontology multi-level association rule mining in the Gene Ontology. PLoS ONE. 2012;7:e47411. doi: 10.1371/journal.pone.0047411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary KL, et al. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc. Natl Acad.Sci. 2010;107:6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oti M, et al. The biological coherence of human phenome databases. Am. J. Hum. Genet. 2009;85:801–808. doi: 10.1016/j.ajhg.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson P, et al. Improved exome prioritization of disease genes through cross species phenotype comparison. Genome Res. 2014;24:340–348. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal N, Brown S. The mouse ascending: perspectives for human-disease models. Nat. Cell Biol. 2007;9:993–999. doi: 10.1038/ncb437. [DOI] [PubMed] [Google Scholar]
- Schofield PN, et al. Mouse genetic and phenotypic resources for human genetics. Hum. Mutat. 2012;33:826–836. doi: 10.1002/humu.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, et al. PhenoDigm: analyzing curated annotations to associate animal models with human diseases. Database J. Biol. Databases Curation. 2013;2013 doi: 10.1093/database/bat025. bat025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Eppig JT. The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009;1:390–399. doi: 10.1002/wsbm.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, et al. Genetic and environmental effects on complex traits in mice. Genetics. 2006;174:959–984. doi: 10.1534/genetics.106.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Driel MA, et al. A text-mining analysis of the human phenome. Eur. J. Hum. Genet. 2006;14:535–542. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
- Washington NL, et al. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7:e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook K, et al. WormBase 2012: more genomes, more data, new website. Nucleic Acids Res. 2012;40:D735–D741. doi: 10.1093/nar/gkr954. [DOI] [PMC free article] [PubMed] [Google Scholar]