Background: Helicase SUV3, polynucleotide phosphorylase (PNPase), or mitochondrial poly(A) polymerases (mtPAP) have individual activity in regulating mitochondrial mRNA (mt-mRNA)-polyadenylated (poly(A)) tails.
Results: SUV3 bridges PNPase and mtPAP to form a transient complex in modulating mt-mRNA poly(A) tail length depending on the mitochondrial matrix Pi level.
Conclusion: mt-mRNA poly(A) tail length is modulated by a SUV3·PNPase·mtPAP complex.
Significance: Mitochondrial energy status affects mt-mRNA poly(A) tail length.
Keywords: Bioenergetics, Mitochondria, Polyadenylation, RNA Helicase, RNA Processing, PNPase, SUV3, Mitochondrial mRNA Poly(A) tails, mtPAP
Abstract
Mammalian mitochondrial mRNA (mt-mRNA) transcripts are polyadenylated at the 3′ end with different lengths. The SUV3·PNPase complex and mtPAP have been shown to degrade and polyadenylate mt mRNA, respectively. How these two opposite actions are coordinated to modulate mt-mRNA poly(A) lengths is of interest to pursue. Here, we demonstrated that a fraction of the SUV3·PNPase complex interacts with mitochondrial polyadenylation polymerase (mtPAP) under low mitochondrial matrix inorganic phosphate (Pi) conditions. In vitro binding experiments using purified proteins suggested that SUV3 binds to mtPAP through the N-terminal region around amino acids 100–104, distinctive from the C-terminal region around amino acids 510–514 of SUV3 for PNPase binding. mtPAP does not interact with PNPase directly, and SUV3 served as a bridge capable of simultaneously binding with mtPAP and PNPase. The complex consists of a SUV3 dimer, a mtPAP dimer, and a PNPase trimer, based on the molecular sizing experiments. Mechanistically, SUV3 provides a robust single strand RNA binding domain to enhance the polyadenylation activity of mtPAP. Furthermore, purified SUV3·PNPase·mtPAP complex is capable of lengthening or shortening the RNA poly(A) tail lengths in low or high Pi/ATP ratios, respectively. Consistently, the poly(A) tail lengths of mt-mRNA transcripts can be lengthened or shortened by altering the mitochondrial matrix Pi levels via selective inhibition of the electron transport chain or ATP synthase, respectively. Taken together, these results suggested that SUV3·PNPase·mtPAP form a transient complex to modulate mt-mRNA poly(A) tail lengths in response to cellular energy changes.
Introduction
RNA polyadenylation is observed in almost all living organisms. In Escherichia coli, plant mitochondria and chloroplasts, transient polyadenylation of RNA transcripts serves as signal for rapid exonucleolytic degradation (1–4). In eukaryotic cells, the nuclear-encoded mRNA requires polyadenylation for stabilization, and subsequent nuclear export and translation initiation in the cytosol (5, 6). The poly(A) tail lengths for nuclear-encoded mRNAs can be as long as 200–300 nucleotides (nt)5 (7). In comparison, human mitochondrial mRNAs (mt-mRNAs) have relatively shorter poly(A) tails (∼50 nt), which are critical for the completion of functional translation stop codons for 7 of 13 mt-mRNAs (8, 9). Polyadenylation has also been shown to be involved in the editing of the 3′-acceptor region in mt-tRNAs (10, 11). Beyond this, the precise role of the human mitochondrial mRNA poly(A) tails remains unclear.
RNA polyadenylation is generally carried out by poly(A) polymerases (PAP), to incorporate ATP into poly(A) tails in a template-independent manner, using primarily ATP as a substrate (12). A mitochondrial specific poly(A) polymerase (mtPAP) has been shown to elongate single-stranded 3′ ends of mt-mRNA transcripts (13–16). Knocking down mtPAP by siRNA results in significant shortenings of the poly(A) tails of the mt-mRNA transcripts. However, the effect on mRNA steady-state levels seems to be transcript-specific (14, 15, 17).
On the other hand, removal of 3′ poly(A) tails of mt-mRNA is thought to be coupled to degradation of the entire transcript as in the E. coli model (18, 19). Polynucleotide phosphorylase (PNPase), a 3′ to 5′ phosphate-dependent exoribonuclease in E. coli RNA degradosome (20), has been implicated to be involved in mt-mRNA degradation and surveillance (21). Intriguingly, PNPase has also been shown to possess 3′ polyadenylation activity via the reverse reaction in E. coli and the chloroplasts (4, 14, 22, 23). PNPase also has multiple functions including RNA import in the mitochondrial intermembrane space (24, 25), and degradations of cytosolic mRNA (26) and microRNA (27). In summary, the roles of mtPAP and PNPase in human mt-mRNA poly(A) length regulation remain controversial and have to be further investigated.
Previously, we demonstrated that human mitochondrial helicase, SUV3, interacts with PNPase to form a heteropentameric complex with a molecular mass of 330 kDa, capable of degrading structured single-stranded RNA (ssRNA) substrates in the presence of Pi in vitro (28, 29). Inactivation of SUV3 in both yeast and human mitochondria leads to accumulation of aberrant RNA molecules (30–32). A recent study showed that both SUV3 and PNPase are essential in degrading mt-mRNAs in vivo, and PNPase is directly involved in deadenylation and degradation of mt-mRNAs (13). Thus, it is likely that 3′ polyadenylation of mt-mRNA may be closely modulated in coordination with the degradation of the transcript, similar to its E. coli counterpart.
In this communication, we demonstrated that small fractions of the SUV3, PNPase, and mtPAP form a complex under low mitochondrial matrix Pi conditions. SUV3 serves as a bridge for binding to mtPAP and PNPase simultaneously. The complex consists of a SUV3 dimer, a mtPAP dimer, and a PNPase trimer based on the molecular sizing experiments. Mechanistically, SUV3 provides robust ssRNA binding ability to enhance the polyadenylation activity of mtPAP. Furthermore, purified SUV3·PNPase·mtPAP complex is capable of lengthening or shortening the RNA poly(A) tail lengths in low or high Pi/ATP ratios, respectively. Consistently, the poly(A) tail lengths of mt-mRNA transcripts can be lengthened or shortened by altering the mitochondrial matrix Pi levels via selective inhibition of the ETC or ATP synthase, respectively. These results suggested that SUV3 bridges PNPase and mtPAP to form a transient complex for modulating mt-mRNA poly(A) tail lengths in response to energy changes.
EXPERIMENTAL PROCEDURES
Tissue Culture and Drug Treatments
Normal mammary epithelial cells, MCF10A, grown to 70% confluence were treated with either 5 μg/ml of oligomycin A or 1 mm 2-d-deoxyglucose (2-DG) and 0.5 mm sodium azide (Az) in the background of 5 μg/ml of actinomycin D unless otherwise specified.
Dithiobis(succinimidyl propionate) Cross-linking
The protocol was adopted from Smith et al. (33). Briefly, MCF10A cells were treated with 0.5 mm dithiobis(succinimidyl propionate) for 30 min and quenched with 1× PBS containing 5 mm Tris (pH 7.4) for 10 min before washing once with 1× ice-cold PBS. Then the cells were scrapped down and subjected to lysis and gel filtration.
Cell Lysate Preparation
Cells were washed 2 times with PBS, trypsinized, and lysed with 500 μl of lysis buffer (50 mm HEPES, pH 7.6, 150 mm NaCl, 2.5 mm EGTA, 10% glycerol, and 0.2% Triton X-100) at 4 °C on a rotator for 15 min for post-drug treatments and/or dithiobis(succinimidyl propionate) cross-linking. The lysate was then clarified at maximum speed for 10 min at 4 °C.
Co-immunoprecipitation
Clarified cell lysate containing 300 μg of protein was incubated with either 1 μg of rabbit polyclonal antibody for SUV3, PNPase, or mtPAP (GeneTex, Irvine, CA), or rabbit IgG at 4 °C on a rotator for 2 h. Subsequently, 50 μl of bovine serum albumin (BSA)-blocked protein G-Sepharose beads (GeneTex, Irvine, CA) was added to the mixture and incubated at 4 °C on a rotator for an additional hour. The beads were washed 3 times by spinning down and replacing the supernatant with 1 ml of fresh lysis buffer. After the wash, 2× SDS-PAGE loading buffer was added to the samples, which was heated in boiling water for 10 min before loading onto a 8% SDS-PAGE gel for immunoblotting.
In Vitro Binding Experiment
The experimental protocol was adopted from Wang et al. (29), except for GST-tagged proteins that were purified using glutathione-Sepharose 4B beads and eluted with 10 mm freshly prepared glutathione solution (GE Healthcare Life Sciences).
SUV3·PNPase·mtPAP Complex Formation Experiment and Analytical Ultracentrifugation
Recombinant His-SUV3, His-PNPase, and His-mtPAP were separately purified with a nickel affinity column, followed by MonoS, MonoQ, and heparin columns (GE Healthcare Life Sciences), respectively. The proteins were mixed at a 2/3/2 molecular ratio and dialyzed in dialysis buffer (20 mm Tris, pH 8.0, 200 mm NaCl, 5% glycerol) at 4 °C for 6 h while stirring at low speed. Subsequently, the protein was concentrated and loaded onto a Superdex 200 gel filtration column (GE Healthcare Life Sciences). Analytical ultracentrifugation was performed as previously described (29).
Polyadenylation Assay and Helicase Assay
12.5 ng/μl of mtPAP was incubated with 40 nm ssRNA substrate (3WRNA) of the sequence 5′-GUUGAGAGAGAGAGAGUUUGAGAGAGAGAG, with or without SUV3 in 100 mm Tris (pH 7.4), 1 mm ATP, 50 μg/μl of BSA, 6 mm DTT, and 2 mm MgCl2 for the denoted times. The reaction was stopped with denaturing loading buffer (1× TBE, 82% formamide, 0.1% bromphenol blue, 0.1% xylene cyanol), and subsequently loaded onto 14% denaturing gel (7 m urea, 1× TBE, 10% formamide). The gel was subsequently dried and visualized using a Storm 320 PhosphorImager (GE Healthcare Life Sciences). Helicase assay was performed as previously described using the same 3WRNA/T20DNA duplex (29).
RNA Poly(A) Processing Assay
50 ng/μl of SUV3·PNPase·mtPAP complex was incubated with 50 nm ssRNA substrate (3WRNA) in 100 mm Tris (pH 7.4), 1 mm ATP, 50 μg/μl of BSA, 3 mm DTT, and 2 mm MgCl2 and sodium phosphate (pH 7.5) of the denoted concentrations for 30 min. The reaction was stopped with formamide loading denaturing loading buffer (1× TBE, 82% formamide, 0.1% bromphenol blue, 0.1% xylene cyanol), and subsequently loaded onto 8% denaturing gel (7 m urea, 1× TBE, 10% formamide). The gel was subsequently dried and visualized using a Storm 320 PhosphorImager (GE Healthcare Life Sciences). Quantifications on the relative abundance of poly(A) tail species that were longer than 50 nt were done by ImageJ.
Circular RT-PCR
The protocol was adopted from Slomvic et al. (34). In brief, 5 μg of total mRNA extracted from purified mitochondria or cells was used for self-ligation overnight using T4 RNA ligase (Ambion, Grand Island, NY). The circularized RNA was purified using Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA), followed by reverse transcription using gene-specific reverse primers. The first round of PCR was performed using forward primer (F1) and reverse primer (R2). The second round of nested PCR was performed using radioactively labeled forward primer (F2) and reverse primer (R2). Primers were ordered from Integrated DNA Technologies (Integrated DNA Technology, San Diego, CA) and sequences were as described in Ref. 34.
Mitochondrial Isolation, Drug Treatment, and RNA Extraction
24 plates of human embryonic kidney (HEK) 293 cells were grown to confluence, washed once with PBS, and trypsinized. Cells were washed once more with PBS, followed by resuspending in 4 ml of mitochondria isolation buffer (70 mm sucrose, 210 mm mannitol, 5 mm HEPES, pH 7.2, 1 mm EGTA, 0.5% (w/v) fatty acid-free BSA). The resuspended cells were then transferred into a Dounce homogenizer and disrupted with 12 passes at 300 rpm. 10 ml of isolation buffer was added to the homogenized cells and centrifuged at 700 × g for 5 min. The supernatant was collected and centrifuged at 700 × g twice more, until very little material was sedimented. Finally, the supernatant was collected and centrifuged at 10,000 × g for 20 min and the mitochondria pellet was suspended in 300 μl of MAS buffer (70 mm sucrose, 210 mm mannitol, 10 mm KH2PO4, 5 mm MgCl2, 25 mm PIPES, pH 6.8, 1 mm EGTA, 0.2% (w/v) fatty acid-free BSA, 0.13 mm NADH). After determining the protein concentration, mitochondria containing 300 mg of protein were transferred to an Eppendorf tube. The isolated mitochondria was treated with either 5 μg/ml (final) of oligomycin A or 5 mm (final) sodium azide in the background of 5 μg/ml (final) of actinomycin D in 100 μl (final) of MAS buffer for 1 h at 37 °C with the lid open. Unless otherwise specified, the entire process was done at 4 °C or on ice.
RESULTS
SUV3, PNPase, and mtPAP Form a Large Molecular Mass Complex in Vivo When the ETC Is Disrupted
To investigate the underlying mechanism that regulates mt-mRNA poly(A) dynamics, we first examined the mitochondrial enzymes that harbor the necessary activities to synthesize and process mt-mRNA poly(A) tails. The potential candidates included mtPAP, which is responsible for the polyadenylation of mt-mRNA poly(A) tails in the presence of ATP (13–16), and the SUV3·PNPase complex, which has been shown to degrade ssRNA in the presence of Pi (29). To test whether these three enzymes can function as a unit to modulate the mt-mRNA poly(A) tail lengths, co-immunoprecipitation experiments were performed. As shown in Fig. 1, A and B, immunoprecipitation of PNPase brought down SUV3 under normal culture conditions, whereas immunoprecipitation of mtPAP under the same conditions failed to bring down any detectable SUV3. Because these enzymatic activities require either ATP or Pi, we then treated cells with oligomycin A (Oligo) or Az/2-DG (a glycolysis inhibitor) to modulate the mitochondrial matrix Pi level. It was noted that import of Pi into the mitochondrial matrix is mediated by a Pi/OH-antiporter at the expense of the proton gradient established by the ETC (35). Inhibiting the ETC by compounds such as Az, will result in disruption of the proton gradient, hence damping the Pi import and decreasing the matrix Pi level (36). Conversely, inhibition of the ATP synthase by oligomycin A would increase the proton gradient across the inner mitochondrial membrane, causing more Pi to be imported into the matrix (37). Intriguingly, mtPAP was coimmunoprecipitated with SUV3 only when the cells were treated with Az, but not oligomycin A (Fig. 1B).
FIGURE 1.
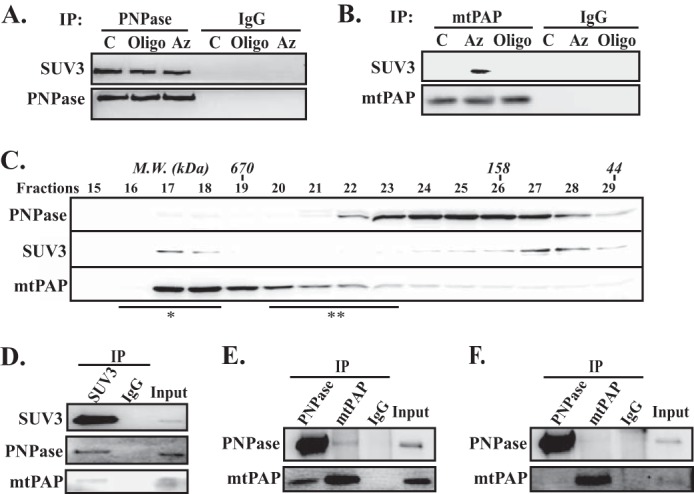
SUV3, PNPase, and mtPAP interact in vivo to form a large molecular mass complex when the ETC is disrupted. A and B, co-immunoprecipitation assays of SUV3 by immunoprecipitating mtPAP or PNPase, respectively, using whole cell lysate post 1 h of oligomycin A (Oligo) or Az and 2-DG treatments. C, control, culture media containing DMSO only. All proteins were visualized by immunoblotting. C, gel filtration (Superdex 200) fractionations of whole cell lysate post 1 h of Az and 2-DG treatments. Approximate molecular masses of gel filtration fractions were determined by calibrating the column with known size markers. SUV3, mtPAP, and PNPase were resolved by SDS-PAGE and visualized by immunoblotting. D, co-immunoprecipitation of mtPAP and PNPase by immunoprecipitating SUV3 with large molecular mass fractions (*). E and F, reciprocal co-immunoprecipitation (IP) of mtPAP and PNPase with large (*) and small (**) gel filtration fractions, respectively.
To further validate this observation, we fractionated the whole cell lysate by size exclusion chromatography post 2-DG/Az treatment (Fig. 1C). Consistent with prior observations, the majority of SUV3 (dimer, calculated molecular mass 166 kDa) and PNPase (trimer, calculated molecular mass 250 kDa) existed in uncomplexed forms (24, 38). Yet, small portions of the two proteins co-fractionated at a peak larger than 670 kDa. In contrast, the majority of mtPAP (dimer, calculated molecular mass 132 kDa (16)) appeared to associate with a large molecular mass complex. Subsequently, the fractions immediately larger than 670 kDa (fractions 16–18) were pooled together for co-immunoprecipitation experiments. As shown in Fig. 1D, immunoprecipitation of SUV3 brought down both PNPase and mtPAP from the pooled large molecular mass fractions. Reciprocal immunoprecipitations of PNPase and mtPAP brought down each other using the same fractions (Fig. 1E), but not the low-molecular weight fractions (fractions 20–23) (Fig. 1F). Together, these results suggested that small portions of SUV3 and PNPase form a stable complex, and only interact with mtPAP under the low matrix Pi level.
SUV3 Serves as a Bridge for PNPase and mtPAP Binding
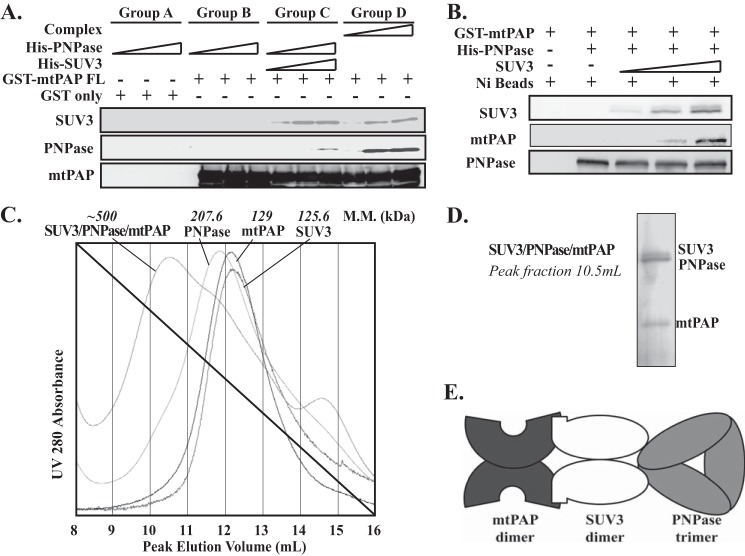
Next, we explored how mtPAP interacts with SUV3 and/or PNPase by in vitro binding experiments using purified recombinant mtPAP, SUV3, and PNPase. As shown in Fig. 2A, when immobilized on resin, mtPAP alone failed to pull down PNPase (Group B). However, mtPAP can pull down PNPase by supplementing SUV3 (Group C). Even more PNPase could be brought down when the pre-formed SUV3·PNPase complex was utilized in lieu of the separated individual proteins (Group D). The same result was obtained in the reverse direction with PNPase immobilized on the resin. Adding SUV3 enhanced the ability of PNPase to bring down mtPAP in a dosage dependent manner (Fig. 2B). To ascertain whether SUV3·PNPase·mtPAP can interact simultaneously using SUV3 as a bridge, we mixed the three purified proteins in vitro. As shown in Fig. 2C, mixing SUV3, mtPAP, and PNPase resulted in the formation of a complex with a molecular mass ∼500 kDa, determined by the gel filtration peak elution volume. The peak elution volume for the SUV3·PNPase·mtPAP complex is 10.5 ml, whereas each individual protein had peak elution volumes around 12.0 ml. Previously, the respective molecular masses of purified SUV3 and PNPase were determined to be 125.6 and 207.6 kDa by an analytical ultracentrifuge (29). The molecular mass of mtPAP was determined by analytical ultracentrifugation to be 129 kDa. Simple stoichiometry suggests the complex is composed of a 1/1/1 ratio of SUV3 dimer, PNPase trimer, and mtPAP dimer, which is confirmed by the silver staining result on the SUV3·PNPase·mtPAP complex peak fraction (Fig. 2D). Collectively, these results demonstrate that mtPAP interacts with the SUV3·PNPase complex through direct binding to SUV3 (Fig. 2E).
FIGURE 2.
SUV3 serves as a bridge for PNPase and mtPAP binding. A, in vitro binding assay. GST-mtPAP full-length (FL) or GST proteins were bound to glutathione-Sepharose 4B beads. Group C was preincubated with increasing amounts of His-SUV3 for 1 h. Subsequently, Groups A, B, and C were incubated with increasing amounts of His-PNPase, whereas Group D was incubated with increasing amounts of preformed SUV3·PNPase complex for 1 h. Post washing, the bound proteins were resolved by SDS-PAGE and visualized by immunoblotting. B, in vitro binding assay. Increasing amounts of un-tagged SUV3 was preincubated with His-PNPase on Ni+ resin for 1 h. Subsequently, equal amounts of GST-mtPAP were added to the mixture and further incubated for 1 h. Post washing, the bound proteins were resolved by SDS-PAGE and visualized by immunoblotting. C, size exclusion chromatography (Superdex 200) elution profiles of the purified recombinant SUV3·PNPase·mtPAP complex and the individual proteins. The molecular masses (M.M.) of the individual proteins were determined by analytical ultracentrifuge. The molecular mass of the SUV3·PNPase·mtPAP complex was approximated from its peak elution volume. D, SDS-PAGE followed by silver staining of the complex. E, schematic representation of the SUV3·PNPase·mtPAP complex.
N-terminal Region of SUV3 Binds to C-terminal Region of mtPAP
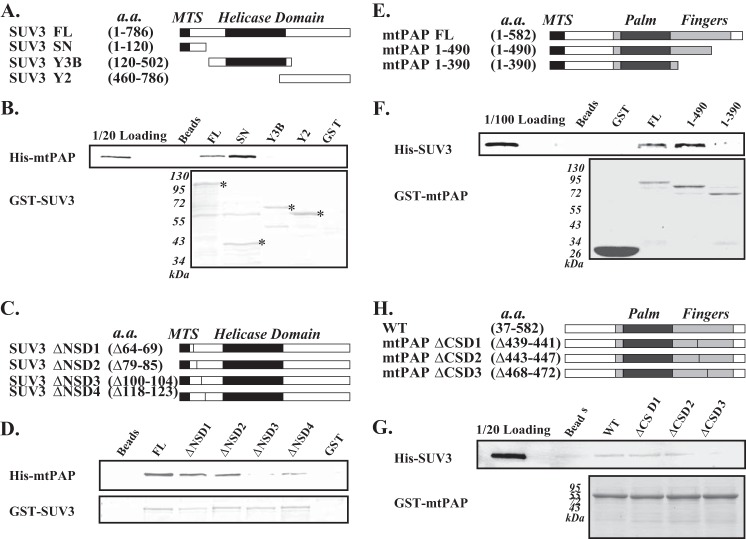
In determining the critical binding regions of the complex, because mtPAP has low binding affinity to PNPase, we focused on interaction between SUV3 and mtPAP. As shown in Fig. 3, A and B, mtPAP was selectively bound to the N-terminal region of SUV3. On the other hand, SUV3 selectively bound to the C-terminal region of mtPAP (Fig. 3, E and F). By systematic deletion analysis within the critical binding regions, we found that a 5-amino acid deletion in either SUV3 or mtPAP can disrupt this interaction. For SUV3, the deletion is at amino acid positions 100–104, designated as ΔNSD3, located upstream of the helicase domain (Fig. 3, C and D). The deletion for mtPAP is at amino acid positions 468–472, designated as ΔCSD3, located in the finger region of mtPAP (Fig. 3, H and G). Interestingly, we have previously shown that PNPase interacts with SUV3 at the region downstream of the helicase domain, as deletion of amino acids 510–514 (designated as ΔSP2) obliterates the PNPase/SUV3 interaction (29). Based on the x-ray crystal structure of SUV3, ΔNSD3 is located in the N-terminal domain A, whereas ΔSP2 is situated at the linker region between domains C and D (39).
FIGURE 3.
N-terminal region of SUV3 binds to the C-terminal region of mtPAP. A and C, schematics of SUV3 constructs used in the in vitro binding assays. B and D, in vitro binding assay. Top panels, immunoblotting of His-tagged mtPAP remained bound to GST-SUV3 fusion proteins on glutathione-Sepharose 4B beads after binding and washing. Bottom panels, protein quantifications of GST-SUV3 fusion proteins by Coomassie Blue staining. *, desired protein species. E and G, schematics of mtPAP constructs used for the in vitro binding assays. F and H, in vitro binding assay. Top panels: immunoblotting of His-tagged SUV3 remained bound to GST-mtPAP fusion proteins on-Sepharose 4B beads after binding and washing. Bottom panels, protein quantifications of GST-mtPAP fusion proteins by Coomassie Blue staining. FL, full-length.
SUV3 Enhances mtPAP Polyadenylation by Providing a Robust ssRNA Binding Domain
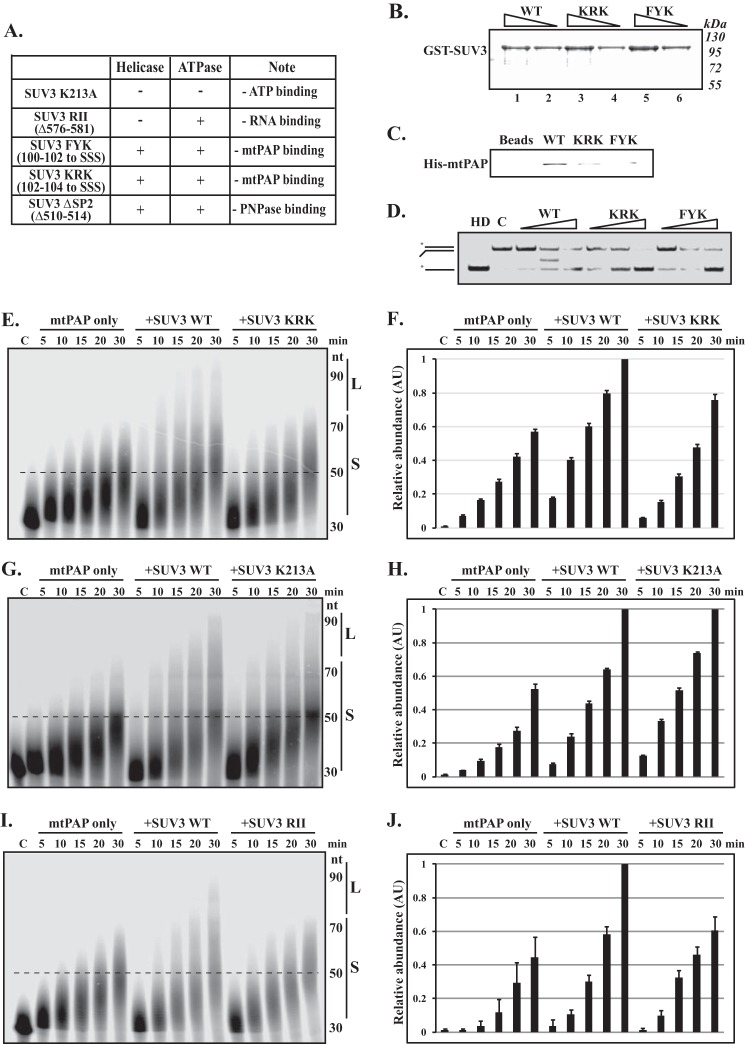
To assess if the interaction with SUV3 has an impact on the polyadenylation activity of mtPAP, we used purified mtPAP and various SUV3 mutants to perform polyadenylation on ssRNA in vitro (Fig. 4A). Consistent with previous studies, mtPAP can carry out 3′ polymerization on ssRNA in the presence of all four nucleotide triphosphates (14, 16), yet the activity is the highest for ATP (Fig. 4B). Interestingly, we observed that SUV3 can enhance the polyadenylation activity of mtPAP (Fig. 4E, left and middle panels). To substantiate the importance of the SUV3/mtPAP protein-protein interaction in the observed cooperative activity, we attempted to employ the mtPAP binding-deficient mutant, SUV3 ΔNSD3, which had the 100FYKRK104 sequence deleted. However, low solubility of the recombinant protein rendered the purification process difficult. Instead, we generated two serine substitution mutants: FYK and KRK to SSS, both of which possessed reduced mtPAP binding activities (Fig. 4, B and C) but intact helicase activities (Fig. 4D). KRK was subsequently chosen for the polyadenylation assay because it had a significantly superior expression and solubility. As shown in Fig. 4, E and F, when compared with SUV3 wild-type (WT), adding SUV3 KRK, even at 2-fold concentrations, did not enhance the activity of mtPAP to the same extent, suggesting a direct interaction between the two proteins is essential for the observed enhanced polyadenylation activity on ssRNA.
FIGURE 4.
SUV3 enhances the activity of mtPAP and PNPase by providing a robust ssRNA binding domain. A, summary table of the various SUV3 mutants pertinent to this study. B, protein quantification of GST-SUV3 WT, KRK, and FYK mutants by Coomassie Blue staining. 10 and 20 μl of protein-bound beads were loaded in lanes 2, 4, and 6, and lanes 1, 3, and 5, respectively. C, in vitro binding assay. Immunoblotting of His-tagged mtPAP remained bound to 20 μl of beads containing GST-SUV3 WT, KRK, and FYK after washing. D, helicase assay. Increasing amounts of SUV3 WT, KRK, and FYK mutants (5, 10, and 20 ng/μl) were incubated with the 3′ overhang helicase substrate at 37 °C for 30 min in the presence of 5 mm ATP. Lane HD, heat denatured; the reaction mixture containing the substrate was heated to 80 °C for 10 min to denature the duplex immediately before loading onto 15% native PAGE. Lane C, control, the size exclusion chromatography buffer containing no protein was added. E, G, and I, polyadenylation assays. mtPAP alone or mtPAP with SUV3 wild-type or the designated mutants were incubated with 30 nt ssRNA (3WRNA) at 37 °C in the presence of 1 mm ATP. In E, the amount of KRK used was twice that of SUV3 WT. In G and I, the respective concentrations of K213A and RII were the same as those of SUV3 WT. F, H, and J, respective quantifications for relative abundance of poly(A) tails that were longer than 50 nt in panels E, G, and I as indicated by dashed lines. AU, arbitrary unit.
To further delineate how SUV3 enhances the activity of mtPAP, we employed two additional SUV3 mutants, K213A and RII, which were previously shown to have disrupted ATPase and RNA binding activity, respectively (28, 29). As shown in Fig. 4, G–J, K213A, but not RII, had comparable enhancing activity for the polyadenylation activity of mtPAP compared with SUV3 WT. Collectively, these results indicated that SUV3 enhances the polyadenylation activity of mtPAP by providing a robust RNA binding domain and this cooperative mechanism does not require helicase activity of SUV3.
SUV3·PNPase·mtPAP Complex Modulates Poly(A) Tail Lengths in Response to Changes in Pi/ATP Ratios
As SUV3·PNPase cooperatively degrade ssRNA (29) and SUV3·mtPAP cooperatively add poly(A) tails to mRNA (Fig. 4), we then examined how the SUV3·PNPase·mtPAP complex modulates RNA poly(A) tail lengths in vitro. As shown in Fig. 5A, the complex polyadenylated the ssRNA substrate in the presence of ATP, and efficiently degraded poly(A) tails upon the addition of Pi. On the other hand, by keeping the ATP concentration constant, the complex preferred to polyadenylate the ssRNA substrate at low Pi levels. As the Pi level rose, the complex degraded the polyadenylated ssRNA instead (Fig. 5B). These results suggested that the complex is capable of lengthening or shortening the lengths of ssRNA poly(A) tails in response to low or high Pi/ATP ratios, respectively. Next, to test whether a change in the mitochondrial matrix Pi level has an effect on altering the mt-mRNA poly(A) tail lengths in cultured cells, we used circular RT-PCR to measure the poly(A) tail lengths of mt-mRNA transcripts, ND3 and ND5, upon manipulating the matrix inorganic phosphate levels as previously described. The entire experiment was done in the presence of actinomycin D, a mitochondrial transcription inhibitor, to avoid the complications of newly synthesized mt-mRNAs. As shown, respectively, in Fig. 5, C and D, ND3 transcripts were primarily oligoadenylated (∼8 nt), whereas ND5 transcripts were polyadenylated (∼50 nt), consistent with a previous report (40). Treatment of Az resulted in 3′ extensions of ND3 transcripts within 1 h (Fig. 5C, lanes 5–7). Conversely, when oligomycin A was administrated to block ATP synthase, most poly(A) tails were shortened to oligo(A) tails over the same time frame (Fig. 5C, lanes 2–4). Similarly, the ratio of poly(A) tails to oligo(A) tails of ND5 was drastically decreased upon oligomycin A treatment (Fig. 5C, compare lane 1 to 3), although increased concentrations of oligomycin A treatments had little effect on oligoadenylation. On the other side, treatment of Az slightly increased ND3 poly(A) tail lengths (Fig. 5D, compare lane 1 to 2). These results suggested that the change of poly(A) tail lengths of both ND3 and ND5 is responding to changes in energetic states of mitochondria.
FIGURE 5.
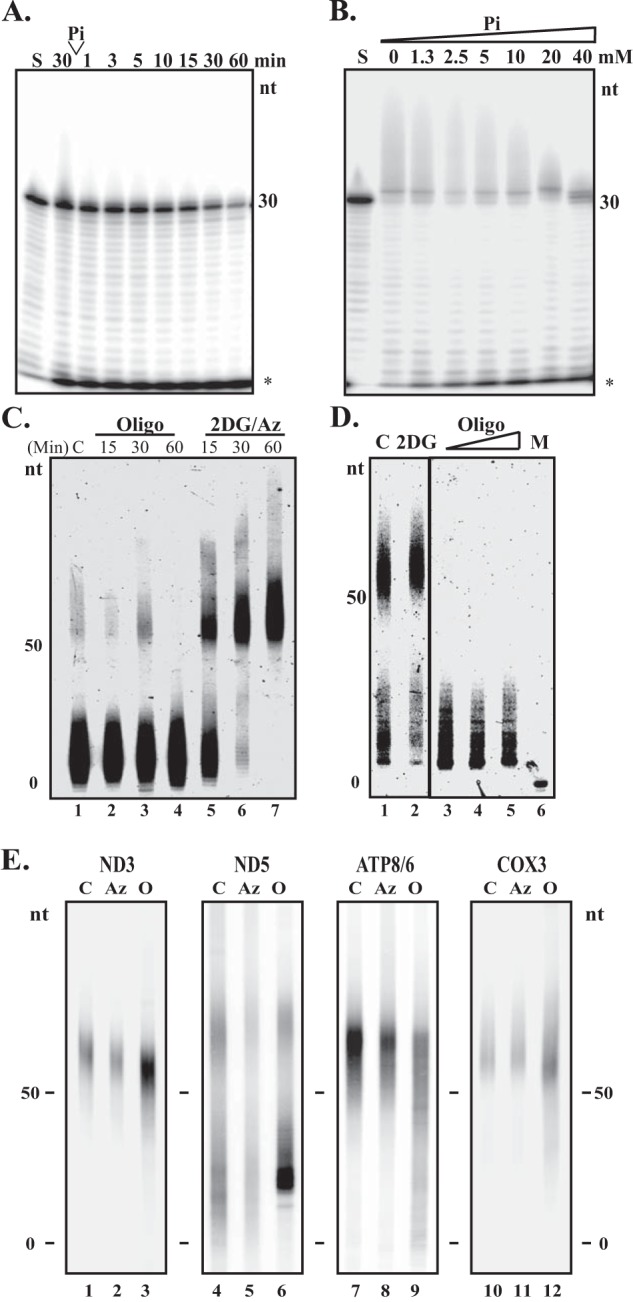
SUV3·PNPase·mtPAP complex modulates poly(A) tail lengths in response to changes in Pi/ATP ratios. A and B, poly(A) tail processing assay. In A the complex was incubated with 3WRNA and 0.1 mm ATP at 37 °C. At 30 min, 10 mm Pi (final) is supplemented to the reaction. In B, the SUV3·PNPase·mtPAP complex was incubated with 3WRNA, 1 mm ATP, and 2-fold increasing concentrations of Pi up to 40 μm (final) at 37 °C for 30 min. S, reaction immediately after all constituents had been added. *, degradation products. C, circular RT-PCR to determine the poly(A) tail lengths of the ND3 RNA transcripts post 15-, 30-, and 60-min treatments of oligomycin A or 2-DG and sodium azide in MCF10A cells. C, control. Cells were treated for 60 min with control medium containing actinomycin D only. D, circular RT-PCR to determine ND5 poly(A) tail lengths post 2.5 h treatment of 1 mm 2-DG only or increasing concentrations of oligomycin A (0.5, 1, and 5 μg/ml) in MCF10A cells. M, marker. Oligo, oligomycin A. E, circular RT-PCR to determine poly(A) tail lengths of mtRNA transcripts post 60-min treatments of oligomycin A or sodium azide in isolated mitochondria. Isolated mitochondria were treated for 60 min with control medium. For C, D, and E, all samples were concurrently treated with actinomycin D to inhibit mitochondrial transcription.
To further substantiate this observation, we isolated metabolically active mitochondria from cultured cells and then treated with the same drugs in the presence of reduced NADH, the electron donor of the ETC, and room air. The majority of mt-mRNA transcripts from the control isolated mitochondria were in the poly(A) states (Fig. 5E, lanes 1, 4, 7, and 10), different from what is observed in the whole cell extract. We attribute that to the fact that we oversaturated the mitochondria with NADH, promoting polyadenylation of mRNA transcripts. However, more than half of the ND5 transcripts were still oligoadenylated compared with other transcripts, consistent with the rare polyadenylation of ND5 in the whole cells extract (Fig. 5E, lane 4). Upon treating the mitochondria with oligomycin A, we observed a shortening of poly(A) tails among the 4 mt-mRNA transcripts tested. On the other hand, azide treatment resulted in no apparent extensions of poly(A) tail lengths, as most transcripts were already in the poly(A) state. These results indicated that altering the energy states via manipulating the Pi levels in mitochondria has an impact on the lengths of mt-mRNA poly(A) tails.
DISCUSSION
In this communication, we demonstrated the existence of a small fraction of SUV3 bridging PNPase and mtPAP to form a complex under low mitochondrial matrix Pi conditions. SUV3 provides a robust ssRNA binding ability to enhance the polyadenylation activity of mtPAP. Furthermore, the purified SUV3·PNPase·mtPAP complex is capable of lengthening or shortening the RNA poly(A) tail lengths in low or high Pi/ATP ratios, respectively. Consistently, the poly(A) tail lengths of mt-mRNA transcripts can be lengthened or shortened by altering the inorganic phosphate (Pi) levels inside the mitochondrial matrix. These results suggested that the transient SUV3·PNPase·mtPAP complex may have a role in modulating mt-mRNA poly(A) tail lengths in response to energetic changes.
PNPase has diverse roles in the mt-mRNA metabolism depending on its localizations. It was demonstrated that the majority of PNPase is localized in the intermembrane space to regulate RNA import, raising doubts on the accessibility of PNPase to other mitochondrial matrix proteins like SUV3 (24, 25). However, using fluorescence microscopy, it was reported that both SUV3 and PNPase co-localized in distinct foci inside the mitochondrial matrix (41), consistent with a previous biochemical study demonstrating that both proteins were associated with or in close vicinity to the mitochondrial DNA nucleoids (42). In addition, a recent study utilizing a genetically targetable peroxidase enzyme that can biotinylate nearby proteins directly confirmed the presence of PNPase in the mitochondrial matrix (43). These observations indicate that a fraction of PNPase, localized in the mitochondrial matrix, may be capable to interact with other mitochondrial matrix proteins, such as SUV3 and mtPAP.
Interestingly, another recent study suggested that the LRPPRC·SLIRP complex suppresses PNPase-mediated mRNA degradation while promoting mtPAP-mediated polyadenylation in vitro, implying multiple layers of control in the regulation of mt-mRNA poly(A) tail lengths by mtPAP and SUV3·PNPase. In the same study, depletion of PNPase, but not PDE12, a 2′-phosphodiesterase identified in human mitochondrial (44, 45), has been shown to significantly extend poly(A) tails and abolish mRNA decay, confirming the role of PNPase as the critical exoribonuclease in mt-mRNA processing (13). Importantly, our data suggested that the majority of mtPAP is associated with a large molecular weight complex besides SUV3·PNPase (Fig. 1). It would be interesting to test if the LRPPRC·SLIRP complex and other mt-mRNA processing complexes co-fractionate with mtPAP. Formation of the SUV3·PNPase·mtPAP complex may require additional signals, such as phosphorylation, acetylation, and other secondary modifications, which demands further investigation.
As aforementioned, both mtPAP and PNPase possess polyadenylation activities, raising the question of which enzyme mediates polyadenylation in human mitochondria. One significant difference between the two enzymes is their substrate specificities: mtPAP has been shown to synthesize homopolymeric poly(A) tails; whereas PNPase is less specific in choosing nucleotides and usually utilizes NDP to generate heteropolymeric tails (7, 23, 46, 47). It has been reported that the chloroplast PNPase is the sole enzyme with polyadenylation activity, and the 3′ RNA extensions were found to contain up to 30% non-adenosine nucleotides (48). On the other hand, sequence analysis of the 3′ ends of human mt-mRNAs showed mainly homopolymeric tails with less than 1% of residues other than adenosine (15), suggesting that the polyadenylation of mt-mRNA is more likely to be performed by mtPAP rather than PNPase. In addition, the requirement of ADP for PNPase to carry out polyadenylation makes it unlikely to be the enzyme in action as the ADP concentration inside the matrix is in a much lower ATP concentration. Last, the discrepancy on the effect of poly(A) tail lengths upon PNPase depletion in prior studies can largely be explained by transient versus stable silencing approaches used in different studies (13, 49).
The observation that the three enzymes, SUV3, PNPase, and mtPAP, form a transient complex to coordinately modulate the poly(A) tail lengths of mt-mRNA is very interesting, as it is known that RNA degradation is coordinated with 3′ polyadenylation in E. coli (19). Mechanistically, it is intriguing that SUV3 provides a robust ssRNA binding domain for both mtPAP and PNPase. In vitro enzymatic assays suggested that the interactions between mtPAP/SUV3 and PNPase/SUV3 are vital for their respective polyadenylase and exoribonuclease activities. Clinically, a A to G missense mutation in exon 9 (N478D) of mtPAP results in shortened mt-mRNA poly(A) tails and mitochondrial dysfunction in an Ohio Amish family (50). Structurally, Asn-478 is located outside of the enzymatic domain, but close to the critical region for SUV3 binding (amino acids 468–472) demonstrated in this study (Fig. 3) (16). It is possible that the clinical manifestation is a direct result of the disruption of the interaction between mtPAP to SUV3 and compromised the complex activity. Further investigation to confirm this conjecture is warranted.
On the other hand, the biological functions of mt-mRNA poly(A) tails and whether the length of poly(A) tails confers mtmRNA stability remain an enigma. Temperley et al. (51) reported that a pathogenic microdeletion (μΔ9205) of human mt-ATP6 mRNA at its 3′ end is associated with the shortened poly(A) tail and decreased stability, favoring the notion that poly(A) conveys stability. However, artificial targeting of cytosolic deadenylase (PARN) to mitochondria or overexpressing PDE12 (45) to force deadenylation of mt-mRNAs (52) showed differential effects on the steady-state levels of mt-mRNAs. Such transcript-specific stabilization or destabilization is also observed in cells depleted with either mtPAP or PNPase (13–15, 49). All in all, the length of the poly(A) does not appear to fully correlate with the stability of mt-mRNA.
Based on the data presented in this endeavor, poly(A) tails of mt-mRNA appear to lengthen and shorten via modulations of the SUV3·PNPase·mtPAP complex in response to changes in cellular energy state. When the mitochondrial matrix Pi level increases, the SUV3·PNPase complex degrades the poly(A) tails of mt-mRNA transcripts. Conversely, as the ATP level rises and matrix Pi level wanes, the SUV3·PNPase complex interacts with mtPAP to incorporate excess ATP in the poly(A) tails of mt-mRNA. This observation may provide an alternative function of mt-mRNA poly(A) tails. Further genetic and biochemical investigations in this regard will be necessary to affirm this postulation.
Acknowledgments
We are grateful to Dr. Yumay Chen, Dr. Chun-Mei Hu, and Bryan Ngo for technical assistance and Dr. Yu-Hou Chen for assistance in analytical ultracentrifuge.
This work was supported, in whole or in part, by National Institutes of Health Grant AG027877 (to W. H. L.).
- nt
- nucleotide
- mt
- mitochondrial
- poly(A)
- polyadenylation
- ssRNA
- single-stranded RNA
- ETC
- electron transport chain
- Az
- sodium azide
- 2-DG
- 2-d-deoxyglucose
- mtPAP
- mitochondrial polyadenylation polymerase
- PNPase
- polynucleotide phosphorylase.
REFERENCES
- 1. Carpousis A. J., Leroy A., Vanzo N., Khemici V. (2001) Escherichia coli RNA degradosome. Methods Enzymol. 342, 333–345 [DOI] [PubMed] [Google Scholar]
- 2. Coburn G. A., Miao X., Briant D. J., Mackie G. A. (1999) Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev. 13, 2594–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hajnsdorf E., Steier O., Coscoy L., Teysset L., Régnier P. (1994) Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams pnp rnb mutant. EMBO J. 13, 3368–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yehudai-Resheff S., Hirsh M., Schuster G. (2001) Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 21, 5408–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dreyfus M., Régnier P. (2002) The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell 111, 611–613 [DOI] [PubMed] [Google Scholar]
- 6. Manley J. L., Proudfoot N. J. (1994) RNA 3′ ends: formation and function, meeting review. Genes Dev. 8, 259–264 [DOI] [PubMed] [Google Scholar]
- 7. Colgan D. F., Manley J. L. (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11, 2755–2766 [DOI] [PubMed] [Google Scholar]
- 8. Ojala D., Montoya J., Attardi G. (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470–474 [DOI] [PubMed] [Google Scholar]
- 9. Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J., Staden R., Young I. G. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 [DOI] [PubMed] [Google Scholar]
- 10. Yokobori S., Pääbo S. (1997) Polyadenylation creates the discriminator nucleotide of chicken mitochondrial tRNA(Tyr). J. Mol. Biol. 265, 95–99 [DOI] [PubMed] [Google Scholar]
- 11. Tomita K., Ueda T., Watanabe K. (1996) RNA editing in the acceptor stem of squid mitochondrial tRNA(Tyr). Nucleic Acids Res. 24, 4987–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jacob S. T., Schindler D. G. (1972) Polyriboadenylate polymerase solubilized from rat liver mitochondria. Biochem. Biophys. Res. Commun. 48, 126–134 [DOI] [PubMed] [Google Scholar]
- 13. Chujo T., Ohira T., Sakaguchi Y., Goshima N., Nomura N., Nagao A., Suzuki T. (2012) LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 40, 8033–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagaike T., Suzuki T., Katoh T., Ueda T. (2005) Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 280, 19721–19727 [DOI] [PubMed] [Google Scholar]
- 15. Tomecki R., Dmochowska A., Gewartowski K., Dziembowski A., Stepien P. P. (2004) Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 32, 6001–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bai Y., Srivastava S. K., Chang J. H., Manley J. L., Tong L. (2011) Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase. Mol Cell 41, 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagao A., Hino-Shigi N., Suzuki T. (2008) Measuring mRNA decay in human mitochondria. Methods Enzymol. 447, 489–499 [DOI] [PubMed] [Google Scholar]
- 18. Borowski L. S., Szczesny R. J., Brzezniak L. K., Stepien P. P. (2010) RNA turnover in human mitochondria: more questions than answers? Biochim. Biophys. Acta 1797, 1066–1070 [DOI] [PubMed] [Google Scholar]
- 19. Xu F., Cohen S. N. (1995) RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature 374, 180–183 [DOI] [PubMed] [Google Scholar]
- 20. Carpousis A. J., Van Houwe G., Ehretsmann C., Krisch H. M. (1994) Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76, 889–900 [DOI] [PubMed] [Google Scholar]
- 21. Carpousis A. J. (2007) The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 61, 71–87 [DOI] [PubMed] [Google Scholar]
- 22. Portnoy V., Palnizky G., Yehudai-Resheff S., Glaser F., Schuster G. (2008) Analysis of the human polynucleotide phosphorylase (PNPase) reveals differences in RNA binding and response to phosphate compared to its bacterial and chloroplast counterparts. RNA 14, 297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mohanty B. K., Kushner S. R. (2000) Polynucleotide phosphorylase functions both as a 3′ → 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 11966–11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang G., Chen H. W., Oktay Y., Zhang J., Allen E. L., Smith G. M., Fan K. C., Hong J. S., French S. W., McCaffery J. M., Lightowlers R. N., Morse H. C., 3rd, Koehler C. M., Teitell M. A. (2010) PNPase regulates RNA import into mitochondria. Cell 142, 456–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen H. W., Rainey R. N., Balatoni C. E., Dawson D. W., Troke J. J., Wasiak S., Hong J. S., McBride H. M., Koehler C. M., Teitell M. A., French S. W. (2006) Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol. Cell. Biol. 26, 8475–8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sarkar D., Fisher P. B. (2006) Human polynucleotide phosphorylase (hPNPase old-35): an RNA degradation enzyme with pleiotrophic biological effects. Cell Cycle 5, 1080–1084 [DOI] [PubMed] [Google Scholar]
- 27. Das S. K., Bhutia S. K., Sokhi U. K., Dash R., Azab B., Sarkar D., Fisher P. B. (2011) Human polynucleotide phosphorylase (hPNPase(old-35)): an evolutionary conserved gene with an expanding repertoire of RNA degradation functions. Oncogene 30, 1733–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shu Z., Vijayakumar S., Chen C. F., Chen P. L., Lee W. H. (2004) Purified human SUV3p exhibits multiple-substrate unwinding activity upon conformational change. Biochemistry 43, 4781–4790 [DOI] [PubMed] [Google Scholar]
- 29. Wang D. D., Shu Z., Lieser S. A., Chen P. L., Lee W. H. (2009) Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J. Biol. Chem. 284, 20812–20821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Khidr L., Wu G., Davila A., Procaccio V., Wallace D., Lee W. H. (2008) Role of SUV3 helicase in maintaining mitochondrial homeostasis in human cells. J. Biol. Chem. 283, 27064–27073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo X. E., Chen C. F., Wang D. D., Modrek A. S., Phan V. H., Lee W. H., Chen P. L. (2011) Uncoupling the roles of the SUV3 helicase in maintenance of mitochondrial genome stability and RNA degradation. J. Biol. Chem. 286, 38783–38794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Margossian S. P., Li H., Zassenhaus H. P., Butow R. A. (1996) The DExH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell 84, 199–209 [DOI] [PubMed] [Google Scholar]
- 33. Smith A. L., Friedman D. B., Yu H., Carnahan R. H., Reynolds A. B. (2011) ReCLIP (reversible cross-link immunoprecipitation): an efficient method for interrogation of labile protein complexes. PLoS One 6, e16206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slomovic S., Laufer D., Geiger D., Schuster G. (2005) Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol. Cell. Biol. 25, 6427–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palmieri F. (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 447, 689–709 [DOI] [PubMed] [Google Scholar]
- 36. Thiaudiere E., Gallis J. L., Dufour S., Rousse N., Canioni P. (1993) Compartmentation of inorganic phosphate in perfused rat liver. Can cytosol be distinguished from mitochondria by 31P NMR? FEBS Lett. 330, 231–235 [DOI] [PubMed] [Google Scholar]
- 37. Vidal G., Gallis J. L., Dufour S., Canioni P. (1997) NMR studies of inorganic phosphate compartmentation in the isolated rat liver during acidic perfusion. Arch. Biochem. Biophys. 337, 317–325 [DOI] [PubMed] [Google Scholar]
- 38. Chen H. W., Koehler C. M., Teitell M. A. (2007) Human polynucleotide phosphorylase: location matters. Trends Cell Biol. 17, 600–608 [DOI] [PubMed] [Google Scholar]
- 39. Jedrzejczak R., Wang J., Dauter M., Szczesny R. J., Stepien P. P., Dauter Z. (2011) Human Suv3 protein reveals unique features among SF2 helicases. Acta Crystallogr. D Biol. Crystallogr. 67, 988–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Temperley R. J., Wydro M., Lightowlers R. N., Chrzanowska-Lightowlers Z. M. (2010) Human mitochondrial mRNAs-like members of all families, similar but different. Biochim. Biophys. Acta 1797, 1081–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Borowski L. S., Dziembowski A., Hejnowicz M. S., Stepien P. P., Szczesny R. J. (2013) Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 41, 1223–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bogenhagen D. F., Rousseau D., Burke S. (2008) The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 283, 3665–3675 [DOI] [PubMed] [Google Scholar]
- 43. Rhee H. W., Zou P., Udeshi N. D., Martell J. D., Mootha V. K., Carr S. A., Ting A. Y. (2013) Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Poulsen J. B., Andersen K. R., Kjær K. H., Durand F., Faou P., Vestergaard A. L., Talbo G. H., Hoogenraad N., Brodersen D. E., Justesen J., Martensen P. M. (2011) Human 2′-phosphodiesterase localizes to the mitochondrial matrix with a putative function in mitochondrial RNA turnover. Nucleic Acids Res. 39, 3754–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rorbach J., Nicholls T. J., Minczuk M. (2011) PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucleic Acids Res. 39, 7750–7763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cao G. J., Sarkar N. (1992) Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl. Acad. Sci. U.S.A. 89, 10380–10384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lin-Chao S., Chiou N. T., Schuster G. (2007) The PNPase, exosome and RNA helicases as the building components of evolutionarily-conserved RNA degradation machines. J. Biomed. Sci. 14, 523–532 [DOI] [PubMed] [Google Scholar]
- 48. Bollenbach T. J., Schuster G., Stern D. B. (2004) Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 78, 305–337 [DOI] [PubMed] [Google Scholar]
- 49. Slomovic S., Schuster G. (2008) Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA. RNA 14, 310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Crosby A. H., Patel H., Chioza B. A., Proukakis C., Gurtz K., Patton M. A., Sharifi R., Harlalka G., Simpson M. A., Dick K., Reed J. A., Al-Memar A., Chrzanowska-Lightowlers Z. M., Cross H. E., Lightowlers R. N. (2010) Defective mitochondrial mRNA maturation is associated with spastic ataxia. Am. J. Hum. Genet. 87, 655–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Temperley R. J., Seneca S. H., Tonska K., Bartnik E., Bindoff L. A., Lightowlers R. N., Chrzanowska-Lightowlers Z. M. (2003) Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Hum. Mol. Genet. 12, 2341–2348 [DOI] [PubMed] [Google Scholar]
- 52. Wydro M., Bobrowicz A., Temperley R. J., Lightowlers R. N., Chrzanowska-Lightowlers Z. M. (2010) Targeting of the cytosolic poly(A) binding protein PABPC1 to mitochondria causes mitochondrial translation inhibition. Nucleic Acids Res. 38, 3732–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]