Abstract
Objective
To investigate whether wild-type TP53 status in high-grade serous ovarian carcinoma is associated with poorer survival.
Methods
Clinical and genomic data of 316 sequenced samples from The Cancer Genome Atlas (TCGA) ovarian high-grade serous carcinoma (HGOSC) study were downloaded from TCGA data portal. Association between wild-type TP53 and survival was analyzed with Kaplan Meier method and Cox regression. The diagnosis of HGOSC was evaluated by reviewing pathological reports and high-resolution hematoxylin and eosin (H&E) images from frozen sections. The authenticity of wild-type TP53 in these tumor samples was assessed by analyzing SNP array data with ASCAT algorithm, reverse phase protein array (RPPA) data and RNAseq data.
Results
Fifteen patients with HGOSCs were identified to have wild-type TP53, which had significantly shorter survival and higher chemoresistance than those with mutated TP53. The authenticity of wild-type TP53 status in these fifteen patients was supported by SNP array, RPPA, and RNAseq data. Except four cases with mixed histology, the classification as high grade serous carcinomas was supported by pathological reports and H&E images. Using RNAseq data, it was found that EDA2R gene, a direct target of wild-type TP53, was highly up-regulated in samples with wild-type TP53 in comparison to samples with either nonsense or missense TP53 mutations.
Conclusion
Patients with wild-type TP53 high grade ovarian serous carcinomas appeared to have a poorer survival and were more chemoresistant than those with mutated TP53. Differentially expressed genes in these TP53 wild-type tumors may provide insight in the molecular mechanism in chemotherapy resistance.
Keywords: Ovarian Cancer, p53 mutation, wild-type p53, EDA2R, Survival Statistics, Collagen VI
Introduction
Many researchers have tried to correlate TP53 mutations with clinical outcomes for patients with ovarian cancer. The results have been conflicting [1–4], likely due to methodological issues, small sample sizes and mixed histological subtypes/grades. The Cancer Genome Atlas consortium has performed whole-genome massively parallel sequencing of selected tumor and nontumor pairs to identify somatic mutations in cancer cells. Recent exome sequencing analysis of 316 high-grade ovarian serous carcinomas (HGOSCs) identified mutations in the tumor repressor protein 53 (TP53) gene in 96% of all cases [5], which supports the idea that TP53 is a driver mutation in ovarian cancer [6]. The use of whole-exome sequencing and subsequent independent validation of TP53 mutations in The Cancer Genome Atlas (TCGA) ovarian cancer samples provide the most reliable data to date. However, it will be stimulating to investigate at the comprehensive TCGA data that are freely available for the 15 cases of HGOSCs that apparently had no detectable TP53 mutation. Here, we interrogated the genomic, pathological and clinical data of the 15 cases with wild-type TP53. The result indicated that these were tumors with authentic wild-type TP53. More interestingly, patients with wild-type TP53 ovarian cancer appeared to have a poorer survival and were more chemoresistant than those with mutated TP53.
Methods
Survival analysis
Clinical data from 316 newly diagnosed patients with ovarian serous adenocarcinoma who had received no prior treatment was retrieved from the supplemental data of the TCGA Nature paper [5] and integrated with TP53 mutation data from the TCGA data portal (S1). Kaplan-Meier survival analysis was performed with SPSS version 19.0 software (IBM, Armonk, New York); Significance was estimated with the log-rank test. The Cox proportional hazards regression model was used to evaluate the relationship between survival time with age and cytoreduction status. Overall survival was defined as the interval from the date of initial surgical resection to the date of last known contact or death. Progression free survival was defined as the interval from the date of initial surgical resection to the date of progression, date of recurrence, or date of last known contact if the patient was alive and has not recurred. The definition of platinum free interval is similar to that of progress free survival but was from the date of last primary platinum treatment. Chemoresistance status was defined as sensitive when the platinum free interval was six months or greater, and there was no evidence of progression or recurrence. Chemoresistance status was defined as resistant when the platinum free interval was less than six months and the patient had progressed or recurred. Chemoresistance status was defined as refractory if the patient had progressed or recurred during the platinum treatment or right after the last primary platinum treatment.
Review of pathological data of wild-type TP53 samples
Pathology Reports were downloaded from The Cancer Genome Atlas data portal control-access directory. Quality control Hematoxylin and Eosin (H&E) images from frozen sections were visualized at TCGA BioSig website hosted at Lawrence Berkeley National by our gynecologic pathologist (Michael Deavers) [7].
Analysis of The Cancer Genome Atlas Data for high-grade ovarian serous cystadenocarcinoma
Level 3 somatic mutations derived from Next Generation exome sequencing and RPPA data were retrieved from The Cancer Genome Atlas data portal open-access directory [8]. Affymetrix SNP6.0 level 1 data was downloaded from controlled-access directory. DNA copy number aberrations were analyzed using level 1 Affymetrix SNP6.0 data processed by Nexus 6.0 software with ASCAT (allele-specific copy number analysis of tumors) algorithm [9]. With the matched normal control DNA, ASCAT can accurately dissect the allele-specific copy number of solid tumors, and simultaneously estimate and adjust for both tumor ploidy and percentage of normal stromal cell contamination. RNAseq data was available from 420 samples and transcript expression level in terms of RPKM (reads per kilobase per million mapped reads), which has been normalized by total count and gene length, were compiled for differential gene expression analysis (S2). RPPA level 3 data for 177 samples were also consolidated for statistical analysis (S3). RPPA data from the TCGA data portal were from MD Anderson Cancer Center. Tumor lysates were two-fold serial diluted for 5 dilutions and arrayed on nitrocellulose coated slides, which were probed with antibodies. Relative protein levels for each sample were determined by interpolation of each dilution curves from the standard curve (“SuperCurve”) as described previously [10]. All the data points were normalized for protein loading and transformed to linear value. Normalized data for RNAseq and RPPA were uploaded to dChip software for identification of differentially expressed genes/proteins by student t-test [11]. The false discovery rate (q-value) was computed using software SAM by randomly permuting the sample labels 200 times [12].
Results
Patients with wild-type TP53 had poorer survival than those with mutated TP53
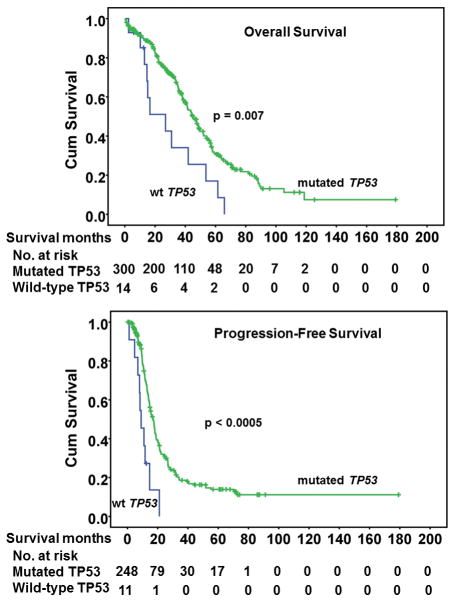
Patients with wild-type TP53 had significantly poorer survival than those with mutated TP53 (Figure 1). The median overall survival for the patients with wild-type TP53 was 27 months, while the median survival for patients with mutated TP53 was 45 months. The mean follow-up time for patients with TP53 wild-type was 57 months and for TP53 mutation was 63 months (p value = 0.272, not significantly different). The median follow-up for patients with TP53 mutation was 62 months which is much longer than the median survival time for patients with TP53 mutation. After adjustment for age and debulking status by Cox regression analysis, the difference in both overall and progression-free survival between mutated TP53 and wild-type TP53 was still significant (p=0.02 and 0.01, respectively). Moreover, seven patients with wild-type TP53 appeared to be resistant to standard chemotherapy (carboplatin/taxane). Four patients had recurrent disease in less than six months and the other three patients had tumors that were refractory to treatment with progressive disease (Table 1).
Figure 1.
Overall (A) and progression-free survival (B) of patients with ovarian tumors with mutated TP53 or wild-type (wt) TP53.
Table 1.
Clinical and Pathological characteristics of TCGA ovarian tumors with wild-type TP53.
| Sample ID | H&E images Characteristics | Pathology Report | Vital status in months (chemoresistance, platinum free interval) |
|---|---|---|---|
| TCGA-9-2056 | High-grade serous | High-grade serous | Living, 12.43 (sensitive, 7.6) |
| TCGA-10-0933 | Poor histology | High-grade with mixed components (serous, endometrioid, mucinous, and sarcomatoid) | Deceased, 14.46 (sensitive, 7.3) |
| TCGA-13-0727 | High-grade serous | N/A | Deceased, 15.15 (resistant, 4.3) |
| TCGA-13-0755 | High-grade serous | N/A | Deceased, 2.46 (refractory, 0) |
| TCGA-13-1408 | High-grade serous | N/A | Living, 5.9 (N/A) |
| TCGA-13-1477 | High grade serous | N/A | Deceased, 53.67 (resistant, 1) |
| TCGA-24-1544 | High-grade serous, with clusters of mucinous neoplastic cells | High-grade serous with minor clear cell component and clusters of mucinous neoplastic cells | Deceased, 26.89 (sensitive, 14.5) |
| TCGA-24-1565 | High-grade serous | High-grade serous | Deceased, 64.1 (N/A) |
| TCGA-24-2038 | Serous borderline tumor (with KRAS mutation) | Serous borderline tumor | Deceased, 44.36 (N/A) |
| TCGA-24-2293 | High-grade serous | Suggestive of primary peritoneal high-grade serous | Deceased, 16.56 (refractory, 0) |
| TCGA-25-1316 | MMMT (with KRAS mutation) | N/A | Deceased, 41.93 (resistant, 1.9) |
| TCGA-25-1328 | Low-grade serous | N/A | Diseased, 68.57 (resistant, 1) |
| TCGA-25-2042 | High-grade serous | N/A | Deceased, 12.98 (refractory, 0) |
| TCGA-25-2408 | Low-grade serous | N/A | Deceased, 30.89 (N/A) |
| TCGA-61-2095 | Low-grade serous | Moderately differentiated serous with focal prominent psammoma body | Deceased, 61.54 (N/A) |
H&E images were visualized at TCGA BioSig website hosted at Lawrence Berkeley National Laboratory. Clinical data was extracted from the supplemental data in the published TCGA ovarian paper in Nature Journal. MMMT, malignant mixed Mullerian tumor; N/A: information not available.
Pathological characteristics of high-grade ovarian serous cystadenocarcinoma with wild-type TP53
Pathology reports were available for seven samples with wild-type TP53. All seven cases had a diagnosis as HGOSC except for TCGA-24-2038, which was diagnosed as serous borderline tumor and was excluded in the survival analysis. Another two cases (TCGA-10-0933 and TCGA-24-1544) had mixed components with serous and mucinous neoplastic cells. High resolution H&E Images from frozen sections were available for all 15 cases from the Biosig website and were retrieved for detail review. In general, the histology images agreed with the available pathology reports. For the eight cases without a pathology report, four had high-grade serous features, one had features of malignant mixed mullerian tumor, and the remaining three had areas of low-grade papillary serous features. The pathological and clinical features were listed in Table 1 and two examples of H&E staining with low-grade features were shown in supplemental data (S4).
Mutational profiles of high-grade ovarian serous cystadenocarcinoma with wild-type TP53
Using somatic mutation level 3 TCGA data (validated mutations) for these 15 tumor samples with wild-type TP53, we identified 354 genes that were mutated (S5). This list of genes was analyzed through the use of IPA (Ingenuity® Systems, www.ingenuity.com). The top 3 biological functions associated with these mutated genes were identified (Table 2). Out of these 354 genes, 88 genes are involved in the pathogenesis of cancer and seven genes in the estrogen receptor signaling, which may be involved in the pathogenesis of these tumors. With the exception of KRAS mutations in two samples (TCGA-24-2038 and TCGA-25-1316), no BRAF, CTNNB1, PIK3CA, or ARIDIA mutations were found in any of these wild-type TP53 samples. Since these mutations were more frequently found in epithelial ovarian cancers of other histological subtypes – BRAF in serous borderline tumors and low-grade serous carcinomas, CTNNB1 in endometrioid ovarian carcinomas, and PIK3CA and ARIDIA in clear cell carcinomas, misclassification of these wild-type TP53 tumors from other histological subtypes as high grade ovarian serous cystadenocarcinomas was less likely.
Table 2.
Top biological functions associated with mutated genes in ovarian high grade serous carcinomas with wild-type TP53
| Top Biological Functions | p-value | Number of Genes |
|---|---|---|
| Top 3 Diseases and Disorders | ||
| Cancer | 0.00015 | 88 |
| Hematological Disease | 0.00024 | 26 |
| Immunological Disease | 0.00024 | 12 |
| Top 3 Molecular and Cellular Functions | ||
| Cellular Development | 0.00015 | 15 |
| Cell Cycle | 0.00015 | 25 |
| Cell Morphology | 0.00015 | 48 |
| Top 3 Canonical Pathways | ||
| Gα12/13 Signaling | 0.00271 | 8 |
| Glioblastoma Multiforme Signaling | 0.00313 | 8 |
| Estrogen Receptor Signaling | 0.00417 | 7 |
Reverse-Phase Protein Array, RNAseq, and SNP Array Data Analysis
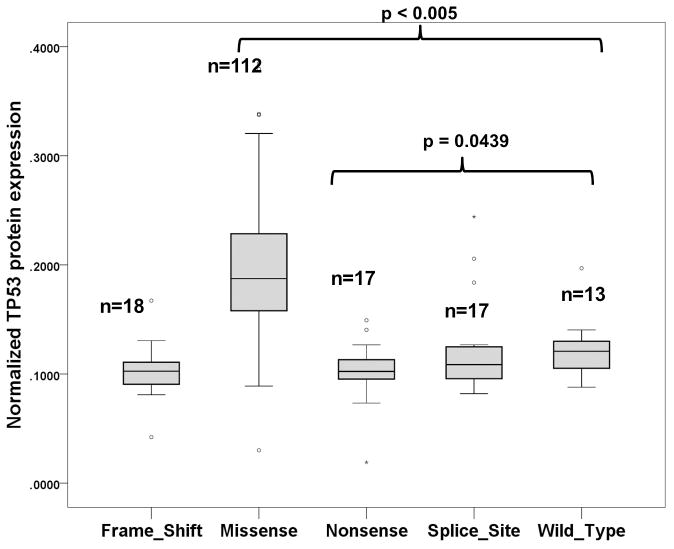
Reverse-phase protein array data for TP53 protein was available for thirteen wild-type TP53 samples and 112 samples with missense TP53 mutations. Wild-type TP53 samples had a significant 1.85-fold (p < 0.005) lower TP53 protein level than those with missense mutated TP53 but 1.22-fold (p=0.0439) higher than that of nonsense mutated TP53 samples (Figure 3). This result is in agreement with the fact that mutated TP53 protein is frequently more stable than wild-type TP53 protein is. Since mammalian cells has an RNA surveillance system (nonsense mediated mRNA decay) to eliminate faulty mRNA transcripts with nonsense mutations [13], we would expect that samples with nonsense TP53 mutation would have lower expression level of TP53 transcript and protein. Using the RNAseq data, we found that the mean TP53 transcript level (41.49 RPKM) of the thirteen wild-type TP53 samples was 4.15 fold higher than the mean TP53 transcript level (9.99 RPKM) of the 35 samples with nonsense TP53 mutation ( p = 0.001397; S4). More interestingly, we also identified that EDA2R gene, a direct target of TP53 [14], was highly up-regulated (>5-fold) in wild-type TP53 samples (S6 and S7). From the RPPA proteomic data, wild-type TP53 samples had a higher expression of collagen VI but lower expressions of B-Raf, Chk2, cyclin E1, GAB2, and Syk (S8 and S9). To determine if the wild-type phenotype could be due to the contamination of stromal component, we applied the ASCAT algorithm on the SNP array data with paired normal tumor samples to determine the percentage of normal stromal contamination and copy number aberrations. The median percentage of normal stromal contamination was 22%, which was not significantly different from the rest of the samples (Table 3). The SNP array data also indicated that these wild-type TP53 samples were genomically more stable with less DNA copy number aberrations than that of missense or nonsense TP53 mutated samples (Table 3, S10). These data support that these tumor samples have authentic wild-type TP53 tumor cells.
Table 3.
Estimation of total DNA copy number aberrations and percentage of normal contamination in TCGA high grade ovarian serous carcinomas by ASCAT algorithm using SNP array data.
| TP53 status | Total DNA Copy Number Aberrations | Percentage of normal contamination | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Mean | Std. Deviation | Range | Median | t-test | Mean | Std. Deviation | Range | Median | t-test | |
| wild-type (n=11) | 132 | 84 | 31–276 | 91 | 34 | 21 | 16–80 | 22 | ||
| Frame_Shift (n=18) | 193 | 112 | 65–560 | 162 | p=0.111 | 33 | 15 | 5–58 | 35 | p=0.940 |
| Missense_Mutation (n=109) | 199 | 122 | 60–670 | 176 | p=0.030 | 31 | 13 | 8–77 | 28 | p=0.633 |
| Nonsense_Mutation (n=17) | 204 | 91 | 74–374 | 208 | p=0.043 | 34 | 13 | 14–62 | 32 | p=0.950 |
| Splice_Site (n=17) | 214 | 122 | 80–556 | 173 | p=0.046 | 23 | 7 | 12–36 | 23 | p=0.135 |
Discussion
In this study, we found that the fifteen patients with wild-type TP53 in the published TCGA high grade ovarian serous carcinoma study (316 patients) had a poor survival than those patients with mutated TP53. To exclude the possibility of pathological misclassification, for these 15 samples we assessed for other gene mutations, reviewed available pathology reports, and reviewed all the images from H& E stained slides with a gynecologic pathologist. Furthermore, the protein and mRNA expression level of TP53 in these wild-type TP53 samples was significantly higher than that of samples with nonsense TP53 mutation. Moreover, there was a significant difference in copy number aberrations between wild-type and missense samples. The differences may be directly related to the TP53 mutation. When the function of TP53 is lost, genomic instability could result from checkpoint loss that leads to aneuploidy, gene amplifications, and other chromosomal aberrations [15]. All these data support that these samples have wild-type TP53.
The existence of HGOSC with wild-type TP53 have previously been demonstrated by sequencing analysis of DNA extracted from purified tumor cells [16] as well as by immunostaining of p21 (a downstream target of wild-type TP53) from these cells [17]. The existence of low-grade features in three of the supposedly HGOSC is interesting since low-grade serous carcinomas are usually more chemoresistant [18]. Although the progression of low-grade serous carcinoma to high-grade serous carcinoma is very rare, Vang et al found no TP53 mutations in a series of six ovarian serous carcinomas with both high-grade and low-grade components [19]. Although no TP53 mutation was detected in these samples, mutations were detected in 354 genes in all these samples together (Table 2, S5). Out of these 354 genes with mutations, 88 genes are involved in the pathogenesis of cancer, seven genes were involved in estrogen receptor signaling (Table 2), and six genes (DSCAML1, KIAA1012, TNN, MUC16, FBXW7, and KRAS) occurred in more than one wild-type TP53 samples (S5). Some of these genes may be involved in the pathogenesis of these tumors. Among these recurrent mutated genes, FBXW7 is a p53-dependent tumor suppressor [20] and down-regulation of FBXW7 expression correlates with poor survival in gastric cancer [21]. In a recent study, FBXW7 mutation was found to be significantly correlated with positive lymph nodes in endometrial cancer [22]. By querying the TCGA uterine cancer study using the cBio Cancer Genomics Portal [23], we found that FBXW7 is more frequently mutated in more aggressive uterine serous carcinomas (33%) than that of uterine endometrioid cancer (12%).
Recently, it was shown that wild-type TP53 promoted ovarian cancer cell survival and invasion by up-regulating genes involved in cell proliferation and metastasis in a mouse model [24]. Wild-type TP53 can also impair the effectiveness of chemotherapy against breast cancer [25]. However, the underlying molecular mechanism is not clear. From the RNAseq data, we found that EDA2R is significantly up-regulated in wild-type TP53 samples. EDA2R is a member of the tumor necrosis factor receptor family, which plays a major role in the activation of the NF-kappaB and JNK pathways [26]. Activation of NF-kappaB can enhance the chemoresistance of ovarian cancer cells through the regulation of MAPK and apoptosis [27]. From the RPPA data, collagen VI was found to be highly up-regulated in wild-type TP53 tumors (S7). Overexpression of collagen VI has previously been shown to contribute to cisplatin resistance in ovarian cancer cells [28]. Thus, the poorer survival of patients with wild-type TP53 tumors may be partly due to their tumors being more chemoresistant than mutated TP53 tumors are. In fact, seven of the ten cases of wild-type TP53 tumors with chemoresponse data were chemoresistant (Table 1). Thus, it may be beneficial to check HGOSC for TP53 mutation status by both DNA sequencing analysis and immunohistochemistry for stratification of patients for future clinical trials since both techniques will provide complementary validation of the TP53 mutational status [29]. Determination of TP53 mutation status will become more important for personalized therapy especially when promising drugs selectively targeting wild-type TP53 or mutant TP53 are emerging [30]. In a recent study, when an ovarian cancer cell line with wild-type TP53 was treated with Nutlin-3a, an MDM2 antagonist, cell cycle arrest and apoptosis were induced through up-regulation of p21 [24]. Nutlin-3a has been shown to have anti-proliferative activity in a number of malignancies with wild-type TP53 [31]. Thus, Nutlin-3a or similar agent could be a possible therapeutic strategy for these rare HGOSC patients with wild-type TP53. This study represents a detailed integrated analysis of 15 wild-type TP53 high grade serous carcinoma samples including RPPA data which was not available in the initial TCGA study. While the TCGA data set is the largest data set available at the moment, it will be important to have another dataset for validation. Since only H&E images from frozen sections for each of the samples were available for pathological review in this study and not all pathology reports are available, we could not eliminate the possibility of misdiagnosis in a few cases. Further investigation of the role of wild-type TP53 in chemoresistance is warranted.
Supplementary Material
S1. Clinical covariates and TP53 mutation status for survival analysis.
S2. Consolidated RNAseq data from 247 samples with TP53 mutation status (transcript expression level is in RPKM).
S3. Consolidated RPPA data from 177 samples (protein expression is in normalized linear values).
S4. Examples of TCGA ovarian high grade serous carcinomas with wild-type TP53 had low-grade serous papillary features.
S5. Gene mutations in TCGA ovarian samples with wild-type TP53.
S6. Differentially expressed genes between thirteen wild-type TP53 samples and 35 nonsense TP53 mutated samples based on RNAseq data.
S7. Differentially expressed genes between thirteen wild-type TP53 samples and 199 missense TP53 mutated samples based on RNAseq data.
S8. Differentially expressed proteins between thirteen wild-type TP53 samples and 17 nonsense TP53 samples based on RPPA data.
S9. Differentially expressed proteins between thirteen TP53 wild-type samples and 112 missense TP53 mutated samples based on RPPA data.
S10. Percentage of stromal contamination and total DNA copy number aberrations estimated by ASCAT algorithm.
Figure 2.
The protein expression level (normalized linear value) of TP53 protein from TCGA samples with wild-type TP53, TP53 missense mutations, and TP53 nonsense mutations. Data were retrieved from TCGA reverse-phase protein assay data. The box is bounded by the 25th and 75th percentile with the median depicted by the line in the box. Outlying values are drawn individually.
Acknowledgments
The results published here are in whole based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutes who constitute the TCGA research network can be found at http://cancergenome.nih.gov. Supported in part by grants from the National Institutes of Health, including The University of Texas MD Anderson Cancer Center Specialized Program of Research Excellence in Ovarian Cancer (P50 CA08369), grant CA133057, and MD Anderson’s Cancer Center Support Grant (CA016672); and the Blanton-Davis Ovarian Cancer Research Program; the Sara Brown Musselman Fund for Serous Ovarian Cancer Research. E.R.K. is supported by the National Cancer Institute–Department of Health and Human Services–National Institutes of Health Training of Academic Oncologists Grant (T32 CA101642).
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest
References
- 1.de Graeff P, Crijns AP, de Jong S, Boezen M, Post WJ, de Vries EG, et al. Modest effect of p53, EGFR and HER-2/neu on prognosis in epithelial ovarian cancer: a meta-analysis. Br J Cancer. 2009;101:149–59. doi: 10.1038/sj.bjc.6605112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hogdall EV, Christensen L, Hogdall CK, Frederiksen K, Gayther S, Blaakaer J, et al. Distribution of p53 expression in tissue from 774 Danish ovarian tumour patients and its prognostic significance in ovarian carcinomas. APMIS. 2008;116:400–9. doi: 10.1111/j.1600-0463.2008.00917.x. [DOI] [PubMed] [Google Scholar]
- 3.Darcy KM, Brady WE, McBroom JW, Bell JG, Young RC, McGuire WP, et al. Associations between p53 overexpression and multiple measures of clinical outcome in high-risk, early stage or suboptimally-resected, advanced stage epithelial ovarian cancers A Gynecologic Oncology Group study. Gynecol Oncol. 2008;111:487–95. doi: 10.1016/j.ygyno.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel F, Jung J, Bohnke A, Gradhand E, Zeng K, Thomssen C, et al. Both germ line and somatic genetics of the p53 pathway affect ovarian cancer incidence and survival. Clin Cancer Res. 2008;14:89–96. doi: 10.1158/1078-0432.CCR-07-1192. [DOI] [PubMed] [Google Scholar]
- 5.Consortium TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biosig T. http://tcga.lbl.gov:8080/biosig/tcgadownload.do.
- 8.Portal TCGAD. https://tcga-data.nci.nih.gov/tcga/
- 9.Van Loo P, Nilsen G, Nordgard SH, Vollan HK, Borresen-Dale AL, Kristensen VN, et al. Analyzing cancer samples with SNP arrays. Methods Mol Biol. 2012;802:57–72. doi: 10.1007/978-1-61779-400-1_4. [DOI] [PubMed] [Google Scholar]
- 10.Neeley ES, Kornblau SM, Coombes KR, Baggerly KA. Variable slope normalization of reverse phase protein arrays. Bioinformatics. 2009;25:1384–9. doi: 10.1093/bioinformatics/btp174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–10. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 14.Tanikawa C, Ri C, Kumar V, Nakamura Y, Matsuda K. Crosstalk of EDA-A2/XEDAR in the p53 signaling pathway. Mol Cancer Res. 2010;8:855–63. doi: 10.1158/1541-7786.MCR-09-0484. [DOI] [PubMed] [Google Scholar]
- 15.Smith ML, Fornace AJ., Jr Genomic instability and the role of p53 mutations in cancer cells. Curr Opin Oncol. 1995;7:69–75. [PubMed] [Google Scholar]
- 16.Salani R, Kurman RJ, Giuntoli R, 2nd, Gardner G, Bristow R, Wang TL, et al. Assessment of TP53 mutation using purified tissue samples of ovarian serous carcinomas reveals a higher mutation rate than previously reported and does not correlate with drug resistance. Int J Gynecol Cancer. 2008;18:487–91. doi: 10.1111/j.1525-1438.2007.01039.x. [DOI] [PubMed] [Google Scholar]
- 17.Cai KQ, Wu H, Klein-Szanto AJ, Xu XX. Acquisition of a second mutation of the Tp53 alleles immediately precedes epithelial morphological transformation in ovarian tumorigenicity. Gynecol Oncol. 2009;114:18–25. doi: 10.1016/j.ygyno.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gershenson DM, Sun CC, Bodurka D, Coleman RL, Lu KH, Sood AK, et al. Recurrent low-grade serous ovarian carcinoma is relatively chemoresistant. Gynecol Oncol. 2009;114:48–52. doi: 10.1016/j.ygyno.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Vang R, Shih Ie M, Kurman RJ. Ovarian low-grade and high-grade serous carcinoma: pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv Anat Pathol. 2009;16:267–82. doi: 10.1097/PAP.0b013e3181b4fffa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–9. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 21.Yokobori T, Mimori K, Iwatsuki M, Ishii H, Onoyama I, Fukagawa T, et al. p53-Altered FBXW7 expression determines poor prognosis in gastric cancer cases. Cancer Res. 2009;69:3788–94. doi: 10.1158/0008-5472.CAN-08-2846. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Dios DA, Lambrechts D, Coenegrachts L, Vandenput I, Capoen A, Webb PM, et al. High-throughput interrogation of PIK3CA, PTEN, KRAS, FBXW7 and TP53 mutations in primary endometrial carcinoma. Gynecol Oncol. 2013;128:327–34. doi: 10.1016/j.ygyno.2012.11.037. [DOI] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullany LK, Liu Z, King ER, Wong KK, Richards JS. Wild-type tumor repressor protein 53 (Trp53) promotes ovarian cancer cell survival. Endocrinology. 153:1638–48. doi: 10.1210/en.2011-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson JG, Pant V, Li Q, Chang LL, Quintas-Cardama A, Garza D, et al. p53-Mediated Senescence Impairs the Apoptotic Response to Chemotherapy and Clinical Outcome in Breast Cancer. Cancer Cell. 21:793–806. doi: 10.1016/j.ccr.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinha SK, Zachariah S, Quinones HI, Shindo M, Chaudhary PM. Role of TRAF3 and -6 in the activation of the NF-kappa B and JNK pathways by X-linked ectodermal dysplasia receptor. J Biol Chem. 2002;277:44953–61. doi: 10.1074/jbc.M207923200. [DOI] [PubMed] [Google Scholar]
- 27.Yang G, Xiao X, Rosen DG, Cheng X, Wu X, Chang B, et al. The biphasic role of NF-kappaB in progression and chemoresistance of ovarian cancer. Clin Cancer Res. 2011;17:2181–94. doi: 10.1158/1078-0432.CCR-10-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman-Baust CA, Weeraratna AT, Rangel LB, Pizer ES, Cho KR, Schwartz DR, et al. Remodeling of the extracellular matrix through overexpression of collagen VI contributes to cisplatin resistance in ovarian cancer cells. Cancer Cell. 2003;3:377–86. doi: 10.1016/s1535-6108(03)00058-8. [DOI] [PubMed] [Google Scholar]
- 29.Yemelyanova A, Vang R, Kshirsagar M, Lu D, Marks MA, Shih Ie M, et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol. 2011;24:1248–53. doi: 10.1038/modpathol.2011.85. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann BD, Pietenpol JA. Targeting mutant p53 in human tumors. J Clin Oncol. 2012;30:3648–50. doi: 10.1200/JCO.2012.44.0412. [DOI] [PubMed] [Google Scholar]
- 31.Kapoor S. The emerging anti-proliferative role of Nutlin-3 in the pathogenesis of systemic malignancies. Cancer Biol Ther. 2013;14:5. doi: 10.4161/cbt.22629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. Clinical covariates and TP53 mutation status for survival analysis.
S2. Consolidated RNAseq data from 247 samples with TP53 mutation status (transcript expression level is in RPKM).
S3. Consolidated RPPA data from 177 samples (protein expression is in normalized linear values).
S4. Examples of TCGA ovarian high grade serous carcinomas with wild-type TP53 had low-grade serous papillary features.
S5. Gene mutations in TCGA ovarian samples with wild-type TP53.
S6. Differentially expressed genes between thirteen wild-type TP53 samples and 35 nonsense TP53 mutated samples based on RNAseq data.
S7. Differentially expressed genes between thirteen wild-type TP53 samples and 199 missense TP53 mutated samples based on RNAseq data.
S8. Differentially expressed proteins between thirteen wild-type TP53 samples and 17 nonsense TP53 samples based on RPPA data.
S9. Differentially expressed proteins between thirteen TP53 wild-type samples and 112 missense TP53 mutated samples based on RPPA data.
S10. Percentage of stromal contamination and total DNA copy number aberrations estimated by ASCAT algorithm.