Abstract
Purpose
Despite aggressive conventional therapy, glioblastoma multiforme (GBM) remains uniformly lethal. Immunotherapy, in which the immune system is harnessed to specifically attack malignant cells, offers a treatment option with less toxicity. The expression of cytomegalovirus (CMV) antigens in GBM presents a unique opportunity to target these viral proteins for tumor immunotherapy. Although the presence of CMV within malignant gliomas has been confirmed by several laboratories, its relevance as an immunological target in GBM has yet to be established. The objective of this study was to explore whether T cells stimulated by CMV pp65 RNA-transfected dendritic cells (DCs) target and eliminate autologous GBM tumor cells in an antigen-specific manner.
Experimental Design
T cells from patients with GBM were stimulated with autologous DCs pulsed with CMV pp65 RNA, and the function of the effector CMV pp65-specific T cells was measured.
Results
In this study, we demonstrate the ability to elicit CMV pp65-specific immune responses in vitro using RNA-pulsed autologous DCs generated from patients with newly diagnosed GBM. Importantly, CMV pp65-specific T cells lyse autologous, primary GBM tumor cells in an antigen-specific manner. Moreover, T cells expanded in vitro using DCs pulsed with total tumor RNA demonstrated a 10–20 fold expansion of CMV pp65-specific T cells as assessed by tetramer analysis and recognition and killing of CMV pp65-expressing target cells.
Conclusion
These data collectively demonstrate that CMV-specific T cells can effectively target glioblastoma tumor cells for immunologic killing and support the rationale for the development of CMV-directed immunotherapy in patients with GBM.
Keywords: Cytomegalovirus, immunotherapy, dendritic cells, gliomas, T cells
Introduction
Despite aggressive surgery, radiation and chemotherapy, treatment for patients with GBM is rarely curative and conventional therapies are inherently non-specific and damaging to surrounding normal tissues (1, 2). Immunotherapy provides a promising alternative due to the intrinsic specificity and potentially long-lasting effects of immune activation. Substantial evidence suggests that T cells can eradicate large, well-established tumors in mice and humans with exquisite precision even when tumors reside within the “immunologically privileged” central nervous system (CNS) (3–5). The barriers that prevent most humoral immune components from entering the brain are not an impediment to T-cell mediated therapy as activated T cells have been shown to have access to the CNS and tumor infiltration of CD8+ T cells in newly-diagnosed GBM has been reported in some studies to be associated with long-term survival (4, 6, 7). The advancement of immunotherapy for patients with GBM is hampered significantly by the lack of identification of tumor rejection antigens that are consistently expressed in the majority of patients. This has prompted the frequent use of unfractionated tumor antigens in the form of lysates, peptides, or total tumor RNA in immunotherapeutic approaches to GBM. Although these methods have shown promise in early phase clinical studies, they are limited by requirement of sufficient tumor tissue for vaccine preparation. The capacity to safely target GBM tumors with an immunogenic and conserved tumor-associated antigen would significantly accelerate the developmental pathway for an effective brain tumor immunotherapy.
The recent identification of antigens unique to CMV being expressed in the majority of GBM tumors, with limited expression in normal brain, brings to the forefront the potential to leverage CMV-directed immunity as a novel approach in the targeting of invasive glial tumors. The role of CMV in gliomagenesis and progression remains under intense study, and the relatively low levels of viral proteins and nucleic acids that require sensitive modalities for reliable detection call into question the physiologic relevance of subclinical CMV expression within these tumors. We explored whether CMV-specific T cell responses could be consistently elicited in vitro using CMV pp65 RNA-pulsed DCs from patients with newly-diagnosed GBM, and whether these T cells were capable of recognition and lytic killing of autologous primary GBM tumor cells expressing endogenous levels of tumor antigens. Mature CMV pp65 RNA-pulsed DCs were reliably generated from patients with GBM and were capable of expanding CMV-specific CD4+ and CD8+ polyfunctional T cells in vitro comparable in function to that from healthy volunteers. Importantly, CMV-specific T cells recognized and lysed autologous primary GBM tumor cells and antigen- pulsed autologous DCs in a CMV-restricted manner in vitro. Influenza-specific T cells generated from patients with GBM were potent in the recognition and killing of influenza expressing target cells but did not recognize and kill autologous tumor cells in vitro, further confirming the antigen-specific killing of GBM tumor cells by CMV-specific T cells. Additional studies using total tumor RNA-pulsed DCs demonstrated the in vitro expansion of CMV-specific T cells when stimulated by DCs expressing total tumor antigens and the killing of CMV pp65 expressing target cells in vitro, providing cross-validation that endogenous CMV antigen expression in GBM tumor cells is sufficient for immunologic recognition and killing.
To our knowledge, this study is the first to demonstrate that CMV-specific T cells are effective in recognition and killing of autologous GBM tumor cells expressing endogenous levels of antigen, and provides compelling support for the development of CMV-directed immunotherapy in patients with malignant gliomas.
Materials and Methods
Peripheral blood mononuclear cells (PBMCs) and tumor cells
PBMCs were obtained by leukapheresis of patients with newly diagnosed GBM post-surgical resection and prior to treatment with radiation or chemotherapy. Cells were obtained from human subjects following written informed consent using protocols approved by the Duke University Institutional Review Board. Patient tumors were rinsed 2–3 times with PBS to remove blood and necrotic tissue and then minced into small fragments. Minced tumor tissue was transferred to a sterile container in Earls Balanced Salt Solution (EBSS). Each gram of tumor was digested with 100 units papain (Worthington) and 1,000 units DNase I (Worthington) with continuous low-speed stirring at 37°C for 20 minutes. The mixture was then triturated for 10 minutes as described by the manufacturer. Cells were harvested and filtered through a 70µm screen into 50ml tubes and centrifuged at 500 × g for 5 minutes. Digested tumor pellets were resuspended in 1 ml Neurobasal medium (Gibco) with DNase (200 Units/ml). After a 5-minute incubation, cells were diluted with PBS and viable cells were harvested over a Ficoll gradient (Sigma). Viable tumor cells at the interface were harvested, washed with PBS, and resuspended in human AB serum with 10% DMSO at 5–10×106 cells/ml. For use as tumor targets, the cells were thawed and cultured in Richter's Zinc Option media with 10% FBS for 7–14 days.
RNA
Generation of pSP73-Sph/A64 was done by adding oligonucleotides containing 64 A-T bp followed by an SpeI restriction site placed between the EcoRI and NarI sites of pGEM4Z (Promega) to create the plasmid pGEM4Z/A64. The HindIII–NdeI fragment of pGEM4Z/A64 was cloned into pSP73 (Promega) digested with HindIII and NdeI to create pSP73/A64. The plasmid pSP73-Sph was created by digesting pSP73/A64 with SphI, filling in the ends with T4 DNA polymerase and re-ligating. pSP73-Sph/A64/Not contains a NotI restriction site adjacent to the SpeI site. The cDNA encoding CMV pp65 in the pBluescript vector (generously provided by Dr. T. Clay, GlaxoSmithKline Biologicals, Rixensart, Belgium) was excised and cloned into the BamHI and SalI sites of pSP73-Sph/A64 (pSP73-Sph/A64/CMVpp65). The cDNA for GFP was derived from pEGFP-N1 (Clontech, Palo Alto, California) and inserted into pGEM4Z/A64 (pGEM4Z/A64/GFP). The gene encoding the full-length Flu M1 matrix protein (generously provided by Dr. A. Steinkasserer, University Hospital Erlangen, Erlangen, Germany) was inserted into the pSP73-Sph/A64 (pSP73-Sph/A64/Flu1). The gene encoding full length survivin was cloned by isolating total RNA from human tumor cells followed by reverse transcription using oligo dT primers. Survivin cDNA was amplified from the first strand using the forward primer 5’-TATATAAGCTTGCCACCATGGGTGCCCCGACGTTG-3’ and the reverse primer 5’-TATATAGAATTCAATCCATGGCAGCCAGC-3’. The resulting fragment was cloned into the HindIII and BamHI sites of pSP73-Sph/A64. All plasmids were digested with SpeI for use as a template for in vitro transcription reactions using the mMESSAGE mMACHINE T7 kit (Ambion, Austin, TX) according to the manufacturer’s protocol. mRNA was purified with the RNeasy mini kit (Qiagen).
Isolation of total cellular RNA from tumor cells
Total RNA was isolated from the autologous tumor cells of patients and autologous PBMCs using RNeasy RNA isolation kits (Qiagen) according to the manufacturer’s protocol.
Generation and pulsing of dendritic cells (DCs)
Cells were thawed, washed in PBS and resuspended at 2×108 cells in 30 ml AIM-V media (Invitrogen) in T-150 tissue-culture flasks. Cells were incubated for 1 hour at 37°C, 5% CO2 in a humidified incubator. Non-adherent cells were harvested by rocking the flask from side-to-side to dislodge them. Adherent cells were replenished with 30 ml AIM-V supplemented with 800 U/ml human GM-CSF and 500 U/ml human IL-4, then incubated at 37°C. DCs were harvested on day 6, by collecting all non-adherent cells, followed by a cold PBS wash. Cells that were still adherent were dissociated with cell dissociation buffer (Invitrogen), 37°C for 20 minutes. DCs were washed, counted and maintained on ice until use. DCs were pulsed in 2-mm cuvettes (200 µl) at 300 V for 500 µs using an Electro Square Porator (ECM 830, BTX, San Diego, CA). Antigen mRNA was used at 3–10 µg/106 DCs. RNA- pulsed DCs were matured for 8–10 hours in AIM-V media containing GM-CSF (800 U/ml), IL-4 (500 U/ml) and the maturation cytokine cocktail of TNF-α (10 ng/ml), IL-1β (10 ng/ml), IL-6 (1000 U/ml), and PGE2 (1 µg/ml). All cytokines were obtained from R&D Systems. PGE2 was purchased from Sigma.
DC phenotype analysis
PE-labeled anti-CD25, anti-CD80, anti-CD86 (BD Biosciences), anti-CD14, anti-CD197/CCR7 (eBioscience), anti-CD40 and anti-CD83 (Immunotech) antibodies were used. DCs were incubated with 20 µl Fc-block (eBioscience) on ice for 20 minutes. Labeled antibodies were added and cells were stained for 20 minutes at 4°C. Cells were washed and kit resuspended in PBS with 1% formaldehyde before being acquired on a FACSCaliber flow cytometer (Becton Dickinson). Data was analyzed using CellQuest (BD Biosciences) or FlowJo (Tree Star, Inc.).
In vitro stimulation of T cells with RNA-transfected DCs
PBMCs were thawed and resuspended in PBS and treated with DNase I (Sigma) at 200 U/ml for 20 minutes at 37°C. DNase I-treated PBMC were incubated for 1 hour at 37°C, 5% CO2 in a humidified incubator. Non-adherent cells (2×106 cells/ml) were harvested and stimulated with RNA-transfected, matured DCs at a responder to stimulator ratio of 10:1 in the presence of 25 ng/ml IL-7. All stimulations were done in RPMI 1640 with 10% FBS, 2 mM L-glutamine, 20 mM HEPES, 1 mM sodium pyruvate, 0.1 mM MEM non-essential amino acids, 100 IU/ml penicillin, 100 µg/ml streptomycin and 5×10−5 M β-mercaptoethanol (CTL stimulation medium). IL-2 was added at 100 U/ml on day 3 and every 4–5 days for 12–14 days. T cells were maintained at 1–2×106 cells/ml in CTL stimulation medium. T cells were harvested on day 12–14, counted and used as effector T cells in a europium-release CTL assay. Autologous DCs transfected with tumor antigen-encoding mRNA were used as targets.
Antibody and tetramer staining of T cells
The following tetramers and fluorochrome-labeled antibodies were used: PE labeled tetramers (Beckman Coulter) loaded with HLA-A*0201 CMV pp65 (amino acids 495–503, NLVPMVATV), HLA-B*0702 CMV pp65 (amino acids 417–426, TPRVTGGGAM), HLA-B*3501 CMV pp65 (amino acids 123–132, IPSINVHHY), HLA-A*2402 CMV pp65 (amino acids 341–349, QYDPVAALF), CD4-APC (BD Biosciences), CD8-FITC (eBioscience), B cell CD19-PE, natural killer (NK) cell CD56-PE, CD25-PE (BD Biosciences). T cells were stained with tetramer and antibodies for 30 minutes at RT. 5×105 in vitro stimulated T cells were incubated with 5 µl of tetramer in 100 µl FACS buffer (PBS with 4% FBS). The cells were washed once with FACS buffer and resuspended in 300 µl PBS with 1% formaldehyde for data acquisition. Isotype controls included the corresponding fluorochrome-conjugated or unconjugated mouse IgG1 or IgG2a. Data was acquired on a FACSCaliber flow cytometer and analyzed using CellQuest or FlowJo.
Analysis of T cell polyfunction
Polyfunctional CMV-specific CD4 and CD8 T cell responses (IFN-γ, IL-2, TNF-α and CD107) were evaluated using a polychromatic intracellular flow cytometry. PBMCs were thawed and rested overnight at 37°C/5% CO2 in RPMI with 10% FBS (R10) for 12–20 hours. PBMCs (2×106/well) were stimulated for 6 hours at 37°C/5% CO2 with CMV pp65 PepMix (1 µg/ml, JPT Peptide Technologies), SEB (Staphylococcal enterotoxin B, 1 µg/ml, Sigma), or no antigen in the presence of co-stimulatory mAbs, anti-CD28 and anti-CD49d (1 µg/ml of each, BD Biosciences), and protein transport inhibitors, monensin (1 µg/ml; Golgistop, BD Biosciences) and Brefeldin A (5 µg/ml, Sigma). Following stimulation, cells were treated with EDTA for 15 minutes at RT (18–22°C). Cells were stained using manufacturer’s protocol for FACSLysing solution and FACSPermeabilization Solution II (BD Biosciences) using the following mAbs: exclusion markers CD14-Pacific Blue (BD Biosciences), CD19-Pacific Blue (Invitrogen), violet LIVE/DEAD fixable dead cell stain kit (Invitrogen); basic subset markers CD3-AmCyan (BD Biosciences), CD4-PerCP-Cy5.5 (BD Biosciences), CD8-APC-Cy7 (BD Biosciences); maturational markers CD45RO-ECD (Beckman Coulter), CD27-APC (BD Biosciences), CD57-FITC (BD Biosciences) and functional markers IFN-γ-PE-Cy7 (BD Biosciences), IL-2-PE (BD Biosciences), TNF-α-Alexa 700 (BD Biosciences), and CD107-PE-Cy5 (BD Biosciences.). Cells were washed in FACS buffer, acquired within 6 hours of staining on a custom 18-parameter LSRII (BD Biosciences) and analyzed using FlowJo.
Alternately, following stimulation (as described above) T cells were only analyzed for phenotype and IFN-γ production using CD4/CD8-FITC, CD69-PE, CD3-PerCP-Cy5.5 and IFN-γ-APC. The cells were washed once with FACS buffer, resuspended in 300 µl PBS with 1% formaldehyde and data acquired on a FACSCaliber.
Gating scheme for polychromatic intracellular flow cytometric assay
Basic subsets for CD4+ and CD8+ T-cells were identified using a series of sequential gates starting with singlets (FSC-W versus FSC-H), viable CD3+ T cells (CD3 versus Exclusion), scatter lymphocytes (FSC-A versus SSC-A), and finally CD8 versus CD4 to identify the three basic subset gates: CD4+CD8-(CD4+ viable T lymphocytes), CD8+CD4- (CD8+ viable T lymphocytes), and double positives (CD4+ CD8+). For each of the 3 basic subsets (total population) 4 cytokine gates were defined: IFN-γ+, TNF-α+, IL-2+, and CD107+. Additionally, maturational subsets were defined as CD45RO-CD27+ (naïve), CD45RO+CD27+ (central memory), CD45RO+CD27- (effector memory), CD45RO+CD57+(effector), CD45RO-CD57+ (terminal effector) were gated off of the 3 basic subsets and functional markers were then gated for each respective maturational subset. The positive and negative controls (SEB and unstimulated) were used at the level of the cytokine gates to maximize the positive signal and minimize the negative using back-gating, or tweaking each of the basic, maturational, and functional gates to improve purity and recovery of the functional population of interest. Cytokine gates were then applied across all test samples.
In vitro cytotoxicity assay
Target cells were harvested, washed to remove all traces of media and labeled with europium (Eu). The Eu-labeling buffer (1 ml/target) contains 1 ml HEPES buffer (50mM HEPES, 93mM NaCl, 5mM KCl, 2mM MgCl2, pH 7.4), 10 µl Eu (10mM EuCl3.6H2O in 0.01N HCl), 5 µl DTPA (100mM diethylenetriamine pentaacetate in HEPES buffer) and 4 µl DS (1% dextran-sulfate) (8). Cells were resuspended in 1 ml of the europium-labeling buffer very gently and incubated on ice for 20 minutes. 30 µl of CaCl2 solution (100mM) was then added to the labeled cells, mixed and the cells were incubated for another 5 minutes on ice. 30 ml of Repair buffer (HEPES buffer with 10mM glucose, 2mM CaCl2) was added to the cells and the cells were centrifuged at 1000 rpm for 10 minutes. Cells were counted and 5×106 cells were washed 4 times with Repair buffer. After the final wash the cells were resuspended in CTL stimulation RPMI-10% FCS medium with no penicillin-streptomycin at 105 cells/ml. 104 Eu-labeled targets and serial dilutions of effector cells were cocultured in 200 µl at 37°C for 4 hours. Eu-release in the supernatant (50 µl) was measured by time- resolved fluorescence (VICTOR3 Multilabel Counter, Perkin Elmer). % specific lysis = [(experimental release - spontaneous release)/(total release -spontaneous release)] × 100. Spontaneous release of the target cells was less than 25% of total release by detergent in all assays for the results of the assay to be valid. Spontaneous release of the target cells was determined by incubating the target cells in medium without T cells. All assays were done in triplicate.
Cytokine Bead Array (CBA) analysis
In addition to measuring cytotoxicity using the Eu-release cytotoxicity assay, an additional 50 µl of supernatant was harvested after 8 hours from the effector + target cocultures described above and used for analysis of cytokines. Supernatant (50 µl) harvested from effector and target cells cultured alone were used as additional controls. Samples were stored frozen at −20°C until the day of the assay. The BD™ CBA Human Th1/Th2 Cytokine Kit II (#551809) was used to measure the presence of the cytokines IL-2, IL-4, IL-6, IL-10, TNF-α and IFN-γ and assayed exactly as described in the kit.
Western Blot
Western blot analysis was performed on de-identified samples of newly diagnosed high-grade glioma (WHO grade IV) from the Duke Medical Center tumor repository. Tumor cells were thawed and lysed in RIPA buffer (Thermo Scientific), separated on a 12.5% PAGE-SDS gel (BioRad), and transferred to nitrocellulose membrane (Bio-Rad) blocked in Protein-Free T20 TBS Blocking Buffer (Thermo Scientific). Blots were probed with monoclonal anti-CMV IE1 (Millipore #MAB810, 1:500 dilution) or monoclonal anti-CMV pp65 (Santa Cruz, #sc-71229, 1:1,000 dilution) and probed with secondary goat anti- mouse IgG-HRP (Thermo Scientific-Pierce, 1:10,000 dilution), developed with SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) and imaged with Alpha InnoTech HD-2. Blots were stripped with Restore Western Blot Stripping Buffer (Thermo Scientific-Pierce) and reprobed with monoclonal anti-β-Actin (clone C4, Santa Cruz, 1:2,500 dilution) and developed with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). Controls include CMV-infected (ViruSys, #CV001-1) and VZV-infected (ViruSys, #AV043-1) cell extract.
Results
Presence of CMV antigens in glioma tissue but not normal healthy brain tissue
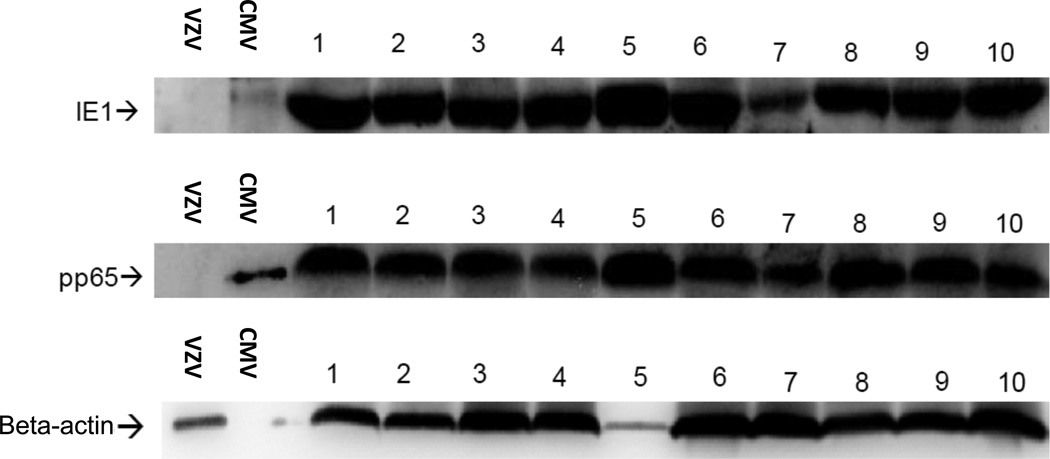
The presence of CMV in glioma tissue was first reported by Cobbs et al. in 2002 (9). Since this first report, seven independent laboratories have confirmed the detection of CMV within the vast majority of malignant gliomas at the DNA, RNA, and protein level (10–16). Most reports to date examining CMV protein expression within GBM have relied on immunohistochemical detection using an optimized and amplified detection protocol. For confirmatory identification of CMV proteins within GBM tumors, we conducted western blot analysis on primary GBM tumors for detection of CMV pp65 and IE1 (Figure 1). Appropriately sized proteins were detected in GBM lysates and lysates from CMV-infected fibroblasts but not in lysates of Varicella Zoster virus-infected fibroblasts (VZV). Additionally, CMV antigens were not detected in normal brain lysates from deceased patients (>50-years old) who died of non-brain tumor related causes (data not shown).
Figure 1. Detection of CMV proteins in primary GBM.
Single cell digests were prepared from de-identified newly diagnosed GBM specimens, washed with PBS and lysates prepared in the presence of proteinase inhibitors. Western blot (WB) analysis was performed using CMV IE1-specific and pp65-specific mAbs on GBM lysates (20 µg per lane). CMV- and VSV-infected fibroblast lysates were used as positive and negative control samples respectively (1 µg per lane). Figure depicts 10 GBM specimens positive for CMV proteins IE1 and pp65. Normal brain lysates obtained from autopsy specimens were negative for detection of CMV proteins (0 out of 5, data not shown).
Generation of mature CMV pp65 RNA-pulsed DCs from patients with newly-diagnosed GBM
To evaluate the T cell stimulation potential of DCs generated from patients with GBM, we harvested peripheral blood mononuclear cells (PBMCs) via leukapheresis from CMV-seropositive patients with newly diagnosed GBM. DCs were generated as described in Methods and then pulsed with antigen-encoding mRNA. Pulsed DCs were matured in a cytokine cocktail containing GM-CSF, IL-4, TNFα, IL-1β, IL-6 and PGE2 after electroporation with CMV pp65 RNA (17). DC maturation is characterized phenotypically by high expression of membrane bound costimulatory molecules, CD80 and CD86, as well as CD83 and MHC class II molecules. Mature DCs also upregulate chemokine receptors, notably CCR7, that enables effective migration to lymph nodes for antigen presentation to T cells (18). Phenotypic analysis of the pulsed, matured DCs revealed a significant increase in surface expression of CD80, CD83, CD25, and CCR7 as compared to pulsed immature DCs (Supplementary Figure S1). Mature DCs also exhibited increase in expression of CD40, MHC class II, and CD86 although the increase in expression of these proteins was modest compared to CD80, CD83, CD25, and CCR7 (Supplementary Figure S2). These results demonstrate that DCs can be consistently generated from the PBMCs of GBM patients using standard protocols, electroporated with RNA and that maturation of these RNA pulsed-DCs using a cytokine cocktail containing TNF-α, IL-1β, IL-6 and PGE2 results in phenotypically mature DCs.
Activation of functional CMV pp65-specific T cells from patients with GBM using CMV pp65 RNA-pulsed autologous DCs
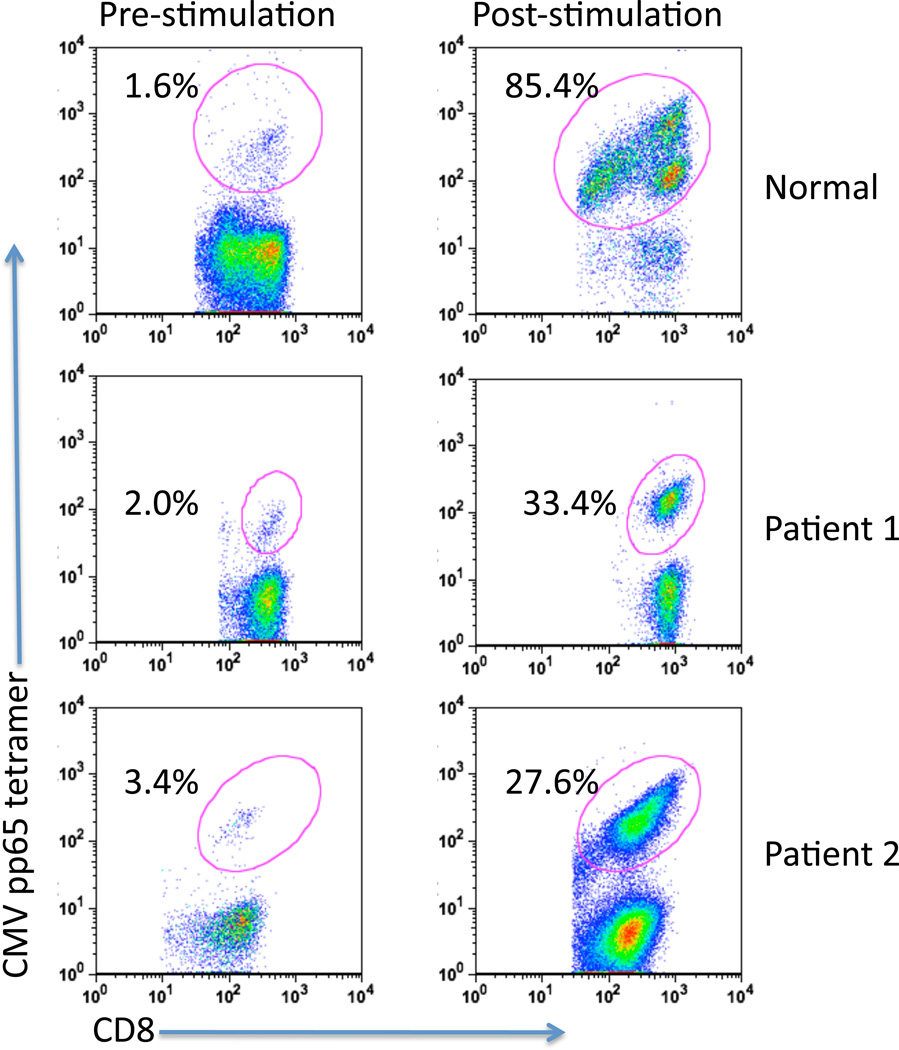
To determine the capacity to induce CMV pp65-specific T cells in vitro, PBMCs from patients with GBM and from a healthy donor were cultured in the presence of autologous CMV pp65 RNA-pulsed matured DCs in CTL stimulation media (Materials and Methods) for 12–14 days. Patient characteristics are presented in Supplementary Table S1. As shown in Figure 2, GBM patient-derived DCs pulsed with CMV pp65 RNA expanded high levels of CD8+ CMV pp65-specific cells, based on the increase of CMV tetramer+ CD8+ T cells (Pre-stimulation versus Post-stimulation). The ability to reliably induce CD8+ CMV pp65-specific cells was further confirmed using cells from additional patients with newly diagnosed GBM (Supplementary Table S2).
Figure 2. CMV pp65 RNA-pulsed mature dendritic cells induce robust activation of CMV pp65-specific CD8+ T cells.
T cells were stimulated ex vivo with autologous CMV pp65 RNA-pulsed DCs using PBMCs from 1 normal donor and 2 GBM patients. T cells were analyzed pre- and post-stimulation for the presence of CD8+ and CMV pp65 tetramer+ cells using flow cytometry as described in Methods. CMV pp65 HLA-A*0201 (normal and Patient 1) and HLA-B*3501 (Patient 2) PE-labeled tetramers were used.
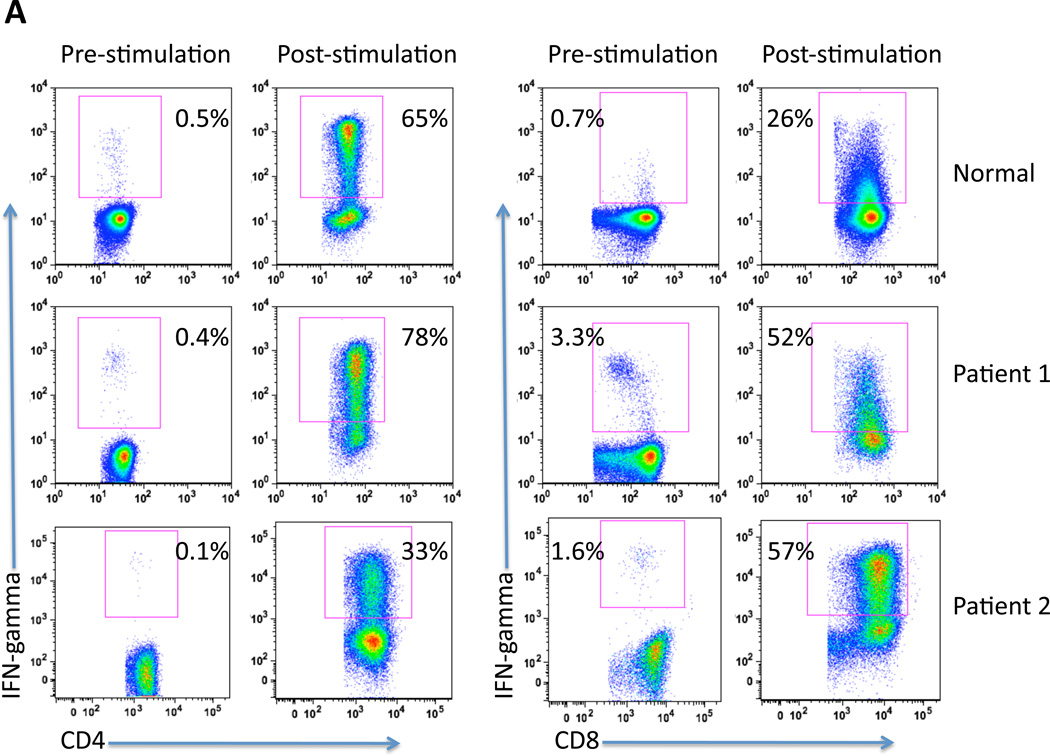
In vitro induced CMV pp65-specific T cells shown in Figure 2 were further characterized for function by measuring IFN-γ production by CD4 and CD8 T cells using intracellular flow cytometry. Effector cells were analyzed from a normal volunteer and 2 patients with GBM, pre- and post-stimulation. As depicted in Figure 3A, there was a substantial increase in IFN-γ+/CD4+ and IFN-γ+/CD8+ cells in all subjects tested post-stimulation with CMV pp65 RNA-loaded DCs. Again, the expansion of activated IFN-γ-secreting T cells was comparable in GBM patients and healthy donors.
Figure 3. CMV pp65 RNA-pulsed mature dendritic cells induce CMV pp65-specific T cell expansion and polyfuntionality.
T cells were stimulated ex vivo with autologous CMV pp65 RNA-pulsed DCs.
A) CD4 and CD8 T cell populations were analyzed for co-expression of IFN-γ pre and post-stimulation. PBMCs from 1 normal donor and 2 GBM patients (Patient 1 and Patient 2) were used.
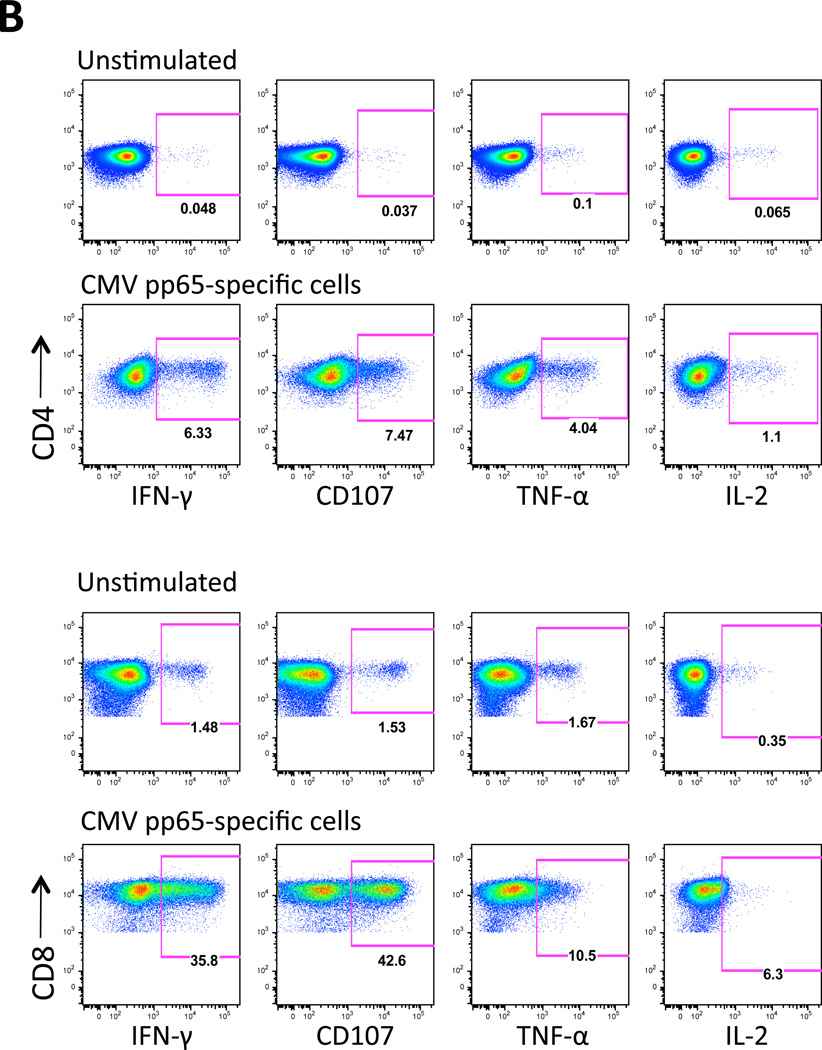
B) GBM patient PBMC-derived CD4 and CD8 T cells were characterized for functionality by measuring IFN-γ, TNF-α, CD107, and IL-2 using flow cytometry as described in Methods. PBMCs from 1 GBM patient (Patient 3) were used.
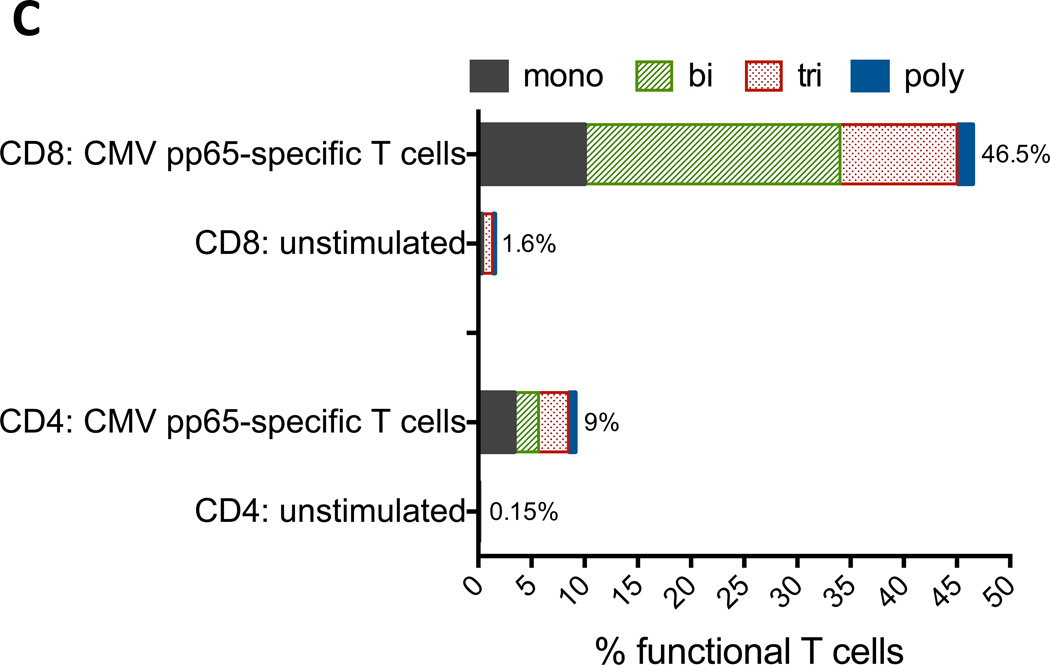
C) Analysis of percent functional T cells from data shown in Figure 3B. Polyfunctional T cells exhibit all four functions. Analysis of PBMCs from 1 GBM patient (Patient 3) is shown.
Multiple studies have reported the importance of polyfunctional T cells, which are characterized as T cells with multiple functions that includes cytotoxicity (expression of CD107) and the secretion of IFN-γ, TNF-α and IL-2 (19). Polyfunctional T cells are associated with protective immunity after vaccination (20–24), control of infections including CMV (25–28), and favorable outcomes in HIV/AIDS (29–32) and tumor regression (33–37).
We therefore evaluated whether CMV pp65-specific T cells generated from patients with GBM exhibited polyfunctional effector responses after in vitro co-culture with CMV pp65 RNA-pulsed DCs. As shown in Figures 3B and 3C, in vitro stimulated T cells demonstrated an increase in functional T cells as measured by expression of CD107 and secretion of IFN-γ, TNF-α and IL-2 compared to unstimulated cells; 1.6% to 46.5% CD8 T cells and 0.15% to 9% activated CD4 T cells respectively (Figure 3C). Moreover, a majority of these T cells (78.5% CD8 and 62.8% CD4) exhibited more than one function (Figure 3C), demonstrating multifunctional capacity.
Collectively, Figures 2 and 3 demonstrate that the functions of the antigen-specific T cells derived from GBM patient PBMCs were comparable to those derived from a healthy donor, indicating the capacity to generate robust CMV pp65-specific T cells from patients with GBM using RNA-pulsed autologous DCs.
CMV pp65-specific T cells recognize and lyse autologous GBM tumors
Figures 2 and 3 demonstrate that CMV pp65-specific T cells can be efficiently induced in vitro using cells from GBM patients. We subsequently tested the ability of CMV pp65-specific T cells to recognize and lyse autologous GBM tumors in vitro in an antigen- specific manner. GBM patient derived T cells were stimulated by culturing them with autologous DCs pulsed with CMV pp65 RNA, and the cytotoxic reactivity of the effector CMV pp65-specific T cells was measured using a europium release CTL assay (38). To demonstrate efficacy, specificity, and off-target effects, autologous DCs electroporated with the following RNA transcripts were used as targets: 1) CMV pp65 RNA (specific target) 2) survivin RNA (non-specific target) 3) Flu M1 RNA (non-specific target) 4) GBM tumor RNA (test surrogate target for autologous tumor) and 5) Total cellular RNA (control target using autologous PBMC-derived total RNA or DC-derived total RNA). To address the most relevant question, namely the ability to immunologically target CMV pp65 as a tumor antigen in GBM tumors, we examined the cytotoxic activity of CMV pp65-specific T cells on autologous patient-derived, primary GBM tumor cells.
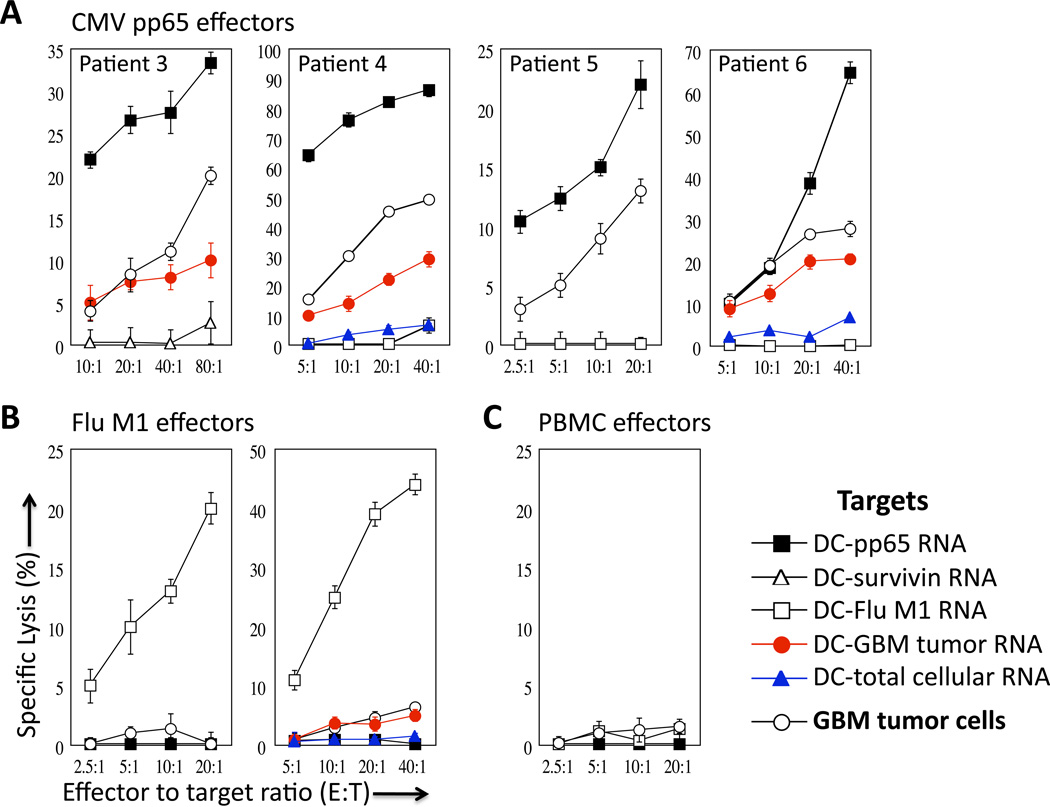
Figure 4A illustrates the CTL reactivity of T cells stimulated with CMV pp65 RNA-transfected DCs from 4 GBM patients following a short co-culture with the aforementioned target cells. All patients’ CMV pp65-specific T cells lysed DC target cells pulsed with pp65 RNA as well as the surrogate tumor target, DCs pulsed with GBM total tumor RNA (not tested in patient 3). Importantly, CMV-specific T cells recognized and killed autologous GBM tumor cells as well, indicating sufficient levels of target antigen expression within tumors for immunologic recognition and susceptibility of GBM tumor cells to T cell-mediated killing. The antigen-specific recognition of these T cells was conclusively demonstrated by the fact that CMV pp65-specific T cells did not lyse the control targets of autologous DCs pulsed with survivin RNA, Flu M1 RNA, or total cellular RNA. Furthermore, Flu M1-specific effectors lysed only Flu M1-expressing targets. (Figure 4B) but did not demonstrate reactivity against DCs pulsed with GBM total tumor RNA or autologous tumor cells. T cells cultured with DCs pulsed with total cellular RNA were incapable of inducing any appreciable level of CTL activity (Figure 4C) further confirming the recognition of ‘non-self’ antigens by the primed CTLs. Cytokine analysis conducted in parallel indicated that vast levels of IFN-γ and appreciable amounts of TNF-α and IL-6 were produced by the CMV pp65-specific T cells upon recognition of the autologous primary GBM tumor cells (Supplementary Figure S3). Collectively, Figures 4A–4C show that in vitro generated CMV pp65-specific T cells are capable of specifically recognizing and lysing autologous GBM tumor cells. These data demonstrate, for the first-time, the immunologic relevance of CMV- specific immunity in the targeting of glial tumors.
Figure 4. CMV pp65-specific T cells lyse autologous GBM tumors.
T cells generated from ex vivo culture with CMV pp65 RNA-pulsed DCs were used as effector cells in a europium release assay to determine CTL activity.
A) Target cells used included DCs pulsed with CMV pp65 RNA, survivin RNA, Flu M1 RNA, GBM tumor RNA, and total cellular RNA (RNA isolated from autologous PBMCs or DCs) as indicated. Patient-derived autologous primary GBM tumor cells were also used as target cells.
B) T cells stimulated with DCs pulsed with Flu M1 RNA were tested against the same targets and demonstrated no activity against patient tumor cells. Flu M1-specific T cells specifically lyse only Flu M1-expressing DC targets. Data using cells from Patient 5 and Patient 6 are depicted.
C) T cells stimulated with DCs pulsed with PBMC RNA (RNA isolated from autologous PBMCs) were incapable of inducing CTL activity against any of the targets tested. Figure represents data using cells from Patient 5.
Total tumor RNA pulsed DCs expand CMV pp65-specific T cells
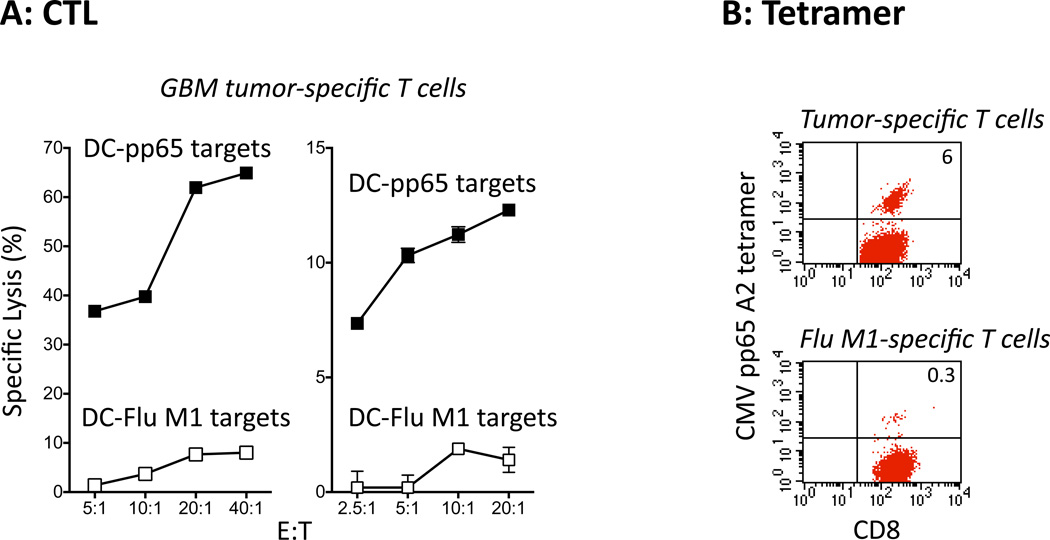
To further explore the significance of targeting CMV in GBM, we used total tumor RNA-transfected DCs from patients with GBM to stimulate autologous T cells and examined the induction of CMV-specific T cells. Figure 5A depicts the results of the europium release CTL assay for two GBM patients in which sufficient tumor RNA for ex vivo T cell stimulation was obtained. T cells stimulated with DCs pulsed with total tumor RNA effectively recognized and lysed DC target cells pulsed with CMV pp65 RNA but not Flu M1 RNA, demonstrating the activation of CMV pp65-specific T cells using total tumor RNA-pulsed DCs. Moreover, one of these two patients, who was HLA-A2-positive, demonstrated the expansion of CMV pp65-specific tetramer-positive CD8+ T cells after stimulation with total tumor RNA-pulsed DCs but no expansion of CMV pp65-specific T cells when stimulated by influenza RNA-pulsed DCs, ruling out a global non-specific expansion of CMV viral memory cells within the DC co-culture (Figure 5B). Taken together, this data provides evidence that CMV pp65 is a viable antigenic target in GBM tumors and supports the rationale for CMV directed immunotherapy for the treatment of GBM.
Figure 5. GBM tumor RNA-pulsed dendritic cells stimulate CMV pp65-specific T cells.
T cells were stimulated with autologous DCs pulsed with GBM tumor RNA (total RNA isolated from autologous primary GBM tumor cells) and tested for CTL activity. DCs pulsed with either CMV pp65 RNA or Flu M1 RNA were used as target cells.
A) GBM tumor-specific T cells specifically lyse CMV pp65-expressing DC targets but not Flu M1-expressing DC targets. Data using cells from Patient 4 and Patient 5 are depicted.
B) T cells stimulated with GBM tumor RNA-pulsed DCs expand CMV pp65-specific CD8+ T cells. T cells from the same patient PBMCs stimulated with Flu M1 RNA-pulsed DCs were used as controls and do not demonstrate the presence of CMV pp65-specific CD8+ T cells.
Figure represents data using cells from Patient 4.
Discussion
Cancer immunotherapy has made great strides in recent years. The ability to harness a patient’s immune system to generate powerful anti-tumor responses with minimal collateral damage to surrounding healthy tissue is essential in any therapeutic setting and is a cornerstone of immunotherapy. This is especially true in patients with malignant brain tumors, as immunologic cross-reactivity with normal brain could lead to severe morbidity. In the present study, we explored the physiologic relevance of targeting human cytomegalovirus as a tumor rejection antigen in GBM. To date, DC targeting of malignant gliomas using unfractionated tumor antigens in the form of tumor lysates, total tumor RNA, and tumor peptides, have shown safety, feasibility, capacity to elicit immune responses and promising efficacy (39–41). However, these approaches are limited by the requirement for acquisition of tumor tissue in a preparation suitable for vaccine preparation and the lack of identity of relevant tumor targets for immune monitoring. Conserved and truly tumor-specific antigens, such as EGFRvIII, can serve as potent antigens for tumor rejection but are rare in example and often limited to only a subset of tumor patients (42–44). The reported detection of CMV antigens in the majority of malignant glioma specimens, the specificity and immunogenicity of well-characterized viral antigens in CMV, and the history of safety in immunologic targeting of CMV antigens even in patients with disseminated CMV disease, potentially highlight CMV antigens as a novel and promising target for immunotherapy in the treatment of CMV-associated gliomas.
In this study, we importantly demonstrate that despite well-described deficits in cell-mediated immunity in patients with GBM, CMV pp65 RNA-pulsed DCs can be reliably generated from the peripheral blood from these patients (Figures 2–5, Supplementary Table S2 and Supplementary Figure S3)and stimulate CMV-specific CD4+ and CD8+ polyfunctional effector cells (Figures 3B and 3C). Most significantly, these CMV pp65-specific T cells were capable of killing autologous tumor cells and autologous DCs pulsed with total tumor RNA (Figure 4). The use of autologous DCs pulsed with RNA as surrogate tumor targets allowed for the exquisite specificity of this recognition to be evaluated in vitro. CMV pp65-specific T cells recognized total tumor antigen-expressing DCs and CMV pp65-expressing DCs, while did not lyse autologous DCs pulsed with a variety of antigen- specific controls, demonstrating a CMV pp65-restricted killing mechanism. While potent influenza-specific T cells could also be stimulated from patients with GBM, these T cells failed to recognize autologous tumor cells or DCs pulsed with total tumor RNA, further validating antigen-specific recognition of CMV antigens. Finally, as an additional demonstration of the relevance of endogenous CMV antigen expression, we observed that stimulation of lymphocytes with total tumor RNA-pulsed DCs led to a 10- to 20-fold expansion of CMV-specific CD8+ T cells by tetramer analysis and the recognition and killing of CMV pp65-expressing targets in vitro (Figure 5). Previous studies have demonstrated the capacity to induce immunologic responses to a variety of GBM-associated antigens, including capacity to expand CMV-specific T cells from patients with GBM (45, 46). However, these studies did not address whether endogenous low levels of CMV antigen expression within these tumors was sufficient to serve as a relevant target for immunotherapy. To our knowledge, this is the first study demonstrating the physiologic relevance of CMV as a therapeutic target in GBM. This study, coupled with demonstrated feasibility for dendritic cell-based vaccination and use of ex vivo expanded T cells for adoptive cellular therapy, strongly support the evaluation of CMV-directed immunotherapy for patients with GBM. A potential limitation of this approach, however, is the well- established heterogeneity in gene expression within GBM tumors that would likely limit the potential efficacy of any antigen-directed therapeutic strategy. Prior studies have demonstrated the frequent detection of CMV antigens in GBM tumors, but the proportion of infected tumor cells has ranged from less than 10% to greater than 80% (11, 47). Our assessment of the proportion of tumor cells expressing sufficient antigenic peptides to serve as targets in cytotoxicity assays revealed that 20–50% of tumor cells could be recognized and killed through immunologic targeting of CMV pp65 (Figure 4). Furthermore, while we have established the capacity to generate functional cytotoxic T cells ex vivo, well-established mechanisms of immunosuppression in vivo, such as elevated proportion of regulatory T cells in patients with GBM may limit potential therapeutic efficacy (48). These potential confounders can be readily addressed through combination antigen targeting approaches, as we and others have shown expression of other CMV antigen targets in GBM tumors, as well as depletion or inhibition of regulatory T cells using clinically available monoclonal antibodies (14, 49, 50). Thus, there is considerable rationale for the clinical evaluation of CMV-directed immunotherapy in patients with GBM.
Supplementary Material
Statement of translation relevance.
Although detection of low levels of human cytomegalovirus (CMV) expression in glioblastoma (GBM) has been reported, the physiologic relevance of this expression remains unclear. Because CMV can serve as a novel immunological target and CMV-directed immunotherapy against primary CMV infection has been shown to be safe and efficacious, we explored the utility of the immunodominant CMV antigen pp65 as a tumor rejection antigen in primary human GBM tumor cells. Despite deficits in cellular immunity in patients with GBM, CMV pp65-specific T cells could be reliably activated and expanded in vitro using CMV pp65 RNA-pulsed autologous dendritic cells. Activated CMV-specific T cells were capable of recognizing and killing of autologous GBM tumor cells expressing endogenous levels of CMV antigens. These studies are the first to demonstrate the relevance of CMV-specific immunity in mediating the killing of glial tumor cells and strongly support the rationale for CMV-targeted immunotherapy in patients with GBM.
Acknowledgements
This work has been supported by the following NIH/NCI grants: 5R01-NS067037 (Mitchell), 5R01-CA134844 (Mitchell), 5P50-CA108786 (Sampson/Bigner), 5R01-CA135272-04 (Sampson) and the CTSA grant UL1RR024128 from the National Center for Research Resources (NCRR), a component of the NIH and NIH Roadmap for Medical Research.
Footnotes
Disclosure of potential conflicts of interest: SKN & DB are co-inventors on the patent describing the use of dendritic cells transfected with tumor antigen encoding RNA that has been licensed by Argos Therapeutics (Durham, NC) through Duke University. SKN & DB have no financial interest in Argos Therapeutics and are not compensated by Argos Therapeutics. JHS & DAM hold a patent related to technologies disclosed in this work. GDL, RS, WX, JS, RL, LAJ, KW and GEA declare no conflicts of interest.
Authors’ Contributions
Conception and design: SKN, DAM
Development of Methodology: SKN, DB, JS, KW, DAM
Acquisition of data: SKN, DB, RS, JS, LAJ, WX, RL
Analysis and interpretation of data: SKN, GDL, DAM, DB, RS, WX, LAJ
Writing, review, and/or revision of manuscript: GDL, SKN, DAM
Administrative, technical, or material support: GEA, KW, JHS
Study supervision: DAM, SKN, JHS
Supplementary data are provided as separate pdf files.
References
- 1.Imperato JP, Paleologos NA, Vick NA. Effects of treatment on long-term survivors with malignant astrocytomas. Ann Neurol. 1990;28:818–822. doi: 10.1002/ana.410280614. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong JJ, Rosenberg SA, Dudley ME, Yang JC, White DE, Butman JA, et al. Successful treatment of melanoma brain metastases with adoptive cell therapy. Clin Cancer Res. 2010;16:4892–4898. doi: 10.1158/1078-0432.CCR-10-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–4557. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang I, Tihan T, Han SJ, Wrensch MR, Wiencke J, Sughrue ME, et al. CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. J Clin Neurosci. 2010;17:1381–1385. doi: 10.1016/j.jocn.2010.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blankenhorn EP, Stranford SA, Smith PD, Hickey WF. Genetic differences in the T cell receptor alleles of LEW rats and their encephalomyelitis-resistant derivative, LER, and their impact on the inheritance of EAE resistance. Eur J Immunol. 1991;21:2033–2041. doi: 10.1002/eji.1830210910. [DOI] [PubMed] [Google Scholar]
- 8.Blomberg K, Granberg C, Hemmila I, Lovgren T. Europium-labelled target cells in an assay of natural killer cell activity. I. A novel non-radioactive method based on time-resolved fluorescence. J Immunol Methods. 1986;86:225–229. doi: 10.1016/0022-1759(86)90457-6. [DOI] [PubMed] [Google Scholar]
- 9.Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH, et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347–3350. [PubMed] [Google Scholar]
- 10.Mitchell DA, Xie W, Schmittling R, Learn C, Friedman A, McLendon RE, et al. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with glioblastoma. Neuro Oncol. 2008;10:10–18. doi: 10.1215/15228517-2007-035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheurer ME, Bondy ML, Aldape KD, Albrecht T, El-Zein R. Detection of human cytomegalovirus in different histological types of gliomas. Acta Neuropathol. 2008;116:79–86. doi: 10.1007/s00401-008-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucas KG, Bao L, Bruggeman R, Dunham K, Specht C. The detection of CMV pp65 and IE1 in glioblastoma multiforme. J Neurooncol. 2011;103:231–238. doi: 10.1007/s11060-010-0383-6. [DOI] [PubMed] [Google Scholar]
- 13.Dziurzynski K, Wei J, Qiao W, Hatiboglu MA, Kong LY, Wu A, et al. Glioma-associated cytomegalovirus mediates subversion of the monocyte lineage to a tumor propagating phenotype. Clin Cancer Res. 2011;17:4642–4649. doi: 10.1158/1078-0432.CCR-11-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soroceanu L, Matlaf L, Bezrookove V, Harkins L, Martinez R, Greene M, et al. Human cytomegalovirus US28 found in glioblastoma promotes an invasive and angiogenic phenotype. Cancer Res. 2011;71:6643–6653. doi: 10.1158/0008-5472.CAN-11-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranganathan P, Clark PA, Kuo JS, Salamat MS, Kalejta RF. Significant association of multiple human cytomegalovirus genomic Loci with glioblastoma multiforme samples. J Virol. 2012;86:854–864. doi: 10.1128/JVI.06097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharjee B, Renzette N, Kowalik TF. Genetic analysis of cytomegalovirus in malignant gliomas. J Virol. 2012;86:6815–6824. doi: 10.1128/JVI.00015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair S, Archer GE, Tedder TF. Isolation and generation of human dendritic cells. Curr Protoc Immunol. 2012;Chapter 7(Unit7):32. doi: 10.1002/0471142735.im0732s99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnis ME, Rooney CM. Enhancement of dendritic cells as vaccines for cancer. Immunotherapy. 2010;2:847–862. doi: 10.2217/imt.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 20.Precopio ML, Betts MR, Parrino J, Price DA, Gostick E, Ambrozak DR, et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med. 2007;204:1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beveridge NE, Price DA, Casazza JP, Pathan AA, Sander CR, Asher TE, et al. Immunisation with BCG and recombinant MVA85A induces long-lasting, polyfunctional Mycobacterium tuberculosis-specific CD4+ memory T lymphocyte populations. Eur J Immunol. 2007;37:3089–3100. doi: 10.1002/eji.200737504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elvang T, Christensen JP, Billeskov R, Thi Kim Thanh Hoang T, Holst P, Thomsen AR, et al. CD4 and CD8 T cell responses to the M. tuberculosis Ag85B-TB10.4 promoted by adjuvanted subunit, adenovector or heterologous prime boost vaccination. PLoS One. 2009;4:e5139. doi: 10.1371/journal.pone.0005139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 25.Lachmann R, Bajwa M, Vita S, Smith H, Cheek E, Akbar A, et al. Polyfunctional T cells accumulate in large human cytomegalovirus-specific T cell responses. J Virol. 2012;86:1001–1009. doi: 10.1128/JVI.00873-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badr G, Bedard N, Abdel-Hakeem MS, Trautmann L, Willems B, Villeneuve JP, et al. Early interferon therapy for hepatitis C virus infection rescues polyfunctional, long-lived CD8+ memory T cells. J Virol. 2008;82:10017–10031. doi: 10.1128/JVI.01083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi JS, Du M, Zajac AJ. A vital role for interleukin-21 in the control of a chronic viral infection. Science. 2009;324:1572–1576. doi: 10.1126/science.1175194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munoz-Cobo B, Solano C, Benet I, Costa E, Remigia MJ, de la Camara R, et al. Functional profile of cytomegalovirus (CMV)-specific CD8+ T cells and kinetics of NKG2C+ NK cells associated with the resolution of CMV DNAemia in allogeneic stem cell transplant recipients. J Med Virol. 2012;84:259–267. doi: 10.1002/jmv.22254. [DOI] [PubMed] [Google Scholar]
- 29.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger CT, Frahm N, Price DA, Mothe B, Ghebremichael M, Hartman KL, et al. High-functional-avidity cytotoxic T lymphocyte responses to HLA-B-restricted Gag-derived epitopes associated with relative HIV control. J Virol. 2011;85:9334–9345. doi: 10.1128/JVI.00460-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duvall MG, Precopio ML, Ambrozak DA, Jaye A, McMichael AJ, Whittle HC, et al. Polyfunctional T cell responses are a hallmark of HIV-2 infection. Eur J Immunol. 2008;38:350–363. doi: 10.1002/eji.200737768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carpenito C, Milone MC, Hassan R, Simonet JC, Lakhal M, Suhoski MM, et al. Control of large, established tumor xenografts with genetically retargeted human T cells containing CD28 and CD137 domains. Proc Natl Acad Sci U S A. 2009;106:3360–3365. doi: 10.1073/pnas.0813101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imai N, Ikeda H, Tawara I, Shiku H. Tumor progression inhibits the induction of multifunctionality in adoptively transferred tumor-specific CD8+ T cells. Eur J Immunol. 2009;39:241–253. doi: 10.1002/eji.200838824. [DOI] [PubMed] [Google Scholar]
- 35.Imai N, Ikeda H, Tawara I, Wang L, Wang L, Nishikawa H, et al. Glucocorticoid-induced tumor necrosis factor receptor stimulation enhances the multifunctionality of adoptively transferred tumor antigen-specific CD8+ T cells with tumor regression. Cancer Sci. 2009;100:1317–1325. doi: 10.1111/j.1349-7006.2009.01179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perales MA, Yuan J, Powel S, Gallardo HF, Rasalan TS, Gonzalez C, et al. Phase I/II study of GM-CSF DNA as an adjuvant for a multipeptide cancer vaccine in patients with advanced melanoma. Mol Ther. 2008;16:2022–2029. doi: 10.1038/mt.2008.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan J, Gnjatic S, Li H, Powel S, Gallardo HF, Ritter E, et al. CTLA-4 blockade enhances polyfunctional NY-ESO-1 specific T cell responses in metastatic melanoma patients with clinical benefit. Proc Natl Acad Sci U S A. 2008;105:20410–20415. doi: 10.1073/pnas.0810114105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volgmann T, Klein-Struckmeier A, Mohr H. A fluorescence-based assay for quantitation of lymphokine-activated killer cell activity. J Immunol Methods. 1989;119:45–51. doi: 10.1016/0022-1759(89)90379-7. [DOI] [PubMed] [Google Scholar]
- 39.Liau LM, Prins RM, Kiertscher SM, Odesa SK, Kremen TJ, Giovannone AJ, et al. Dendritic cell vaccination in glioblastoma patients induces systemic and intracranial T-cell responses modulated by the local central nervous system tumor microenvironment. Clin Cancer Res. 2005;11:5515–5525. doi: 10.1158/1078-0432.CCR-05-0464. [DOI] [PubMed] [Google Scholar]
- 40.Prins RM, Soto H, Konkankit V, Odesa SK, Eskin A, Yong WH, et al. Gene expression profile correlates with T-cell infiltration and relative survival in glioblastoma patients vaccinated with dendritic cell immunotherapy. Clin Cancer Res. 2011;17:1603–1615. doi: 10.1158/1078-0432.CCR-10-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su Z, Dannull J, Heiser A, Yancey D, Pruitt S, Madden J, et al. Immunological and clinical responses in metastatic renal cancer patients vaccinated with tumor RNA-transfected dendritic cells. Cancer Res. 2003;63:2127–2133. [PubMed] [Google Scholar]
- 42.Sampson JH, Aldape KD, Archer GE, Coan A, Desjardins A, Friedman AH, et al. Greater chemotherapy-induced lymphopenia enhances tumor-specific immune responses that eliminate EGFRvIII-expressing tumor cells in patients with glioblastoma. Neuro Oncol. 2011;13:324–333. doi: 10.1093/neuonc/noq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sampson JH, Heimberger AB, Archer GE, Aldape KD, Friedman AH, Friedman HS, et al. Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28:4722–4729. doi: 10.1200/JCO.2010.28.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sampson JH, Archer GE, Mitchell DA, Heimberger AB, Herndon JE, 2nd, Lally-Goss D, et al. An epidermal growth factor receptor variant III-targeted vaccine is safe and immunogenic in patients with glioblastoma multiforme. Mol Cancer Ther. 2009;8:2773–2779. doi: 10.1158/1535-7163.MCT-09-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghazi A, Ashoori A, Hanley PJ, Brawley VS, Shaffer DR, Kew Y, et al. Generation of polyclonal CMV-specific T cells for the adoptive immunotherapy of glioblastoma. J Immunother. 2012;35:159–168. doi: 10.1097/CJI.0b013e318247642f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crough T, Beagley L, Smith C, Jones L, Walker DG, Khanna R. Ex vivo functional analysis, expansion and adoptive transfer of cytomegalovirus-specific T-cells in patients with glioblastoma multiforme. Immunol Cell Biol. 2012;90:872–880. doi: 10.1038/icb.2012.19. [DOI] [PubMed] [Google Scholar]
- 47.Dziurzynski K, Chang SM, Heimberger AB, Kalejta RF, McGregor Dallas SR, Smit M, et al. Consensus on the role of human cytomegalovirus in glioblastoma. Neuro Oncol. 2012;14:246–255. doi: 10.1093/neuonc/nor227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fecci PE, Mitchell DA, Whitesides JF, Xie W, Friedman AH, Archer GE, et al. Increased regulatory T-cell fraction amidst a diminished CD4 compartment explains cellular immune defects in patients with malignant glioma. Cancer Res. 2006;66:3294–3302. doi: 10.1158/0008-5472.CAN-05-3773. [DOI] [PubMed] [Google Scholar]
- 49.Matlaf LA, Harkins LE, Bezrookove V, Cobbs CS, Soroceanu L. Cytomegalovirus pp71 protein is expressed in human glioblastoma and promotes pro-angiogenic signaling by activation of stem cell factor. PLoS One. 2013;8:e68176. doi: 10.1371/journal.pone.0068176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell DA, Cui X, Schmittling RJ, Sanchez-Perez L, Snyder DJ, Congdon KL, et al. Monoclonal antibody blockade of IL-2 receptor alpha during lymphopenia selectively depletes regulatory T cells in mice and humans. Blood. 2011;118:3003–3012. doi: 10.1182/blood-2011-02-334565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.