Abstract
Identifying the factors governing the maintenance of genetic variation is a central challenge in evolutionary biology. New genomic data, methods and conceptual advances provide increasing evidence that balancing selection, mediated by antagonistic species interactions, maintains functionally-important genetic variation within species and natural populations. Because diverse interactions between plants and herbivorous insects dominate terrestrial communities, they provide excellent systems to address this hypothesis. Population genomic studies of Arabidopsis thaliana and its relatives suggest spatial variation in herbivory maintains adaptive genetic variation controlling defense phenotypes, both within and among populations. Conversely, inter-species variation in plant defenses promotes adaptive genetic variation in herbivores. Emerging genomic model herbivores of Arabidopsis could illuminate how genetic variation in herbivores and plants interact simultaneously.
Introduction
Understanding the maintenance of genetic variation within species and populations is a fundamental goal in evolutionary biology. Balancing selection, a suite of adaptive evolutionary processes that maintain greater genetic or phenotypic diversity in a population or species than expected under a neutral evolutionary model, was once regarded as the primary force maintaining functional genetic variation. However, until recently, a paucity of genomic signatures of balancing selection suggested that polymorphisms maintained by balancing selection may be rare [1]. Advances in population genomics (e.g. [2]) and in linking genotype to fitness in nature (e.g. [3]) have provided new support for widespread balancing selection acting on genes underlying ecologically important traits.
Despite the ecological ubiquity of plant-herbivore interactions, the extent to which they maintain genetic variation in plants and insects is not well understood. Here, we highlight empirical examples and theoretical predictions related to how plant-herbivore interactions could maintain genetic variation through balancing selection. Non-exclusive forms of balancing selection include fitness advantages for heterozygotes, frequency dependent selection favoring rare alleles, and antagonistic selection across temporally and spatially variable environments (reviewed in [1,4]). We focus on the role of spatially varying selection (SVS) because of its rich theoretical framework and testability with modern genomic resources. We suggest major questions that future studies might address, and highlight experimental techniques and genetically-enabled model systems well suited to answer these questions.
Why should plant-herbivore interactions maintain genetic diversity?
Host-pathogen interactions are among the most important selective forces known to maintain genetic variation in both hosts [5–7] and pathogens [8], and SVS plays a key role in this process. For example, geographic variation in pathogen communities may be the strongest selective force maintaining non-neutral genetic variation across human populations [6]. Spatial variation in plant and herbivore populations and communities is likely to produce a similar effect. SVS may be particularly important for herbivores, as plants comprise a large fraction of an herbivore’s environment and may be more important than abiotic factors in determining herbivore fitness [9].
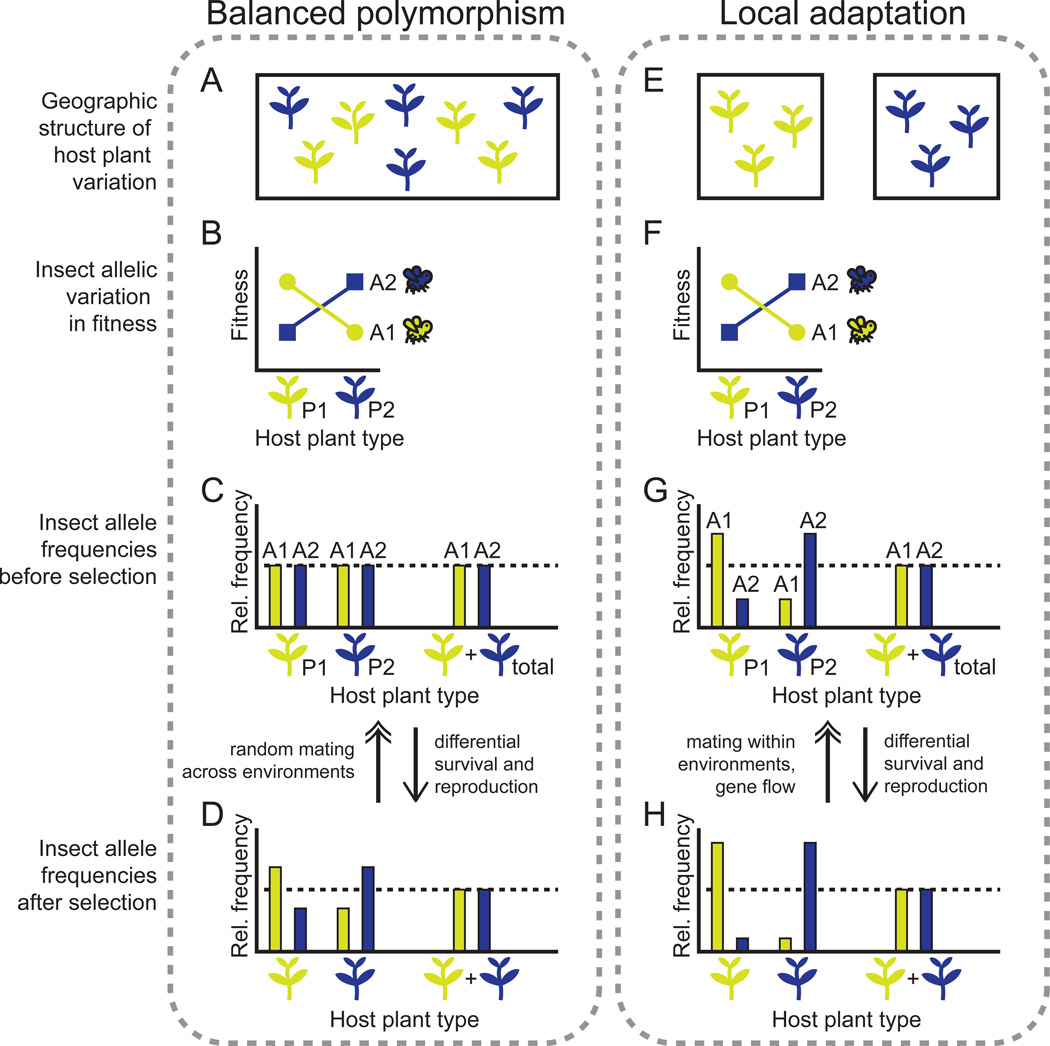
Under SVS, selective advantages or disadvantages of alleles at a locus differ between environments that individuals of a species occupy (Fig. 1B,F; Supplementary Table 1). A simple model of populations inhabiting multiple environments, connected by varying levels of gene flow, forms the foundation of theoretical models of polymorphism maintenance within populations [1,10] and among locally adapted populations and host races [11,12]. Figure 1 illustrates an application of this model to plant-herbivore interactions: spatial variation in a plant defensive trait (e.g. [13]) – which may arise through complex biotic interactions, abiotic interactions, or genetic drift – is expected to maintain genetic variation within or among insect populations.
Figure 1. Two simplified models for spatially varying selection (SVS) due to spatial heterogeneity in host plant characteristics.
Two alleles (“A1” and “A2”) in an herbivorous insect have opposing effects on insect fitness on two host plant types (“P1” and “P2”), which may represent plant genotypes or species that differ in a defensive trait. When a single insect population feeds on both sympatric host types (A), the two alleles can be maintained at intermediate frequency within the population (C,D). When host plant types are spatially separated so that insect gene flow between host types is low (E), allele frequencies will diverge between populations feeding on each host type, and the two allele polymorphism will be maintained at the species level (G,H) by migration-selection balance [12]. Both novel mutations and standing genetic variation can be driven by SVS to the intermediate frequencies depicted in the figure. The two models presented above are simplified extremes of situations in nature, which can fall along a continuum of high (right column) versus low (left column) host plant segregation and insect gene flow among host types.
Levene [10] first demonstrated mathematically that SVS can maintain multiple alleles at stable equilibrium frequencies in a single, randomly-mating population. Subsequent studies revealed that when gene flow across environments is low, maintenance of polymorphism becomes more favorable [1,10,14]. Further, when the costs of host resistance and enemy virulence vary between environments [15], global polymorphisms in interacting host and enemy genes are even more likely; for plants, geographically variable components of the environment can alter the cost of defense [16]. A final important insight is that the maintenance of polymorphism is more favorable as environment-specific fitness advantages or disadvantages of an allele increase [10,17]. Thus, alleles maintained by SVS are likely to have large phenotypic effects and contribute a disproportionately large amount to fitness [17,18]. These predictions are consistent with the finding that traits under biotic selection are controlled by loci with larger effects than traits under abiotic selection in plants [19].
Do herbivores maintain genetic variation in plants?
Population genomic analyses, enabled by whole-genome resequencing of natural Arabidopsis thaliana (Arabidopsis) accessions ([20], http://www.1001genomes.org/), suggest abundant adaptive variation exists for defense-related traits. Loci underlying defense-related traits [21] are highly differentiated between populations compared to the genome overall [22]. The same loci showed little evidence for selective sweeps, inconsistent with an arms race model in which repetitive sweeps reduce diversity [22]. Instead, plant enemies maintain species-wide defense polymorphisms over broad geographic scales. These polymorphisms manifest through both protein structure and gene expression: genes controlling defense traits, such as glucosinolate production, show high levels of genetic polymorphism and high variation in expression between individuals [23]. However, the extent to which anti-herbivore or anti pathogen defense genes each contribute to these patterns in Arabidopsis is unclear.
Climate-responsive genes in Arabidopsis show elevated polymorphism [24] and predict fitness in common gardens [3,25], suggesting climatic variation maintains ecologically important genetic variation in Arabidopsis. However, heterogeneity in biotic interactions (e.g., herbivory) may also contribute to these patterns if biotic and climatic variables co-vary. In fact, allele frequencies at genes involved in defense varied with climate more often than expected by chance [25]. Observational data of herbivore distributions, integrated with common garden experiments to identify genetic variation underlying fitness trade-offs in the presence/absence of herbivores in different geographic contexts, may help link genetic variation to spatially varying herbivory.
Additional studies connecting genotype to phenotype at individual loci have also illuminated functionally important variation maintained by herbivores. In Arabidopsis, geographic variation at a locus controlling variation in glucosinolate profiles correlated with the relative long-term abundance of two specialist aphids across Europe, consistent with the direction of differential selection imposed by these species in the laboratory [13]. Similarly, variation in glucosinolate biosynthetic genes in Boechera stricta, a close relative of Arabidopsis, explained geographic variation in herbivore damage and fitness in common gardens [26]. Herbivory may also contribute to the maintenance of defense variation at finer spatial scales: amino acid polymorphisms underlying a trade-off between growth or defense against biotrophic pathogens and aphids are maintained at intermediate frequencies across populations in ACD6, a gene controlling leaf necrosis [27].
Do plants maintain genetic variation in herbivores?
The strongest evidence that plant diversity drives genetic variation in herbivorous insects exists for host races – sympatric insect populations that use different hosts and are genetically differentiated, despite gene flow among populations [11]. Antagonistic selection when feeding on different hosts, a form of SVS, is hypothesized to generate and maintain genetic divergence at loci affecting preference and performance on different host plants, reducing inter-race gene flow and creating more subtle divergence in nearby genomic regions [11]. Genome-wide scans revealed that regions with loci affecting preference for, and population growth rate on, different hosts have diverged in pea aphid (Acyrthosiphon pisum) host races [28,29]. Meanwhile, divergent genomic regions in apple maggot races (Rhagoletis pomonella) control diapause timing [30]. These findings highlight that adaptations to host-specific defenses can maintain genetic differences between host races, but other differences between host plants (e.g. phenology) can also be important.
At present, there is little evidence from the literature for host plant variation maintaining polymorphism within herbivores in the absence of host race formation (but see [31,32]). However, differential performance of different spider mite (Tetranychus urticae) genotypes across hosts suggests spatial mosaics of host plants can maintain significant phenotypic variation in generalist herbivores [33]. Similar patterns may occur even in relatively specialized insects as well: though geography explains patterns of relatedness among populations of the large pine weevil (Hylobius abietis), allele frequencies at a few loci of unknown function differ between individuals feeding on spruce or pine [34].
Detecting plant-driven balancing selection in herbivorous insects
Illuminating signatures of balancing selection in herbivorous insects driven by plant variation requires three phases: (1) identifying plant genes or traits affecting insect fitness, (2) identifying insect genes interacting with plant genes or traits that mediate effects on insect fitness, and (3) using population genetic tests for balancing selection with appropriate null hypotheses. The ability to rapidly generate genomic sequence data from many individuals within natural populations, and to conduct experimental evolution and common garden experiments using completely sequenced plant accessions, now makes achieving these criteria feasible at the scale of genomic analyses. Cost-effective methods relying on pooled sequencing are particularly promising (Box 1).
Box 1: The utility of pooled sequencing experiments for mapping targets of balancing selection in herbivorous insects.
Genome sequences can be generated easily for non-model insects. However, generating, sequencing, and maintaining inbred lines required for traditional quantitative trait locus (QTL) mapping or genome wide association (GWA) studies remains laborious and expensive. Conversely, short read, next-generation sequencing of pooled individuals, in which allele frequencies can be compared between biologically meaningful groups of individuals, offers a desirable alternative in the following contexts:
Extreme-QTL mapping requires the generation of large populations that exhibit segregating variation for a trait, isolation of many progeny with extreme trait values, and estimation of allele frequencies in phenotypically extreme individuals through pooled sequencing [52]. Accuracy of the approach is similar to GWA in Drosophila melanogaster [53], though pooled X-QTL approaches preclude estimation of trait heritability, epistasis, and locus effect sizes. High-throughput phenotyping of herbivore weight gain or development time on different plant mutant genotypes, ecotypes, or species offers an avenue to identify herbivore genetic variation maintained by variation in plant defenses. Experimental populations for phenotyping could be generated by crossing phenotypically divergent parents from a single population or across locally adapted populations, or derived from directly sampling wild individuals to take advantage of natural, low levels of linkage disequilibrium.
Evolve-and-resequence approaches [54] involve altering phenotypes of experimental populations through artificial selection or divergent growth conditions, followed by pooled resequencing of experimental populations to uncover causal genetic variants. Replicate selection for high and low performance insect populations on different plant types could directly uncover loci with antagonistic effects that depend on host plant characteristics.
Allelic distributions in nature have been used to infer local adaptation [55]. Alleles underlying preferential feeding or high survival on particular, sympatric plants should be at higher frequencies in insects consuming those plants, and the distributions of locally adaptive alleles across populations should be explained more by habitat characteristics (e.g. common host plant species, genotypes, or chemotypes) than population structure.
False positive and negative results stemming from confounding factors, such as population structure, are a major obstacle to mapping loci through genome-wide association studies [35]. Meanwhile, many population genetic tests for balancing selection (Supplementary Table 2) suffer high rates of false positives resulting from genetic drift. Further, simulations [36] reveal that when SVS favors multiple alleles within a population, partial selective sweeps during which a new mutation rises to intermediate frequency proceed extremely slowly. As a result, recombination during the sweep limits hitchhiking of neutral polymorphism to narrow windows near the selected site, and signatures of balancing selection are difficult to detect. Reduced-representation sequencing strategies such as RAD-tag sequencing [37], therefore, may not generate sequence data within small genomic regions showing signatures of balancing selection, particularly in species with low levels of linkage disequilibrium. Integrating genetic mapping studies with population analysis using whole genome sequences (e.g., re-sequencing) is therefore necessary to reveal highly informative, genome-scale patterns.
Model systems for testing if plant-herbivore interactions maintain variation
Studying the maintenance of variation by reciprocal plant-insect interactions requires model systems that are experimentally tractable, genomically characterized and interact in nature. While features affecting the ability to perform experiments and generate genomic data – such as genome size or the ease of rearing and manipulating the organism in the laboratory or field – are important to consider, ecological and evolutionary inferences require knowledge of the distributions and ecologies of the interacting species. Ideally, herbivore species amenable to addressing these questions would be nested evolutionarily among a set of species with genomic resources, have low linkage disequilibrium and a large effective population size, and feed naturally on model plant species with sequenced accessions and functional mutants.
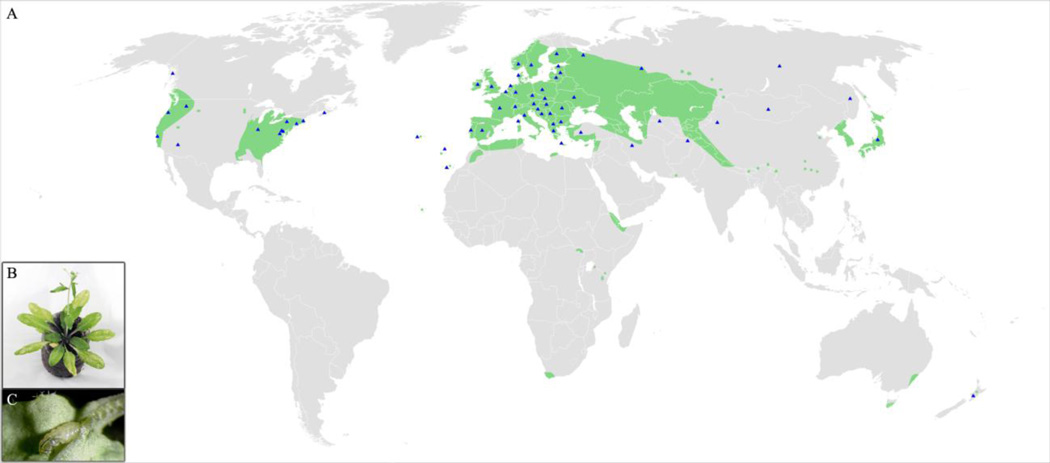
Given the available genomic resources and detailed insight into the phenotypic and genetic basis of important defense traits, Arabidopsis and its close relatives are excellent models to study genetic variation under balancing selection. A number of herbivorous arthropod species that attack Arabidopsis have genomic resources in various stages of development; and these species differ in feeding mode (piercing-sucking or chewing), host breadth (specialist or generalist), and mode of reproduction (Figure 2). New genomic resources for chewing herbivores such as the genome sequence of the diamondback moth (Plutella xylostella) and the two-spotted spider mite (T. urticae) have facilitated novel insight into the evolution of herbivory and resistance to mustard defenses [38,39]. Other species are also emerging as useful models for addressing if and how plant-herbivore interactions maintain genetic variation. For example, the leaf-mining drosophilid fly Scaptomyza flava specializes on Arabidopsis and relatives (Brassicaceae) in the wild [40] (Fig. 3). S. flava is closely related to the many Drosophila species with completely sequenced genomes and has a relatively small genome (290 Mb) [41]. Remarkably, this species recently evolved from within the microbe-feeding Drosophila species [42]. S. flava, like other chewing herbivores, is sensitive to canonical jasmonate-dependent defenses, including glucosinolates, and exhibits variation in performance across Arabidopsis accessions [41,43]. Genes that are functionally important in detoxification against glucosinolates have been shown to be under positive selection [Gloss et al., in review]. Because each generation is sexual, it promises to be a good candidate for use in laboratory selection\ experiments. A sequenced transcriptome [43] and genome [R. Lapoint et al., unpublished] will allow this species to be leveraged in a population genomics context to complement the important lessons learned from other systems.
Figure 2. Comparison of some herbivorous insects of Arabidopsis thaliana with emerging genomic resources.
Draft genome sequences of two M. persicae clones are available for BLAST searches (AphidBase; URL: http://www.aphidbase.com/, http://tools.genouest.org/tools/myzus/login).
Figure 3. Arabidopsis thaliana and Scaptomyza flava are globally distributed and share much of their range.
(A) Distribution of A. thaliana modified from [56] in green. Blue triangles mark presence of S. flava within a country, region, or island group. (B) A. thaliana with adult S. flava oviposition damage on leaves. (C) Leaf-mining larva of S. flava partially removed from A. thaliana leaf. Distribution references are provided in Supplementary Table 3.
In addition to Arabidopsis, population and comparative genomic resources are rapidly accumulating for species of economic and ecological importance, particularly crop plants and their wild relatives [44–46] and forest trees [47]. Importantly, these species span a wide range of taxonomic diversity and vary in defensive traits and life history strategies. Emerging genomic resources for herbivores in these systems will complement those already available (e.g. [48,49]), enabling identification of common patterns underlying the maintenance of variation through plant-herbivore interactions across diverse model systems.
Outstanding questions/future directions
Together, population genomic analysis of natural populations coupled with genetic mapping and experimental evolution (Box 1) can address whether local adaptation or balanced polymorphisms explain much of the adaptive nucleotide variation found in genomic datasets [50]. Outstanding questions include:
Is balancing selection, particularly through SVS, an important force maintaining variation in plant-herbivore systems?
How important are different sources of plant variation – intraspecific, interspecific, and non-genetic – for the maintenance of genetic variation in herbivores, and vice versa?
How does the amount of variation maintained by balancing selection differ between systems with specific vs. diffuse species interactions?
What are the spatial and temporal scales over which plant-herbivore interactions can maintain balanced polymorphism?
How often do genes under balancing selection within populations diverge adaptively among populations or species, given that selective sweeps favoring new mutations erode polymorphism?
Do genes under balancing selection through plant-herbivore interactions provide standing variation co-opted for other adaptations, such as pesticide resistance?
Answers to these questions may differ between plants and arthropod herbivores, primarily because many plant defense traits can be constrained by the multitude of diverse herbivores attacking each host plant species [51]. Systems in which one, highly specialized herbivore heavily influences plant fitness are ideal for studying both sides of the interaction (plant and herbivore) simultaneously, but may be less generalizable (e.g. [9]).
Conclusions
Established evolutionary theory indicates that adaptive processes can facilitate the maintenance, rather than simply the erosion, of genetic variation within and among populations of plants and herbivores. Rapid progress on the development of genomic resources for model plant species with wild relatives has facilitated the illumination of the genes and alleles underlying natural trait variation, as well as how genomes are shaped by adaptive and neutral processes. Arabidopsis has been a key model in this regard, and the promise of >1000 completely sequenced genomes, an active research community investigating all facets of its biology, and emerging model herbivores will enable studies linking genetic variation in plants to variation within herbivore species and communities. The extent to which balancing selection sensu lato can account for the large amount of genetic variation present in plant and herbivorous insect populations is a general one considering that most named species of life are herbivorous insects and the plants on which they feed.
Supplementary Material
Highlights.
Genomic studies reveal variation at genes mediating plant-herbivore interactions
Theory suggests spatially varying selection (SVS) maintains this variation
Genetic mapping and population genetics are complementary for investigating SVS
Studying model species can functionally link interacting plant and insect variation
Acknowledgments
A.D.G. and B.G.H. were each supported by Graduate Research Fellowships from the National Science Foundation (DGE-0646147). A.C.N.D. was supported by a PERT fellowship from the National Institutes of Health (5K12GM000708-13). N.K.W. was supported by grants from the National Science Foundation (DEB-1256758), the National Geographic Society (#9097-12), and the University of Arizona (Faculty Seed Grant).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hedrick PW. Genetic polymorphism in heterogeneous environments: The age of genomics. Annu Rev Ecol Evol Syst. 2006;37:67–93. [Google Scholar]
- 2.Sellis D, Callahan BJ, Petrov DA, Messer PW. Heterozygote advantage as a natural consequence of adaptation in diploids. Proceedings of the National Academy of Sciences. 2011;108:2066–2071. doi: 10.1073/pnas.1114573108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334:86–89. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- 4.Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergelson J, Kreitman M, Stahl EA, Tian D. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- 6.Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admettla A, Pattini L, Nielsen R. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7:e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leffler EM, Gao Z, Pfeifer S, Ségurel L, Auton A, Venn O, Bowden R, Bontrop R, Wall JD, Sella G, et al. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 2013 doi: 10.1126/science.1234070. published online 14 Feb 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Amambua-Ngwa A, Tetteh KKA, Manske M, Gomez-Escobar N, Stewart LB, Deerhake ME, Cheeseman IH, Newbold CI, Holder AA, Knuepfer E, et al. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet. 2012;8:e1002992. doi: 10.1371/journal.pgen.1002992. • The authors identified genes showing balancing selection across 65 fully sequenced isolates of the malaria parasite, Plasmodium falciparum. Similar population genomic scans, which are being embraced in model organisms and in studies of human pathogens, could help reveal the extent of balancing selection on counter-defense genes in insect herbivores.
- 9.Garrido E, Andraca-Gómez G, Fornoni J. Local adaptation: simultaneously considering herbivores and their host plants. New Phytol. 2012;193:445–453. doi: 10.1111/j.1469-8137.2011.03923.x. [DOI] [PubMed] [Google Scholar]
- 10.Levene H. Genetic equilibrium when more than one ecological niche is available. Amer Nat. 1953;87:331–333. [Google Scholar]
- 11.Drès M, Mallet J. Host races in plant–feeding insects and their importance in sympatric speciation. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences. 2002;357:471–492. doi: 10.1098/rstb.2002.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7:1225–1241. [Google Scholar]
- 13. Zust T, Heichinger C, Grossniklaus U, Harrington R, Kliebenstein DJ, Turnbull LA. Natural enemies drive geographic variation in plant defenses. Science. 2012;338:116–119. doi: 10.1126/science.1226397. • In A. thaliana, geographic variation in a polymorphic locus controlling glucosinolate profiles was correlated with the relative long-term abundance of two specialist aphids across Europe, consistent with the direction of differential selection imposed by these two species in laboratory experiments.
- 14.Yeaman S, Otto SP. Establishment and maintenance of adaptive genetic divergence under migration, selection, and drift. Evolution. 2011;65:2123–2129. doi: 10.1111/j.1558-5646.2011.01277.x. [DOI] [PubMed] [Google Scholar]
- 15.Tellier A, Brown JK. Spatial heterogeneity, frequency-dependent selection and polymorphism in host-parasite interactions. BMC evolutionary biology. 2011;11:319. doi: 10.1186/1471-2148-11-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cipollini D, Heil M. Costs and benefits of induced resistance to herbivores and pathogens in plants. Plant Sciences Reviews. 2010;5:1–5. [Google Scholar]
- 17.Connallon T, Clark AG. A general population genetic framework for antagonistic selection that accounts for demography and recurrent mutation. Genetics. 2012;190:1477–1489. doi: 10.1534/genetics.111.137117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charlesworth B, Hughes KA. The maintenance of genetic variation in life-history traits. In: Singh RS, Krimbas CB, editors. Evolutionary genetics: from molecules to morphology. Cambridge University Press; 1999. pp. 369–392. [Google Scholar]
- 19.Louthan AM, Kay KM. Comparing the adaptive landscape across trait types: larger QTL effect size in traits under biotic selection. BMC Evol Biol. 2011;11 doi: 10.1186/1471-2148-11-60. 60-2148-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 21.Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjalmsson BJ, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet. 2012;44:212–216. doi: 10.1038/ng.1042. • 1,307 worldwide accessions of A. thaliana, genotyped at 250,000 SNPs, provide a high-resolution picture of genetic variation and natural selection. Previously-identified SNPs underlying defense phenotypes show a unique evolutionary history relative to SNPs controlling flowering time, development, and mineral accumulation.
- 23.Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CR, Mitchell-Olds T. Environmental adaptation contributes to gene polymorphism across the Arabidopsis thaliana genome. Mol Biol Evol. 2012;29:3721–3728. doi: 10.1093/molbev/mss174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, Toomajian C, Roux F, Bergelson J. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334:83–86. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- 26. Prasad KV, Song BH, Olson-Manning C, Anderson JT, Lee CR, Schranz ME, Windsor AJ, Clauss MJ, Manzaneda AJ, Naqvi I, et al. A gain-of-function polymorphism controlling complex traits and fitness in nature. Science. 2012;337:1081–1084. doi: 10.1126/science.1221636. • Using an elegant combination of experiments using genetic mapping populations in the field and transgenic plants in the lab, the authors characterized a locally adaptive quantitative trait locus controlling glucosinolate chemistry, herbivory damage, survival, and reproduction in a wild relative of A. thaliana. The study thoroughly illustrates how complex geographic variation in herbivory can drive adaptive evolution and maintenance of polymorphism in plant defense genes.
- 27.Todesco M, Balasubramanian S, Hu TT, Traw MB, Horton M, Epple P, Kuhns C, Sureshkumar S, Schwartz C, Lanz C, et al. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature. 2010;465:632–636. doi: 10.1038/nature09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jaquiery J, Stoeckel S, Nouhaud P, Mieuzet L, Maheo F, Legeai F, Bernard N, Bonvoisin A, Vitalis R, Simon JC. Genome scans reveal candidate regions involved in the adaptation to host plant in the pea aphid complex. Mol Ecol. 2012;21:5251–5264. doi: 10.1111/mec.12048. • Hierarchical, population genomic analysis of pea aphid races identified genes encoding salivary proteins and olfactory receptors as candidates driving host-specific divergence in the face of gene flow.
- 29.Via S, Conte G, Mason-Foley C, Mills K. Localizing FST outliers on a QTL map reveals evidence for large genomic regions of reduced gene exchange during speciation-with-gene-flow. Mol Ecol. 2012;21:5546–5560. doi: 10.1111/mec.12021. [DOI] [PubMed] [Google Scholar]
- 30.Michel AP, Sim S, Powell THQ, Taylor MS, Nosil P, Feder JL. Widespread genomic divergence during sympatric speciation. Proceedings of the National Academy of Sciences. 2010;107:9724–9729. doi: 10.1073/pnas.1000939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berenbaum MR, Zangerl AR. Chemical phenotype matching between a plant and its insect herbivore. Proceedings of the National Academy of Sciences. 1998;95:13743–13748. doi: 10.1073/pnas.95.23.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heidel-Fischer HM, Vogel H, Heckel DG, Wheat CW. Microevolutionary dynamics of a macroevolutionary key innovation in a Lepidopteran herbivore. BMC Evol Biol. 2010;10 doi: 10.1186/1471-2148-10-60. 60-2148-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kant MR, Sabelis MW, Haring MA, Schuurink RC. Intraspecific variation in a generalist herbivore accounts for differential induction and impact of host plant defences. Proc Biol Sci. 2008;275:443–452. doi: 10.1098/rspb.2007.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manel S, Conord C, Després L. Genome scan to assess the respective role of host-plant and environmental constraints on the adaptation of a widespread insect. BMC Evolutionary Biology. 2009;9:288. doi: 10.1186/1471-2148-9-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segura V, Vilhjálmsson BJ, Platt A, Korte A, Seren Ü, Long Q, Nordborg M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet. 2012;44:825–830. doi: 10.1038/ng.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connallon T, Clark AG. Antagonistic versus nonantagonistic models of balancing selection: characterizing the relative timescales and hitchhiking effects of partial selective sweeps. Evolution. 2013;67:908–917. doi: 10.1111/j.1558-5646.2012.01800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP Discovery and Genetic Mapping Using Sequenced RAD Markers. PLoS ONE. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grbic M, Van Leeuwen T, Clark RM, Rombauts S, Rouze P, Grbic V, Osborne EJ, Dermauw W, Ngoc PC, Ortego F, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You M, Yue Z, He W, Yang X, Yang G, Xie M, Zhan D, Baxter SW, Vasseur L, Gurr GM, et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet. 2013;45:220–225. doi: 10.1038/ng.2524. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell-Olds T. Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends in Ecology & Evolution. 2001;16:693–700. [Google Scholar]
- 41.Whiteman NK, Groen SC, Chevasco D, Bear A, Beckwith N, Gregory TR, Denoux C, Mammarella N, Ausubel FM, Pierce NE. Mining the plant-herbivore interface with a leafmining Drosophila of Arabidopsis. Mol Ecol. 2011;20:995–1014. doi: 10.1111/j.1365-294X.2010.04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lapoint RT, O'Grady PM, Whiteman NK. Diversification and Dispersal of the Hawaiian Drosophilidae: the evolution of Scaptomyza. Mol Phylogenet Evol. doi: 10.1016/j.ympev.2013.04.032. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whiteman NK, Gloss AD, Sackton TB, Groen SC, Humphrey PT, Lapoint RT, Sonderby IE, Halkier BA, Kocks C, Ausubel FM, Pierce NE. Genes involved in the evolution of herbivory by a leaf-mining, drosophilid fly. Genome Biol Evol. 2012;4:788–804. doi: 10.1093/gbe/evs063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zouine M, Latché A, Rousseau C, Regad F, Pech J, Philippot M, Bouzayen M, Delalande C, Frasse P, Schiex T. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hufford MB, Bilinski P, Pyhäjärvi T, Ross-Ibarra J. Teosinte as a model system for population and ecological genomics. Trends in Genetics. 2012;28:606–615. doi: 10.1016/j.tig.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Huang X, Lu T, Han B. Resequencing rice genomes: an emerging new era of rice genomics. Trends in Genetics. 2013;29:225–232. doi: 10.1016/j.tig.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Neale DB, Kremer A. Forest tree genomics: growing resources and applications. Nature Reviews Genetics. 2011;12:111–122. doi: 10.1038/nrg2931. [DOI] [PubMed] [Google Scholar]
- 48.Keeling CI, Yuen MM, Liao NY, Docking TR, Chan SK, Taylor GA, Palmquist DL, Jackman SD, Nguyen A, Li M. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 2013;14:R27. doi: 10.1186/gb-2013-14-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giron D, Huguet E. A genomically tractable and ecologically relevant model herbivore for a model plant: new insights into the mechanisms of insect–plant interactions and evolution. Mol Ecol. 2011;20:990–994. [Google Scholar]
- 50.Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- 51.Wise MJ, Rausher MD. Evolution of resistance to a multiple-herbivore community: Genetic correlations, diffuse coevolution, and constraints on the plant's response to selection. Evolution. 2013 doi: 10.1111/evo.12061. published online 13 Feb 2013. [DOI] [PubMed] [Google Scholar]
- 52.Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, Gresham D, Caudy AA, Kruglyak L. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464:1039–1042. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swarup S, Huang W, Mackay TF, Anholt RR. Analysis of natural variation reveals neurogenetic networks for Drosophila olfactory behavior. Proc Natl Acad Sci U S A. 2013;110:1017–1022. doi: 10.1073/pnas.1220168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner TL, Stewart AD, Fields AT, Rice WR, Tarone AM. Population-based resequencing of experimentally evolved populations reveals the genetic basis of body size variation in Drosophila melanogaster. PLoS Genet. 2011;7:e1001336. doi: 10.1371/journal.pgen.1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kolaczkowski B, Kern AD, Holloway AK, Begun DJ. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics. 2011;187:245–260. doi: 10.1534/genetics.110.123059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- 57.Ramsey JS, Wilson AC, de Vos M, Sun Q, Tamborindeguy C, Winfield A, Malloch G, Smith DM, Fenton B, Gray SM, Jander G. Genomic resources for Myzus persicae : EST sequencing, SNP identification, and microarray design. BMC Genomics. 2007;8:423. doi: 10.1186/1471-2164-8-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang XW, Luan JB, Li JM, Bao YY, Zhang CX, Liu SS. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genomics. 2010;11 doi: 10.1186/1471-2164-11-400. 400-2164-11-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rotenberg D, Whitfield AE. Analysis of expressed sequence tags for Frankliniella occidentalis, the western flower thrips. Insect Mol Biol. 2010;19:537–551. doi: 10.1111/j.1365-2583.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- 60.Negre V, Hotelier T, Volkoff AN, Gimenez S, Cousserans F, Mita K, Sabau X, Rocher J, Lopez-Ferber M, d'Alencon E, et al. SPODOBASE: an EST database for the lepidopteran crop pest Spodoptera. BMC Bioinformatics. 2006;7:322. doi: 10.1186/1471-2105-7-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.