Abstract
Recent years have provided us with spectacular insights into the biology of the plant hormone auxin, leaving the impression of a highly versatile molecule involved in virtually every aspect of plant development. A combination of genetics, biochemistry, and cell biology has established auxin signaling pathways, leading to the identification of two distinct modes of auxin perception and downstream regulatory cascades. Major targets of these signaling modules are components of the polar auxin transport machinery, mediating directional distribution of the phytohormone throughout the plant body, and decisively affecting plant development. Alterations in auxin transport, metabolism, or signaling that occur as a result of intrinsic as well as environmental stimuli, control adjustments in morphogenetic programs, giving rise to defined growth responses attributed to the activity of the phytohormone. Some of the results obtained from the analysis of auxin, however, do not fit coherently into a picture of highly specific signaling events, but rather suggest mutual interactions between auxin and fundamental cellular pathways, like the control of intracellular protein sorting or translation. Crosstalk between auxin and these basic determinants of cellular activity and how they might shape auxin effects in the control of morphogenesis are the subject of this review.
Keywords: Auxin, Protein sorting, Plasma membrane, Translational control, Cytoskeleton
Introduction
Plant growth regulators, also referred to as phytohormones, decisively influence plant development and environmental interactions. Similar to hormones in metazoans, the activity of these compounds is tightly controlled, with sophisticated pathways modulating activity and specificity of phytohormones. Auxin, for example, (indole-3-acetic acid, IAA, representing the predominant compound found in plants), had been implicated in the regulation of plant growth long before its chemical structure was identified (Kögl and Haagen-Smits 1931). Even Charles Darwin speculated about the participation of a mobile signal in the directional growth of plant organs (Darwin and Darwin 1881). Half a century later, Frits Went and Nikolai Cholodny independently demonstrated that auxin functions as a mediator of such tropic growth responses (Went 1926; Cholodny 1927). However, it took another half a century until the first molecular determinants mediating auxin responses were identified.
As with other plant hormones or secondary messengers, the auxin signals first need to be perceived by specific receptor molecules, and the initial signal consequently needs to be transmitted to ultimately promote hormonal responses. This might be reflected in, for example, adjustments in the transcription of target genes leading to defined cellular responses. Auxin however, is quite unique among phytohormones, as plants have evolved complex machineries mediating intra- as well as intercellular polar auxin transport (PAT). This enables the establishment of hormonal concentration and—hence—activity gradients, thus shaping plant growth and development by active hormone transport mechanisms, which is unparalleled among plant growth regulators (Leyser 2011; Peer et al. 2011; Wabnik et al. 2011; Lau et al. 2012; Barbez and Kleine-Vehn 2013; Ljung 2013; Spalding 2013). In the first sections of this review, we provide an overview, summarizing general aspects of auxin transport and signaling. Further on, we summarize recent work, which has revealed novel insights into auxin-signaling pathways, thereby connecting auxin responses to global cellular activities and thus raising questions about the specificity of auxin-mediated growth control. These novel findings and scenarios, integrating fundamental activities in the regulation of auxin signaling are the main subject of this review.
Essentials of auxin perception and signaling
The search for proteins that might act as cellular receptors for auxin was already initiated by biochemical approaches in the 1970s. It was straightforward genetics though, that led to identification of TRANSPORT INHIBITOR RESPONSE 1 (TIR1), the so far best-characterized auxin receptor in the model plant Arabidopsis (Ruegger et al. 1998; Gray et al. 1999; Dharmasiri et al. 2005a, b; Kepinski and Leyser 2005). TIR1 represents one of numerous F-box proteins that constitute part of a heteromeric F-box E3 ubiquitin ligase complex, which mediates the reversible covalent attachment of the small protein ubiquitin onto target proteins (Jackson and Eldridge 2002; Lechner et al. 2006). In the case of SCFTIR1/AFB-type E3 ligases, this reaction depends on auxin binding by TIR1 or by closely related AUXIN SIGNALING F-BOX (AFB) proteins, which in turn enhances an interaction with short-lived AUXIN/INDOLE ACETIC ACID (Aux/IAA) proteins that function as auxin co-receptors (Theologis et al. 1985; Dharmasiri et al. 2003; Tan et al. 2007; Calderon Villalobos et al. 2012) (Fig. 1). Ubiquitylation of Aux/IAA proteins likely occurs as a result of auxin binding, leading to the subsequent degradation of Aux/IAA via the proteasomal pathway (Gray et al. 2001; Yang et al. 2004; Maraschin et al. 2009). Control of Aux/IAA degradation is crucial for plant responses to auxin, as it links hormone perception to the other essential function of Aux/IAA proteins in transcriptional regulation. Aux/IAA forms heterodimers with AUXIN RESPONSE FACTOR (ARF) transcriptional regulators via protein interaction domains located in the C-terminal portion of ARFs and Aux/IAAs. Upon auxin-induced degradation of Aux/IAAs, ARF activity is no longer be repressed by heteromerization thus allowing transmission of auxin signals into downstream transcriptional networks (Kim et al. 1997; Ulmasov et al. 1997a, b; Tiwari et al. 2001) (Fig. 1). This, remarkably simple, mechanism of transcriptional de-repression appears to be sufficient for the integration of a highly diverse range of signaling events controlled by auxin. To some extent, this might explain the considerable number of Aux/IAA genes found in the genomes of higher plants (Liscum and Reed 2002), all of which in principle could interact with the different SCFTIR1/AFB receptors (Calderon Villalobos et al. 2012). Variations in auxin-binding affinities, arising as a result of such variable interactions, might impact on the proteolytic turnover of Aux/IAAs, thereby specifying downstream ARF activities in transcriptional control (Calderon Villalobos et al. 2012; Yu et al. 2013). Moreover, plant genomes are characterized by extensive ARF gene families, whose members exhibit highly diverse expression profiles and activities. This allows for efficient fine-tuning and coordination of auxin-controlled transcriptional responses (Okushima et al. 2005; Overvoorde et al. 2005).
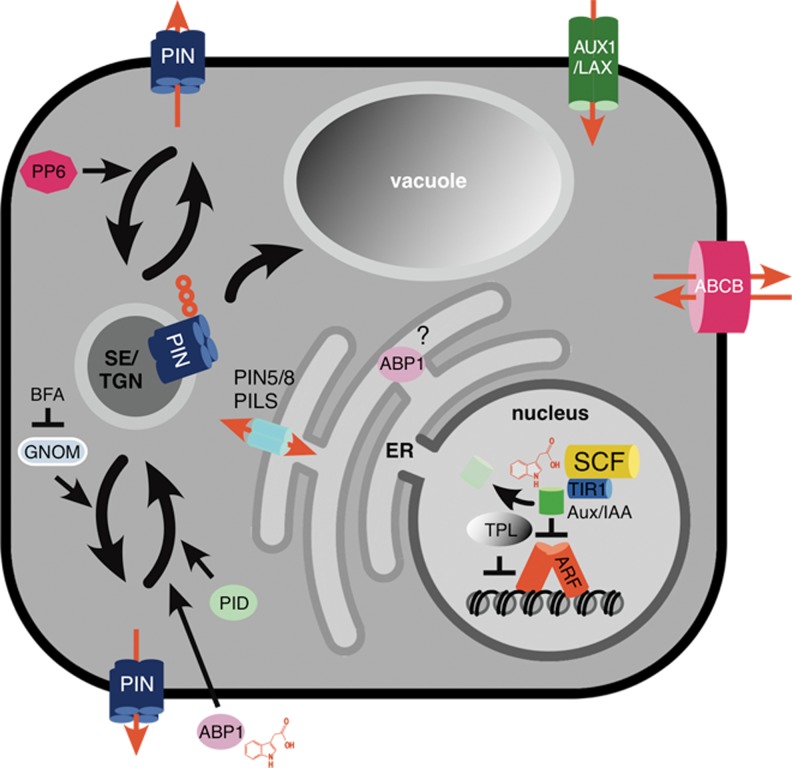
Fig. 1.
Essential elements of auxin signaling and transport in plant cells. Auxin perception by nuclear SCFTIR1/AFB (blue, yellow) triggers ubiquitination and proteasome-mediated degradation of Aux/IAA co-receptors (green). As a result transcriptional repression via TPL (grey) activity and via Aux/IAA-mediated inhibition of ARF transcriptional regulators (red) is relieved. Auxin binding by extracellular ABP1 (lilac) impacts on endocytic protein sorting (see also Fig. 2a), whereas the role of ER-resident ABP1 is still not known. Cellular uptake of auxin is mediated by AUX1/LAX-type permeases (green) and by selected ABCB-type transporters (pink). Cellular auxin efflux requires PIN- (blue) and ABCB-type (pink) transport proteins. Auxin sequestration between cytoplasm and ER involves activity of PILS and PIN proteins (turquois) lacking the central hydrophilic loop that is characteristic for plasma membrane-resident PINs. Auxin transport proteins are subject to intracellular sorting from the plasma membrane to sorting endosomes (SE)/trans Golgi network (TGN). Recycling from SE/TGN to the plasma membrane involves activity of BFA-sensitive ARF-GEF GNOM, specifically for sorting at the basal cellular domain. PIN sorting and transcytosis is controlled by its phosphorylation status. PID (light green) kinase activity promotes sorting of phosphorylated PINs preferentially to the apical plasma membrane domain, a response that is antagonized by PP6 (purple) phosphatase activity triggering sorting to the basal domain. Apart from protein recycling and transcytosis between plasma membrane domains, PINs are also subject to endocytic sorting via late endosomes/multivesicular bodies into the lytic vacuole for their irreversible degradation. Sorting via this pathway is controlled by ubiquitination of PINs (red circles)
Further cellular activities, implicated in shaping auxin effects on transcription, are related to the control of chromatin architecture and histone modifications (Sablowski 2011; Yoshida et al. 2013). Topless (TPL) and TPL-related (TPR) proteins represent transcriptional co-repressors, down-regulating gene expression via interaction with diverse transcriptional regulators (Long et al. 2006; Smith and Long 2010; Wang et al. 2013b). Specifically, TPL/TPR appear to be involved in the recruitment of histone deacetylase activity, thus promoting establishment of a chromatin status, which is less accessible for the transcriptional machinery (Long et al. 2006; Wang et al. 2013b). TPL was demonstrated to physically interact with Aux/IAA proteins, suggesting that apart from sequestering ARFs, Aux/IAA-mediated down-regulation of auxin responses involves TPL-mediated establishment of a repressive chromatin status (Szemenyei et al. 2008) (Fig. 1). Clear-cut genetic evidence linking the regulation of chromatin architecture to established auxin signaling pathways came from screens for suppressors of slr-1, a dominant mutation in SOLITARY-ROOT (SLR) /IAA14. Slr-1 is entirely defective in auxin-regulated lateral root formation, due to a point mutation stabilizing the Aux/IAA protein (Fukaki et al. 2002). Loss of chromatin remodeling activity mediated by PICKLE (PKL) /GYMNOS, a CHD subfamily II ATP-dependent chromatin remodeling factor, suppresses slr-1 lateral root formation defects. Such suppression of slr-1 phenotypes by pkl requires ARF transcription factors that act in conjunction with SLR/IAA14, a finding that placed PKL together with ARFs in transcriptional regulation of lateral root development (Fukaki et al. 2006). Nevertheless, whilst PKL-mediated chromatin remodeling activity was found to determine histone H3 lysine 27 trimethylation levels at repressed loci (Zhang et al. 2012), it is still not resolved, whether auxin itself could influence PKL activities on chromatin. Additional studies support scenarios in which auxin directly impacts on chromatin architecture and histone modifications. Chromatin in the promoter region of SKP2B, an Arabidopsis F-box protein essential for cell cycle progression during early stages of lateral root formation, undergoes reversible adjustments in the composition of nucleosomes as well as histone acetylation in an auxin-dependent manner (Manzano et al. 2012). This was suggested to modulate SKB2B expression in response to the growth regulator, being essential for transmission of hormonal signals on cell proliferation (Manzano et al. 2012). In another report, PROPORZ1 (PRZ1)/AtADA2b, an Arabidopsis ortholog of ADA2 transcriptional adaptor protein (Sieberer et al. 2003; Vlachonasios et al. 2003), was demonstrated to channel auxin effects on histone acetylation in the promoter region of auxin-controlled loci, which coincided with alterations in auxin-responsive gene expression (Anzola et al. 2010). Similar to ADA2 homologs in other organisms, PRZ1 most likely represents a component of multi-subunit chromatin remodeling complexes that are required for the recruitment of histone acetyl transferase GCN5 to chromatin (Mao et al. 2006). Prz1 shortcomings in controlling variations in histone acetylation thus could result from very general defects in chromatin remodeling activities (Anzola et al. 2010). Detailed insights into mechanisms of an epigenetic control that could transmit auxin signals via reversible histone modifications have however, not been provided so far.
Identification of another type of auxin receptor was originally based on characterization of auxin-binding activities in cellular fractions (Ray 1977; Cross and Briggs 1978; Jones and Venis 1989). Such experiments resulted in the identification of AUXIN-BINDING PROTEIN 1 (ABP1), which localizes predominantly to the ER (Hertel et al. 1972; Löbler and Klämbt 1985a, b; Shimomura et al. 1986) (Fig. 1). For several decades however, it was not clear, how ABP1 might participate in auxin-controlled processes, and only recently a role for ABP1 in post-transcriptional regulation of hormonal responses has been revealed (Robert et al. 2010; Xu et al. 2010). This work indicates that ABP1 functions as a positive regulator of clathrin-mediated endocytosis from the plasma membrane. ABP1 activity appears to be blocked upon auxin binding, resulting in inhibition of endocytosis (Robert et al. 2010). Furthermore, ABP1 function has been linked to polarity establishment in cells, which involves ABP1-mediated activation of downstream Rho-GTPases (ROP) to affect cytoskeleton organization (Xu et al. 2010). Thus unlike TIR1/AFB pathways, ABP1-mediated transmission of auxin signals primarily affects post-translational responses. To complicate matters further, a recent report has demonstrated additional ABP1 activities in the regulation of Aux/IAA protein via pathways antagonizing SCFTIR1/AFB activities (Tromas et al. 2013). This places ABP1 function in transcriptional regulation of responses to auxin, apart from its role in protein sorting. However, it remains a mystery as to how ABP1 might fulfill these quite different tasks. In particular, we still have no clear clue about mechanisms that would connect a predominantly ER-localized protein to activities at the plasma membrane (Löbler and Klämbt 1985b; Inohara et al. 1989). It appears that at least a minor portion of ABP1 escapes from the ER, and gets released into the extracellular apoplast, where upon auxin binding downstream signaling events might be initiated (Jones and Herman 1993) (Fig. 1). It is still unclear how ABP1 might enter the secretory pathway, as the specific trans-acting determinants involved have not yet been characterized. In addition, signaling from the extracellular apoplast to the cytosolic face of the plasma membrane would require docking to and signal transmission across the plasma membrane (Shi and Yang 2011). Determinants that could be involved in these aspects of ABP1-dependent auxin signaling events are under investigation, but still await functional characterization (Shi and Yang 2011; Sauer et al. 2013).
Key determinants of polar auxin transport
Ever since Darwin’s work, an active relocation of endogenous signals has been implicated in the regulation of plant growth responses (Darwin and Darwin 1881; Boysen Jensen 1910). Subsequent experiments repeatedly demonstrated directional, polar transport of auxin along the main plant body axes, and a model has been put forward, integrating early observations and suggesting a remarkably simple mechanism for the transport of the hormone (Went and Thimann 1937; Rubery and Sheldrake 1974; Raven 1975). According to the chemiosmotic model, PAT along transport routes would involve cellular uptake of apoplastic, protonated IAAH either via uptake carriers or via diffusion across the plasma membrane. Once inside the cell, further export of deprotonated cytosolic IAA− into the apoplast would require activity of specific efflux carriers, and an asymmetric, polar distribution of such carrier proteins could pre-determine the direction of hormone transport within a given cell file (Rubery and Sheldrake 1974; Raven 1975). By making use of the model plant Arabidopsis, a number of auxin transport proteins have been identified, which essentially belong to three classes: (1) a family of intrinsic membrane proteins with resemblance to tryptophan permeases implicated in cellular uptake of the growth regulator; (2) a group of ATP-binding cassette membrane proteins involved in cellular efflux and uptake of auxin; and (3) a plant-specific class of membrane proteins involved in auxin cellular efflux as well as in intracellular compartmentalization of the growth regulator. Genetic and cell biological analysis of selected members of these classes of auxin transport proteins has provided substantial insights into the biology of auxin and has essentially confirmed the models for PAT proposed about 40 years ago (Luschnig 2002; Kramer 2004; Paponov et al. 2005; Geisler and Murphy 2006; Bandyopadhyay et al. 2007; Benjamins and Scheres 2008; Zazimalova et al. 2010) (Fig. 1).
AUXIN1 (AUX1) and LIKE AUX1 (LAX1) function in cellular uptake of the hormone, and analysis of mutants deficient in these loci demonstrated their involvement in auxin-regulated processes like organ tropism, control of lateral root formation as well as phyllotactic patterning of leaves (Bennett et al. 1996; Marchant et al. 1999; Reinhardt et al. 2003; Swarup et al. 2005; Bainbridge et al. 2008). When viewing the localization of AUX1 in root meristems, a polar distribution was revealed in cells along major auxin transport routes. This presumably optimizes cellular uptake of the growth regulator and could be of particular importance for the establishment of transient auxin maxima in the regulation of growth responses (Swarup et al. 2005). Heterologous expression of AUX1 in Xenopus oocytes and in Schizosaccharomyces pombe indicated auxin uptake activity in a saturable manner at physiological pH, thus providing strong evidence for AUX1 function as an auxin uptake facilitator (Yang et al. 2006; Yang and Murphy 2009).
Among the plant ABC- or multidrug-resistant-type carrier proteins characterized to date, members of a subclade have been found to be involved in cellular efflux and uptake of auxin (Rea 2007). ABCB1, 4, 19, and 21 (ATP-BINDING CASSETTE SUBFAMILY B-TYPE) function in cellular auxin efflux (Gaedeke et al. 2001; Noh et al. 2001; Geisler et al. 2005; Terasaka et al. 2005; Kamimoto et al. 2012), and additional experimental evidence has demonstrated facultative auxin uptake activities for ABCB4 and ABCB21 which depended on cytosolic auxin concentrations (Terasaka et al. 2005; Kamimoto et al. 2012). Expression and subcellular localization of ABCB proteins, together with phenotypes of abcb loss-of-function mutants, and mutant combinations, led to models suggesting rather general functions in long-distance auxin transport and loading into such transport highways (Bandyopadhyay et al. 2007). In addition, ABCB auxin transport protein function has been linked to highly defined growth responses, like root hair formation, adventitious root formation, or phototropism (Cho et al. 2007; Christie et al. 2011; Sukumar et al. 2013). Modulation of some of these growth responses involves interaction between distinct auxin transport activities, specifying auxin distribution via mutual regulation of auxin transport proteins (Bouchard et al. 2006; Blakeslee et al. 2007; Mravec et al. 2008; Titapiwatanakun et al. 2009).
Mutants in some members of the PIN-FORMED (PIN) family, representing a plant-specific family of membrane proteins (Zazimalova et al. 2010), produce phenotypes that are suggestive of severe aberrations in auxin-controlled growth responses. Pin1 mutant alleles, for example, exhibit broad defects in embryogenesis as well as in development of inflorescences and floral organs, whereas pin2 alleles have initially been characterized as agravitropic root growth mutants (Bell and Maher 1990; Okada et al. 1991; Chen et al. 1998; Gälweiler et al. 1998; Luschnig et al. 1998; Utsuno et al. 1998). Additional phenotypes affecting diverse aspects of plant development have been identified in mutant combinations deficient in multiple PINs, underlining functional redundancies within this protein family (Blilou et al. 2005; Vieten et al. 2005). Most importantly however, PIN localization studies revealed asymmetric, polar signals restricted to domains at the plasma membrane, a distribution that conforms to the chemiosmotic model for PAT. PIN1 for example accumulates at the basal end of vascular stele cells where it participates in rootward auxin transport towards the primary root meristem (Gälweiler et al. 1998). Similar results have been obtained for further PIN proteins, which exhibit a polar distribution at the plasma membrane, concurrent with the direction of auxin transport routes in different tissues and cell files (Müller et al. 1998; Friml et al. 2002, 2003; Benkova et al. 2003; Reinhardt et al. 2003; Sorefan et al. 2009). These observations linked PIN function to cellular auxin efflux, and results from auxin transport assays, performed either in plants or heterologous hosts, further supported this notion (Petrasek et al. 2006; Yang and Murphy 2009).
Owing to their central role in plant development, myriads of reports connecting PINs to a large variety of biological processes have been published since their initial discovery about 15 years ago. In that respect it is surprising that we still lack clear mechanistic insights into catalytic activities of PINs, as no transport assay in an entirely defined experimental system has been established. This is also true for PIN topology: no structural model as based on protein crystallization data has so far been provided for the PINs. Thus, for the time being, we need to deal with the fact that we still do not fully understand the function of PINs in auxin efflux in mechanistic terms (Peer et al. 2011). This might explain the rather vague term “auxin transport facilitator” that is frequently used in the literature when referring to PINs.
Apart from conventional PIN proteins characterized by two transmembrane domain regions and separated by a large hydrophilic loop, additional PIN proteins share the transmembrane domains but encode only a small central loop region (Krecek et al. 2009). These proteins appear evolutionary older than conventional PINs and were found to localize to ER membranes, where they have been implicated in the control of intracellular auxin homeostasis (Mravec et al. 2009; Dal Bosco et al. 2012; Ding et al. 2012b; Bender et al. 2013; Cazzonelli et al. 2013; Sawchuk et al. 2013). The purpose of auxin translocation across ER membranes is not fully understood, but it is evident that auxin redistribution between different intracellular compartments could influence a wide range of auxin-controlled signaling events (Barbez and Kleine-Vehn 2013) (Fig. 1). A biological significance of this activity is underlined further by characterization of another class of auxin transport proteins, exhibiting limited similarity to PINs, and therefore termed PILS (for PIN-LIKES) genes (Barbez et al. 2012). PILS proteins are found throughout the entire plant lineage and, by analogy to ER-resident PINs, appear to modulate intracellular auxin homeostasis (Barbez et al. 2012; Feraru et al. 2012).
While still not fully understood in terms of function, characterization of these different groups of PIN and PILS proteins changed our view on the evolution of auxin transport and signaling (Zazimalova et al. 2010; Feraru et al. 2012). The occurrence of ancestral ER-resident auxin transport proteins suggests that a common ancestor of auxin transport proteins first developed activities, mediating intracellular hormone transport. In the course of acquisition of morphological complexity, plants might have evolved modified PIN proteins that get sorted to the plasma membrane, allowing for directional, intercellular auxin transport. Perhaps, this was one of the prerequisites for orchestrating sophisticated developmental programs upon plants’ conquest of land.
Protein sorting and auxin
The key to the establishment of directional intercellular auxin transport is a highly polar distribution of auxin transport proteins at distinct plasma membrane domains. Four major domains, comprising an apical and a basal domain together with proximal (inner) and distal (outer) lateral domains as well as additional sub-domains have so far been described in plant cells (Dettmer and Friml 2011; Yang and Lavagi 2012). Auxin transport proteins have been found at these domains and have been described to undergo dynamic rearrangements in their localization (Friml et al. 2002, 2003, 2004). The functional significance of such a polar distribution, and variations therein, was demonstrated by in planta expression of constitutively mis-localized PIN proteins. This coincided with alterations in auxin distribution and the occurrence of auxin-related growth defects, highlighting additionally a role for dynamic adjustments in PIN localization as a mediator of directional auxin transport (Wisniewska et al. 2006; Dhonukshe et al. 2010; Huang et al. 2010; Zhang et al. 2010).
Characterization of the machinery controlling the distribution of auxin transport proteins has revealed pathways that modulate sorting to and from distinct domains at the plasma membrane (Geldner 2009; Titapiwatanakun and Murphy 2009; Grunewald and Friml 2010; Richter et al. 2010; Robinson et al. 2012; Löfke et al. 2013). Plasma membrane proteins are sorted along the secretory pathway and reach their destination at the plasma membrane after passage through Golgi compartments and the trans Golgi network/early endosomes (TGN/EE) (Jürgens and Geldner 2007; Marti et al. 2010; Zarsky and Potocky 2010; Cai et al. 2011; Ding et al. 2012a). This is followed by re-internalization via endocytic sorting into TGN/EE, and recycling back to the plasma membrane, a response implicated in modulation of plasma membrane protein function (Geldner et al. 2001) (Fig. 2a). Alternatively, plasma membrane proteins are sorted into multivesicular bodies/prevacuolar compartment for degradation in the lytic vacuole (Richter et al. 2009; Schellmann and Pimpl 2009; Reyes et al. 2011) (Fig. 2b). Recycling of plasma membrane proteins has been linked to ADP-ribosylation factor (ARF) GTPase activity that is controlled by ARF guanine-nucleotide exchange factors (ARF-GEF). Detailed analysis of PIN1 sorting revealed a Brefeldin A-(BFA)-sensitive ARF-GEF GNOM as an essential regulator for its recycling and transcytosis between distinct plasma membrane domains (Steinmann et al. 1999; Geldner et al. 2003; Kleine-Vehn et al. 2008a, 2010). In addition, protein phosphorylation, mediated by PINOID (PID) and related serine/threonine kinases, was found to affect PIN targeting and plant growth (Friml et al. 2004; Santner and Watson 2006; Michniewicz et al. 2007; Dhonukshe et al. 2010) (Fig. 2a). Specifically, phosphorylation of conserved TPRXS(N/S) motifs and further sites within the PIN central hydrophilic loop domain, appears to promote sorting to apical plasma membrane domains (Dhonukshe et al. 2010; Huang et al. 2010; Zhang et al. 2010), whereas PP6-type heterotrimeric phosphatase antagonizes such activity, favoring PIN sorting to basal cellular domains (Michniewicz et al. 2007; Dai et al. 2012; Ballesteros et al. 2013) (Fig. 2a). Next to intracellular protein cycling, proteolytic degradation via sorting to lytic vacuoles represents another means to adjust auxin transport activities (Abas et al. 2006; Kleine-Vehn et al. 2008b; Laxmi et al. 2008; Marhavy et al. 2011). Plants, by analogy to other eukaryotes, utilize reversible cargo ubiquitination as a recognition signal for protein sorting from the plasma membrane to the vacuole (Reyes et al. 2011; Korbei and Luschnig 2013), which is consistent with observations demonstrating that ubiquitination of Arabidopsis PIN2 functions as a vacuolar targeting signal (Leitner et al. 2012) (Fig. 2b).
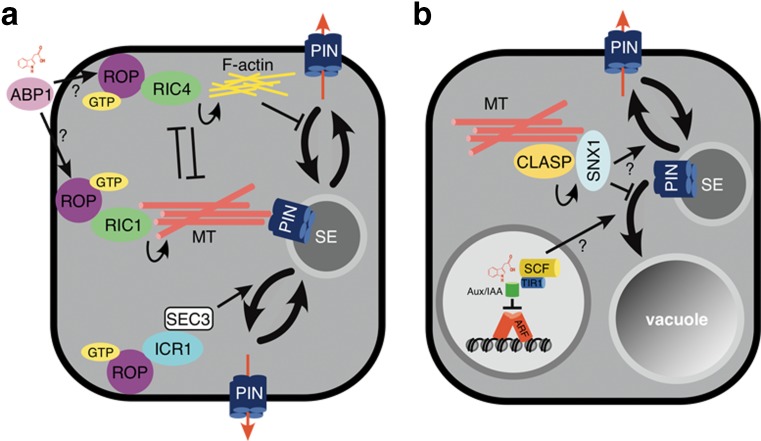
Fig. 2.
Auxin and its role in protein sorting. a Auxin binding by extracellular ABP1 (lilac) causes activation of ROP GTPases (purple) and downstream effectors RIC1 and RIC4 (green). ROP2-RIC4 stimulates F-actin (yellow) accumulation at localized domains, which is suggested to antagonize clathrin-mediated endocytosis of plasma membrane proteins like PINs. This pathway is antagonized by ROP6-RIC1 activity promoting local formation of cortical MT arrays (pink), which interferes with activation of ROP2. ROP2 on the other hand sequesters RIC1, thereby blocking formation of ordered MT arrays. ICR1, another ROP-interacting protein (light blue) is essential for polarized distribution of PIN proteins at the plasma membrane. This involves interaction with exocyst vesicle-tethering complex subunit SEC3 (white). b Decisions about endocytic sorting and further targeting on PIN proteins into the lytic vacuole depend on putative retromer subunit SNX1 (light blue), which functions as a gating factor, promoting protein recycling to the plasma membrane and thereby antagonizing vacuolar sorting. CLASP-(yellow)-mediated tethering of SNX1 to MTs (pink), links PIN sorting to cytoskeleton components and possibly ABP1-mediated signaling. Variations in auxin homeostasis sensed by nuclear SCFTIR1/AFB auxin receptor induce vacuolar sorting of PIN proteins. Mechanisms and pathways involved are still elusive
Recent years have seen a systematic characterization of evolutionary conserved pathways for protein sorting in plants. It turns out that auxin decisively influences these stereotypic sorting processes, which has now established the phytohormone as a plant-specific regulator of universal cellular activities. Paciorek and colleagues were the first to demonstrate that treatment with potent auxin analogs, as well as alterations in intracellular auxin homeostasis, interfere with endocytic sorting of PIN proteins. This is hypothesized to modulate auxin flow as part of a regulatory feedback switch (Paciorek et al. 2005). Auxin effects on plasma membrane protein sorting, however, are quite general and not restricted to PIN proteins, acting via inhibition of clathrin-mediated endocytosis (Paciorek et al. 2005; Robert et al. 2010). In plants, experimental evidence for clathrin sorting has been provided only recently, controlling variations in protein abundance at the plasma membrane via endocyosis, with far-reaching implications for a range of metabolic pathways and signaling cascades (Dhonukshe et al. 2007). Clathrin represents a triskelion-shaped complex, comprising a structural backbone of three clathrin heavy chain (CHC) molecules, and three clathrin light chain (CLC) molecules, which upon interaction form a polyhedral clathrin lattice (Kirchhausen and Harrison 1981; Kirchhausen et al. 1986; Royle 2006). For endocytic cargo sorting, clathrin needs to associate with the plasma membrane, where it interacts with cargo, adaptor protein complex-2 and further accessory proteins, resulting in formation of clathrin-coated pits. In animals, this is followed by maturation into cargo-loaded clathrin-coated vesicles (CCV) upon internalization from the plasma membrane and analogous processes appear to be present in plants (Royle 2006; Kitakura et al. 2011; Ito et al. 2012; Di Rubbo et al. 2013; Fan et al. 2013; Kim et al. 2013; Wang et al. 2013a; Yamaoka et al. 2013).
Auxin's effect on endocytic sorting has been linked to the control of clathrin recruitment at the plasma membrane. In fact, synthetic auxin analogs and IAA were found to interfere with recruitment of Arabidopsis CLC and CHC isoforms to the plasma membrane, thereby interfering with CCV assembly and endocytic sorting (Robert et al. 2010; Wang et al. 2013a). Inhibitory effects of auxin on CLC recruitment to the plasma membrane appear to be mediated by auxin-binding by ABP1, whereas SCFTIR1/AFB-controlled auxin perception seems dispensable thus supporting models in which ABP1 acts as positive regulator of clathrin-dependent endocytosis (Kitakura et al. 2011; Wang et al. 2013a). Downstream components linking ABP1 signaling to endocytic sorting comprise ROP-type small GTPases, which were demonstrated to be activated by auxin in an ABP1-dependent manner (Xu et al. 2010) (Fig. 2a). ROPs represent the only small GTPase family found in plants, with several of its members implicated in intracellular cargo sorting via regulating actin and microtubule components of the cytoskeleton (Molendijk et al. 2001; Gu et al. 2003; Vernoud et al. 2003; Fu et al. 2009; Craddock et al. 2012) and a number of elegant studies have now established their role in auxin responses via organization of the cytoskeleton. Arabidopsis ROP2 was shown to affect PIN1 endocytosis in emerging lobe regions of interdigitated leaf pavement cells (Nagawa et al. 2012). This involves the activity of the ROP2 effector RIC4 (ROP-INTERACTIVE CRIB MOTIF-CONTAINING PROTEIN), which stimulates localized accumulation of F-actin and is suggested to antagonize clathrin-dependent endocytosis of PIN1 (Nagawa et al. 2012) (Fig. 2a). A related pathway has been proposed for PIN2 sorting in root meristem cells, with the SPK1-(for SPIKE1, an activator of ROP6)-ROP6-RIC1 regulatory module promoting actin filament stabilization and supposedly interfering with clathrin-dependent PIN2 endocytosis (Chen et al. 2012; Lin et al. 2012). PIN retention at the plasma membrane might cause elevated auxin flow into the apoplast, which in turn could induce auxin signaling via apoplastic ABP1, altogether constituting a robust feed-forward loop essential for auxin-driven growth responses (Yang and Lavagi 2012).
Auxin-controlled ROP activity has been connected to microtubules (MT) as well, seemingly antagonizing auxin effects mediated via control of the actin cytoskeleton. ROP6 was found to bind and activate RIC1 a MT-associated protein (MAP) in an ABP1-dependent manner (Fu et al. 2005, 2009; Xu et al. 2010). This in turn would promote localized accumulation of cortical MT arrays that are aligned perpendicular to the axis of cell expansion and are implicated in guidance of cellulose synthase activity (Crowell et al. 2009; Bringmann et al. 2012) (Fig. 2a). Variations in ROP6 activity could thus modulate cell expansion via differential cellulose deposition at the cell wall. Activated ROP2, on the other hand, locally sequesters RIC1 to interfere with ordered MT alignment, whereas RIC1-mediated MT array organization suppresses activation of ROP2 (Xu et al. 2010). Thus ROP2 and ROP6 pathways mutually inhibit each other in the organization of the cytoskeleton, which presumably feeds back on the sorting of PINs and corresponding auxin transport activities in the control of morphogenesis (Yang and Lavagi 2012) (Fig. 2a). Another link between ROP activity and PIN sorting was established by Hazak and colleagues, who demonstrated an involvement of the ROP effector ICR1 (INTERACTOR OF CONSTITUTIVE ACTIVE ROP1) in the control of polar PIN distribution at the plasma membrane (Hazak et al. 2010). Notably, an earlier report demonstrated interaction between ICR1 and SEC3, the latter representing a subunit of an Arabidopsis vesicle-tethering exocyst protein complex (Lavy et al. 2007; Zarsky et al. 2009; Zhang et al. 2013) (Fig. 2a). Such interaction would suggest a role for ROP-ICR1 in secretory protein sorting, potentially modulating exocyst activity and its effects on PIN distribution at the plasma membrane (Drdova et al. 2013).
While it seems evident that the organization of the cytoskeleton feeds back on auxin distribution via sorting of auxin transport proteins, mechanisms linking, e.g., PIN sorting to the cytoskeleton remain to be resolved (Geldner 2009; Heisler et al. 2010; Brandizzi and Wasteneys 2013). A recent report has come up with an intriguing model, linking MTs, the predicted Arabidopsis retromer component SNX1 (SORTING NEXIN 1) and PIN sorting (Ambrose et al. 2013). The authors demonstrated that a MAP termed CLASP (CYTOPLASMIC LINKER-ASSOCIATED PROTEIN) interacts with SNX1 to tether retromer-associated endosomes to MTs (Fig. 2b). SNX1 has been implicated in PIN2 sorting, presumably acting as a gating factor that promotes recycling of endocytosed PINs back to the plasma membrane (Jaillais et al. 2006, 2008; Kleine-Vehn et al. 2008b). In the absence of either SNX1 or CLASP, PIN2 no longer efficiently recycles to the plasma membrane but relocates to the vacuole for its degradation, underlining a role for both proteins as positive effectors of PIN recycling (Kleine-Vehn et al. 2008b; Ambrose et al. 2013). While conflicting evidence for CLASP function in PIN sorting has been published elsewhere (Kakar et al. 2013), data presented by Ambrose and colleagues strongly support a function for MTs in PIN recycling, presumably controlled by SNX1/CLASP-mediated tethering of sorting endosomes to MTs (Ambrose et al. 2013). Perhaps, ROP effects on microtubule organization modulate such interaction, indirectly affecting sorting and recycling of PIN proteins.
Experiments and results, linking auxin-binding by ABP1 to cytoskeleton organization and PIN endocytic sorting, suggest that variations in PIN retention time at the plasma membrane modulate auxin efflux rates that are further translated into developmental cues. Notably, another link between actin and auxin transport has been established, highlighting effects of auxin transport inhibitors (ATI) on the dynamic distribution of the auxin transport machinery. Specifically, 2,3,5-triiodobenzoic acid and 2-(1-pyrenoyl) benzoic acid, which both efficiently block cellular auxin efflux, were found to interfere with organization of the actin cytoskeleton with drastic consequences for protein endocytosis and vesicle motility, including the sorting of PINs (Geldner et al. 2001; Dhonukshe et al. 2008). Whether or not treatment with ATIs affects pathways overlapping with those controlled by ABP1 is currently not known. Nevertheless, ATI effects on the actin cytoskeleton underline the significant role for intracellular vesicle dynamics in the regulation of PAT.
Crosstalk between ABP1 and clathrin has established a plausible function for this auxin receptor. However, auxin signaling via SCFTIR1/AFB-controlled pathways contributes to the regulation of endocytic sorting as well. Apart from auxin-induced inhibition of PIN endocytosis, the opposite effect, namely enhanced degradation of PINs, has been observed upon alterations in auxin homeostasis (Sieberer et al. 2000; Vieten et al. 2005; Abas et al. 2006; Baster et al. 2013). PIN2 in particular, undergoes vacuolar sorting in response to altered auxin levels, presumably mediated by increased protein ubiquitination, which signals enhanced turnover of the auxin carrier protein (Leitner et al. 2012). It seems possible that down-regulation of PINs in response to auxin antagonizes inhibitory auxin effects on plasma membrane protein endocytosis, resetting cellular PIN protein levels back to default. This could be of particular importance for the control of differential growth responses that involve transient establishment of auxin maxima (Band et al. 2012). When trying to elucidate pathways involved, it turned out that SCFTIR1/AFB-controlled auxin signaling is essential for PIN vacuolar sorting, while ABP1 is not, demonstrating that both major auxin-signaling pathways are involved in overlapping but distinct aspects of intracellular sorting of PIN proteins (Sieberer et al. 2000; Baster et al. 2013) (Fig. 2a,b).
Insights into pathways transmitting effects of auxin perception by ABP1 or SCFTIR1/AFB to the sorting of membrane proteins in general, or PIN proteins in particular, are still rudimentary. However, given the prominent phenotypes that can be observed, when disturbing such signaling, it seems clear that auxin-mediated regulation of protein sorting is one of the key regulators of plant morphogenesis. Consistently, genetic or pharmacological interference with cytoskeleton organization and/or clathrin-mediated sorting processes results in phenotypic alterations, characteristic for auxin signaling and/or transport mutants (Dhonukshe et al. 2008; Kitakura et al. 2011; Wang et al. 2013a). While this underlines essential roles for auxin in a multitude of developmental processes, it seems quite unlikely that all the classical “auxin-related” phenotypes that can be observed upon affecting, e.g., endocytic sorting processes, arise as an exclusive consequence of altered auxin transport or signaling. Rather it appears that phenotypes observed in auxin signaling and transport mutants reflect widespread, combinatorial consequences of altered protein sorting, not necessarily directly related to a disturbance of PAT. Perhaps, it will be necessary to reevaluate causes and consequences of the broad phenotypic responses that arise as result of highly pleiotropic activities of this universal plant growth regulator.
Auxin and translational control
Auxin-clathrin crosstalk underlines the hormone's role in an evolutionary conserved pathway, whereas analysis of mutants defective in ribosomal function have linked auxin signaling to translational regulation, another fundamental feature of gene expression control. Mutants affected in ribosomal protein functionality exhibit growth defects reminiscent of phenotypes observed upon disturbing auxin transport or signaling (Van Lijsebettens et al. 1994; Ito et al. 2000; Weijers et al. 2001; Degenhardt and Bonham-Smith 2008; Szakonyi and Byrne 2011). Similar auxin-related defects are caused by a mutation in the nucleolin PARL1 (PARALLEL1) likely required for ribosomal maturation (Petricka and Nelson 2007). It remains unclear though, as to how defects in such general determinants of cellular metabolism could result in rather specific auxin-related phenotypes. A mechanistic link between auxin, translational control, and protein sorting was established when screening mutagenized Arabidopsis seedlings for defects in vacuolar cargo sorting (Rosado et al. 2010). In this report the authors identified mutations in the ribosomal proteins RPL4A and RPL4D, and found that both loci are required for efficient vacuolar sorting of synthetic reporter proteins (Miao et al. 2008; Rosado et al. 2010). In addition, rpl4a turned out to be deficient in vacuolar targeting of PIN2, and mutant vacuolar sorting defects could be phenocopied by application of high concentrations of auxin, causing secretion of reporter proteins, instead of sorting to the vacuole (Rosado et al. 2010). Intriguingly, the authors observed that expression of RPL4A is responsive to auxin and to inhibition of auxin transport, which led them to suggest that auxin could modulate ribosomal biogenesis, linking ribosome function to auxin signaling and protein sorting (Rosado et al. 2010).
Similar to characterized ribosomal protein or ribosome biogenesis mutants, rpl4 alleles exhibit developmental aberrations that appear related to altered auxin responses, and models have been put forward in an attempt to explain these rather specific growth defects. Formation of heterogeneous populations of ribosomes characterized by incorporation of different ribosomal protein isoforms has been proposed to cause variations in the ribosomes’ specificity for different transcripts or transcript groups. As an alternative, a role for ribosomal gene dosage, rather than variations in ribosome composition has been suggested to modulate efficiency of protein translation, which might affect cellular processes primarily related to auxin signaling (Giavalisco et al. 2005; Komili et al. 2007; Carroll et al. 2008; Rosado et al. 2010). However, all these models fall short when trying to explain the specific defects of ribosomal protein mutants, without further knowledge about downstream targets modulated by such translational control.
A study by Nishimura and colleagues provided some insights into mechanisms that could link translational control to the regulation of auxin responses (Nishimura et al. 2005). The authors identified a mutant they termed short valve 1 (stv1), exhibiting apical-basal patterning defects in the gynoceum that are reminiscent of mutants defective in the ARF transcriptional regulators ETTIN (ETT/ARF3) and MONOPTEROS (MP/ARF5) (Sessions et al. 1997; Hardtke and Berleth 1998). Cloning of STV1 revealed that it encodes RPL24B, a ribosomal protein found in archaebacterial and eukaryotic genomes and implicated in enhancing translational efficiency (Dresios et al. 2000). On top of that, Arabidopsis RPL24 was found to interact with a viral transactivator protein, and to stimulate translational efficiency via translation reinitiation of polycistronic viral RNA (Park et al. 2001). In eukaryotes, translation reinitiation has been shown to modulate translation of ORFs that are flanked by an upstream, translatable ORF located in the 5′-leader region of the mature mRNA (Kozak 1999; Roy and von Arnim 2013). Upon initiation of translation, such uORFs are recognized by the scanning translation initiation complex that moves along the mRNA, prompting translation of these—often times very small—ORFs (Rajkowitsch et al. 2004). Subsequent translation of the “main” ORF, located downstream of the uORF requires efficient translation reinitiation, ensuring that the ribsome resumes scanning to recognize downstream Start AUGs (Park et al. 2001; Zhou et al. 2010). Transcripts of both, ETT and MP encode uORFs, which might suggest that stv1 patterning defects could reflect deficiencies in the translational control of these ARF genes. Indeed, when expressing an engineered ETT transcript lacking its 5′ uORF, stv1 patterning defects were partially suppressed, supporting scenarios, in which RPL24B modulates auxin responsive organ development via controlling translational reinitiation of ARF genes (Nishimura et al. 2005). Related observations have been made with rpl4d and rpl5a ribosomal protein mutants and with a mutant in translational initiation factor eIF3h, seemingly affected in translation of ARF loci that all are characterized by uORFs in their 5′-regions (Zhou et al. 2010; Rosado et al. 2012) (Fig. 3).
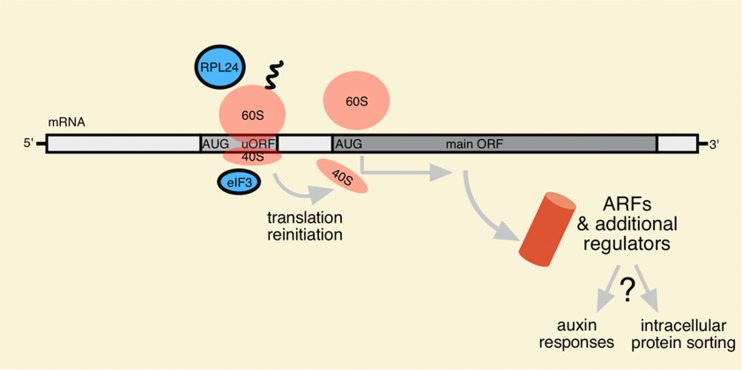
Fig. 3.
Translational control as a mediator of auxin signaling. Translation of mRNAs comprising uORFs commences with initiation of translation at the AUG of the uORF (ribosomal subunits in pink). Further translation of the main ORF positioned downstream of the uORF, depends on efficient translation reinitiation, which is stimulated by ribosomal proteins like RPL24 or translation initiation complex subunit eIF3 (blue). Several mRNAs encoding ARF transcription factors were found to contain uORFs. Variations in availability and/or functionality of rate-limiting determinants of translation reinitiation like RPL24 could define amounts of ARF protein and additional regulatory proteins to control auxin responses and intracellular protein sorting
Control of ARF expression via reinitiation of translation might represent an integral part of auxin signaling cascades, controlling intracellular protein sorting via adjustments in transcriptional control of auxin responses (Rosado et al. 2010, 2012). However, about one third of Arabidopsis mRNAs appears to contain uORFs (Kim et al. 2007), all potentially subject to expression control via translation reinitiation. Assuming that RPL24B and additional determinants of translational control are involved in expression control of all these loci, it appears questionable that auxin-related growth defects arise as an exclusive consequence of diminished expression of such a huge proportion of the Arabidopsis proteome. Analogous to the situation described for crosstalk between auxin and components of the cytoskeleton, phenotypes of ribosomal mutants clearly reflect the outstanding importance of auxin signaling and transport. However, a closer look on phenotypes and the proteome of these mutants is definitely required to characterize the role of translational control in higher plants.
Another even more puzzling link between translational control and auxin signaling was established when characterizing a dominant, auxin-resistant Arabidopsis mutant derived from a screen performed in the presence of sirtinol (Perry et al. 2005). Apart from being less responsive to externally applied auxin, mutant plants exhibited defects in apical hook formation upon germination in the dark and developed less lateral roots, both representing hallmark features of auxin-response mutants. The mutation responsible for these phenotypes was found in the anticodon loop of a tRNAAla, changing its anticodon from 5′-CGC-3′ to 5′-CAC-3′, which no longer recognizes alanine but valine codons, instead (Perry et al. 2005). Given that further tRNA identity determinants are not affected by this mutation, it seems likely that, although charged with alanine, the mutant tRNA will correctly associate with ribosomes and valine triplett codons, resulting in translation of mutant proteins. The Arabidopsis genome is predicted to contain 630 tRNA loci, amongst which 33 will recognize a codon for alanine, whereas 30 will bind to valine codons. When not considering wobbling, seven tRNAAla will recognize 5′-GCG-3′ codons, whereas 8 tRNAVal will interact with 5′-GUG-3′ codons, suggestive of a substantial degree of functional redundancy within these tRNA families ((Lowe and Eddy 1997); http://gtrnadb.ucsc.edu/Athal/). In that respect it is quite surprising that a mutation in a single tRNA locus is sufficient to cause such dramatic effects on plant development. Perhaps even more surprising is the observation that defects in auxin-controlled growth responses appear to represent the major consequence of this mutation. Unfortunately, we still lack detailed information on tRNA expression control in plants (Lin et al. 1992; Stange et al. 1992; Carneiro et al. 1994; Ulmasov and Folk 1995). So, it remains a mystery as to how a single tRNA anticodon mutation could produce highly defined growth defects, as those observed by Perry and colleagues (Perry et al. 2005).
Outlook
In recent years, the plant community experienced an almost inflationary increase of data, linking auxin to essentially every aspect of plant growth and development. By pursuing a range of experimental approaches, substantial progress has been made in the molecular characterization of auxin signaling cascades, metabolism as well as transport processes. However, in-depth analysis and integration of all these findings into coherent networks lags behind. This is particularly true for some key components of the auxin network like the PIN proteins and ABP1, as their actual function is still not entirely resolved. Also, new approaches are needed to elucidate crosstalk between auxin signals and general cellular activities like cytoskeleton organization or translational control. In the light of these far-reaching auxin effects, a systematic characterization of the underlying mechanisms and targets involved should be given highest priority. This will greatly improve our understanding of plant growth in general, and will also provide a solid foundation for further studies aiming at an integration of auxin responses into a multidimensional network of cellular activities.
Acknowledgments
The authors are indebted to Jürgen Kleine-Vehn (BOKU, Vienna) for valuable comments on the manuscript. Work in the lab of C.L. is supported by the Austrian Science Fund FWF (P25931), K.R. is supported by a Doc fforte fellowship from the Austrian Academy of Sciences, B.K. holds a Hertha Firnberg fellowship from the FWF (T477), and H.B. was supported by a fellowship from the Pakistani Commission for Higher Education (HEC).
Abbreviations
- ABC
Atp-binding cassette
- ABCB
ATP-BINDING CASSETTE SUBFAMILY B-TYPE
- ABP1
AUXIN BINDING PROTEIN 1
- ADA2
TRANSCRIPTIONAL ADAPTOR 2
- AFB
AUXIN SIGNALING F-BOX
- ARF
ADP-ribosylation factor
- ARF
AUXIN RESPONSE FACTOR
- ATI
Auxin transport inhibitor
- ATP
Adenosine triphosphate
- Aux/IAA
AUXIN/INDOLE ACETIC ACID
- AUX1
AUXIN 1
- BFA
Brefeldin A
- CCP
Clathrin-coated pits
- CCV
Clathrin-coated vesicles
- CHC
Clathrin heavy chain
- CHD
Chromodomain helicase DNA
- CLASP
CYTOPLASMIC LINKER-ASSOCIATED PROTEIN
- CLC
Clathrin light chain
- eIF3h
Eukaryotic translation initiation factor 3 subunit H
- ER
Endoplasmic reticulum
- ETT
ETTIN
- GCN5
GENERAL CONTROL NONDEPRESSIBLE 5
- GEF
Guanine-nucleotide exchange factor
- HAT
Histone acetyltransferase
- HDAC
Histone deacetylase
- IAA
Indole-3-acetic acid
- ICR1
INTERACTOR OF CONSTITUTIVE ROP 1
- LAX
LIKE AUXIN 1
- MAP
Microtubule-associated protein
- MP
MONOPTEROS
- MT
Microtubule
- MVB
Multivesicular bodies
- ORF
Open reading frame
- PARL1
PARALLEL 1
- PAT
Polar auxin transport
- PBA
2-(1-pyrenoyl) benzoic acid
- PILS
PIN-LIKES
- PIN
PIN-FORMED
- PKL
PICKLE
- PP6
PROTEIN PHOSPHATASE 6
- PRZ1
PROPORZ 1
- PVC
Prevacuolar compartment
- RIC
ROP-INTERACTIVE CRIB MOTIF-CONTAINING PROTEIN
- ROP
Rho-type
- RPL
Ribosomal protein of the large subunit
- SCF
SKP, CULLIN, F-BOX
- SEC3
SECRETORY 3
- SKP2B
S-PHASE KINASE-ASSOCIATED PROTEIN 2B
- SLR
SOLITARY-ROOT
- SNX1
SORTING NEXIN 1
- SPK1
SPIKE 1
- STV1
SHORT VALVE 1
- TGN/EE
Trans golgi network/early endosomes
- TIBA
2,3,5-triiodobenzoic acid
- TIR1
TRANSPORT INHIBITOR RESPONSE 1
- TPL
TOPLESS
- TPR
TPL-RELATED
- uORF
Upstream ORF
References
- Abas L, Benjamins R, Malenica N, Paciorek T, Wisniewska J, Moulinier-Anzola JC, Sieberer T, Friml J, Luschnig C. Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat Cell Biol. 2006;8(3):249–256. doi: 10.1038/ncb1369. [DOI] [PubMed] [Google Scholar]
- Ambrose C, Ruan Y, Gardiner J, Tamblyn LM, Catching A, Kirik V, Marc J, Overall R, Wasteneys GO. CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev Cell. 2013;24(6):649–659. doi: 10.1016/j.devcel.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Anzola JM, Sieberer T, Ortbauer M, Butt H, Korbei B, Weinhofer I, Mullner AE, Luschnig C. Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc Natl Acad Sci U S A. 2010;107(22):10308–10313. doi: 10.1073/pnas.0913918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge K, Guyomarc'h S, Bayer E, Swarup R, Bennett M, Mandel T, Kuhlemeier C. Auxin influx carriers stabilize phyllotactic patterning. Genes Dev. 2008;22(6):810–823. doi: 10.1101/gad.462608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros I, Dominguez T, Sauer M, Paredes P, Duprat A, Rojo E, Sanmartin M, Sanchez-Serrano JJ. Specialized functions of the PP2A subfamily II catalytic subunits PP2A-C3 and PP2A-C4 in the distribution of auxin fluxes and development in Arabidopsis. Plant J. 2013;73(5):862–872. doi: 10.1111/tpj.12078. [DOI] [PubMed] [Google Scholar]
- Band LR, Wells DM, Larrieu A, Sun JY, Middleton AM, French AP, Brunoud G, Sato EM, Wilson MH, Peret B, Oliva M, Swarup R, Sairanen I, Parry G, Ljung K, Beeckman T, Garibaldi JM, Estelle M, Owen MR, Vissenberg K, Hodgman TC, Pridmore TP, King JR, Vernoux T, Bennett MJ. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc Natl Acad Sci U S A. 2012;109(12):4668–4673. doi: 10.1073/pnas.1201498109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Blakeslee JJ, Lee OR, Mravec J, Sauer M, Titapiwatanakun B, Makam SN, Bouchard R, Geisler M, Martinoia E, Friml J, Peer WA, Murphy AS. Interactions of PIN and PGP auxin transport mechanisms. Biochem Soc Trans. 2007;35(Pt 1):137–141. doi: 10.1042/BST0350137. [DOI] [PubMed] [Google Scholar]
- Barbez E, Kleine-Vehn J. Divide Et Impera–cellular auxin compartmentalization. Curr Opin Plant Biol. 2013;16(1):78–84. doi: 10.1016/j.pbi.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Barbez E, Kubes M, Rolcik J, Beziat C, Pencik A, Wang B, Rosquete MR, Zhu J, Dobrev PI, Lee Y, Zazimalova E, Petrasek J, Geisler M, Friml J, Kleine-Vehn J. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature. 2012;485(7396):119–122. doi: 10.1038/nature11001. [DOI] [PubMed] [Google Scholar]
- Baster P, Robert S, Kleine-Vehn J, Vanneste S, Kania U, Grunewald W, De Rybel B, Beeckman T, Friml J. SCF(TIR1/AFB)-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J. 2013;32(2):260–274. doi: 10.1038/emboj.2012.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Maher EP. Mutants of Arabidopsis thaliana with abnormal gravitropic responses. Mol Gen Genet. 1990;220(2):289–293. doi: 10.1007/BF00260496. [DOI] [Google Scholar]
- Bender RL, Fekete ML, Klinkenberg PM, Hampton M, Bauer B, Malecha M, Lindgren K, Maki J, Perera MA, Nikolau BJ, Carter CJ. PIN6 is required for nectary auxin response and short stamen development. Plant J. 2013;74(6):893–904. doi: 10.1111/tpj.12184. [DOI] [PubMed] [Google Scholar]
- Benjamins R, Scheres B. Auxin: the looping star in plant development. Annu Rev Plant Biol. 2008;59:443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115(5):591–602. doi: 10.1016/S0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science. 1996;273(5277):948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, Geisler M, Nagashima A, Sakai T, Martinoia E, Friml J, Peer WA, Murphy AS. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell. 2007;19(1):131–147. doi: 10.1105/tpc.106.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433(7021):39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Bouchard R, Bailly A, Blakeslee JJ, Oehring SC, Vincenzetti V, Lee OR, Paponov I, Palme K, Mancuso S, Murphy AS, Schulz B, Geisler M. Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J Biol Chem. 2006;281(41):30603–30612. doi: 10.1074/jbc.M604604200. [DOI] [PubMed] [Google Scholar]
- Boysen Jensen P. Über die Leitung des phototropischen Reizes in Avenakeimpflanzen. Ber Deut Bot Ges. 1910;28:118–120. [Google Scholar]
- Brandizzi F, Wasteneys GO (2013) Cytoskeleton-dependent endomembrane organization in plant cells: an emerging role for microtubules. Plant J 75(2):339–349 [DOI] [PubMed]
- Bringmann M, Landrein B, Schudoma C, Hamant O, Hauser MT, Persson S. Cracking the elusive alignment hypothesis: the microtubule-cellulose synthase nexus unraveled. Trends Plant Sci. 2012;17(11):666–674. doi: 10.1016/j.tplants.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jia T, Lam SK, Ding Y, Gao C, San MW, Pimpl P, Jiang L. Multiple cytosolic and transmembrane determinants are required for the trafficking of SCAMP1 via an ER-Golgi-TGN-PM pathway. Plant J. 2011;65(6):882–896. doi: 10.1111/j.1365-313X.2010.04469.x. [DOI] [PubMed] [Google Scholar]
- Calderon Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, Zheng N, Napier R, Kepinski S, Estelle M. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol. 2012;8(5):477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro VT, Dietrich A, Marechal-Drouard L, Cosset A, Pelletier G, Small I. Characterization of some major identity elements in plant alanine and phenylalanine transfer RNAs. Plant Mol Biol. 1994;26(6):1843–1853. doi: 10.1007/BF00019497. [DOI] [PubMed] [Google Scholar]
- Carroll AJ, Heazlewood JL, Ito J, Millar AH. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol Cell Proteomics. 2008;7(2):347–369. doi: 10.1074/mcp.M700052-MCP200. [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Vanstraelen M, Simon S, Yin K, Carron-Arthur A, Nisar N, Tarle G, Cuttriss AJ, Searle IR, Benkova E, Mathesius U, Masle J, Friml J, Pogson BJ. Role of the Arabidopsis PIN6 auxin transporter in auxin homeostasis and auxin-mediated development. PLoS One. 2013;8(7):e70069. doi: 10.1371/journal.pone.0070069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH. The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Natl Acad Sci U S A. 1998;95(25):15112–15117. doi: 10.1073/pnas.95.25.15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Naramoto S, Robert S, Tejos R, Lofke C, Lin D, Yang Z, Friml J. ABP1 and ROP6 GTPase signaling regulate clathrin-mediated endocytosis in Arabidopsis roots. Curr Biol. 2012;22(14):1326–1332. doi: 10.1016/j.cub.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Cho M, Lee SH, Cho HT. P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell. 2007;19(12):3930–3943. doi: 10.1105/tpc.107.054288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholodny N. Wuchshormone und Tropismem bei den Pflanzen. Biol Zent. 1927;47:604–626. [Google Scholar]
- Christie JM, Yang H, Richter GL, Sullivan S, Thomson CE, Lin J, Titapiwatanakun B, Ennis M, Kaiserli E, Lee OR, Adamec J, Peer WA, Murphy AS (2011) phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol 9 (6):e1001076. [DOI] [PMC free article] [PubMed]
- Craddock C, Lavagi I, Yang Z. New insights into Rho signaling from plant ROP/Rac GTPases. Trends Cell Biol. 2012;22(9):492–501. doi: 10.1016/j.tcb.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JW, Briggs WR. Properties of a solubilized microsomal auxin-binding protein from coleoptiles and primary leaves of Zea mays. Plant Physiol. 1978;62(1):152–157. doi: 10.1104/pp.62.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Hofte H, Vernhettes S. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell. 2009;21(4):1141–1154. doi: 10.1105/tpc.108.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Zhang C, Kania U, Chen F, Xue Q, McCray T, Li G, Qin G, Wakeley M, Terzaghi W, Wan J, Zhao Y, Xu J, Friml J, Deng XW, Wang H. A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell. 2012;24(6):2497–2514. doi: 10.1105/tpc.112.098905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Bosco C, Dovzhenko A, Liu X, Woerner N, Rensch T, Eismann M, Eimer S, Hegermann J, Paponov IA, Ruperti B, Heberle-Bors E, Touraev A, Cohen JD, Palme K (2012) The endoplasmic reticulum localized PIN8 is a pollen-specific auxin carrier involved in intracellular auxin homeostasis. Plant J 71(5):860–870 [DOI] [PubMed]
- Darwin C, Darwin FE. The power of movement in plants. New York: Appleton; 1881. [Google Scholar]
- Degenhardt RF, Bonham-Smith PC. Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol. 2008;147(1):128–142. doi: 10.1104/pp.107.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Friml J. Cell polarity in plants: when two do the same, it is not the same. Curr Opin Cell Biol. 2011;23(6):686–696. doi: 10.1016/j.ceb.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Jones AM, Estelle M. Auxin action in a cell-free system. Curr Biol. 2003;13(16):1418–1422. doi: 10.1016/S0960-9822(03)00536-0. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9(1):109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol. 2007;17(6):520–527. doi: 10.1016/j.cub.2007.01.052. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Grigoriev I, Fischer R, Tominaga M, Robinson DG, Hasek J, Paciorek T, Petrasek J, Seifertova D, Tejos R, Meisel LA, Zazimalova E, Gadella TW, Jr, Stierhof YD, Ueda T, Oiwa K, Akhmanova A, Brock R, Spang A, Friml J. Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proc Natl Acad Sci U S A. 2008;105(11):4489–4494. doi: 10.1073/pnas.0711414105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Huang F, Galvan-Ampudia CS, Mahonen AP, Kleine-Vehn J, Xu J, Quint A, Prasad K, Friml J, Scheres B, Offringa R. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development. 2010;137(19):3245–3255. doi: 10.1242/dev.052456. [DOI] [PubMed] [Google Scholar]
- Di Rubbo S, Irani NG, Kim SY, Xu ZY, Gadeyne A, Dejonghe W, Vanhoutte I, Persiau G, Eeckhout D, Simon S, Song K, Kleine-Vehn J, Friml J, De Jaeger G, Van Damme D, Hwang I, Russinova E. The clathrin adaptor complex AP-2 mediates endocytosis of BRASSINOSTEROID INSENSITIVE1 in Arabidopsis. Plant Cell. 2013;25(8):2986–2997. doi: 10.1105/tpc.113.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang J, Stierhof YD, Robinson DG, Jiang L. Unconventional protein secretion. Trends Plant Sci. 2012;17(10):606–615. doi: 10.1016/j.tplants.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Ding Z, Wang B, Moreno I, Duplakova N, Simon S, Carraro N, Reemmer J, Pencik A, Chen X, Tejos R, Skupa P, Pollmann S, Mravec J, Petrasek J, Zazimalova E, Honys D, Rolcik J, Murphy A, Orellana A, Geisler M, Friml J. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat Commun. 2012;3:941. doi: 10.1038/ncomms1941. [DOI] [PubMed] [Google Scholar]
- Drdova EJ, Synek L, Pecenkova T, Hala M, Kulich I, Fowler JE, Murphy AS, Zarsky V (2013) The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. Plant J 73(5):709–719 [DOI] [PubMed]
- Dresios J, Derkatch IL, Liebman SW, Synetos D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000;39(24):7236–7244. doi: 10.1021/bi9925266. [DOI] [PubMed] [Google Scholar]
- Fan L, Hao H, Xue Y, Zhang L, Song K, Ding Z, Botella MA, Wang H, Lin J. Dynamic analysis of Arabidopsis AP2 sigma subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development. 2013;140(18):3826–3837. doi: 10.1242/dev.095711. [DOI] [PubMed] [Google Scholar]
- Feraru E, Vosolsobe S, Feraru MI, Petrasek J, Kleine-Vehn J. Evolution and structural diversification of PILS putative auxin carriers in plants. Front Plant Sci. 2012;3:227. doi: 10.3389/fpls.2012.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2002;415(6873):806–809. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426(6963):147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, Hooykaas PJ, Palme K, Offringa R. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306(5697):862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z. Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell. 2005;120(5):687–700. doi: 10.1016/j.cell.2004.12.026. [DOI] [PubMed] [Google Scholar]
- Fu Y, Xu T, Zhu L, Wen M, Yang Z. A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr Biol. 2009;19(21):1827–1832. doi: 10.1016/j.cub.2009.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29(2):153–168 [DOI] [PubMed]
- Fukaki H, Taniguchi N, Tasaka M (2006) PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J 48(3):380–389 [DOI] [PubMed]
- Gaedeke N, Klein M, Kolukisaoglu U, Forestier C, Muller A, Ansorge M, Becker D, Mamnun Y, Kuchler K, Schulz B, Mueller-Roeber B, Martinoia E. The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J. 2001;20(8):1875–1887. doi: 10.1093/emboj/20.8.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282(5397):2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Geisler M, Murphy AS. The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett. 2006;580(4):1094–1102. doi: 10.1016/j.febslet.2005.11.054. [DOI] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, Ejendal KF, Smith AP, Baroux C, Grossniklaus U, Muller A, Hrycyna CA, Dudler R, Murphy AS, Martinoia E. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J : Cell Mol Biol. 2005;44(2):179–194. doi: 10.1111/j.1365-313X.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- Geldner N. Cell polarity in plants: a PARspective on PINs. Curr Opin Plant Biol. 2009;12(1):42–48. doi: 10.1016/j.pbi.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jurgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413(6854):425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jurgens G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112(2):219–230. doi: 10.1016/S0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Wilson D, Kreitler T, Lehrach H, Klose J, Gobom J, Fucini P. High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol Biol. 2005;57(4):577–591. doi: 10.1007/s11103-005-0699-3. [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999;13(13):1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414(6861):271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Grunewald W, Friml J. The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 2010;29(16):2700–2714. doi: 10.1038/emboj.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Vernoud V, Fu Y, Yang Z. ROP GTPase regulation of pollen tube growth through the dynamics of tip-localized F-actin. J Exp Bot. 2003;54(380):93–101. doi: 10.1093/jxb/erg035. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17(5):1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazak O, Bloch D, Poraty L, Sternberg H, Zhang J, Friml J, Yalovsky S. A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol. 2010;8(1):e1000282. doi: 10.1371/journal.pbio.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jonsson H, Traas J, Meyerowitz EM. Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol. 2010;8(10):e1000516. doi: 10.1371/journal.pbio.1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel R, Russo VEA, Thomson KS (1972) In-vitro auxin binding to particulate cell fractions from corn coleoptiles. Planta 107 (4):325-&. [DOI] [PubMed]
- Huang F, Zago MK, Abas L, van Marion A, Galvan-Ampudia CS, Offringa R. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell. 2010;22(4):1129–1142. doi: 10.1105/tpc.109.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Shimomura S, Fukui T, Futai M. Auxin-binding protein located in the endoplasmic reticulum of maize shoots: molecular cloning and complete primary structure. Proc Natl Acad Sci U S A. 1989;86(10):3564–3568. doi: 10.1073/pnas.86.10.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Kim GT, Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000;22(3):257–264. doi: 10.1046/j.1365-313x.2000.00728.x. [DOI] [PubMed] [Google Scholar]
- Ito E, Fujimoto M, Ebine K, Uemura T, Ueda T, Nakano A (2012) Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J 69(2):204–216 [DOI] [PubMed]
- Jackson PK, Eldridge AG. The SCF ubiquitin ligase: an extended look. Mol Cell. 2002;9(5):923–925. doi: 10.1016/S1097-2765(02)00538-5. [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miege C, Rollin C, Gaude T. AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature. 2006;443(7107):106–109. doi: 10.1038/nature05046. [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miege C, Gaude T (2008) Evidence for a sorting endosome in Arabidopsis root cells. Plant J 53(2):237–247 [DOI] [PubMed]
- Jones AM, Herman EM. KDEL-containing auxin-binding protein is secreted to the plasma membrane and cell wall. Plant Physiol. 1993;101(2):595–606. doi: 10.1104/pp.101.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Venis MA. Photoaffinity labeling of indole-3-acetic acid-binding proteins in maize. Proc Natl Acad Sci U S A. 1989;86(16):6153–6156. doi: 10.1073/pnas.86.16.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G, Geldner N. The high road and the low road: trafficking choices in plants. Cell. 2007;130(6):977–979. doi: 10.1016/j.cell.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Kakar K, Zhang H, Scheres B, Dhonukshe P. CLASP-mediated cortical microtubule organization guides PIN polarization axis. Nature. 2013;495(7442):529–533. doi: 10.1038/nature11980. [DOI] [PubMed] [Google Scholar]
- Kamimoto Y, Terasaka K, Hamamoto M, Takanashi K, Fukuda S, Shitan N, Sugiyama A, Suzuki H, Shibata D, Wang B, Pollmann S, Geisler M, Yazaki K. Arabidopsis ABCB21 is a facultative auxin importer/exporter regulated by cytoplasmic auxin concentration. Plant Cell Physiol. 2012;53(12):2090–2100. doi: 10.1093/pcp/pcs149. [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. Protein–protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci U S A. 1997;94(22):11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Cai X, Vaughn JN, von Arnim AG. On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 2007;8(4):R60. doi: 10.1186/gb-2007-8-4-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Xu ZY, Song K, Kim DH, Kang H, Reichardt I, Sohn EJ, Friml J, Juergens G, Hwang I. Adaptor protein complex 2-mediated endocytosis is crucial for male reproductive organ development in Arabidopsis. Plant Cell. 2013;25(8):2970–2985. doi: 10.1105/tpc.113.114264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC. Protein organization in clathrin trimers. Cell. 1981;23(3):755–761. doi: 10.1016/0092-8674(81)90439-6. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC, Heuser J. Configuration of clathrin trimers: evidence from electron microscopy. J Ultrastruct Mol Struct Res. 1986;94(3):199–208. doi: 10.1016/0889-1605(86)90067-4. [DOI] [PubMed] [Google Scholar]
- Kitakura S, Vanneste S, Robert S, Lofke C, Teichmann T, Tanaka H, Friml J. Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell. 2011;23(5):1920–1931. doi: 10.1105/tpc.111.083030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Dhonukshe P, Sauer M, Brewer PB, Wisniewska J, Paciorek T, Benkova E, Friml J. ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr Biol. 2008;18(7):526–531. doi: 10.1016/j.cub.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Leitner J, Zwiewka M, Sauer M, Abas L, Luschnig C, Friml J. Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc Natl Acad Sci U S A. 2008;105(46):17812–17817. doi: 10.1073/pnas.0808073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Ding Z, Jones AR, Tasaka M, Morita MT, Friml J. Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc Natl Acad Sci U S A. 2010;107(51):22344–22349. doi: 10.1073/pnas.1013145107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kögl F, Haagen-Smits AJ. Über die Chemie des Wuchsstoffs. Proc K Ned Akad Wet. 1931;34:1411–1416. [Google Scholar]
- Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131(3):557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbei B, Luschnig C. Plasma membrane protein ubiquitylation and degradation as determinants of positional growth in plants. J Integr Plant Biol. 2013;55(9):809–823. doi: 10.1111/jipb.12059. [DOI] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234(2):187–208. doi: 10.1016/S0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Kramer EM. PIN and AUX/LAX proteins: their role in auxin accumulation. Trends Plant Sci. 2004;9(12):578–582. doi: 10.1016/j.tplants.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Krecek P, Skupa P, Libus J, Naramoto S, Tejos R, Friml J, Zazimalova E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009;10(12):249. doi: 10.1186/gb-2009-10-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S, Slane D, Herud O, Kong J, Jurgens G. Early embryogenesis in flowering plants: setting up the basic body pattern. Annu Rev Plant Biol. 2012;63:483–506. doi: 10.1146/annurev-arplant-042811-105507. [DOI] [PubMed] [Google Scholar]
- Lavy M, Bloch D, Hazak O, Gutman I, Poraty L, Sorek N, Sternberg H, Yalovsky S. A novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr Biol. 2007;17(11):947–952. doi: 10.1016/j.cub.2007.04.038. [DOI] [PubMed] [Google Scholar]
- Laxmi A, Pan J, Morsy M, Chen R. Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS One. 2008;3(1):e1510. doi: 10.1371/journal.pone.0001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner E, Achard P, Vansiri A, Potuschak T, Genschik P. F-box proteins everywhere. Curr Opin Plant Biol. 2006;9(6):631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Leitner J, Petrasek J, Tomanov K, Retzer K, Parezova M, Korbei B, Bachmair A, Zazimalova E, Luschnig C. Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc Natl Acad Sci U S A. 2012;109(21):8322–8327. doi: 10.1073/pnas.1200824109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. Auxin, self-organisation, and the colonial nature of plants. Curr Biol. 2011;21(9):R331–R337. doi: 10.1016/j.cub.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Lin TY, March R, Scanlon SR, Folk WR. Isolation and transcriptional competence of three tRNA(Trp) genes from Arabidopsis thaliana L. Plant Mol Biol. 1992;18(1):159–160. doi: 10.1007/BF00018472. [DOI] [PubMed] [Google Scholar]
- Lin D, Nagawa S, Chen J, Cao L, Chen X, Xu T, Li H, Dhonukshe P, Yamamuro C, Friml J, Scheres B, Fu Y, Yang Z. A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr Biol. 2012;22(14):1319–1325. doi: 10.1016/j.cub.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49(3–4):387–400. doi: 10.1023/A:1015255030047. [DOI] [PubMed] [Google Scholar]
- Ljung K. Auxin metabolism and homeostasis during plant development. Development. 2013;140(5):943–950. doi: 10.1242/dev.086363. [DOI] [PubMed] [Google Scholar]
- Löbler M, Klämbt D. Auxin-binding protein from coleoptile membranes of corn (Zea mays L.). I. Purification by immunological methods and characterization. J Biol Chem. 1985;260(17):9848–9853. [PubMed] [Google Scholar]
- Löbler M, Klämbt D. Auxin-binding protein from coleoptile membranes of corn (Zea mays L.). II. Localization of a putative auxin receptor. J Biol Chem. 1985;260(17):9854–9859. [PubMed] [Google Scholar]
- Löfke C, Luschnig C, Kleine-Vehn J. Posttranslational modification and trafficking of PIN auxin efflux carriers. Mech Dev. 2013;130(1):82–94. doi: 10.1016/j.mod.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312(5779):1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C. Auxin transport: ABC proteins join the club. Trends Plant Sci. 2002;7(8):329–332. doi: 10.1016/S1360-1385(02)02292-6. [DOI] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 1998;12(14):2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano C, Ramirez-Parra E, Casimiro I, Otero S, Desvoyes B, De Rybel B, Beeckman T, Casero P, Gutierrez C, Del Pozo CJ. Auxin and epigenetic regulation of SKP2B, an F-box that represses lateral root formation. Plant Physiol. 2012;160(2):749–762. doi: 10.1104/pp.112.198341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Pavangadkar KA, Thomashow MF, Triezenberg SJ. Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim Biophys Acta. 2006;1759(1–2):69–79. doi: 10.1016/j.bbaexp.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Maraschin F, Memelink J, Offringa R (2009) Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 59(1):100–109 [DOI] [PubMed]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999;18(8):2066–2073. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavy P, Bielach A, Abas L, Abuzeineh A, Duclercq J, Tanaka H, Parezova M, Petrasek J, Friml J, Kleine-Vehn J, Benkova E. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell. 2011;21(4):796–804. doi: 10.1016/j.devcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Marti L, Fornaciari S, Renna L, Stefano G, Brandizzi F. COPII-mediated traffic in plants. Trends Plant Sci. 2010;15(9):522–528. doi: 10.1016/j.tplants.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Miao Y, Li KY, Li HY, Yao X, Jiang L (2008) The vacuolar transport of aleurain-GFP and 2S albumin-GFP fusions is mediated by the same pre-vacuolar compartments in tobacco BY-2 and Arabidopsis suspension cultured cells. Plant J 56(5):824–839 [DOI] [PubMed]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, Schwab R, Weigel D, Meyerowitz EM, Luschnig C, Offringa R, Friml J. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130(6):1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- Molendijk AJ, Bischoff F, Rajendrakumar CS, Friml J, Braun M, Gilroy S, Palme K. Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 2001;20(11):2779–2788. doi: 10.1093/emboj/20.11.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Kubes M, Bielach A, Gaykova V, Petrasek J, Skupa P, Chand S, Benkova E, Zazimalova E, Friml J. Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development. 2008;135(20):3345–3354. doi: 10.1242/dev.021071. [DOI] [PubMed] [Google Scholar]
- Mravec J, Skupa P, Bailly A, Hoyerova K, Krecek P, Bielach A, Petrasek J, Zhang J, Gaykova V, Stierhof YD, Dobrev PI, Schwarzerova K, Rolcik J, Seifertova D, Luschnig C, Benkova E, Zazimalova E, Geisler M, Friml J. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459(7250):1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tanzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998;17(23):6903–6911. doi: 10.1093/emboj/17.23.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa S, Xu T, Lin D, Dhonukshe P, Zhang X, Friml J, Scheres B, Fu Y, Yang Z. ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol. 2012;10(4):e1001299. doi: 10.1371/journal.pbio.1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell. 2005;17(11):2940–2953. doi: 10.1105/tpc.105.036533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell. 2001;13(11):2441–2454. doi: 10.1105/tpc.13.11.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the auxin polar transport-system in early stages of Arabidopsis floral bud formation. Plant Cell. 1991;3(7):677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17(2):444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, Smith A, Yu G, Theologis A. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell. 2005;17(12):3282–3300. doi: 10.1105/tpc.105.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazimalova E, Ruthardt N, Petrasek J, Stierhof YD, Kleine-Vehn J, Morris DA, Emans N, Jurgens G, Geldner N, Friml J. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature. 2005;435(7046):1251–1256. doi: 10.1038/nature03633. [DOI] [PubMed] [Google Scholar]
- Paponov IA, Teale WD, Trebar M, Blilou I, Palme K. The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci. 2005;10(4):170–177. doi: 10.1016/j.tplants.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell. 2001;106(6):723–733. doi: 10.1016/S0092-8674(01)00487-1. [DOI] [PubMed] [Google Scholar]
- Peer WA, Blakeslee JJ, Yang H, Murphy AS. Seven things we think we know about auxin transport. Mol Plant. 2011;4(3):487–504. doi: 10.1093/mp/ssr034. [DOI] [PubMed] [Google Scholar]
- Perry J, Dai X, Zhao Y. A mutation in the anticodon of a single tRNAala is sufficient to confer auxin resistance in Arabidopsis. Plant Physiol. 2005;139(3):1284–1290. doi: 10.1104/pp.105.068700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, Dhonukshe P, Skupa P, Benkova E, Perry L, Krecek P, Lee OR, Fink GR, Geisler M, Murphy AS, Luschnig C, Zazimalova E, Friml J. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312(5775):914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Nelson TM. Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 2007;144(1):173–186. doi: 10.1104/pp.106.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L, Vilela C, Berthelot K, Ramirez CV, McCarthy JE. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J Mol Biol. 2004;335(1):71–85. doi: 10.1016/j.jmb.2003.10.049. [DOI] [PubMed] [Google Scholar]
- Raven JA. Transport of indoleacetic-acid in plant–cells in relation to ph and electrical potential gradients, and its significance for polar Iaa transport. New Phytol. 1975;74(2):163–172. doi: 10.1111/j.1469-8137.1975.tb02602.x. [DOI] [Google Scholar]
- Ray PM. Auxin-binding sites of maize coleoptiles are localized on membranes of the endoplasmic reticulum. Plant Physiol. 1977;59(4):594–599. doi: 10.1104/pp.59.4.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea PA. Plant ATP-binding cassette transporters. Annu Rev Plant Biol. 2007;58:347–375. doi: 10.1146/annurev.arplant.57.032905.105406. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426(6964):255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- Reyes FC, Buono R, Otegui MS. Plant endosomal trafficking pathways. Curr Opin Plant Biol. 2011;14(6):666–673. doi: 10.1016/j.pbi.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Richter S, Voss U, Jurgens G. Post-Golgi traffic in plants. Traffic. 2009;10(7):819–828. doi: 10.1111/j.1600-0854.2009.00916.x. [DOI] [PubMed] [Google Scholar]
- Richter S, Anders N, Wolters H, Beckmann H, Thomann A, Heinrich R, Schrader J, Singh MK, Geldner N, Mayer U, Jurgens G. Role of the GNOM gene in Arabidopsis apical-basal patterning—from mutant phenotype to cellular mechanism of protein action. Eur J Cell Biol. 2010;89(2–3):138–144. doi: 10.1016/j.ejcb.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Covanova M, Hayashi K, Dhonukshe P, Yang Z, Bednarek SY, Jones AM, Luschnig C, Aniento F, Zazimalova E, Friml J. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell. 2010;143(1):111–121. doi: 10.1016/j.cell.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Pimpl P, Scheuring D, Stierhof YD, Sturm S, Viotti C. Trying to make sense of retromer. Trends Plant Sci. 2012;17(7):431–439. doi: 10.1016/j.tplants.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Rosado A, Sohn EJ, Drakakaki G, Pan S, Swidergal A, Xiong Y, Kang BH, Bressan RA, Raikhel NV. Auxin-mediated ribosomal biogenesis regulates vacuolar trafficking in Arabidopsis. Plant Cell. 2010;22(1):143–158. doi: 10.1105/tpc.109.068320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A, Li R, van de Ven W, Hsu E, Raikhel NV. Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc Natl Acad Sci U S A. 2012;109(48):19537–19544. doi: 10.1073/pnas.1214774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, von Arnim AG. Translational regulation of cytoplasmic mRNAs. Arabidopsis Book. 2013;11:e0165. doi: 10.1199/tab.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle SJ. The cellular functions of clathrin. Cell Mol Life Sci. 2006;63(16):1823–1832. doi: 10.1007/s00018-005-5587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubery PH, Sheldrake A. Carrier-mediated auxin transport. Planta. 1974;118(2):101–121. doi: 10.1007/BF00388387. [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 1998;12(2):198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R. Plant stem cell niches: from signalling to execution. Curr Opin Plant Biol. 2011;14(1):4–9. doi: 10.1016/j.pbi.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Santner AA, Watson JC. The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. Plant J. 2006;45(5):752–764. doi: 10.1111/j.1365-313X.2005.02641.x. [DOI] [PubMed] [Google Scholar]
- Sauer M, Robert S, Kleine-Vehn J. Auxin: simply complicated. J Exp Bot. 2013;64(9):2565–2577. doi: 10.1093/jxb/ert139. [DOI] [PubMed] [Google Scholar]
- Sawchuk MG, Edgar A, Scarpella E. Patterning of leaf vein networks by convergent auxin transport pathways. PLoS Genet. 2013;9(2):e1003294. doi: 10.1371/journal.pgen.1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S, Pimpl P. Coats of endosomal protein sorting: retromer and ESCRT. Curr Opin Plant Biol. 2009;12(6):670–676. doi: 10.1016/j.pbi.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124(22):4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- Shi JH, Yang ZB. Is ABP1 an auxin receptor yet? Mol Plant. 2011;4(4):635–640. doi: 10.1093/mp/ssr050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura S, Sotobayashi T, Futai M, Fukui T. Purification and properties of an auxin-binding protein from maize shoot membranes. J Biochem. 1986;99(5):1513–1524. doi: 10.1093/oxfordjournals.jbchem.a135621. [DOI] [PubMed] [Google Scholar]
- Sieberer T, Seifert GJ, Hauser MT, Grisafi P, Fink GR, Luschnig C. Post-transcriptional control of the Arabidopsis auxin efflux carrier EIR1 requires AXR1. Curr Biol. 2000;10(24):1595–1598. doi: 10.1016/S0960-9822(00)00861-7. [DOI] [PubMed] [Google Scholar]
- Sieberer T, Hauser MT, Seifert GJ, Luschnig C. PROPORZ1, a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr Biol. 2003;13(10):837–842. doi: 10.1016/S0960-9822(03)00327-0. [DOI] [PubMed] [Google Scholar]
- Smith ZR, Long JA. Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature. 2010;464(7287):423–426. doi: 10.1038/nature08843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galvan-Ampudia CS, Offringa R, Friml J, Yanofsky MF, Ostergaard L. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature. 2009;459(7246):583–586. doi: 10.1038/nature07875. [DOI] [PubMed] [Google Scholar]
- Spalding EP. Diverting the downhill flow of auxin to steer growth during tropisms. Am J Bot. 2013;100(1):203–214. doi: 10.3732/ajb.1200420. [DOI] [PubMed] [Google Scholar]
- Stange N, Beier D, Beier H. Intron excision from tRNA precursors by plant splicing endonuclease requires unique features of the mature tRNA domain. Eur J Biochem. 1992;210(1):193–203. doi: 10.1111/j.1432-1033.1992.tb17408.x. [DOI] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Galweiler L, Palme K, Jurgens G. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286(5438):316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Sukumar P, Maloney GS, Muday GK. Localized induction of the ATP-binding cassette B19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol. 2013;162(3):1392–1405. doi: 10.1104/pp.113.217174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol. 2005;7(11):1057–1065. doi: 10.1038/ncb1316. [DOI] [PubMed] [Google Scholar]
- Szakonyi D, Byrne ME. Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J. 2011;65(2):269–281. doi: 10.1111/j.1365-313X.2010.04422.x. [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319(5868):1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446(7136):640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Terasaka K, Blakeslee JJ, Titapiwatanakun B, Peer WA, Bandyopadhyay A, Makam SN, Lee OR, Richards EL, Murphy AS, Sato F, Yazaki K. PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell. 2005;17(11):2922–2939. doi: 10.1105/tpc.105.035816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A, Huynh TV, Davis RW. Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J Mol Biol. 1985;183(1):53–68. doi: 10.1016/0022-2836(85)90280-3. [DOI] [PubMed] [Google Scholar]
- Titapiwatanakun B, Murphy AS. Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J Exp Bot. 2009;60(4):1093–1107. doi: 10.1093/jxb/ern240. [DOI] [PubMed] [Google Scholar]
- Titapiwatanakun B, Blakeslee JJ, Bandyopadhyay A, Yang H, Mravec J, Sauer M, Cheng Y, Adamec J, Nagashima A, Geisler M, Sakai T, Friml J, Peer WA, Murphy AS (2009) ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J 57(1):27–44 [DOI] [PubMed]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13(12):2809–2822. doi: 10.1105/tpc.13.12.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A, Paque S, Stierle V, Quettier AL, Muller P, Lechner E, Genschik P, Perrot-Rechenmann C. Auxin-binding protein 1 is a negative regulator of the SCF(TIR1/AFB) pathway. Nat Commun. 2013;4:2496. doi: 10.1038/ncomms3496. [DOI] [PubMed] [Google Scholar]
- Ulmasov B, Folk W. Analysis of the role of 5′ and 3′ flanking sequence elements upon in vivo expression of the plant tRNATrp genes. Plant Cell. 1995;7(10):1723–1734. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276(5320):1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9(11):1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsuno K, Shikanai T, Yamada Y, Hashimoto T. Agr, an agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant Cell Physiol. 1998;39(10):1111–1118. doi: 10.1093/oxfordjournals.pcp.a029310. [DOI] [PubMed] [Google Scholar]
- Van Lijsebettens M, Vanderhaeghen R, Deblock M, Bauw G, Villarroel R, Vanmontagu M. An S18 ribosomal-protein gene copy at the Arabidopsis Pfl locus affects plant development by its specific expression in meristems. EMBO J. 1994;13(14):3378–3388. doi: 10.1002/j.1460-2075.1994.tb06640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131(3):1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benkova E, Benjamins R, Beeckman T, Luschnig C, Friml J. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development. 2005;132(20):4521–4531. doi: 10.1242/dev.02027. [DOI] [PubMed] [Google Scholar]
- Vlachonasios KE, Thomashow MF, Triezenberg SJ. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell. 2003;15(3):626–638. doi: 10.1105/tpc.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wabnik K, Govaerts W, Friml J, Kleine-Vehn J. Feedback models for polarized auxin transport: an emerging trend. Mol BioSyst. 2011;7(8):2352–2359. doi: 10.1039/c1mb05109a. [DOI] [PubMed] [Google Scholar]
- Wang C, Yan X, Chen Q, Jiang N, Fu W, Ma B, Liu J, Li C, Bednarek SY, Pan J. Clathrin light chains regulate clathrin-mediated trafficking, auxin signaling, and development in Arabidopsis. Plant Cell. 2013;25(2):499–516. doi: 10.1105/tpc.112.108373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci U S A. 2013;110(2):761–766. doi: 10.1073/pnas.1215010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R. An Arabidopsis minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development. 2001;128(21):4289–4299. doi: 10.1242/dev.128.21.4289. [DOI] [PubMed] [Google Scholar]
- Went FW. On growth accelerating substances in the coleoptile of Avena sativa. Proc K Ned Akad Wet. 1926;30:10–19. [Google Scholar]
- Went FW, Thimann KV. Phytohormones. New York: Macmillan Company; 1937. [Google Scholar]
- Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, Blilou I, Rouquie D, Benkova E, Scheres B, Friml J. Polar PIN localization directs auxin flow in plants. Science. 2006;312(5775):883. doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell. 2010;143(1):99–110. doi: 10.1016/j.cell.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka S, Shimono Y, Shirakawa M, Fukao Y, Kawase T, Hatsugai N, Tamura K, Shimada T, Hara-Nishimura I. Identification and dynamics of Arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell. 2013;25(8):2958–2969. doi: 10.1105/tpc.113.114082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lavagi I. Spatial control of plasma membrane domains: ROP GTPase-based symmetry breaking. Curr Opin Plant Biol. 2012;15(6):601–607. doi: 10.1016/j.pbi.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Murphy AS (2009) Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J 59(1):179–191 [DOI] [PubMed]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1). Plant J 40(5):772–782 [DOI] [PubMed]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol. 2006;16(11):1123–1127. doi: 10.1016/j.cub.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Saiga S, Weijers D. Auxin regulation of embryonic root formation. Plant Cell Physiol. 2013;54(3):325–332. doi: 10.1093/pcp/pcs170. [DOI] [PubMed] [Google Scholar]
- Yu H, Moss BL, Jang SS, Prigge M, Klavins E, Nemhauser JL, Estelle M. Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol. 2013;162(1):295–303. doi: 10.1104/pp.113.215582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarsky V, Potocky M. Recycling domains in plant cell morphogenesis: small GTPase effectors, plasma membrane signalling and the exocyst. Biochem Soc Trans. 2010;38(2):723–728. doi: 10.1042/BST0380723. [DOI] [PubMed] [Google Scholar]
- Zarsky V, Cvrckova F, Potocky M, Hala M. Exocytosis and cell polarity in plants—exocyst and recycling domains. New Phytol. 2009;183(2):255–272. doi: 10.1111/j.1469-8137.2009.02880.x. [DOI] [PubMed] [Google Scholar]
- Zazimalova E, Murphy AS, Yang H, Hoyerova K, Hosek P. Auxin transporters—why so many? Cold Spring Harb Perspect Biol. 2010;2(3):a001552. doi: 10.1101/cshperspect.a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nodzynski T, Pencik A, Rolcik J, Friml J. PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc Natl Acad Sci U S A. 2010;107(2):918–922. doi: 10.1073/pnas.0909460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bishop B, Ringenberg W, Muir WM, Ogas J. The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 2012;159(1):418–432. doi: 10.1104/pp.112.194878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Immink R, Liu CM, Emons AM, Ketelaar T. The Arabidopsis exocyst subunit SEC3A is essential for embryo development and accumulates in transient puncta at the plasma membrane. New Phytol. 2013;199(1):74–88. doi: 10.1111/nph.12236. [DOI] [PubMed] [Google Scholar]
- Zhou F, Roy B, von Arnim AG. Translation reinitiation and development are compromised in similar ways by mutations in translation initiation factor eIF3h and the ribosomal protein RPL24. BMC Plant Biol. 2010;10:193. doi: 10.1186/1471-2229-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]