Abstract
Microglia, the immunocompetent cells of the CNS, are rapidly activated in response to injury and microglia migration towards and homing at damaged tissue plays a key role in CNS regeneration. Lysophosphatidic acid (LPA) is involved in signaling events evoking microglia responses through cognate G protein-coupled receptors. Here we show that human immortalized C13NJ microglia express LPA receptor subtypes LPA1, LPA2, and LPA3 on mRNA and protein level. LPA activation of C13NJ cells induced Rho and extracellular signal-regulated kinase activation and enhanced cellular ATP production. In addition, LPA induced process retraction, cell spreading, led to pronounced changes of the actin cytoskeleton and reduced cell motility, which could be reversed by inhibition of Rho activity. To get an indication about LPA-induced global alterations in protein expression patterns a 2-D DIGE/LC-ESI-MS proteomic approach was applied. On the proteome level the most prominent changes in response to LPA were observed for glycolytic enzymes and proteins regulating cell motility and/or cytoskeletal dynamics. The present findings suggest that naturally occurring LPA is a potent regulator of microglia biology. This might be of particular relevance in the pathophysiological context of neurodegenerative disorders where LPA concentrations can be significantly elevated in the CNS.
Keywords: 2-D DIGE, Cell biology, Lipids, Migration, MS, Rho
1 Introduction
Microglia are both supportive glia and immunocompetent defense cells. This hybrid character enables microglia not only to support but also to protect neurons within the CNS [1]. Microglia are activated in response to extracellular stimuli, leading to morphological transformation (increased size of cell bodies, thickening of proximal processes, decreased ramification of distal branches), proliferation, migration, phagocytosis, and the production of bioactive molecules [2]. These events are presumably the first steps that mobilize the cellular and molecular defense machinery in the CNS leading to microglia “activation”. The highly locomotive and morphologically flexible features of activated microglia [3] are supported by dynamic remodeling of the cytoskeleton and concomitant changes in adhesion molecule expression and antigen presentation during activation [4].
Lysophosphatidic acid (LPA) is a bioactive lysophospholipid that is synthesized by activated platelets, in response to growth factor stimulation or after cell injury [5]. The brain contains significant levels of LPA, which increase in response to CNS injury [6, 7]. In vitro and in vivo data demonstrate that LPA receptors play a prominent role in the CNS (reviewed in [8, 9]). The effects of LPA are mediated through activation of cognate G protein-coupled receptors that are encoded by LPA receptors (LPA1–3), which are the “classical” members of the endothelial cell differentiation gene family, and LPA4–7, belonging to the P2Y subgroup [10].
In the CNS LPA displays profound effects on brain capillary endothelial cells, neurons, and glia cells. At the level of brain endothelial cells that form the physical basis of the blood–brain barrier, LPA induces the disruption of junctional complexes [11]. In mature neurons, LPA induces an increase of cytosolic Ca2+ levels ([Ca2+]cyto), rapid growth cone collapse, and neurite retraction [12]. Mice lacking the lpa1 gene show craniofacial defects and perinatal lethality due to impaired suckling behavior [13]. LPA and subsequent LPA receptor engagement were also shown to be responsible for neuropathic pain and hyperalgesia [14, 15]. Among glia cells, astrocytes respond to LPA treatment with [Ca2+]cyto increases, DNA synthesis, and cell rounding [16, 17]. Oligodendrocytes have been shown to express LPA1 mRNA in vivo [18] and respond to LPA with a rise of [Ca2+]cyto [19].
Microglia express LPA receptors in a species-specific manner [20]. These differences are also reflected on the cellular level since in rat microglia a rise in [Ca2+]cyto in response to LPA is due to an activation of Ca2+ entry, while in mouse microglia LPA triggers Ca2+ mobilization from intracellular Ca2+ stores [20]. In murine BV-2 microglia LPA elicits membrane hyperpolarization due to an activation of Ca2+-dependent K+ currents [21]. Moreover it was suggested that Ca2+-activated K+ channels are a requirement for LPA-dependent induction of microglia migration [22]. However, it is still unclear how LPA transmits these signals, since no LPA-receptor transcripts were detected in murine BV-2 cells [22]. In contrast, quantitative real-time PCR (qPCR) profiling in primary rat microglia revealed predominant expression of LPA1 and LPA3 with the expression levels being closely coupled to the cellular activation status [23]. Taken together, these data indicate that activation of LPA-dependent signaling cascades via corresponding LPA receptor subtypes results in altered physiological properties of microglia that are central to their protective role in the CNS.
A thorough understanding of extracellular signals regulating microglia function as well as a characterization of microglia responses to these signals is important in the healthy and diseased CNS. Therefore in the current study we aimed to identify expression of the classical LPA1–3 by human microglia (using human immortalized C13NJ cells as a model system). In the first set of experiments specific cellular responses towards LPA activation were characterized. The second part of our study aimed to investigate global alterations in protein expression patterns in response to LPA by a combination of 2-D DIGE and LC-ESI-MS. A number of proteins showing differential expression in LPA-treated cells could be potent regulators of microglia biology (i.e. energy homeostasis, cytoskeletal dynamics, and migrational properties).
2 Materials and methods
2.1 Materials
Cell culture supplies were from Gibco (Invitrogen, Vienna, Austria), PAA Laboratories (Linz, Austria) and Costar (Vienna, Austria). LPA (1-oleoyl-2-hydroxy-sn-glycero-3-phosphate) was from Avanti Polar Lipids (Alabaster, AL, USA). DIGE cyanine dyes (Cy3 and Cy5), immobilized pH gradient strips (IPG strips, pH 3–10), and ECL Plus Western Blotting Detection Reagents were from Amersham Biosciences (Vienna; Austria). Urea, thiourea, acrylamide, acetic acid, CHAPS, glycerol, iodacetamide, DTT, NH4HCO3, Tris, SDS, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were from Sigma (Vienna, Austria). ACN (HPLC grade) and CaCl2 were from Merck (Vienna, Austria). Modified trypsin (sequencing grade) was from Promega (Vienna, Austria). The primary rabbit polyclonal anti-LPA1 antibody (raised against a peptide corresponding to amino acids 342–359 of human LPA1) was purchased from Cayman Chemical (Ann Arbor, MI, USA), the polyclonal rabbit anti-LPA2 antibody (raised against the N-terminus of human LPA2) was from Abgent (San Diego, CA, USA), and the polyclonal rabbit anti-LPA3 antibody (raised against a peptide corresponding to the N-terminal amino acids 1–12 of mouse LPA3) was from Cayman (Biozol, Eching, Germany). HRP-conjugated secondary goat-anti-rabbit antibody was from Sigma. Antibodies against extracellular signal-regulated kinases 1/2 (ERK1/2) and phosphorylated ERK1/2 (pERK1/2) were from Promega (Mannheim, Germany) and Cell Signaling (Frankfurt, Germany), respectively. The monoclonal anti-β-tubulin antibody was from Sigma. Rhodamine-labeled phalloidin was from Molecular Probes (Invitrogen), 4′,6-diamidino-2-phenylindol (DAPI) was from Partec (Munster, Germany), the EZ-Detect Rho activation Kit from Pierce (Rockford, IL, USA). Exoenzyme C3 Transferase from Clostridium botulinum used to selectively inactivate the GTPases RhoA, RhoB, and RhoC was from Cytoskeleton (tebu-bio, Germany). Cell transfection supplies were from Amaxa (Cologne, Germany). The LPA1/3 antagonist 3-(4-[4-([1-(2-Chlorophenyl)ethoxy] carbonylamino)-3-methyl-5-isoxazolyl]benzylthio) propanoic acid (Ki16425) was from Sigma.
2.2 Methods
2.2.1 Cell culture and LPA treatment
In this study we have used the human C13NJ microglia as model system. These immortalized cells possess the macrophage characteristics of adherence and phagocytosis and express several macrophage antigens [24, 25]. C13NJ cells were maintained in DMEM/high glucose supplemented with 10% FCS (PAA Laboratories), pyruvate (1 mL/500 mL medium) 50 μg/mL gentamycin at 371C under 5% CO2. For LPA treatment, cells were incubated in DMEM with 0.5% FCS and 0.1% BSA in the absence or presence of LPA (aqueous oleoyl-LPA stock solutions, 10 mM, stored at −70°C, and used within 2 wk) at indicated concentrations and times.
2.2.2 LPA receptor expression analysis by RT-PCR
To generate a cDNA template, total RNA was isolated using RNeasy Mini Kit (Qiagen, Hilgen, Germany) and 3 μg of DNase treated (Ambion, Brunn am Gebirge, Austria) RNA was reverse transcribed using 200 U SuperScript II Reverse Transcriptase and 500 ng Oligo(dT)12–18 according to the manufacturer’s instructions (Invitrogen). During the present study we focused on LPA1–3, which are the classical members of the endothelial differentiation gene family receptors. For primer combinations for RT-PCR detection of LPA receptor genes see Table 1.
Table 1.
Human LPA receptor primers
| Target gene | Primers pos. | Sequence | Amplicon size (bp) |
|---|---|---|---|
| LPA1, NM_001401 | Forward 968 | 5′-GTCTTCTGGGCCATTTTCAAC-3′ | 332 |
| Reverse 1319 | 3′-TGCCTAAAGGTGGCGCTCAT-5′ | ||
| LPA2, NM_004720 | Forward 257 | 5′-CCTACCTCTTCCTCATGTTC-3′ | 784 |
| Reverse 1061 | 3′-TAAAGGGTGGAGTCCATCAG-5′ | ||
| LPA3, NM_012152 | Forward 541 | 5′-GGAATTGCCTCTGCAACATC-3′ | 382 |
| Reverse 923 | 3′-GAGTAGATGATGGGGTTCA-5′ |
2.2.3 Western blot analysis of LPA receptor expression
SDS-PAGE was performed on protein extracts from C13NJ cells (50 mM Tris-HCl pH 7.4, 1% NP-40, 150 mM NaCl, 1 mM Na3VO4, 1 mM NaF, 1 mM EDTA, 10 μM PMSF, aprotinin, leupeptin, pepstatin: 1 μg/mL each). Separation was performed by linear SDS-PAGE (12%) under reducing conditions and proteins were electrophoretically transferred to PVDF membranes (150 mA). Immunochemical detection was performed using polyclonal rabbit anti-human LPA1 (1:250), anti-human LPA2 (1:500), and anti-mouse LPA3 (1:400) anti-serum as primary antibodies. Immunoreactive bands were visualized using HRP-conjugated goat anti-rabbit or rabbit anti-mouse IgG (1:5000) and subsequent ECL development. Alternatively, cell lysates were subjected to enzymatic deglycosylation with Endo HF as described in the manufacturer’s (New England Biolabs, Frankfurt, Germany) protocol prior to immunoblotting.
2.2.4 Rho and ERK1/2 activation
Prior to measurement of Rho family member activation, C13NJ cells were serum-starved for 24 h. Cells were incubated in the absence or presence of LPA for the indicated times and concentrations. Analysis of active Rho was performed using the EZ-Detect Rho Activation Kit (Pierce), which is based on affinity precipitation of GTP-bound RhoA/B/C by a mouse Rhotekin-RBD GST fusion protein. Cells were processed rapidly on ice and were snap-frozen in liquid nitrogen (according to our experience this is an essential step to avoid GTP hydrolysis) until further processing. Protein lysates were separated by SDS-PAGE (15%; 500 μg protein/lane) and transferred to PVDF membranes. Immunoreactive Rho was detected with a mouse monoclonal anti-Rho IgG (1:1000; recognizing all three Rho family members; i.e. RhoA, B, and C) as primary antibody, followed by HRP-conjugated goat anti-mouse IgG (1:2000) and ECL detection.
Immunochemical detection of ERK1/2, and pERK1/2 was performed with sequence-specific antibodies as described previously [26]. Immunoreactive bands were visualized with HRP-conjugated goat anti-rabbit or goat anti-mouse IgG (both 1:2500) and the ECL detection system. To inhibit LPA receptor engagement the LPA1/3-specific antagonist Ki16425 (10 μM) was used. Bands were quantified by densitometry of films with a Herolab EASY RH densitometer (Herolab, Wiesloch, Germany) and the EASY Win32 software.
2.2.5 Adenine nucleotide analysis
Adenine nucleotides were analyzed as reported previously [27] with some modifications: separation was performed on a Hypersil ODS column (5μm, 250 × mm id), using a L2200 autosampler, two L-2130 HTA pumps, and a L2450 diode array detector (all from VWR Hitachi). EZchrom Elite (VWR) was used for data acquisition and analysis. Cellular proteins of C13NJ cells were precipitated with 250μL of perchloric acid (0.4 M). After centrifugation (12 000 × g), 100μL of the supernatant were neutralized with 20–25 μL of potassium carbonate (2M, 4°C). The supernatant (40 μL) obtained after centrifugation was used for HPLC analysis. The pellets of the acid extract were dissolved in 1 mL of sodium hydroxide (0.1 M) and used for protein determination at a dilution of 1:10 (BCA Assay; Pierce).
2.2.6 Mitochondrial activity assay
To test effects of LPA on mitochondrial activity and C13NJ viability, the MTT assay was used [28]. Yellow MTT is reduced to purple formazan. This reduction takes place only when mitochondrial reductase enzymes are active, and therefore conversion is related to mitochondrial activity. Briefly, after incubating cells with LPA (2 μM) for the indicated times, 50 μL of the MTT solution were added to cells (in 500 μL medium) and incubated for 1 h as described previously [28].
2.2.7 Mitochondrial membrane potential
To examine the mitochondrial membrane potential, the JC-1 assay was used. In viable cells with high (negative) mitochondrial membrane potential, JC-1 spontaneously forms complexes known as J-aggregates with intense red fluorescence. In apoptotic or compromised cells with low (depolarized) membrane potential, JC-1 remains in the monomeric form, which shows only green fluorescence. Briefly, C13NJ cells were plated in black 96-well plates (10 000 cells per well) and allowed to grow 24 h before addition of LPA (2 μM; 1 and 5 h) in serum-deprived medium at indicated concentrations and indicated time periods. Cells were then incubated with JC-1 (2.5 μM, dissolved in serum-free medium, 100 μL per well) at 37°C in the dark for 30 min followed by two washing steps with 100 μL PBS. Aliquots of 50 μL PBS were then added to each well and the fluorescence intensities read at 484/540 (excitation/emission) for detection of green fluorescence, and 544/590 (excitation/emission) for detection of red fluorescence on a Victor 1420 multilabel counter.
2.2.8 Time-lapse video microscopy and image quantification
For time-lapse video microscopy the cells were seeded on polystyrol cover slips and mounted in a micro-incubation chamber on the microscope stage (Axiovert 35 microscope, Zeiss) at 37°C. After 18 h, LPA (2 μM) was added directly into the incubation chamber. Images were acquired every 5 min at five different positions of the chamber using a digital camera (Axiocam HRc 1300 × 1030 pixel with a 10 × Plan Neoflour objective). For automatically focusing and data recording KS300 software (Zeiss) was used. Data analysis was carried out using ImageJ (NIH). Image intensity correction was achieved by correcting each image median value to 125 (8 bit images have a resolution of 255 grey levels). Image stabilization was achieved using the Lucas-Kanade method with a macro for ImageJ (K. Li, “The image stabilizer plugin for ImageJ”; http://www.cs.cmu.edu/~kangli/code/Image_Stabilizer.html; ®Kang Li) to correct for changes in position due to mechanical tolerances in the microscope stage, which resulted in residual position changes that make positional information difficult to interpret. Equalization of low-frequency variations in the background signal of the image using an FFT band pass filtering and reducing low and high frequency changes in the images (low frequency filter set to 40 pixels and high frequency filter set to 6 pixels) enabled simple thresholding of the images. After thresholding, which allowed object selection, a Gaussian blur was applied, 4 pixels in size to enable object closing with an additional thresholding step. The resulting binary images were analyzed using ImageJ inherent functions. Measurements included the direct parameters area (pixel), perimeter (pixels), object number, and the calculated parameters average object size (pixels) and circularity (4π(area)/perimeter2; a circle corresponds to 1).
2.2.9 Phalloidin and DAPI stains
C13NJ cells were cultured on Permanox chamberslides (Nalge Nunc International, Rochester, NY, USA) in the absence or presence of LPA (2 μM). For F-actin staining a one-step fixation, permeabilization, and labeling procedure was used. Living cells were incubated with a lyso-palmi–toylphosphatidylcholine (50 μg/mL)/formaldehyde (3.7% v/v) solution, containing 10 U of rhodamine phalloidin (Molecular Probes) for 20 min at 4°C. Laser scanning microscopy (LSM) was performed on a Leitz/Leica TCSSP2 microscope (Leica Lasertechnik, Heidelberg, Germany). Rho inhibition was achieved by a pre-incubation of cells for 8 h in the presence of exoenzyme C3 transferase (1 μg/mL). Nuclei were stained with DAPI (10 min, 1:1000, 22°C).
2.2.10 Expression and visualization of YFP-tagged β-tubulin
C13NJ microglia were electroporated using the Amaxa Mouse Astrocyte Nucleofector Kit (Amaxa). Cells were transiently transfected with 3 μg of a purified yellow fluorescent protein (YFP)-tagged β-tubulin plasmid and seeded on polystyrol cover slips. Forty-eight hours post transfection, cells were mounted in an incubation chamber on an inverted array confocal laser scanning microscope (Axiovert 200 M, Zeiss Microsystems, Jena, Germany) equipped with VoxCell Scan® (VisiTech, Sunderland, UK), a 150 mW Ar laser (Laser Physics; West Jordan, UT, USA), and controlled by Metamorph 6.2r6 (Universal Imaging, Visitron Systems, Puchheim, Germany). Cells were visualized before and 50 min after addition of LPA (2 μM).
2.2.11 β-tubulin immunofluorescence
C13NJ cells were cultured on Permanox chamberslides and incubated with 2 μM LPA for 60 min in serum-free medium (containing 0.1% BSA). Coverslips were washed twice in PBS, fixed with Cytoskelfix (Cytoskeleton; 4 min at −20°C), rehydrated in PBS (10 min), and treated with protein blocking solution for 20 min. Coverslips were incubated with primary tubulin antibody [monoclonal anti-β-tubulin (1:100)] for 60 min, rinsed with PBS, and incubated with Cy-5-conjugated secondary antibody (1:300; 60 min). Cell nuclei were stained with DAPI (1:1000) for 10 min at room temperature; then the slides were rinsed again and mounted with Mowiol.
2.2.12 Cell migration
Migration of C13NJ microglia was assessed in 12-well modified Boyden Transwell microchambers containing 8 μm pore size polyethylene terephthalate membranes. For each assay, 200 000 cells were resuspended in 100 μL of DMEM supplemented with 0.5% FCS and plated on the upper chamber, and lower wells were filled with either DMEM containing 0.5% FCS and 0.1% BSA (nonspecific migration) or with DMEM containing 0.5% FCS, 0.1% BSA, and LPA (2 μM) and incubated at 37°C for 2 h. Subsequent to stimulation cells of the bottom wells were transferred to 1.1 mL microtubes (Bioquote; UK) and pelleted by centrifugation for 7 min at 400 × g. Supernatants were discarded and the cells were resuspended in 150 μL ice-cold fixative solution (CellFix, Becton Dickinson, Schwechat, Austria) diluted 1:10 in distilled water and 1:4 in FacsFlow (Becton Dickinson) to preserve cell shape until analysis. Determination of cell counts was performed using a FACSCalibur flow cytometer from Becton Dickinson by measuring the amount of migrated cells for 60 s. Inhibition of RhoA, B, and C was achieved by a pre-incubation of cells for 6 h in the presence of exoenzyme C3 transferase (1 μg/mL), which was also present during the migration assay.
2.2.13 Sample preparation for 2-D DIGE
Confluent C13NJ cell monolayers (12 cm Petri dishes) were incubated in the absence or presence of LPA (2 μM; 6 h). Cells were washed with sucrose (250 μM in 10 mM Tris/HCl, pH 7.4), then scraped in 50 μL lysis buffer (5 M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris) and processed for 2-D DIGE. Cell lysates were sonicated (5 × 15 s on ice) and centrifuged (5 min, 4°C, 13 000 rpm). Lysates from control and LPA-treated cells were labeled using Cy5 and Cy3 dyes according to the manufacturer’s (Amersham Biosciences) recommendations. The protein content was determined using the Bradford assay. Protein (50 μg) from cell lysates was labeled with 400 pmol of amine-reactive Cy3 or Cy5 N-hydroxysuccinamide ester DIGE dyes (Amersham Biosciences), freshly dissolved in anhydrous dimethyl formamide. The labeling mixture was incubated on ice in the dark for 30 min. The reaction was quenched by addition of lysine (10 nmol) followed by incubation on ice for another 10 min. Equal volumes of 2 × sample buffer containing 7 M urea, 2 M thiourea, 4% w/v CHAPS, 65 mM DTT, and 2% v/v IPG buffer containing 450 μg of the corresponding unlabeled protein lysates were added to each of the labeled protein samples, and the two samples were mixed prior to IEF on Immobiline IPG strips.
2.2.14 2-D DIGE and image analysis
The IPG strips (24 cm, linear pH range of 3–10) were rehydrated overnight in ceramic strip holders in 450 μL sample buffer (containing 1 mg of protein lysates from control and LPA-treated cells; see above). IEF was carried out at 18°C with a maximum current set at 50 μA/strip using an Ettan IPGphor unit (Amersham Biosciences). After IEF (3 h at 150 V, 3 h at 300 V, 3 h at 600 V, remaining time at 8000 V to reach a total of 50 000 Vh) strips were equilibrated in SDS equilibration buffer (50 mM Tris/HCl, 6 M urea, 30% glycerol, 2% SDS, 0.5% DTT, 4.5% iodoacetamide, pH 8.8). IPG strips were directly applied to self-cast SDS gels (12%). SDS-PAGE was carried out at 10°C (Ettan Dalt, Amersham Biosciences; 15 mA/gel, 17 h). Fluorescence imaging was performed on a Typhoon 9400 scanner (Amersham Biosciences; Ex 520/620 nm, Em 590/680 nm for Cy3 and Cy5, respectively). Statistical analysis and quantification of protein spots was carried out using the DeCyder-DIA software (Amersham Biosciences). Fold regulation is expressed as mean from two different gels run from two independent LPA stimulations. Differentially expressed protein spots were picked using an Ettan spot picker (Amersham Biosciences).
2.2.15 LC-MS/MS analysis
Protein spots were digested with trypsin (20 μg in 200 μL 1 mM HCl) as described previously [29]. Peptide extracts were dissolved in 0.1% formic acid and separated on a nano-HPLC system (Ultimate 3000™, LC Packings, Amsterdam, The Netherlands). Samples of 70 μL were injected and concentrated on the loading column (LC Packings C18 Pep-Map™, 5 μm, 100Å, 300 μm inner diameter × 1 mm) for 5 min using 0.1% formic acid as isocratic solvent at a flow rate of 20 μL/min. The column was then switched into the nanoflow circuit, and the sample was loaded on the nano-column (LC-Packings C18 PepMap™, 75 μm inner diameter × 150 mm) at a flow rate of 300 nL/min and separated using the following gradient: solvent A: water, 0.3% formic acid, solvent B: ACN/water 80/20 v/v, 0.3% formic acid; 0–5 min: 4% B, after 40 min 55% B, then for 5 min 90% B and 47 min re-equilibration at 4% B. The sample was ionized in a Finnigan nano-ESI source equipped with NanoSpray tips (PicoTip™ Emitter, New Objective, Woburn, MA, USA) and analyzed in a Thermo-Finnigan LTQ linear iontrap mass-spectrometer (Thermo, San Jose, CA, USA). Raw data files were processed using the “Data Extractor” of the Agilent-Spectrum Mill Software (Rev. A. 03.03.078).
2.2.16 Protein identification
Processed data were searched against the mammalian protein subdatabase (downloaded from “ftp://ftp.ncbi.nih.gov/blast/db/FASTA/” on 18.6.2006; 3717264 total entries). Database searching was performed with the MS/MS Search feature of the Agilent-Spectrum Mill Software (Rev. 03.03.078). The precursor mass tolerance was set to 2.0 Da, the product mass tolerance to 0.7 Da. The following search parameters were used: variable modification due to methionine oxidation, fixed cysteine modification due to carbamidomethylation, N-terminal pyro glutamic acid allowed, and two missed cleavage sites in case of incomplete trypsin hydrolysis. Acceptance parameters were minimum number of peptides ≥3, peptide score of ≥10, and an SPI% (the percentage of assigned spectrum intensity of total spectrum intensity) of ≥70. Only Homo sapiens proteins regulated in both independent experiments are shown.
2.2.17 qPCR
Cells were incubated in the absence or presence of LPA (2 μM) for 1, 5, and 12 h in triplicate dishes. Total RNA was isolated according to the RNeasy (Qiagen, Hilgen, Germany) protocol. Aliquots of 3 μg of total RNA were reverse transcribed according to the manufacturer’s instructions using random hexamer primers (Amersham Biosciences). Real-time PCRs were performed with an Applied Biosystems 7900HT Fast Real Time PCR System, the QuantiFast SYBR Green PCR kit and QuantiTect Primer Assays (Qiagen, Hilgen, Germany). The following QuantiTect primer assays were used: Hydroxymethylbilane synthase (Hs_HMBS_ 1_SG; internal standard), lactate dehydrogenase B (Hs_ LDHB_1-SG), methionine adenosyltransferase 2 subunit β (Hs_MAT2B_1_SG), Annexin A1 (Hs-ANXA1_1_SG; ANXA1, Annexin A1), tubulin folding cofactor B (TBCB, Hs_TBCB_1_SG), 78 kDa glucose-regulated protein (GRP78) (Hs_HSPA5_1_SG), and Hsp90 co-chaperone Cdc37 (CDC37) (Hs_CDC37_2_SG).
2.2.18 Statistical analysis
Data are presented as mean±SD. Either Student’s t-test (using the GraphPad Prism package) or the Mann–Whitney U-test (video time lapse microscopy) was used for analysis of statistical significance. All values of p<0.05 were considered significant. For proteome analyses only spots that were regulated ≥2-fold (up or down; DeCyder software) were picked and further processed for identification.
Statistical significance of differences in mRNA expression levels was analyzed using the relative expression software tool (REST©, http://www.gene-quantification.de/rest.html) using a pair-wise fixed reallocation test [30].
3 Results
3.1 Characterization of LPA receptor expression
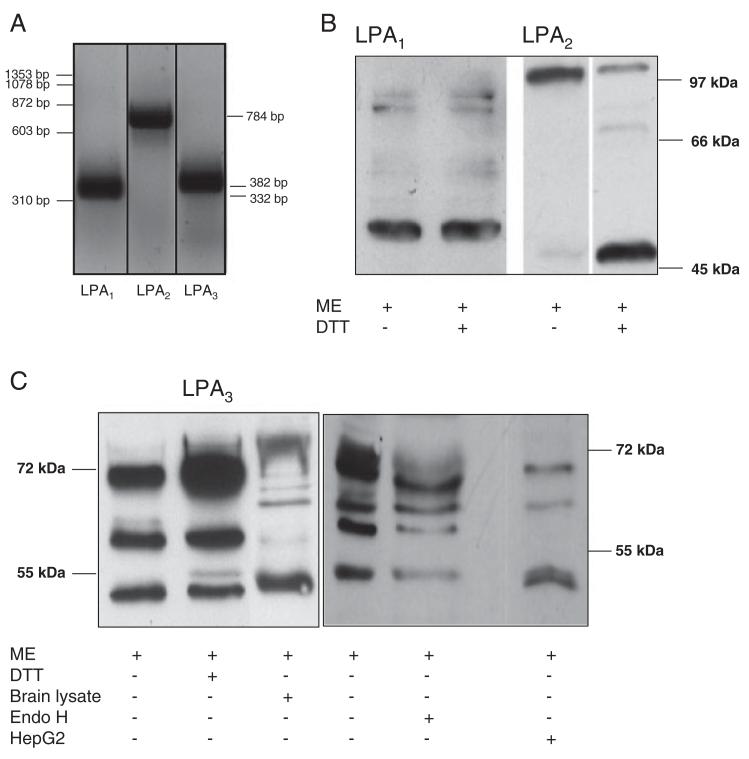
LPA receptor expression analysis was performed on mRNA and protein level. Using RT-PCR, the presence of three classical LPA receptor transcripts was revealed (Fig. 1A) and identity was confirmed by sequencing the corresponding PCR products (data not shown). Western blot analysis confirmed LPA1–3 expression on protein level. The major immunoreactive band for LPA1 was detected at an apparent molecular mass of 53 kDa (Fig. 1B). In contrast, the majority of immunoreactive LPA2 was detected at approx. 100 kDa. Under more drastic conditions using DTT as additional reducing agent, the bulk of the 100 kDa band was shifted to an apparent molecular mass of 48 kDa. Similar observations of mercaptoethanol-resistant dimers were reported for sphingosine-1-phosphate specific lysophospholipid receptors [31]. Immunoreactivity for LPA3 was detected in three bands of 52, 60, and 72 kDa, from which the 52 kDa band comigrated with LPA3 detected in mouse brain protein lysates (Fig. 1C). Reduction by mercaptoethanol in the presence of DTT had no effects on molecular mass distribution of the three bands. Deglycosylation of cellular proteins by Endo H treatment prior to immunoblotting shifted the 71 kDa band to approx. 65 kDa, in line with N15 and N172 representing two potential N-glycosylation sites (LPAR3_HUMAN, http://www.uniprot.org/uniprot/Q9UBY5). HepG2 cell lysates were used as additional controls for LPA3 expression and revealed three immunoreactive bands detected at apparent molecular weights of 51, 63, and 70 kDa comigrating with three of the major bands detected in C13NJ lysates (Fig. 1C).
Figure 1.
LPA receptor expression by C13NJ microglia. (A) Expression levels of LPA1–3 transcripts were determined by RT-PCR using the primers and conditions described in Table 1. Lane 1: 100 bp standard. (B and C) Western blot analysis of LPA receptor expression. Fifty micrograms of protein of total cell lysates were reduced under standard conditions (5% mercaptoethanol; ME) or treated additionally with DTT (100 mM) or EndoH followed by SDS-PAGE separation. Blots were probed with polyclonal rabbit anti-human LPA1, anti-human LPA2, or anti-mouse LPA3 anti-serum and HRP-conjugated goat-anti-rabbit antibody as indicated. LPA2 samples were run on two different gels. Immunoreactive bands were visualized using the ECL-detection system. Mouse brain and HepG2 cell lysates were used as control for LPA3 identification.
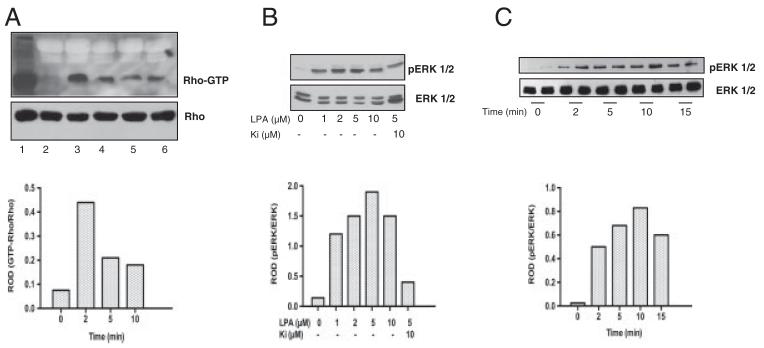
LPA receptor activation leads to activation of the Rho (via G12/13) and ERK1/2 (via Gi/o) pathway [32], both of which can lead to enhanced glycolysis and elevated intracellular ATP levels. To test whether the Rho pathway is induced in C13NJ microglia in response to LPA, Rho activation was studied in a time-dependent manner using a Rhotekin-GST pull-down kit (Fig. 2A). These experiments revealed that Rho was rapidly activated upon stimulation with LPA reaching maximum activity at 5 min, while the amount of total Rho protein (determined by Western blotting of cell lysates) remained unchanged. LPA-induced signaling resulted also in time- and concentration-dependent phosphorylation of ERK1/2, with maximum phosphorylation rates at 5 μM (Fig. 2B). Importantly the LPA1 and LPA3 specific antagonist Ki16425 resulted in significantly attenuated pERK1/2 signals, indicating LPA1/3-dependent signal transduction to the mitogen activated protein kinase pathway in C13NJ microglia (Fig. 2B). ERK1/2 phosphorylation was maximal at an incubation time of approx. 10 min (Fig. 2C).
Figure 2.
Rho and ERK1/2 activation in C13NJ microglia in response to LPA receptor engagement. (A) Serum-starved cells were incubated in the presence of LPA. Cells were lysed and GST-Rhotekin-RBD and SwellGel Immobilized Glutathione Discs were added to the lysate (700 μg) to pull down GTP-bound Rho. Samples were analyzed by Western blotting using anti-Rho antibody followed by HRP-conjugated goat-anti-mouse antibody. Bands were visualized with the ECL system. Total RhoA was immunoblotted to ensure comparable loading of cell lysates. 1, total lysate; 2, control; 3, positive control as provided by supplier; 4–6, LPA (2 μM) 2, 5, and 10 min, respectively. Bar graph shows relative optical densities (ROD) of lanes 2 and 4–6 (Rho-GTP/Rho). (B) Cell lysates of control or cells stimulated with LPA in the indicated concentrations in the absence or presence of Ki16425 were separated by SDS-PAGE and transferred to PVDF membranes. pERK1/2 and total ERK1/2 were detected as described in Section 2. The bar graph shows densitometric analysis of ERK1/2 phosphorylation from the Western blots. Data are presented as ROD (pERK1/2/ERK1/2). (C) Cells were incubated with LPA (2 μM) for the indicated periods of time. Protein lysates were separated by SDS-PAGE, blotted to PVDF membranes, and pERK1/2 and ERK1/2 was detected as described in Section 2. The bar graph shows densitometric analysis of ERK1/2 phosphorylation from the Western blots. Data are presented as mean ROD (pERK1/2/ERK1/2).
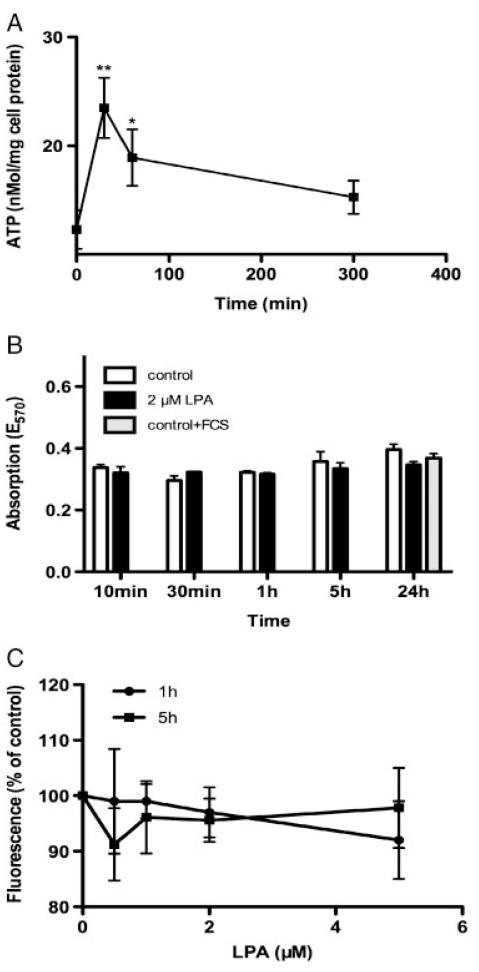
In line with induction of the Rho signaling pathway (Fig. 2A), LPA treatment also led to significantly elevated ATP concentrations after 30 and 60 min (1.9- and 1.5-fold over controls, respectively) approaching baseline levels after 300 min (Fig. 3A). Stimulation of C13NJ microglia by LPA was without measurable effects on mitochondrial metabolic function and membrane potential, as determined by MTT (Fig. 3B) and JC-1 assays (Fig. 3C). This indicates that LPA neither compromises nor enhances mitochondrial metabolic function. Thus, increased ATP levels are most likely the result of increased glycolysis (see below).
Figure 3.
ATP content, viability, and mitochondrial membrane potential of untreated and LPA-treated C13NJ microglia. (A) ATP content of cells incubated in the presence of LPA (2 μM) for the indicated times. ATP levels were analyzed by HPLC as described in Section 2. Results represent mean±SD from triplicate dishes. p<0.001, p<0.05 versus baseline. (B) Mitochondrial activity of cells was analyzed using the MTT assay. Cells were incubated in the presence of LPA (2 μM) for the indicated times and processed as described in Section 2. Results represent mean±SD (n = 4). (C) Measurement of mitochondrial membrane potential by JC-1 staining assay. Cells were cultured on 96-well plates and incubated in the presence of the indicated LPA concentrations for 1 or 5 h. Staining with JC-1 and fluorescence measurements were performed as described in Section 2. Results (fluorescence intensities) are expressed as percentage of controls (no LPA added) and shown as mean±SD (n = 6).
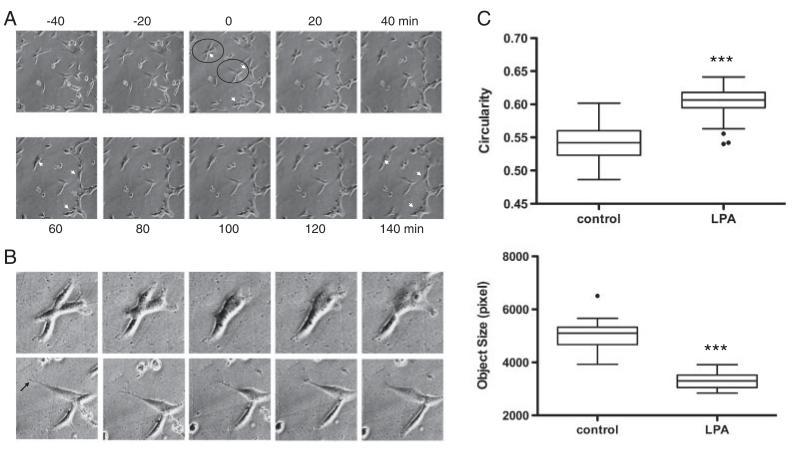
LPA-induced morphological alterations were investigated by a time-lapse video microscopy approach. Upon LPA stimulation the cells responded with morphological changes including retraction and thickening of processes as well as thickened morphology of cell bodies, an effect that was evident already 40 min after stimulation (Fig. 4A). Cell rounding became evident after 120 and 140 min incubation in the presence of LPA. The cells encircled in Fig. 4A are shown at higher magnification in Fig. 4B. From these micrographs the transformation to a more amoeboid morphology in response to LPA is evident (upper panel). The cell shown in the lower panel reacts to LPA by retraction of a much thinner, branched process (arrow), which is almost completed after 60 min. Also, this cell transforms into a more flattened, amoeboid type, morphological transformations indicative of microglia activation. Image quantification (Fig. 4C) revealed that LPA treatment resulted in increased circularity (0.55 versus 0.61; controls versus LPA) and decreased cellular area (4992 versus 3301 pixel; controls versus LPA).
Figure 4.
Effects of LPA on C13NJ microglia morphology. (A) Cells were cultured on polystyrol cover slips and were photographed by time-lapse video microscopy at 20 min intervals. The numbers refer to time relative to the addition of LPA (2 μM; time 0). Representative micrographs are shown. Arrowheads indicate cells that significantly change cell morphology in response to LPA supplementation. (B) The cells enclosed by circles in (A) are shown at higher magnification. An arrow indicates the fine protrusion visible in one cell in the lower panel that is retracted in response to LPA. (C) Image quantification and calculation of circularity and object size was performed as described in Section 2. Results for circularity (a shape factor; 1 corresponds to a circle) and object size are shown. Circularity and object size were evaluated in 70 images (control or LPA-treated cells) where a minimum of 50 objects/image was assessed for pixel size. These measurements were performed at five different x/y-positions of the microscope stage, ***p<0.001.
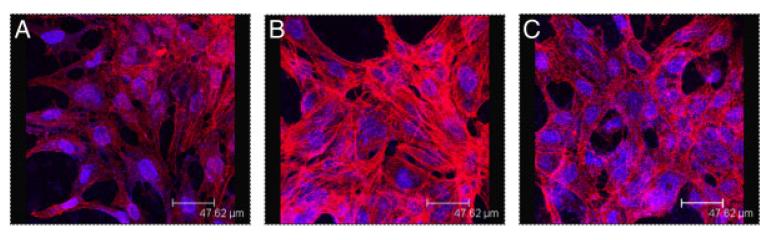
To reveal effects of LPA on cytoskeletal dynamics, the actin cytoskeleton of control and LPA-treated cells was visualized by phalloidin staining (Fig. 5). In control cells F-actin staining was observed along well-defined bundles, which are oriented along the cell axis (Fig. 5A). In the presence of LPA the formation of condensed, thickened stress fibers of F-actin was observed (Fig. 5B). The F-actin stains in LPA-treated cells were reminiscent of LPA-induced cell spreading (as observed in Fig. 6), resulting in a rounder and more flattened cell type (Fig. 5B). To study the involvement of Rho GTPases during LPA-induced actin stress fiber formation, C13NJ cells were incubated with C3 exoenzyme, which selectively inactivates all three Rho-family members by ADP-ribosylation at Asn41 [33]. These experiments revealed that LPA-induced stress fiber formation was almost completely abolished by C3 exoenzyme (Fig. 5C). All alterations in cell morphology described above (i.e. rounding, process retraction and stress fiber formation) are compatible with LPA signaling via G12/13 and subsequent Rho activation.
Figure 5.
Effects of LPA on the actin cytoskeleton of C13NJ microglia. Cells were incubated in the absence (A) or presence of LPA (2 μM, 30 min) preincubated without (B) or with C3 exoenzyme (C) (1 μg/mL, added 6 h prior to and present during LPA stimulation). The actin cytoskeleton was visualized by fluorescence microscopy after rhodamine-phalloidin staining (red), nuclear staining was performed with DAPI (blue).
Figure 6.
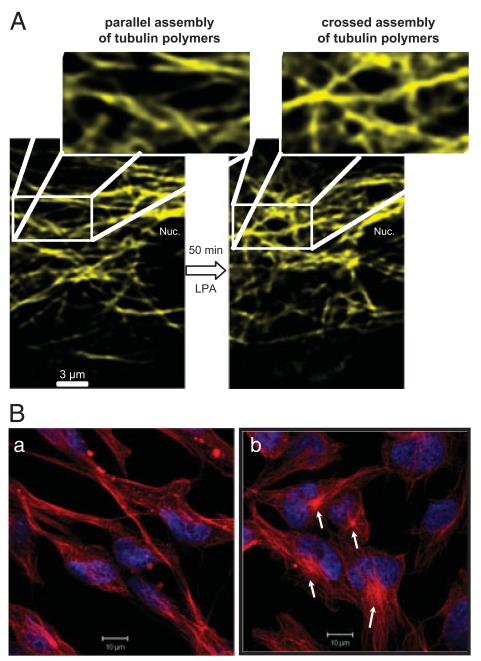
Analysis of β-tubulin organization of C13NJ microglia. (A) Cells were transiently transfected with an YFP-tagged β-tubulin plasmid. Forty-eight hours after transfection, β-tubulin organization was analyzed by LSM in untreated and LPA-treated (2 μM; 50min) cells. The indicated areas are shown at higher magnification. (B) Cells on chamberslides were incubated with LPA (2 μM for 60min, serum-free medium, 0.1% BSA). After washing, incubation with primary (monoclonal anti-β-tubulin) and secondary (Cy-5 conjugated) antibody, cells were analyzed by confocal LSM. Nuclei were stained with DAPI. Red = β-tubulin, blue = DAPI.
To get an indication about potential effects of LPA on microtubule morphology, YFP-β-tubulin was transiently expressed in C13NJ cells and visualized by LSM. These experiments indicate that LPA treatment (50 min) leads to a remodeling of the microtubule network, transforming a parallel assembly of tubulin polymers into a more cross-linked structure (Fig. 6A). Immunofluorescence studies revealed more pronounced β-tubulin staining at cellular structures resembling centromers of LPA-stimulated cells as compared with control cells (Fig. 6B).
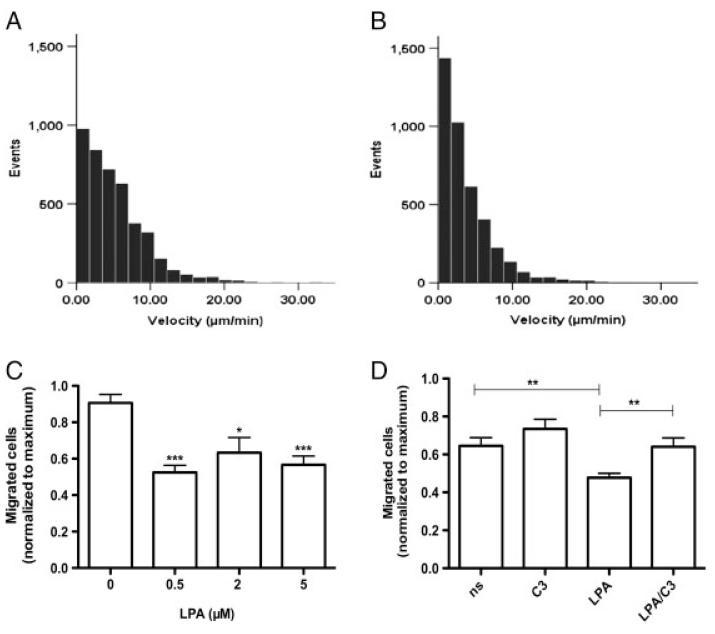
LPA is a potent effector of cell motility; therefore, cell migration of LPA-treated microglia was analyzed by two independent techniques. First, time-lapse video microscopy revealed pronounced impairment of cell motility of LPA-treated cells: While control cells migrated a mean distance of 5.3 μm/min, LPA-treated cells showed a statistically significant (p<0.005; Mann–Whitney U-test) lower migration (3.8 μm/min; Figs. 7A and B). Second, in Boyden chamber assays quantitatively comparable observations were made: Under the experimental conditions applied, the number of transmigrated cells in the presence of LPA (≤10 μM) was significantly lower (between 20 and 35%) as compared with controls (Fig. 7C). The migrational block was released in response to Rho family inactivation by C3 exoenzyme (Fig. 7D). In summary these findings indicate that LPA receptor engagement in C13NJ microglia leads to altered cellular energy homeostasis, cytoskeleton arrangement, and cell motility.
Figure 7.
Effect of LPA on C13NJ microglia motility and migration. (A and B) Cell motility was recorded using time-lapse microscopy. Photographs were taken every 5 min and the migratory paths of cells were evaluated in μm/min using Image J. The histograms show the velocity distribution of >60 individual cells monitored for 5 h in the absence (A) or presence (B) of LPA (2 μM). Data were analyzed for statistical significance using the Mann–Whitney U-Test. (C) Migration assays were carried out using Boyden chambers as described in Section 2. Results represent mean±SD (n = 10) and are expressed as percentage of migrated cells under control conditions. ***p<0.001, *p<0.05. (D) Migration assays were carried out in Boyden chambers in serum-free medium (ns), containing C3 exoenzyme (1 μg/mL), LPA (2 μM), or LPA plus C3 exoenzyme (2 μM and 1 μg/mL, respectively). Results represent mean±SD (n = 12); **p<0.005.
3.2 Effects of LPA on microglia protein expression patterns
To investigate changes on the proteome level in response to LPA, a 2-D DIGE proteomics approach was used. Protein lysates from control (labeled with Cy5, Supporting Information Fig. IA) and LPA-treated C13NJ microglia (labeled with Cy3, Supporting Information Fig. IB) were run on the same gel. Expression analysis was performed using the DeCyder software package, by which the gel images were normalized and compared to detect differences in spot volume. During these analyses 762 spots were detected, out of which 110 were regulated ≥2-fold (71 up, 39 down). Differentially expressed proteins were picked and processed for identification by MS and 55 spots corresponding to 42 different proteins were identified. Five proteins were detected in two or more spots, most probably indicative of posttranslational modifications. A gray scale image of an overlay with spot labeling is shown in Supporting Information Fig. IC.
The identified proteins were grouped in five different functional classes. The complete list of regulated proteins is given in Table 2. Within these five groups enzymes involved in metabolism/glycolysis (group 1) revealed the most pronounced regulations in response to LPA (consistent upregulation between 2.1- and 7.9-fold). The second group comprises proteins that are potent regulators of cell motility and/or cytoskeletal dynamics (regulation −2.3 to +3.9-fold). Within the third (chaperones), fourth (protein processing), and fifth (RNA processing) group we identified proteins that were regulated between −6.3- and +6.4-fold.
Table 2.
Protein identification
| Spot no. | Full protein name | Protein name | Swiss-Prot accession no. |
Mw theor. | Pl theor. | Mw exp. | pl exp. | Fold-regulation | No. of peptides matched |
Sequence coverage (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Groupl: metabolism | ||||||||||
| Upregulated | ||||||||||
| 1551, 1672, 1673,1682, 1684, 1691, 1729, 1849 | α-Enolase | ENOA | P06733 | 47 169 | 7.01 | 49 200–55 600 | 5.8–7.5 | ↑2.2–7.9 | 6–23 | 22–64 |
| 2186, 2189 | Fructose-bisphosphate aldolase A | ALDOA | P04075 | 39 420 | 8.30 | 41 200 | 8.7 | ↑2.6/2.5 | 6/7 | 19/25 |
| 2405, 2418, 2423, 2435 | Glyceraldehyde-3-phosphate dehyd rogenase | GAPDH | P04406 | 36 053 | 8.57 | 36 500 | 6.7–9.7 | ↑2.1–2.8 | 3–6 | 13–21 |
| 1433 | Pyruvate kinase isozymes M1/M2 | KPYM | P14618 | 57 937 | 7.96 | 61900 | 7.2 | ↑2.5 | 10 | 24 |
| 3013 | Triosephosphate isomerase | TPIS | P60174 | 26 669 | 6.45 | 22 700 | 7.0 | ↑2.4 | 3 | 14 |
| 2422 | l-lactate dehydrogenase B chain | LDHB | P07195 | 36 638 | 5.71 | 35800 | 5.5 | ↑5.6 | 9 | 31 |
| 1850 | Fumarate hydratase, mitochondrial | FUMH | P07954 | 54 637 | 8.85 | 49 200 | 7.1 | ↑3.9 | 6 | 17 |
| 1574 | GARS protein | Q15374 | 45 987 | 6.34 | 56000 | 7.2 | ↑5.1 | 11 | 41 | |
| 2136 | Gastric-associated differentially lexpressed protein YA61P (drug-sensitive protein 1) | Q9NZ23 | 14 867 | 6.83 | 42 700 | 5.8 | ↑2.8 | 3 | 33 | |
| Downregulated | ||||||||||
| 2184 | Cytosolic acyl coenzyme A thioester hydrolase | BACH | 000154 | 41 796 | 8.85 | 46 200 | 7.1 | ↓5.1 | 3 | 10 |
| 1038 | Guanine monophosphate synthetase variant | Q53F90 | 76 696 | 6.24 | 72 600 | 6.7 | ↓2.0 | 6 | 12 | |
| 2405 | Methionine adenosyltransferase 2 subunit β | MAT2B | Q9NZL9 | 37 552 | 6.9 | 36 400 | 6.7 | ↓4.9 | 4 | 18 |
| Group 2: cytoskeleton/motility | ||||||||||
| Upregulated | ||||||||||
| 2485 | Annexin A1 | ANXA1 | P04083 | 38 714 | 6.57 | 34 600 | 6.7 | ↑2.2 | 16 | 60 |
| 3368 | Galectin-7 | LEG7 | P47929 | 15 075 | 7.02 | 13800 | 6.9 | ↑2.1 | 3 | 30 |
| 3295 | Nucleoside diphosphate kinase B | NDKB | P22392 | 17 298 | 8.52 | 15 600 | 6.7 | ↑3.9 | 4 | 14 |
| 2148 | Twinfilin-2 | TWF2 | Q6IBS0 | 39 548 | 6.37 | 41700 | 6.3 | ↑3.7 | 5 | 24 |
| 1130 | WD repeat-containing protein 1 | WDR1 | 075083 | 66194 | 6.17 | 70000 | 6.6 | ↑2.2 | 9 | 29 |
| 2535 | WD repeat-containing protein 61 | WDR61 | Q9GZS3 | 33 581 | 5.16 | 33300 | 6.5 | ↑4.1 | 4 | 24 |
| Downregulated | ||||||||||
| 2839 | Tubulin folding cofactor B | TBCB | Q99426 | 27 326 | 5.06 | 27 000 | 5.0 | ↓2.4 | 3 | 17 |
| 1383 | Vimentin | VIME | P08670 | 53 652 | 5.06 | 60800 | 5.0 | ↓2.3 | 24 | 54 |
| Group 3: chaperones/unfolded protein response | ||||||||||
| Upregulated | ||||||||||
| 3056 | BAG family molecular chaperone regulator 2 | BAG 2 | 095816 | 23 772 | 6.25 | 22 200 | 6.5 | ↑6.4 | 6 | 30 |
| 477 | Heat shock protein 105 kDa | HS105 | Q92598 | 96 865 | 5.28 | 92 900 | 5 | ↑2.8 | 33 | 42 |
| 904 | 78kDa glucose-regulated protein (immunoglobulin heavy chain-binding protein) | GRP78 | P11021 | 72 333 | 5.07 | 80 500 | 4.7 | ↑3.3 | 22 | 43 |
| 584, 591 | Endoplasmin [Precursor] | ENPL | P14625 | 92 469 | 4.76 | 89 500 | 5.3 | ↑2.1/2.6 | 18/6 | 33/10 |
| Downregulated | ||||||||||
| 1660 | Hsp90 co-chaperone Cdc37 | CDC37 | Q16543 | 44 468 | 5.17 | 53700 | 4.6 | ↓2.1 | 6 | 22 |
| Group 4: protein binding, synthesis, modification, and secretion | ||||||||||
| Upregulated | ||||||||||
| 3227 | Calmodulin-like protein 5 | CALL5 | Q9NZT1 | 15921 | 4.34 | 17 400 | 7.9 | ↑2.4 | 5 | 35 |
| 1786 | Septin-2 | SEPT2 | Q15019 | 41 487 | 6.15 | 51 200 | 6.3 | ↑2.9 | 9 | 34 |
| 1786 | Septin-5 | SEPT5 | Q99719 | 42 111 | 6.21 | 51 200 | 6.3 | ↑2.9 | 3 | 11 |
| 2485 | Vacuolar protein sorting-associated protein 37B | VP37B | Q9H9H4 | 31 307 | 6.78 | 34 600 | 6.7 | ↑2.2 | 3 | 19 |
| 2919 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | UCHL1 | P09936 | 24 824 | 5.33 | 24700 | 5.3 | ↑2.8 | 11 | 51 |
| Downregulated | ||||||||||
| 1041 | Annexin A6 | ANXA6 | P08133 | 75 873 | 5.42 | 118900 | 7.5 | ↓2.1 | 4 | 9 |
| 2787 | Calcyclin-binding protein | Q6NVY0 | 26182 | 7.63 | 28000 | 8.6 | ↓2.5 | 7 | 36 | |
| 2270 | Calmodulin-like skin protein variant [Fragment] | Q53H37 | 15 876 | 4.34 | 39 000 | 6.7 | ↓6.3 | 3 | 19 | |
| 2692 | Proteasome activator complex subunit 3 | PSME3 | P61289 | 29 506 | 5.69 | 30000 | 5.5 | ↓2.3 | 7 | 29 |
| 2150 | Protein disulfide-isomerase [Precursor] | PDIA1 | P07237 | 57 116 | 4.76 | 41 200 | 4.7 | ↓2.0 | 5 | 12 |
| 2839 | Tumor protein D52-like 2 | Q5JWU8 | 21 450 | 5.71 | 27 000 | 5.0 | ↓2.4 | 4 | 27 | |
| Group 5: RNA synthesis, binding, turnover | ||||||||||
| Upregulated | ||||||||||
| 1679 | Lupus La protein (La autoantigen) | LA | P05455 | 46 837 | 6.68 | 53700 | 6.8 | ↑6.2 | 5 | 15 |
| 2307 | Heterogeneous nuclear ribonucleoproteins C1/C2 | HNRPC | P07910 | 33 670 | 4.95 | 37 800 | 5.3 | ↑2.7 | 5 | 23 |
| Downregulated | ||||||||||
| 2424 | Heterogeneous nuclear | ROA1 | P09651 | 38 846 | 9.26 | 35800 | 9.4 | ↓2.5 | 3 | 12 |
| ribonucleoprotein A1 | ||||||||||
| 2288 | Heterogeneous nuclear | HNRH3 | P31942 | 36 926 | 6.27 | 37 800 | 6.9 | ↓2.5 | 3 | 12 |
| ribonucleoprotein H3 | ||||||||||
| 830, 849 | Far upstream element-binding protein 2 | FUBP2 | Q92945 | 73146 | 6.84 | 80 500 | 7.0 | ↓2.1/2.2 | 13/16 | 24/27 |
| 1989 | TAR DNA-binding protein 43 | TADBP | Q13148 | 44 740 | 5.85 | 46700 | 5.6 | ↓3.7 | 8 | 29 |
Information on protein name, the Swiss-Prot accession number, theoretical Mw and pl, experimental Mw and pl, fold-regulation, number of identified peptide sequences, and % sequence coverage are included. Protein identifications are based on multiple peptide sequences with a minimum number of three distinct peptides.
The most pronounced and consistent effects of LPA treatment were observed for expression levels of glycolytic enzymes (enolase A (ENOA), aldolase A (ALDOA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), pyruvate kinase isozymes M1/M2, and triosephosphate isomerase (TPIS), which were upregulated between 2.1- and 7.9-fold (Table 2). Three of the enzymes (ENOA, ALDOA, and GAPDH) were detected in two or more spots, being indicative for posttranslational modification. This phenomenon was most pronounced for ENOA, which was detected in eight consecutive spots. In line, the Swiss-Prot entry (P06733) lists several S, T, and Y residues that are potential phosphorylation sites and one K-residue that is subject to acetylation. However, posttranslational modifications were not verified analytically.
The next set of proteins affected by LPA is involved in the regulation of cell motility and/or cytoskeletal dynamics (Table 2, group 2). Within this group six proteins (ANXA1, LEG7, NDKB, TWF2, WDR1 (WDR, WD repeat-containing protein) and WDR61 were upregulated (2.2- to 4.1-fold) while two (TBCB, VIME) were downregulated (2.3- and 2.4- fold). Findings of these proteome analyses are entirely consistent with pronounced effects of LPA on cellular ATP production, effects on cell size and circularity, alterations in the actin and tubulin cytoskeleton, and altered migrational properties of C13NJ microglia.
Proteome data obtained for groups 3 (chaperones), 4 (protein binding, synthesis, modification, and secretion) and 5 (RNA synthesis, binding, and turnover) were not characterized on a functional level. However, altered expression levels of proteins classified in these groups could significantly impact on microglia biology. Within group 3, BAG family molecular chaperone regulator 2 (BAG2), HSP105, endoplasmin (ENPL, also called 94 kDa glucose-regulated protein; GRP94), and 78 kDa glucose-regulated protein (GRP78) were upregulated in response to LPA. BAG2 is a cochaperone that shuttles microtubule-associated Tau protein that is prone to misfolding to a ubiquitin-independent degradation pathway, thus preventing accumulation of ubiquinated Tau-filaments as observed in many neurodegenerative diseases [34]. GRP78 and GRP94 are coordinately regulated by common trans-acting factors and it was suggested that upregulation of these ER chaperones is a protective mechanism to prevent amyloid beta deposition [35]. In group 4 (among other proteins) two members of the septin protein family were upregulated in response to LPA – namely septin-2 and -5. This might be of relevance for microglia biology as septins coordinate cytokinesis with chromosome congression and segregation [36] and could thus be regulators of microglia cell division. Among the downregulated proteins in this group is protein disulfide-isomerase (PDI), a finding, which, in light of upregulated chaperone expression (group 3), is somewhat surprising. Among the proteins involved in RNA synthesis/binding, Lupus La protein was upregulated. La protein is involved in diverse aspects of RNA metabolism, including binding and protecting 3-prime UUU elements of newly transcribed RNA, processing 5- and 3-prime ends of pre-tRNA precursors, acting as an RNA chaperone, and binding viral RNAs associated with hepatitis C virus [37]. Three identified proteins are members of the heterogeneous nuclear ribonucleoprotein family of proteins, which share common structural domains and play central roles in DNA repair, telomere biogenesis, signaling events, and are regulators of gene expression at both transcriptional and translational levels [38].
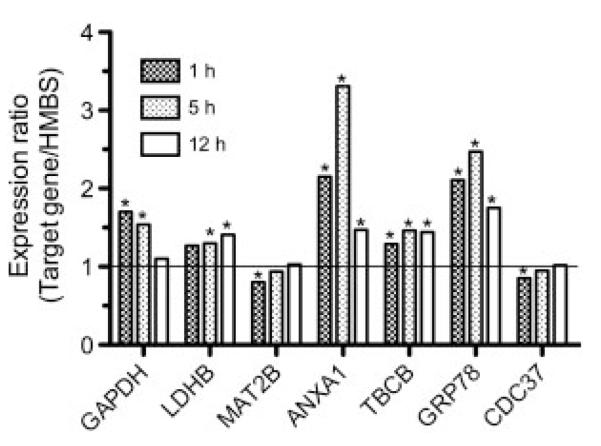
To verify the proteome data, expression levels of seven genes arbitrarily chosen from up- and downregulated members in groups 1–3 (Table 2) were quantitated by qPCR (Fig. 8A). Upregulated mRNA levels consistently accompanied upregulated protein levels for the genes evaluated (GAPDH, LDHB, ANXA1, and GRP78). With regard to downregulated candidates, protein and mRNA expression correlated for MAT2B and CDC37 (at 1 and 5 h), but not for TBCB. TBCB was downregulated on protein but upregulated on mRNA level. Whether this is a result of post-translational or proteasomal processing is currently not clear.
Figure 8.
qPCR analysis of C13NJ gene expression in response to LPA treatment. Cells were cultured on 6-well trays. Cells were incubated in the absence or presence of LPA (2 μM; triplicate dishes per time point) and mRNA levels for GAPDH, LDHB, MAT2B, ANXA1, TBCB, GRP78, and CDC37 were determined by qPCR. At the indicated time points total RNA was isolated and reverse transcribed. cDNA was amplified using the QuantiTect primer assays described in Section 2. Relative gene expression of target genes is presented in relation to hydroxymethylbilane synthase. Gene expression ratios were calculated by REST© as described in Section 2; *p<0.05.
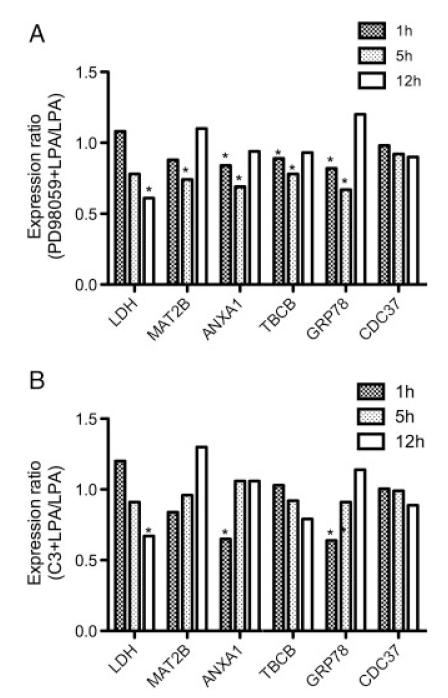
Finally, the effects of ERK and Rho inhibition on mRNA expression levels were analyzed by qPCR (Fig. 9). Preincubation of microglia with the MEK1 inhibitor PD98059 resulted in downregulated mRNA levels of LDH, MAT2B, ANXA1, TBCB, and GRP78. Except for LDH the effects of PD98059 on mRNA levels were transient and detectable only at 1 and 5 h (Fig. 9A). Inhibition of Rho with C3 exoenzyme resulted in significantly lower mRNA levels of LDH (12 h), ANXA1 (1 h), and GRP78 (1 h) (Fig. 9B). These findings are in line with published data demonstrating ERK1/2- and Rho-dependent crosstalk of ANXA1 [39, 40] and GRP78 [41, 42].
Figure 9.
Effect of ERK- and Rho inhibition on C13NJ gene expression. Cells were cultured on 6-well trays. Cells were incubated in the presence of LPA (2 μM; triplicate dishes per time point) with or without PD98059 (25 μM, 1 h preincubation; (A) or C3 exoenzyme (1 μg/mL, 4 h preincubation; (B) mRNA levels of LDHB, MAT2B, ANXA1, TBCB, GRP78, and CDC37 were determined by qPCR as described in Fig. 8; *p<0.05.
4 Discussion
In the present study the cellular outcome of LPA receptor stimulation in human C13NJ microglia was characterized on the proteome level and in a physiological context. On the proteomic level the most pronounced alterations were observed for glycolytic enzymes and for proteins that are regulators of cell motility and/or cytoskeletal dynamics. These findings could be verified on the functional level: (i) LPA-induced Rho and ERK1/2 activation and increased cellular ATP production, (ii) LPA-induced process retraction and cell spreading, (iii) the cytoskeletal architecture was significantly altered after LPA treatment, and, (iv) cell motility was significantly reduced in response to LPA. Many of our observations are compatible with activation of the Rho signaling pathway, which is induced in response to LPA receptor engagement [43]. This was further corroborated by the inhibitory effect of C3 exoenzyme on LPA-triggered stress fiber formation and reversal of the migrational block.
4.1 Glycolysis
Most pronounced alterations in protein expression were detected for glycolytic enzymes in LPA-treated microglia coinciding with time- and concentration-dependent phosphorylation of ERK1/2. This is one likely explanation for the strong induction of glycolytic enzymes: In cardiomyocytes α-enolase is induced in response to ERK1/2 activation during ischemic hypoxia and reoxygenation [44]. Moreover, activation of ERK1/2 resulted in elevated cellular ATP levels (similar as observed during the present study; Fig. 3A), an effect that was alleviated in the presence of the mitogen activated protein kinase inhibitor PD98059 [44]. In line, Fujita et al. [45] have demonstrated increased ATP-release from primary rat microglia in response to LPA. With regard to Rho activation Jung et al. [46] reported the assembly of a glycolytic enzyme minicomplex in response to LPA-mediated activation of the Rho/ROCK/LIM kinase pathway in HeLa cells. Subsequent cofilin phosphorylation and translocation of a phospho-cofilin/triose-phosphate isomerase complex to the plasma membrane forms a glycolytic enzyme minicomplex providing energy support for ion pump activity. This would be in line with the observation that glycolysis-derived ATP is mainly used for fuelling plasma membrane located ion pumps, whereas ATP derived from oxidative metabolism is used for contractile function [47]. Thus, upregulation of glycolytic enzymes in response to LPA treatment of C13NJ cells could serve two purposes, (i) as a survival signal by generating sufficient amounts of ATP and (ii) as a source of cellular energy to fuel plasma membrane located, ATP-dependent ion pumps.
4.2 Migration, cytoskeleton
Engagement of LPA1 and LPA2 leads to activation of the Rho pathway, followed by activation of the Rho-associated kinase known to induce formation of stress fibers, retraction and cell rounding [48], and inhibition of neuronal migration [12]. In line, the proteomic approach applied here revealed pronounced changes in microglial expression levels of proteins, which are regulators of the cytoskeletal architecture and cell migration. Microglia and macrophages rely on dynamic changes in membrane cytoskeleton for their functions since in response to activation microglia undergo dramatic changes in cell shape, migration, and phagocytic properties [49].
ANXA1 is a member of the annexin protein superfamily, which binds to phospholipids in a Ca2+-dependent manner, thereby providing a link between Ca2+ signaling and membrane function (reviewed in [50]). ANXA1 is induced by glucocorticoids in inflammatory cells and mediates many of the anti-inflammatory actions of these drugs (reviewed in [51, 52]). In LPS-stimulated microglia, ANXA1 potently inhibits induction of inducible nitric oxide synthase and COX-2, which are major players in neurodegenerative disorders with an inflammatory component [53]. Of note, ANXA1 displays pronounced effects on migratory properties of immunocompetent cells. In a murine model of peritonitis full length ANXA1 and an N-terminal fragment inhibited leukocyte migration [54]. Reduced migration in response to ANXA1 binding was also reported for neutrophils [55] and monocytes in vitro [56] and in vivo [57]. Thus, LPA-mediated upregulation of ANXA1 levels could serve as a homing signal and blunt the inflammatory response of microglia commonly thought to drive neurodegeneration.
The nucleoside diphosphate kinase family is encoded by eight different genes in humans (nm23-H1 to nm23-H8) (reviewed in [58]). Here we show that NDKB (NM23-H2) is upregulated (3.9-fold) in response to LPA. NDKB was identified as an interacting partner of ICAP-1 and β1 integrins in spreading fibroblasts, an event that could (via Rac1 inhibition) increase Rho activity, favoring focal adhesion assembly and cell spreading [59], as observed in the present study (Fig. 4A and Fig. 6). Also, in human breast cancer cells (MCF-7) overexpression of NDKB resulted in a significant reduction of cell migration in Boyden chamber assays [60]. Thus, upregulation of NDKB would be compatible with reduced migration of microglia as observed here.
The assembly and disassembly of actin filaments and their organization in functional 3-D networks is under tight control of actin-binding proteins. In line, two potential candidates namely twinfilin-2 and WD repeat-containing protein 1 (WDR1) that could be involved in these processes were identified herein. Twinfilin is an evolutionary conserved actin-binding protein involved in motile and morphological processes [61]. In line with identification of twinfilin-2 in human microglia during the present study, it was demonstrated that twinfilin-2 (like twinfilin-1) mRNA is expressed in brain and both proteins are present at elevated levels in the mouse embryo brain around E14 [62].
Another pivotal mediator of actin dynamics is cofilin, which promotes actin filament severing and depolymerization acting in concert with actin interacting protein 1 (AIP1). The mammalian homologue of AIP1 is WDR1, which was upregulated by LPA in C13NJ cells (Table 2). Knockdown of WDR1 by RNA interference leads to pronounced alterations of cell morphology, resulting in mitotic cell flattening [63]. Most importantly functional WDR1 is critical for development and function of megakaryocytes and neutrophils [64].
Two proteins within group 2 (Table 2) were found to be downregulated, namely TBCB and vimentin. Over-expression of TBCB leads to depolymerization of microtubules, which is a result of a TBCB/TBCE interaction that greatly enhances the efficacy of the latter cofactor for tubulin dissociation [65]. It was also suggested that TBCB expression levels control microtubule densities in microglia where high TBCB levels are associated with a low density of microtubules, thus increasing actin-based cell migration [66]. This would be in line with findings obtained during the present study: LPA downregulated TBCB expression, resulting in higher β-tubulin immunofluorescence at centromers (Fig. 6B) and decreased cell migration (Fig. 7).
Several of the proteins identified within group 3 (Table 2) contribute to cell survival by preventing cell injury and/or death. Although not further investigated here, increasing evidence suggests that molecular chaperones provide a first line of defense against neurodegenerative disorders that involve defect protein folding.
Our findings suggest that LPA is a potent regulator of microglia protein expression and morphological plasticity. On the cellular level LPA induced Rho and ERK activation, which triggered increased cellular ATP production, alterations in the actin and tubulin cytoskeleton, and decreased migration. In agreement with increased LPA concentrations at sites of neuronal damage [15] our observations can be interpreted as a protective microglia response to CNS injury [58].
Supplementary Material
Acknowledgments
The authors are grateful to Drs. J. Mazella (Institut de Pharmacologie Moléculaire et Cellulaire, Valbonne, France) and M. Tardieu (Université Paris sud, le Kremlin-Bicétre, France) for providing C13NJ cells. Financial support was provided by the Austrian Science Fund (P19074-B05, P19424-B05, F3007, F3010) and the Austrian Research Promotion Agency (Bridge P810994).
Abbreviations
- ALDOA
aldolase A
- ANXA1
annexin A1
- BAG2
BAG family molecular chaperone regulator 2
- CDC37
Hsp90 co-chaperone Cdc37
- DAPI
4′,6-diamidino-2-phenyl-indol
- ENOA
enolase A
- ENPL
endoplasmin
- ERK1/2
extracellular signal-regulated kinase1/2
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GRP78
78 kDa glucose-regulated protein
- LPA
lysophosphatidic acid
- LPA1-3
lysophosphatidic acid receptors 1–3
- LSM
laser scanning microscopy
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PDI
protein disulfide-isomerase
- pERK
phosphorylated ERK
- qPCR
quantitative real-time PCR
- TBCB
tubulin folding cofactor B
- TPIS
triosephosphate isomerase
- WDR
WD repeat-containing protein
- YFP
yellow fluorescent protein
Footnotes
The authors have declared no conflict of interest.
References
- [1].Streit WJ. Microglia as neuroprotective, immunocompetent cells of the CNS. Glia. 2002;40:133–139. doi: 10.1002/glia.10154. [DOI] [PubMed] [Google Scholar]
- [2].Aldskogius H. Regulation of microglia-potential new drug targets in the CNS. Expert Opin. Ther. Targets. 2001;5:655–668. doi: 10.1517/14728222.5.6.655. [DOI] [PubMed] [Google Scholar]
- [3].Rezaie P, Trillo-Pazos G, Greenwood J, Everall IP, Male DK. Motility and ramification of human fetal microglia in culture: an investigation using time-lapse video microscopy and image analysis. Exp. Cell Res. 2002;274:68–82. doi: 10.1006/excr.2001.5431. [DOI] [PubMed] [Google Scholar]
- [4].Raivich G, Bohatschek M, Kloss CU, Werner A, et al. Neuroglial activation repertoire in the injured brain: graded response, molecular mechanisms and cues to physiological function. Brain Res. Brain Res. Rev. 1999;30:77–105. doi: 10.1016/s0165-0173(99)00007-7. [DOI] [PubMed] [Google Scholar]
- [5].Anliker B, Chun J. Lysophospholipid G protein-coupled receptors. J. Biol. Chem. 2004;279:20555–20558. doi: 10.1074/jbc.R400013200. [DOI] [PubMed] [Google Scholar]
- [6].Tigyi G, Hong L, Yakubu M, Parfenova H, et al. Lysophosphatidic acid alters cerebrovascular reactivity in piglets. Am. J. Physiol. 1995;268:H2048–H2055. doi: 10.1152/ajpheart.1995.268.5.H2048. [DOI] [PubMed] [Google Scholar]
- [7].Sun GY, Lu FL, Lin SE, Ko MR. Decapitation ischemia-induced release of free fatty acids in mouse brain Relationship with diacylglycerols and lysophospholipids. Mol. Chem. Neuropathol. 1992;17:39–50. doi: 10.1007/BF03159980. [DOI] [PubMed] [Google Scholar]
- [8].Ye X, Fukushima N, Kingsbury MA, Chun J. Lysophosphatidic acid in neural signaling. Neuroreport. 2002;13:2169–2175. doi: 10.1097/00001756-200212030-00002. [DOI] [PubMed] [Google Scholar]
- [9].Birgbauer E, Chun J. New developments in the biological functions of lysophospholipids. Cell. Mol. Life Sci. 2006;63:2695–2701. doi: 10.1007/s00018-006-6155-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Aoki J, Inoue A, Okudaira S. Two pathways for lysophosphatidic acid production. Biochim. Biophys. Acta. 2008;1781:513–518. doi: 10.1016/j.bbalip.2008.06.005. [DOI] [PubMed] [Google Scholar]
- [11].Schulze C, Smales C, Rubin LL, Staddon JM. Lysophosphatidic acid increases tight junction permeability in cultured brain endothelial cells. J. Neurochem. 1997;68:991–1000. doi: 10.1046/j.1471-4159.1997.68030991.x. [DOI] [PubMed] [Google Scholar]
- [12].Fukushima N, Weiner JA, Kaushal D, Contos JJ, et al. Lysophosphatidic acid influences the morphology and motility of young, postmitotic cortical neurons. Mol. Cell. Neurosci. 2002;20:271–282. doi: 10.1006/mcne.2002.1123. [DOI] [PubMed] [Google Scholar]
- [13].Contos JJ, Fukushima N, Weiner JA, Kaushal D, Chun J. Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior. Proc. Natl. Acad. Sci. USA. 2000;97:13384–13389. doi: 10.1073/pnas.97.24.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Inoue M, Rashid MH, Fujita R, Contos JJ, et al. Initiation of neuropathic pain requires lysophosphatidic acid receptor signaling. Nat. Med. 2004;10:712–718. doi: 10.1038/nm1060. [DOI] [PubMed] [Google Scholar]
- [15].Ueda H. Peripheral mechanisms of neuropathic pain – involvement of lysophosphatidic acid receptor-mediated demyelination. Mol. Pain. 2008;4:11. doi: 10.1186/1744-8069-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rao TS, Lariosa-Willingham KD, Lin FF, Palfreyman EL, et al. Pharmacological characterization of lysophospholipid receptor signal transduction pathways in rat cerebrocortical astrocytes. Brain Res. 2003;990:182–194. doi: 10.1016/s0006-8993(03)03527-3. [DOI] [PubMed] [Google Scholar]
- [17].Keller JN, Steiner MR, Holtsberg FW, Mattson MP, Steiner SM. Lysophosphatidic acid-induced proliferation-related signals in astrocytes. J. Neurochem. 1997;69:1073–1084. doi: 10.1046/j.1471-4159.1997.69031073.x. [DOI] [PubMed] [Google Scholar]
- [18].Weiner JA, Hecht JH, Chun J. Lysophosphatidic acid receptor gene vzg-1/lpA1/edg-2 is expressed by mature oligodendrocytes during myelination in the postnatal murine brain. J. Comp. Neurol. 1998;398:587–598. [PubMed] [Google Scholar]
- [19].Moller T, Musante DB, Ransom BR. Lysophosphatidic acid-induced calcium signals in cultured rat oligodendrocytes. Neuroreport. 1999;10:2929–2932. doi: 10.1097/00001756-199909290-00010. [DOI] [PubMed] [Google Scholar]
- [20].Moller T, Contos JJ, Musante DB, Chun J, Ransom BR. Expression and function of lysophosphatidic acid receptors in cultured rodent microglial cells. J. Biol. Chem. 2001;276:25946–25952. doi: 10.1074/jbc.M102691200. [DOI] [PubMed] [Google Scholar]
- [21].Schilling T, Repp H, Richter H, Koschinski A, et al. Lysophospholipids induce membrane hyperpolarization in microglia by activation of IKCa1 Ca(2+)-dependent K(+) channels. Neuroscience. 2002;109:827–835. doi: 10.1016/s0306-4522(01)00534-6. [DOI] [PubMed] [Google Scholar]
- [22].Schilling T, Stock C, Schwab A, Eder C. Functional importance of Ca2+-activated K+channels for lysophosphatidic acid-induced microglial migration. Eur. J. Neurosci. 2004;19:1469–1474. doi: 10.1111/j.1460-9568.2004.03265.x. [DOI] [PubMed] [Google Scholar]
- [23].Tham CS, Lin FF, Rao TS, Yu N, Webb M. Microglial activation state and lysophospholipid acid receptor expression. Int. J. Dev. Neurosci. 2003;21:431–443. doi: 10.1016/j.ijdevneu.2003.09.003. [DOI] [PubMed] [Google Scholar]
- [24].Janabi N, Peudenier S, Heron B, Ng KH, Tardieu M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci. Lett. 1995;195:105–108. doi: 10.1016/0304-3940(94)11792-h. [DOI] [PubMed] [Google Scholar]
- [25].Martin S, Vincent JP, Mazella J. Involvement of the neurotensin receptor-3 in the neurotensin-induced migration of human microglia. J. Neurosci. 2003;23:1198–1205. doi: 10.1523/JNEUROSCI.23-04-01198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Egger T, Schuligoi R, Wintersperger A, Amann R, et al. Vitamin E (αTocopherol) attenuates cyclooxygenase-2 transcription and synthesis in immortalized murine BV-2 microglia. Biochem. J. 2003;370:459–467. doi: 10.1042/BJ20021358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Furst W, Hallstrom S. Simultaneous determination of myocardial nucleotides, nucleosides, purine bases and creatine phosphate by ion-pair high-performance liquid chromatography. J. Chromatogr. 1992;578:39–44. doi: 10.1016/0378-4347(92)80222-c. [DOI] [PubMed] [Google Scholar]
- [28].Kratzer I, Wernig K, Panzenboeck U, Bernhart E, et al. Apolipoprotein A-I coating of protamine-oligonucleotide nanoparticles increases particle uptake and transcytosis in an in vitro model of the blood-brain barrier. J. Control. Release. 2007;117:301–311. doi: 10.1016/j.jconrel.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- [30].Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Van Brocklyn JR, Behbahani B, Lee NH. Homo-dimerization and heterodimerization of S1P/EDG sphingosine-1-phosphate receptors. Biochim. Biophys. Acta. 2002;1582:89–93. doi: 10.1016/s1388-1981(02)00141-5. [DOI] [PubMed] [Google Scholar]
- [32].Gardell SE, Dubin AE, Chun J. Emerging medicinal roles for lysophospholipid signaling. Trends Mol. Med. 2006;12:65–75. doi: 10.1016/j.molmed.2005.12.001. [DOI] [PubMed] [Google Scholar]
- [33].Wilde C, Genth H, Aktories K, Just I. Recognition of RhoA by Clostridium botulinum C3 exoenzyme. J. Biol. Chem. 2000;275:16478–16483. doi: 10.1074/jbc.M910362199. [DOI] [PubMed] [Google Scholar]
- [34].Carrettiero DC, Hernandez I, Neveu P, Papagiannakopoulos T, Kosik KS. The cochaperone BAG2 sweeps paired helical filament-insoluble tau from the microtubule. J. Neurosci. 2009;29:2151–2161. doi: 10.1523/JNEUROSCI.4660-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hoshino T, Nakaya T, Araki W, Suzuki K, et al. Endoplasmic reticulum chaperones inhibit the production of amyloid-beta peptides. Biochem. J. 2007;402:581–589. doi: 10.1042/BJ20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Spiliotis ET, Kinoshita M, Nelson WJ. A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science. 2005;307:1781–1785. doi: 10.1126/science.1106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Maraia RJ, Bayfield MA. The La protein-RNA complex surfaces. Mol. Cell. 2006;21:149–152. doi: 10.1016/j.molcel.2006.01.004. [DOI] [PubMed] [Google Scholar]
- [38].Carpenter B, MacKay C, Alnabulsi A, MacKay M, et al. The roles of heterogeneous nuclear ribonucleoproteins in tumour development and progression. Biochim. Biophys. Acta. 2006;1765:85–100. doi: 10.1016/j.bbcan.2005.10.002. [DOI] [PubMed] [Google Scholar]
- [39].Kim JY, Kim DY, Ro JY. Granule formation in NGF-cultured mast cells is associated with expressions of pyruvate kinase type M2 and annexin I proteins. Int. Arch. Allergy Immunol. 2008;146:287–297. doi: 10.1159/000121463. [DOI] [PubMed] [Google Scholar]
- [40].McArthur S, Yazid S, Christian H, Sirha R, et al. Annexin A1 regulates hormone exocytosis through a mechanism involving actin reorganization. FASEB J. 2009;23:4000–4010. doi: 10.1096/fj.09-131391. [DOI] [PubMed] [Google Scholar]
- [41].Song MS, Park YK, Lee JH, Park K. Induction of glucose-regulated protein 78 by chronic hypoxia in human gastric tumor cells through a protein kinase C-epsilon/ERK/AP-1 signaling cascade. Cancer Res. 2001;61:8322–8330. [PubMed] [Google Scholar]
- [42].Xu W, Liu L, Charles IG, Moncada S. Nitric oxide induces coupling of mitochondrial signalling with the endoplasmic reticulum stress response. Nat. Cell Biol. 2004;6:1129–1134. doi: 10.1038/ncb1188. [DOI] [PubMed] [Google Scholar]
- [43].Zhang H, Wang D, Sun H, Hall RA, Yun CC. MAGI-3 regulates LPA-induced activation of Erk and RhoA. Cell. Signal. 2007;19:261–268. doi: 10.1016/j.cellsig.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mizukami Y, Iwamatsu A, Aki T, Kimura M, et al. ERK1/2 regulates intracellular ATP levels through alpha-enolase expression in cardiomyocytes exposed to ischemic hypoxia and reoxygenation. J. Biol. Chem. 2004;279:50120–50131. doi: 10.1074/jbc.M402299200. [DOI] [PubMed] [Google Scholar]
- [45].Fujita R, Ma Y, Ueda H. Lysophosphatidic acid-induced membrane ruffling and brain-derived neurotrophic factor gene expression are mediated by ATP release in primary microglia. J. Neurochem. 2008;107:152–160. doi: 10.1111/j.1471-4159.2008.05599.x. [DOI] [PubMed] [Google Scholar]
- [46].Jung J, Yoon T, Choi EC, Lee K. Interaction of cofilin with triose-phosphate isomerase contributes glycolytic fuel for Na,K-ATPase via Rho-mediated signaling pathway. J. Biol. Chem. 2002;277:48931–48937. doi: 10.1074/jbc.M208806200. [DOI] [PubMed] [Google Scholar]
- [47].Weiss J, Hiltbrand B. Functional compartmentation of glycolytic versus oxidative metabolism in isolated rabbit heart. J. Clin. Invest. 1985;75:436–447. doi: 10.1172/JCI111718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jalink K, van Corven EJ, Hengeveld T, Morii N, et al. Inhibition of lysophosphatidate- and thrombin-induced neurite retraction and neuronal cell rounding by ADP ribosylation of the small GTP-binding protein Rho. J. Cell Biol. 1994;126:801–810. doi: 10.1083/jcb.126.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kreutzberg GW. Microglia: a sensor for pathological events in the CNS. Trends Neurosci. 1996;19:312–318. doi: 10.1016/0166-2236(96)10049-7. [DOI] [PubMed] [Google Scholar]
- [50].Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- [51].Parente L, Solito E. Annexin 1: more than an anti-phospholipase protein. Inflamm. Res. 2004;53:125–132. doi: 10.1007/s00011-003-1235-z. [DOI] [PubMed] [Google Scholar]
- [52].Buckingham JC, John CD, Solito E, Tierney T, et al. Annexin 1, glucocorticoids, and the neuroendocrine-immune interface. Ann. N. Y. Acad. Sci. 2006;1088:396–409. doi: 10.1196/annals.1366.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Minghetti L, Nicolini A, Polazzi E, Greco A, et al. Down-regulation of microglial cyclo-oxygenase-2 and inducible nitric oxide synthase expression by lipocortin 1. Br. J. Pharmacol. 1999;126:1307–1314. doi: 10.1038/sj.bjp.0702423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Getting SJ, Flower RJ, Perretti M. Inhibition of neutrophil and monocyte recruitment by endogenous and exogenous lipocortin 1. Br. J. Pharmacol. 1997;120:1075–1082. doi: 10.1038/sj.bjp.0701029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lim LH, Solito E, Russo-Marie F, Flower RJ, Perretti M. Promoting detachment of neutrophils adherent to murine postcapillary venules to control inflammation: effect of lipocortin 1. Proc. Natl. Acad. Sci. USA. 1998;95:14535–14539. doi: 10.1073/pnas.95.24.14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Solito E, Romero IA, Marullo S, Russo-Marie F, Weksler BB. Annexin 1 binds to U937 monocytic cells and inhibits their adhesion to microvascular endothelium: involvement of the alpha 4 beta 1 integrin. J. Immunol. 2000;165:1573–1581. doi: 10.4049/jimmunol.165.3.1573. [DOI] [PubMed] [Google Scholar]
- [57].Perretti M, Ingegnoli F, Wheller SK, Blades MC, et al. Annexin 1 modulates monocyte-endothelial cell interaction in vitro and cell migration in vivo in the human SCID mouse transplantation model. J. Immunol. 2002;169:2085–2092. doi: 10.4049/jimmunol.169.4.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Fournier HN, Albiges-Rizo C, Block MR. New insights into Nm23 control of cell adhesion and migration. J. Bioenerg. Biomembr. 2003;35:81–87. doi: 10.1023/a:1023450008347. [DOI] [PubMed] [Google Scholar]
- [59].Fournier HN, Dupe-Manet S, Bouvard D, Lacombe ML, et al. Integrin cytoplasmic domain-associated protein 1α (ICAP-1α) interacts directly with the metastasis suppressor nm23-H2, and both proteins are targeted to newly formed cell adhesion sites upon integrin engagement. J. Biol. Chem. 2002;277:20895–20902. doi: 10.1074/jbc.M200200200. [DOI] [PubMed] [Google Scholar]
- [60].Rayner K, Chen YX, Hibbert B, White D, et al. Discovery of NM23-H2 as an estrogen receptor beta-associated protein: Role in estrogen-induced gene transcription and cell migration. J. Steroid Biochem. Mol. Biol. 2008;108:72–81. doi: 10.1016/j.jsbmb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- [61].Paavilainen VO, Bertling E, Falck S, Lappalainen P. Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 2004;14:386–394. doi: 10.1016/j.tcb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- [62].Vartiainen MK, Sarkkinen EM, Matilainen T, Salminen M, Lappalainen P. Mammals have two twinfilin isoforms whose subcellular localizations and tissue distributions are differentially regulated. J. Biol. Chem. 2003;278:34347–34355. doi: 10.1074/jbc.M303642200. [DOI] [PubMed] [Google Scholar]
- [63].Fujibuchi T, Abe Y, Takeuchi T, Imai Y, et al. AIP1/WDR1 supports mitotic cell rounding. Biochem. Biophys. Res. Commun. 2005;327:268–275. doi: 10.1016/j.bbrc.2004.11.156. [DOI] [PubMed] [Google Scholar]
- [64].Kile BT, Panopoulos AD, Stirzaker RA, Hacking DF, et al. Mutations in the cofilin partner Aip1/Wdr1 cause autoinflammatory disease and macrothrombocytopenia. Blood. 2007;110:2371–2380. doi: 10.1182/blood-2006-10-055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kortazar D, Fanarraga ML, Carranza G, Bellido J, et al. Role of cofactors B (TBCB) and E (TBCE) in tubulin heterodimer dissociation. Exp. Cell Res. 2007;313:425–436. doi: 10.1016/j.yexcr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- [66].Fanarraga ML, Villegas JC, Carranza G, Castano R, Zabala JC. Tubulin cofactor B regulates microtubule densities during microglia transition to the reactive states. Exp. Cell Res. 2009;315:535–541. doi: 10.1016/j.yexcr.2008.10.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.