Significance
Interleukin (IL)-15 plays a role in diverse functions of cells involved in the protection against cancer and infections. IL-15 signal is delivered via a mechanism called trans-presentation, in which IL-15 expressed at the surface of presenting cells via the membrane-bound IL-15Rα chain, is presented during the interaction with responding cells expressing the transducing receptor. IL-15Rα can be cleaved from the surface of presenting cells and we wondered whether such a mechanism could participate in the regulation of IL-15 trans-presentation. By generating an uncleavable form of IL-15Rα, we found an unprecedented cytokine pathway in which the IL-15.IL-15Rα complex cleaved from presenting cells allows responding cells to internalize, store and use IL-15.IL-15Rα complex for their own proliferation and survival.
Keywords: autocrine, IL-2Rβ, cell–cell interaction, synapse, lymphocyte
Abstract
Interleukin (IL)-15 and its specific receptor chain, IL-15Rα, support the development of various effector cells, including NK and CD8 T cells via a mechanism called trans-presentation. Whereas the dynamic of trans-presentation has been shown to involve the recycling of IL-15Rα by presenting cells, the way responding cells integrate, or take advantage of this process has not been evaluated yet. To address this question, we set up a trans-presentation model using a membrane-bound IL-15.IL-15Rα fusion protein, and found that IL-15 is detectable within responding cells following IL-15 trans-presentation. The role of the proteolytic cleavage of IL-15Rα in this process was investigated by generating an uncleavable form of IL-15Rα. We showed that IL-15 entry into responding cells necessitates the cleavage of IL-15.IL-15Rα complex from the surface of IL-15 presenting cells, and observed that IL-15Rα cleavage is associated with a decrease of the duration of Stat5 signaling. Once separated from presenting cells, responding cells are able to recycle IL-15.IL-15Rα complexes via intracellular compartments, for residual proliferation in a time-limited manner. These studies define an unprecedented cytokine pathway in which the IL-15.IL-15Rα complex cleaved from presenting cells allows responding cells to internalize, store and use IL-15.IL-15Rα complex for their own proliferation and survival.
Cytokines are key molecules involved in a wide variety of lymphocyte functions, including differentiation, proliferation, activation and survival. The way they are regulated impacts on the cell responses and its consequences on the immune system. Interleukin (IL)-15 belongs to the IL-2 cytokine family, which uses in common the gamma chain receptor (CD132). IL-2 and IL-15 are closely related cytokines sharing the same heterodimeric transducing receptor made of IL-2/15Rβ (CD122) and CD132 (1). The specificity of action of each cytokine is conferred by their private alpha chain, IL-2Rα and IL-15Rα, respectively (2). Even structurally related, these two receptor chains differ in their potency to bind their cytokine. Whereas IL-2 binds to IL-2Rα-chain with a low affinity (Kd = 10 nM), IL-15 retains a high affinity for IL-15Rα (Kd = 50 pM). This high affinity allows IL-15 to act in trans-presentation, a mechanism in which membrane-bound IL-15.IL-15Rα complex interacts and delivers IL-15 signal efficiently to cells expressing the dimeric CD122/CD132 receptor (3). Trans-presentation has been shown to be the dominant mechanism delivering IL-15 signal in vivo, to support the development and the survival of NK cells and CD8 T cells (4, 5). Prior studies have described the mechanism of establishment of IL-15 trans-presentation in which IL-15 binds to IL-15Rα, playing a role of chaperone, within presenting cells prior from emerging to the cell surface (6). In this study, to mimic IL-15 and IL-15Rα preassociation, we created a fusion molecule that comprises the total IL-15Rα chain, including the transmembrane domain, covalently linked to IL-15 (wt.ILR). That molecule was routed to the cell surface allowing IL-15 to be trans-presented in the context of membrane anchored IL-15Rα, allowing us to study the fate of the membrane anchored IL-15.IL-15Rα complex following trans-presentation.
Numerous studies have described the high degree of regulation of IL-15 expression at transcriptional, translational as well as posttranslational levels. Our laboratory showed that soluble IL-15Rα (sIL-15Rα) could be generated by proteolytic cleavage, acting as a potent antagonist on cell expressing the trimeric IL-15Rα/CD122/CD132 receptor (7). In contrast, the soluble IL-15Rα could also potentiate IL-15 action, by reinforcing IL-15 binding capacity on the CD122/CD132 dimeric receptor (8–10). Thus, we asked whether such a mechanism could participate in the regulation of IL-15 trans-presentation. To address this question, we have identified amino acid residues critical for the cleavage of human IL-15Rα and generated an uncleavable form of this receptor. We have used this uncleavable receptor to examine the role of this proteolytic cleavage in IL-15 signaling.
Results and Discussion
IL-15 Trans-Presentation Is Followed by IL-15 Internalization by Responding Cells.
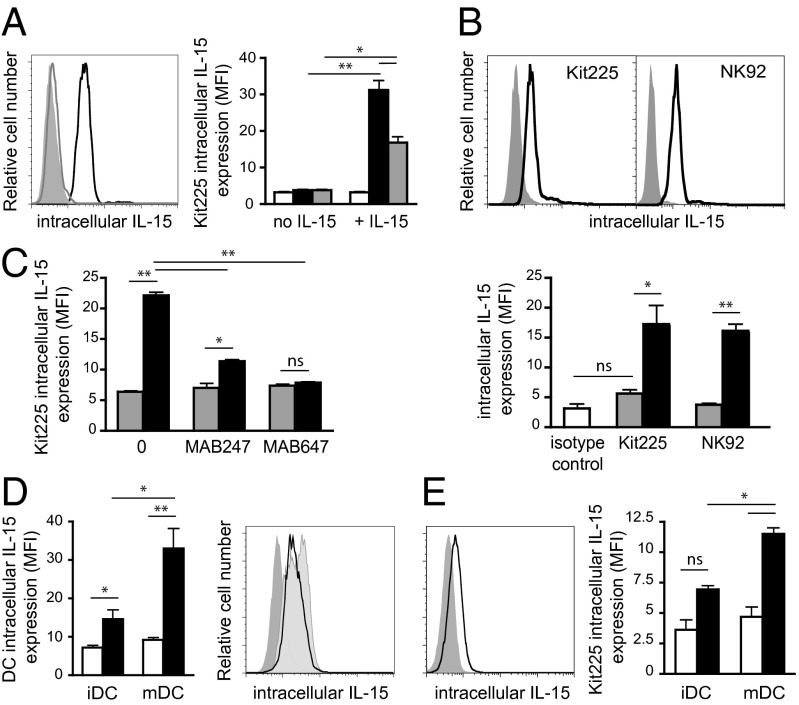
To investigate the fate of the IL-15.IL-15Rα complex following trans-presentation, stable transfected cell lines (HEK-293) were generated that expressed the membrane-bound IL-15Rα chain. These cells that do not express IL-15 mRNA (Fig. S1A) were cultured with IL-15 overnight to allow IL-15 binding to IL-15Rα and recycling of the complex to the cell surface (3). As expected, IL-15 was detectable by flow cytometry at their surface only after IL-15 loading (Fig. S1B). To study IL-15 trans-presentation, IL-15 loaded IL-15Rα transfected cells were washed to remove soluble IL-15 and cocultured with Kit225 cells for 24 h. Interestingly, intracellular IL-15 was detected by flow cytometry inside Kit225 cells and the intensity of fluorescence decreased when increasing the ratio of responding over IL-15 presenting cells, suggesting that Kit225 cells were competing for trans-presented IL-15. No IL-15 could be detected inside Kit225 cells in the absence of IL-15 loading (Fig. 1A). Kit225 cells also failed to express IL-15 mRNA (Fig. S1A).
Fig. 1.
IL-15 cytokine is detectable within responding cells following trans-presentation by IL-15 presenting cells. (A Left) Representative flow cytometric analysis of intracellular IL-15 expression within Kit225 responding cells following 24 h coculture with overnight IL-15 (100 pM) loaded IL-15Rα (black histogram) or unloaded IL-15Rα (gray histogram) HEK-293 presenting cells. Isotype control staining is shown as gray filled histogram. (A Right) Graph showing in MFI intracellular IL-15 staining within Kit225 responding cells at ratios of responding over presenting cells of 1:1 (black bars) or 10:1 (gray bars). White bars represent isotype control staining. (B) Representative flow cytometric analyses of intracellular IL-15 expression within Kit225 and NK92 responding cells following 24 h coculture with wt.ILR (black histogram) or wt.IL-15Rα (gray histogram) HEK-293 cells. Graph below shows intracellular staining of IL-15 in MFI, within annotated cells cocultured with wt.IL-15Rα cells (gray bars) or wt.ILR (black bars). (C) Flow cytometric analysis of intracellular IL-15 expression (MFI) within Kit225 responding cells following 24 h coculture with wt.ILR (black histogram) or wt.IL-15Rα (gray histogram) HEK-293 expressing cells in the presence or the absence of MAB247 or MAB647 anti–IL-15 blocking antibody. (D) Graph shows intracellular expression of IL-15 in MFI within immature and mature DCs (black bars). White bars represent the isotype control staining. (Right) Representative flow cytometric analysis of IL-15 intracellular expression within LPS matured DCs cultured for 24 h in the absence (pale gray filled histogram) or in the presence of Kit225 (black histogram). Isotype staining control is shown in gray filled histogram. (E) Representative flow cytometric analysis of IL-15 intracellular expression within Kit225 cocultured 24 h with mature DC. Isotype staining control is shown in gray filled histogram. Graph shows intracellular staining of IL-15 in MFI within Kit225 cocultured with immature and mature DC (black bars). White bars represent the isotype control staining. Graphs show a pool of at least three independent experiments. ns, not significant; **P < 0.01; ***P < 0.001.
IL-15 and IL-15Rα are preformed as complexes in presenting cells in response to stimulation before emerging to the cell surface (6). To mimic the process of IL-15.IL-15Rα preassociation within the presenting cells, we generated a membrane-bound fusion molecule on which IL-15 is covalently attached via a linker peptide to the N terminus of IL-15Rα. This IL-15-linker-IL-15Rα fusion molecule (herein referred as wt.ILR) was created also to rule out any dissociation of IL-15 as a soluble entity that could act through cis-presentation. IL-15 expression at the surface of transfected cells was checked by flow cytometry (Fig. S2B). Using a similar assay as below, wt.ILR transfected cells were cocultured with Kit225 or NK92 cells (ratio 1:1) and the location of IL-15 was analyzed by flow cytometry. We controlled that NK92 cells, like Kit225, did not express IL-15 mRNA (Fig. S1A). Surprisingly, after coculture with ILR expressing cells, both Kit225 and NK92 responding cells expressed intracellular IL-15 (Fig. 1B), suggesting that IL-15 was internalized together with its receptor. This IL-15 entry was strongly diminished by anti–IL-15 antibodies that block IL-15 binding to either the CD122 (MAB247) or CD132 (MAB647) IL-15 receptor chains (Fig. 1C). To investigate whether these observations could be reproduced with primary cells, we generated monocyte derived immature dendritic cells (iDCs) that were subsequently matured to mDCs by LPS. LPS treatment led to a significant increase of IL-15 expression (Fig. 1D) that could potentially be presented by the diverse spliced IL-15Rα isoforms recently described (11). Immature DCs and mDCs were cocultured for 24 h with Kit225 cells. No significant IL-15 could be detected on Kit225 cells after culture with iDCs, whereas significant intracellular IL-15 was observed after culture with mDCs (Fig. 1E). Moreover, the level of IL-15 expression on mDCs was simultaneously decreased, reinforcing the idea of a transfer of IL-15 from DCs to Kit225 cells (Fig. 1D, Right). Altogether, the results obtained with primary DCs validate the physiological relevance of our IL-15 trans-presentation model using IL-15Rα and ILR transfected cells.
IL-15 Entry Within Responding Cells Requires Cell–Cell Contact Rather Than Soluble IL-15.IL-15Rα Complexes Released by Proteolytic Cleavage.
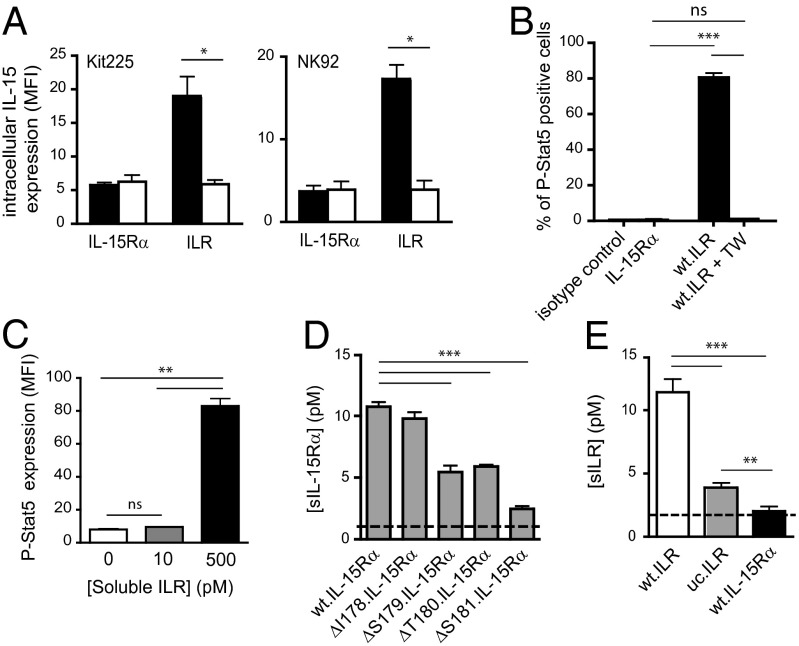
We next investigated whether the IL-15 protein detected inside responding cells was derived from soluble ILR (sILR) released into the culture medium after proteolytic cleavage of its membrane-bound counterpart, or from membrane-bound ILR during cell–cell interaction. To distinguish between these two alternatives, IL-15 presenting cells were physically separated from Kit225 or NK92 responding cells by a transwell insert (TW; Boyden chamber). No IL-15 was detected within either Kit225 or NK92 cells after 24 h of coculture with ILR expressing cells (Fig. 2A), minimizing the importance of the soluble IL-15.IL-15Rα complex released during the coculture. These results are in agreement with our previous studies (ref. 6 and Fig. S1C). IL-15 signaling following trans-presentation was then assessed by measuring Stat5 phosphorylation (12) within responding cells by flow cytometry. Whereas p-Stat5 was induced in Kit225 by coculturing with wt.ILR cells, no signal was detected within Kit225 when separated from wt.ILR using a TW insert, or when cocultured with IL-15Rα cells (Fig. 2B). This finding indicates that both IL-15 internalization and IL-15 signaling in this in vitro trans-presentation model rely predominantly on membrane-bound ILR.
Fig. 2.
Soluble IL-15 component released is not sufficient for IL-15 action: generation of an uncleavable form of IL-15Rα. (A) Flow cytometric analyses of intracellular IL-15 expression (MFI) within Kit225 and NK92 cells following 24 h coculture separated from wt.ILR or wt.IL-15Rα HEK-293 cells by an insert (white bars) or not (black bars). (B) Flow cytometric analysis of the percentage of p-Stat5 positive Kit225 cells following 1 h coculture with wt.ILR HEK-293 cells separated by an insert (white bar) or not (black bar). (C) Flow cytometric analysis of p-Stat5 expression (MFI) within Kit225 cells following 1 h stimulation with 10 pM and 500 pM of sILR. (D) ELISA detection of sIL-15Rα released from the different annotated point-mutated IL-15Rα HEK-293 transfected cells after 24 h of culture. Dashed line indicates the ELISA detection threshold (1 pM). (E) ELISA measurement of sILR released in the culture supernatant from wt.ILR, uc.ILR, and wt.IL-15Rα HEK-293 transfected cells after 24 h of culture. Dashed line indicates the ELISA detection threshold (2 pM). All data are representative of at least three separate experiments. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
In parallel, wt.ILR cell culture supernatants were checked for their ability to release soluble IL-15.IL-15Rα complex. These soluble components were detected in the supernatants at low concentrations (10 pM range) that were unable to induce Stat5 phosphorylation within Kit225 cells (Fig. 2E), compared with that induced by 500 pM sILR (Fig. 2C). Together, our data indicate that IL-15 trans-presentation leading to IL-15 internalization by responding cells requires cell–cell contact and is not mimicked by soluble IL-15.IL-15Rα complexes released in culture supernatant (3, 6).
To better understand the physiological importance of IL-15Rα shedding, we attempted to locate the sequence of IL-15Rα responsible for its proteolytic cleavage. Our previous studies revealed the existence of a soluble IL-15Rα protein, and the apparent molecular mass of this fragment suggested that the proteolytic cleavage site was located in IL-15Rα’s extracellular domain close to the plasma membrane (7). To precisely locate the proteolytic cleavage site, we assayed single amino acid deletions on IL-15Rα. IL-15Rα constructs were generated in which I178, S179, T180, and S181 were, respectively, deleted and transfected into HEK-293 cell line. Culture supernatants were harvested and sIL-15Rα concentrations were measured by radioactive ELISA. Whereas IL-15Rα shedding was not significantly affected by the I178 deletion, it was progressively reduced to reach a marked inhibition when S181 was deleted, without affecting IL-15Rα surface expression (Fig. 2D and Fig. S2A). This finding is comparable to that described for the cleavage of IL-6R (13). Recently, Chertova et al. identified G170 as the C terminus residue of the naturally cleaved IL-15Rα (14). However, G170 belongs to domain 5 of IL-15Rα, and deletion of that domain does not affect the release of soluble IL-15Rα (7). Interestingly, Lorenzen et al. (15) demonstrated that the membrane-proximal domain of ADAM17, involved in the shedding of several membrane-bound protein including IL-6R and IL-15Rα, is essential for recognition of the transmembrane proteins substrates. Altogether, these findings suggest that S181 could belong to a preferential anchoring site on IL-15Rα for its protease or that its deletion induces a conformational change in the membrane proximal domain of IL-15Rα that affects protease anchoring.
IL-15Rα Shedding Is Responsible for the Entry of IL-15 into Responding Cells.
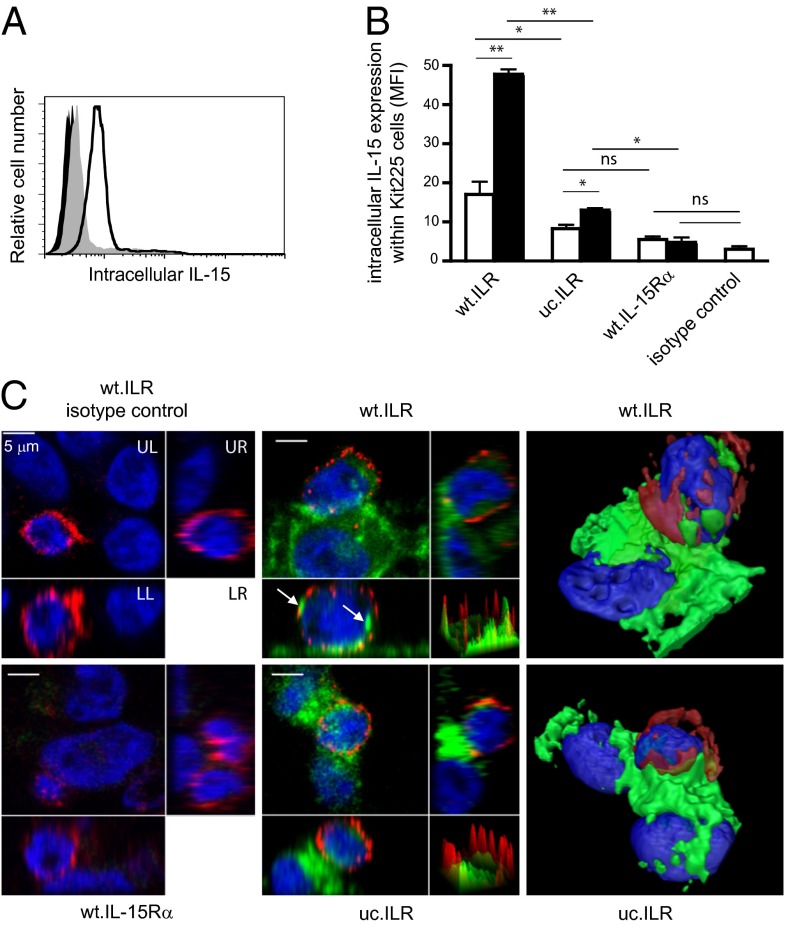
Our identification of S181 as a critical residue for supporting IL-15Rα cleavage provided a unique opportunity to address the role of IL-15Rα shedding in IL-15 internalization by responding cells during trans-presentation. We generated stably transfected cell lines expressing an uncleavable form of membrane-anchored ILR (uc.ILR). Whereas wt.ILR and uc.ILR cells expressed similar amounts of IL-15 at their surface (Fig. S2B), the S181 deletion strongly reduced the release of sILR (Fig. 2E). Using a radio-immunoassay measuring only free soluble IL-15Rα (7), no sIL-15Rα was detected in wt.ILR cell culture supernatant indicating that, even after cleavage of ILR from the membrane, IL-15 stays bound to IL-15Rα (Fig. S2C). In the trans-presentation assay, whereas a clear intracellular IL-15 signal was detected by flow cytometry inside Kit225 or NK92 cocultured with wt.ILR, it was significantly decreased by threefold at 24 h and fivefold at 48 h when cocultured with uc.ILR (Fig. 3 A and B and Fig. S2E). The same observation was made using IL-15 loaded wt.IL-15Rα and uc.IL-15Rα (Fig. S2D). However, IL-15 detection within responding cells cocultured with uc.ILR or IL-15 loaded uc.IL-15Rα was significantly higher compared with wt.IL-15Rα control (Fig. 3B and Fig. S2 D and E), suggesting that IL-15 entry is not exclusively dependent on IL-15Rα cleavage. This was confirmed by confocal microscopy. When wt.ILR cells were cocultured with CD4 positive Kit225, intracellular IL-15 was detected inside both cell types. Moreover, IL-15 stored into Kit225 seems gathered into clusters (Fig. 3C). By contrast, IL-15 protein expression inside Kit225 cocultured with uc.ILR cells was strongly reduced (Fig. 3C), indicating that IL-15Rα cleavage is crucial for IL-15 entry into responding cells. In contrast, polarization of IL-15 at the interface between presenting and responding cells (16, 17) did not seem to be affected by the deletion of the S181 residue. No signal was detected using isotypic control nor inside Kit225 cocultured with wt.IL-15Rα. Altogether, these data indicate that the proteolytic cleavage of IL-15Rα is not involved in IL-15 polarization but is essential for the entry of IL-15 inside responding cells.
Fig. 3.
Role of IL-15Rα shedding during IL-15 trans-presentation. (A) Representative flow cytometric analysis of intracellular IL-15 expression within Kit225 responding cells following 24 h coculture with wt.ILR (black histogram), uc.ILR (gray filled histogram), or wt.IL-15Rα (black filled histogram) HEK-293 presenting cells. (B) Graph shows a pool of three independent experiments of intracellular expression of IL-15 (MFI) within Kit225 cocultured for 24 h (white bars) and 48 h (black bars) with annotated cells. (C) Confocal microscopy analysis of intracellular IL-15 expression within Kit225 responding cells following 1-h coculture with wt.ILR, uc.ILR, or wt.IL-15Rα HEK-293 cells. Cells were stained with an Alexa-647–conjugated anti-CD4 Ab, fixed, permeablilzed and stained with a FITC–conjugated anti–IL-15 Ab. Each picture is divided in four quadrants: UL shows Z-project XY view; orthogonal views are shown on UR (YZ plane) and LL (XZ plane); and surface plot of XY view on LR (peak = pixel IF). Arrows show IL-15 clusters within Kit225. The two right panels show pictures from a 3D reconstitution. ns, not significant; *P < 0.05; **P < 0.01.
Abrogation of IL-15Rα Shedding Leads to Prolonged Stat5 Signaling.
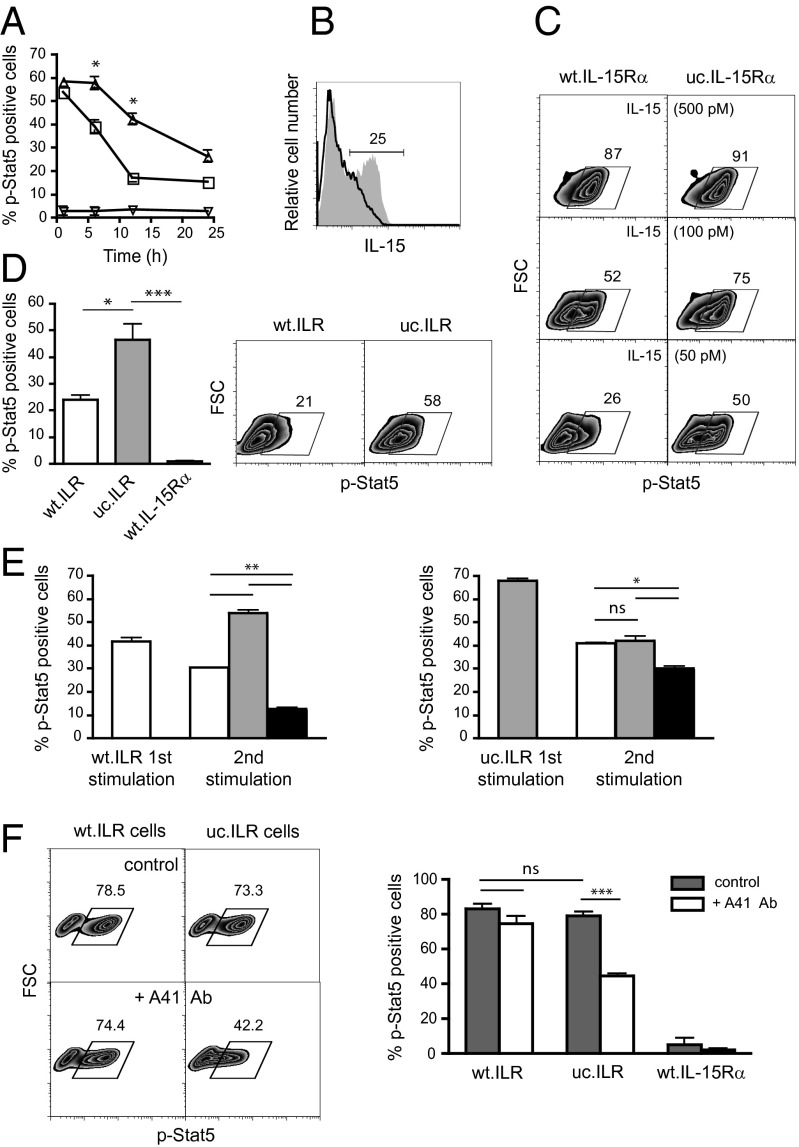
IL-15Rα cleavage defective cells were further used to investigate the role of this cleavage in trans-presentation associated signaling. Kit225 cells were added to wt.IL-15Rα or uc.IL-15Rα previously loaded with 100 pM IL-15 and their Stat5 phosphorylation was analyzed by flow cytometry over a 24 h period. P-Stat5 was maintained at a significantly higher level under IL-15 trans-presentation by uc.IL-15Rα than by wt.IL-15Rα (Fig. 4A). As a negative control, no Stat5 phosphorylation was detectable when Kit225 were cultured with wt.IL-15Rα. That observation was concomitant with a higher maintenance at 24 h of IL-15 expression at the surface of uc.IL-15Rα than of wt.IL-15Rα (Fig. 4B). To further test the impact of the concentration of IL-15 at the surface of presenting cells, wt.IL-15Rα or uc.IL-15Rα were loaded with increasing concentrations of IL-15. At high (500 pM) IL-15 loading concentration, the percentage of p-Stat5 positive Kit225 induced by wt.IL-15Rα was similar to that induced by uc.IL-15Rα, whereas it was lower by 30% and 50%, respectively, when lower (100 pM and 50 pM) IL-15 loading concentrations were used (Fig. 4C). This finding suggests that IL-15Rα proteolytic cleavage leads to a limitation of Stat5 phosphorylation caused by an impoverishment of IL-15 at the surface of presenting cells as a result of IL-15 internalization by responding cells. Then, we investigated the impact of the proteolytic cleavage in the context of ILR constructs. Kit225 cells were cocultured with wt.ILR or uc.ILR cells for 1 h and analyzed for p-Stat5 by flow cytometry. No difference in the level of p-Stat5 was observed (Fig. 4F), suggesting that internalization of IL-15 is not necessary for its signaling. However, 45% of Kit225 remained positive for Stat5 phosphorylation after 24 h of coculture with uc.ILR cells compared with 23% with wt.ILR cells (Fig. 4D, Left). As a negative control, no Stat5 phosphorylation was detectable when Kit225 were cultured 1 h or 24 h with wt.IL-15Rα cells. Thus, IL-15Rα cleavage leads to an attenuation of IL-15 signaling with time.
Fig. 4.
Differential dynamics of activation. (A) Flow cytometric analysis of p-Stat5 within Kit225 cells cocultured for indicated time points with transiently transfected wt.IL-15Rα (open squares) or uc.IL-15Rα (open triangles) HeLa cells loaded with 100 pM IL-15 or not (open inverted triangles). (B) Representative flow cytometric analysis of IL-15 expression at the surface of transiently transfected wt.IL-15Rα or uc.IL-15Rα HeLa cells loaded with 100 pM IL-15 after 24 h. (C and D) Representative flow cytometric analysis of p-Stat5 within Kit225 cells cocultured for 24 h with wt.IL-15Rα or uc.IL-15Rα loaded with 50, 100, and 500 pM IL-15 (C) or with wt.ILR, uc.ILR and IL-15Rα HeLa cells (D). Graph in D, Left shows a pool of four independent experiments. (E) Flow cytometric analysis of p-Stat5 within Kit225 cells prestimulated for 3 h with wt.ILR (Left) or uc.ILR HeLa cells (Right) and restimulated for 1 h with wt.ILR (white bars), uc.ILR (gray bars) or wt.IL-15Rα (black bars) HeLa cells. (F Left) Representative flow cytometric analysis of p-Stat5 within Kit225 responding cells, pretreated with or without A41 anti-CD122 antibody, then washed to remove unbound antibody, and cocultured for 1 h with wt.ILR, uc.ILR or wt.IL-15Rα HEK-293 presenting cells. (F Right) Graph shows a pool of at least three independent experiments. Numbers shown in zebra plots indicate the percentage of cells in the indicated gates. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
To further examine the impact of IL-15Rα cleavage on the kinetics of IL-15 signaling, we set up a reactivation assay in which Kit225 were stimulated for 3 h with wt.ILR or uc.ILR, and after washing, restimulated with wt.ILR, uc.ILR, or wt.IL-15Rα for 1 h. A higher Stat5 phosphorylation level was confirmed when Kit225 cells were stimulated with uc.ILR compared with wt.ILR (68 vs. 42%, respectively) (Fig. 4E). When stimulated with wt.ILR and restimulated in the absence of IL-15 (wt.IL-15Rα cells), the percentage of p-Stat5 positive Kit225 dropped from 42% to 12%. As expected, when restimulation was done with wt.ILR, this percentage was maintained to 30%. However, if the cells were restimulated with uc.ILR, it was increased to 54%, indicating again that uc.ILR was able to mobilize more efficiently p-Stat5 than does wt.ILR. In the case of initial stimulation with uc.ILR, restimulation with either wt.ILR or uc.ILR sustained similarly p-Stat5 level to 40%. Altogether, these data indicate that proteolytic cleavage of IL-15Rα during trans-presentation leads to a rapid decrease of Stat5 phosphorylation within responding cells.
The role of CD122 on IL-15 trans-presentation was investigated using the A41 anti-CD122 monoclonal antibody that has been shown to block soluble IL-15 signaling through the CD122/CD132 heterodimeric receptor, but not IL-15 cis-presentation through the IL-15Rα/CD122/CD132 trimeric receptor (18). Kit225 cells were pretreated with A41, then washed to remove unbound antibody and cocultured with wt.ILR, uc.ILR, or wt.IL-15Rα cells for 1 h before being stained for p-Stat5. Surprisingly, the A41 antibody was unable to block signaling by wt.ILR, whereas it significantly reduced signaling when IL-15 was trans-presented by uc.ILR (Fig. 4F). This finding suggests that in the absence of IL-15Rα cleavage, IL-15 trans-presentation is acting through the CD122/CD132 heterodimeric receptor, whereas the proteolytic cleavage of IL-15Rα makes IL-15 trans-presentation mimics IL-15 cis-presentation through IL-15Rα/CD122/CD132. In the case of the IL-2 system, a sequential assembly of the cytokine to its different receptor chains has been depicted (19, 20), where initial IL-2 binding to IL-2Rα induces a local conformational change in IL-2 that increases its affinity for CD122. Similarly, IL-15 binds first to IL-15Rα, that association however occurring within the cells prior from emerging at the cell surface, and undergoes an increased affinity for CD122 (6). This allosteric effect, due to a conformational change in the structure of the IL-15.IL-15Rα complex, was shown to be required for efficient IL-15 trans-presentation (21). Our results suggest that following IL-15 trans-presentation, IL-15Rα cleavage would allow a conformational change in the positioning of the cleaved IL-15Rα within the high-affinity quaternary complex resembling the cis-presentation mode and rendering the A41 binding epitope unavailable.
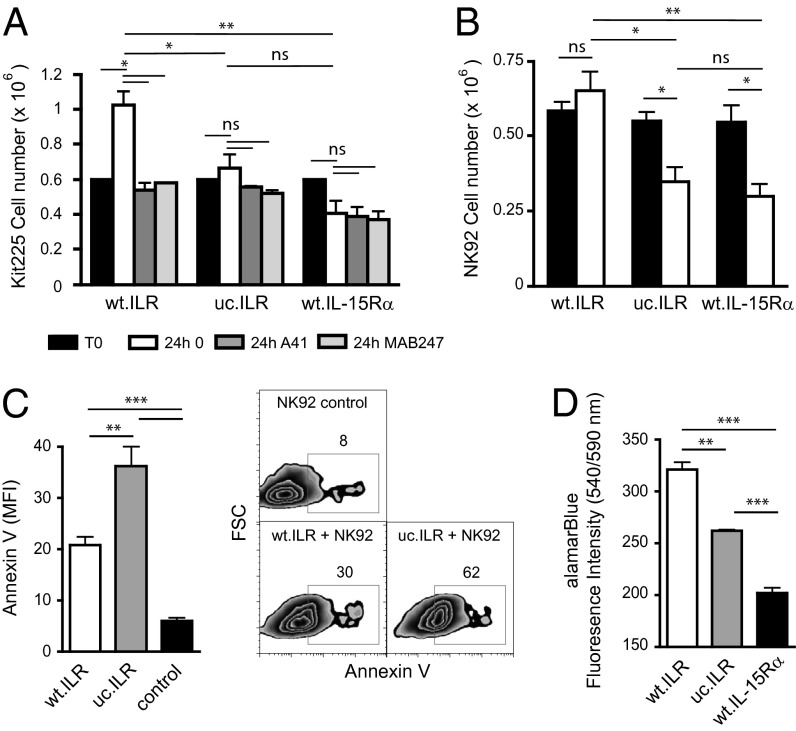
IL-15 Within Responding Cells Is Essential for Prolonged Cell Survival by Maintaining a Residual Proliferation.
We next tested whether and how IL-15 that entered into responding cells following trans-presentation could impact the behavior of these cells. Kit225 or NK92 cells were cocultured with wt.ILR, uc.ILR, or wt.IL-15Rα overnight to allow them to store IL-15.IL-15Rα complexes (Fig. S2F). They were then separated from the adherent presenting cells, cultured alone in new medium, and counted 24 h after separation. The number of Kit225 cells increased by twofold when previously cocultured in the presence of wt.ILR cells, whereas it did not change significantly when cultured with either uc.ILR or wt.IL-15Rα cells (Fig. 5A). In the case of NK92 cells, their number was maintained when precultured with wt.ILR, whereas it decreased significantly by around twofold after uc.ILR or IL-15Rα cells preculture (Fig. 5B). The same tendency was observed when Kit225 or NK92 were cultured overnight in the presence of sILR, washed, and quantitated at different time points to follow proliferation (Fig. S3A and ref. 22). Thus, the process of IL-15Rα cleavage linked to trans-presentation and subsequent IL-15.IL-15Rα internalization supports the proliferation and survival of responding cells after their separation from presenting cells. This effect was fully abrogated by A41 Ab (Fig. 5A), suggesting that it involved an extracellular factor operating through CD122. Given the fact that it was also inhibited by an anti–IL-15 Ab, it is likely due to soluble ILR released by responding cells. Together, these data suggest that once IL-15 responding cells are separated from presenting cells, they switch to another mechanism in which they recycle IL-15.IL-15Rα complexes, previously collected during trans-presentation to continue to proliferate and survive. This recycling process was further documented at the signaling level (Fig. S3 B and C). The percentage of p-Stat5 positive Kit225 cells observed at 1 h after Kit225/wt.ILR separation was completely abrogated by the A41 Ab added after cell separation, further indicating that the IL-15.IL-15Rα complex recycling process leading to residual proliferation involves CD122 and Stat5 phosphorylation. This recycling process was also shown to improve cell survival. Indeed, the number of Annexin V-positive responding cells was increased by twofold when precultured in the presence of uc.ILR, compared with wt.ILR (Fig. 5C). Moreover, as assessed by alamarBlue coloration, NK92 precultured with wt.ILR cells were almost twofold more metabolically active than when precultured with uc.ILR cells (Fig. 5D). Thus, IL-15.IL-15Rα complexes stored during physical contact lead to improved survival as well as proliferation of responding cells once separated from an IL-15 presenting source. This mechanism could allow the cells to migrate and establish multiple contacts with other IL-15 presenting cells such as mDCs and macrophages (23). During trans-presentation, as discussed above, a physical contact between presenting and responding cells is the dominant mechanism, and this recycling autocrine process probably plays a secondary role. However, the advantage for responding cells of an IL-15Rα cleavage process leading to such a time controlled autocrine IL-15.IL-15Rα pathway could be to protect them from overexposure to IL-15 signaling. Indeed, overexpression of IL-15 in transgenic mice could lead to fatal leukemia with a T and NK phenotype (24). Uncontrolled expression of IL-15 was described in numerous pathological conditions involving IL-15 autocrine loops (25–30). Moreover, soluble IL-15Rα, detected in patients suffering from T-LGL leukemia, could participate to the pathogenesis by decreasing IL-15 response threshold (31). Altogether, this reinforces the idea of a highly regulated system to protect from such malignant consequences.
Fig. 5.
Intracellular IL-15 collected by responding cells is essential for a prolonged effect on cell proliferation and survival after cytokine withdrawal. (A and B) Kit225 cells (A) or NK92 (B) were cocultured overnight with wt.ILR, uc.ILR or wt.IL-15Rα transiently transfected HeLa cells. Then, responding cells were separated from IL-15 presenting cells and cultured with or without A41 anti-CD122 or MAB247 anti–IL-15 antibodies. Cells were counted 24 h later. (C) NK92 were cocultured overnight with wt.ILR or uc.ILR transiently transfected HeLa cells. Then, NK92 cells were isolated from IL-15 presenting cells using CD56 Ab positive selection and cultured for 24 h before being stained for Annexin V and analyzed by flow cytometry. Numbers indicate the percentage of cells in the indicated gates. (D) After NK92 isolation, alamarBlue dye was added to culture media for 24 h. Cell health was analyzed by measuring fluorescence intensity. Graph shows a pool of at least two independent experiments. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001.
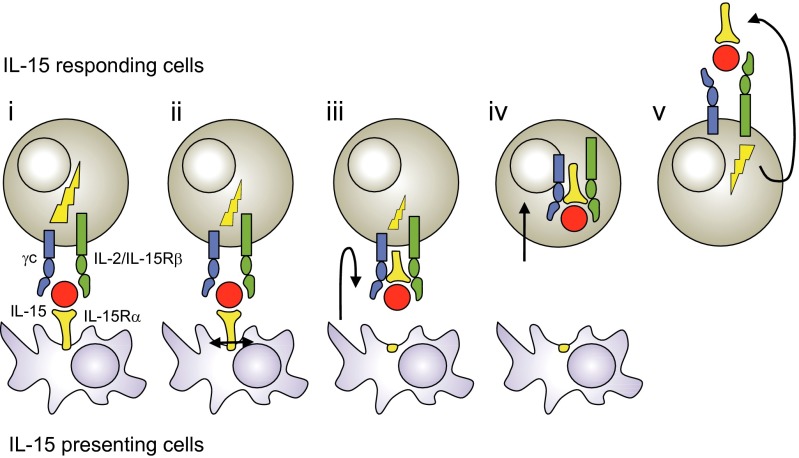
In summary, we have identified an unprecedented cytokine pathway by which IL-15.IL-15Rα complexes expressed by presenting cells are internalized following trans-presentation within responding cells. That process is dependent on the proteolytic cleavage of IL-15Rα, which allows IL-15.IL-15Rα complexes to separate from presenting cells and enter into responding cells via the CD122/CD132 receptor. Once inside the responding cells, the IL-15.IL-15Rα complexes accumulate during the time of cell–cell interaction. This process allows responding cells, when separated from presenting cells, to recycle temporarily IL-15.IL-15Rα complexes, previously stored, for residual proliferation (Fig. 6). These findings extend our understanding of the complex mode of action of IL-15, and highlight how the immune system can regulate the delivery of a cytokine signal in a highly compartmentalized and time-regulated fashion.
Fig. 6.
Proposed model of IL-15 trans-presentation in five steps. IL-15 and IL-15Rα are preassociated within presenting cells prior from emerging to the cell surface. (i) IL-15 is trans-presented by IL-15Rα at the surface of presenting cells to IL-15 responding cells expressing CD122/CD132 dimeric receptor. (ii) Membrane-bound IL-15.IL-15Rα complexes are cleaved following trans-presentation. (iii) The IL-15 receptor undergoes conformational change. (iv) IL-15.IL-15Rα complexes are internalized inside responding cells (v) and recycled for residual proliferation of responding cells. Signaling during IL-15 trans-presentation (lightning symbol) decreases following IL-15Rα cleavage.
Materials and Methods
Cell Lines, Cytokines, Antibodies.
The Kit225 and NK92 cell lines were used in that study. A41 anti-human CD122 antibody was reported previously (32). See SI Materials and Methods.
Molecular Constructs and Transfections.
For mutagenesis, deletions were realized using sequences containing the desired deletion and Quick Change Site-Directed Mutagenesis kit (Stratagene) protocol. After amplification, the sequences were ligated to pcDNA3.1/myc-His mammalian expression vector (Invitrogen) and sequenced. The ILR molecule that comprises the IL-15Rα linked to IL-15 was generated as described (8). See SI Materials and Methods.
For transfection, cells were transfected following a standard PEI Polyplus transfection protocol. For stable transfections, cells were cloned using a FACS Aria III cytometer (BD Biosciences).
Soluble IL-15Rα and ILR Quantification.
The quantification of sIL-15Rα and sILR was determined by Sandwich Radio-Immuno-Assay as described (7). See SI Materials and Methods.
Flow Cytometry and Immunofluorescence.
Intracellular detection of IL-15, p-Stat5 and annexin V staining (BD Biosciences) were assessed according to manufacturer instructions. Cells were analyzed with a Calibur cytometer (BD Biosciences) and FlowJo software. Fluorescent images were acquired using a Nikon A1 RS confocal microscope and analyzed with FIJI and Amira softwares. See SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Averil Ma and Dr. Nidia Alvarez Rueda for critically reading this manuscript, Philippe Hulin for technical assistance, and PiCell and CytoCell facilities. This work was supported by Inserm, Centre National de la Recherche Scientifique, and Agence Nationale de la Recherche. Y.J. and E.M. are supported by the Ligue Nationale Contre le Cancer (LNCC) as an Equipe Labellisée. E.M. was supported by the Association pour la Recherche sur le Cancer, and F.T. was supported by a fellowship from the LNCC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405514111/-/DCSupplemental.
References
- 1.Giri JG, et al. IL-15, a novel T cell growth factor that shares activities and receptor components with IL-2. J Leukoc Biol. 1995;57(5):763–766. doi: 10.1002/jlb.57.5.763. [DOI] [PubMed] [Google Scholar]
- 2.Giri JG, et al. Identification and cloning of a novel IL-15 binding protein that is structurally related to the α chain of the IL-2 receptor. EMBO J. 1995;14(15):3654–3663. doi: 10.1002/j.1460-2075.1995.tb00035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubois S, Mariner J, Waldmann TA, Tagaya Y. IL-15Ralpha recycles and presents IL-15 In trans to neighboring cells. Immunity. 2002;17(5):537–547. doi: 10.1016/s1074-7613(02)00429-6. [DOI] [PubMed] [Google Scholar]
- 4.Burkett PR, et al. Coordinate expression and trans presentation of interleukin (IL)-15Ralpha and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. J Exp Med. 2004;200(7):825–834. doi: 10.1084/jem.20041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mortier E, et al. Macrophage- and dendritic-cell-derived interleukin-15 receptor alpha supports homeostasis of distinct CD8+ T cell subsets. Immunity. 2009;31(5):811–822. doi: 10.1016/j.immuni.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 6.Mortier E, Woo T, Advincula R, Gozalo S, Ma A. IL-15Ralpha chaperones IL-15 to stable dendritic cell membrane complexes that activate NK cells via trans presentation. J Exp Med. 2008;205(5):1213–1225. doi: 10.1084/jem.20071913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortier E, Bernard J, Plet A, Jacques Y. Natural, proteolytic release of a soluble form of human IL-15 receptor alpha-chain that behaves as a specific, high affinity IL-15 antagonist. J Immunol. 2004;173(3):1681–1688. doi: 10.4049/jimmunol.173.3.1681. [DOI] [PubMed] [Google Scholar]
- 8.Mortier E, et al. Soluble interleukin-15 receptor alpha (IL-15R alpha)-sushi as a selective and potent agonist of IL-15 action through IL-15R beta/gamma. Hyperagonist IL-15 x IL-15R alpha fusion proteins. J Biol Chem. 2006;281(3):1612–1619. doi: 10.1074/jbc.M508624200. [DOI] [PubMed] [Google Scholar]
- 9.Rubinstein MP, et al. Converting IL-15 to a superagonist by binding to soluble IL-15Ralpha. Proc Natl Acad Sci USA. 2006;103(24):9166–9171. doi: 10.1073/pnas.0600240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoklasek TA, Schluns KS, Lefrançois L. Combined IL-15/IL-15Ralpha immunotherapy maximizes IL-15 activity in vivo. J Immunol. 2006;177(9):6072–6080. doi: 10.4049/jimmunol.177.9.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller JR, Waldmann TA, Kruhlak MJ, Dubois S. Paracrine and transpresentation functions of IL-15 are mediated by diverse splice versions of IL-15Rα in human monocytes and dendritic cells. J Biol Chem. 2012;287(48):40328–40338. doi: 10.1074/jbc.M112.378612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by γ(c) family cytokines. Nat Rev Immunol. 2009;9(7):480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müllberg J, et al. The soluble human IL-6 receptor. Mutational characterization of the proteolytic cleavage site. J Immunol. 1994;152(10):4958–4968. [PubMed] [Google Scholar]
- 14.Chertova E, et al. Characterization and favorable in vivo properties of heterodimeric soluble IL-15·IL-15Rα cytokine compared to IL-15 monomer. J Biol Chem. 2013;288(25):18093–18103. doi: 10.1074/jbc.M113.461756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenzen I, et al. The membrane-proximal domain of A Disintegrin and Metalloprotease 17 (ADAM17) is responsible for recognition of the interleukin-6 receptor and interleukin-1 receptor II. FEBS Lett. 2012;586(8):1093–1100. doi: 10.1016/j.febslet.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 16.Barreira da Silva R, Graf C, Münz C. Cytoskeletal stabilization of inhibitory interactions in immunologic synapses of mature human dendritic cells with natural killer cells. Blood. 2011;118(25):6487–6498. doi: 10.1182/blood-2011-07-366328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brilot F, Strowig T, Roberts SM, Arrey F, Münz C. NK cell survival mediated through the regulatory synapse with human DCs requires IL-15Ralpha. J Clin Invest. 2007;117(11):3316–3329. doi: 10.1172/JCI31751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehours P, et al. Subunit structure of the high and low affinity human interleukin-15 receptors. Eur Cytokine Netw. 2000;11(2):207–215. [PubMed] [Google Scholar]
- 19.Rickert M, Wang X, Boulanger MJ, Goriatcheva N, Garcia KC. The structure of interleukin-2 complexed with its alpha receptor. Science. 2005;308(5727):1477–1480. doi: 10.1126/science.1109745. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Rickert M, Garcia KC. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 2005;310(5751):1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 21.Ring AM, et al. Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat Immunol. 2012;13(12):1187–1195. doi: 10.1038/ni.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perdreau H, et al. Different dynamics of IL-15R activation following IL-15 cis- or trans-presentation. Eur Cytokine Netw. 2010;21(4):297–307. doi: 10.1684/ecn.2010.0207. [DOI] [PubMed] [Google Scholar]
- 23.Beuneu H, et al. Dynamic behavior of NK cells during activation in lymph nodes. Blood. 2009;114(15):3227–3234. doi: 10.1182/blood-2009-06-228759. [DOI] [PubMed] [Google Scholar]
- 24.Fehniger TA, et al. Fatal leukemia in interleukin 15 transgenic mice follows early expansions in natural killer and memory phenotype CD8+ T cells. J Exp Med. 2001;193(2):219–231. doi: 10.1084/jem.193.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burton JD, et al. A lymphokine, provisionally designated interleukin T and produced by a human adult T-cell leukemia line, stimulates T-cell proliferation and the induction of lymphokine-activated killer cells. Proc Natl Acad Sci USA. 1994;91(11):4935–4939. doi: 10.1073/pnas.91.11.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azimi N, et al. Human T cell lymphotropic virus type I Tax protein trans-activates interleukin 15 gene transcription through an NF-kappaB site. Proc Natl Acad Sci USA. 1998;95(5):2452–2457. doi: 10.1073/pnas.95.5.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azimi N, Jacobson S, Leist T, Waldmann TA. Involvement of IL-15 in the pathogenesis of human T lymphotropic virus type I-associated myelopathy/tropical spastic paraparesis: Implications for therapy with a monoclonal antibody directed to the IL-2/15R beta receptor. J Immunol. 1999;163(7):4064–4072. [PubMed] [Google Scholar]
- 28.Sato N, et al. Development of an IL-15-autocrine CD8 T-cell leukemia in IL-15-transgenic mice requires the cis expression of IL-15Rα. Blood. 2011;117(15):4032–4040. doi: 10.1182/blood-2010-09-307504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kukita T, et al. Autocrine and/or paracrine growth of adult T-cell leukaemia tumour cells by interleukin 15. Br J Haematol. 2002;119(2):467–474. doi: 10.1046/j.1365-2141.2002.03813.x. [DOI] [PubMed] [Google Scholar]
- 30.Tinhofer I, Marschitz I, Henn T, Egle A, Greil R. Expression of functional interleukin-15 receptor and autocrine production of interleukin-15 as mechanisms of tumor propagation in multiple myeloma. Blood. 2000;95(2):610–618. [PubMed] [Google Scholar]
- 31.Chen J, et al. Increased serum soluble IL-15Rα levels in T-cell large granular lymphocyte leukemia. Blood. 2012;119(1):137–143. doi: 10.1182/blood-2011-04-346759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaluza B, Betzl G, Shao H, Diamantstein T, Weidle UH. A general method for chimerization of monoclonal antibodies by inverse polymerase chain reaction which conserves authentic N-terminal sequences. Gene. 1992;122(2):321–328. doi: 10.1016/0378-1119(92)90221-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.