Abstract
Western European coastal sand dunes are highly important for nature conservation. Communities of the creeping willow (Salix repens) represent one of the most characteristic and diverse vegetation types in the dunes. We report here the results of the first kingdom-wide fungal diversity assessment in S. repens coastal dune vegetation. We carried out massively parallel pyrosequencing of ITS rDNA from soil samples taken at ten sites in an extended area of joined nature reserves located along the North Sea coast of the Netherlands, representing habitats with varying soil pH and moisture levels. Fungal communities in Salix repens beds are highly diverse and we detected 1211 non-singleton fungal 97% sequence similarity OTUs after analyzing 688,434 ITS2 rDNA sequences. Our comparison along a north-south transect indicated strong correlation between soil pH and fungal community composition. The total fungal richness and the number OTUs of most fungal taxonomic groups negatively correlated with higher soil pH, with some exceptions. With regard to ecological groups, dark-septate endophytic fungi were more diverse in acidic soils, ectomycorrhizal fungi were represented by more OTUs in calcareous sites, while detected arbuscular mycorrhizal genera fungi showed opposing trends regarding pH. Furthermore, we detected numerous red listed species in our samples often from previously unknown locations, indicating that some of the fungal species currently considered rare may be more abundant in Dutch S. repens communities than previously thought.
Introduction
Fungi represent one of the largest groups of living organisms. However, fungi are still poorly understood and appreciated compared to plants and animals and our knowledge of fungal diversity lags far behind [1]. Approximately 100,000 species of Eumycota (the true Fungi) have been described. Already well before the routine use of DNA sequencing in fungal biodiversity assessments, their true diversity was estimated to be around 1.5 million species [2], while more recent estimates suggests there may be 0.7 to 5 million species [3],[4]. Despite the differences among various estimates, it is clear that we currently know only a fraction of the total fungal biodiversity.
The task of discovering a significant portion of yet unknown species has only recently become possible with the advent of high-throughput DNA sequencing of environmental samples that has enormous potential to further boost data acquisition in biodiversity research [5],[6],[7],[8],[9],[10],[11],[12],[13],[14],[15],[16]. This methodology involves identification of multiple species from environmental samples using targeted loci specifically selected with the purpose of identification. These loci are typically the same ones that are used in specimen-based large-scale DNA barcoding efforts (e.g., [17],[18]), hence providing both appropriate power of resolution for the group of interest and maximum amount of reference sequences available from vouchered specimens.
The fungal diversity assessment presented here focuses on Salix repens L. sand dune communities along the North Sea coast, because these areas are highly important for nature conservation, water resource management, and recreational purposes. Coastal sand dunes include an outstanding ecological diversity in terms of environmental heterogeneity and species composition [19]. In the Netherlands, they are home to more than half of all higher plant species, in spite of covering only 1% of the land area [20],[21]. In Europe, coastal dune areas have been reduced by ca. 70% in the last century [22], and biodiversity in the remaining dune ecosystems is highly endangered [23]. In the Netherlands, however, coastal dunes have been relatively well preserved, with less than 10% area loss, largely due to their roles in drinking water management and as barriers against the sea [21]. These dunes represent one of the last nutrient-poor habitats in the Netherlands and most plant species featured on the Dutch Red List now have their main distribution in these coastal areas. Moreover, the reserves featured in this manuscript are part of Natura 2000, an ecological network of protected areas in the territory of the European Union (http://www.natura2000.nl/pages/kaartpagina.aspx).
The creeping willow (Salix repens) is a widespread shrub in western Europe and inhabits a wide range of vegetation types. It forms both ectomycorrhiza and arbuscular mycorrhiza [24],[25], often supporting a diverse community of symbiotic fungi in its extensive root system [26]. It is a particularly characteristic component of coastal sand dune ecosystems where it is often the only ectomycorrhizal (ECM) host plant [27],[26]. Therefore, it has been repeatedly noted that the presence of S. repens adds wealth to the overall fungal diversity in sand dune ecosystems [25],[26]. Somewhat surprisingly, species composition of fungi in S. repens communities have not been previously investigated using molecular methods to our knowledge. To date, characterizations of fungi in S. repens assemblages have been based on morphology-based identification of fruitbodies and ECM root structures. For example, in one of the early studies comparing above- and below-ground diversity of ECM fungi, Van der Heijden et al. [25] found fruitbodies of 78 ECM taxa belonging to 12 genera and identified fifteen ECM root morphotypes in S. repens communities in the Netherlands. Watling [26] reported almost 80 species of macrofungi from S. repens beds of the northern Scottish islands based on fruiting records. Despite these outstanding pioneer works, important gaps remain in our knowledge about total fungal diversity in this important ecosystem.
In this study, we collected soil samples at multiple locations in strictly protected coastal areas and conducted a large-scale biodiversity assessment of soil fungi inhabiting the rhizosphere of S. repens using pyrosequencing techniques. The aims of this work were 1) to characterize fungal communities found in S. repens vegetation; 2) compare the effects of environmental factors (e.g., soil pH, geological district) on communities through multiple sites; and 3) to supplement sporocarp-based mapping data points of rare fungi with soil-based data points. Our results provide the first kingdom-wide fungal diversity assessment in coastal sand dune communities and demonstrate the potential use of high-throughput DNA sequencing of environmental samples to fungal conservation.
Materials and Methods
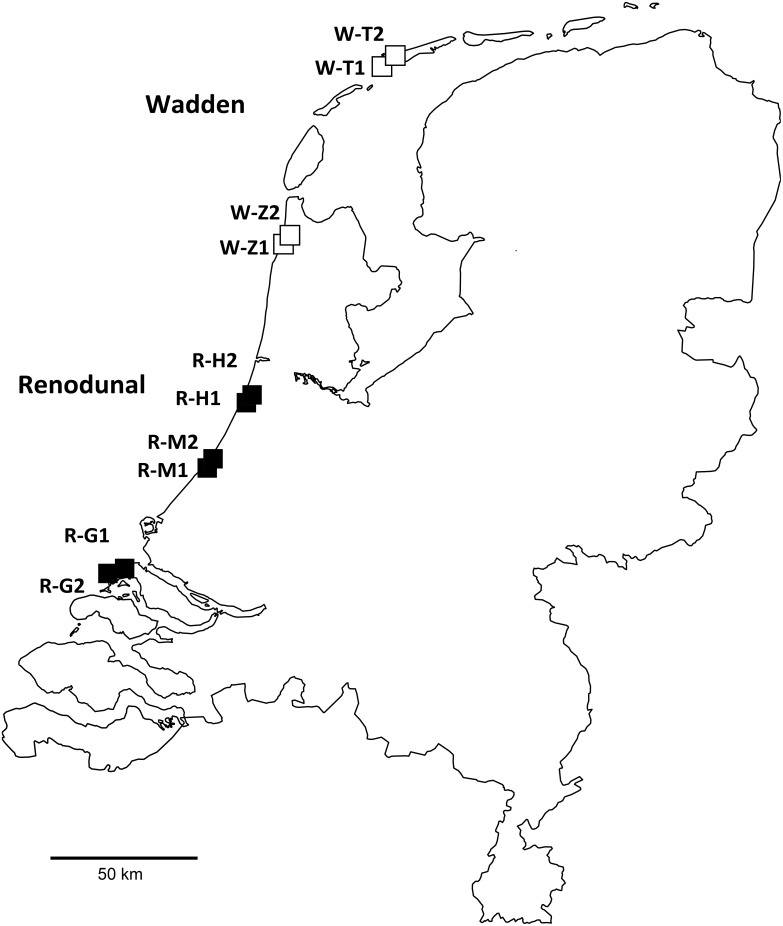
The Study Region
The following environmental agencies granted permission for the fieldwork (with sampled field sites): Amsterdamse Waterleidingduinen (Haarlem-Bentveld), Dunea (Wassenaar-Meijendel), Natuurmonumenten (Goeree, Callantsoog-Zwanenwater), and Staatsbosbeheer (Terschelling). GPS coordinates of the sampling sites are listed in Table 1. The field sampling only included soil and did not involve endangered or protected species. The selected sites represented sand dune S. repens beds (European Environment Agency habitat code: 2170) in an extended area of joined nature reserves located along the North Sea coast of the Netherlands, representing a south-north transect (Figure 1, Table 1). This coastal dune system is characterized by an oceanic climate with mean annual temperature of 9–10°C (July, 16–17°C; January, 2–3°C) and precipitation of 675–800 mm [21]. Sand-lime content varies between 0.1 and 10% [21], with generally calcareous soils in the southern Renodunal district (represented by the sampling sites Goeree, Meijendel, and Haarlem-Bentveld) and more acidic soils in the northern Wadden district (Zwanenwater, Terschelling). In most areas, the height of the dunes reaches 15–25 m [28]. Current management policies are directed toward multiple purposes, such as drinking water supply, defense against storm floods, recreation, and nature conservation [29]. Soil samples were taken at sites monitored by Natuurmonumenten (Goeree and Zwanenwater), Dunea (Meijendel), Staatsbosbeheer (Terschelling), and the Amsterdamse Waterleidingduinen (Haarlem-Bentveld). In March of 2010, 20 soil cores, 2 cm in diameter and ca. 20 cm deep, were taken at each site in a way that cores were at least 2 m from each other to minimize the probability of sampling the same genet repeatedly. For every plot, the 20 cores were pooled, resulting in a composite soil sample for each plot. Soils were deposited in 50 mL Falcon tubes and stored at –80°C until lyophilization.
Table 1. Sampling sites of coastal Salix repens communities used in this study.
| Site | Code | Geological district, habitat | Latitude, longitude | pH | OTUs |
| Goeree: Middelduinen | R-G1 | Renodunal, wet | 51.827238; 3.943405 | 6.01 | 971 |
| Goeree: Westduinen | R-G2 | Renodunal, wet | 51.810850; 3.861609 | 6.73 | 934 |
| Wassenaar: Meijendel 1 | R-M1 | Renodunal, dry | 52.137439; 4.341960 | 7.3 | 691 |
| Wassenaar: Meijendel 2 | R-M2 | Renodunal, dry | 52.142707; 4.333034 | 7.42 | 790 |
| Haarlem-Bentveld: Amsterdamse Waterleidingduinen 1 | R-H1 | Renodunal, wet | 52.338001; 4.531978 | 7.11 | 727 |
| Haarlem-Bentveld: Amsterdamse Waterleidingduinen 2 | R-H2 | Renodunal, dry | 52.337312; 4.533910 | 6.56 | 660 |
| Callantsoog: Zwanenwater 1 | W-Z1 | Wadden, dry | 52.822073; 4.709229 | 5.71 | 867 |
| Callantsoog: Zwanenwater 2 | W-Z2 | Wadden, wet | 52.821516; 4.708264 | 5.32 | 762 |
| Terschelling: Groene Strand | W-T1 | Wadden, wet | 53.364741; 5.201769 | 6.39 | 718 |
| Terschelling: West aan Zee | W-T2 | Wadden, dry | 53.398326; 5.269489 | 5.78 | 804 |
Code, geological district, habitat type (wet dune slack vs. dry dune side and top), geographic coordinates, soil pH, number of quality-filtered sequences and the number of 97% ITS2 sequence similarity OTUs are displayed for each site.
Figure 1. Sampling sites along the coast of the Netherlands.
Sites corresponding to the Renodunal and Wadden geological districts are marked with black and white squares, respectively.
DNA Extraction, PCR, and Pyrosequencing
We follow the recently proposed guidelines for standardizing the description of next-generation sequencing datasets in publications as proposed by Nilsson et al. [30]. Genomic DNA was extracted from 8 g of lyophilized soil from each composite sample using the Mo Bio Powersoil kit following the manufacturer’s instructions. We used high-throughput tag-encoded 454 GS-FLX amplicon sequencing to assess fungal diversity in the selected plant communities. Amplicon libraries were performed using a combination of tagged primers designed for the variable ITS region, as recommended for the tag-encoded pyrosequencing method [31]. Genomic DNA samples were amplified using the fungal-specific primer pair ITS1F (5′-XCTTGGTCATTTAGAGGAAGTAA) and ITS4 (5′-YxxxxxTCCTCCGCTTATTGATATGC), where X and Y represent the two pyrosequencing primers (CCTATCCCCTGTGTGCCTTGGCAGTCTCAGT and CCATCTCATCCCTGCGTGTCTCCGACTCAGA) and xxxxx represents the barcodes designed for sample identification. Barcodes differed from all other barcodes by at least two nucleotides. Four PCR reactions were performed per sample using the following temperature program: 95°C/2 min, 25 cycles of 95°C/30 s, 54°C/1 min, 72°C/2 min, and 72°C/10 min. One µl of DNA template was used for the 40 µl PCR reaction containing 25.6 µl of MQ water, 4 µl of 10x buffer, 1.5 µl dNTP’s (2.5 mM), 1.5 µl of reverse and forward primers (10 mM), 4 µl MgCl2 (50 mM), 0.5 µl BSA (10 mg/ml) and 0.4 µl BIOTAQ polymerase (5 U/µl). PCR products were pooled for each sample, purified and sequencing-adapters/tags were integrated via MegaPrimers (PCR). The amplicon length and concentration were estimated, and an equimolar mix of all amplicon libraries was used for pyrosequencing (with the ITS4 primer). Tagged samples were pooled and sequenced using the Genome Sequencer GS FLX 454 System (454 Life Sciences/Roche Applied Biosystems, Nutley, NJ, USA) at GATC Biotech AG (Konstanz, Germany) and the Leiden Genome Technology Center (Leiden, the Netherlands).
Bioinformatic Work
Pyrosequencing resulted in 688 434 reads following quality checks and trimming, with median read length of 284 bp. The raw data have been submitted to the European Nucleotide Archive (ERP001713). We used a parallel version of MOTHUR v. 1.32.1 [32] installed at the University of Alaska Life Sciences Informatics Portal for sequence analyses. We filtered sequences by quality score with a sliding window approach as in Brown et al. [33]. The FASTQ files were converted to FASTA and QUAL files, and the sequences were subjected to quality filtering whereby each sequence was screened for thresholds for average Phred score of Q≥25 in a sliding window of 50 bp (qwindowaverage = 25, qwindowsize = 50), no ambiguous bases (maxambig = 0) and homopolymers no longer than 8 bp (maxhomop = 8). Sequences shorter than 150 bp or longer than 400 bp were omitted from further analyses (mintlength = 150, maxlength = 400). A total of 369 181 sequences remained after quality filtering and trimming. Because next-generation sequencing libraries generally vary in size, we normalized the number of sequences for all samples prior to OTU clustering, as recommended by Gihring et al. [34], to ensure that estimators across all samples are comparable. For this purpose, we randomly subsampled the number of trimmed and quality-filtered reads to the size of the smallest library (30 497).
The resulting 304 970 sequences served as input for operational taxonomic unit (OTU) clustering using OTUPIPE [35] based on 97% sequence similarity. Such 97% sequence similarity cut-off value is routinely used in fungal community studies as a proxy for species, accounting for intraspecific variation and errors generated by PCR (e.g., [36],[37],[38],[7],[39],[40],[9],[12],[15],[41]. We simultaneously removed 13 279 putatively chimeric sequences, using a curated dataset of fungal ITS sequences as reference dataset [42]. Pyrosequencing datasets comprising orders of magnitude more sequences per taxa than traditional methods tend to contain a higher proportion of artifactual OTUs, because sequencing errors tend to be unique, while the accumulation of real taxa starts to level off [43],[12]. Because most pyrosequencing singletons tend to be artifactual and such sequencing errors can overestimate the diversity of ‘rare taxa’ (e.g., [44],[12]), we opted to be conservative and excluded all singletons from further analyses. For identification, sequences were subjected to USEARCH [45] against the latest release of quality-checked UNITE+INSD fungal ITS sequence database containing both identified and unidentified sequences, many of which are assigned to Species Hypothesis groups as defined by experts using phylogenetic evaluations [46]. OTUs that did not have at least 80% similarity over at least 100 bp to any fungal sequence were excluded from further analyses. Taxonomic affiliations were based on the current Index Fungorum classification as implemented in UNITE.
Comparing Fungal Communities among Sampling Sites
We used the 97% ITS sequence similarity OTUs inferred earlier as input data for the various ordination methods. To visualize the variation in the occurrence of OTUs across sites, we carried out ordination using non-metric multidimensional scaling (NMDS) with the quantitative version of Sorenson similarity (Bray-Curtis index) with 500 iterations in PC-Ord [47]. Read abundance in 454 sequencing has been shown to be approximately quantitative only within species, while between-species comparisons can be biased by innate sequence structure [39]. Because of such uncertainties regarding the reliability of read abundance, we carried out the ordination analyses with square-root transformed abundance to moderate the influence of OTUs with high sequence counts, while maintaining some approximation of abundance that often reflects ecological significance. The solution with the lowest stress was derived from 250 runs using real data and was then subjected to 250 randomized runs using a Monte Carlo test to evaluate the probability of final the NMDS patterns being greater than chance occurrences. Orthogonal rotation of the resulting NMDS solution was used to maximize correlation between the strongest environmental variables (as indicated by Pearson r values) and major axes. The solution with the lowest number of dimensions was selected when the decrease in the final stress was <5 by adding another dimension [47]. The Pearson correlation coefficient (r) values between environmental and fungal community variables and axes 1 and 2 were calculated. Soil pH and the number of OTUs representing various taxonomic groups per site were included in a site environmental factor matrix as quantitative variables. For this latter, we used only taxonomic orders and genera represented by at least 3 OTUs were included for a site as quantitative variables (see Table 2 for full list). Geological district (Renodunal = 1, Wadden = 0) was included as categorical variable. Following NMDS ordination of sites, we examined the Pearson correlation values between the community ordination axes and these variables. We also tested whether fungal communities were statistically different across the sites using a multiresponse permutation procedure (MRPP), also in PC-Ord. For the MRPP analyses, three categorical variables were tested: geological district, as described above, habitat type (wet dune slack vs. dry dune side and top), and soil pH (acidic = 0, non-acidic = 1, based on a pH = 6.5 threshold following the USDA soil classification for acidic and non-acidic soils). Finally, we determined any preferences of individual OTUs for pH categories using indicator species analyses.
Table 2. Pearson’s correlation values (r) for variables in the NMDS analyses.
| Axis 1 | Axis 2 | Axis 1 | Axis 2 | ||||||
| r | r2 | r | r2 | r | r2 | r | r2 | ||
| pH | 0.718 | 0.516 | 0.094 | 0.009 | Ascomycete genera | ||||
| OTU richness | −0.263 | 0.069 | −0.29 | 0.084 | Acremonium | −0.229 | 0.052 | 0.11 | 0.012 |
| Ascomycota | 0.028 | 0.001 | 0.306 | 0.093 | Alternaria | 0.594 | 0.353 | −0.144 | 0.021 |
| Basidiomycota | 0.321 | 0.103 | 0.503 | 0.253 | Archaeorhizomyces | −0.704 | 0.495 | 0.479 | 0.229 |
| Glomeromycota | 0.33 | 0.109 | 0.197 | 0.039 | Cadophora | 0.655 | 0.429 | −0.142 | 0.02 |
| Mucoromycotina | −0.318 | 0.101 | 0.529 | 0.28 | Cladonia | 0.854 | 0.729 | −0.527 | 0.278 |
| Chytridiomycota | −0.585 | 0.342 | 0.159 | 0.025 | Devriesia | −0.198 | 0.039 | 0.027 | 0.001 |
| Exophiala | 0.881 | 0.777 | −0.406 | 0.165 | |||||
| Ascomycete orders | Fusarium | −0.026 | 0.001 | −0.006 | 0 | ||||
| Archaeorhizomycetales | −0.704 | 0.495 | 0.479 | 0.229 | Geoglossum | 0.062 | 0.004 | 0.176 | 0.031 |
| Capnodiales | −0.232 | 0.054 | 0.222 | 0.049 | Geomyces | 0.424 | 0.179 | −0.463 | 0.214 |
| Chaetosphaeriales | −0.371 | 0.138 | 0.223 | 0.05 | Geopora | 0.73 | 0.532 | −0.092 | 0.008 |
| Chaetothyriales | 0.242 | 0.059 | −0.282 | 0.079 | Ilyonectria | 0.344 | 0.119 | −0.213 | 0.046 |
| Diaporthales | 0.202 | 0.041 | −0.47 | 0.221 | Lachnum | −0.854 | 0.729 | 0.527 | 0.278 |
| Eurotiales | −0.599 | 0.359 | −0.07 | 0.005 | Lophiostoma | 0.154 | 0.024 | 0.199 | 0.04 |
| Geoglossales | 0.012 | 0 | 0.258 | 0.067 | Massarina | 0.02 | 0 | 0.107 | 0.011 |
| Glomerales | 0.418 | 0.175 | −0.254 | 0.065 | Meliniomyces | −0.392 | 0.154 | 0.195 | 0.038 |
| Helotiales | −0.624 | 0.389 | −0.058 | 0.003 | Metarhizium | 0.437 | 0.191 | −0.59 | 0.348 |
| Hypocreales | −0.274 | 0.075 | −0.177 | 0.031 | Myrothecium | 0.287 | 0.083 | 0.155 | 0.024 |
| Lecanorales | 0.829 | 0.687 | −0.441 | 0.194 | Penicillium | −0.63 | 0.397 | 0.013 | 0 |
| Microascales | 0.195 | 0.038 | −0.257 | 0.066 | Phialophora | 0.17 | 0.029 | −0.26 | 0.068 |
| Onygenales | 0.643 | 0.414 | −0.749 | 0.561 | Phoma | 0.899 | 0.808 | −0.309 | 0.096 |
| Orbiliales | −0.396 | 0.157 | 0.17 | 0.029 | Pochonia | −0.359 | 0.129 | −0.332 | 0.11 |
| Pezizales | 0.921 | 0.848 | −0.15 | 0.023 | Podospora | 0.873 | 0.761 | −0.566 | 0.32 |
| Phyllachorales | −0.419 | 0.176 | 0.014 | 0 | Preussia | 0.04 | 0.002 | 0.167 | 0.028 |
| Pleosporales | 0.759 | 0.577 | −0.184 | 0.034 | Scolecobasidium | 0.588 | 0.346 | −0.414 | 0.171 |
| Saccharomycetales | −0.563 | 0.317 | −0.272 | 0.074 | Teratosphaeria | 0.817 | 0.667 | −0.211 | 0.044 |
| Sordariales | 0.883 | 0.779 | −0.492 | 0.242 | Trichoderma | −0.378 | 0.143 | 0.051 | 0.003 |
| Verrucariales | −0.535 | 0.286 | 0.485 | 0.235 | Verticillium | −0.551 | 0.303 | −0.435 | 0.189 |
| Xylariales | −0.037 | 0.001 | 0.57 | 0.325 | |||||
| Basidiomycete orders | Basidiomycete genera | ||||||||
| Agaricales | 0.479 | 0.229 | −0.137 | 0.019 | Ceratobasidium | −0.255 | 0.065 | −0.331 | 0.11 |
| Auriculariales | −0.796 | 0.634 | 0.122 | 0.015 | Clavicorona | −0.445 | 0.198 | −0.118 | 0.014 |
| Boletales | 0.015 | 0 | 0.213 | 0.045 | Clavulina | −0.63 | 0.397 | 0.052 | 0.003 |
| Cantharellales | −0.179 | 0.032 | −0.292 | 0.085 | Clavulinopsis | −0.564 | 0.318 | 0.125 | 0.016 |
| Corticiales | −0.186 | 0.035 | −0.19 | 0.036 | Cortinarius | −0.62 | 0.385 | 0.594 | 0.353 |
| Filobasidiales | −0.398 | 0.158 | −0.023 | 0.001 | Cryptococcus | −0.183 | 0.034 | −0.32 | 0.102 |
| Hymenochaetales | −0.465 | 0.216 | −0.208 | 0.043 | Entoloma | −0.453 | 0.205 | 0.672 | 0.451 |
| Polyporales | −0.212 | 0.045 | −0.376 | 0.142 | Hebeloma | −0.089 | 0.008 | 0.503 | 0.253 |
| Russulales | −0.447 | 0.2 | 0.026 | 0.001 | Hygrocybe | 0.166 | 0.028 | −0.126 | 0.016 |
| Sebacinales | 0.39 | 0.152 | −0.13 | 0.017 | Inocybe | 0.836 | 0.698 | −0.164 | 0.027 |
| Sporidiobolales | −0.372 | 0.138 | 0.122 | 0.015 | Laccaria | −0.489 | 0.239 | 0.52 | 0.27 |
| Thelephorales | 0.824 | 0.679 | −0.456 | 0.208 | Mycena | −0.56 | 0.314 | −0.165 | 0.027 |
| Trechisporales | −0.752 | 0.566 | −0.129 | 0.017 | Panaeolus | 0.745 | 0.555 | −0.216 | 0.047 |
| Tremellales | 0.069 | 0.005 | 0.106 | 0.011 | Paxillus | −0.114 | 0.013 | 0.255 | 0.065 |
| Trichosporonales | −0.853 | 0.727 | 0.243 | 0.059 | Rhodotorula | −0.416 | 0.173 | −0.036 | 0.001 |
| Rickenella | −0.465 | 0.216 | −0.208 | 0.043 | |||||
| Russula | −0.477 | 0.227 | 0.471 | 0.221 | |||||
| Sebacina | −0.115 | 0.013 | 0.288 | 0.083 | |||||
| Sporobolomyces | −0.004 | 0 | 0.265 | 0.07 | |||||
| Stropharia | 0.356 | 0.127 | −0.026 | 0.001 | |||||
| Thanatephorus | 0.515 | 0.265 | −0.237 | 0.056 | |||||
| Thelephora | −0.633 | 0.4 | −0.223 | 0.05 | |||||
| Tomentella | 0.939 | 0.882 | −0.403 | 0.163 | |||||
| Trichosporon | −0.853 | 0.727 | 0.243 | 0.059 |
Ordinations were performed with square-root transformed abundance in the OTU vs. site matrix. Final stress value for the 2-dimensional NMDS solution was 0.03062. Variables with |r| ≥0.5 values are shown in bold and are displayed in the NMDS ordinations in Figure 3.
Results
OTU Diversity and Taxonomic Affiliations
Out of the 688 434 original reads, 402 500 sequences passed the various filtering steps. After normalizing the library size across all samples, 304 970 sequences were assembled into 7156 OTUs of which 1788 were singletons. After excluding all singletons and OTUs with <80% similarity or <100 bp pairwise alignment length to a fungal sequence as performed by USEARCH, 3611 OTUs were retained for subsequent analyses. The number of OTUs at a given site ranged from 660 (Amsterdamse Waterleidingduinen, R-H2) to 971 (Goeree: Middelduinen, R-G1) (Table 1).
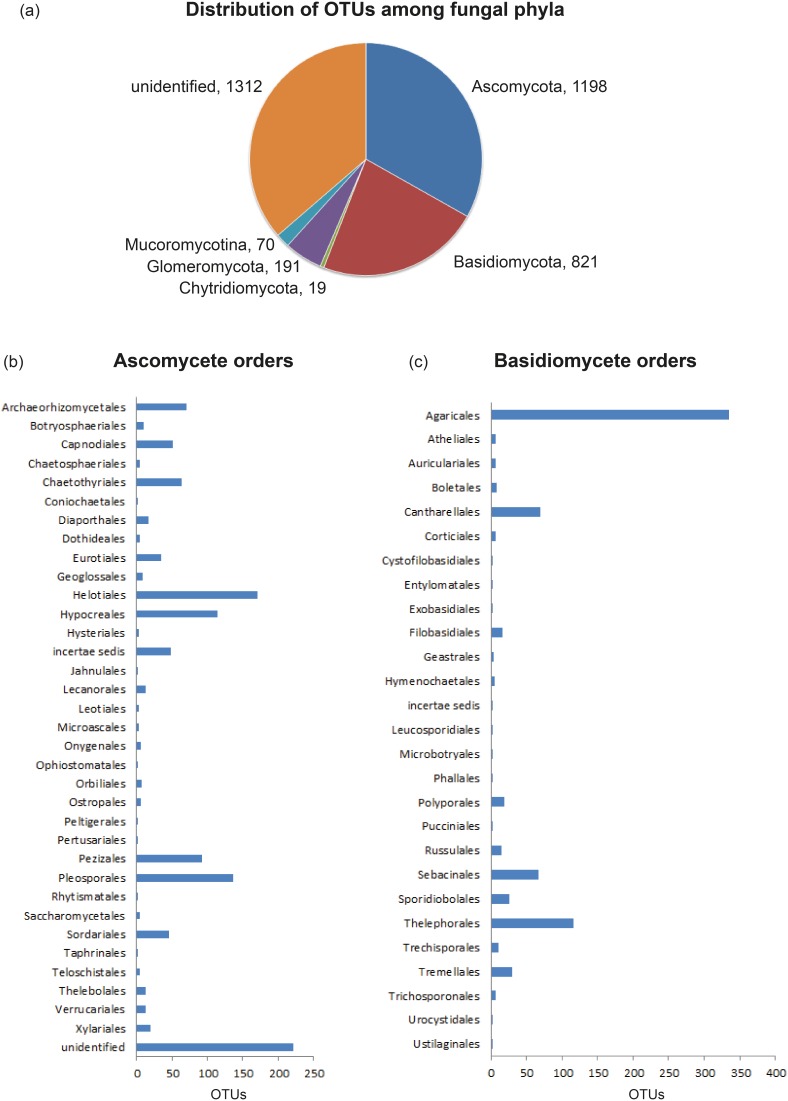
Ascomycota was the dominant phylum and accounted for 33.18% of the OTUs, followed by Basidiomycota (22.73%) Glomeromycota (5.29%), Mucoromycotina, incertae sedis (1.94%), and Chytridiomycota (0.53%), while unidentified fungal OTUs with most similar sequences to other environmental sequences without assignment to phylum accounted for 36.33% (Figure 2a). In Ascomycota, the ranking of taxonomic orders based on the number of representative OTUs was as follows: Helotiales (171), Pleosporales (136), Hypocreales (114), Pezizales (92), Archaeorhizomycetales (71), Chaetothyriales (64), Capnodiales (51), Sordariales (45), Eurotiales (34), Xylariales (19), Diaporthales (17), Verrucariales (13), Lecanorales (13), Thelebolales (12), Botryopshpaeriales (10), followed by numerous other orders with <10 OTUs. In addition, there were 49 ascomycete OTUs with incertae sedis classification, representing mostly mitosporic, asexual fungi (Figure 2b). In Basidiomycota, Agaricales was by far the most species-rich order in the sample with 335 OTUs, followed by Thelephorales (116), Cantharellales (69), Sebacinales (67), Tremellales (30), Sporidiobolales (26), Polyporales (19), Filobasidiales (16), Russulales (15), Trechisporales (11) and numerous other orders with <10 OTUs each, while there were only 2 basidiomycete OTUs with uncertain taxonomic placement (i.e, incertae sedis) (Figure 2c).
Figure 2. Taxonomic groupings of fungi retrieved from the samples.
Proportional distribution of the 1022 unique BLASTN hits among different phyla and taxonomic orders in Basidiomycota and Ascomycota across all sampling sites based on sequences most closely related to 97% ITS OTUs in our sample.
There were 354 OTUs with >97% sequence similarity to publicly available sequences with species identification (Table S1). Although most of these represented microfungi, we also detected many species that form macroscopic fruitbodies (e.g., mushrooms, bracket fungi, cup fungi, earth tongues, truffles etc.), among which we found numerous red listed species in our samples, often from previously unknown locations, based on mapping data and red list categories obtained from the Fungal Mapping website of the Dutch Mycological Society (http://www.verspreidingsatlas.nl/paddenstoelen).
Comparing Fungal Communities among Sampling Sites
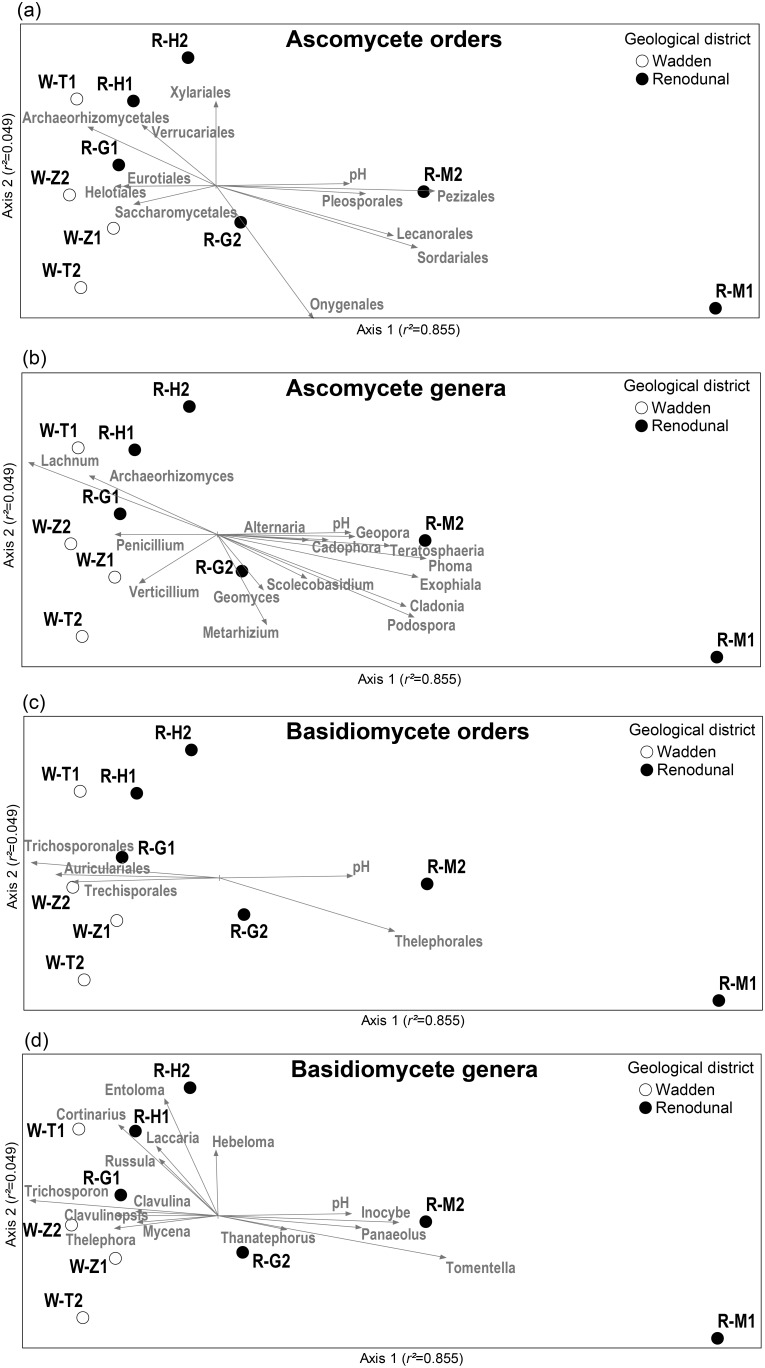
NMDS analyses resulted in a 2-dimensional solution with a final stress of 0.03062 and a final instability <0.00001. The Monte Carlo test results indicated that this 2-dimensional solution using the real data was significantly better than chance occurrences (p = 0.008). The two axes explained the majority of variability in the sampled fungal communities (axis 1: r2 = 0.855; axis 2: r2 = 0.049; total r2 = 0.904; orthogonality = 79.5%). The NMDS ordination plot was orthogonally rotated by pH to visualize correlations between pH and fungal community composition in general and the taxonomic groups in particular. The Pearson correlation coefficient (r) values between all fungal community variables and axes 1 and 2 are displayed in Table 2. As in the fungal community ecology paper of Rogers et al. [48], we considered variables with |r| ≥0.5 values for axis 1 important for characterizing changes in fungal community structure along a pH gradient and these variables were superimposed on the ordination plot as direction and strength vectors (Figure 3).
Figure 3. Non-metric multidimensional scaling (NMDS) ordination plot for fungal communities from Salix repens sites.
Ordinations were based on the square-root transformed abundance of 97% ITS sequence similarity OTUs. Labels, localities and descriptions of the sampling sites are given in Table 1. Vectors are shown for soil pH, total OTU richness, and for the most diverse taxonomic orders (A) and genera (B) that correlated with the ordination axes at r2>0.200.
There was a strong structuring of the occurrence of fungal OTUs according to soil pH, that co-correlated with the geological district. Sites belonging to the Wadden district were clustered on the left side of the ordination plot with strongly negative axis 1 values. Even though the Renodunal district sites were more scattered, the Wadden and Renodunal sites formed non-overlapping clusters (Figure 3). However, one Renodunal site (Goeree: Middelduinen, R-G1), that had an acidic soil pH value uncharacteristic of the Renodunal sites, was appeared to be more similar to the Wadden communities than to other Renodunal sites. MRPP tests confirmed the importance of soil pH in shaping fungal community composition (effect size A = 0.04259, probability p = 0.00463), while the effect of geological district was somewhat weaker (A = 0.02786, p = 0.03157). Habitat type (wet dune slack vs. dry dune side and top) did not seem to have a significant influence on community composition (A = 0.01113, p = 0.17961).
The joint plots illustrate the strength and direction of correlation of variables to each ordination axis (Figure 3). The correlation of soil pH with axis 1 was strong (r = 0.718). At the phylum level, only Chytridiomycota showed correlation with axis1 (r = −0.585) and was more diverse in the acidic sites, while diversity in other phyla did not show significant trends. At the taxonomic order level, the ascomycete Archaeorhizomycetales, Helotiales, Eurotiales, Saccharomycetales, and Verrucariales and the basidiomycete Trichosporonales, Auriculariales, Trechisporales were found to be more diverse in the acidic sites (Figure 3, Table 2). Orders showing greater richness in the alkaline soil samples included the ascomycete Pezizales, Sordariales, Lecanorales, Pleosporales, Onygenales and the basidiomycete Thelephorales. At the genus level, the diversity in ascomycete genera Archaeorhizomyces, Lachnum, Penicillium, and Verticillium and basidiomycete genera Trichosporon, Thelephora, Clavulinopsis, Cortinarius, Clavulina, and Mycena were negatively correlated with pH, while the ascomycete Phoma, Exophiala, Podospora, Cladonia, Teratospora, Geopora, Cadophora, Alternaria, Scolecobasidium and the basidiomycete Tomentella, Inocybe, Panaeolus, and Tanatephorus were more diverse in the alkaline samples (Figure 3, Table 2).
There were 96 OTUs identified as significant (p<0.05) indicators for a certain pH category. Of these, 82 were indicators for acidic and 14 for alkaline sites. Several indicators belonged to root-associated fungi. For example, an unidentified root-endophytic Phialocephala and the ECM Russula pascua (SH117152.05FU), Thelephora albomarginata, Thelephora sp. (SH112629.05FU), and Tulasnella sp. (SH117022.05FU) were associated with acidic sites, while the ECM Inocybe dunensis (SH110718.05FU) and an unidentified Tomentella were characteristic of the alkaline sites. In addition, there were several saprobic that were found almost exclusively in the acidic soil samples, such as Cryptococcus sp. (SH117277.05FU), Fusarium sp. (SH106152.05FU), various Mortierella species, Mycena simian (SH102569.05FU), Penicillium adametzii, Rickenella fibula (SH109302.05FU), Schizoblastosporion starkeyihenricii, Umbelopsis ramanniana (SH100505.05FU), and a Verticillium sp. (SH111596.05FU), some of which can be potentially pathogenic. An unidentified species of the nematode pathogen Pochonia genus (SH106625.05FU) also showed strong preference for acidic sites. There were only two saprobic species identified as indicators for the alkaline soils: Fusarium torulosum and a Podospora species. The full list of indicator OTUs are shown in Table 3.
Table 3. OTUs considered as significant indicators of the altitudinal vegetation zones with corresponding p-values, and with accession numbers, sequence similarity, pairwise alignment length, name, and taxonomic classification of the most similar sequence in the UNITE+INSD database.
| OTU | pH | p | Accession no. | % | bp | SH | Name | Phylum | Order |
| 1573 | acidic | 0.0402 | HQ212048 | 99.1 | 230 | SH114088.05FU | Agaricomycotina sp. | Basidiomycota | - |
| 1200 | acidic | 0.0084 | EF445994 | 80.4 | 337 | SH113104.05FU | Arthrobotrys eudermata | Ascomycota | Orbiliales |
| 339 | acidic | 0.017 | JQ272358 | 98.1 | 324 | - | Ascomycota sp. | Ascomycota | - |
| 3541 | acidic | 0.0426 | JQ272358 | 96.5 | 198 | - | Ascomycota sp. | Ascomycota | - |
| 747 | acidic | 0.0306 | JQ272358 | 99.1 | 324 | - | Ascomycota sp. | Ascomycota | - |
| 1385 | acidic | 0.047 | HF947900 | 97.1 | 314 | - | Ascomycota sp. | Ascomycota | - |
| 1687 | acidic | 0.0382 | HF947896 | 95.3 | 215 | - | Ascomycota sp. | Ascomycota | - |
| 397 | acidic | 0.047 | HF947899 | 100 | 322 | - | Ascomycota sp. | Ascomycota | - |
| 535 | acidic | 0.0468 | HM230880 | 95.3 | 257 | - | Chaetothyriales | Ascomycota | Chaetothyriales |
| 361 | acidic | 0.0084 | KF060235 | 100 | 350 | - | Cryptococcus sp. | Basidiomycota | Incertae sedis |
| 6307 | acidic | 0.0084 | FJ553137 | 96.5 | 201 | SH117277.05FU | Cryptococcus sp. | Basidiomycota | Incertae sedis |
| 474 | acidic | 0.027 | JQ437607 | 100 | 321 | SH106152.05FU | Fusarium sp. | Ascomycota | Hypocreales |
| 4597 | alkaline | 0.023 | JX534258 | 99.2 | 258 | SH107221.05FU | Fusarium torulosum | Ascomycota | Hypocreales |
| 3605 | acidic | 0.0376 | HE794041 | 97.2 | 181 | SH113008.05FU | Glomeraceae sp. | Glomeromycota | Glomerales |
| 347 | alkaline | 0.0084 | JN859279 | 100 | 303 | SH167718.05FU | Helotiales | Ascomycota | Helotiales |
| 395 | acidic | 0.0468 | HM190115 | 99.7 | 327 | SH115846.05FU | Helotiales sp. | Ascomycota | Helotiales |
| 4961 | acidic | 0.0244 | JN655622 | 98.7 | 227 | SH115846.05FU | Helotiales sp. | Ascomycota | Helotiales |
| 686 | acidic | 0.0486 | JN655578 | 100 | 320 | SH114192.05FU | Helotiales sp. | Ascomycota | Helotiales |
| 746 | acidic | 0.0476 | JN655598 | 97.9 | 338 | SH117370.05FU | Helotiales sp. | Ascomycota | Helotiales |
| 2760 | acidic | 0.044 | HQ021773 | 83.6 | 238 | - | Hyaloriaceae sp. | Basidiomycota | Auriculariales |
| 773 | acidic | 0.044 | HQ021773 | 83.6 | 281 | - | Hyaloriaceae sp. | Basidiomycota | Auriculariales |
| 338 | alkaline | 0.0426 | UDB017616 | 100 | 249 | SH110718.05FU | Inocybe dunensis | Basidiomycota | Agaricales |
| 475 | acidic | 0.047 | JQ272448 | 100 | 256 | - | Mortierella macrocystis | Mucoromycotina | Mortierellales |
| 608 | acidic | 0.0494 | JX976005 | 99.7 | 331 | - | Mortierella parvispora | Mucoromycotina | Mortierellales |
| 577 | acidic | 0.0494 | KC018246 | 100 | 198 | - | Mortierella sp. | Mucoromycotina | Mortierellales |
| 6109 | acidic | 0.0242 | JN943795 | 85.8 | 225 | SH117207.05FU | Mortierella verticillata | Mucoromycotina | Mortierellales |
| 4807 | acidic | 0.044 | GU234138 | 95 | 179 | SH102569.05FU | Mycena simia | Basidiomycota | Agaricales |
| 812 | acidic | 0.0084 | GU234138 | 98.5 | 205 | SH102569.05FU | Mycena simia | Basidiomycota | Agaricales |
| 379 | acidic | 0.0162 | KC773822 | 99.6 | 259 | - | Penicillium adametzii | Ascomycota | Eurotiales |
| 1127 | acidic | 0.0084 | FN678832 | 99.6 | 226 | SH116036.05FU | Pezizomycetes sp. | Ascomycota | - |
| 781 | acidic | 0.0084 | JQ272412 | 100 | 264 | - | Pezizomycotina sp. | Ascomycota | - |
| 1249 | acidic | 0.0188 | JQ346810 | 99.1 | 338 | - | Phialocephala sp. | Ascomycota | Helotiales |
| 844 | acidic | 0.047 | EU726301 | 99.7 | 322 | - | Phyllachorales sp. | Ascomycota | Phyllachorales |
| 821 | alkaline | 0.0166 | KC218448 | 100 | 244 | - | Plectosphaerella sp. | Ascomycota | Incertaesedis |
| 1469 | acidic | 0.047 | DQ516079 | 98.9 | 187 | SH106625.05FU | Pochonia sp. | Ascomycota | Hypocreales |
| 158 | alkaline | 0.0084 | KC180725 | 100 | 154 | - | Podospora sp. | Ascomycota | Sordariales |
| 859 | acidic | 0.047 | UDB015270 | 99.3 | 272 | SH109302.05FU | Rickenella fibula | Basidiomycota | Hymenochaetales |
| 1456 | acidic | 0.0468 | FN687605 | 95.7 | 161 | - | Russula pascua | Basidiomycota | Russulales |
| 627 | acidic | 0.0468 | UDB017659 | 98.5 | 197 | SH117152.05FU | Russula pascua | Basidiomycota | Russulales |
| 429 | acidic | 0.0294 | HF558658 | 99.6 | 262 | - | Schizoblastosporion starkeyihenricii | Ascomycota | Saccharomycetales |
| 1618 | alkaline | 0.0294 | JN802314 | 98.1 | 210 | SH113075.05FU | Sordariales sp. | Ascomycota | Sordariales |
| 346 | acidic | 0.0084 | JQ761571 | 99 | 196 | SH112969.05FU | Sordariomycetes sp. | Ascomycota | - |
| 555 | acidic | 0.023 | HQ211703 | 91.6 | 323 | SH100328.05FU | Sordariomycetes sp. | Ascomycota | - |
| 560 | acidic | 0.047 | UDB017379 | 100 | 216 | - | Thelephora albomarginata | Basidiomycota | Thelephorales |
| 514 | acidic | 0.047 | JN858076 | 100 | 192 | SH112629.05FU | Thelephora sp. | Basidiomycota | Thelephorales |
| 1281 | alkaline | 0.0458 | KC840637 | 99.1 | 222 | - | Tomentella sp. | Basidiomycota | Thelephorales |
| 568 | acidic | 0.0494 | GQ268679 | 88.4 | 258 | - | Trechisporales | Basidiomycota | Trechisporales |
| 1126 | acidic | 0.0376 | JF691365 | 96.2 | 213 | SH110459.05FU | Trechisporales sp. | Basidiomycota | Trechisporales |
| 420 | acidic | 0.0426 | JF519135 | 100 | 195 | SH114084.05FU | Trechisporales sp. | Basidiomycota | Trechisporales |
| 5679 | acidic | 0.044 | JF519135 | 95.5 | 156 | SH114084.05FU | Trechisporales sp. | Basidiomycota | Trechisporales |
| 4735 | acidic | 0.0234 | JQ272445 | 96.5 | 173 | - | Tremellomycetes sp. | Basidiomycota | - |
| 1929 | acidic | 0.047 | JN655663 | 99.2 | 242 | SH117022.05FU | Tulasnella sp. | Basidiomycota | Cantharellales |
| 2522 | acidic | 0.0494 | EU715662 | 97.4 | 193 | SH100505.05FU | Umbelopsis ramanniana | Mucoromycotina | Mucorales |
| 540 | acidic | 0.0252 | KC489481 | 96.4 | 195 | - | Umbelopsis sp. | Mucoromycotina | Mucorales |
| 561 | acidic | 0.0166 | HQ157863 | 98.4 | 253 | SH102024.05FU | Umbelopsis sp. | Mucoromycotina | Mucorales |
| 731 | acidic | 0.033 | KC489481 | 99 | 204 | - | Umbelopsis sp. | Mucoromycotina | Mucorales |
| 1205 | acidic | 0.047 | DQ914739 | 97.4 | 229 | SH111596.05FU | Verticillium sp. | Ascomycota | Incertae sedis |
| 1037 | acidic | 0.044 | KC588535 | 89.4 | 227 | - | uncultured fungus | - | - |
| 1076 | acidic | 0.0084 | KC588535 | 86.5 | 296 | - | uncultured fungus | - | - |
| 1211 | alkaline | 0.049 | AJ875364 | 99.7 | 343 | SH100236.05FU | uncultured fungus | - | - |
| 1459 | acidic | 0.0084 | JQ312779 | 100 | 209 | - | uncultured fungus | - | - |
| 1651 | acidic | 0.047 | HM439535 | 96.8 | 220 | SH101693.05FU | uncultured fungus | - | - |
| 1845 | acidic | 0.0084 | FJ197879 | 93.1 | 232 | - | uncultured fungus | - | - |
| 1913 | acidic | 0.047 | EF521253 | 80.6 | 345 | - | uncultured fungus | - | - |
| 1969 | acidic | 0.047 | JQ313102 | 99.6 | 226 | - | uncultured fungus | - | - |
| 2667 | alkaline | 0.049 | EU516756 | 89.6 | 328 | - | uncultured fungus | - | - |
| 328 | acidic | 0.0464 | JQ312820 | 98.5 | 259 | - | uncultured fungus | - | - |
| 358 | alkaline | 0.0458 | JN889968 | 99.4 | 315 | SH102623.05FU | uncultured fungus | - | - |
| 363 | acidic | 0.0084 | KC588722 | 100 | 261 | - | uncultured fungus | - | - |
| 367 | alkaline | 0.0084 | JX316640 | 99 | 306 | - | uncultured fungus | - | - |
| 401 | acidic | 0.0244 | KC588704 | 100 | 320 | - | uncultured fungus | - | - |
| 4316 | alkaline | 0.0188 | GU308427 | 99.6 | 255 | - | uncultured fungus | - | - |
| 4368 | acidic | 0.0224 | KC416188 | 97.5 | 158 | - | uncultured fungus | - | - |
| 4423 | acidic | 0.044 | KC416184 | 95 | 161 | - | uncultured fungus | - | - |
| 451 | acidic | 0.0084 | AF504839 | 99.7 | 364 | SH109673.05FU | uncultured fungus | - | - |
| 466 | alkaline | 0.034 | FN397289 | 100 | 322 | SH112415.05FU | uncultured fungus | - | - |
| 4887 | acidic | 0.0396 | JQ312820 | 91.7 | 229 | - | uncultured fungus | - | - |
| 502 | acidic | 0.0084 | DQ309106 | 97.6 | 328 | SH114572.05FU | uncultured fungus | - | - |
| 573 | acidic | 0.047 | JQ312957 | 94.5 | 218 | - | uncultured fungus | - | - |
| 6263 | acidic | 0.047 | JQ312957 | 96.1 | 207 | - | uncultured fungus | - | - |
| 630 | acidic | 0.047 | AM260886 | 97.8 | 274 | SH110947.05FU | uncultured fungus | - | - |
| 643 | acidic | 0.0084 | KC588567 | 99.3 | 283 | - | uncultured fungus | - | - |
| 6519 | alkaline | 0.049 | JX316640 | 95.7 | 207 | - | uncultured fungus | - | - |
| 6520 | acidic | 0.048 | KC588554 | 95.9 | 217 | - | uncultured fungus | - | - |
| 665 | acidic | 0.047 | DQ309136 | 99.6 | 271 | - | uncultured fungus | - | - |
| 711 | acidic | 0.047 | HM136622 | 99.2 | 355 | - | uncultured fungus | - | - |
| 713 | acidic | 0.047 | JQ312796 | 99.5 | 199 | - | uncultured fungus | - | - |
| 723 | acidic | 0.036 | DQ093782 | 97.6 | 329 | SH099908.05FU | uncultured fungus | - | - |
| 762 | acidic | 0.0458 | JQ312820 | 96.4 | 193 | - | uncultured fungus | - | - |
| 787 | acidic | 0.0084 | JF300531 | 90.2 | 368 | - | uncultured fungus | - | - |
| 803 | acidic | 0.0084 | JN890270 | 96.3 | 327 | - | uncultured fungus | - | - |
| 838 | acidic | 0.0434 | FN298702 | 93.3 | 326 | SH104632.05FU | uncultured fungus | - | - |
| 852 | acidic | 0.0454 | JQ312820 | 97.8 | 230 | - | uncultured fungus | - | - |
| 857 | acidic | 0.0162 | HM037687 | 99 | 196 | SH110459.05FU | uncultured fungus | - | - |
| 899 | acidic | 0.047 | HM208720 | 99.4 | 334 | SH099545.05FU | uncultured fungus | - | - |
| 828 | acidic | 0.0308 | HQ021815 | 99.7 | 320 | SH108513.05FU | uncultured fungus | - | - |
Where available, the Species Hypothesis (SH) numbers are given for the corresponding sequence as published by Köljalg et al. (2013).
Discussion
Acidic vs. Non-acidic Sand Dune Fungal Communities
While environmental factors influencing the distribution of plants and animals have been thoroughly studied, environmental factors controlling the spatial patterns and community composition of soil microorganisms are still poorly understood [49]. Recent molecular studies on various microbial groups (mostly bacteria) have started to explore the distributional patterns of soil communities, however, many of them applied DNA-fingerprinting techniques or phospholipid analyses that do not permit the taxonomic surveys and comparisons of different communities [50],[51]. Soil pH is generally implicitly considered important for shaping fungal communities with some supporting evidence from experimental studies mostly with AM fungi [52],[53] and, in recent years, from joint pyrosequencing analyses of bacterial and fungal communities in agricultural soils and land use types [54],[49]. Unfortunately, both of these latter sequencing studies relied on nuclear small-subunit (SSU) ribosomal genes, meaning that their power of taxonomic resolution was at family-level groups at best. Therefore, information on the effect of soil pH on the distribution of specific fungal taxa and on the composition of their communities in natural habitats remains surprisingly scarce.
Our large-scale comparison of various acidic and non-acidic coastal dune Salix repens beds clearly indicates that soil pH strongly correlates with the composition of fungal communities. In NMDS ordinations, soil pH had high Pearson correlation values with the dominant ordination axis and overrode the effect of the geological district (Wadden vs. Renodunal) on community composition in both the NMDS and the MRPP analyses (Figures 3 and 4). This is in agreement with the findings of Van der Heijden et al. [25] that soil pH significantly contributed to the variation explained in ECM morphotype composition of communities associated with S. repens. In our NMDS ordinations and MRPP analyses, Inocybe and Tomentella showed strong preference for alkaline soils, while Clavulina, Cortinarius, Thelephora, and to some extent Laccaria, were more diverse in acidic sites (Figure 3, Table 2). It is worth noting, that because our study focuses on natural systems, it is impossible to determine based on our current data whether the communities are structured directly by pH or indirectly via the interaction of pH with soil communities and various environmental factors (e.g., nutrient availability, organic C characteristics, vegetation differences etc.). Because results from previous studies using a pH gradient with controlled variables suggest no or only weak effect of pH on community structure [54],[49] and because many fungal species have a relatively wide pH optimum (e.g. [55],[56]), it is likely that the observed correlation of pH with community composition is mainly indirect, e.g., via nutrient availability and altered competitive interactions between soil fungi and bacteria [57], and other soil biota. While our data suggest that the effect of soil pH on fungal community composition may be stronger in relatively undisturbed sites than in agricultural settings, disentangling causal relationships is beyond the scope of our work.
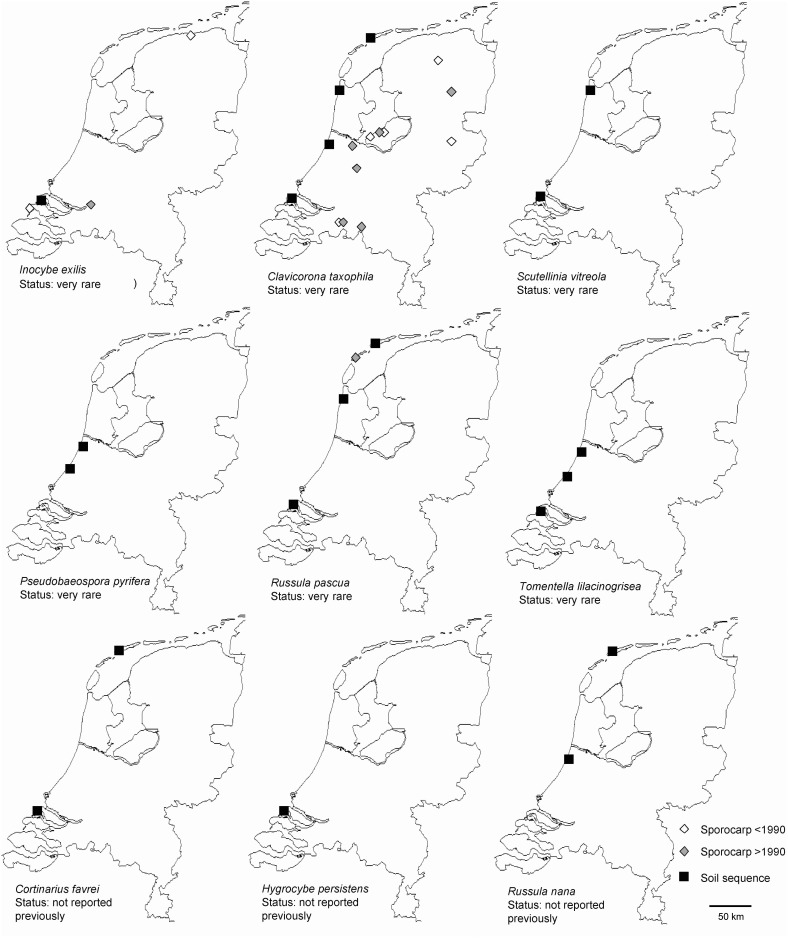
Figure 4. Examples for OTUs found in our soil samples showing high sequence similarity with rare, red-listed fungi.
Sporocarp records (before and after 1990) are based on the mapping database of the Dutch Mycological Society (http://www.verspreidingsatlas.nl/paddenstoelen).
Root-associated Fungi
Although taxa detected by our sequencing efforts featured groups from a wide variety of ecological guilds, including many saprobic and/or plant pathogenic fungi (e.g., Fusarium cereale, Gibberella zeae, Phoma multirostrata) as well as animal or fungal pathogens (e.g., Hypomyces chrysospermus, Metarhizium anisopliae, M. flavoviride and other Hypocreales), we focus our discussion on groups that are most relevant to current conservation priorities, such as fungi with macroscopic fruitbodies and/or with symbiotic associations with S. repens or with red listed plants (e.g. orchids) in these habitats.
Previous studies based on sporocarps and ECM root structures have shown that Salix repens communities generally harbour numerous ectomycorrhizal basidiomycetes [25],[26]. This was confirmed by our large-scale soil sequence data, as we found 418 ECM OTUs. The majority of these showed similarity to basidiomycete genera known to form associations with willows (e.g. [58],[59],[15]), such as Inocybe (95 OTUs), Thelephora/Tomentella (83), Cortinarius (36), Hebeloma (10), Russula (7), Laccaria (7), Clavulina (6), Lactarius (5), Paxillus (2). Also, there were 4 OTUs (Amanita pantherina, Hydnum rufescens, Suillus luteus and Tulasnella sp.) that represent species that are known to form ECM associations with other host trees found near the sampling sites, such as oak, pine or poplar. In addition, we found 52 OTUs belonging to the Sebacinaceae, a taxonomically difficult group with diverse ecological roles, including various forms of mycorrhizae (ECM, orchid, ericoid, arbutoid) and other associations with plant roots [60]. We also identified several ectomycorrhizal ascomycetes: Cenococcum geophilum, one of the most widespread and abundant ECM fungus with a wide host range [59], Geopora arenicola and G. cervina, a genus known to occur in sand dunes [61], Helvella compressa and H. maculata, and an unidentified Trichophaea species. In addition, we found four truffles that are likely symbionts of Quercus that is widespread in the dunes: Tuber anniae, and three unidentified Tuber species.
Salix repens is one of the very few plants known to form both ECM and AM [24],[62]. In dry coastal foredunes, where phosphorus is the main limiting nutrient, AM fungi and their hosts generally predominate, while nitrogen is the main limiting element in dune slacks, resulting in the dominance of ECM taxa [62]. With the ability to form both ECM and AM associations, Salix repens is particularly well-suited to survive in the highly dynamic dune ecosystems and is able to colonize habitats that range from dry to wet and from acidic to calcareous [62]. We detected 191 glomeromycete OTUs in our samples, representing Glomus (53), Rhizophagus (10), Archaeospora (6), Acaulospora (4), Claroideoglomus (4), Scutellospora (4), Cetraspora (3), Paraglomus (3), Entrophospora (2), Diversispora (1), Racocetra (1), and 100 unidentified glomeromycete OTUs. Because the majority of land plants are known to form AM, it is likely that the majority of these taxa are also associated with a wide range of plants beside Salix repens.
We also found numerous OTUs with very high sequence similarity to known dark septate root endophytes (DSE) (e.g., Cadophora, Cladophialophora, Exophiala, Leptodontidium, Meliniomyces, Phialocephala, Phialophora spp.). Although DSE colonization and diversity have previously been shown to be higher in acidic conditions in general [63],[64], our data suggest that different genera may prefer different pH conditions. For example, Cadophora and Exophiala were represented by more OTUs in the alkaline sites, while Meliniomyces was somewhat more diverse in samples with lower pH (Table 2).
The sampling areas included in our study harbor some of the richest orchid populations in the Netherlands. The diversity of orchid mycorrhizae may be particularly relevant for nature conservation as they provide symbioses to this unique group of plants with high public appeal and conservation value. The majority of orchid mycorrhizal symbionts belong to the Ceratobasidiaceae, Sebacinaceae and Tulasnellaceae [65],[66], although ECM fungi have also been reported to be associated with orchids, albeit mainly with non-photosynthetic, mycoheterotrophic species that often use the ECM fungus as a channel to obtain carbohydrates from the ECM host tree [65],[67]. Among the first group of orchid symbionts that are typically associated with photosynthetic orchids, we found 21 Ceratobasidiaceae and 12 Tulasnellaceae OTUs in addition to the 52 Sebacinaceae OTUs mentioned above. Among other ECM lineages, the basidiomycete Inocybe, Tomentella, and the ascomycete Tuber species have been found to form mycorrhizal associations with the mycoheterotrophic orchid Epipactis helleborine [67],[68]. This orchid is represented by two subspecies in the Netherlands, among these, E. h. ssp. helleborine is widespread in a variety of habitats, while E. h. ssp. neerlandica is restricted to the dunes and is endangered [68]. This latter rare subspecies is present in most of our sampling sites and we found multiple species of all three ECM genera reportedly associated with E. helleborine. Tomentella and Inocybe were the most species-rich genera in our samples and are important symbionts for Salix repens as well, as specified earlier. Because E. h. ssp. neerlandica is mostly found near Salix repens [68],[69], it is highly likely that willow-associated ECM fungi are crucial for the survival of this endangered orchid. The same might apply to two other rare orchid species in the Netherlands, Platanthera bifolia and Dactylorhiza incarnata ssp. coccinea of which the associated ECM fungi are still largely unexplored but which are also mostly found near Salix repens [69].
Contribution of Soil Data to Mapping Rare Fungi
The Netherlands boasts one of the most efficient programs in the world for gathering distribution data on macrofungi, where the Netherlands Mushroom Mapping Working Group (Werkgroep Paddenstoelenkartering Nederland, WPN) has been producing distribution maps and fruiting statistics since 1980 with a nation-wide network of ca. 600 volunteers that produce appr. 60,000 data points yearly [70],[71],[72]. The long-term mapping program of the WPN has been crucially important in compiling a near complete checklist of macrofungi [73], in highlighting high diversity sites [70], and in drawing attention to the serious decline of macrofungi in the Netherlands [72]. As a result, an official national Red List of fungi was published in 1996, followed by a revision in 2008 [74]. On the other hand, due to inherent limitations of the data collecting method, data on the diversity and distribution of microfungi and taxa with inconspicuous fruitbodies have remained scarce. Building on the methodological advances provided by next-generation sequencing techniques, we can still greatly improve the exhaustiveness of species inventories by providing complementary information, particularly with regard to asexual fungi and species with microscopic and/or inconspicuous fruiting bodies, in addition to providing new spatial data points for macrofungi already featured in the species lists.
The massive amount of sequence data we generated provide the first kingdom-wide fungal diversity assessment for this coastal ecosystem, offering a complementary view on species diversity with a particularly large contribution on asexual fungi and species with microscopic and/or inconspicuous fruiting bodies that fall beyond the scope of the above mapping project. Even for macrofungi, despite the obvious spatial limitations of the methodology we used, we detected numerous red listed species in our samples, often from previously unknown locations. Several taxa with scarce (<5) previous sporocarp records, such as Inocybe exilis, Pseudobaeospora pyrifera, Russula pascua, Scutellinia vitreola and several Tomentella and Sebacina species to name a few, were found in multiple locations, indicating that they may be more widespread in the Netherlands than previously thought (Figure 4). This may be particularly true for Tomentella, a species-rich genus with often inconspicuous sporocarps, that is regularly shown to be much more diverse and abundant in soil and root samples than in sporocarp-based diversity assessments (e.g., [15],[75],[76]). We also provide valuable data for additional taxa that are considered either rare or sporadic, often with “vulnerable” or “endangered” red list categories, e.g., Clavicorona taxophila, Clavulinopsis corniculata, C. luteoalba, Conocybe subxerophytica, Cortinarius alnetorum, C. casimiri, C. cinnamomeus, C. diasemospermus, C. parvannulatus, Entoloma asprellum, E. clandestinum, E. longistriatum, E. turbidum, Geastrum striatum, Geoglossum fallax, G. umbratile, Geopora arenicola, Hebeloma collariatum, H. leucosarx, H. nigellum, Hemimycena tortuosa, Hydnum rufescens, Hygrocybe helobia, H. vitellina, Inocybe agardhii, I. decipiens, I. dunensis, I. griseovelata, I. jacobi, I. nitidiuscula, I. serotina, I. vulpinella, I. whitei, Lactarius aurantiacus, Mycena albidolilacea, Panaeolus papilionaceus, Peziza ampliata, Pleurotus eryngii, Russula foetens, Russula puellaris, Squamanita odorata, Tephrocybe tylicolor, Thanatephorus cucumeris, Tricholoma lascivum, Tulostoma brumale, Xerocomus riperiellus etc.
Highly similar matching sequences from species not formerly reported from the Netherlands represent a special case and confirming the presence of some species will require additional evidence. Some such examples among ascomycetes are the truffle Tuber anniae, the earth-tongue Geoglossum glabrum, the cup fungus Geopora cervina, and the false-morel Verpa digitaliformis. In Basidiomycota, the list of taxa formerly not reported from the Netherlands and found among the highly similar (>98%) matching sequences of our OTUs include: Clavaria californica, Cortinarius favrei, C. sertipes, Heterobasidion parviporum, Hygrocybe persistens, Inocybe giacomi, Mycena simia, Pluteus satur, Russula integra, R. nana, Sphaerobolus ingoldii, and Tubaria minima. Occurrence data regarding species that usually occur in arctic-alpine regions, such as Cortinarius favrei and Russula nana, are particularly interesting, given the geographic position and the lack of mountains in the Netherlands. In the Arctic, these species are symbionts of various dwarf willow species (e.g. Salix polaris, S. reticulata) [77],[Geml pers. obs.], while in the Netherlands, they appear to grow with Salix repens. Both Cortinarius favrei and Russula nana have been found associated with Salix repens in the oceanic archipelago north of Scotland [26]. It has to be noted that Russula laccata, a species originally described from the Netherlands albeit with no current mapping data, is very closely related to R. nana and whether or not they are truly distinct species is currently uncertain. Therefore, detailed taxonomic work is needed to test whether or not the two taxa are conspecific and whether R. laccata refer to the same taxon as our OTU (no. 695), in which case the name R. nana will be preserved based on priority.
Conclusions
Fungal communities are notoriously difficult to fully characterize for ecological and biodiversity studies and for conservation purposes. Even for macrofungi (e.g., such as mushrooms, true and false truffles etc.), that have the longest history of diversity studies among fungi [1],[70],[78], basic questions about the number of species at a given location or differences in species richness among vegetation types have generally remained unanswered due to taxonomic problems and the scarcity of long-term sporocarp-monitoring projects [78].
Our work provides a wealth of information on the kingdom-wide diversity of fungal communities along the protected coastal dune habitats in the Netherlands, extending both the list of species likely occurring in the Netherlands as well as providing new data points for many rare species. Furthermore, our work highlights that fungal communities found in the rhizosphere of S. repens can have very different species composition due to edaphic factors and that the protection of areas that harbour such varied S. repens beds is vital for the conservation of co-habiting fungi, including many red-listed species. Future fruitbody collecting projects should utilize our data to concentrate sampling and monitoring efforts on selected localities in order to confirm species occurrences that are currently only based on soil sequence data. Although more detailed, taxon-specific studies will follow, this project already provides examples for the potential contribution of high-throughput soil sequencing studies to fungal mapping and conservation.
Data Accessability
The raw data have been submitted to the European Nucleotide Archive (ERP001713).
Supporting Information
List of OTUs that showed >97% ITS2 sequence similarity to fully identified fungi in coastal Salix repens communities used in this study with best identified match (GenBank or UNITE accession number, Species Hypothesis, name), similarity (%), pairwise alignment length (bp), sequence counts in the sampled sites, phylum, and order classifications.
(XLS)
Acknowledgments
The authors also thank Marcel Eurlings for support with initial labwork, and Shawn Houston and Rob Cermak at the University of Alaska Fairbanks for bioinformatics support. Finally, we are grateful to the Dutch Mycological Society (Nederlandse Mycologische Vereniging) for providing access to mapping data and to the environmental agencies that granted permission for the fieldwork: Amsterdamse Waterleidingduinen (Haarlem-Bentveld), Dunea, Natuurmonumenten, and Staatsbosbeheer. (Terschelling).
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Data are available from the European Nucleotide Archive under the accession number: ERP001713.
Funding Statement
This research was sponsored by a Next-Generation Sequencing grant from the Naturalis Biodiversity Center to JG and MEN and by the Kits van Waveren Postdoctoral Fellowship to JG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Perini C, Dahlberg A, Fraiture A (2008) Conservation and mapping of macrofungi in Europe – Advancement during the last decade. Coolia 51: 181–195. [Google Scholar]
- 2. Hawksworth DL (1991) The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycol Res 95: 641–655. [Google Scholar]
- 3. Blackwell M (2011) The Fungi: 1, 2, 3 … 5.1 million species? Am J Bot 98: 426–438. [DOI] [PubMed] [Google Scholar]
- 4. Schmit JP, Mueller GM (2007) An estimate of the lower limit of global fungal diversity. Biodivers Conserv 16: 99–111. [Google Scholar]
- 5. Schadt CW, Martin AP, Lipson DA, Schmidt SK (2003) Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301: 1359–1361. [DOI] [PubMed] [Google Scholar]
- 6. Porter TM, Skillman JE, Moncalvo JM (2008) Fruiting body and soil rDNA sampling detects complementary assemblage of Agaricomycotina (Basidiomycota, Fungi) in a hemlock-dominated forest plot in southern Ontario. Mol Ecol 17: 3037–3050. [DOI] [PubMed] [Google Scholar]
- 7. Geml J, Laursen GA, Timling I, McFarland JM, Booth MG, et al. (2009) Molecular phylogenetic biodiversity assessment of arctic and boreal Lactarius Pers. (Russulales; Basidiomycota) in Alaska, based on soil and sporocarp DNA. Mol Ecol 18: 2213–2227. [DOI] [PubMed] [Google Scholar]
- 8. Abarenkov K, Nilsson RH, Larsson K-H, Alexander IJ, Eberhardt U, et al. (2010) The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytol 186: 281–285. [DOI] [PubMed] [Google Scholar]
- 9. Geml J, Laursen GA, Herriott I, McFarland JM, Booth MG, et al. (2010) Phylogenetic and ecological analyses of soil and sporocarp DNA sequences reveal high diversity and strong habitat partitioning in the boreal ectomycorrhizal genus Russula Pers. (Russulales; Basidiomycota). New Phytol 187: 494–507. [DOI] [PubMed] [Google Scholar]
- 10. Jumpponen A, Jones KL, Mattox JD, Yaege C (2010) Massively parallel 454-sequencing of fungal communities in Quercus spp. ectomycorrhizas indicates seasonal dynamics in urban and rural sites. Mol Ecol 19: S41–S53. [DOI] [PubMed] [Google Scholar]
- 11. Taylor DL, Herriott IC, Stone KE, McFarland JW, Booth MG, et al. (2010) Structure and resilience of fungal communities in Alaskan boreal forest soils. Can J Forest Res 40: 1288–1301. [Google Scholar]
- 12. Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Sadam A, et al. (2010) 454 pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol 188: 291–301. [DOI] [PubMed] [Google Scholar]
- 13. Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk P, et al. (2011) Progress in molecular and morphological taxon discovery in Fungi and options for formal classification of environmental sequences. Fungal Biol Rev 25: 38–47. [Google Scholar]
- 14. Blaalid R, Carlsen T, Kumar S, Halvorsen R, Ugland KI, et al. (2012) Changes in the root-associated fungal communities along a primary succession gradient analysed by 454 pyrosequencing. Mol Ecol 21: 1897–1908. [DOI] [PubMed] [Google Scholar]
- 15. Geml J, Timling I, Robinson CH, Lennon N, Nusbaum HC, et al. (2012) An arctic community of symbiotic fungi assembled by long-distance dispersers: phylogenetic diversity of ectomycorrhizal basidiomycetes in Svalbard based on soil and sporocarp DNA. J Biogeogr 39: 74–88. [Google Scholar]
- 16. Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E (2012) Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol 21: 2045–2050. [DOI] [PubMed] [Google Scholar]
- 17. Hebert PDN, Cywinska A, Ball SL, de WaardJR (2003) Biological identification through DNA barcodes. P Roy Soc Lond B Bio 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seifert KA (2008) Integrating DNA barcoding into the mycological sciences. Persoonia 21: 162–166. [Google Scholar]
- 19. Van der Maarel E (2003) Some remarks on the functions of European coastal ecosystems. Phytocoenologia 33: 187–202. [Google Scholar]
- 20.Van Zadelhoff FJ (1981) Nederlandse kustduinen: geobotanie. Pudoc, Wageningen. 121 p. [Google Scholar]
- 21. Bakker TWM, Doing H (1987) De Nederlandse kustduinen en hun internationale betekenis. Duin 10: 34–41. [Google Scholar]
- 22. Brown AC, McLachlan A (2002) Sandy shore ecosystems and the threats facing them: some predictions for the year 2025. Environ Conserv 29: 62–77. [Google Scholar]
- 23.Van der Meulen F, Udo de Haes HA (1996) Nature conservation and integrated coastal.
- 24.Harley JL, Harley EL (1987) A checklist of mycorrhiza in the British flora. New Phytol 105 (suppl.): 1–102.
- 25. Van der Heijden EW, de Vries FW, Kuyper TW (1999) Mycorrhizal associations of Salix repens L. communities in succession of dune ecosystems. I. Above-ground and below-ground views of ectomycorrhizal fungi in relation to soil chemistry Can J Bot 77: 1821–1832. [Google Scholar]
- 26. Watling R (2005) Fungal associates of Salix repens in northern oceanic Britain and their conservation significance. Mycol Res 109: 1418–1424. [DOI] [PubMed] [Google Scholar]
- 28.Salman A (1991) Sand dune inventory, Netherlands. In: Doody JP, editor. Sand dune inventory of Europe. Joint Nature Conservation Committee (UK) and EUCC. 28–32.
- 29. Feola S, Carranza ML, Schaminée JHJ, Janssen JAM, Acosta ATR (2011) EU habitats of interest: an insight into Atlantic and Mediterranean beach and foredunes. Biodivers Conserv 20: 1457–1468. [Google Scholar]
- 30. Nilsson RH, Tedersoo L, Lindahl BD, Kjøller R, Carlsen T, et al. (2011) Towards standardization of the description and publication of next-generation sequencing datasets of fungal communities. New Phytol 191: 314–318. [DOI] [PubMed] [Google Scholar]
- 31. Acosta-Martinez V, Dowd S, Sun Y, Allen V (2008) Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol Bioch 40: 2762–2770. [Google Scholar]
- 32. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. (2009) Introducing mother: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown SP, Callaham MA, Oliver AK, Jumpponen A (2013) Deep Ion Torrent sequencing identifies soil fungal community shifts after frequent prescribed fires in a southeastern US forest ecosystem. FEMS Microbiol Ecol doi: 10.1111/1574-6941.12181 [DOI] [PubMed] [Google Scholar]
- 34. Gihring TM, Green SJ, Schadt CW (2012) Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ Microbiol 14: 285–290. [DOI] [PubMed] [Google Scholar]
- 35. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O’Brien H, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R (2005) Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microb 71: 5544–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arnold AE, Lutzoni F (2007) Diversity and host range of foliar fungal endophytes: Are tropical leaves biodiversity hotspots? Ecology 88: 541–549. [DOI] [PubMed] [Google Scholar]
- 38. Higgins KL, Arnold AE, Miadlikowska J, Sarvate SD, Lutzoni F (2007) Phylogenetic relationships, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Mol Phyl Evol 42: 543–555. [DOI] [PubMed] [Google Scholar]
- 39. Amend AS, Seifert KA, Bruns TD (2010) Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol Ecol 19: 5555–5565. [DOI] [PubMed] [Google Scholar]
- 40. Bjorbækmo MFM, Carlsen T, Brysting A, Vrålstad T, Høiland K, et al. (2010) High diversity of root associated fungi in both alpine and arctic Dryas octopetala . BMC Plant Biol 10: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kauserud H, Kumar S, Brysting AK, Nordén J, Carlsen T (2012) High consistency between replicate 454 pyrosequencing analyses of ectomycorrhizal plant root samples. Mycorrhiza 22: 309–315. [DOI] [PubMed] [Google Scholar]
- 42. Nilsson RH, Abarenkov K, Larsson K-H, Köljalg U (2011a) Molecular identification of fungi: rationale, philosophical concerns, and the UNITE database. TOAINFOJ 5: 81–86. [Google Scholar]
- 43. Reeder J, Knight R (2009) The ‘rare biosphere’: a reality check. Nat Methods 6: 636–637. [DOI] [PubMed] [Google Scholar]
- 44. Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12: 118–123. [DOI] [PubMed] [Google Scholar]
- 45. Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- 46. Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, et al. (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22: 5271–5277. [DOI] [PubMed] [Google Scholar]
- 47.McCune B, Grace JB (2002) Analysis of Ecological Communities. MjM Software, Gleneden Beach, Oregon, U.S.A.
- 48. Rogers PC, Moore KD, Ryel RJ (2009) Aspen succession and nitrogen loading: a case for epiphytic lichens as bioindicators in the Rocky Mountains, USA. J Veg Sci 20: 498–510. [Google Scholar]
- 49. Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, et al. (2010) Soil bacterial and fungal communities across a pH gradient in an arable soil. The ISME J 4: 1340–1351. [DOI] [PubMed] [Google Scholar]
- 50. Bååth E, Anderson TH (2003) Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol Biochem 35: 955–963. [Google Scholar]
- 51. Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. P Natl Acad Sci USA 103: 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Porter WM, Robson AD, Abbott LK (1987) Field survey of the distribution of Vesicular-arbuscular mycorrhizal fungi in relation to soil pH. J Appl Ecol 24: 659–662. [Google Scholar]
- 53. Coughlan AP, Dalpé Y, Lapoint L, Piché Y (2000) Soil pH-induced changes in root colonization, diversity, and reproduction of symbiotic arbuscular mycorrhizal fungi from healthy and declining maple forests. Can J Forest Res 30: 1543–1554. [Google Scholar]
- 54. Lauber CL, Strickland MS, Bradford MA, Fierer N (2008) The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40: 2407–2415. [Google Scholar]
- 55. Nevarez L, Vasseur V, Le Madec L, Le Bras L, Coroller L, et al. (2009) Physiological traits of Penicillium glabrum strain LCP 08.5568, a filamentous fungus isolated from bottled aromatised mineral water. Int J Food Microbiol 130: 166–171. [DOI] [PubMed] [Google Scholar]
- 56. Wheeler KA, Hurdman BF, Pitt JI (1991) Influence of pH on the growth of some toxigenic species of Aspergillus, Penicillium and Fusarium . Int J Food Microbiol 12: 141–150. [DOI] [PubMed] [Google Scholar]
- 57. Rousk J, Aldén Demoling L, Bahr A, Bååth E (2008) Examining the fungal and bacterial niche overlap using selective inhibitors in soil. FEMS Microbiol Ecol 63: 350–358. [DOI] [PubMed] [Google Scholar]
- 58.Molina R, Massicotte H, Trappe JM (1992) Specificity Phenomena in Mycorrhizal Symbioses: Community-Ecological Consequences and Practical Implications. In: Routledge AMF, editor. Mycorrhizal Functioning: An Integrative Plant-Fungal Process. Chapman and Hall, New York. 357–423.
- 59. De Roman M, Claveria V, De Miguel AM (2005) A revision of the descriptions of ectomycorrhizas published since 1961. Mycol Res 109: 1063–1104. [DOI] [PubMed] [Google Scholar]
- 60. Weiss M, Selosse M-A, Rexer K-H, Urban A, Oberwinkler F (2004) Sebacinales: a hitherto overlooked cosm of heterobasidiomycetes with a broad mycorrhizal potential. Mycol Res 108: 1003–1010. [DOI] [PubMed] [Google Scholar]
- 61. Tamm H, Pöldmaa K, Kullman B (2010) Phylogenetic relationships in genus Geopora (Pyrenomycetaceae, Pezizales). Mycol Prog 9: 509–522. [Google Scholar]
- 62. Van der Heijden EW (2001) Differential benefits of arbuscular mycorrhizal and ectomycorrhizal infection of Salix repens . Mycorrhiza 10: 185–193. [Google Scholar]
- 63. Postma JWM, Olsson PA, Falkengren-Grerup U (2006) Root colonization by arbuscular mycorrhizal, fine endophytic and dark septate fungi across a pH gradient in acid beech forests. Soil Biol Biochem 39: 400–408. [Google Scholar]
- 64. Geml J, Pastor N, Fernandez L, Pacheco S, Semenova TA, Becerra AG, Wicaksono CY, Nouhra ER (2014) Large-scale fungal diversity assessment in the Andean Yungas forests reveals strong community turnover among forest types along an altitudinal gradient. Mol Ecol 23: 2452–2472. [DOI] [PubMed] [Google Scholar]
- 65. Taylor DL, Bruns TD (1997) Independent, specialized invasions of ectomycorrhizal mutualism by two nonphotosynthetic orchids. P Natl Acad Sci USA 94: 4510–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Taylor DL, McCormick MK (2008) Internal transcribed spacer primers and sequences for improved characterization of basidiomycetous orchid mycorrhizas. New Phytol 177: 1020–1033. [DOI] [PubMed] [Google Scholar]
- 67. Bidartondo MI, Burghardt B, Gebauer G, Bruns TD, Read DJ (2004) Changing partners in the dark: isotopic and molecular evidence of ectomycorrhizal liaisons between forest orchids and trees. P Roy Soc Lond B Bio 271: 1799–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kuiper M, Oostermeijer JGB, Gravendeel B (2009) Duinwespenorchis: standplaatsvariatie of soort in wording? Holland’s Duinen 54: 16–21. [Google Scholar]
- 69.Delforge P (2007) Guide des orchidées de France, de Suisse et du Benelux. Delachaux et Niestlé, Paris. 288 p. [Google Scholar]
- 70.Jalink LM, Nauta MM (2001) Recognising and managing mycologically valuable sites in The Netherlands. In: Moore D, Nauta MM, Evans SE, Rotheroe M, editors. Fungal conservation – Issues and solutions. Cambridge University Press, Cambridge. 89–94.
- 71.Nauta MM, Jalink LM (2001) The contribution of national mycological societies: the Dutch Mycological Society and its Committee for Fungi and Nature Conservation. In: Moore D, Nauta MM, Evans SE, Rotheroe M, editors. Fungal conservation – Issues and solutions. Cambridge University Press, Cambridge. 242–246.
- 72.Nauta MM, Vellinga EC (1995) Atlas van Nederlandse paddestoelen. A.A. Balkema Uitgevers B.V., Rotterdam. 352 p. [Google Scholar]
- 73.Arnolds E, Kuyper TW, Noordeloos ME (1995) Overzicht van de paddenstoelen in Nederland. Nederlandse Mycologische Vereniging, Wijster. 872 p. [Google Scholar]
- 74.Arnolds E, Veerkamp M (2008) Rode Lijst Paddenstoelen. Basisrapport. Nederlandse Mycologische Vereniging. 295 p. [Google Scholar]
- 75. Gardes M, Bruns TD (1996) Community structure of ectomycorrizal fungi in a Pinus muricata forest: above- and below-ground views. Can J Bot 74: 1572–1583. [Google Scholar]
- 76. Kõljalg U, Dahlberg A, Taylor AFS, larsson E, Hallenberg N, et al. (2000) Diversity and abundance of resupinate thelephoroid fungi as ectomycorrhizal symbionts in Swedish boreal forests. Mol Ecol 9: 1985–1996. [DOI] [PubMed] [Google Scholar]
- 77. Väre H, Vestberg M, Eurola S (1992) Mycorrhiza and root-associated fungi in Spitsbergen. Mycorrhiza 1: 93–104. [Google Scholar]
- 78.Lodge J, Ammirati JF, O’Dell TO, Mueller GM, Huhndorf SM, et al.. (2004) Terrestrial and lignicolous macrofungi. In: Mueller GM, Bills GF, Foster MS, editors. Biodiversity of Fungi – Inventory and monitoring methods. Elsevier Academic Press. 128–172.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of OTUs that showed >97% ITS2 sequence similarity to fully identified fungi in coastal Salix repens communities used in this study with best identified match (GenBank or UNITE accession number, Species Hypothesis, name), similarity (%), pairwise alignment length (bp), sequence counts in the sampled sites, phylum, and order classifications.
(XLS)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Data are available from the European Nucleotide Archive under the accession number: ERP001713.