Abstract
The size and shape of the nucleus are tightly regulated, indicating the physiological significance of proper nuclear morphology, yet the mechanisms and functions of nuclear size and shape regulation remain poorly understood. Correlations between altered nuclear morphology and certain disease states have long been observed, most notably many cancers are diagnosed and staged based on graded increases in nuclear size. Here we review recent studies investigating the mechanisms regulating nuclear size and shape, how mitotic events influence nuclear morphology, and the role of nuclear size and shape in subnuclear chromatin organization and cancer progression.
Introduction
Many structural components of the nucleus control nuclear size and shape. The nuclear envelope (NE) is a double lipid bilayer consisting of the outer nuclear membrane (ONM), continuous with the endoplasmic reticulum (ER), and inner nuclear membrane (INM). The nuclear pore complex (NPC) embedded in the NE mediates nucleocytoplasmic transport. The nucleoplasmic face of the INM is lined by the nuclear lamina, a meshwork of intermediate lamin filaments that structurally supports the NE and mediates connections with chromatin. Linker of nucleoskeleton and cytoskeleton (LINC) complexes connect the nuclear lamina with the cytoskeleton through the NE, mediated by interactions between INM SUN-domain proteins and ONM KASH-domain proteins (reviewed in [1,2]).
The nucleus is a dynamic organelle, particularly during mitosis in metazoans when the NE breaks down to facilitate mitotic spindle assembly. Reassembly of the NE, nuclear lamina, and NPCs occurs after chromosome segregation [1], and recent studies show that these post-mitotic events are important in determining proper nuclear morphology in the subsequent interphase. Yeast studies have also elucidated the regulation of nuclear size and shape, however in contrast to the open mitosis of animal cells, many yeasts undergo a closed mitosis that necessitates dramatic cell cycle regulated changes in nuclear morphology [3-6].
Changes in nuclear size and shape are associated with cell differentiation, development, and disease. Of note, nuclear morphology is frequently altered in cancer cells [7,8]. By and large the physiological consequences of altered nuclear size and shape are not known but could potentially impact chromatin organization and gene expression, particularly in the context of tumor development and cancer progression. Therefore, it is important to understand the mechanisms that regulate nuclear size and shape as well as the function of proper nuclear morphology control.
In this review we focus on recent studies addressing mechanisms of nuclear size and shape regulation, in particular the roles of nuclear structural elements, the cytoskeleton, membrane, and the extracellular matrix (ECM). We then discuss how mitotic events impact nuclear morphology and how nuclear size and shape might impact subnuclear structure and function. We conclude with recent studies investigating the contributions of nuclear morphology to cancer and some future directions.
Mechanisms of nuclear size regulation
Nucleocytoplasmic transport, nuclear structural components, and post-mitotic nuclear assembly can all impact nuclear size. Although genome size scales with nuclear size across a wide range of species, DNA content tends to be a less important contributor to nuclear size regulation in a variety of experimental systems, primarily establishing a minimum nuclear size (reviewed in [9-11]). Here we will integrate results from older studies with newer findings on the roles of the nuclear lamina, LINC complexes, and NPCs in the regulation of nuclear size (Table 1).
Table 1.
Nuclear envelope structural elements that regulate nuclear morphology.
| Protein | Organism/ System |
Function | Alteration and nuclear phenotype |
|---|---|---|---|
| Lamin A/C | Vertebrates | Structural support to the nucleus, roles in DNA replication and gene expression |
Mutations in LMNA gene cause diseases with misshaped nuclei [34- 40] |
| Lamin A/C | Neutrophil- differentiated HL-60 cells |
As above | Overexpression causes hypolobulated nuclei [30,31] |
| Lamin B1 | Mammals Cortical neurons |
As above | Deficiency causes misshaped nuclei, nuclear blebs [32] |
| Lamin B2 | As above | As above | Deficiency leads to elongated nuclei [32] |
| Lamin B2 | Zebrafish embryo |
As above | Overexpression causes lobulated nuclei with intranuclear membranes [16] |
| Lamin B3 |
Xenopus egg extract |
As above | Depletion decreases nuclear size [13] Ectopic addition increases nuclear size [14] |
| Lamin-like nuclear proteins |
Arabidopsis
thaliana |
Should have similar functions to vertebrate lamina |
Deletions of LINC1/2 genes result in smaller and more round nuclei [17] |
| LBR | Neutrophil- differentiated HL-60 cells |
Binds lamins and chromatin |
Depletion causes hypolobulated nuclei [30,31] |
| Lap2β |
Xenopus
egg extract |
Binds lamins and chromatin |
Addition of truncated Lap2β causes small scalloped nuclei [19] |
| LEM4 |
Caenorhabditis
elegans |
Interacts with lamina and chromatin |
Depletion causes misshapen, multilobed nuclei [58] |
| AtSun1/2 |
Arabidopsis
thaliana |
Components of LINC complex |
Depletion causes nuclear rounding [46] |
| AtWIPs |
Arabidopsis
thaliana |
Plant specific KASH domain proteins, components of LINC complex |
Mutations that disrupt LINC interactions cause nuclear rounding [47] |
| Nesprin2 | HaCaT cells | KASH domain protein; binds to actin cytoskeleton and SUN proteins; part of LINC complex |
Nes2ΔABD overexpression increases nuclear size; Nes2-mini overexpression decreases nuclear size [21,22] |
| Pom121 |
Xenopus egg extract |
Nucleoporin, structural component of NPC |
Addition of dominant negative fragment blocks NE growth [26] |
| Nup188 |
Xenopus egg extract |
As above | Depletion increases nuclear size [27] |
| AtNup136 |
Arabidopsis
thaliana |
As above | Overexpression increases nuclear size and nuclear elongation; Depletion decreases nuclear size and causes nuclear rounding [44,45] |
Several studies support a role for nuclear lamins in nuclear size regulation. In Xenopus egg extracts, the lamin Ig-fold motif was required for post-mitotic lamina assembly and NE growth [12], lamin B3 depletion resulted in small nuclei that failed to expand [13], and ectopic addition of lamin B3 increased the rate of nuclear growth [14] (Fig. 1a). In tissue culture cells and Xenopus oocytes, NE growth was promoted by the C-terminal domain from B-type lamins, which contains a farnesylated CaaX motif required for lamin interaction with the INM [15,16]. Lamin B overexpression in zebrafish embryos and tissue culture cells resulted in extranuclear cisternae-like lamin/membrane arrays, dependent on farnesylation [16]. Furthermore, in Arabidopsis thaliana, deletion of genes encoding lamin-like nuclear matrix proteins, LITTLE NUCLEI 1/2, resulted in decreased nuclear size and altered nuclear morphology [17].
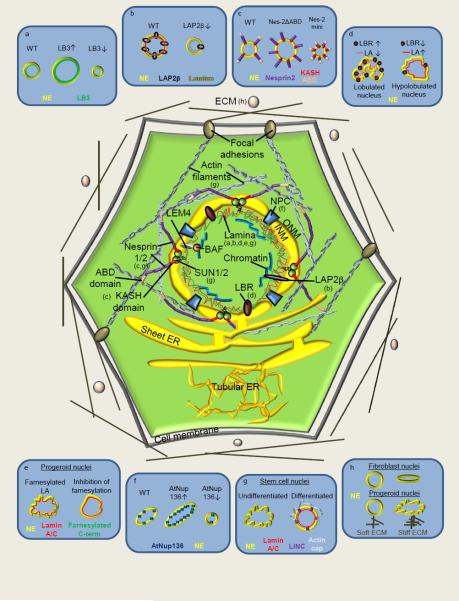
Figure 1. Mechanisms of nuclear size and shape regulation.
The central diagram depicts the major cellular components involved in regulating nuclear morphology. The blue boxes around the edge depict specific examples where mechanisms determining nuclear size and/or shape have been identified. (a) In Xenopus egg extracts, lamin B3 (LB3) depletion reduces nuclear size [13], while supplementing extract with LB3 increases the rate of NE expansion [14]. (b) Mislocalization of LAP2 or addition of a dominant negative fragment of LAP2 to Xenopus egg extract inhibits nuclear growth [19,20]. (c) Expression of nesprin-2 lacking the ABD increases nuclear size, while expression of nesprin-2-mini decreases nuclear size [21,22]. (d) Altered LBR and lamin A (LA) expression in neutrophils affects nuclear lobulation [30,31]. (e) Progerin expression leads to the formation of misshapen nuclei that can be rescued with farnesylation inhibitors [34]. (f) Altered expression of Arabidopsis thaliana Nup136 affects both nuclear size and elongation [44,45]. (g) Stem cell differentiation is associated with acquisition of a perinuclear actin cap that regulates nuclear morphology through LINC and lamina interactions [49]. (h) ECM stiffness modulates nuclear shape [53].
The lamina-associated polypeptides (LAPs) establish connections between the lamins and chromatin [18]. Addition of the nucleoplasmic chromatin-binding domain of human LAP2β to Xenopus extract blocked nuclear lamina assembly, inhibited nuclear growth, and resulted in a scalloped NE phenotype, demonstrating a role for LAP2 in postmitotic nuclear size determination [19]. Additionally, LAP2 was mislocalized upon depletion of TPX2, an important regulator of spindle assembly, resulting in dramatically smaller, but functional, interphase nuclei [20] (Fig. 1b).
LINC complexes also contribute to the regulation of nuclear size. In HaCaT cells, F-actin depolymerization resulted in small, highly dysmorphic nuclei, while microtubule depolymerization increased nuclear size. Notably, depolymerization of both cytoskeletal components decreased nuclear size, suggesting that nuclear connections with the actin cytoskeleton may be dominant [21]. These effects may be mediated through the actin-binding domain (ABD) of the ONM KASH-domain protein nesprin-2. Expression of nesprin-2ΔABD, the ABD alone, or the KASH domain alone increased nuclear area. Conversely, expression of nesprin-2-mini, a fusion protein consisting of only the KASH domain and ABD and lacking the large central spectrin-repeat rod domain, resulted in reduced nuclear size [21,22] (Fig. 1c).
During normal interphase, nuclear volume and NPC number nearly double, however distinct mechanisms seem to regulate these two processes because disruption of interphase NPC assembly in HeLa cells by cyclin-dependent kinase inhibition negligibly affected nuclear growth [23,24]. Nonetheless, altered NPC composition can affect nuclear size. In budding yeast, a RSC chromatin remodeling complex mutant exhibited NPC mislocalization, accumulation of nuclear membrane sheets, and altered nuclear morphology [25]. In Xenopus egg extract, a dominant-negative fragment of the nucleoporin POM121 blocked NE growth when added to intact nuclei [26], while depletion of Nup188 led to the formation of enlarged nuclei with intact NPCs that exhibited increased import of INM proteins [27].
Mechanisms of nuclear shape regulation
Recent studies support previous research demonstrating that nuclear lamins modulate nuclear shape [11,28,29]. During granulopoesis, neutrophils developing lobulated nuclei increase expression of lamin B receptor (LBR) and downregulate lamin A. Reducing LBR in neutrophil-differentiated HL-60 cells resulted in hypolobulated nuclei, while lamin A overexpression caused both nuclear hypolobulation and impaired neutrophil migration [30,31] (Fig. 1d). Cortical neurons in lamin B1 knock-out mice exhibited abnormally shaped nuclei with blebs and irregular lamin B2 distribution, while lamin B2 deficiency resulted in neurons with elongated nuclei accompanied by severe defects in brain development [32]. Abnormal nuclear shape and premature senescence of Ataxia telangiectasia cells were rescued by reducing lamin B1 levels [33].
Diseases caused by mutations in nuclear lamin genes, collectively termed laminopathies, are frequently associated with altered nuclear shape [34]. Lamin A mutations give rise to muscular dystrophies, familial partial lipodystrophy, dilated cardiomyopathy, and Hutchinson-Gilford progeria syndrome (HGPS). HGPS misshapen nuclei are caused by an inappropriately farnesylated form of lamin A, called progerin, that improperly incorporates into the lamina (Fig. 1e). Inhibition of farnesyltransferase or farnesylcysteine methylation improved nuclear morphology and phenotypes of progeroid mice [35,36]. Similarly, inhibiting prelamin A farnesylation significantly reduced nuclear shape abnormalities in HGPS patient fibroblasts [37]. Strikingly, reducing levels of the INM protein Sun1 in laminopathy mouse models and HGPS patient cells rescued defects in nuclear shape and cellular senescence [38], as did inhibition of the mTOR pathway which reduced progerin levels and nuclear blebbing, detected by a novel automated and high-throughput image analysis method that quantifies NE curvature [39,40].
NE components other than the lamina also influence nuclear shape [11,28]. Mutations in budding yeast nucleoporins that cause NPC clustering are frequently associated with altered nuclear shape [41,42]. Deletion of yeast proteins Mlp1p and Mlp2p, structural components of the NPC basket thought to link and properly distribute neighboring NPCs, led to increased NPC mobility and clustering and the formation of fragile, misshapen nuclei that frequently exhibited NE blebs [43]. Arabidopsis thaliana expresses a plant-specific nucleoporin, Nup136, a functional homolog of vertebrate Nup153. Nup136 overexpression increased nuclear size and elongation in many tissues, whereas reducing Nup136 expression resulted in smaller, more spherical nuclei [44,45] (Fig. 1f). Arabidopsis also expresses LINC complex proteins that regulate nuclear shape. Knockdown of AtSUN1 and AtSUN2 produced round nuclei in root hairs, compared to highly elongated wild-type nuclei [46]. Furthermore, Arabidopsis WIPs, plant-specific KASH-domain proteins, interact with SUN1/2 proteins, and disrupting these interactions reduced nuclear elongation [47].
Recent studies highlight an important role for perinuclear actin caps in controlling nuclear shape and positioning [48]. The multi-lobed, elongated nuclei of embryonic stem cells lack a perinuclear actin cap. During differentiation, these nuclei were observed to take on a more smooth, rounded morphology concomitantly with the appearance of a perinuclear actin cap, which wrapped around the nuclear surface making contacts across the NE with lamin A/C through LINC complex proteins [49] (Fig. 1g). Intriguingly, in mouse models of progeria and muscular dystrophy, the perinuclear actin cap was disrupted or absent and nuclei assumed deformed shapes [50].
There is growing evidence that physical properties of the ECM modulate nuclear shape. The actin-myosin cytoskeleton transmits mechanical force from focal adhesions at the cell membrane/ECM junction to nuclear LINC complexes and the lamina (reviewed in [29]), and the actin cytoskeleton was also shown to be important in coordinating nuclear shape with cell shape [51]. Rigidity of the substrate on which NIH 3T3 cells were grown modulated nuclear shape, such that soft substrates produced cells with round nuclei while stiff substrates led to flattened nuclei [52]. Dermal fibroblasts from laminopathy patients exhibited round nuclei on soft substrates but misshapen or ruptured nuclei on stiff substrates [53] (Fig. 1h). Intriguing interactions between lamin A levels, ECM stiffness, and cell differentiation have also recently emerged [54], as well as a novel role for keratin filaments in regulating nuclear shape [55]. Taken together, nuclear shape is determined by structural elements of the nucleus (Table 1), cytoplasmic and extracellular structures, and cytoskeletal tension transduced from the ECM. In the case of disease, weakened nuclei may contribute to abnormal nuclear morphology.
Cell cycle events that influence nuclear morphology
Events that occur during mitosis are important in establishing normal interphase nuclear morphology [1]. Depletion of microtubule-binding ER proteins REEP3/4 caused inappropriate ER accumulation on metaphase chromosomes, leading to NE defects during interphase [56] (Fig. 2a). In addition to clearance of ER membrane from metaphase chromosomes, mitotic microtubule dynamics also influence interphase nuclear morphology. In Xenopus egg extract, chromatin-binding protein Dppa2 inhibits post-mitotic microtubule polymerization, and depletion of Dppa2 led to the formation of small, misshapen nuclei with decreased nuclear lamin and NPC assembly. Strikingly, these nuclear morphology defects could be rescued by ectopically depolymerizing microtubules, suggesting that precisely tuned microtubule dynamics are required for proper nuclear assembly [57] (Fig. 2b). Also important for establishing correct nuclear shape are interactions between chromatin-bound BAF and LEM family INM proteins that mediate nuclear assembly (Fig. 1). LEM4 depletion or mutation caused misshapen, multi-lobed nuclei in C. elegans [58] (Fig. 2c), and in Schizosaccharomyces japonicus, the conserved LEM-domain protein Man1 was required for equal partitioning of nuclear membrane and NPCs to daughter nuclei [59]. During the closed mitoses of many yeasts, spindle pole bodies inserted into the NE drive intranuclear spindle formation. Elongation of the internally-forming spindle profoundly alters nuclear shape, as the intact nucleus expands along the mother-daughter axis prior to cytokinesis [3-6].
Figure 2. Cell cycle events that influence nuclear morphology.
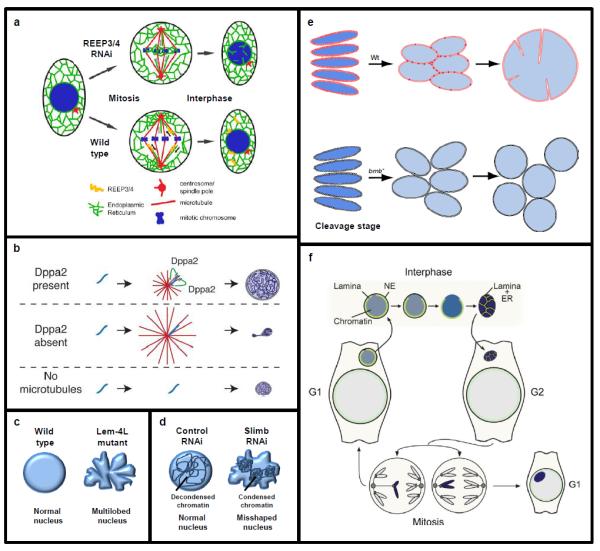
(a) REEP3/4 are required to clear ER membrane from metaphase chromatin. Failure to do so leads to intranuclear membrane invaginations extending into the interphase nucleus. Image adapted with permission from [56]. (b) Post-mitotic suppression of microtubule (red) polymerization by Dppa2 (green) is required for the formation of a nucleus with normal morphology. Chromatin is shown in blue. Image adapted with permission from [57]. (c) Depletion of LEM4 in C. elegans leads to misshapen, multi-lobed nuclei [58]. (d) Depletion of the ubiquitin ligase SCFSlimb leads to increased condensin II activity in interphase, chromatin compaction, and deformed nuclear morphology [70]. (e) The brambleberry protein (red) is required for karyomere fusion during early zebrafish development. In the absence of brambleberry (bmb) multiple micronuclei form. Image adapted with permission from [71]. (f) The process of micronuclear formation and disruption is depicted. A micronucleus forms around a lagging chromosome at the end of mitosis. During interphase, disorganization of the nuclear lamina leads to NE collapse, chromatin compaction, and intercalation of tubular ER. Image adapted with permission from [104].
The regulation of phospholipid biosynthesis is important in maintaining normal nuclear structure and dynamics. In fission yeast, a temperature-sensitive RanGEF mutant exhibited asymmetric cell divisions, reduced post-mitotic nuclear growth, and frequent NE breakage, phenotypes that could be rescued by slowing spindle elongation, increasing proliferation of ER membrane, or increasing the relative proportion of ER sheets [60]. In budding yeast, expression of a dominant negative SUN-domain protein, Mps3, led to over-proliferation of the INM that could be rescued by altering lipid homeostasis [61]. Upregulating lipid biosynthesis in yeast led to ER and NE membrane proliferation and the formation of misshapen nuclei in which the NE expanded specifically in the region adjacent to the nucleolus, forming a structure termed a "flare" [28,62,63]. More irregular nuclear shapes were observed if endosome to late Golgi trafficking was also disrupted, but a normal nuclear/cell volume ratio was maintained [64]. Mitotic delay induced similar nucleolar flares that could be rescued by inhibition of phospholipid synthesis [65], and ER-NE lipid partitioning controlled NE assembly and growth in higher eukaryotes as well [66,67]. In C. elegans, downregulation of lipin homolog LPIN-1 altered ER composition, NE breakdown, chromosome segregation, and nuclear morphology [68]. Mutations affecting trafficking through the ER-Golgi intermediate compartment also affected NE structure, disrupting transport of NE proteins and nucleoporins [69].
During interphase different mechanisms promote the maintenance of proper nuclear morphology. Inappropriate condensin II activity in interphase, caused by depletion of the ubiquitin ligase SCFSlimb, resulted in chromatin compaction and deformed, ruffled nuclei [70] (Fig. 2d). Early in development in some fish and amphibian embryos, post-mitotic NE assembly is initiated around individual chromosomes, forming structures called karyomeres that eventually fuse to form an intact nucleus. The zebrafish protein brambleberry was shown to be required for fusion of karyomeres into a mononucleus and for the regulation of normal nuclear morphology in early development [71] (Fig. 2e).
Nuclear morphology, chromatin organization, and gene expression
An important function of nuclear architecture is to organize chromatin and regulate gene expression. Some factors that structure and modify chromatin also influence nuclear morphology and size. Whether nuclear size directly impacts subnuclear function remains an open question. Recent studies have quantified chromatin positioning and gene expression during development and differentiation, processes associated with changes in nuclear size (reviewed in [72,73]), and modeling studies suggest that nuclear volume could drive chromatin organization [74,75].
In immortalized mammary epithelial cells, knockdown of a chromatin-remodeling enzyme, BRG1, resulted in nuclear periphery grooves [76]. During neuronal cell maturation, the chromatin-associated protein MeCP2 was required for normal developmental nuclear growth and gene transcription [77]. RNAi of soybean GmFWL1 resulted in decreased nuclear size, likely due to altered heterochromatization [78]. During C. elegans embryogenesis, reductions in nuclear size were shown to correlate with increased mitotic chromosome condensation [79] and interphase genome reorganization resulting in activated genes shifting towards the nuclear lumen and silenced genes localizing to the NE [80]. Interestingly, increasing the size of Xenopus embryonic nuclei did not result in increased mitotic chromosome length or width [81]. During T-cell activation, both actin-mediated nuclear elongation and activation of signaling intermediates were required to alter gene expression [82].
Changes in histone modifications have also been linked to nuclear size and shape. Bone marrow mesenchymal stem cells placed under mechanical strain by microtopographic patterning exhibited elongated nuclei and increased histone acetylation and gene transcription [83,84]. During myotube formation in human primary myoblasts, nuclei became smaller and more flattened, accompanied by altered histone modifications, chromatin remodeling, and gene expression silencing [85]. In mouse myoblasts, increased histone H3 acetylation correlated with increased nuclear size and F-actin cytoskeleton content [86], which has been shown to be required in large nuclei to stabilize subnuclear organization against gravitational forces [87]. Null mutations in the mouse cannabinoid receptor type 1 resulted in spermatozoa with elongated nuclei, reduced histone retention, and poor chromatin quality, which could be rescued by estrogen treatment [88].
Osmotic stress is known to induce global changes in gene expression. Articular chondrocytes under hyper-osmotic conditions showed increased nucleocytoplasmic transport, decreased nuclear size, and a more convoluted NE morphology. Conversely, under hypo-osmotic stress, nuclei swelled, assuming a smooth spherical shape limited by NE stiffness, with no effect on nucleocytoplasmic transport [89,90]. Osmotic stress may be a useful system for elucidating the relationship between nuclear morphology and gene expression [91].
Some structural components of the NE that regulate nuclear size have also been shown to affect chromatin structure. During Drosophila embryogenesis, the NE protein Kugelkern and dynamic microtubules were necessary to maintain normal NE morphology and chromatin dynamics and to activate zygotic gene transcription [92,93]. During differentiation of human embryonic stem cells, increased expression of lamin A/C and emerin were associated with increased nuclear size, the appearance of nuclear invaginations or lobes, and large-scale chromatin reorganization [94]. Overexpression of budding yeast Esc1, a NE protein with roles in chromatin organization and gene expression, led to NE extensions into the cytoplasm [95]. In Arabidopsis thaliana, seed germination increased expression of lamin-like nuclear matrix proteins, resulting in independently regulated nuclear growth and chromatin decondensation [96].
Nuclear morphology in cancer
Nuclear size differs between normal and cancer cells, and nuclear atypia is a common diagnostic and prognostic marker [7,8]. In lung cancer cells and adenocarcinomas, increased p53 expression and decreased expression of p16INK4A, a regulator of Rb, correlated with increased nuclear size and chromatin density as well as distortion of the NE [97]. In mucinous ovarian cancer, LINE1 DNA hypomethylation and increased nuclear area correlated with greater cell proliferation rates, aneuploidy, and reduced survival probability [98]. Lamin A was identified as a potential new biomarker for prostate cancer progression, which is associated with altered nuclear size, shape, and heterochromatin organization. Compared to benign samples, lamin A was downregulated in low grade tumors and upregulated in high grade tumors [99,100]. Tumor regression in response to treatment of breast cancer with anti-estrogen therapy was associated with decreased nuclear size in tumor cells, suggesting that reductions in nuclear size might be used to assess treatment efficacy [101].
Micronuclei, extranuclear structures generated by chromosome missegregation during mitosis, are common in cancer cells and may be degraded by nucleophagy [102]. Micronuclei were observed to generate extensively fragmented chromosomes that could be distributed to daughter nuclei, potentially contributing to chromosomal rearrangements and aneuploidy associated with cancer progression [103]. Micronuclear disruption, triggered by NE collapse and lamina disorganization, was frequently observed in cancer cells and may represent a general characteristic of genomic instability useful in diagnosis or prognosis [104,105] (Fig. 2f). Nuclear blebs are also common pathological features in cancers and laminopathies. Modeling studies indicated that separation of lamin fibers within the meshwork of the lamina is required for bleb formation, suggesting a possible approach to preventing these NE deformations in disease [106,107].
Recent diagnostic advances relevant to altered nuclear size in cancer have been reported. High grade urothelial carcinomas exhibited increased nuclear size, relative to low grade cases, that correlated with increased numbers of centromeres, detected by FISH using centromere enumeration probes (CEPs). Such CEPs may be applied to diagnosis of bladder carcinomas [108]. Tomographic imaging performed on fibrocystic and malignant mammary epithelial cells revealed abnormal nuclear shape, increased nuclear volume, greater numbers of nucleoli, and increased chromatin density and clumping, compared to normal cells [109]. Digital image analysis performed on melanocytic lesions comparing 62 features, including nuclear area, shape, and texture, allowed for effective differentiation of melanoma stages and subtypes [110]. Taken together, automated quantitative 3D nuclear morphometry could be useful as a novel diagnostic tool. These and future advancements in the analysis of digital histological images promise to improve diagnosis and provide for more individualized therapeutic interventions.
Conclusions
Some common mechanistic themes in the regulation of nuclear morphology are beginning to emerge. NE structural components, especially nuclear lamins, lamin-associated and lamin-like proteins, LINC complex proteins, and NPCs, are frequently involved, as is regulated nuclear import of these components. Perinuclear elements also influence nuclear morphology, such as ER structure and mechanical forces transduced by the actin and microtubule cytoskeletons, sometimes through ECM associations. In many systems, DNA amount appears to not be a primary determinant of nuclear size, while chromatin structure and modification can affect nuclear morphology. It is also becoming evident that mitotic events determine interphase nuclear morphology, where clearance of ER and microtubules from chromosomes and proper lipid homeostasis are important.
Open questions remain regarding the functional significance of nuclear morphology and how steady-state nuclear morphology is determined. The regulation of nuclear size and shape may be intimately linked, for instance it was recently proposed that changes in nuclear shape that maintain a constant karyoplasmic ratio may in fact be manifestations of altered nuclear size [28,64]. Although factors that influence nuclear size and shape are known, an integrated model of the mechanisms controlling nuclear morphology has yet to emerge, and upstream regulatory determinants and/or signals of nuclear size regulation remain to be identified. Emerging technologies, such as microfluidics [111], cell encapsulation [112], advances in microscopy [113], and 3D cell culture systems [114], broaden the possibilities for studying mechanisms of organelle scaling. In one recent example, microfluidic devices were used to encapsulate mitotic Xenopus extract within microdroplets of defined and tunable size, demonstrating how cytoplasmic volume contributes to mitotic spindle length scaling [115,116] and offering evidence for limiting-component models of organelle size regulation [117-119]. Similar approaches may be applied to study mechanisms regulating nuclear size and shape. Coupled with emerging technologies, Xenopus has been, and will continue to be, a powerful system to investigate intrinsic mechanisms of organelle size regulation [120].
Higher order chromatin organization is usually perturbed in cancers and can affect mutation frequencies [121,122], but the cause and effect relationships between increased nuclear size and altered subnuclear organization remain to be elucidated. One hypothesis is that nuclear size directly affects chromatin organization and gene expression. Enlarged nuclei are often observed in cells adjacent to a tumor, and these cells appear otherwise normal by histology. An explanation for this “field effect” is that genetic alterations leading to cancer occur in a stepwise fashion and cells in the field around the tumor represent a clonal population arising from an early genetic change that was a precursor to carcinogenesis [123]. By this model, precancerous alterations in nuclear size might disrupt chromatin positioning, thereby influencing transcriptional profiles and priming cancer development or promoting additional genetic alterations that contribute to cancer progression.
Development, differentiation, and disease progression are associated with changes in nuclear size, nuclear morphology, chromatin organization, and gene expression. Determining how these parameters relate to one another can now be accomplished using new techniques for mapping global chromatin organization coupled with massively parallel sequencing technologies (reviewed in [72,73,124]). Elucidating how developmentally regulated changes in nuclear size affect gene expression will help to clarify the relationship between aberrant nuclear morphology and diverse disease states such as cancer, progeria and other laminopathies, and neuronal disorders [77,125]. Answering fundamental questions about nuclear size and shape regulation promises to provide novel approaches to disease diagnosis, prevention, and treatment.
Highlights.
Mechanisms of nuclear size and shape regulation
Cell cycle events that influence nuclear morphology
Nuclear morphology, chromatin organization, and gene expression
Nuclear morphology in cancer
Acknowledgements
We thank Karen White for critical reading of the manuscript. P.J. is supported by the University of Wyoming Agricultural Experiment Station’s Competitive Grants Program. L.J.E. and L.D.V. are supported by Wyoming IDeA Networks for Biomedical Research Excellence (INBRE) graduate assistantships. Work in the Levy lab is supported by the NIGMS (R15GM106318).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
•• of outstanding interest
- 1.Schooley A, Vollmer B, Antonin W. Building a nuclear envelope at the end of mitosis: coordinating membrane reorganization, nuclear pore complex assembly, and chromatin de-condensation. Chromosoma. 2012;121:539–554. doi: 10.1007/s00412-012-0388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tapley EC, Starr DA. Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr Opin Cell Biol. 2013;25:57–62. doi: 10.1016/j.ceb.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang D, Oliferenko S. Remodeling the nuclear membrane during closed mitosis. Curr Opin Cell Biol. 2013;25:142–148. doi: 10.1016/j.ceb.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Martinez J, Rout MP. Nuclear pore complex biogenesis. Curr Opin Cell Biol. 2009;21:603–612. doi: 10.1016/j.ceb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smoyer CJ, Jaspersen SL. Breaking down the wall: the nuclear envelope during mitosis. Curr Opin Cell Biol. 2014;26:1–9. doi: 10.1016/j.ceb.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Winey M, Bloom K. Mitotic spindle form and function. Genetics. 2012;190:1197–1224. doi: 10.1534/genetics.111.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jevtic P, Levy DL. Mechanisms of nuclear size regulation in model systems and cancer. In: Schirmer EC, de Las Heras J, editors. In Cancer Biology and the Nuclear Envelope. Springer Science+Business; 2013. In Press. [Google Scholar]
- 8.Chow KH, Factor RE, Ullman KS. The nuclear envelope environment and its cancer connections. Nat Rev Cancer. 2012;12:196–209. doi: 10.1038/nrc3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edens LJ, White KH, Jevtic P, Li X, Levy DL. Nuclear size regulation: from single cells to development and disease. Trends Cell Biol. 2013;23:151–159. doi: 10.1016/j.tcb.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Levy DL, Heald R. Mechanisms of intracellular scaling. Annu Rev Cell Dev Biol. 2012;28:113–135. doi: 10.1146/annurev-cellbio-092910-154158. [DOI] [PubMed] [Google Scholar]
- 11.Webster M, Witkin KL, Cohen-Fix O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J Cell Sci. 2009;122:1477–1486. doi: 10.1242/jcs.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shumaker DK, Lopez-Soler RI, Adam SA, Herrmann H, Moir RD, Spann TP, Goldman RD. Functions and dysfunctions of the nuclear lamin Ig-fold domain in nuclear assembly, growth, and Emery-Dreifuss muscular dystrophy. Proc Natl Acad Sci U S A. 2005;102:15494–15499. doi: 10.1073/pnas.0507612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newport JW, Wilson KL, Dunphy WG. A lamin-independent pathway for nuclear envelope assembly. J Cell Biol. 1990;111:2247–2259. doi: 10.1083/jcb.111.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy DL, Heald R. Nuclear size is regulated by importin alpha and Ntf2 in Xenopus. Cell. 2010;143:288–298. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ralle T, Grund C, Franke WW, Stick R. Intranuclear membrane structure formations by CaaX-containing nuclear proteins. J Cell Sci. 2004;117:6095–6104. doi: 10.1242/jcs.01528. [DOI] [PubMed] [Google Scholar]
- 16.Prufert K, Vogel A, Krohne G. The lamin CxxM motif promotes nuclear membrane growth. J Cell Sci. 2004;117:6105–6116. doi: 10.1242/jcs.01532. [DOI] [PubMed] [Google Scholar]
- 17.Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell. 2007;19:2793–2803. doi: 10.1105/tpc.107.053231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chmielewska M, Dubinska-Magiera M, Sopel M, Rzepecka D, Hutchison CJ, Goldberg MW, Rzepecki R. Embryonic and adult isoforms of XLAP2 form microdomains associated with chromatin and the nuclear envelope. Cell Tissue Res. 2011;344:97–110. doi: 10.1007/s00441-011-1129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gant TM, Harris CA, Wilson KL. Roles of LAP2 proteins in nuclear assembly and DNA replication: truncated LAP2beta proteins alter lamina assembly, envelope formation, nuclear size, and DNA replication efficiency in Xenopus laevis extracts. J Cell Biol. 1999;144:1083–1096. doi: 10.1083/jcb.144.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Brien LL, Wiese C. TPX2 is required for postmitotic nuclear assembly in cell-free Xenopus laevis egg extracts. J Cell Biol. 2006;173:685–694. doi: 10.1083/jcb.200512107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu W, Schneider M, Neumann S, Jaeger VM, Taranum S, Munck M, Cartwright S, Richardson C, Carthew J, Noh K, et al. Nesprin interchain associations control nuclear size. Cell Mol Life Sci. 2012;69:3493–3509. doi: 10.1007/s00018-012-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.The actin binding domain of nesprin-2, interaction of nesprin-2 with nesprin-3, and actin cytoskeletal connections through LINC complexes are required for proper nuclear size determination in human and COS-7 cell culture.
- 22.Luke Y, Zaim H, Karakesisoglou I, Jaeger VM, Sellin L, Lu W, Schneider M, Neumann S, Beijer A, Munck M, et al. Nesprin-2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J Cell Sci. 2008;121:1887–1898. doi: 10.1242/jcs.019075. [DOI] [PubMed] [Google Scholar]
- 23.Maeshima K, Iino H, Hihara S, Funakoshi T, Watanabe A, Nishimura M, Nakatomi R, Yahata K, Imamoto F, Hashikawa T, et al. Nuclear pore formation but not nuclear growth is governed by cyclin-dependent kinases (Cdks) during interphase. Nat Struct Mol Biol. 2010;17:1065–1071. doi: 10.1038/nsmb.1878. [DOI] [PubMed] [Google Scholar]
- 24.Maeshima K, Iino H, Hihara S, Imamoto N. Nuclear size, nuclear pore number and cell cycle. Nucleus. 2011;2:113–118. doi: 10.4161/nucl.2.2.15446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.The doubling of nuclear volume and NPC number during interphase are regulated through distinct mechanisms, and Cdk activity is required during interphase in human cells for early nuclear pore generation.
- 25.Titus LC, Dawson TR, Rexer DJ, Ryan KJ, Wente SR. Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol Biol Cell. 2010;21:1072–1087. doi: 10.1091/mbc.E09-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaulov L, Gruber R, Cohen I, Harel A. A dominant-negative form of POM121 binds chromatin and disrupts the two separate modes of nuclear pore assembly. J Cell Sci. 2011;124:3822–3834. doi: 10.1242/jcs.086660. [DOI] [PubMed] [Google Scholar]
- 27.Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol. 2010;189:1129–1142. doi: 10.1083/jcb.200912045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walters AD, Bommakanti A, Cohen-Fix O. Shaping the nucleus: Factors and forces. J Cell Biochem. 2012;113:2813–2821. doi: 10.1002/jcb.24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, Olins AL, Herrmann H, Weitz DA, Lammerding J. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J Biol Chem. 2013;288:8610–8618. doi: 10.1074/jbc.M112.441535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.Differentiated neutrophils are characterized by lobulated nuclear shape and ability to traverse constrictions smaller than their own diameter. Lamin A levels significantly affect the ability of differentiated neutrophils to pass through micron-sized pores, while the multilobulated shape of the nuclei plays a lesser role.
- 31.Olins AL, Ernst A, Zwerger M, Herrmann H, Olins DE. An in vitro model for Pelger-Huet anomaly: stable knockdown of lamin B receptor in HL-60 cells. Nucleus. 2010;1:506–512. doi: 10.4161/nucl.1.6.13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coffinier C, Jung HJ, Nobumori C, Chang S, Tu Y, Barnes RH, 2nd, Yoshinaga Y, de Jong PJ, Vergnes L, Reue K, et al. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol Biol Cell. 2011;22:4683–4693. doi: 10.1091/mbc.E11-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barascu A, Le Chalony C, Pennarun G, Genet D, Imam N, Lopez B, Bertrand P. Oxidative stress induces an ATM-independent senescence pathway through p38 MAPK-mediated lamin B1 accumulation. Embo J. 2012;31:1080–1094. doi: 10.1038/emboj.2011.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar DZ, Barkan R, Meshorer E, Gruenbaum Y. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med. 2009;13:1059–1085. doi: 10.1111/j.1582-4934.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Ostlund C, Worman HJ. Blocking protein farnesylation improves nuclear shape abnormalities in keratinocytes of mice expressing the prelamin A variant in Hutchinson-Gilford progeria syndrome. Nucleus. 2010;1:432–439. doi: 10.4161/nucl.1.5.12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ibrahim MX, Sayin VI, Akula MK, Liu M, Fong LG, Young SG, Bergo MO. Targeting isoprenylcysteine methylation ameliorates disease in a mouse model of progeria. Science. 2013;340:1330–1333. doi: 10.1126/science.1238880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bifulco M, D'Alessandro A, Paladino S, Malfitano AM, Notarnicola M, Caruso MG, Laezza C. N6-isopentenyladenosine improves nuclear shape in fibroblasts from humans with progeroid syndromes by inhibiting the farnesylation of prelamin A. FEBS J. 2013;280:6223–6232. doi: 10.1111/febs.12544. [DOI] [PubMed] [Google Scholar]
- ••.Inhibition of farnesylation with an inhibitor of farnesyl diphosphate synthase, N6-isopentenyladenosine, decreases farnesylation of prelamin A, and in combination with lovastatin significantly improves nuclear morphology in fibroblasts isolated from atypical progeria patients.
- 38.Chen CY, Chi YH, Mutalif RA, Starost MF, Myers TG, Anderson SA, Stewart CL, Jeang KT. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell. 2012;149:565–577. doi: 10.1016/j.cell.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao K, Graziotto JJ, Blair CD, Mazzulli JR, Erdos MR, Krainc D, Collins FS. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci Transl Med. 2011;3:89ra58. doi: 10.1126/scitranslmed.3002346. [DOI] [PubMed] [Google Scholar]
- 40.Driscoll MK, Albanese JL, Xiong ZM, Mailman M, Losert W, Cao K. Automated image analysis of nuclear shape: what can we learn from a prematurely aged cell? Aging (Albany NY) 2012;4:119–132. doi: 10.18632/aging.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wente SR, Blobel G. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol. 1994;125:955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pemberton LF, Rout MP, Blobel G. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc Natl Acad Sci U S A. 1995;92:1187–1191. doi: 10.1073/pnas.92.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, Strambio-De-Castillia C. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell. 2013;24:3920–3938. doi: 10.1091/mbc.E13-07-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2010;22:4084–4097. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Hara-Nishimura I. Involvement of the nuclear pore complex in morphology of the plant nucleus. Nucleus. 2011;2:168–172. doi: 10.4161/nucl.2.3.16175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oda Y, Fukuda H. Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J. 2011;66:629–641. doi: 10.1111/j.1365-313X.2011.04523.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhou X, Graumann K, Evans DE, Meier I. Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol. 2012;196:203–211. doi: 10.1083/jcb.201108098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gay O, Gilquin B, Nakamura F, Jenkins ZA, McCartney R, Krakow D, Deshiere A, Assard N, Hartwig JH, Robertson SP, et al. RefilinB (FAM101B) targets filamin A to organize perinuclear actin networks and regulates nuclear shape. Proc Natl Acad Sci U S A. 2011;108:11464–11469. doi: 10.1073/pnas.1104211108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khatau SB, Kusuma S, Hanjaya-Putra D, Mali P, Cheng L, Lee JS, Gerecht S, Wirtz D. The differential formation of the LINC-mediated perinuclear actin cap in pluripotent and somatic cells. PLoS One. 2012;7:e36689. doi: 10.1371/journal.pone.0036689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.Nuclear envelope morphology changes during the differentiation of stem cells, correlating with formation of a perinuclear actin cap, expression and proper localization of lamin A and LINC complexes, and establishment of intranuclear connections with actin cap filaments that contribute to the modulation of nuclear shape.
- 50.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci U S A. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Versaevel M, Grevesse T, Gabriele S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat Commun. 2012;3:671. doi: 10.1038/ncomms1668. [DOI] [PubMed] [Google Scholar]
- 52.Lovett DB, Shekhar N, Nickerson JA, Roux KJ, Lele TP. Modulation of Nuclear Shape by Substrate Rigidity. Cell Mol Bioeng. 2013;6:230–238. doi: 10.1007/s12195-013-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.To test how mechanical properties of the ECM influence nuclear morphology, NIH 3T3 fibroblasts were grown on polyacrylamide gels of differing stiffness and nuclei became more elongated and flattened with increasing matrix stiffness.
- 53.Tamiello C, Kamps MA, van den Wijngaard A, Verstraeten VL, Baaijens FP, Broers JL, Bouten CC. Soft substrates normalize nuclear morphology and prevent nuclear rupture in fibroblasts from a laminopathy patient with compound heterozygous LMNA mutations. Nucleus. 2013;4:61–73. doi: 10.4161/nucl.23388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee CH, Kim MS, Chung BM, Leahy DJ, Coulombe PA. Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat Struct Mol Biol. 2012;19:707–715. doi: 10.1038/nsmb.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlaitz AL, Thompson J, Wong CC, Yates JR, 3rd, Heald R. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev Cell. 2013;26:315–323. doi: 10.1016/j.devcel.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.A biochemical screen to detect proteins that mediate interactions between membrane-bound organelles and microtubules identified REEP4, an ER binding protein. REEP3/4 redundantly regulate ER membrane clearance from post-mitotic chromosomes, important for ensuring normal nuclear morphology in the subsequent interphase.
- 57.Xue JZ, Woo EM, Postow L, Chait BT, Funabiki H. Chromatin-bound Xenopus dppa2 shapes the nucleus by locally inhibiting microtubule assembly. Dev Cell. 2013;27:47–59. doi: 10.1016/j.devcel.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.Proper spatiotemporal regulation of microtubule dynamics is important for nuclear assembly and regulation of normal nuclear size and shape. The Dppa2 protein inhibits microtubule polymerization around chromatin during NE reassembly, and excessive microtubules resulting from Dppa2 depletion led to the formation of misshaped nuclei and altered DNA replication.
- 58.Asencio C, Davidson IF, Santarella-Mellwig R, Ly-Hartig TB, Mall M, Wallenfang MR, Mattaj IW, Gorjanacz M. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell. 2012;150:122–135. doi: 10.1016/j.cell.2012.04.043. [DOI] [PubMed] [Google Scholar]
- 59.Yam C, Gu Y, Oliferenko S. Partitioning and Remodeling of the Schizosaccharomyces japonicus Mitotic Nucleus Require Chromosome Tethers. Curr Biol. 2013;23:2303–2310. doi: 10.1016/j.cub.2013.09.057. [DOI] [PubMed] [Google Scholar]
- 60.Gonzalez Y, Meerbrey K, Chong J, Torii Y, Padte NN, Sazer S. Nuclear shape, growth and integrity in the closed mitosis of fission yeast depend on the Ran-GTPase system, the spindle pole body and the endoplasmic reticulum. J Cell Sci. 2009;122:2464–2472. doi: 10.1242/jcs.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Friederichs JM, Ghosh S, Smoyer CJ, McCroskey S, Miller BD, Weaver KJ, Delventhal KM, Unruh J, Slaughter BD, Jaspersen SL. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet. 2011;7:e1002365. doi: 10.1371/journal.pgen.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell JL, Lorenz A, Witkin KL, Hays T, Loidl J, Cohen-Fix O. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol Biol Cell. 2006;17:1768–1778. doi: 10.1091/mbc.E05-09-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siniossoglou S. Lipins, lipids and nuclear envelope structure. Traffic. 2009;10:1181–1187. doi: 10.1111/j.1600-0854.2009.00923.x. [DOI] [PubMed] [Google Scholar]
- 64.Webster MT, McCaffery JM, Cohen-Fix O. Vesicle trafficking maintains nuclear shape in Saccharomyces cerevisiae during membrane proliferation. J Cell Biol. 2010;191:1079–1088. doi: 10.1083/jcb.201006083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Witkin KL, Chong Y, Shao S, Webster MT, Lahiri S, Walters AD, Lee B, Koh JL, Prinz WA, Andrews BJ, et al. The Budding Yeast Nuclear Envelope Adjacent to the Nucleolus Serves as a Membrane Sink during Mitotic Delay. Curr Biol. 2012;22:1128–1133. doi: 10.1016/j.cub.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.In a screen for budding yeast mutants with abnormal nuclear morphology, mutations causing cell cycle delays were frequently associated with the appearance of nuclear extensions, that could be rescued by inhibition of phospholipid synthesis.
- 66.Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol. 2008;182:911–924. doi: 10.1083/jcb.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kiseleva E, Morozova KN, Voeltz GK, Allen TD, Goldberg MW. Reticulon 4a/NogoA locates to regions of high membrane curvature and may have a role in nuclear envelope growth. J Struct Biol. 2007;160:224–235. doi: 10.1016/j.jsb.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Golden A, Liu J, Cohen-Fix O. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Sci. 2009;122:1970–1978. doi: 10.1242/jcs.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tran D, Chalhoub A, Schooley A, Zhang W, Ngsee JK. A mutation in VAPB that causes amyotrophic lateral sclerosis also causes a nuclear envelope defect. J Cell Sci. 2012;125:2831–2836. doi: 10.1242/jcs.102111. [DOI] [PubMed] [Google Scholar]
- 70.Buster DW, Daniel SG, Nguyen HQ, Windler SL, Skwarek LC, Peterson M, Roberts M, Meserve JH, Hartl T, Klebba JE, et al. SCFSlimb ubiquitin ligase suppresses condensin II-mediated nuclear reorganization by degrading Cap-H2. J Cell Biol. 2013;201:49–63. doi: 10.1083/jcb.201207183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abrams EW, Zhang H, Marlow FL, Kapp L, Lu S, Mullins MC. Dynamic assembly of brambleberry mediates nuclear envelope fusion during early development. Cell. 2012;150:521–532. doi: 10.1016/j.cell.2012.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.The fidelity of rapid cell divisions during early fish and amphibian embryogenesis is ensured by the envelopment of individual segregated chromosomes with nuclear membrane, called karyomeres. In zebrafish, brambleberry localizes to regions where individual karyomeres contact each other and regulates NE fusion, as brambleberry mutations caused formation of multiple micronuclei.
- 72.Naumova N, Dekker J. Integrating one-dimensional and three-dimensional maps of genomes. J Cell Sci. 2010;123:1979–1988. doi: 10.1242/jcs.051631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 74.Kang J, Xu B, Yao Y, Lin W, Hennessy C, Fraser P, Feng J. A dynamical model reveals gene co-localizations in nucleus. PLoS Comput Biol. 2011;7:e1002094. doi: 10.1371/journal.pcbi.1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Albert B, Mathon J, Shukla A, Saad H, Normand C, Leger-Silvestre I, Villa D, Kamgoue A, Mozziconacci J, Wong H, et al. Systematic characterization of the conformation and dynamics of budding yeast chromosome XII. J Cell Biol. 2013;202:201–210. doi: 10.1083/jcb.201208186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Imbalzano KM, Cohet N, Wu Q, Underwood JM, Imbalzano AN, Nickerson JA. Nuclear shape changes are induced by knockdown of the SWI/SNF ATPase BRG1 and are independent of cytoskeletal connections. PLoS One. 2013;8:e55628. doi: 10.1371/journal.pone.0055628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yazdani M, Deogracias R, Guy J, Poot RA, Bird A, Barde YA. Disease modeling using embryonic stem cells: MeCP2 regulates nuclear size and RNA synthesis in neurons. Stem Cells. 2012;30:2128–2139. doi: 10.1002/stem.1180. [DOI] [PubMed] [Google Scholar]
- 78.Libault M, Zhang XC, Govindarajulu M, Qiu J, Ong YT, Brechenmacher L, Berg RH, Hurley-Sommer A, Taylor CG, Stacey G. A member of the highly conserved FWL (tomato FW2 -like) gene family is essential for soybean nodule organogenesis. Plant J. 2010;62:852–864. doi: 10.1111/j.1365-313X.2010.04201.x. [DOI] [PubMed] [Google Scholar]
- 79.Hara Y, Iwabuchi M, Ohsumi K, Kimura A. Intranuclear DNA density affects chromosome condensation in metazoans. Mol Biol Cell. 2013;24:2442–2453. doi: 10.1091/mbc.E13-01-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.Mitotic chromosome size directly correlates with interphase nuclear size during early C. elegans development, and altering the DNA amount leads to changes in chromatin condensation. In a similar fashion, altering nuclear size in Xenopus extracts changes chromosome size at mitosis, further suggesting that the degree of metaphase chromosome condensation is dynamically determined by interphase nuclear size.
- 80.Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 2010;24:766–782. doi: 10.1101/gad.559610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kieserman EK, Heald R. Mitotic chromosome size scaling in Xenopus. Cell Cycle. 2011;10:3863–3870. doi: 10.4161/cc.10.22.17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gupta S, Marcel N, Sarin A, Shivashankar GV. Role of actin dependent nuclear deformation in regulating early gene expression. PLoS One. 2012;7:e53031. doi: 10.1371/journal.pone.0053031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Y, Chu JS, Kurpinski K, Li X, Bautista DM, Yang L, Sung KL, Li S. Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys J. 2011;100:1902–1909. doi: 10.1016/j.bpj.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.Micropatterning of bone marrow mesenchymal stem cells and induced mechanical strain induce changes in nuclear matrix composition, nuclear morphology, and histone acetylation, indicating that microtransduction of the physical environment of the cell can alter chromatin structure and gene transcription.
- 84.Jain N, Iyer KV, Kumar A, Shivashankar GV. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci U S A. 2013;110:11349–11354. doi: 10.1073/pnas.1300801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rozwadowska N, Kolanowski T, Wiland E, Siatkowski M, Pawlak P, Malcher A, Mietkiewski T, Olszewska M, Kurpisz M. Characterisation of nuclear architectural alterations during in vitro differentiation of human stem cells of myogenic origin. PLoS One. 2013;8:e73231. doi: 10.1371/journal.pone.0073231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chambliss AB, Wu PH, Chen WC, Sun SX, Wirtz D. Simultaneously defining cell phenotypes, cell cycle, and chromatin modifications at single-cell resolution. FASEB J. 2013;27:2667–2676. doi: 10.1096/fj.12-227108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feric M, Brangwynne CP. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat Cell Biol. 2013;15:1253–1259. doi: 10.1038/ncb2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.Nuclei larger than ~10μm experience gravitational forces that can affect internal structures, in particular an F-actin scaffold within Xenopus germinal vesicles prevents gravitational sedimentation and fusion of subnuclear structures, including nucleoli and histone locus bodies.
- 88.Cacciola G, Chioccarelli T, Altucci L, Viggiano A, Fasano S, Pierantoni R, Cobellis G. Nuclear size as estrogen-responsive chromatin quality parameter of mouse spermatozoa. Gen Comp Endocrinol. 2013;193:201–209. doi: 10.1016/j.ygcen.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 89.Finan JD, Guilak F. The effects of osmotic stress on the structure and function of the cell nucleus. J Cell Biochem. 2010;109:460–467. doi: 10.1002/jcb.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Finan JD, Leddy HA, Guilak F. Osmotic stress alters chromatin condensation and nucleocytoplasmic transport. Biochem Biophys Res Commun. 2011;408:230–235. doi: 10.1016/j.bbrc.2011.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martins RP, Finan JD, Guilak F, Lee DA. Mechanical regulation of nuclear structure and function. Annu Rev Biomed Eng. 2012;14:431–455. doi: 10.1146/annurev-bioeng-071910-124638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hampoelz B, Azou-Gros Y, Fabre R, Markova O, Puech PH, Lecuit T. Microtubule-induced nuclear envelope fluctuations control chromatin dynamics in Drosophila embryos. Development. 2011;138:3377–3386. doi: 10.1242/dev.065706. [DOI] [PubMed] [Google Scholar]
- 93.Polychronidou M, Hellwig A, Grosshans J. Farnesylated nuclear proteins Kugelkern and lamin Dm0 affect nuclear morphology by directly interacting with the nuclear membrane. Mol Biol Cell. 2010;21:3409–3420. doi: 10.1091/mbc.E10-03-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Butler JT, Hall LL, Smith KP, Lawrence JB. Changing nuclear landscape and unique PML structures during early epigenetic transitions of human embryonic stem cells. J Cell Biochem. 2009;107:609–621. doi: 10.1002/jcb.22183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hattier T, Andrulis ED, Tartakoff AM. Immobility, inheritance and plasticity of shape of the yeast nucleus. BMC Cell Biol. 2007;8:47. doi: 10.1186/1471-2121-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van Zanten M, Koini MA, Geyer R, Liu Y, Brambilla V, Bartels D, Koornneef M, Fransz P, Soppe WJ. Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc Natl Acad Sci U S A. 2011;108:20219–20224. doi: 10.1073/pnas.1117726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Okudela K. An association between nuclear morphology and immunohistochemical expression of p53 and p16INK4A in lung cancer cells. Med Mol Morphol. 2013 doi: 10.1007/s00795-013-0052-x. In press. [DOI] [PubMed] [Google Scholar]
- ••.Increased p53 expression and decreased p16INK4A expression are shown to correlate with larger nuclei, increased chromatin density, and nuclear membrane distortion in lung cancer and adenocarcinomas. Different factors may underlie nuclear atypia in different cancers.
- 98.Zeimet AG, Fiegl H, Goebel G, Kopp F, Allasia C, Reimer D, Steppan I, Mueller-Holzner E, Ehrlich M, Marth C. DNA ploidy, nuclear size, proliferation index and DNA-hypomethylation in ovarian cancer. Gynecol Oncol. 2011;121:24–31. doi: 10.1016/j.ygyno.2010.12.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Skvortsov S, Schafer G, Stasyk T, Fuchsberger C, Bonn GK, Bartsch G, Klocker H, Huber LA. Proteomics profiling of microdissected low- and high-grade prostate tumors identifies Lamin A as a discriminatory biomarker. J Proteome Res. 2011;10:259–268. doi: 10.1021/pr100921j. [DOI] [PubMed] [Google Scholar]
- 100.Veltri RW, Christudass CS, Isharwal S. Nuclear morphometry, nucleomics and prostate cancer progression. Asian J Androl. 2012;14:375–384. doi: 10.1038/aja.2011.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Samarnthai N, Elledge R, Prihoda TJ, Huang J, Massarweh S, Yeh IT. Pathologic changes in breast cancer after anti-estrogen therapy. Breast J. 2012;18:362–366. doi: 10.1111/j.1524-4741.2012.01251.x. [DOI] [PubMed] [Google Scholar]
- 102.Mijaljica D, Devenish RJ. Nucleophagy at a glance. J Cell Sci. 2013;126:4325–4330. doi: 10.1242/jcs.133090. [DOI] [PubMed] [Google Scholar]
- 103.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.Nuclear envelope collapse, triggered by lamin B1 loss and lamina disorganization, causes micronuclear disruption in cancer cells.
- 105.Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 2012;3:88–100. doi: 10.4161/nucl.18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Funkhouser CM, Sknepnek R, Shimi T, Goldman AE, Goldman RD, Olvera de la Cruz M. Mechanical model of blebbing in nuclear lamin meshworks. Proc Natl Acad Sci U S A. 2013;110:3248–3253. doi: 10.1073/pnas.1300215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••.Nuclear blebs, frequently observed in cancer and progeria, are shown by modeling to preferentially form at sites where fibers in the lamina meshwork separate.
- 107.Wren NS, Zhong Z, Schwartz RS, Dahl KN. Modeling nuclear blebs in a nucleoskeleton of independent filament networks. Cell Mol Bioeng. 2012;5:73–81. doi: 10.1007/s12195-011-0196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shuto M, Seyama A, Gotoh Y, Kamada K, Nakamura M, Warigaya K, Watanabe H, Ueno M, Shimizu M, Fukuda T, et al. Significant Correlation between Chromosomal Aberration and Nuclear Morphology in Urothelial Carcinoma. Acta Histochem Cytochem. 2012;45:25–33. doi: 10.1267/ahc.11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nandakumar V, Kelbauskas L, Hernandez KF, Lintecum KM, Senechal P, Bussey KJ, Davies PC, Johnson RH, Meldrum DR. Isotropic 3D nuclear morphometry of normal, fibrocystic and malignant breast epithelial cells reveals new structural alterations. PLoS One. 2012;7:e29230. doi: 10.1371/journal.pone.0029230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miedema J, Marron JS, Niethammer M, Borland D, Woosley J, Coposky J, Wei S, Reisner H, Thomas NE. Image and statistical analysis of melanocytic histology. Histopathology. 2012;61:436–444. doi: 10.1111/j.1365-2559.2012.04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Y, Singh AK. Microfluidic platforms for single-cell protein analysis. J Lab Autom. 2013;18:446–454. doi: 10.1177/2211068213494389. [DOI] [PubMed] [Google Scholar]
- 112.Wang S, Guo Z. Bio-inspired encapsulation and functionalization of living cells with artificial shells. Colloids Surf B Biointerfaces. 2014;113:483–500. doi: 10.1016/j.colsurfb.2013.09.024. [DOI] [PubMed] [Google Scholar]
- 113.Zumbusch A, Langbein W, Borri P. Nonlinear vibrational microscopy applied to lipid biology. Prog Lipid Res. 2013;52:615–632. doi: 10.1016/j.plipres.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 114.Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science. 2013;342:853–856. doi: 10.1126/science.1243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. Cytoplasmic volume modulates spindle size during embryogenesis. Science. 2013;342:856–860. doi: 10.1126/science.1243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goehring NW, Hyman AA. Organelle growth control through limiting pools of cytoplasmic components. Curr Biol. 2012;22:R330–339. doi: 10.1016/j.cub.2012.03.046. [DOI] [PubMed] [Google Scholar]
- 118.Chan YH, Marshall WF. How cells know the size of their organelles. Science. 2012;337:1186–1189. doi: 10.1126/science.1223539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hara Y, Kimura A. Cell-size-dependent control of organelle sizes during development. Results Probl Cell Differ. 2011;53:93–108. doi: 10.1007/978-3-642-19065-0_5. [DOI] [PubMed] [Google Scholar]
- 120.Edens LJ, Levy DL. Size scaling of subcellular organelles and structures in X. laevis and X. tropicalis. In: Kubiak J, Kloc M, editors. In Xenopus Development. Life Science Books, Wiley-Blackwell; 2013. In press. [Google Scholar]
- 121.Reddy KL, Feinberg AP. Higher order chromatin organization in cancer. Semin Cancer Biol. 2013;23:109–115. doi: 10.1016/j.semcancer.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 123.Chai H, Brown RE. Field effect in cancer-an update. Ann Clin Lab Sci. 2009;39:331–337. [PubMed] [Google Scholar]
- 124.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zwerger M, Ho CY, Lammerding J. Nuclear mechanics in disease. Annu Rev Biomed Eng. 2011;13:397–428. doi: 10.1146/annurev-bioeng-071910-124736. [DOI] [PMC free article] [PubMed] [Google Scholar]