Abstract
Nuclear-cytoskeletal connections are central to fundamental cellular processes, including nuclear positioning and chromosome movements in meiosis. The cytoskeleton is coupled to the nucleoskeleton through conserved KASH-SUN bridges, or LINC complexes, that span the nuclear envelope. KASH proteins localize to the outer nuclear membrane where they connect the nucleus to the cytoskeleton. New findings have expanded the functional diversity of KASH proteins, showing that they interact with microtubule motors, actin, intermediate filaments, a nonconventional myosin, RanGAP, and each other. The role of KASH proteins in cellular mechanics is discussed. Genetic mutations in KASH proteins are associated with autism, hearing loss, cancer, muscular dystrophy and other diseases.
Introduction
SUN and KASH proteins form a bridge across the nuclear envelope, often referred to as the LINC complex, connecting the nucleoskeleton to the cytoskeleton [1]. KASH proteins are named after the founding members of the family, Drosophila Klarsicht, C. elegans ANC-1, and mammalian SYNE-1 and -2 (nesprin-1 and -2) [2]. All KASH proteins contain a C-terminal trans-membrane domain followed by a short (~10–32 residues), conserved luminal KASH domain that is necessary and sufficient to target the large, unconserved cytoplasmic domains to the outer surface of the nuclear envelope [1,3–5]. KASH proteins are targeted to the outer nuclear membrane through a direct physical interaction between the KASH domain and SUN proteins in the peri-nuclear space of the nuclear envelope. The KASH-SUN interaction was recently described at the structural level [6,7] and thoroughly reviewed [8–11]. There are many excellent comprehensive reviews on KASH and SUN proteins [1,3–5]. Here we focus on recent developments on the diverse array of functions that KASH proteins play at the cytoplasmic surface of the nucleus (Figure 1). KASH proteins function in transmitting mechanical forces from the cytoplasm to the nucleus. During meiosis, KASH proteins transmit forces generated in the cytoplasm that move telomeres inside the nucleus [12]. Given the wide variety of cell and developmental functions KASH proteins play, it is not surprising that defects in KASH proteins have been linked to an ever-growing list of human diseases (Table 1).
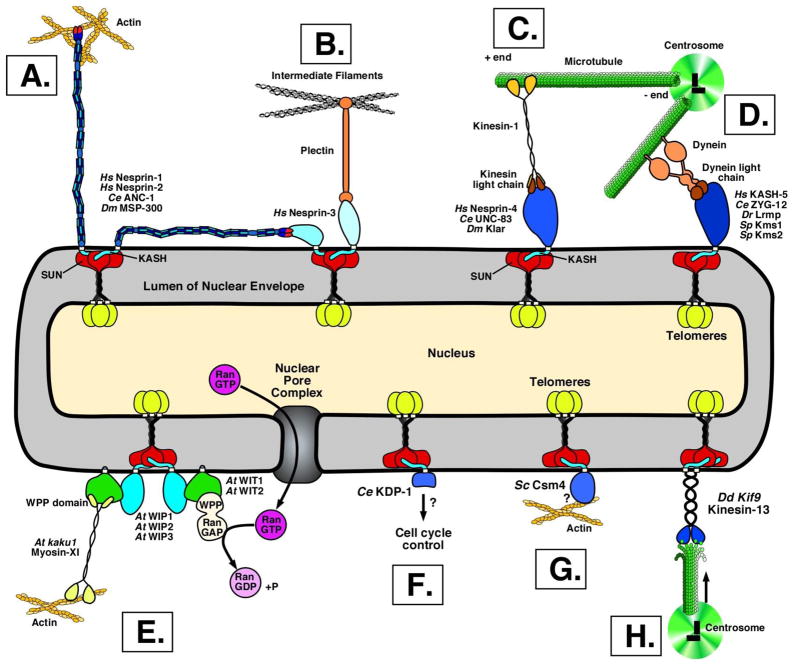
Figure 1. Functions of KASH proteins at the cytoplasmic face of the nucleus.
SUN proteins form trimers in the inner nuclear membrane with their conserved SUN domains (red) and coiled domains in the lumen of the nuclear envelope and nucleoplasmic domains (yellow). SUN domains interact with the KASH domain (light blue) in the lumen of the nuclear envelope. The cytoplasmic domains of KASH proteins (various shades of blue) are on the outer surface of the nucleus. SUN and KASH proteins are thought to interact in a three-to-three ratio. Only one or two KASH proteins are shown for each complex for simplicity. (A) Giant KASH proteins made of spectrin repeats tether the nucleus to actin networks. (A–B) Giant KASH proteins also interact with nesprin-3 to form a cage around the nucleus. (B) Nesprin-3 interacts with intermediate filaments through plectin. (C) KASH proteins recruit kinesin-1 to move nuclei. (D) KASH proteins recruit dynein to move nuclei, telomeres (or pairing centers in worms) or to connect centrosomes. (E) The plant KASH proteins WIP1, 2, and 3 interact with WITs (gren) to recruit a myosin-XI to move nuclei, and a RanGAP to catalyze the hydrolosis of GTP in Ran (pink) as it exits the nucleus. (F) The worm KASH protein KDP-1 regulates the cell cycle through unknown mechanisms. (G) Yeast Csm4 fits the definition of a KASH protein used to move telomeres along actin. (H) A novel kinesin-13 is a KASH protein that functions to attach the centrosome to nuclei. The names of KASH proteins from various systems, including humans (Hs), roundworms (Ce), fruit flies (Dm), zebrafish (Dr), fission yeast (Sp), budding yeast (Sc), slime molds (Dd), and angiosperms (At) are indicated.
Table 1.
Human diseases associated with genetic mutations in KASH proteins
| KASH protein | Disease | Reference |
|---|---|---|
| Nesprin-1 | Emery-Dreifuss muscular dystrophy | [70] |
| Myogenic autosomal recessive arthrogyroposis | [71] | |
| Autosomal recessive cerebellar ataxia | [72,73] | |
| Colorectal cancer | [74] | |
| Ovarian cancer | [75] | |
| Bipolar disorder | [76,77] | |
| Recurrent major depression | [77] | |
| Familial autism spectrum disorders | [78,79] | |
| Nesprin-2 | Emery-Dreifuss muscular dystrophy | [70] |
| Breast cancer | [74] | |
| Gastrointesitnal stromal tumors | [80] | |
| Nesprin-4 | High-frequency hearing loss | [19] |
KASH proteins form a complex nuclear scaffold
The canonical KASH proteins are mammalian nesprin-1 and nesprin-2, and their worm and fly orthologs ANC-1 and MSP-300 (Figure 1A). These giant (800–1000 kDa) proteins tether nuclei to the actin cytoskeleton, and play important roles in muscle and neuronal development [1]. They consist of N-terminal actin-binding domains and C-terminal KASH domains separated by a long rod consisting of over 70 spectrin repeats in the case of nesprin-1 [13]. Adding to the difficulties of studying nesprin-1 and -2 is the abundance of isoforms; 16 5′ start sites and 14 3′UTR in the nesprin-1 locus alone encode countless isoforms, many of which are tissue specific [14]. It was recently shown that the N-termini of nesprin-1 and -2 interact with the cytoplasmic domains of other KASH proteins, including the intermediate filament-associated KASH protein nesprin-3 [15] (Figure 1A–B). Together, these data suggest a model where various isoforms of nesprin-1 and -2 form a scaffold around the nucleus [15,16]. Furthermore, this nuclear scaffold likely plays a role in regulating the size of the nucleus [15].
Hearing and KASH protein-mediated recruitment of kinesin to the nuclear envelope
Another class of KASH proteins including mammalian nesprin-4, C. elegans UNC-83, and Drosophila Klarsicht, interact with a kinesin light chain to target kinesin-1 to the surface of the nucleus [17,18] (Figure 1C). Recently, nesprin-4 was found to function in ear development and hearing. Human geneticists identified a family with a mutation in nesprin-4 that caused hearing loss, and nesprin-4 knockout mice were shown to be normal except for loss of hearing [19]. Major structural defects were observed in the cochlea of nesprin-4, or the SUN partner sun-1, knockout mice. By postnatal day 30, the outer hair cells in these mice exhibited degenerated stereocilia and nuclei that were mispositioned to the apical cellular surface [19].
Dynein-KASH protein interactions
There is a large class of KASH proteins that recruit dynein to the surface of the nuclear envelope (Figure 1D). Mouse Sun1 functions in homologous chromosome pairing in meiosis to attach telomeres to the inner surface of the nuclear envelope [20]. KASH5 was recently identified as a meiosis-specific protein with a conserved KASH domain that localizes to the nuclear envelope in a Sun1-dependent manner [21,22]. Ectopic expression of KASH5 recruits dynein to the nuclear envelope, KASH5 co-immunoprecipitates with dynein and dynactin, and KASH5 co-localizes with SUN proteins and telomeres during meiosis [22]. Furthermore, KASH5 knockout mice are infertile, have a severe meiotic chromosome pairing defect, fail to repair double-strand breaks, and fail to recruit dynein to telomere attachment sites [22]. The KASH5 homolog in zebrafish, Lrmp, functions in the zygote to attach centrosomes to the male pronucleus and is required for female pronuclear migration [23]. In C. elegans, ZYG-12 and SUN-1 recruit dynein to sub-telomeric regions (pairing centers in worms) that serve as attachment sites in meiosis. Dynein then is required for the intranuclear movements of chromosomes, which aid in homolog pairing and synapsis [24]. ZYG-12 also mediates the nuclear envelope localization of dynein during pro-nuclear migration [25], making ZYG-12 a likely functional ortholog of KASH5 and Lrmp. During meiosis in S. pombe, the KASH proteins Kms1 and Kms2 recruit dynein to specific spots on the nuclear envelope that associate with telomeres attached to the inside of the nuclear envelope. The γ-TuRC complex is then recruited to these spots resulting in the formation of mini microtubule organizing centers, or telocentrosomes, which move the telomeres toward the spindle pole body [26]. The KASH proteins UNC-83, Klarsicht, nesprin-1 and nesprin-2 also function, in part, to recruit dynein to the nuclear envelope [27–30]. Therefore, dynein is recruited to the surface of nuclei by SUN-KASH bridges. At the surface of the nucleus, dynein movements function to move nuclei, connect microtubules or centrosomes to nuclei, and to move chromosomes inside the nucleus during meiotic homolog pairing.
KASH proteins in plants
SUN and KASH proteins were recently described in plants [31–33] and have been hypothesized to mediate actin-dependent nuclear migrations[34]. Three Arabidopsis WIP (for WPP domain-Interacting Proteins; Figure 1E) proteins are the first identified KASH proteins in plants [32,35]. WIP proteins interact with WIT1 and 2 (WPP-Interacting Tail-anchored proteins) in the outer nuclear membrane [36]. AtWIP1, 2, and 3 fail to localize to the nuclear envelope in SUN double mutants and all three co-immunoprecipitate with AtSUN1 and AtSUN2 in a KASH-domain-dependent manner [32]. Arabidopsis mutants lacking the SUN, WIP, or WIT proteins lead to abnormally round nuclei, suggesting that nuclei are no longer attached to the actin cytoskeleton [32,37]. Forward genetic screens for similar mutants identified the Arabidopsis gene kaku1, which encodes a nonconventional myosin, myosin-XI [37]. Myosin-XI contains a WPP domain that interacts with WIT2 and then the WIP/SUN bridges to recruit it to the outer nuclear membrane to stretch and move nuclei [37] (Figure 1E). WIP and WIT proteins also recruit RanGAP1 to the cytoplasmic surface of the nuclear envelope [32,35,36,38]. In animal cells, RanGAP interacts with the cytoplasmic filaments of nuclear pore complexes to induce the hydrolysis of RanGTP to RanGDP as it exits the nucleoplasm [39]. Alternatively, in plants, RanGAP has a novel WPP domain that interacts with WITs at the outer nuclear membrane [32,36,38] (Figure 1E). Other plant KASH proteins likely remain unidentified since SUN proteins appear to play a similar role in maize meiosis as they do in yeast and animals [31], but their KASH partners remain unknown.
Novel KASH proteins and their functions
Novel functions for new KASH proteins continue to be found. C. elegans KDP-1 is a KASH protein that regulates progression of the cell cycle through unknown mechanisms [40] (Figure 1F). Genetic phenotypes, the presence of a tail-anchored domain, and the interaction with the SUN protein Mps3 [41,42] are consistent with Csm4 being a KASH protein that connects actin to meiotic telomeres in S. cerevisiae (Figure 1G). Kif9, a Dictyostelium kinesin-13, is a KASH protein that functions to connect nuclei to centrosomes [43,44]. Kif9 localizes to the nuclear envelope near the centrosome with Sun1; once at the nuclear envelope, it is thought that Kif9 depolymerizes microtubules, pulling the centrosome toward the nucleus [43] (Figure 1H). It is likely that additional KASH proteins remain to be discovered.
KASH proteins and cellular mechanics
Mechanical stimuli have been known for sometime to be communicated into the nuclear interior through the cytoskeleton during various fundamental cellular processes such as cell adhesion, migration, and differentiation [45–47]. However, the molecular mechanism responsible for this communication remained unclear until recently. KASH proteins, through their participation in LINC complexes, have been hypothesized to be responsible for this force transmission [48,49].
Four lines of experimental evidence where the over-expression of dominant negative KASH or SUN protein constructs was used to disrupt the formation of endogenous LINC complexes have supported this hypothesis. First, the lack of functional LINC complexes was shown to cause a loss of cytoskeletal mechanical stiffness in fibroblasts [50]. Second, cells containing disrupted LINC complexes had defective nuclear-cytoskeletal coupling as measured by the deformation of the nucleus caused by a microneedle pulling on the cytoskeleton [51]. These cells also displayed an altered organization of the actin and intermediate filaments. Similar results were obtained in experiments where shear forces transmitted through magnetic beads adhered onto the plasma membrane of cells [52]. Third, the actin-dependent rotation of nuclei observed in cells exposed to cyclic stretch requires intact LINC complexes [53]. Fourth, the rigidity dependence of nuclear height was disrupted in cells lacking functional LINC complexes [54]. While collectively these results implicate LINC complexes in intracellular force transmission, they do not identify specific KASH or SUN proteins.
Specific KASH proteins are necessary for three examples of nuclear-cytoskeletal force transmission. First, nesprin-1 is required for the reorientation of endothelial cells in response to applied uniaxial cyclic strain perpendicular to the direction of mechanical strain, which is important during proper angiogenesis [55]. Nesprin-1-depletion also resulted in an increase in focal adhesion assembly, cell traction, and nuclear height. Since a similar change in nuclear height was observed in cells where myosin-II activity was inhibited, it was suggested that the actomyosin tension is balanced in part by the nucleus. However, in the absence of nesprin-1, actomyosin tension was balanced by the increase in focal adhesions [55]. In a separate study, nesprin-1-depleted endothelial cells subjected to uniaxial stretching exhibited increased nuclear strain, which is an indicator of force transmission to the nucleus [56]. Second, nesprin-3 was found to be required for the ability of endothelial cells to polarize in response to shear stress such that their centrosomes are positioned on the side of the nucleus facing the source of stress [57]. Third, the ability of shear flow-stimulated myoblasts to assemble a specialized subset of actin cables above their nuclei was shown to require nesprin-3 and to a lesser extent, nesprin-2G [58]. The formation of these cables, which have been referred to as the PeriNuclear Actin Cap (PNAC), occurs in response to sheer stresses that are 50-fold lower than those required to form the actin cables found underneath the nucleus. PNAC cables terminate in a specialized subset of focal adhesions that are important for the mechano-sensing of matrix stiffness [59]. Interestingly, the focal adhesion protein zyxin was specifically required for the fast assembly of the PNAC, suggesting that extracellular mechanical stimuli can be sensed by the PNAC and quickly relayed to the LINC complex and into the nucleus [58]. Taken together, these results are consistent with a model where the LINC complex mediates nuclear-cytoskeletal force transmission.
A comparison between the PNAC and TAN lines
The PNAC is a subset of highly organized and dynamic actin cables that form over the apical surface of interphase nuclei in adherent cells [59–61]. In addition to mechano-sensation, several functions have been attributed to the PNAC including maintenance of nuclear shape, control of cellular differentiation, and three-dimensional cell migration {Khatau:2012il, Bui:2013in, Kihara:2011it}. A similar structure to the PNAC, known as Transmembrane Actin-associated Nuclear (TAN) lines, has also been identified and demonstrated to be required for nuclear movement in migrating fibroblasts {Luxton:2010eq, Luxton:2011ft, Luxton:2011jk}. TAN lines are linear arrays of nesprin-2G/Sun2 LINC complexes that form on the dorsal nuclear surface of fibroblasts that are preparing to migrate. Through their anchorage by A-type lamins, TAN lines are able to harness the forces generated by retrograde flow of the perinuclear actin cables, resulting in the movement of the nucleus to the rear of the cell, which is required for efficient cell migration [65–67]. The inner nuclear membrane protein Samp1 is also a TAN line component [68]. Samp1 associates with both SUN2 and A-type lamins, but requires the later for its localization to the nuclear envelope, suggesting that Samp1 assists in anchoring TAN lines. While linear arrays of LINC complexes have not been visualized in the PNAC, it is possible that these structures are related to one another. For instance, TAN lines may only form during active movement of the nucleus in migrating cells. It will be important to further understand the similarities and differences between the PNAC and TAN lines.
Conclusions
Our understanding of the cellular, developmental, and disease-related roles of KASH proteins is growing at an accelerating rate. It has become clear that KASH proteins, as part of LINC complexes, are critical mediators of nuclear-cytoskeletal force transmission events and cellular mechanics. KASH proteins also function in other events including cell cycle regulation and nuclear transport. The ability of KASH proteins to participate in such a wide variety of cellular functions stems from the ever-growing list of KASH proteins and their functions at the surface of the nucleus. Unsurprisingly, genetic mutations in KASH proteins are associated with various human diseases including Emery-Dreifuss muscular dystrophy, mental disorders, several cancers, and hearing loss [3,69] (Table 1 and references within). Much remains to be studied about the regulation of KASH proteins. It is not know when KASH proteins are assembled onto SUN proteins, or how KASH proteins are regulated during important developmental switches such as between nuclear migration and anchorage. Most importantly, how mutations in KASH proteins contribute to the pathologies of various human diseases remains to be determined.
Highlights.
KASH proteins localize to the cytoplasmic surface of the nucleus
New functions for KASH proteins interacting with the cytoskeleton are discussed
KASH proteins play a central role in cellular mechanics
Genetics mutations in KASH proteins are associated with a variety of human diseases
Acknowledgments
DAS thanks David Fay for hosting him at the University of Wyoming, Laramie while on sabbatical. Studies in the Starr lab are supported by grant R01 GM073874 from the National Institutes of Health NIGMS. GWGL thanks the members of his laboratory, Meg Titus, and Melissa Gardner for insightful discussions. Studies in the Luxton lab are supported by start up funding from the University of Minnesota and P30 Pilot and Feasibility Grant from the Paul and Sheila Wellstone Muscular Dystrophy Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
G. W. Gant Luxton, Email: gwgluxton@umn.edu.
Daniel A. Starr, Email: dastarr@ucdavis.edu.
References
- 1.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Starr DA. KASH and SUN proteins. Curr Biol. 2011;21:R414–5. doi: 10.1016/j.cub.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152:1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Developmental Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC Complexes Form by Binding of Three KASH Peptides to Domain Interfaces of Trimeric SUN Proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, Shi Z, Jiao S, Chen C, Wang H, Liu G, Wang Q, Zhao Y, Greene MI, Zhou Z. Structural insights into SUN-KASH complexes across the nuclear envelope. [Internet] Cell Res. 2012 doi: 10.1038/cr.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tapley EC, Starr DA. Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Current opinion in cell biology. 2012 doi: 10.1016/j.ceb.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sosa BA, Kutay U, Schwartz TU. Structural insights into LINC complexes. Curr Opin Struct Biol. 2013;23:285–291. doi: 10.1016/j.sbi.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothballer A, Schwartz TU, Kutay U. LINCing complex functions at the nuclear envelope: What the molecular architecture of the LINC complex can reveal about its function. Nucleus. 2013;4:29–36. doi: 10.4161/nucl.23387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke B. It takes KASH to hitch to the SUN. Cell. 2012;149:961–963. doi: 10.1016/j.cell.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Developmental Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Autore F, Pfuhl M, Quan X, Williams A, Roberts RG, Shanahan CM, Fraternali F. Large-scale modelling of the divergent spectrin repeats in nesprins: giant modular proteins. PLoS ONE. 2013;8:e63633. doi: 10.1371/journal.pone.0063633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Rajgor D, Mellad JA, Autore F, Zhang Q, Shanahan CM. Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. PLoS ONE. 2012;7:e40098. doi: 10.1371/journal.pone.0040098. Nesprin-1 and -2 are encoded by large, complicated loci. This study is the most complete describing the existence of dozens of isoforms of nesprin-1 and -2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu W, Schneider M, Neumann S, Jaeger V-M, Taranum S, Munck M, Cartwright S, Richardson C, Carthew J, Noh K, et al. Nesprin interchain associations control nuclear size. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajgor D, Shanahan CM. Nesprins: from the nuclear envelope and beyond. Expert Rev Mol Med. 2013;15:e5. doi: 10.1017/erm.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 2009;136:2725–2733. doi: 10.1242/dev.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci US A. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Horn HF, Brownstein Z, Lenz DR, Shivatzki S, Dror AA, Dagan-Rosenfeld O, Friedman LM, Roux KJ, Kozlov S, Jeang K-T, et al. The LINC complex is essential for hearing. Journal of Clinical Investigation. 2013 doi: 10.1172/JCI66911. A novel role for nesprin-4 is described. Nesprin-4 was found linked to families with hearing loss. Nesprin-4 knockout mice have misplaced nuclei and defective stereocilia in the ear hair cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Developmental Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K-I, Han M, Watanabe Y. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol. 2012;198:165–172. doi: 10.1083/jcb.201204085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Horn HF, Kim DI, Wright GD, Wong ESM, Stewart CL, Burke B, Roux KJ. A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J Cell Biol. 2013;202:1023–1039. doi: 10.1083/jcb.201304004. The identification of a new KASH protein, KASH 5, is reported. KASH5 is the missing link connecting SUN proteins to dynein during mammalian male and female meiosis. Dynein, through KASH5, then moves meiotic telomeres to assist in homolog pairing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindeman RE, Pelegri F. Localized Products of futile cycle/lrmp Promote Centrosome-Nucleus Attachment in the Zebrafish Zygote. Curr Biol. 2012;22:843–851. doi: 10.1016/j.cub.2012.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907–919. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malone CJ, Misner L, Le Bot N, Tsai M-C, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 26*.Yoshida M, Katsuyama S, Tateho K, Nakamura H, Miyoshi J, Ohba T, Matsuhara H, Miki F, Okazaki K, Haraguchi T, et al. Microtubule-organizing center formation at telomeres induces meiotic telomere clustering. J Cell Biol. 2013 doi: 10.1083/jcb.201207168. In this paper, the role of the KASH protein Kms1 is shown in the formation of new microtubule-organizing centers termed telocentrosomes, because they form across the nuclear envelope from a telomere. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fridolfsson HN, Ly N, Meyerzon M, Starr DA. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol. 2010;338:237–250. doi: 10.1016/j.ydbio.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer JA, Acosta S, Kenny A, Cater C, Robinson C, Hook J. Drosophila klarsicht has distinct subcellular localization domains for nuclear envelope and microtubule localization in the eye. Genetics. 2004;168:1385–1393. doi: 10.1534/genetics.104.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu J, Lei K, Zhou M, Craft CM, Xu G, Xu T, Zhuang Y, Xu R, Han M. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Human molecular genetics. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy SP, Bass HW. The maize (Zea mays L.) desynaptic (dy) mutation defines a pathway for meiotic chromosome segregation linking nuclear morphology, telomere distribution, and synapsis. J Cell Sci. 2012 doi: 10.1242/jcs.108290. [DOI] [PubMed] [Google Scholar]
- 32**.Zhou X, Graumann K, Evans DE, Meier I. Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol. 2012;196:203–211. doi: 10.1083/jcb.201108098. This paper shows for the first time that plants have KASH proteins called WIPs. The WIPs function to recruit RanGAP to the outer nuclear membrane where it functions to hydrolyze GTP in Ran as Ran exits the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graumann K, Runions J, Evans DE. Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 2009;61:134–144. doi: 10.1111/j.1365-313X.2009.04038.x. [DOI] [PubMed] [Google Scholar]
- 34.Chytilova E, Macas J, Sliwinska E, Rafelski SM, Lambert GM, Galbraith DW. Nuclear dynamics in Arabidopsis thaliana. Mol Biol Cell. 2000;11:2733–2741. doi: 10.1091/mbc.11.8.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu XM, Meulia T, Meier I. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol. 2007;17:1157–1163. doi: 10.1016/j.cub.2007.05.076. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Q, Brkljacic J, Meier I. Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell. 2008;20:1639–1651. doi: 10.1105/tpc.108.059220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I. Myosin XI-i Links the Nuclear Membrane to the Cytoskeleton to Control Nuclear Movement and Shape in Arabidopsis. Curr Biol. 2013 doi: 10.1016/j.cub.2013.07.035. By studying a mutant with abnormal nuclear shapes and movements, a myosin XI was found that is recruited to the surface of nuclei by Arabidopsis KASH proteins. [DOI] [PubMed] [Google Scholar]
- 38.Zhou X, Meier I. How plants LINC the SUN to KASH [Internet] Nucleus. 2013;4:206–215. doi: 10.4161/nucl.24088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutten S, Flotho A, Melchior F, Kehlenbach RH. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Mol Biol Cell. 2008;19:2300–2310. doi: 10.1091/mbc.E07-12-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGee MD, Stagljar I, Starr DA. KDP-1 is a nuclear envelope KASH protein required for cell-cycle progression. J Cell Sci. 2009;122:2895–2905. doi: 10.1242/jcs.051607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133:1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- 42.Lee C-Y, Conrad MN, Dresser ME. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet. 2012;8:e1002730. doi: 10.1371/journal.pgen.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Tikhonenko I, Magidson V, Gräf R, Khodjakov A, Koonce MP. A kinesin-mediated mechanism that couples centrosomes to nuclei. Cell Mol Life Sci. 2013;70:1285–1296. doi: 10.1007/s00018-012-1205-0. For the first time, a microtubule motor that contains its own KASH domain is identified. A kinesin-13 is targeted directly to the outer nuclear membrane through its KASH domain. The kinesin-13 then plays a role in coupling the centrosome to the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tikhonenko I, Nag DK, Robinson DN, Koonce MP. Microtubule-nucleus interactions in Dictyostelium discoideum mediated by central motor kinesins. Eukaryotic Cell. 2009;8:723–731. doi: 10.1128/EC.00018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci US A. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martins RP, Finan JD, Guilak F, Lee DA. Mechanical regulation of nuclear structure and function. Annu Rev Biomed Eng. 2012;14:431–455. doi: 10.1146/annurev-bioeng-071910-124638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nature reviews. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- 48.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nature reviews. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 49.Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nature reviews. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. Journal of Biological Chemistry. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iyer KV, Pulford S, Mogilner A, Shivashankar GV. Mechanical Activation of Cells Induces Chromatin Remodeling Preceding MKL Nuclear Transport. Biophys J. 2012;103:1416–1428. doi: 10.1016/j.bpj.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brosig M, Ferralli J, Gelman L, Chiquet M, Chiquet-Ehrismann R. Interfering with the connection between the nucleus and the cytoskeleton affects nuclear rotation, mechanotransduction and myogenesis. Int J Biochem Cell Biol. 2010;42:1717–1728. doi: 10.1016/j.biocel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 54.Lovett DB, Shekhar N, Nickerson JA, Roux KJ, Lele TP. Modulation of Nuclear Shape by Substrate Rigidity. Cell Mol Bioeng. 2013;6:230–238. doi: 10.1007/s12195-013-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chancellor TJ, Lee J, Thodeti CK, Lele T. Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys J. 2010;99:115–123. doi: 10.1016/j.bpj.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anno T, Sakamoto N, Sato M. Role of nesprin-1 in nuclear deformation in endothelial cells under static and uniaxial stretching conditions. Biochem Biophys Res Commun. 2012;424:94–99. doi: 10.1016/j.bbrc.2012.06.073. [DOI] [PubMed] [Google Scholar]
- 57.Morgan JT, Pfeiffer ER, Thirkill TL, Peng G, Fridolfsson HN, Douglas GC, Starr DA, Barakat AI. Nesprin-3 Regulates Endothelial Cell Morphology, Perinuclear Cytoskeletal Architecture, and Flow-Induced Polarization. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-04-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58*.Chambliss AB, Khatau SB, Erdenberger N, Robinson DK, Hodzic D, Longmore GD, Wirtz D. The LINC-anchored actin cap connects the extracellular milieu to the nucleus for ultrafast mechanotransduction. Sci Rep. 2013;3:1087. doi: 10.1038/srep01087. Low shear stresses were shown to induce the nesprin-2G and -3-dependent assembly of the PNAC in fibroblasts at rates faster than those required for biochemical stimulation. Actin cables in the PNAC terminated in focal adhesions containing zyxin, which was required for this process suggesting that mechanotransduction in cells can quickly be relayed across the cytoskeleton into the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim D-H, Chambliss AB, Wirtz D. The multi-faceted role of the actin cap in cellular mechanosensation and mechanotransduction. Soft Matter. 2013;9:5516–5523. doi: 10.1039/C3SM50798J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci US A. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khatau SB, Kim D-H, Hale CM, Bloom RJ, Wirtz D. The perinuclear actin cap in health and disease. Nucleus. 2010;1:337–342. doi: 10.4161/nucl.1.4.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62*.Khatau SB, Kusuma S, Hanjaya-Putra D, Mali P, Cheng L, Lee JSH, Gerecht S, Wirtz D. The differential formation of the LINC-mediated perinuclear actin cap in pluripotent and somatic cells. PLoS ONE. 2012;7:e36689. doi: 10.1371/journal.pone.0036689. Both nesprin-2G and -3, which are required for PNAC formation, were found to critical for three-dimensional cell migration. In the absence of the PNAC, cells did not form pseudopodial protrusions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bui KH, Appen von A, Diguilio AL, Ori A, Sparks L, Mackmull M-T, Bock T, Hagen W, Andrés-Pons A, Glavy JS, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- 64.Kihara T, Haghparast SMA, Shimizu Y, Yuba S, Miyake J. Physical properties of mesenchymal stem cells are coordinated by the perinuclear actin cap. Biochem Biophys Res Commun. 2011;409:1–6. doi: 10.1016/j.bbrc.2011.04.022. [DOI] [PubMed] [Google Scholar]
- 65.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luxton GWG, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Current opinion in cell biology. 2011;23:579–588. doi: 10.1016/j.ceb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Folker ES, Östlund C, Luxton GWG, Worman HJ, Gundersen GG. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci US A. 2011;108:131–136. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Borrego-Pinto J, Jegou T, Osorio DS, Aurade F, Gorjanacz M, Koch B, Mattaj IW, Gomes ER. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci. 2012;125:1099–1105. doi: 10.1242/jcs.087049. [DOI] [PubMed] [Google Scholar]
- 69.Worman HJ. Nuclear lamins and laminopathies. [Internet] J Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Human molecular genetics. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- 71.Attali R, Warwar N, Israel A, Gurt I, McNally E, Puckelwartz M, Glick B, Nevo Y, Ben-Neriah Z, Melki J. Mutation of SYNE-1, encoding an essential component of the nuclear lamina, is responsible for autosomal recessive arthrogryposis. Human molecular genetics. 2009;18:3462–3469. doi: 10.1093/hmg/ddp290. [DOI] [PubMed] [Google Scholar]
- 72.Noreau A, Bourassa CV, Szuto A, Levert A, Dobrzeniecka S, Gauthier J, Forlani S, Durr A, Anheim M, Stevanin G, et al. SYNE1 Mutations in Autosomal Recessive Cerebellar Ataxia. JAMA Neurol. 2013 doi: 10.1001/jamaneurol.2013.3268. [DOI] [PubMed] [Google Scholar]
- 73.Gros-Louis F, Dupré N, Dion P, Fox MA, Laurent S, Verreault S, Sanes JR, Bouchard J-P, Rouleau GA. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nature genetics. 2007;39:80–85. doi: 10.1038/ng1927. [DOI] [PubMed] [Google Scholar]
- 74.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 75.Doherty JA, Rossing MA, Cushing-Haugen KL, Chen C, Van Den Berg DJ, Wu AH, Pike MC, Ness RB, Moysich K, Chenevix-Trench G, et al. ESR1/SYNE1 polymorphism and invasive epithelial ovarian cancer risk: an Ovarian Cancer Association Consortium study. Cancer Epidemiology Biomarkers & Prevention. 2010;19:245–250. doi: 10.1158/1055-9965.EPI-09-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature genetics. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Green EK, Grozeva D, Forty L, Gordon-Smith K, Russell E, Farmer A, Hamshere M, Jones IR, Jones L, McGuffin P, et al. Association at SYNE1 in both bipolar disorder and recurrent major depression. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.48. [DOI] [PubMed] [Google Scholar]
- 78.Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, Schmitz-Abe K, Harmin DA, Adli M, Malik AN, et al. Using Whole-Exome Sequencing to Identify Inherited Causes of Autism. Neuron. 2013;77:259–273. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, MacKenzie AP, Ng SB, Baker C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature genetics. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schoppmann SF, Vinatzer U, Popitsch N, Mittlböck M, Liebmann-Reindl S, Jomrich G, Streubel B, Birner P. Novel clinically relevant genes in gastrointestinal stromal tumors identified by exome sequencing. Clin Cancer Res. 2013;19:5329–5339. doi: 10.1158/1078-0432.CCR-12-3863. [DOI] [PubMed] [Google Scholar]