Graphical abstract
Keywords: Fusion peptide, Membrane fusion, Viral entry, Peptide-lipid interaction
Highlights
-
•
The presence of a fusion peptide (FP) is a hallmark of viral fusion glycoproteins.
-
•
Structure–function relationships underlying FP conservation remain greatly unknown.
-
•
FPs establish interactions satisfying their folding within pre-fusion glycoproteins.
-
•
Upon fusion activation FPs insert into and restructure target membranes.
-
•
FPs can finally combine with transmembrane domains to form integral membrane bundles.
Abstract
Fusion peptides comprise conserved hydrophobic domains absolutely required for the fusogenic activity of glycoproteins from divergent virus families. After 30 years of intensive research efforts, the structures and functions underlying their high degree of sequence conservation are not fully elucidated. The long-hydrophobic viral fusion peptide (VFP) sequences are structurally constrained to access three successive states after biogenesis. Firstly, the VFP sequence must fulfill the set of native interactions required for (meta) stable folding within the globular ectodomains of glycoprotein complexes. Secondly, at the onset of the fusion process, they get transferred into the target cell membrane and adopt specific conformations therein. According to commonly accepted mechanistic models, membrane-bound states of the VFP might promote the lipid bilayer remodeling required for virus-cell membrane merger. Finally, at least in some instances, several VFPs co-assemble with transmembrane anchors into membrane integral helical bundles, following a locking movement hypothetically coupled to fusion-pore expansion. Here we review different aspects of the three major states of the VFPs, including the functional assistance by other membrane-transferring glycoprotein regions, and discuss briefly their potential as targets for clinical intervention.
1. Introduction: viral glycoprotein-induced membrane fusion
Membrane fusion, i.e., the merging of two initially separate and apposed lipid bilayers with the result of the mixing of two initially distinct aqueous compartments, is ubiquitous to cell life. This unfavorable event typically advances at the expense of protein refolding energy, and under the control of mechanisms that ensure its evolvement at defined cell locations and physiological stages. Despite the variety of physiological conditions involving membrane fusion events, the same basic principles underlying lipid bilayer remodeling seem to apply to all the expressions of this phenomenon (see Chernomordik and Kozlov, 2003, Cohen and Melikyan, 2004, Frolov and Zimmerberg, 2010, Kozlov et al., 2010, Zimmerberg et al., 1993) and references therein for comprehensive reviews on this issue).
The protein-assisted membrane fusion reaction has been co-opted by lipid-enveloped pathogens, including enveloped viruses (White, 1992, White et al., 2008). Thus, enveloped viruses comprising highly relevant human pathogens such as Influenza virus (IFV), human immunodeficiency virus (HIV) or Ebola virus (EBOV), make use of membrane glycoproteins for selecting host cells, induce membrane fusion and gain access to internal compartments, a sequence of processes collectively known as “viral entry”(White et al., 2008). Given the dynamic nature of viral genomes and the vast number of replication cycles taking place during infection of a single host, viral entry machineries are subject to intense molecular evolution. Thus, the themes of glycoprotein structure–function common to all enveloped viruses are highly significant for the understanding of the general mechanism of protein-mediated fusion (Blumenthal et al., 2012, Chernomordik and Kozlov, 2003, Cohen and Melikyan, 2004, Harrison, 2008, Hernandez et al., 1996, Kozlov et al., 2010, Lentz et al., 2000, Melikyan, 2008). In addition, the conserved elements of these machineries, transiently or permanently exposed on the virion surface, provide clinical targets for development of inhibitors (antivirals) and immunogens (vaccines) (Blumenthal and Dimitrov, 2007, Doms and Moore, 2000, Eckert and Kim, 2001, Forssmann et al., 2010, Huarte et al., 2011, Munch et al., 2007).
Fig. 1A displays a generally accepted mechanism of membrane fusion induced by prototypical Class I viral glycoproteins, such as those of IFV, HIV or EBOV. The model is supported by the available structural and functional evidence (reviewed in references Blumenthal et al., 2012, Eckert and Kim, 2001, Harrison, 2008, Melikyan, 2008, Skehel and Wiley, 2000, Wiley and Skehel, 1987). The mature envelope glycoproteins in those viruses are organized as trimers of non-covalently associated heterodimers. Each heterodimer is composed of a surface and a trans-membrane subunit, which mediate receptor binding and virus-cell fusion, respectively (Karlsson Hedestam et al., 2008, Roux and Taylor, 2007, Skehel and Wiley, 2000, Wiley and Skehel, 1987). Upon fusion activation, the ectodomain of the membrane-anchored subunit undergoes a series of conformational changes conducive to membrane merger. First a “pre-hairpin” intermediate forms to anchor viral and cell membranes through the transmembrane (TMD) and fusion peptide (FP) domains, respectively. The presence of the FP within the ectodomain exposed to the aqueous phase constitutes a feature shared by all viral fusion proteins, and represents an absolute requirement for their fusogenic function (previously reviewed in: Durell et al., 1997, Epand, 2003, Nieva and Agirre, 2003, Tamm et al., 2002). These hydrophobic and conserved sequences usually located at the N-terminal end of the fusogenic subunit, or close to it (Table 1 ), are thought to be involved in driving the initial partitioning of the fusion protein into the target membrane (Nieva and Suarez, 2000). In this first stage, formation (IFV hemagglutinin (HA)) or completion (HIV-Envelope protein (Env)) of an extended coiled coil (red rods in Fig. 1A) by N-terminal helices (or N-terminal heptad repeats, NHRs) would be the propelling force that brings about exposition and translocation of the initially cryptic FP into close vicinity of the target membrane.
Fig. 1.
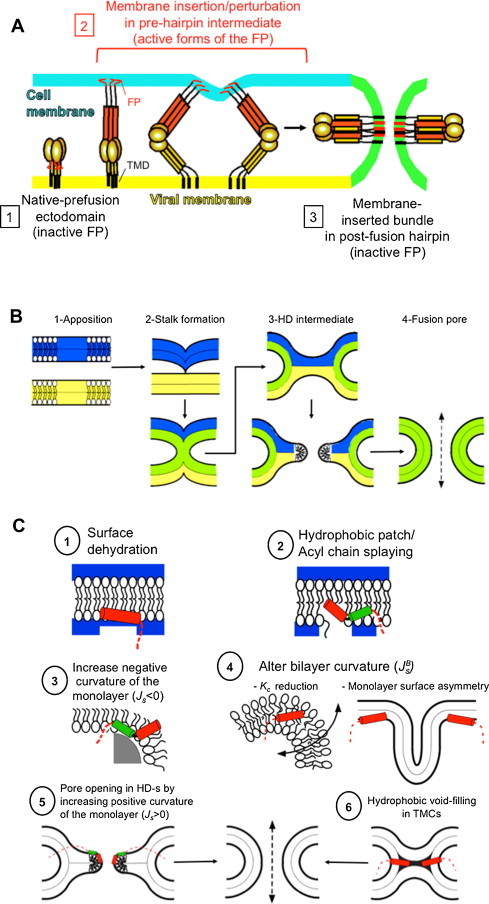
Viral glycoprotein-induced membrane fusion and proposed functions for the FP in the process. (A) General model of membrane fusion promoted by Class I viral fusion glycoproteins. (B) Stages of the process from a lipid-centric perspective (see text). (C) Putative effects of FPs on target membrane rupture and deformation during fusion (see text).
Table 1.
VFPs of Class I fusion proteins.
| Family | Speciesa | Sequencea | Position | FYWb | ΔG Interfacialc | Ala Percentd | Gly Percentd |
|---|---|---|---|---|---|---|---|
| Retroviridae | HTLV-1 | AVPVAVWLVSALAMGAGVAGGITGS | N-ter | Yes | –1.24 | 24/9 | 20/8 |
| HIV-1 | AVGIGALFLGFLGAAGSTMGARS | N-ter | Yes | –2.29 | 22/7 | 26/7 | |
| ASLV | GPTARIFASILAPGVAAAQALREIERLA | Internal | Yes | 5.94 | 29/8 | 7/7 | |
| SIV | GVFVLGFLGFLATAGSAMGAAS | N-ter | Yes | –3.86 | 23/7 | 23/7 | |
| BLV | SPVAALTLGLALSVGLTGINVAVS | N-ter | No | -0.58 | 17/7 | 13/3 | |
| Filoviridae | EBOV | GAAIGLAWIPYFGPAAE | Internal | Yes | –1.3 | 29/5 | 18/6 |
| MARV | LAAGLSWIPFFGPGI | Internal | Yes | -4.45 | 13/5 | 20/7 | |
| Coronaviridae | SARS-CoV | MYKTPTLKYFGGFNFSQIL | N-ter | Yes | –3.07 | 0/8 | 11/7 |
| GAALQIPFAMQMAYRF | Internal | Yes | –1.42 | 25/8 | 6/7 | ||
| Orthomyxoviridae | IFV-A | GLFGAIAGFIENGWEGMIDGWYG | N-ter | Yes | –2.52 | 9/8 | 30/6 |
| Paramyxoviridae | PIV5 | FAGVVIGLAALGVATAAQVTAAVALV | N-ter | Yes | 0.04 | 35/11 | 13/4 |
| NDV | FIGAIIGSVALGVATAAQITAA | N-ter | Yes | –0.45 | 30/8 | 13/8 | |
| HeV | LAGVVMAGIAIGIATAAQITAGV | N-ter | No | 0.27 | 30/6 | 17/6 |
HTLV-1: Human T-Cell Leukemia Virus Type 1, HIV-1: Human Immunodeficiency Virus Type 1, ASLV: Avian Sarcoma and Leucosis Virus, SIV: Simian Immunodeficiency Virus, BLV: Bovine Leukemia Virus, EBOV: Ebola Virus, MARV: Marburg Virus SARS-CoV: Severe Acute Respiratory Syndrome-Associated Coronavirus, IFV-A: Influenza A Virus, PIV5: Parainfluenza Virus 5 NDV: Newcastle Disease Virus and HeV: Hendra Virus.
Presence of aromatic residues within the fusion peptide sequence.
Computed according to the Wimley-White scale of water-membrane partitioning energies.
Mole percent of alanine and glycine in fusion peptide over whole protein sequence.
The filamentous structure of the “pre-hairpin” subsequently collapses into the low-energy trimeric “hairpin”. In the trimeric hairpin structure, the ectodomain amino- and carboxy-termini are placed at the same end of the molecule (Eckert and Kim, 2001, Weissenhorn et al., 1997). In several instances, this second step might ensue due to the fact that the extended N-terminal coiled coil creates highly conserved grooves into which C-terminal helices (or C-terminal heptad repeats, CHRs) may hydrophobically pack in an antiparallel orientation (yellow rods in Fig. 1A). It is assumed that the conformational energy released during the trimeric hairpin formation can be used to pull membranes together and induce their merger, while its completion upon full zippering of the CHR would stabilize an open state of the fusion pore and likely contribute to its expansion (Melikyan, 2008).
Hence, insertion of the hydrophobic FP into the target membrane and formation of the low-energy 6-HB structure are common themes to all fusogenic Class I glycoproteins, which otherwise may vary in receptor specificity, size, sequence and activation pathways (Melikyan, 2008, White et al., 2008). A great deal of experimental work produced in the course of the last 25 years supports this view. In one hand, convincing evidence for the insertion of the VFPs into target membranes was early provided by Brunner's group using hydrophobic photolabeling approaches (Durrer et al., 1996, Harter et al., 1989, Tsurudome et al., 1992, Weber et al., 1994). The IFV glycoprotein HA is synthesized as a single polypeptide, which is posttranslationally cleaved into two disulfide-linked chains, HA1 and HA2. The hydrophobic N terminus of HA2 generated after cleavage embodies the IFV-FP. Brunner and co-workers demonstrated that, upon incubation at conditions that activate IFV fusion (i.e., pH 5.0 and 37 °C), the hydrophobic interactions of isolated BHA2 (the bromelain-solubilized form of hemagglutinin) or IFV virions were mediated solely by the N-terminal segment of the HA2 subunit, which corresponds to the FP (Harter et al., 1989, Tsurudome et al., 1992). They found that the predominant sites of labeling within this segment were spaced in average 3–4-residues, thereby suggesting that the FP inserted into the target membrane adopting a helical structure with an amphiphilic character (see also Fig. 3B below). From asymmetric hydrophobic photolabeling of membranes, evidence was obtained indicating that HA2-FP penetrates only the external leaflet of the bilayer in the fusion pH conformation (Brunner, 1989). In addition these authors provided evidence to sustain a mechanism of IFV inactivation at acidic pH, according to which the HA2 FP irreversibly inserted into the viral membrane (Weber et al., 1994).
Fig. 3.
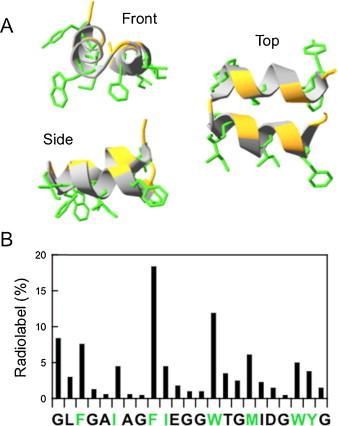
Helical hairpin model for IFV-FP. (A) NMR structure of the synthetic HA-FP1–23 peptide in DPC micelles (PDB entry code: 2KXA). Side-chains of hydrophobic residues are displayed in green, while orange stretches in the ribbon mark positions of Gly residues. (B) Residue labeling in the sequence covered by HA-FP1–23 upon hydrophobic photolabeling of BHA incubated at low-pH in the presence of membranes. Percentages of label in the residues were calculated based on the data reported by (Harter et al., 1989). Side-chains of residues in green characters are depicted in the previous panel.
On the other hand, crystallographic evidence for the formation of low-energy trimeric hairpins by divergent virus glycoproteins has been accumulated during the last two decades (Eckert and Kim, 2001, Harrison, 2008, Weissenhorn et al., 1997, White et al., 2008, Yin et al., 2006). The fact that Class II glycoproteins make use of trimeric hairpins to induce fusion underpins the functional relevance of these structures (Harrison, 2008, Vaney and Rey, 2011). However, in contrast to the helical domains implied in Class I fusion, the Class II glycoprotein subunits employ articulated β-barrel domains to assemble membrane-pulling trimeric hairpins (Harrison, 2008, Vaney and Rey, 2011, White et al., 2008). Yet another class of fusion glycoproteins combine structural features of Class I and Class II fusion proteins, and have been designated as Class III (Backovic and Jardetzky, 2009, Baquero et al., 2013, Weissenhorn et al., 2007, White et al., 2008). Fusogenic subunits of Class II and Class III proteins possess conserved hydrophobic loops that connect extended strands at the tip of elongated β-barrel domains. These “fusion loops” (FLs) are thought to insert into the target membrane and function during fusion in a way reminiscent of Class I FPs. Thus, the fact that Class I/II/III glycoproteins make use of similar elements, but different structural scaffolds, highlights the functional convergence underlying cell-virus membrane fusion.
Structure–function relationships of VFPs derived from diverse viruses with relevance as human pathogens, such as IFV (Han et al., 2001, Lear and DeGrado, 1987, Rafalski et al., 1991), HIV-1 (Agirre et al., 2000, Kliger et al., 1997, Li and Tamm, 2007, Mobley et al., 1999, Nieva et al., 1994, Pritsker et al., 1999, Rafalski et al., 1990, Saez-Cirion and Nieva, 2002, Shnaper et al., 2004, Yang et al., 2004b), and EBOV (Ruiz-Arguello et al., 1998) have been extensively studied in the course of the last decades. We previously reviewed many aspects of VFP-membrane interactions, paying particular attention to the physiological relevance of studies based on synthetic peptides and membrane models (Nieva and Agirre, 2003). For excellent, contemporary reviews on VFP-related issues the reader is referred to: (Durell et al., 1997, Epand, 2003, Epand et al., 1992, Peisajovich and Shai, 2003, Tamm and Han, 2000). Here, we first give a brief overview on our current understanding of the structure and function of the VFPs, and then update some case studies (Sections 2, 3, respectively). This review primarily focuses on Class I VFPs, as they conform more closely to the notion of FP domain (Epand et al., 1992, Gallaher, 1987). Nonetheless, Section 4 will be devoted to the description of Class II/III FLs. Section 5 will deal with the possible assistance by other hydrophobic-conserved regions present in glycoprotein sequences, particularly the “membrane-proximal external regions” known as MPER domains. Finally in Section 6, based on the HIV-1 case, we will succinctly discuss the potential implications of this knowledge for the development of unprecedented anti-viral compounds.
2. Structure–function analyses of VFPs
2.1. Primary structure
Table 1 summarizes information relative to the primary structures of VFPs derived from most prominent Class I enveloped viruses. One feature shared by these sequences is that, despite the phylogenetic divergence, amino acid homology between FPs from Class I viral families can be as high as 90% (Gallaher, 1987). VFPs also gather unusual amounts of Ala and Gly residues, and, in particular when located internally, they contain invariable Pro residues (Gomara et al., 2004). Such a degree of residue conservation and Ala/Gly content are uncommon for hydrophobic protein domains as signal sequences or transmembrane anchors. The occurrence of these residues further suggests an intrinsic conformational flexibility that cannot be explained by the simple requirement of membrane insertion. Thus, FP sequences have evolved constrained by selective pressures beyond the thresholds imposed by mere hydrophobicity or amphipathicity.
Aromatic residues are also present in VFP sequences, although exceptions to this rule may be found (Table 1). These residues may help overcome the energy cost of peptide bond partitioning into membranes, particularly when this cost cannot be counterbalanced by the adoption of a defined secondary structure (Nieva and Suarez, 2000, White and Wimley, 1999). In this regard, aromatics’ contribution maybe relevant for enabling membrane insertion of internal FPs (Table 1). In these instances, the absence of a free N-terminus constraints conformational transitions in contact with the membrane surface (Gomara et al., 2004, Suarez et al., 2003). The favorable interactions of aromatic side chains with phospholipid moieties located at the membrane interface region may in addition contribute to stabilize the insertion of the FP sequence into just one bilayer leaflet (White and Wimley, 1999). An initial interaction restricted to one monolayer has been proposed to generate elastic stresses conducive to bilayer fusion (see below).
2.2. Native structures of VFPs
The most stringent constraint imposed onto VFP sequences is probably the requirement of folding after synthesis as a constituent of the glycoprotein's globular ectodomain. This would represent the first stage in the life of VFPs (Fig. 1A-(1)). The IFV-FP solved within the trimeric HA complex was for many years the only available VFP atomic structure in a native, pre-fusion state (Wiley and Skehel, 1987). The native IFV-FP folds as a loop partly buried into a pocket of ionizable residues, and with the hydrophobic N-terminus occluded at interfaces between HA2 transmembrane subunits. However, the IFV-FP model is not generalizable to all Class I viral fusion glycoproteins, as evidenced by the native structures available later on (Lee et al., 2008, Lyumkis et al., 2013, Yin et al., 2006). Thus, EBOV-FP and Parainfluenza virus-5 (PIV5)-FP locate in the periphery of the pre-fusion glycoprotein complex, partially accessible from solution, and interacting extensively with the surface subunits (Lee et al., 2008, Yin et al., 2006). A similar pattern has been recently suggested for the HIV-FP (Julien et al., 2013, Lyumkis et al., 2013). Thus, the occurrence of distinct native structures suggest that the VFP sequence homology between divergent virus families probably sustains common functional roles beyond folding after synthesis.
2.3. Active VFP structures and membrane destabilization
After fusion activation the VFPs must be exposed in the vicinity of the target membrane. Especially in the case of N-terminal sequences, it is assumed that membrane partitioning, initially driven by favorable hydrophobic interactions between FP monomers and the membrane interface (Table 1), will result in the partitioning-coupled structuring of the FP sequence as an autonomous domain. Thus, structures adopted within the target membrane milieu might represent a second functional state common to all VFPs (Fig. 1A-(2)). From that perspective, experimental approaches based on synthetic surrogates and model membranes have been applied to establish physiologically relevant, functional VFP states (see for earlier discussions on this issue Durell et al., 1997, Epand et al., 1992, Nieva and Agirre, 2003). Insertion patterns and conformations adopted by synthetic versions of the VFPs in model membranes and mimics have been characterized by a plethora of biophysical and structural techniques including Nuclear Magnetic Resonance (NMR) (Chang et al., 1997, Han et al., 2001, Qiang et al., 2008), Electron Paramagnetic Resonance (EPR) (Macosko et al., 1997, Tamm et al., 2007), and Infrared spectroscopy (IR) (Buzon et al., 2005, Castano and Desbat, 2005, Epand et al., 2001, Gray et al., 1996, Haque et al., 2005, Li and Tamm, 2007, Luneberg et al., 1995, Martin et al., 1993, Mobley et al., 1999, Pereira et al., 1995, Pereira et al., 1997). These studies reveal that the main conformation adopted by VFPs in membranes, insertion depth and insertion angle may vary depending on factors like the membrane lipid composition (particularly the presence of cholesterol or anionic phospholipids) or peptide length (Kliger et al., 1997, Lai et al., 2012, Wexler-Cohen et al., 2006). When physiologically relevant, as it is the case of single sequence substitutions that recreate mutations interfering with the fusogenic activity of the full glycoprotein, these factors allow delineating structure–function relationships shared by VFPs from distinct origin (Nieva and Agirre, 2003).
The IFV-FP sequence is probably the most widely studied VFP following this approach. The HA2 post-fusion structure reveals N- and C-terminal residues that form an N cap terminating both the N-terminal α-helix and the central coiled coil (Chen et al., 1999). This structure implies that continuous α helices are not required for membrane fusion at either the N or C termini of the trimeric hairpin. In addition, residue 34 marks the end of the capping structure at the N-terminus, thereby suggesting that the functional structures and dynamics in membranes of the upstream IFV-FP (residues 1–23) are likely to be barely affected by the rest of the HA2 sequence. A main helical structure adopted by a synthetic IFV-FP in contact with membranes and its mimics was put forward by early studies (Chang et al., 2000, Dubovskii et al., 2000, Gray et al., 1996, Lear and DeGrado, 1987, Luneberg et al., 1995). Additional studies revealed that insertion of this element could lead to lipid bilayer destabilization as evidenced by the induction of leakage of aqueous contents and lipid-mixing when added to liposomes (Colotto and Epand, 1997, Epand et al., 2001, Lear and DeGrado, 1987, Rafalski et al., 1991). A further line of evidence suggested that an oblique angle of insertion into the lipid bilayer might be required to sustain those membrane destabilization phenomena (Epand et al., 2001, Gray et al., 1996, Luneberg et al., 1995).
The effects of replacing N-terminal glycine, which may cause hemifusion or abolish fusion activity in the full protein, were found to affect both the adopted structure and the capacity for destabilizing lipid vesicles by the IFV-FP (reviewed in: Nieva and Agirre, 2003, see also next section). In one study, the main α-helical conformation was observed in all the analyzed variants (Gray et al., 1996). However, the nonfusogenic peptides exhibited larger amounts of β-like conformers. A later work related the absence of activity of the HA2 bearing the G1V mutation to a change in the angle of insertion of the helical FP into membranes as measured by polarized IR (Epand et al., 2001).
Similarly, the polar V2E substitution at the N-terminus of the HIV-FP was known to interfere with gp41-induced fusion (Nieva and Agirre, 2003). The V2E change impaired insertion of synthetic HIV-1-FP into membrane monolayers and caused a reduction of membrane activity measured in liposomes. However, in contrast to what is observed for IFV-FP, the HIV-FP may adopt a functional β-strand structure in contact with membranes (Buzon et al., 2005, Castano and Desbat, 2005, Lai et al., 2012, Mobley et al., 1999, Sackett and Shai, 2005, Yang et al., 2001), and the V2E substitution was shown to interfere with the adoption of that conformation (Gabrys et al., 2013, Pereira et al., 1995). A series of Attenuated Total Reflectance (ATR)-IR studies also suggested an oblique angle of insertion for the membrane-bound functional HIV-FP (Martin et al., 1993, Martin et al., 1996), which was confirmed by later structural determinations (Castano and Desbat, 2005, Morris et al., 2004). Thus, insertion in an oblique angle and the capability of alternating α/β conformations depending on membrane dose and lipid composition emerged as common structural patterns from those analyses. However, the membrane-bound conformation most active at generating membrane destabilization differed in the IFV-FP or HIV-FP.
The functional meaning of the conformational polymorphism of membrane-bound VFPs is yet unclear. Some authors argue that VFPs function as inert anchors, i.e., “secondary” transmembrane domains (TMDs) inserted into the target membrane (Donald et al., 2011, Skehel and Wiley, 2000). According to this view, conformational polymorphism of the membrane-inserted FP might contribute to the flexibility of the fusogenic complex during the fusion reaction cycle (Saez-Cirion and Nieva, 2002, Suarez et al., 2003). Following the “hairpin” model (Fig. 1A), the rod-shaped glycoprotein ectodomain must alternate a configuration with its axis perpendicular to the target bilayer normal (inserted prefusion state) with a configuration mainly parallel to the membrane interface supposedly adopted within the trimeric hairpin structure. Thus, the presence of jointed kinks to enable backbone reorientation and the capacity for alternating FP insertion levels might be required for the final stages of the fusion cascade to evolve.
An alternative option is that it is the membrane structure, rather than peptide's structure, what changes upon VFP insertion. Under this view, VFPs would be membrane-active agents, while the hairpins would behave as mechanical devices that bring into apposition membranes poised for fusion (Fig. 1A). Accordingly, the functional link with conformational polymorphism has been established by combining peptide structural studies, membrane stability assays and determinations of changes in structure and material properties of the bilayer, both in liposome and bulk lipid systems (Colotto and Epand, 1997, Epand et al., 1992, Epand and Epand, 1994, Epand et al., 2001, Haque et al., 2011, Pereira et al., 1999, Siegel and Epand, 2000, Tristram-Nagle et al., 2010).
Fig. 1B briefly describes the remodeling processes undergone by the lipid bilayer along the proposed fusion pathway (Chernomordik and Kozlov, 2003, Cohen and Melikyan, 2004, Kozlov et al., 2010). Bilayers must be first brought into close apposition, a process that requires overcoming the highly repulsive hydration and electrostatic forces operating at the membrane surface (Fig. 1B-1). Repulsion can be minimized within pointlike membrane protrusions created by focal application of curvature/tension stress (Fig. 1B-2-top). Formation of these structures might lead to closer apposition of the contacting monolayers and generation of small hydrophobic patches. The most widely accepted model, based on a continuous approach of membranes in terms of their elastic properties, posits that an hourglass-shaped lipid structure termed “stalk”, with mean negative curvature, subsequently emerges at the point where interbilayer contact has been established (Fig. 1B-2-bottom). The stalk allows mixing of the lipids from the contacting monolayers (cis-monolayers), but not of those from the distal ones (trans-monolayers). According to the original proposal of the stalk theory, distal monolayers collapse into a single bilayer referred to as a “hemifusion diaphragm” (HD) (Fig. 1B-3-top). Increasing tension at the HD leads to the formation of a small hydrophilic pore, a process facilitated by positive curvature of the HD monolayers (Fig. 1B-3-bottom). A “fusion pore” is eventually formed, which connects the internal aqueous volumes enclosed by the originally separated membranes (Fig. 1B-4).
Fig. 1C summarizes the VFP effects putatively related to the promotion of the membrane rupture and/or deformation processes along that pathway. In a first instance insertion into one monolayer of an amphipatic VFP structure might help overcoming the hydration repulsion force between approaching bilayers by orienting a poorly solvated face toward the external medium (Fig. 1C (1)). Pre-stalk fusion intermediates maybe subsequently elicited by breaching lipid continuity and generating focal hydrophobic patches (Fig. 1C (2)). Recent molecular dynamic simulations (MDS) suggest that hydrophobic defects generated by IFV-FP in the “boomerang” conformation (see below) might cause lipid tail protrusions (Larsson and Kasson, 2013). It has been argued that a lipid molecule splaying its two acyl chains may suffice to build an initial lipid bridge between contacting bilayers (Frolov and Zimmerberg, 2010, Kinnunen and Holopainen, 2000, Kozlov et al., 2010).
VFP-induced monolayer negative curvature may also minimize the stalk formation energy (Fig. 1C (3)). Indirect proof consistent with this effect comes from the capacity of VFPs for lowering the temperatures of lamellar-to-non-lamellar (HII) phase transitions. Accordingly, the ability of HIV-1-FP to induce lipid mixing in large unilamellar liposomes was correlated with its ability for modulating the lipid polymorphism in bulk systems (Pereira et al., 1999). This concept was also supported by studies addressing the effects of point mutations at the N-terminal amino acid, Gly, of IFV-FP (Epand and Epand, 1994, Epand et al., 2001). Only the wild-type peptide could substantially decrease the temperature of the lamellar-to-non-lamellar (HII) phase transition at pH 5. In addition a correlation existed between the fusogenic activities of the full protein and the effect of the FPs on monolayer curvature, as monitored by a shift in the temperature of the polymorphic phase transition.
More recent theoretical developments stipulate that asymmetric insertion of hydrophobic protein domains into one membrane monolayer may promote expansion of the polar head region and impart curvature stress onto the overall lipid bilayer (reviewed in Kozlov et al., 2010). In the case of the HIV-FP, it was hypothesized that insertion into the external membrane monolayer may induce lateral stress in the hydrocarbon region and/or selectively induce an increment of its surface (Agirre et al., 2000). As a consequence bulges (or dimples) that protrude from the membrane plane would be created (Fig. 1C (4) right panel). The tops of these structures are highly curved, an arrangement that can facilitate the formation of lipid contacts between fusing bilayers (Kozlov et al., 2010). This mode of VFP action would be also consistent with an alternative mechanism of fusion, which posits that the process can be mediated by activated glycoproteins with their FPs inserted into the viral membrane (Chernomordik and Kozlov, 2003). Bilayer deformation can be additionally facilitated by compounds that lower the bending rigidity of the bilayer. Thus membrane-embedded VFPs may behave as additives that disorder and soften the lipid bilayer matrix making it more prone for bending or expansion (Fig. 1C (4) left panel). Recent low-angle x-ray scattering studies reveal that the inserted HIV-FP indeed reduces the bending modulus (K C) of the lipid bilayer (Tristram-Nagle et al., 2010, Tristram-Nagle and Nagle, 2007).
Finally, VFPs may sustain the aperture of lipid fusion pores with distinctive architectures. In one hand, VFPs may promote the opening of hydrophilic pores within the expanding HD (Fig. 1C (5)). Such mechanism would be supported by the finding that functional VFPs bear a pore-formation activity below the membrane rupture tensions (Lau et al., 2004, Longo et al., 1997, Longo et al., 1998), and by their capacity for inducing positive curvature put forward by recent MDS (Fuhrmans and Marrink, 2012). However, it is difficult to envision how the FP attached to the glycoprotein (Fig. 1A) would get access to the expanding HD, an element confined to a topologically distinct compartment.
On the other hand, under the assumption that the stalk does not expand, stalk formation and its evolution into a fusion pore are restrained by the energy of hydrophobic interstices (voids) emerging between contacting monolayers (Lentz et al., 2000, Siegel and Epand, 1997). Thus, VFPs might contribute to lower the energy of those voids, thereby facilitating lipid mixing of cis-monolayers and/or establishment of trans-monolayer contacts (TMCs) (Fig. 1C (6)). Evidence supporting this VFP role mainly derives from experiments of PEG-mediated fusion (Chakraborty et al., 2013, Haque et al., 2011). The physiological relevance of those studies has been put forward by the comparison of wild-type IFV-FP and two derived mutants, G1E and G13L. The wild-type peptide enhanced PEG-induced fusion, increased the bilayer interior packing and filled the membrane free volume, while the mutants caused no change in PEG-induced fusion or interior packing. It was inferred that the functional IFV-FP promotes conversion of the stalk to an expanded TMC intermediate through its ability to fill the voids within this latter structure (Fig. 1C (6)).
In addition, the ability for minimizing interstice energy can be indirectly inferred from the effects on the thermotropic lamellar-to-non-lamellar (HII) phase transition. VFPs have been shown to drive the process toward the formation of bicontinuous inverted cubic phases (QII) (Harroun et al., 2003). Small-angle X-ray diffraction results showed that the action of the IFV-FP on lipid polymorphism was strongly pH-dependent (Colotto and Epand, 1997). In particular, the ability of peptide to promote cubic phases exhibited the same dependence on the pH as viral fusion. Since bilayers arranged into bicontinuous QII phases bear curvatures reminiscent of those purportedly existing within unexpanded fusion pores, it has been proposed that a plausible mechanism of fusion-promotion by the IFV-FP could be that this element may help destabilizing bilayer diaphragms at TMCs, or favor bilayer states of negative Gaussian curvature (Siegel and Epand, 2000, Tenchov et al., 2013).
2.4. Membrane-inserted inactive VFP forms and fusion pore expansion
The neck in the early fusion pore structures can be sufficiently narrow as to impede diffusion of large solutes. Thus, driven by forces favoring membrane unbending, the final stage of the viral fusion process comprises the irreversible expansion of the initial fusion pore (Cohen and Melikyan, 2004), a phenomenon required for the translocation of the macromolecular nucleocapsid complex into the cytosol of the host cell. Formation of the fully zipped trimer-of-hairpins ectodomain has been shown to be required for this step (Blumenthal et al., 2012, Melikyan, 2008). In turn, 6-HB completion may lead to the formation of complexes between the membrane-embedded N-terminal FP and the C-terminal TMD (Armstrong et al., 2000, Tamm, 2003). These integral membrane oligomers might represent the third and last stage in the life of the VFPs (Fig. 1A-(3)). In contrast to forms inserted into single monolayers, FP-TMD transbilayer bundles would favor bilayers with 0 net curvature. Thus, membrane remodeling upon this locking movement might be coupled to fusion pore enlargement.
Although the participation of FP during the pre-hairpin intermediate state has been widely studied, only a few studies have focused on its possible co-assembly with TMDs and the involvement of the process in the late stages of fusion. Since FP and TMD domains lie in close proximity in the final trimeric hairpin structure of IFV-HA (Chen et al., 1999), and being this final conformation necessary to induce pore enlargement, Cohen and co-workers proposed that both domains approach each other to their maximum extent during the late stages of fusion (Borrego-Diaz et al., 2003, Cohen and Melikyan, 2004).
Following this idea, Chang et al. explored the existence of actual interactions between HA FP and TMD and its involvement in the IFV fusion process (Chang et al., 2008). They used fluorescently labeled peptides and model membranes, to assay formation of complexes between FP and TMD domains. The assembly occurred in an antiparallel manner, the TMD being at the core of the structure. In addition, as compared to homo-oligomers, the hetero-oligomers showed an enhanced membrane disrupting ability, likely due to a deeper membrane penetration of the FPs. These authors concluded that the assembly of the FP-TMD structure and its recruitment to the fusion site could cause the rupture of the membrane inner leaflets in a post-stalk state, resulting in initiation and enlargement of the fusion pore.
In a later contribution, Donald et al. addressed the existence of FP-TMD interactions in the case of the Class I fusion protein F of PIV5 (Donald et al., 2011). These authors observed that homo-oligomerization of isolated PIV5 FPs into hexameric bundles occurred in DPC micelles. According to ATR-IR results, FPs would adopt a transmembrane orientation in lipid bilayers. Supporting this idea, MDS of the FP six-helix bundle in lipid bilayers predicted a parallel orientation of the constituent monomers, with its periodically conserved small residues localized at the helix-helix interfaces, and allowing permeation of water into the hexamer core. Tight hetero-association of FP with TMD peptides also occurred in the membrane mimetic DPC. Based on their experimental data and the outcome of the simulations, the authors proposed that FP hexamers may create holes (“pinpricks”) within the target membrane. Further tilting of the helices within approaching bilayers enable the formation of FP-TMD bundles, which eventually provides a low-energy pathway to direct fusion.
Finally, a more recent study of Reuven et al. has shown that the HIV-1 gp41 FP may also functionally associate with the C-terminal TMD (Reuven et al., 2012). These authors made use of an in vivo fluorescence assay to determine FP hetero-assembly with the TMD (probably via its GLRI motif) within the membrane milieu, and the ability of the latter to self-assemble via its GxxxG motif. Besides, heteroassociation between FP and TMD peptides led to enhanced lipid mixing of model membranes. Furthermore, peptides derived from the TMD and FP could inhibit virus-cell fusion probably by interfering with the functional association of the parental endogenous regions. In conjunction these studies suggest that conserved Gly-Ala residues at regularly spaced intervals might sustain VFP homo- and hetero-oligomerization in membranes.
3. Updated case studies
3.1. Influenza HA
Combining NMR-solved structures in micelles with EPR distance constraints measured in LUV, Tamm and co-workers proposed an “inverted V” or “boomerang” conformation for the functional IFV-FP in membranes (Han et al., 2001). As compared to the neutral pH structure, the low pH structure of a 20-mer synthetic peptide inserts deeper while the C-terminal arm transitions from an extended structure to a shorter 310-helix (Fig. 2 B-left). This low-pH-induced structural change was proposed to intensify the FP capacity to distort bilayer architecture (see also (Larsson and Kasson, 2013)). The “inverted V” model was subsequently tested by assessing the effects of IFV-FP mutations that interfere with fusion activity of the full HA protein. In particular, the effects of G1S and G1V mutations, which respectively cause hemifusion and abolish fusion, were first examined (Li et al., 2005). The NMR structures disclosed a V with a disrupted “glycine edge” on its N-terminal arm and a slightly tilted linear helical structure, for the G1S and G1V mutants, repectively. Abolishment of the kink in G1V resulted in reduced hydrophobic penetration of the lipid bilayer and an increased propensity to form β-structures at the membrane surface (Fig. 2B-left).
Fig. 2.
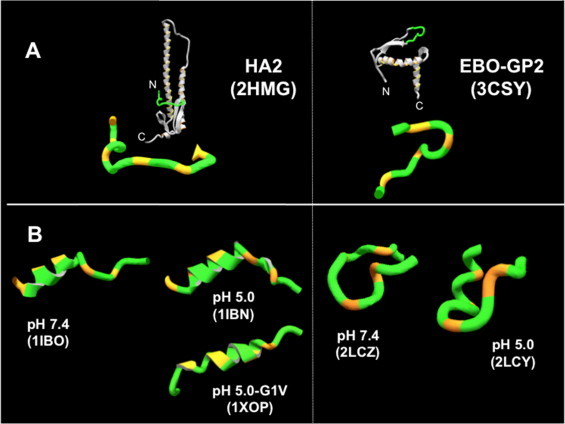
Native and membrane-inserted structures of IFV and EBOV fusion peptides. (A) X-ray structures disclosing several turns in the native fusogenic subunits. Structures on top disclose their position within the ectodomain. (B) NMR structures of peptides in DPC micelles reflecting features of α-helix. In both panels the orange stretches mark the positions of Gly/Pro residues. Structures with the PDB codes in brackets have been used to render the Figure.
The preservation of the low pH-induced, double-arm, “inverted V” structure as a requirement for IFV-FP function was further supported by the outcome of MDS in detergent micelles and lipid bilayers (Lague et al., 2005), and a series of structure–function studies (Lai et al., 2006, Lai and Tamm, 2007). The role of a specific fixed angle “boomerang” structure in fusion was underpinned by the effect of changing W14, a residue involved in kink stabilization (Lai et al., 2006). The W14A substitution was shown to cause inactivation of HA, while the structure of a synthetic IFV-FP bearing the mutation disclosed a flexible kink with an inverted orientation. Thus, the mutant pointed out of the membrane, in contrast to the ordered boomerang of the wild-type, which pointed into the membrane. The W14A fusion phenotype and FP structures were later reproduced by a double substitution F9A/I10A, an observation that underscores the functional importance of membrane-oriented large hydrophobic residues on both sides of the kink region of IFV-FP to fix the angle of the boomerang structure (Lai and Tamm, 2007). Recently published MDS analyses give further support to this idea (Legare and Lague, 2012).
The importance of selecting FP sequence boundaries in structural studies was emphasized by the findings reported by Lorieau et al. (Lorieau et al., 2010). These authors included the conserved residues Trp21-Tyr22-Gly23 at the C-terminus of the IFV-FP, and analyzed the NMR structure in DPC micelles of the resulting specimen, which was designated as HA-FP1–23. The NMR structure of HA-FP1–23 dissolved in DPC disclosed an α-helix-turn-α-helix motif where N- and C-terminal helices packed tightly together in anti-parallel fashion (Fig. 3 A). Interactions between C-terminally added Trp21-Tyr22-Gly23 and the N-terminal residues of HA-FP1–23 were fundamental in the stabilization of this hairpin-like structure. The tight packing was in addition sustained by four interhelical aliphatic H-bonds. The hairpin was predominantly maintained even when lowering the pH below the pKa value of its acidic side chains. A charge–dipole interaction between the positive charge of the Gly1 α-amino group and the dipole moment of the second helix was subsequently described as an additional stabilizing force (Lorieau et al., 2011).
As shown in Fig. 3A, the HA-FP1–23 hairpin segregates into one surface of the structure the hydrophobic residues Phe-3, Ile-6, Phe-9, Trp-14, Met-17, Trp-21 and Tyr-22. Interestingly, early assays performed by Brunner and coworkers (Harter et al., 1989) had identified in the radioactivity profiles these same residues (i.e., including Trp-21 and, to a lower extent, Tyr-22) as the main sites of labeling by membrane-residing photoactivable probes after low pH-induced interaction of BHA with liposomes (Fig. 3B). This arrangement of the hydrophobic and hydrophilic residues relative to the hairpin surface generates an amphipatic structure predicted to stably associate with the water-lipid interface of a membrane leaflet. Thus, the hairpin structure would be also consistent with previous asymmetric hydrophobic photolabeling assays indicating that BHA-membrane interaction induced by low pH involves HA-FP insertion into the external membrane monolayer (Brunner, 1989).
However, these authors later reported that at low pH, where the fusion process is triggered, HA-FP1–23 transiently visits activated states consisting of two stable helices switching between L-shape and extended arrangements, where the L-shape provides an intermediate step in the transition from the hairpin to a totally extended structure (Lorieau et al., 2012). They hypothesized that extension of the hairpin would elongate the IFV-FP to traverse the lipid bilayer, and therefore allow it assembling into bundles with C-terminal transmembrane helices (see preceding Section 2.4). Recent Solid State (SS)-NMR analyses confirm the adoption of closed hairpin structures in membranes by the 20-mer IFV-FP (Sun and Weliky, 2009), although the fractional populations of hairpin and extended structures may be different in the lipid-bilayer context, as compared to micelles (Ghosh et al., 2013). In this regard SS-NMR results confirm the presence of “boomerang” structures in detergent micelles, but support the dominance of closed and semi-closed structures for membrane-associated IFV-FP at pH 5.1.
Thus it appears that segmentation into two helical arms joined by a kink, and fixation of the angle between both arms depending on sequence, pH and lipid environment, may encompass functional requirements for IFV-FP activity along the fusion pathway. It is possible that favorable IFV-FP insertion is driven by the compact hairpin structure, while subsequent target membrane destabilization would be promoted by its expanded structures. As mentioned above, a recent MDS study suggests that the “boomerang” structure might be involved in pre-stalk stages of the fusion process (Larsson and Kasson, 2013).
3.2. HIV-1 gp41
Similarly to the IFV-FP, high-resolution studies carried out in the course of the last decade reiterate the existence of conformationally polymorphic HIV-FP structures (Fig. 4 ), which are likely required to sustain the processes of folding into the globular ectodomain (Julien et al., 2013, Lyumkis et al., 2013), complex formation with inhibitors (Forssmann et al., 2010, Munch et al., 2007) or adoption of alternating structures in a membrane milieu (Gordon et al., 2002, Gordon et al., 2004, Jaroniec et al., 2005, Li and Tamm, 2007, Qiang et al., 2009). In addition, MDS studies have provided supporting evidence for this idea (Barz et al., 2008, Grasnick et al., 2011, Promsri et al., 2012).
Fig. 4.
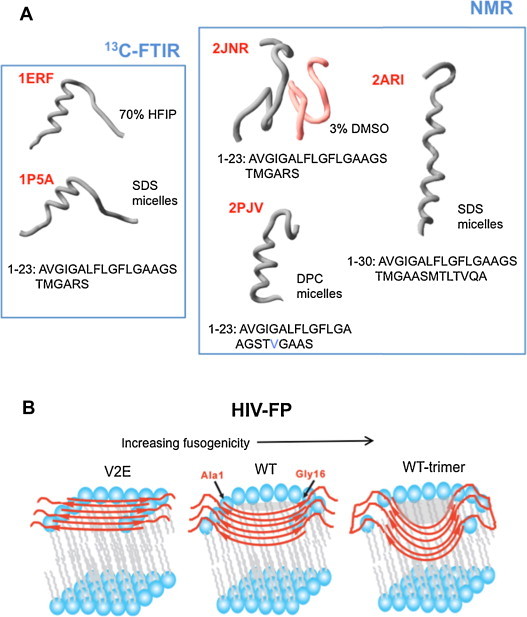
Conformational plasticity of HIV-FP. (A) Gallery of structures of synthetic peptides solved by IR (Gordon et al., 2002, Gordon et al., 2004) and NMR (Jaroniec et al., 2005, Li and Tamm, 2007, Munch et al., 2007). Corresponding PDB entry codes are displayed in red. 2JNR corresponds to the FP in complex with VIRIP (reddish chain). (B) Models for the FP inserted into Chol-containing membranes adopting 6-strand, anti-parallel β-sheet structures, as inferred from SS-NMR determinations. The most fusion-active version inserts deeper (see text). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Source: Adapted from Qiang et al. (2009), © 2009 by National Academy of Sciences, USA.
However, these studies also confirm early observations indicating that membrane-active structures of IFV-FP and HIV-FP are different. After some debate at the time the proposal was first formulated (Mobley et al., 1999, Nieva et al., 1994, Pereira et al., 1995, Pereira et al., 1997), considerable experimental evidence has been accumulated, which ascribes a fusogenic role to β-like extended HIV-FP structures (Buzon et al., 2005, Castano and Desbat, 2005, Haque et al., 2005, Lai et al., 2012, Sackett et al., 2010, Sackett and Shai, 2005, Saez-Cirion and Nieva, 2002). Most prominently, SS-NMR data reported by Weliky's group has provided in conjunction unambiguous evidence for the adoption of oligomeric β strand structure by the HIV-FP inserted into lipid bilayers (Qiang et al., 2008, Yang et al., 2001, Yang et al., 2003, Yang et al., 2004a). In particular, the work by Qiang et al. was key to determine the relevance of this structure under fusion-promoting conditions (Qiang et al., 2009) (see also Fig. 4B). In that work the authors compared structure, fusogenicity and insertion depth of wild-type HIV-FP, a V2E mutant and a cross-linked trimeric HIV-FP that emulates the putative oligomerization state of HIV gp41. All peptides adopted a β-sheet structure in membranes with physiologically relevant cholesterol content. However, in liposome fusion assays the V2E peptide was not active, while trimeric HIV-FP displayed higher potency than the monomeric peptide. The membrane locations as inferred from SS-NMR measurements indicated subtle insertion of the V2E mutant at the membrane surface, deeper insertion into a single membrane leaflet for the monomeric wt, and the deepest level of insertion for the trimeric HIV-FP, which contacted with the center of the membrane. Thus, for a fixed β-sheet secondary structure, a deeper level of insertion correlated with higher fusogenicity. As depicted in the model of Fig. 4B, an intriguing possibility is that the deeply inserted, distorted β-sheets might induce both local dehydration and hydrophobic defects in the monolayer (shadowed areas), thereby increasing the probability of hydrophobic tail flipping upon membrane adhesion (Fig. 1B-(2)).
Thus, attending to the data derived from IFV-FP and HIV-FP studies, it appears that, irrespective of the main conformation, the level of insertion is key for inducing membrane remodeling. This concept has been further elaborated by Lai et al. in a more recent contribution (Lai et al., 2012). These authors report structure–function experiments consistent with the fusogenicity of α- and β-structures adopted by the HIV-FP in Chol-depleted or Chol-containing membranes, respectively (see also the discussion by Epand, 2012). According to these authors, it is possible that fusion domains first assemble as β-sheets on membrane surfaces but later convert into α-helices to complete fusion.
In the last years, particular attention has been also paid to the effect of additional gp41 sequences on the structure and fusogenicity of HIV-FP. In a series of works initiated by Sackett and Shai (Sackett and Shai, 2003), the structure–function of the FP within constructs encompassing this region plus downstream gp41 helical domains has been investigated (Sackett et al., 2009, Sackett et al., 2010, Sackett and Shai, 2005, Sackett et al., 2011). The early studies in this series demonstrated that in trimeric constructs containing the NHR oligomerization domain to model the pre-hairpin intermediate, the FP sequence remained active while retaining its tendency to assemble into β-sheet structures in the membrane (Sackett and Shai, 2003, Sackett and Shai, 2005). Later analyses have revealed that fusogenicity of the FP is hampered within a construct representing post-fusion 6-HB structures, even though the membrane-associated extended structure still existed within these constructs (Sackett et al., 2010, Sackett et al., 2009). In conjunction, these data underscore the fusogenic function of the FP sequence during the fusion stages that precede trimeric hairpin formation, and support the hypothesis that the FP of HIV constitutes an autonomous folding domain.
3.3. Paramyxovirus F protein
The native structure of the PIV5-FP combines features of β-sheet and α-helical conformations (Yin et al., 2006). The C-terminal helix of the FP extends into the F protein N-terminal helical domain, and the joint connecting both domains likely remains helical upon fusion activation. Thus, in contrast to IFV-FP, the post-fusion PIV5-FP would extend the N-terminal helix into the membrane to enable its assembly with the TMD into an integral 6-helix bundle (Donald et al., 2011). Consistent with the functional role of the α-helical conformation, Gly substitutions in the FP exhibited increase in fusion activity in transfected cells (Horvath and Lamb, 1992), and augmented the helical content of the cognate synthetic peptide (Peisajovich et al., 2002).
Some structure–function studies combining synthetic Paramyxovirus FPs and model membranes have been added in recent years to the works already consigned in our previous review (Nieva and Agirre, 2003). Smith and coworkers identified a double mutation VM114/115AA in Hendra F protein that blocked fusion activity (Smith et al., 2012). These authors subsequently examined changes in membrane interactions, using synthetic peptides representative of both wild-type and VM114/115AA mutant F proteins. Consistent with an alteration of the membrane interaction pattern, incubation with the VM114/115AA peptide resulted in a 75% reduction of hemolysis. Moreover, the wild-type peptide exhibited greater helical content as estimated from circular dichroism measurements. Together, these data support that the helical FP structure is important to drive efficient paramyxovirus membrane fusion.
In a more recent study Yao and Hong have analyzed the lipid interactions of the PIV5-FP by means of SS-NMR (Yao and Hong, 2013). These studies reveal a conformational polymorphism for PIV5-FP as a function of the composition of the phospholipid membrane in a way reminiscent of the HIV-FP. Thus, PIV5-FP was found to adopt α-helical structure in anionic membranes, but it was mostly β-sheet in neutral ones. The latter structure caused membrane surface dehydration and curvature. Accordingly the authors ascribe a role for the PIV5-FP β-sheet structure in promoting/stabilizing the hemifusion intermediate. In contrast, the α-helical state would be associated with the extended prehairpin state and the post-fusion state (Donald et al., 2011).
3.4. Ebola GP
The crystal structure of the EBOV-FP was solved in the trimeric, pre-fusion conformation of the envelope glycoprotein (GP) (Lee et al., 2008). In contrast to the previously reported IFV-FP or PIV5-FP structures, the EBOV-FP embodied an internal fusion loop lacking a free N terminus (Fig. 2A-right and Table 1). The hydrophobic EBOV-FP was displayed onto an antiparallel β-stranded scaffold, which was stabilized by a disulphide bond at the base. However, similarly to PIV5-FP, the side chains of the most hydrophobic residues at the tip of the EBOV-FP packed into a neighboring GP surface subunit in the trimer, while the rest of the sequence sat in the periphery of the complex, partly accessible from solution.
Prior to the X-ray structure resolution of the native structure, several studies addressed the interactions of the internal EBOV-FP with membranes. These initial reports suggested that anionic phospholipids might be required for the efficient insertion of EBOV-FP into membranes (Ruiz-Arguello et al., 1998, Suarez et al., 2003). This requirement was not based on the establishment of strong peptide-lipid electrostatic interactions, but rather on the easier access of the FP sequence to the hydrophobic region in the anionic bilayers, and the favorable contribution of its partial structuring therein to the overall energy of partitioning. A helix inserted in a tilted orientation was subsequently posited as the effective fusogenic structure of the sequence (Adam et al., 2004). Somehow contrasting this proposal, it was further suggested that stabilization of a loop structure within membranes by a Pro residue was required for the EBOV-FP membrane activity (Gomara et al., 2004).
Both possibilities need not be mutually exclusive as put forward by later high-resolution NMR studies (Freitas et al., 2007, Gregory et al., 2011) (Fig. 2B-right). In accordance with previous low-resolution studies, the EBOV-FP sequence was shown to get partially structured in contact with SDS micelles at pH 7.0 by adopting a 310-helix in the central part (Freitas et al., 2007). Furthermore, spectroscopic measurements and MDS suggested the participation of the central aromatic residues in interactions with rigid membranes (see also (Freitas et al., 2011)). Gregory and co-workers further analyzed the effect of the β-sheet scaffold and pH on the structure adopted by EBOV-FP in membrane mimics (Gregory et al., 2011). Their analysis yielded a model for the structure–function of this domain (Fig. 2B-right). According to these authors, a conformational change from a relatively flat extended loop structure at pH 7.0 to a bent structure at pH 5.5 would reorient the hydrophobic patch at the tip of the FP. The latter structure display helical features within a reversing turn inserting into the membrane. It is tempting to speculate that the energy contribution from structuring one portion of the Cα chain is required for the favorable insertion of the full loop. This mode of interaction would facilitate disruption of lipids in a way reminiscent to that of N-terminal VFPs.
4. Fusion loops in Class II and Class III glycoproteins
Based on sequence similarities, it has been proposed that Class II/III FLs and internal Class I FPs share functional and structural features (Table 2 ). However, the available experimental evidence is not fully compatible with that assumption. In one hand, some of the crystallographically solved Class II glycoproteins allow inferring the structure adopted by FL-containing regions in pre-fusion, fusion-activated and post-fusion states (Baquero et al., 2013, Gibbons et al., 2004, Klein et al., 2013). All those structures reveal that the exposed hydrophobic FLs at the tip of elongated β-barrel domains (DII-s) are usually shorter than the predicted sequences (Table 2). Aliphatic, aromatic as well as Gly residues are commonly found in β-strands, which, together with the unusual length of those elements in DII domains, obscured initial Class II/III FP detection based on sequence length and hydrophobicity distribution (Nieva and Suarez, 2000).
Table 2.
VFPs of Class II and III fusion proteins.
| Class | Family | Speciesa | Sequencea | Position | Prefusion State | ΔG Interfacialb | Ala Percentc | Gly Percentc |
|---|---|---|---|---|---|---|---|---|
| Class II | Flaviviridae | TBEV | DRGWHNGCGLFGKGSI | Internal | Homodimer | 0.5 | 0/8 | 31/9 |
| DENV-1 | DRGWGNGCGLFGKGSL | Internal | Homodimer | –0.7 | 0/7 | 38/10 | ||
| Togaviridae | SFV | VYTGVYPFMWGGAYCFCDS | Internal | Heterodimer | –3.86 | 4/8 | 13/7 | |
| CHIKV | VYPFMWGGAYCFCDTENT | Internal | Heterodimer | –2.11 | 6/11 | 12/6 | ||
| Class III | Rhabdoviridae | VSV | WY/YA | Bipartite | Trimeric | –3.56 | 25/5 | 0/7 |
| Herpesviridae | HSV | VWFGHRY/RVEAFHRY | Bipartite | Trimeric | 0.7 | 7/11 | 7/7 | |
| Baculoviridae | AcMNPV | YAYNGGSLDPNTRV/VKRQNNNHFAHHTCNK | Bipartite | Trimeric | 8.36 | 7/5 | 7/5 |
TEBV: Tick-Borne Encephalitis Virus, DENV-1: Dengue Virus type 1, SFV: Semliki Forest Virus, CHIKV: Chikungunya Virus, VSV: Vesicular Stomatitis Virus, HSV: Herpes Simplex Virus and AcMNPV: Autographa californica Multiple Nucleopolyhedrovirus.
Computed according to the Wimley-White scale of water-membrane partitioning energies.
Mole percent of alanine and glycine in fusion peptide over whole protein sequence.
On the other hand, the FLs adopt almost invariable conformations in pre- and post-fusion states (Fig. 5 ). This is because the globular structures of the FL-containing DII-s behave as rigid bodies along the fusion pathway, i.e., they do not undergo significant structural rearrangements during the process besides their spatial reorientation. It is thus inferred that, in contrast to Class I internal FPs, short Class II FLs would retain their short-loop conformation upon insertion into the target membrane, an assumption supported by structural studies based on synthetic sequences (Melo et al., 2009, Mendes et al., 2012, Mohanram et al., 2012).
Fig. 5.
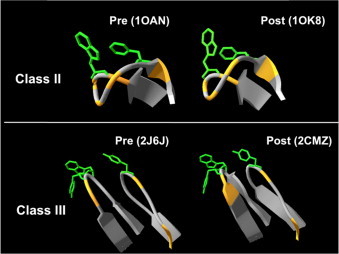
Fusion loops derived from Class II and Class III glycoproteins. X-ray structures of FLs in pre- and post-fusion states of Class II DENV-E and Class III VSV-G glycoproteins are displayed (top and bottom panels, respectively). In both panels the exposed side-chains of aromatic residues are shown in green, while orange stretches mark the positions of Gly/Pro residues. Structures with the PDB codes in brackets have been used to render the figure.
The interaction involving just few hydrophobic residues, and the absence of autonomous folding within the membrane milieu suggest a subtle FL insertion into the outer leaflet of the membrane, which is likely insufficient to compromise the lipid bilayer architecture. This notion is further substantiated by the findings reported in the case of the Class III VSV-G protein (Roche et al., 2006). VSV-G contains at the tip of domain IV a bipartite FL composed of 2 short loops, which together contribute just three aromatic residues to the interaction with the target membrane (Fig. 5 and Table 2). The fact that they may remain inserted into the viral membrane in the pre-fusion state further emphasizes that insertion of those residues does not by itself destabilize membranes (Roche et al., 2007). This is in contrast to Class I IFV, which shows dramatic changes in envelope morphology, fused virions and disorganized HA spikes upon low-pH-induced FP insertion into the envelope (Fontana et al., 2012).
Thus, a cooperative effect attained upon fusion activation has been invoked to explain the induction of target membrane deformations by Class II/III glycoproteins. In vitro assays based on synthetic peptides and liposomes suggest that the context of the trimer can be critical for DENV E FL-induced membrane destabilization (Pan et al., 2010). Moreover, contacts between FLs inserted into the target membrane have been shown to render rings made of several SFV E1 trimers (Gibbons et al., 2004). The particular architecture of those rings would generate nipples within the target membrane, reminiscent of pre-stalk fusion intermediates.
5. Other glycoprotein hydrophobic regions: MPER domains
Besides the canonical VFPs and VFLs discussed above, other regions within viral fusion proteins have been proposed to participate in fusion (reviewed in Peisajovich and Shai, 2003). Among these regions, the NHRs of Class I glycoproteins were found to interact with membranes and postulated to cooperate in FP-induced fusion. This finding applied to several glycoproteins including influenza HA2 (Epand et al., 1999), Sendai virus F1 (Ghosh and Shai, 1999) and HIV-1 gp41 (Sackett and Shai, 2002). In this latter case, substitution of the NHR by the coiled-coil promoting GCN4 sequence, rendered a less fusogenic construct (although still more active than the 33-mer FP) indicating that not only the coiled-coil conformation but also the sequence of NHR is essential for an increased fusion efficiency (Wexler-Cohen et al., 2005).
The presence of additional sequences featuring homology to the canonical FPs was found within the Sendai F1 (Peisajovich et al., 2000), Measles F1 (Samuel and Shai, 2001) and SARS-CoV S2 (Guillen et al., 2005) proteins. Based on their potential capacity for interacting with lipid bilayers multiple regions have been additionally identified within diverse fusion glycoproteins such as Hepatitis C virus E1 and E2 (Perez-Berna et al., 2006), Dengue E (Nemesio et al., 2011) or Herpex Simplex virus gB and gH (Galdiero et al., 2005, Galdiero et al., 2008).
All in all, this evidence suggests that the classical view of a single FP being solely responsible for the membrane interacting/perturbing activity of viral fusion proteins should be qualified by assuming a concerted action of different membrane-transferring regions. In support of this notion, the membrane proximal external region (MPER or preTM) that in Class I glycoproteins connects the CHR with the TMD, has been proposed to act as a C-terminal FP in several viral proteins (Lorizate et al., 2008, Peisajovich and Shai, 2003). A feature shared by MPER/preTM domains is the enrichment in aromatic residues, which are likely to enable their insertion into the viral membrane-interface (Suarez et al., 2000a). The presence of MPER domains has been extended to several Class I and II glycoproteins (Garry and Dash, 2003, Lorizate et al., 2008).
Since its discovery a great deal of effort has been expended on dissecting the structure–function of the most paradigmatic MPER: the one derived from HIV-1 gp41. The first hint of the implication of this region in fusion was the discovery of the DP-178 peptide, now designated as T-20 (brand name: Fuzeon, generic name: enfuvirtide) (Wild et al., 1994). It was the first fusion inhibitor approved by the FDA in 2003 and the only currently approved drug targeting gp41. Its sequence (amino acids 643–678) overlaps partially that of the MPER (amino acids 662–683). Moreover, gp41-targeting antibodies that block HIV-1 entry into cells bind to epitopes within MPER (Lorizate et al., 2008).
Direct confirmation of MPER involvement on fusion was provided by mutagenesis studies reported a few years later (Munoz-Barroso et al., 1999, Salzwedel et al., 1999). Deletions, substitutions, and insertions in the MPER, abrogated the ability of HIV-1 Env to mediate both cell-cell fusion and virus entry, without affecting the normal maturation, transport, or CD4-binding ability of the protein. Interestingly, substitution of three of the five highly conserved tryptophan residues by alanines sufficed to block cell-cell fusion and virus entry (Salzwedel et al., 1999). Further analysis of the W(1-5)A mutant revealed that fusion was abrogated at the non-expanding fusion pore stage (Munoz-Barroso et al., 1999). However, the precise mechanism by which this region participated in fusion was unclear.
Almost in parallel, Suarez et al. reported that a region spanning residues 664–683 showed high tendency to partition into the membrane interface, as revealed by the Wimley-White interfacial hydrophobicity scale (Suarez et al., 2000a). Confirming this prediction, a synthetic peptide representing this region bound, permeabilized and fused model membranes made of neutral phospholipids and cholesterol, even more potently than a peptide representing the FP. Moreover, replacement in this peptide of the first three Trp residues by Ala, reported by Salzwedel et al. to render a fusion defective gp41 phenotype, abrogated its ability to induce membrane fusion, supporting the physiological relevance of the in vitro approach (Suarez et al., 2000a, Suarez et al., 2000b). Further development of the WW analysis suggested that MPER could be segmented into two segregated interfacial subdomains: C-terminal residues (673–683) are hydrophobic-at-interface and N-terminal residues (664–672) include a stretch whose face shows strong affinity for membranes if folded into an α-helix, i.e., an amphipathic-at-interface helix (Lorizate et al., 2006a, Saez-Cirion et al., 2003). Supporting the functional relevance of this organization, a number of fusion-blocking deletions described by Salzwedel et al. correlated with the ablation and/or merging of interfacial sub-domains (Saez-Cirion et al., 2003, Salzwedel et al., 1999). Later reported NMR structures confirmed the predicted MPER segmentation into two short helical subdomains (Sun et al., 2008).
To further establish a correlation between MPER insertion into and perturbation of the membrane and fusion, Vishwanathan and Hunter carried out a mutagenesis study (Vishwanathan and Hunter, 2008) where MPER was replaced by the membrane-perturbing tryptophan-rich cationic peptide indolicidin, and revealed that some of the mutant Envs maintained fusogenic activity. This indicates that MPER insertion into membranes is not only a structure-related pattern, but also a requirement for the fulfillment of a membrane disrupting function during fusion. In addition, a number of studies employing model membranes and MPER-based synthetic peptides have shown that these membrane-perturbing activities are finely modulated by the lipid composition and the MPER segmentation into hydrophobic subdomains (Apellaniz et al., 2009, Apellaniz et al., 2010, Apellaniz et al., 2011a, Apellaniz et al., 2011b, Ivankin et al., 2012, Saez-Cirion et al., 2002, Shnaper et al., 2004, Suarez et al., 2000b). Taken together, these data point to a fusion model where MPER interfacial activity might help remodeling of the merging membranes along the fusion pathway by sustaining transient disruption of lipid continuity (Saez-Cirion et al., 2002, Shnaper et al., 2004) (Fig. 1B-(2)). Alternatively, it has been suggested that MPER insertion might generate the bulging out of the viral membrane, while the curved end-caps of such bulges would be highly fusogenic (Buzon et al., 2010, Kozlov et al., 2010) (Fig. 1B-(4)).
Finally, although high-resolution structures for the region are not yet available, an interesting possibility put forward by experimental evidence (Bellamy-McIntyre et al., 2007, Lorizate et al., 2006a, Lorizate et al., 2006b), is that both gp41 hydrophobic domains, MPER and FP, combine in the pre-fusion structure to render an overall soluble complex. The recently published SOSIP structure does not exclude such possibility (Julien et al., 2013). The fact that the FP stabilizes relevant structures of the MPER epitope targeted by the broadly neutralizing 2F5 antibody supports the interaction between both regions, and might as well have implications for the design of peptide immunogens (Huarte et al., 2012, Julien et al., 2008).
6. Prospects as therapeutic targets
Self-oligomerization of membrane-embedded synthetic FPs has been claimed to reflect a physiologically meaningful phenomenon (Chang et al., 2005, Kliger et al., 1997, Lau et al., 2004, Yang et al., 2004a). In the most amply studied case of the HIV, interference with formation of structurally defined oligomeric complexes was invoked to explain the inhibitory effect exerted by short synthetic FPs in cell systems (Kliger et al., 1997, Owens et al., 1990, Pritsker et al., 1998, Slepushkin et al., 1993). Gomara and co-workers followed this idea to develop a functional screening assay (Gomara et al., 2006). These authors took the advantage that in-membrane self-oligomerization of HIV-FP is required to establish permeating pores in vesicles made of anionic phospholipid (Nieva et al., 1994). Thus, they used leakage of aqueous contents induced by the synthetic HIV-FP as a surrogate assay for self-oligomerization in membranes. Subsequently, a D-hexapeptide library was screened for its capacity to block FP-induced leakage (for a description of this assay see also (Huarte et al., 2011)). Using this approach two hexapeptides were identified that blocked gp41-induced fusion, thereby supporting the FP region as a potential target for inhibitor development.
Such statement was soon confirmed by the finding of VIRIP, the sole HIV-FP inhibitor tested so far in clinical studies (Forssmann et al., 2010, Munch et al., 2007). Münch and coworkers purified an oligopeptide from hemofiltrate obtained from patients suffering from chronic renal failure, which was capable of blocking HIV-1 infection by targeting gp41 FP (Munch et al., 2007). This peptide, termed VIRIP (virus-inhibitory peptide), was later identified as a derivative from the C-terminal sequence of circulating serine protease inhibitor α1-antitrypsin. Optimized versions of VIRIP resulted as potent as T-20 in blocking viral infection and were devoid of cellular toxicity.
The VIRIP's molecular target was evidenced by its capacity for inhibiting synthetic FP-promoted hemolysis (Mobley et al., 2001). VIRIP derivatives showing improved anti-HIV capacity were also more potent interfering with FP-induced hemolysis. Consistent with a direct FP-VIRIP interaction, structural determinations by NMR revealed the formation of HIV-FP/VIRIP complexes, in which both peptides bracketed together via hydrophobic interactions of the amino acid side chains (Fig. 4A).
The outcome of a 10-day monotherapy clinical trial enrolling 18 HIV-1-infected patients was subsequently reported (Forssmann et al., 2010). The trial was meant to evaluate the safety, pharmacokinetics and antiviral efficacy of VIR-576, an optimized derivative of VIRIP. Treatment with VIR-576 was well tolerated by patients, and moderately reduced their plasma viral load. Together, VIRIP identification and its clinical evaluation, constituted a first proof of concept that FP inhibitors could suppress viral replication in infected individuals, and, therefore, support their clinical potential.
Finally, an intriguing possibility put forward by Shai's group is that the HIV-FP might bind the TMD of the T-cell receptor (TCR) α subunit, and interfere with the assembly of the TCR complex (Quintana et al., 2005). Further MDS analyses supported the establishment of specific β-sheet/α-helix interactions between both sequences within the membrane (Bloch et al., 2007). Thus, hetero-association with the TCRα TMD and ensuing immunosuppression may reflect that, behind the conventional membrane fusion, a hidden HIV-FP life might exist, also of relevance under pathological conditions.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Transparency document
Acknowledgements
We thank current financial support by the Spanish MINECO (BIO2011-29792), the Basque Government (IT838-13) and the National Institutes of Health (USA) (1R01AI097051-01). The authors acknowledge the contribution to their present understanding of VFP structure–function of many works not cited in this review due to space limitations.
References
- Adam B., Lins L., Stroobant V., Thomas A., Brasseur R. Distribution of hydrophobic residues is crucial for the fusogenic properties of the Ebola virus GP2 fusion peptide. J. Virol. 2004;78:2131–2136. doi: 10.1128/JVI.78.4.2131-2136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre A., Flach C., Goni F.M., Mendelsohn R., Valpuesta J.M., Wu F., Nieva J.L. Interactions of the HIV-1 fusion peptide with large unilamellar vesicles and monolayers. A cryo-TEM and spectroscopic study. Biochim. Biophys. Acta. 2000;1467:153–164. doi: 10.1016/s0005-2736(00)00214-5. [DOI] [PubMed] [Google Scholar]
- Apellaniz B., Garcia-Saez A., Nir S., Nieva J.L. Destabilization exerted by peptides derived from the membrane-proximal external region of HIV-1 gp41 in lipid vesicles supporting fluid phase coexistence. Biochim. Biophys. Acta. 2011;1808:1797–1805. doi: 10.1016/j.bbamem.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Apellaniz B., Ivankin A., Nir S., Gidalevitz D., Nieva J.L. Membrane-proximal external HIV-1 gp41 motif adapted for destabilizing the highly rigid viral envelope. Biophys. J. 2011;101:2426–2435. doi: 10.1016/j.bpj.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apellaniz B., Nieva J.L., Schwille P., Garcia-Saez A.J. All-or-none versus graded: single-vesicle analysis reveals lipid composition effects on membrane permeabilization. Biophys. J. 2010;99:3619–3628. doi: 10.1016/j.bpj.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apellaniz B., Nir S., Nieva J.L. Distinct mechanisms of lipid bilayer perturbation induced by peptides derived from the membrane-proximal external region of HIV-1 gp41. Biochemistry. 2009;48:5320–5331. doi: 10.1021/bi900504t. [DOI] [PubMed] [Google Scholar]
- Armstrong R.T., Kushnir A.S., White J.M. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 2000;151:425–437. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backovic M., Jardetzky T.S. Class III viral membrane fusion proteins. Curr. Opin. Struct. Biol. 2009;19:189–196. doi: 10.1016/j.sbi.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero E., Albertini A.A., Vachette P., Lepault J., Bressanelli S., Gaudin Y. Intermediate conformations during viral fusion glycoprotein structural transition. Curr. Opin. Virol. 2013;3:143–150. doi: 10.1016/j.coviro.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barz B., Wong T.C., Kosztin I. Membrane curvature and surface area per lipid affect the conformation and oligomeric state of HIV-1 fusion peptide: a combined FTIR and MD simulation study. Biochim. Biophys. Acta. 2008;1778:945–953. doi: 10.1016/j.bbamem.2007.11.014. [DOI] [PubMed] [Google Scholar]
- Bellamy-McIntyre A.K., Lay C.S., Baar S., Maerz A.L., Talbo G.H., Drummer H.E., Poumbourios P. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J. Biol. Chem. 2007;282:23104–23116. doi: 10.1074/jbc.M703485200. [DOI] [PubMed] [Google Scholar]
- Bloch I., Quintana F.J., Gerber D., Cohen T., Cohen I.R., Shai Y. T-cell inactivation and immunosuppressive activity induced by HIV gp41 via novel interacting motif. FASEB J. 2007;21:393–401. doi: 10.1096/fj.06-7061com. [DOI] [PubMed] [Google Scholar]
- Blumenthal R., Dimitrov D.S. Targeting the sticky fingers of HIV-1. Cell. 2007;129:243–245. doi: 10.1016/j.cell.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Blumenthal R., Durell S., Viard M. HIV entry and envelope glycoprotein-mediated fusion. J. Biol. Chem. 2012;287:40841–40849. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrego-Diaz E., Peeples M.E., Markosyan R.M., Melikyan G.B., Cohen F.S. Completion of trimeric hairpin formation of influenza virus hemagglutinin promotes fusion pore opening and enlargement. Virology. 2003;316:234–244. doi: 10.1016/j.virol.2003.07.006. [DOI] [PubMed] [Google Scholar]
- Brunner J. Testing topological models for the membrane penetration of the fusion peptide of influenza virus hemagglutinin. FEBS Lett. 1989;257:369–372. doi: 10.1016/0014-5793(89)81574-1. [DOI] [PubMed] [Google Scholar]
- Buzon V., Natrajan G., Schibli D., Campelo F., Kozlov M.M., Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6:e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon V., Padros E., Cladera J. Interaction of fusion peptides from HIV gp41 with membranes: a time-resolved membrane binding, lipid mixing, and structural study. Biochemistry. 2005;44:13354–13364. doi: 10.1021/bi050382r. [DOI] [PubMed] [Google Scholar]
- Castano S., Desbat B. Structure and orientation study of fusion peptide FP23 of gp41 from HIV-1 alone or inserted into various lipid membrane models (mono-, bi- and multibi-layers) by FT-IR spectroscopies and Brewster angle microscopy. Biochim. Biophys. Acta. 2005;1715:81–95. doi: 10.1016/j.bbamem.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Chakraborty H., Tarafdar P.K., Klapper D.G., Lentz B.R. Wild-type and mutant hemagglutinin fusion peptides alter bilayer structure as well as kinetics and activation thermodynamics of stalk and pore formation differently: mechanistic implications. Biophys. J. 2013;105:2495–2506. doi: 10.1016/j.bpj.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.K., Cheng S.F., Chien W.J. The amino-terminal fusion domain peptide of human immunodeficiency virus type 1 gp41 inserts into the sodium dodecyl sulfate micelle primarily as a helix with a conserved glycine at the micelle-water interface. J. Virol. 1997;71:6593–6602. doi: 10.1128/jvi.71.9.6593-6602.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.K., Cheng S.F., Deo Trivedi V., Yang S.H. The amino-terminal region of the fusion peptide of influenza virus hemagglutinin HA2 inserts into sodium dodecyl sulfate micelle with residues 16–18 at the aqueous boundary at acidic pH. Oligomerization and the conformational flexibility. J. Biol. Chem. 2000;275:19150–19158. doi: 10.1074/jbc.M907148199. [DOI] [PubMed] [Google Scholar]
- Chang D.K., Cheng S.F., Kantchev E.A., Lin C.H., Liu Y.T. Membrane interaction and structure of the transmembrane domain of influenza hemagglutinin and its fusion peptide complex. BMC Biol. 2008;6:2. doi: 10.1186/1741-7007-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.K., Cheng S.F., Lin C.H., Kantchev E.B., Wu C.W. Self-association of glutamic acid-rich fusion peptide analogs of influenza hemagglutinin in the membrane-mimic environments: effects of positional difference of glutamic acids on side chain ionization constant and intra- and inter-peptide interactions deduced from NMR and gel electrophoresis measurements. Biochim. Biophys. Acta. 2005;1712:37–51. doi: 10.1016/j.bbamem.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Chen J., Skehel J.J., Wiley D.C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L.V., Kozlov M.M. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- Cohen F.S., Melikyan G.B. The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J. Membr. Biol. 2004;199:1–14. doi: 10.1007/s00232-004-0669-8. [DOI] [PubMed] [Google Scholar]
- Colotto A., Epand R.M. Structural study of the relationship between the rate of membrane fusion and the ability of the fusion peptide of influenza virus to perturb bilayers. Biochemistry. 1997;36:7644–7651. doi: 10.1021/bi970382u. [DOI] [PubMed] [Google Scholar]
- Doms R.W., Moore J.P. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 2000;151:F9–F14. doi: 10.1083/jcb.151.2.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald J.E., Zhang Y., Fiorin G., Carnevale V., Slochower D.R., Gai F., Klein M.L., DeGrado W.F. Transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3958–3963. doi: 10.1073/pnas.1019668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubovskii P.V., Li H., Takahashi S., Arseniev A.S., Akasaka K. Structure of an analog of fusion peptide from hemagglutinin. Protein Sci. 2000;9:786–798. doi: 10.1110/ps.9.4.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durell S.R., Martin I., Ruysschaert J.M., Shai Y., Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion (review) Mol. Membr. Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- Durrer P., Galli C., Hoenke S., Corti C., Gluck R., Vorherr T., Brunner J. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J. Biol. Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Epand R.F., Macosko J.C., Russell C.J., Shin Y.K., Epand R.M. The ectodomain of HA2 of influenza virus promotes rapid pH dependent membrane fusion. J. Mol. Biol. 1999;286:489–503. doi: 10.1006/jmbi.1998.2500. [DOI] [PubMed] [Google Scholar]
- Epand R.M. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta. 2003;1614:116–121. doi: 10.1016/s0005-2736(03)00169-x. [DOI] [PubMed] [Google Scholar]
- Epand R.M. Further insights into the properties of the HIV gp41 fusion domain: commentary on the article by A.L. Lai et al. J. Mol. Biol. 2012;418:1–2. doi: 10.1016/j.jmb.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Epand R.M., Cheetham J.J., Epand R.F., Yeagle P.L., Richardson C.D., Rockwell A., Degrado W.F. Peptide models for the membrane destabilizing actions of viral fusion proteins. Biopolymers. 1992;32:309–314. doi: 10.1002/bip.360320403. [DOI] [PubMed] [Google Scholar]
- Epand R.M., Epand R.F. Relationship between the infectivity of influenza virus and the ability of its fusion peptide to perturb bilayers. Biochem. Biophys. Res. Commun. 1994;202:1420–1425. doi: 10.1006/bbrc.1994.2089. [DOI] [PubMed] [Google Scholar]
- Epand R.M., Epand R.F., Martin I., Ruysschaert J.M. Membrane interactions of mutated forms of the influenza fusion peptide. Biochemistry. 2001;40:8800–8807. doi: 10.1021/bi0107187. [DOI] [PubMed] [Google Scholar]
- Fontana J., Cardone G., Heymann J.B., Winkler D.C., Steven A.C. Structural changes in Influenza virus at low pH characterized by cryo-electron tomography. J. Virol. 2012;86:2919–2929. doi: 10.1128/JVI.06698-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forssmann W.G., The Y.H., Stoll M., Adermann K., Albrecht U., Barlos K., Busmann A., Canales-Mayordomo A., Gimenez-Gallego G., Hirsch J., Jimenez-Barbero J., Meyer-Olson D., Munch J., Perez-Castells J., Standker L., Kirchhoff F., Schmidt R.E. Short-term monotherapy in HIV-infected patients with a virus entry inhibitor against the gp41 fusion peptide. Sci. Transl. Med. 2010;2:63re63. doi: 10.1126/scitranslmed.3001697. [DOI] [PubMed] [Google Scholar]
- Freitas M.S., Follmer C., Costa L.T., Vilani C., Bianconi M.L., Achete C.A., Silva J.L. Measuring the strength of interaction between the Ebola fusion peptide and lipid rafts: implications for membrane fusion and virus infection. PLoS ONE. 2011;6:e15756. doi: 10.1371/journal.pone.0015756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas M.S., Gaspar L.P., Lorenzoni M., Almeida F.C., Tinoco L.W., Almeida M.S., Maia L.F., Degreve L., Valente A.P., Silva J.L. Structure of the Ebola fusion peptide in a membrane-mimetic environment and the interaction with lipid rafts. J. Biol. Chem. 2007;282:27306–27314. doi: 10.1074/jbc.M611864200. [DOI] [PubMed] [Google Scholar]
- Frolov V.A., Zimmerberg J. Cooperative elastic stresses, the hydrophobic effect, and lipid tilt in membrane remodeling. FEBS Lett. 2010;584:1824–1829. doi: 10.1016/j.febslet.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmans M., Marrink S.J. Molecular view of the role of fusion peptides in promoting positive membrane curvature. J. Am. Chem. Soc. 2012;134:1543–1552. doi: 10.1021/ja207290b. [DOI] [PubMed] [Google Scholar]
- Gabrys C.M., Qiang W., Sun Y., Xie L., Schmick S.D., Weliky D.P. Solid-state nuclear magnetic resonance measurements of HIV fusion peptide CO to lipid P proximities support similar partially inserted membrane locations of the alpha helical and beta sheet peptide Structures. J. Phys. Chem. A. 2013 doi: 10.1021/jp312845w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero S., Falanga A., Vitiello M., Browne H., Pedone C., Galdiero M. Fusogenic domains in herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 2005;280:28632–28643. doi: 10.1074/jbc.M505196200. [DOI] [PubMed] [Google Scholar]
- Galdiero S., Vitiello M., D’Isanto M., Falanga A., Cantisani M., Browne H., Pedone C., Galdiero M. The identification and characterization of fusogenic domains in herpes virus glycoprotein B molecules. Chembi: Eur. J. Chem. Biol. 2008;9:758–767. doi: 10.1002/cbic.200700457. [DOI] [PubMed] [Google Scholar]
- Gallaher W.R. Detection of a fusion peptide sequence in the transmembrane protein of human immunodeficiency virus. Cell. 1987;50:327–328. doi: 10.1016/0092-8674(87)90485-5. [DOI] [PubMed] [Google Scholar]
- Garry R.F., Dash S. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology. 2003;307:255–265. doi: 10.1016/s0042-6822(02)00065-x. [DOI] [PubMed] [Google Scholar]
- Ghosh J.K., Shai Y. Direct evidence that the N-terminal heptad repeat of Sendai virus fusion protein participates in membrane fusion. J. Mol. Biol. 1999;292:531–546. doi: 10.1006/jmbi.1999.3097. [DOI] [PubMed] [Google Scholar]
- Ghosh U., Xie L., Weliky D.P. Detection of closed influenza virus hemagglutinin fusion peptide structures in membranes by backbone (13)CO- (15)N rotational-echo double-resonance solid-state NMR. J. Biomol. NMR. 2013;55:139–146. doi: 10.1007/s10858-013-9709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons D.L., Vaney M.C., Roussel A., Vigouroux A., Reilly B., Lepault J., Kielian M., Rey F.A. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- Gomara M.J., Lorizate M., Huarte N., Mingarro I., Perez-Paya E., Nieva J.L. Hexapeptides that interfere with HIV-1 fusion peptide activity in liposomes block GP41-mediated membrane fusion. FEBS Lett. 2006;580:2561–2566. doi: 10.1016/j.febslet.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Gomara M.J., Mora P., Mingarro I., Nieva J.L. Roles of a conserved proline in the internal fusion peptide of Ebola glycoprotein. FEBS Lett. 2004;569:261–266. doi: 10.1016/j.febslet.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Gordon L.M., Mobley P.W., Lee W., Eskandari S., Kaznessis Y.N., Sherman M.A., Waring A.J. Conformational mapping of the N-terminal peptide of HIV-1 gp41 in lipid detergent and aqueous environments using 13C-enhanced Fourier transform infrared spectroscopy. Protein Sci. 2004;13:1012–1030. doi: 10.1110/ps.03407704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon L.M., Mobley P.W., Pilpa R., Sherman M.A., Waring A.J. Conformational mapping of the N-terminal peptide of HIV-1 gp41 in membrane environments using (13)C-enhanced Fourier transform infrared spectroscopy. Biochim. Biophys. Acta. 2002;1559:96–120. doi: 10.1016/s0005-2736(01)00443-6. [DOI] [PubMed] [Google Scholar]
- Grasnick D., Sternberg U., Strandberg E., Wadhwani P., Ulrich A.S. Irregular structure of the HIV fusion peptide in membranes demonstrated by solid-state NMR and MD simulations. Eur. Biophys. J. 2011;40:529–543. doi: 10.1007/s00249-011-0676-5. [DOI] [PubMed] [Google Scholar]
- Gray C., Tatulian S.A., Wharton S.A., Tamm L.K. Effect of the N-terminal glycine on the secondary structure, orientation, and interaction of the influenza hemagglutinin fusion peptide with lipid bilayers. Biophys. J. 1996;70:2275–2286. doi: 10.1016/S0006-3495(96)79793-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory S.M., Harada E., Liang B., Delos S.E., White J.M., Tamm L.K. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11211–11216. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen J., Perez-Berna A.J., Moreno M.R., Villalain J. Identification of the membrane-active regions of the severe acute respiratory syndrome coronavirus spike membrane glycoprotein using a 16/18-mer peptide scan: implications for the viral fusion mechanism. J. Virol. 2005;79:1743–1752. doi: 10.1128/JVI.79.3.1743-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Bushweller J.H., Cafiso D.S., Tamm L.K. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat. Struct. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- Haque M.E., Chakraborty H., Koklic T., Komatsu H., Axelsen P.H., Lentz B.R. Hemagglutinin fusion peptide mutants in model membranes: structural properties, membrane physical properties, and PEG-mediated fusion. Biophys. J. 2011;101:1095–1104. doi: 10.1016/j.bpj.2011.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque M.E., Koppaka V., Axelsen P.H., Lentz B.R. Properties and structures of the influenza and HIV fusion peptides on lipid membranes: implications for a role in fusion. Biophys. J. 2005;89:3183–3194. doi: 10.1529/biophysj.105.063032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harroun T.A., Balali-Mood K., Gourlay I., Bradshaw J.P. The fusion peptide of simian immunodeficiency virus and the phase behaviour of N-methylated dioleoylphosphatidylethanolamine. Biochim. Biophys. Acta. 2003;1617:62–68. doi: 10.1016/j.bbamem.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Harter C., James P., Bachi T., Semenza G., Brunner J. Hydrophobic binding of the ectodomain of influenza hemagglutinin to membranes occurs through the fusion peptide. J. Biol. Chem. 1989;264:6459–6464. [PubMed] [Google Scholar]
- Hernandez L.D., Hoffman L.R., Wolfsberg T.G., White J.M. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- Horvath C.M., Lamb R.A. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J. Virol. 1992;66:2443–2455. doi: 10.1128/jvi.66.4.2443-2455.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte N., Araujo A., Arranz R., Lorizate M., Quendler H., Kunert R., Valpuesta J.M., Nieva J.L. Recognition of membrane-bound fusion-peptide/MPER complexes by the HIV-1 neutralizing 2F5 antibody: implications for anti-2F5 immunogenicity. PLoS ONE. 2012;7:e52740. doi: 10.1371/journal.pone.0052740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte N., Lorizate M., Perez-Paya E., Nieva J.L. Membrane-transferring regions of gp41 as targets for HIV-1 fusion inhibition and viral neutralization. Curr. Top. Med. Chem. 2011;11:2985–2996. doi: 10.2174/156802611798808460. [DOI] [PubMed] [Google Scholar]
- Ivankin A., Apellaniz B., Gidalevitz D., Nieva J.L. Mechanism of membrane perturbation by the HIV-1 gp41 membrane-proximal external region and its modulation by cholesterol. Biochim. Biophys. Acta. 2012;1818:2521–2528. doi: 10.1016/j.bbamem.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroniec C.P., Kaufman J.D., Stahl S.J., Viard M., Blumenthal R., Wingfield P.T., Bax A. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- Julien J.P., Bryson S., Nieva J.L., Pai E.F. Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site. J. Mol. Biol. 2008;384:377–392. doi: 10.1016/j.jmb.2008.09.024. [DOI] [PubMed] [Google Scholar]
- Julien J.P., Cupo A., Sok D., Stanfield R.L., Lyumkis D., Deller M.C., Klasse P.J., Burton D.R., Sanders R.W., Moore J.P., Ward A.B., Wilson I.A. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson Hedestam G.B., Fouchier R.A., Phogat S., Burton D.R., Sodroski J., Wyatt R.T. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 2008;6:143–155. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- Kinnunen P.K., Holopainen J.M. Mechanisms of initiation of membrane fusion: role of lipids. Biosci. Rep. 2000;20:465–482. doi: 10.1023/a:1010402819509. [DOI] [PubMed] [Google Scholar]
- Klein D.E., Choi J.L., Harrison S.C. Structure of a dengue virus envelope protein late-stage fusion intermediate. J. Virol. 2013;87:2287–2293. doi: 10.1128/JVI.02957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliger Y., Aharoni A., Rapaport D., Jones P., Blumenthal R., Shai Y. Fusion peptides derived from the HIV type 1 glycoprotein 41 associate within phospholipid membranes and inhibit cell-cell Fusion. Structure–function study. J. Biol. Chem. 1997;272:13496–13505. doi: 10.1074/jbc.272.21.13496. [DOI] [PubMed] [Google Scholar]
- Kozlov M.M., McMahon H.T., Chernomordik L.V. Protein-driven membrane stresses in fusion and fission. Trends Biochem. Sci. 2010;35:699–706. doi: 10.1016/j.tibs.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lague P., Roux B., Pastor R.W. Molecular dynamics simulations of the influenza hemagglutinin fusion peptide in micelles and bilayers: conformational analysis of peptide and lipids. J. Mol. Biol. 2005;354:1129–1141. doi: 10.1016/j.jmb.2005.10.038. [DOI] [PubMed] [Google Scholar]
- Lai A.L., Moorthy A.E., Li Y., Tamm L.K. Fusion activity of HIV gp41 fusion domain is related to its secondary structure and depth of membrane insertion in a cholesterol-dependent fashion. J. Mol. Biol. 2012;418:3–15. doi: 10.1016/j.jmb.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A.L., Park H., White J.M., Tamm L.K. Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J. Biol. Chem. 2006;281:5760–5770. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- Lai A.L., Tamm L.K. Locking the kink in the influenza hemagglutinin fusion domain structure. J. Biol. Chem. 2007;282:23946–23956. doi: 10.1074/jbc.M704008200. [DOI] [PubMed] [Google Scholar]
- Larsson P., Kasson P.M. Lipid tail protrusion in simulations predicts fusogenic activity of influenza fusion peptide mutants and conformational models. PLoS Comp. Biol. 2013;9:e1002950. doi: 10.1371/journal.pcbi.1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau W.L., Ege D.S., Lear J.D., Hammer D.A., DeGrado W.F. Oligomerization of fusogenic peptides promotes membrane fusion by enhancing membrane destabilization. Biophys. J. 2004;86:272–284. doi: 10.1016/S0006-3495(04)74103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear J.D., DeGrado W.F. Membrane binding and conformational properties of peptides representing the NH2 terminus of influenza HA-2. J. Biol. Chem. 1987;262:6500–6505. [PubMed] [Google Scholar]
- Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legare S., Lague P. The influenza fusion peptide adopts a flexible flat V conformation in membranes. Biophys. J. 2012;102:2270–2278. doi: 10.1016/j.bpj.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz B.R., Malinin V., Haque M.E., Evans K. Protein machines and lipid assemblies: current views of cell membrane fusion. Curr. Opin. Struct. Biol. 2000;10:607–615. doi: 10.1016/s0959-440x(00)00138-x. [DOI] [PubMed] [Google Scholar]
- Li Y., Han X., Lai A.L., Bushweller J.H., Cafiso D.S., Tamm L.K. Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion. J. Virol. 2005;79:12065–12076. doi: 10.1128/JVI.79.18.12065-12076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.L., Tamm L.K. Structure and plasticity of the human immunodeficiency virus gp41 fusion domain in lipid micelles and bilayers. Biophys. J. 2007;93:876–885. doi: 10.1529/biophysj.106.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo M.L., Waring A.J., Gordon L.M., Hammer D.A. Area expansion and permeation of phospholipid membrane bilayers by influenza fusion peptides and melittin. Langmuir. 1998;14:2385–2395. [Google Scholar]
- Longo M.L., Waring A.J., Hammer D.A. Interaction of the influenza hemagglutinin fusion peptide with lipid bilayers: area expansion and permeation. Biophys. J. 1997;73:1430–1439. doi: 10.1016/S0006-3495(97)78175-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieau J.L., Louis J.M., Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieau J.L., Louis J.M., Bax A. Helical hairpin structure of influenza hemagglutinin fusion peptide stabilized by charge-dipole interactions between the N-terminal amino group and the second helix. J. Am. Chem. Soc. 2011;133:2824–2827. doi: 10.1021/ja1099775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieau J.L., Louis J.M., Schwieters C.D., Bax A. pH-triggered, activated-state conformations of the influenza hemagglutinin fusion peptide revealed by NMR. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19994–19999. doi: 10.1073/pnas.1213801109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorizate M., de la Arada I., Huarte N., Sanchez-Martinez S., de la Torre B.G., Andreu D., Arrondo J.L., Nieva J.L. Structural analysis and assembly of the HIV-1 Gp41 amino-terminal fusion peptide and the pretransmembrane amphipathic-at-interface sequence. Biochemistry. 2006;45:14337–14346. doi: 10.1021/bi0612521. [DOI] [PubMed] [Google Scholar]
- Lorizate M., Gomara M.J., de la Torre B.G., Andreu D., Nieva J.L. Membrane-transferring sequences of the HIV-1 Gp41 ectodomain assemble into an immunogenic complex. J. Mol. Biol. 2006;360:45–55. doi: 10.1016/j.jmb.2006.04.056. [DOI] [PubMed] [Google Scholar]
- Lorizate M., Huarte N., Saez-Cirion A., Nieva J.L. Interfacial pre-transmembrane domains in viral proteins promoting membrane fusion and fission. Biochim. Biophys. Acta. 2008;1778:1624–1639. doi: 10.1016/j.bbamem.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luneberg J., Martin I., Nussler F., Ruysschaert J.M., Herrmann A. Structure and topology of the influenza virus fusion peptide in lipid bilayers. J. Biol. Chem. 1995;270:27606–27614. doi: 10.1074/jbc.270.46.27606. [DOI] [PubMed] [Google Scholar]
- Lyumkis D., Julien J.P., de Val N., Cupo A., Potter C.S., Klasse P.J., Burton D.R., Sanders R.W., Moore J.P., Carragher B., Wilson I.A., Ward A.B. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko J.C., Kim C.H., Shin Y.K. The membrane topology of the fusion peptide region of influenza hemagglutinin determined by spin-labeling EPR. J. Mol. Biol. 1997;267:1139–1148. doi: 10.1006/jmbi.1997.0931. [DOI] [PubMed] [Google Scholar]
- Martin I., Defrise-Quertain F., Decroly E., Vandenbranden M., Brasseur R., Ruysschaert J.M. Orientation and structure of the NH2-terminal HIV-1 gp41 peptide in fused and aggregated liposomes. Biochim. Biophys. Acta. 1993;1145:124–133. doi: 10.1016/0005-2736(93)90389-h. [DOI] [PubMed] [Google Scholar]
- Martin I., Schaal H., Scheid A., Ruysschaert J.M. Lipid membrane fusion induced by the human immunodeficiency virus type 1 gp41 N-terminal extremity is determined by its orientation in the lipid bilayer. J. Virol. 1996;70:298–304. doi: 10.1128/jvi.70.1.298-304.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan G.B. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo M.N., Sousa F.J., Carneiro F.A., Castanho M.A., Valente A.P., Almeida F.C., Da Poian A.T., Mohana-Borges R. Interaction of the Dengue virus fusion peptide with membranes assessed by NMR: the essential role of the envelope protein Trp101 for membrane fusion. J. Mol. Biol. 2009;392:736–746. doi: 10.1016/j.jmb.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes Y.S., Alves N.S., Souza T.L., Sousa I.P., Jr., Bianconi M.L., Bernardi R.C., Pascutti P.G., Silva J.L., Gomes A.M., Oliveira A.C. The structural dynamics of the flavivirus fusion peptide-membrane interaction. PLoS ONE. 2012;7:e47596. doi: 10.1371/journal.pone.0047596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley P.W., Pilpa R., Brown C., Waring A.J., Gordon L.M. Membrane-perturbing domains of HIV type 1 glycoprotein 41. AIDS Res. Hum. Retroviruses. 2001;17:311–327. doi: 10.1089/08892220150503681. [DOI] [PubMed] [Google Scholar]
- Mobley P.W., Waring A.J., Sherman M.A., Gordon L.M. Membrane interactions of the synthetic N-terminal peptide of HIV-1 gp41 and its structural analogs. Biochim. Biophys. Acta. 1999;1418:1–18. doi: 10.1016/s0005-2736(99)00014-0. [DOI] [PubMed] [Google Scholar]
- Mohanram H., Nip A., Domadia P.N., Bhunia A., Bhattacharjya S. NMR structure, localization, and vesicle fusion of Chikungunya virus fusion peptide. Biochemistry. 2012;51:7863–7872. doi: 10.1021/bi300901f. [DOI] [PubMed] [Google Scholar]
- Morris K.F., Gao X., Wong T.C. The interactions of the HIV gp41 fusion peptides with zwitterionic membrane mimics determined by NMR spectroscopy. Biochim. Biophys. Acta. 2004;1667:67–81. doi: 10.1016/j.bbamem.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Munch J., Standker L., Adermann K., Schulz A., Schindler M., Chinnadurai R., Pohlmann S., Chaipan C., Biet T., Peters T., Meyer B., Wilhelm D., Lu H., Jing W., Jiang S., Forssmann W.G., Kirchhoff F. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell. 2007;129:263–275. doi: 10.1016/j.cell.2007.02.042. [DOI] [PubMed] [Google Scholar]
- Munoz-Barroso I., Salzwedel K., Hunter E., Blumenthal R. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 1999;73:6089–6092. doi: 10.1128/jvi.73.7.6089-6092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemesio H., Palomares-Jerez F., Villalain J. The membrane-active regions of the dengue virus proteins C and E. Biochim. Biophys. Acta. 2011;1808:2390–2402. doi: 10.1016/j.bbamem.2011.06.019. [DOI] [PubMed] [Google Scholar]
- Nieva J.L., Agirre A. Are fusion peptides a good model to study viral cell fusion? Biochim. Biophys. Acta. 2003;1614:104–115. doi: 10.1016/s0005-2736(03)00168-8. [DOI] [PubMed] [Google Scholar]
- Nieva J.L., Nir S., Muga A., Goni F.M., Wilschut J. Interaction of the HIV-1 fusion peptide with phospholipid vesicles: different structural requirements for fusion and leakage. Biochemistry. 1994;33:3201–3209. doi: 10.1021/bi00177a009. [DOI] [PubMed] [Google Scholar]
- Nieva J.L., Suarez T. Hydrophobic-at-interface regions in viral fusion protein ectodomains. Biosci. Rep. 2000;20:519–533. doi: 10.1023/a:1010458904487. [DOI] [PubMed] [Google Scholar]
- Owens R.J., Tanner C.C., Mulligan M.J., Srinivas R.V., Compans R.W. Oligopeptide inhibitors of HIV-induced syncytium formation. AIDS Res. Hum. Retroviruses. 1990;6:1289–1296. doi: 10.1089/aid.1990.6.1289. [DOI] [PubMed] [Google Scholar]
- Pan J., Lai C.B., Scott W.R., Straus S.K. Synthetic fusion peptides of tick-borne encephalitis virus as models for membrane fusion. Biochemistry. 2010;49:287–296. doi: 10.1021/bi9017895. [DOI] [PubMed] [Google Scholar]
- Peisajovich S.G., Epand R.F., Epand R.M., Shai Y. Sendai virus N-terminal fusion peptide consists of two similar repeats, both of which contribute to membrane fusion. Eur. J. Biochem./FEBS. 2002;269:4342–4350. doi: 10.1046/j.1432-1033.2002.03132.x. [DOI] [PubMed] [Google Scholar]
- Peisajovich S.G., Samuel O., Shai Y. Paramyxovirus F1 protein has two fusion peptides: implications for the mechanism of membrane fusion. J. Mol. Biol. 2000;296:1353–1365. doi: 10.1006/jmbi.2000.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisajovich S.G., Shai Y. Viral fusion proteins: multiple regions contribute to membrane fusion. Biochim. Biophys. Acta. 2003;1614:122–129. doi: 10.1016/s0005-2736(03)00170-6. [DOI] [PubMed] [Google Scholar]
- Pereira F.B., Goni F.M., Muga A., Nieva J.L. Permeabilization and fusion of uncharged lipid vesicles induced by the HIV-1 fusion peptide adopting an extended conformation: dose and sequence effects. Biophys. J. 1997;73:1977–1986. doi: 10.1016/S0006-3495(97)78228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F.B., Goni F.M., Nieva J.L. Liposome destabilization induced by the HIV-1 fusion peptide effect of a single amino acid substitution. FEBS Lett. 1995;362:243–246. doi: 10.1016/0014-5793(95)00257-a. [DOI] [PubMed] [Google Scholar]
- Pereira F.B., Valpuesta J.M., Basanez G., Goni F.M., Nieva J.L. Interbilayer lipid mixing induced by the human immunodeficiency virus type-1 fusion peptide on large unilamellar vesicles: the nature of the nonlamellar intermediates. Chem. Phys. Lipids. 1999;103:11–20. doi: 10.1016/s0009-3084(99)00087-0. [DOI] [PubMed] [Google Scholar]
- Perez-Berna A.J., Moreno M.R., Guillen J., Bernabeu A., Villalain J. The membrane-active regions of the hepatitis C virus E1 and E2 envelope glycoproteins. Biochemistry. 2006;45:3755–3768. doi: 10.1021/bi0523963. [DOI] [PubMed] [Google Scholar]
- Pritsker M., Jones P., Blumenthal R., Shai Y. A synthetic all D-amino acid peptide corresponding to the N-terminal sequence of HIV-1 gp41 recognizes the wild-type fusion peptide in the membrane and inhibits HIV-1 envelope glycoprotein-mediated cell fusion. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7287–7292. doi: 10.1073/pnas.95.13.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritsker M., Rucker J., Hoffman T.L., Doms R.W., Shai Y. Effect of nonpolar substitutions of the conserved Phe11 in the fusion peptide of HIV-1 gp41 on its function, structure, and organization in membranes. Biochemistry. 1999;38:11359–11371. doi: 10.1021/bi990232e. [DOI] [PubMed] [Google Scholar]
- Promsri S., Ullmann G.M., Hannongbua S. Molecular dynamics simulation of HIV-1 fusion domain-membrane complexes: Insight into the N-terminal gp41 fusion mechanism. Biophys. Chem. 2012;170:9–16. doi: 10.1016/j.bpc.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Qiang W., Bodner M.L., Weliky D.P. Solid-state NMR spectroscopy of human immunodeficiency virus fusion peptides associated with host-cell-like membranes: 2D correlation spectra and distance measurements support a fully extended conformation and models for specific antiparallel strand registries. J. Am. Chem. Soc. 2008;130:5459–5471. doi: 10.1021/ja077302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang W., Sun Y., Weliky D.P. A strong correlation between fusogenicity and membrane insertion depth of the HIV fusion peptide. Proc. Natl. Acad. Sci. U.S.A. 2009;106:15314–15319. doi: 10.1073/pnas.0907360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana F.J., Gerber D., Kent S.C., Cohen I.R., Shai Y. HIV-1 fusion peptide targets the TCR and inhibits antigen-specific T cell activation. J. Clin. Invest. 2005;115:2149–2158. doi: 10.1172/JCI23956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski M., Lear J.D., DeGrado W.F. Phospholipid interactions of synthetic peptides representing the N-terminus of HIV gp41. Biochemistry. 1990;29:7917–7922. doi: 10.1021/bi00486a020. [DOI] [PubMed] [Google Scholar]
- Rafalski M., Ortiz A., Rockwell A., van Ginkel L.C., Lear J.D., DeGrado W.F., Wilschut J. Membrane fusion activity of the influenza virus hemagglutinin: interaction of HA2 N-terminal peptides with phospholipid vesicles. Biochemistry. 1991;30:10211–10220. doi: 10.1021/bi00106a020. [DOI] [PubMed] [Google Scholar]
- Reuven E.M., Dadon Y., Viard M., Manukovsky N., Blumenthal R., Shai Y. HIV-1 gp41 transmembrane domain interacts with the fusion peptide: implication in lipid mixing and inhibition of virus-cell fusion. Biochemistry. 2012;51:2867–2878. doi: 10.1021/bi201721r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S., Bressanelli S., Rey F.A., Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- Roche S., Rey F.A., Gaudin Y., Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- Roux K.H., Taylor K.A. AIDS virus envelope spike structure. Curr. Opin. Struct. Biol. 2007;17:244–252. doi: 10.1016/j.sbi.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Ruiz-Arguello M.B., Goni F.M., Pereira F.B., Nieva J.L. Phosphatidylinositol-dependent membrane fusion induced by a putative fusogenic sequence of Ebola virus. J. Virol. 1998;72:1775–1781. doi: 10.1128/jvi.72.3.1775-1781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackett K., Nethercott M.J., Epand R.F., Epand R.M., Kindra D.R., Shai Y., Weliky D.P. Comparative analysis of membrane-associated fusion peptide secondary structure and lipid mixing function of HIV gp41 constructs that model the early pre-hairpin intermediate and final hairpin conformations. J. Mol. Biol. 2010;397:301–315. doi: 10.1016/j.jmb.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackett K., Nethercott M.J., Shai Y., Weliky D.P. Hairpin folding of HIV gp41 abrogates lipid mixing function at physiologic pH and inhibits lipid mixing by exposed gp41 constructs. Biochemistry. 2009;48:2714–2722. doi: 10.1021/bi8019492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackett K., Shai Y. The HIV-1 gp41 N-terminal heptad repeat plays an essential role in membrane fusion. Biochemistry. 2002;41:4678–4685. doi: 10.1021/bi0255322. [DOI] [PubMed] [Google Scholar]
- Sackett K., Shai Y. How structure correlates to function for membrane associated HIV-1 gp41 constructs corresponding to the N-terminal half of the ectodomain. J. Mol. Biol. 2003;333:47–58. doi: 10.1016/j.jmb.2003.07.008. [DOI] [PubMed] [Google Scholar]
- Sackett K., Shai Y. The HIV fusion peptide adopts intermolecular parallel beta-sheet structure in membranes when stabilized by the adjacent N-terminal heptad repeat: a 13C FTIR study. J. Mol. Biol. 2005;350:790–805. doi: 10.1016/j.jmb.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Sackett K., TerBush A., Weliky D.P. HIV gp41 six-helix bundle constructs induce rapid vesicle fusion at pH 3.5 and little fusion at pH 7.0: understanding pH dependence of protein aggregation, membrane binding, and electrostatics, and implications for HIV-host cell fusion. Eur. Biophys. J. 2011;40:489–502. doi: 10.1007/s00249-010-0662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Cirion A., Arrondo J.L., Gomara M.J., Lorizate M., Iloro I., Melikyan G., Nieva J.L. Structural and functional roles of HIV-1 gp41 pretransmembrane sequence segmentation. Biophys. J. 2003;85:3769–3780. doi: 10.1016/S0006-3495(03)74792-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Cirion A., Nieva J.L. Conformational transitions of membrane-bound HIV-1 fusion peptide. Biochim. Biophys. Acta. 2002;1564:57–65. doi: 10.1016/s0005-2736(02)00400-5. [DOI] [PubMed] [Google Scholar]
- Saez-Cirion A., Nir S., Lorizate M., Agirre A., Cruz A., Perez-Gil J., Nieva J.L. Sphingomyelin and cholesterol promote HIV-1 gp41 pretransmembrane sequence surface aggregation and membrane restructuring. J. Biol. Chem. 2002;277:21776–21785. doi: 10.1074/jbc.M202255200. [DOI] [PubMed] [Google Scholar]
- Salzwedel K., West J.T., Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel O., Shai Y. Participation of two fusion peptides in measles virus-induced membrane fusion: emerging similarity with other paramyxoviruses. Biochemistry. 2001;40:1340–1349. doi: 10.1021/bi001533n. [DOI] [PubMed] [Google Scholar]
- Shnaper S., Sackett K., Gallo S.A., Blumenthal R., Shai Y. The C- and the N-terminal regions of glycoprotein 41 ectodomain fuse membranes enriched and not enriched with cholesterol, respectively. J. Biol. Chem. 2004;279:18526–18534. doi: 10.1074/jbc.M304950200. [DOI] [PubMed] [Google Scholar]
- Siegel D.P., Epand R.M. The mechanism of lamellar-to-inverted hexagonal phase transitions in phosphatidylethanolamine: implications for membrane fusion mechanisms. Biophys. J. 1997;73:3089–3111. doi: 10.1016/S0006-3495(97)78336-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D.P., Epand R.M. Effect of influenza hemagglutinin fusion peptide on lamellar/inverted phase transitions in dipalmitoleoylphosphatidylethanolamine: implications for membrane fusion mechanisms. Biochim. Biophys. Acta. 2000;1468:87–98. doi: 10.1016/s0005-2736(00)00246-7. [DOI] [PubMed] [Google Scholar]
- Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- Slepushkin V.A., Kornilaeva G.V., Andreev S.M., Sidorova M.V., Petrukhina A.O., Matsevich G.R., Raduk S.V., Grigoriev V.B., Makarova T.V., Lukashov V.V. Inhibition of human immunodeficiency virus type 1 (HIV-1) penetration into target cells by synthetic peptides mimicking the N-terminus of the HIV-1 transmembrane glycoprotein. Virology. 1993;194:294–301. doi: 10.1006/viro.1993.1260. [DOI] [PubMed] [Google Scholar]
- Smith E.C., Gregory S.M., Tamm L.K., Creamer T.P., Dutch R.E. Role of sequence and structure of the Hendra fusion protein fusion peptide in membrane fusion. J. Biol. Chem. 2012;287:30035–30048. doi: 10.1074/jbc.M112.367862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez T., Gallaher W.R., Agirre A., Goni F.M., Nieva J.L. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 2000;74:8038–8047. doi: 10.1128/jvi.74.17.8038-8047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez T., Gomara M.J., Goni F.M., Mingarro I., Muga A., Perez-Paya E., Nieva J.L. Calcium-dependent conformational changes of membrane-bound Ebola fusion peptide drive vesicle fusion. FEBS Lett. 2003;535:23–28. doi: 10.1016/s0014-5793(02)03847-4. [DOI] [PubMed] [Google Scholar]
- Suarez T., Nir S., Goni F.M., Saez-Cirion A., Nieva J.L. The pre-transmembrane region of the human immunodeficiency virus type-1 glycoprotein: a novel fusogenic sequence. FEBS Lett. 2000;477:145–149. doi: 10.1016/s0014-5793(00)01785-3. [DOI] [PubMed] [Google Scholar]
- Sun Y., Weliky D.P. 13C-13C correlation spectroscopy of membrane-associated influenza virus fusion peptide strongly supports a helix-turn-helix motif and two turn conformations. J. Am. Chem. Soc. 2009;131:13228–13229. doi: 10.1021/ja905198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.Y., Oh K.J., Kim M., Yu J., Brusic V., Song L., Qiao Z., Wang J.H., Wagner G., Reinherz E.L. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Tamm L.K. Hypothesis: spring-loaded boomerang mechanism of influenza hemagglutinin-mediated membrane fusion. Biochim. Biophys. Acta. 2003;1614:14–23. doi: 10.1016/s0005-2736(03)00159-7. [DOI] [PubMed] [Google Scholar]
- Tamm L.K., Han X. Viral fusion peptides: a tool set to disrupt and connect biological membranes. Biosci. Rep. 2000;20:501–518. doi: 10.1023/a:1010406920417. [DOI] [PubMed] [Google Scholar]
- Tamm L.K., Han X., Li Y., Lai A.L. Structure and function of membrane fusion peptides. Biopolymers. 2002;66:249–260. doi: 10.1002/bip.10261. [DOI] [PubMed] [Google Scholar]
- Tamm L.K., Lai A.L., Li Y. Combined NMR and EPR spectroscopy to determine structures of viral fusion domains in membranes. Biochim. Biophys. Acta. 2007;1768:3052–3060. doi: 10.1016/j.bbamem.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenchov B.G., MacDonald R.C., Lentz B.R. Fusion peptides promote formation of bilayer cubic phases in lipid dispersions. An x-ray diffraction study. Biophys. J. 2013;104:1029–1037. doi: 10.1016/j.bpj.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristram-Nagle S., Chan R., Kooijman E., Uppamoochikkal P., Qiang W., Weliky D.P., Nagle J.F. HIV fusion peptide penetrates, disorders, and softens T-cell membrane mimics. J. Mol. Biol. 2010;402:139–153. doi: 10.1016/j.jmb.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristram-Nagle S., Nagle J.F. HIV-1 fusion peptide decreases bending energy and promotes curved fusion intermediates. Biophys. J. 2007;93:2048–2055. doi: 10.1529/biophysj.107.109181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurudome M., Gluck R., Graf R., Falchetto R., Schaller U., Brunner J. Lipid interactions of the hemagglutinin HA2 NH2-terminal segment during influenza virus-induced membrane fusion. J. Biol. Chem. 1992;267:20225–20232. [PubMed] [Google Scholar]
- Vaney M.C., Rey F.A. Class II enveloped viruses. Cell. Microbiol. 2011;13:1451–1459. doi: 10.1111/j.1462-5822.2011.01653.x. [DOI] [PubMed] [Google Scholar]
- Vishwanathan S.A., Hunter E. Importance of the membrane-perturbing properties of the membrane-proximal external region of human immunodeficiency virus type 1 gp41 to viral fusion. J. Virol. 2008;82:5118–5126. doi: 10.1128/JVI.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Paesold G., Galli C., Mischler R., Semenza G., Brunner J. Evidence for H(+)-induced insertion of influenza hemagglutinin HA2 N-terminal segment into viral membrane. J. Biol. Chem. 1994;269:18353–18358. [PubMed] [Google Scholar]
- Weissenhorn W., Dessen A., Harrison S.C., Skehel J.J., Wiley D.C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W., Hinz A., Gaudin Y. Virus membrane fusion. FEBS Lett. 2007;581:2150–2155. doi: 10.1016/j.febslet.2007.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler-Cohen Y., Johnson B.T., Puri A., Blumenthal R., Shai Y. Structurally altered peptides reveal an important role for N-terminal heptad repeat binding and stability in the inhibitory action of HIV-1 peptide DP178. J. Biol. Chem. 2006;281:9005–9010. doi: 10.1074/jbc.M512475200. [DOI] [PubMed] [Google Scholar]
- Wexler-Cohen Y., Sackett K., Shai Y. The role of the N-terminal heptad repeat of HIV-1 in the actual lipid mixing step as revealed by its substitution with distant coiled coils. Biochemistry. 2005;44:5853–5861. doi: 10.1021/bi047666g. [DOI] [PubMed] [Google Scholar]
- White J.M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White S.H., Wimley W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- Wild C.T., Shugars D.C., Greenwell T.K., McDanal C.B., Matthews T.J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D.C., Skehel J.J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- Yang J., Gabrys C.M., Weliky D.P. Solid-state nuclear magnetic resonance evidence for an extended beta strand conformation of the membrane-bound HIV-1 fusion peptide. Biochemistry. 2001;40:8126–8137. doi: 10.1021/bi0100283. [DOI] [PubMed] [Google Scholar]
- Yang J., Prorok M., Castellino F.J., Weliky D.P. Oligomeric beta-structure of the membrane-bound HIV-1 fusion peptide formed from soluble monomers. Biophys. J. 2004;87:1951–1963. doi: 10.1529/biophysj.103.028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Prorok M., Castellino F.J., Weliky D.P. A trimeric HIV-1 fusion peptide construct which does not self-associate in aqueous solution and which has 15-fold higher membrane fusion rate. J. Am. Chem. Soc. 2004;126:14722–14723. doi: 10.1021/ja045612o. [DOI] [PubMed] [Google Scholar]
- Yang R., Yang J., Weliky D.P. Synthesis, enhanced fusogenicity, and solid state NMR measurements of cross-linked HIV-1 fusion peptides. Biochemistry. 2003;42:3527–3535. doi: 10.1021/bi027052g. [DOI] [PubMed] [Google Scholar]
- Yao H., Hong M. Membrane-dependent conformation, dynamics, and lipid interactions of the fusion peptide of the paramyxovirus PIV5 from solid-state NMR. J. Mol. Biol. 2013;425:563–576. doi: 10.1016/j.jmb.2012.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H.S., Wen X., Paterson R.G., Lamb R.A., Jardetzky T.S. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J., Vogel S.S., Chernomordik L.V. Mechanisms of membrane fusion. Annu. Rev. Biophys. Biomol. Struct. 1993;22:433–466. doi: 10.1146/annurev.bb.22.060193.002245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


