Abstract
Antibiotic-resistant bacterial pathogens threaten public health. Because many anti-biotics target specific bacterial enzymes or reactions, corresponding genes may mutate under selection and lead to antibiotic resistance. Accordingly, antimicrobials that selectively target overall microbial cell integrity may offer alternative approaches to therapeutic design. Naturally occurring mammalian α- and θ-defensins are potent, non-toxic microbicides that may be useful for treating infections by antibiotic-resistant pathogens, because certain defensin peptides disrupt bacterial but not mammalian cell membranes. To test this concept, clinical isolates of methicillin-resistant Staphylococcus aureus (MRSA), including vancomycin heteroresistant strains, and ciprofloxacin-resistant Pseudomonas aeruginosa (CipR-PA) were tested for sensitivity to α-defensins Crp-4, RMAD-4, and HNPs 1–3, and to RTD-1, a macaque θ-defensin-1. In vitro, 3 µM Crp-4, RMAD-4, and RTD-1 reduced MRSA cell survival by 99%, regardless of vancomycin susceptibility. For PA clinical isolates that differ in fluoroquinolone resistance and virulence phenotype, peptide efficacy was independent of strain ciprofloxacin resistance, site of isolation, or virulence factor expression. Thus, Crp-4, RMAD-4, and RTD-1 are effective in vitro antimicrobials against clinical isolates of MRSA and CipR-PA, perhaps providing templates for development of α- and θ-defensin-based microbicides against antibiotic resistant or virulent infectious agents.
Keywords: Antimicrobial peptides, ExoU, ExoS, clinical isolates, peptide therapeutics, bacteremia
INTRODUCTION
The economic costs of antimicrobial resistant infections has reached nearly $30 billion dollars per year in the US1. In 2002, of health care–associated infections that resulted in 99,000 deaths, approximately 16% were reported to the CDC as resistant to antibiotics2–5. The increase in antibiotic resistance narrows the options for the treatment of infections caused by multidrug-resistant bacteria5, 6.
The Gram-negative bacterium Pseudomonas aeruginosa (PA) is a leading cause of nosocomial infections for which the fluoroquinolone antibiotics, e.g. ciprofloxacin, are commonly prescribed. Over the past decade, the prevalence of ciprofloxacin-resistant (CipR) PA strains has increased 3-fold in parallel to the trend of prescribing this antibiotic class7. Upwards of 30% of clinical PA strains are multidrug-resistant, and some are resistant to all available antibiotics5, 8–10. In addition, PA strains have an arsenal of virulence factors, including a type III secretion system (TTSS) that induces cytotoxicity and expression of ExoU or ExoS effector proteins which are virulence factors that influence disease severity by phagocyte evasion during acute infections11.
Methicillin-resistant Staphylococcus aureus (MRSA) are Gram-positive bacteria with resistance to all β-lactam compounds except one, accounting for nearly 60% of all clinical isolates from ICU patients12. Limited to the healthcare setting in the past, new, more virulent MRSA strains have emerged in the community, and they are now responsible for infections across both the community and healthcare settings. In addition, MRSA strains increasingly have developed varying degrees of resistance to vancomycin, the accepted treatment standard13, and hospital-associated health care costs for patients with MRSA-related infections are nearly double those of patients with methicillin-sensitive S. aureus14.
Multi-drug resistant PA and MRSA are treated with antibiotics that target specific bacterial reactions or enzymes to inhibit cell replication, cell wall biosynthesis, or they may kill bacteria directly. Antibiotic exposure enables bacteria to acquire resistance through mutations that allow for target drug degradation, reduced drug affinity to target sites, altered metabolic pathways, and/or reduced drug accumulation15–17. For example, ciprofloxacin inhibits bacterial replication by inhibiting DNA gyrase and topoisomerase IV, thereby blocking bacterial cell division18. CipR PA acquire mutations to DNA gyrase and or topoisomerase to escape its lethal antibiotic effects and by active removal of ciprofloxacin via broad substrate efflux pumps to expel the drug and prevent its accumulation within cells17. On the other hand, vancomycin inhibits bacterial cell wall biosynthesis by binding to the C-terminal D-Ala-D-Ala of pentapeptide peptidoglycan precursors to block transpeptidation19. Heteroresistance to vancomycin in MRSA reportedly results from repeated vancomycin exposure selecting for a thickened cell wall that blocks vancomycin from its target site20, and this resistant phenotype has accounted in part for persistence of bacteremia and increased mortality21, 22. One approach to facilitate the treatment of anti-biotic-resistant infections may be to combine current therapies with broad-spectrum peptide microbicides that kill bacteria by general, independent mechanisms such as membrane disruption.
Mammalian defensins are 2–5 kDa, broad spectrum, cationic antimicrobial peptides, with structures that are defined by specific tridisulfide arrays23. The α-, β-, and θ- defensins compose the mammalian defensin family, and each subfamily differs with respect to structural features that are imposed by specific disulfide connectivities, and they have distinct primary sites of expression24. For example, the α-defensin tridisulfide array is characterized by conserved CI–VI, CII–IV, CIII–V cysteine pairings, and β-defensins are characterized by a CI–V, CII–IV, CIII–VI disulfide connectivity25. α-Defensin genes are expressed by promyelocytes mainly accumulate in neutrophil azurophil granules, and those expressed by Paneth cells are secreted into the small intestinal lumen. Certain β-defensins, on the other hand, are expressed widely by epithelia at diverse mucosal barrier interfaces26, 27. θ-Defensins are the only macrocyclic peptides known in the animal kingdom and are expressed in the bone marrow of only Old World monkeys. The peptides derive from truncated α-defensin genes, assemble from two hemi-precursors, and contain a tridisulfide array that is arranged in the form of a parallel ladder28.
The mechanisms of defensin microbicidal activity have been investigated extensively29. Based on in vitro studies, micromolar levels of α-defensins may disrupt microbial membranes selectively by inducing either stable or transient defects of variable size in model membranes composed of microbial phospholipids30, 31. Induction of membrane defects leads to target cell permeabilization, K+ efflux, depolarization, dissipation of electrochemical gradients, leakage, and eventual cell death32–35. Analyses of defensin-bilayer in5 teractions by small angle X-ray scatter, showed that mouse Paneth cell α-defensin cryptdin-4 (Crp-4), rhesus myeloid α-defensin RMAD-4, and the rhesus θ-defensin RTD-1 induce negative Gaussian, or saddle splay, curvature to create pores in model membranes and facilitating membrane disruption36. On the other hand, at lower peptide concentrations defensins can also inhibit bacterial peptidoglycan synthesis by lipid II binding37, 38, and defensins may use a lipid II binding mechanism to exert antimicrobial effects.
Because α-, and θ-defensins kill bacteria by these general mechanisms, which differ from those of ciprofloxacin and vancomycin, we reasoned that vancomycin-heteroresistant MRSA and CipR PA may be susceptible to the microbicidal effects of exposure to these defensin peptides. To test this hypothesis, the survival of clinical isolates of vancomycinheteroresistant MRSA and CipR PA exposed to Crp4, RMAD-4, RTD-1, and human neutrophil α-defensins HNPs 1–3 were determined. Under the conditions of the in vitro bactericidal assays, nearly all MRSA and PA strains were sensitive to all peptides except for HNPs, irrespective of antibiotic resistance. In addition, PA sensitivity to α-defensins was not related to site of isolation, degree of ciprofloxacin resistance, TTSS effector genotype, or cytotoxic potential. Because Crp-4, RMAD-4, and RTD-1 are non-hemolytic, resistant to proteolytic degradation, and among the most potent known defensins, they may offer promise for development of novel antimicrobial therapeutics.
MATERIALS AND METHODS
Peptide Preparation
Peptides (Figure 1) were purified to homogeneity by reverse phase high performance liquid chromatography (RP-HPLC), and their identities were confirmed by MALDI-TOF MS and by acid-urea polyacrylamide gel electrophoresis (AU-PAGE) as described39, 40. Recombinant Crp-4 and RMAD-4 peptides were expressed in Escherichia coli as N-terminal His6-tagged fusion proteins using the pET28a expression system (Novagen, Inc. Madison, WI)32, 41, 42. Crp-4 and RMAD-4 templates were cloned in pCR-2.1 TOPO, verified by DNA sequencing, subcloned into pET28a plasmid DNA (Novagen, Inc., Madison, WI), and transformed into E. coli BL21(DE3)-CodonPlus-RIL cells (Stratagene) for recombinant expression32, 41. His6-tagged Crp-4 fusion peptides were purified using nickel-nitrilotriacetic acid (Ni-NTA, Qiagen) resin affinity chromatography43. After CNBr cleavage of the His6 tag, peptides were purified by sequential C18 reverse-phase high performance liquid chromatography (RP-HPLC), and molecular masses of purified peptides were determined using MALDI-TOF MS on a Bruker Microflex LRF (Bruker, Fremont, CA). Solution structures of recombinant peptides prepared by this approach have been determined by NMR44, 45.
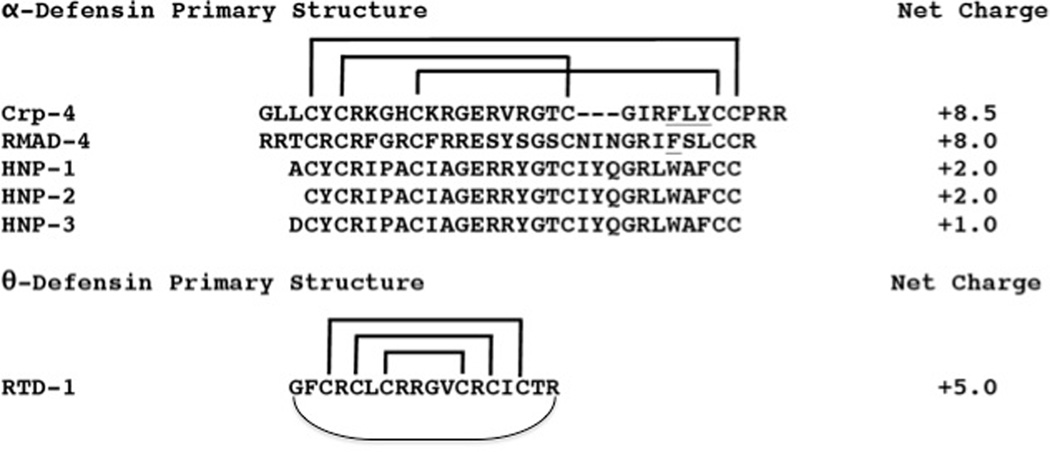
Figure 1. Primary structures of α- and θ-defensins.
The primary structures of peptides investigated are aligned; net overall charge is shown at right. α-Defensins are aligned by their conserved Cys residues (in bold), and their CI-CVI, CII-CIV, and CIII-CV disulfide connectivities are depicted by brackets. The α-defensins from mouse Paneth cells and rhesus neutrophils are highly cationic as compared to the human myeloid peptides, HNPs 1–3. The octadecapeptide RTD-1, a θ-defensin, is a macrocyclic peptide identified in rhesus macaque neutrophils and found only in Old World monkeys.
Human neutrophil peptides (HNPs) were isolated from samples enriched in neutrophils granules prepared from peripheral blood leukocytes46, 47. Briefly, after removing platelets from human blood by centrifugation at 200 × g for 10 min and hypotonic lysis of red blood cells, the leukocyte-enriched cell fraction was resuspended in 0.34 M sucrose, pH 7.4, homogenized, and cellular debris deposited by centrifugation at 200 × g for 10 min, leaving a granule-rich supernatant. Granules deposited by 30 min centrifugation at 27,000 × g at 4°C were extracted for 18 h at 4°C with 10% acetic acid, and protein extracts were clarified by centrifugation at 27,000 × g for 30 min at 4°C. A mixture of natural HNPs 1–3 was purified by gel permeation chromatography using BioGel P10 (Bio-Rad Laboratories, Hercules, CA). HNP-2 was purified further from pooled HNPs 1–3 by C18 RP-HPLC.
RTD-1 was synthesized as previously reported48. Synthetic RTD-1 is structurally and biologically indistinguishable from peptide isolated from rhesus monkey neutrophils28.
Clinical Isolates
Bacterial isolates were grown from culture specimens obtained from hospitalized patients at Huntington Hospital, Pasadena, CA as part of a longitudinal epidemiologic surveillance study of resistant pathogens at the institution and were stored at −80°C until testing. All data were analyzed anonymously. Susceptibility of MRSA isolates to vancomycin was determined by Etest-based method according to manufacturer’s instructions (bio- Meriéux, Durham, NC). Specifically, vancomycin heteroresistant phenotype was determined using the Etest Glycopeptide Resistance Detection (GRD) method49. PA susceptibility to ciprofloxacin was performed by broth microdilution method as recommended by the Clinical Laboratory Standards Institute, with ciprofloxacin resistance defined by mini7 mum inhibitory concentration of 2 µg/ml or greater. As for PA strains, genes encoding the TTSS effector proteins ExoU and ExoS were assayed by polymerase chain reaction as before50. In vitro experiments were performed to determine PA cytotoxicity by infecting A549 lung epithelial cells at a multiplicity of infection (MOI) of 10 and measuring LDH release at 3 h post-infection using the CytoTox96 assay kit (Promega, Madison, WI).
MRSA bloodstream isolates and PA strains that caused pneumonia, as well as bacteremia, wound and urinary tract infections were selected for this study to represent clinical isolates that exhibit varying degrees of resistance to vancomycin (MRSA) and ciprofloxacin (PA), respectively. The MRSA cohort included strains that caused persistent blood-stream infection and different molecular epidemiologic characteristics; PA strains included those with different virulence potential based on TTSS effector genotype and the rate and extent of cytotoxicity observed in A549 cells. Two additional reference MRSA strains with heteroresistance (Mu3) and intermediate resistance (Mu50) to vancomycin also were studied.
In vitro Bactericidal Assays
The α- and θ-defensins were tested for bactericidal activity against clinical isolates of MRSA and PA in in vitro cell suspension assays32, 41. Bacteria grown to midexponential phase in trypticase soy broth were deposited by centrifugation at 10,000 × g for 3 min, and washed 3 times with 10 mM piperazine-1,4-bis(2-ethanesulfonic acid) (PIPES), pH 7.4, supplemented with 1% (vol/vol) of trypticase soy broth (10 mM PIPES-TSB, pH 7.4). In triplicate, 1–5 × 106 bacterial colony forming units (CFU)/ml were exposed to peptides in 50 µl 10 mM PIPES-TSB in 96-well polystyrene plates. Samples were incubated at 37°C with shaking for 1 h, diluted 1:100 in 10 mM PIPES (pH 7.4), and plated on TSB agar plates using an Autoplate 4000 (Spiral Biotech Inc., Bethesda, MD). Bacterial cell survival as a function of peptide exposure was determined by counting CFU after overnight growth at 37°C. In these assays, after 1 h of peptide exposure replicate peptide-bacterial mixtures are plated with a plating stylus onto the agar plate surface. The assay enables bacteria that are not exposed or affected by peptide exposure to be enumerated, rather than produce lawns that cannot be counted. On the other hand, sample dilution limits the lower end of the assay, i.e., when >99.9% of exposed bacteria die. Because of the dilution factors involved, plates on which no CFU were detected after overnight incubation actually may have had between 1 to 999 viable CFU in the peptide-bacterial mixture before dilution and plating. Accordingly, the limit of detection is ≤ 103 CFU/ml, when no CFU are detected, and the ordinates of bacterial survival curves are labeled in that manner.
RESULTS
Defensin sensitivity of vancomycin-heteroresistant MRSA strains
MRSA clinical isolates with varied resistance to vancomycin were exposed to Crp-4, RMAD-4, and RTD-1 to determine their sensitivities to the peptides. Against standard laboratory strains of S. aureus, Crp-4, RMAD-4, and RTD-1 are potent microbicides, reducing cell viability 1000-fold at low micromolar peptide concentrations51, 52. Here, Crp-4, RMAD-4, and RTD-1 similarly reduced viability of vancomycin-intermediate S. aureus (VISA) control strain Mu50 and heteroresistant VISA (hVISA) control strain Mu3 with MBC values between 1.5 and 3 µM peptide (Table 1, Figs. 2A and B). Clinical isolates characterized as hVISA by the Etest GRD method (Materials and Methods) were highly sensitive to all defensins tested (Figs. 2C and D), with RMAD-4 had the greatest bactericidal activity against every VISA/hVISA strain tested with 1.5 µM MBC values for all strains (Table 1, Figs. 2A–D).
Table 1.
Defensin MBC Values against VISA and hVISA MRSA Strains
| MBC Value (µM) | |||
|---|---|---|---|
| MRSA Strain | Crp-4 | RMAD-4 | RTD-1 |
| Mu50 (VISA) | 3 | 1.5 | 3 |
| Mu3 (hVISA) | 1.5 | 3 | 1.5 |
| DH 290 (hVISA) | 3 | 1.5 | 3 |
| DH280 (hVISA) | 3 | 1.5 | 1.5 |
| EB151§ | 1.5 | 1.5 | 1.5 |
| EB123§ | 3 | 1.5 | 3 |
| EB135§ | 3 | 1.5 | 3 |
| JJ593‡ | 1.5 | 1.5 | 1.5 |
| EB77‡ | 3 | 1.5 | 3 |
| EB378‡ | 3 | 1.5 | 3 |
Infection cleared by vancomycin administration.
Persistent infection with vancomycin administration.
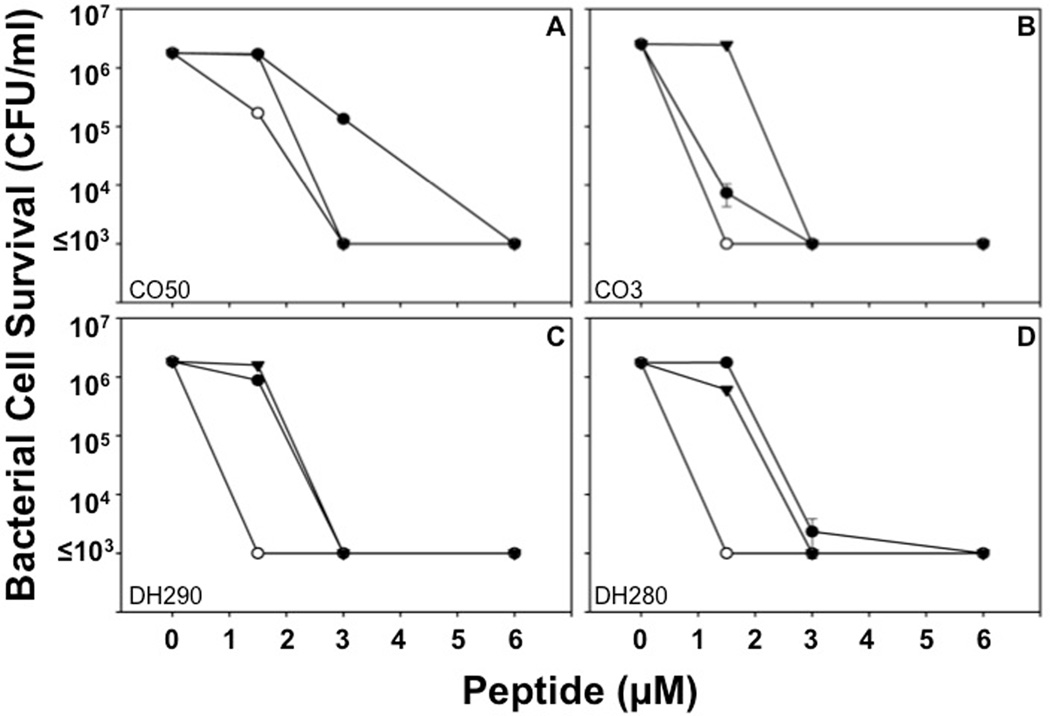
Figure 2. Sensitivity of control MRSA strains to RMAD-4, Crp-4, and RTD-1 is independent of antibiotic resistance.
Bactericidal peptide assays were performed in triplicate as described (see Materials and Methods). Briefly, MRSA strains were exposed to peptides for 1 h, and surviving bacteria were counted as CFU/ml at each peptide concentration (see Materials and Methods). Values ≤ 1 × 103 CFU/ml signify that no colonies were detected on plates after overnight growth. Strain Mu50 (A) is a vancomycin-intermediate S. aureus (VISA) control, and Mu3 (B), DH290 (C), and DH280 (D) are classified as heteroresistant VISA (hVISA) strains. Each condition was performed in triplicate, with error bars denoting standard deviations from the mean. Symbols: Crp-4 (-●-), RMAD-4 (-○-), and RTD-1 (-▼-).
The potent microbicidal effects of Crp-4, RMAD-4, and RTD-1 against VI-SA/hVISA control strains prompted us to test whether MRSA that had persisted in bacteremic patients after extensive vancomycin treatment would be sensitive to these defensins. Clinical blood isolates of MRSA that were not cleared by vancomycin treatment were assessed for survival after Crp-4, RMAD-4, and RTD-1 exposure in vitro, and all MRSA clinical isolates were sensitive to Crp-4, RMAD-4, and RTD-1 a peptide concentration dependent manner, regardless of response to vancomycin treatment (Table 1, Figs. 3A–F). For example, the MBC for RMAD-4 was 1.5 µM against vancomycin resistant and sensitive MRSA strains (Table 1, Fig. 3), and no association existed between antibiotic resistance and defensin susceptibility. Also, vancomycin-heteroresistant MRSA isolated from bacteremic patients that failed to resolve with antibiotic therapy were susceptible to the three defensins and as sensitive as culture adapted S. aureus strains (Table 1).
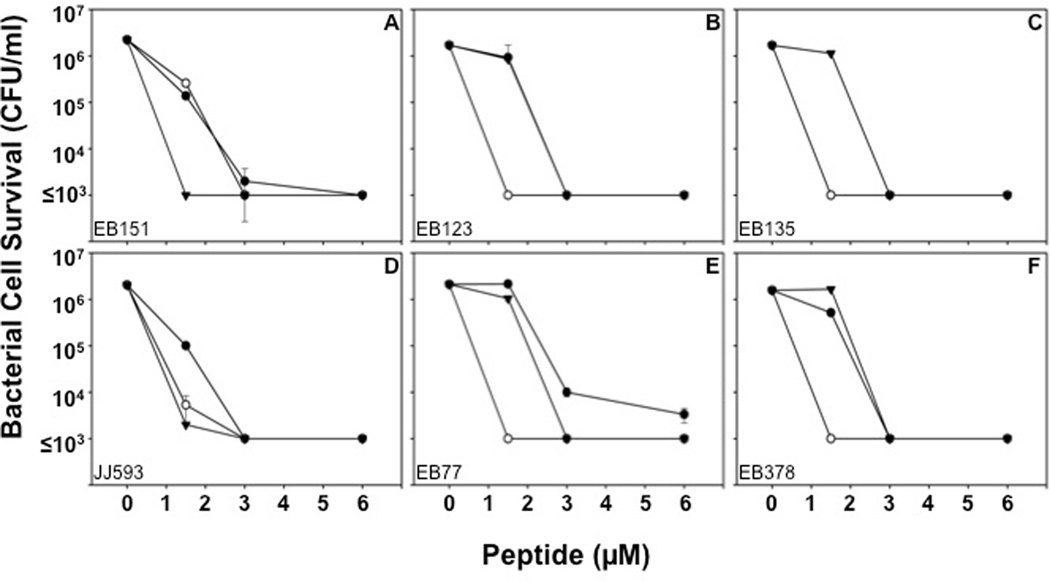
Figure 3. MRSA sensitivity to RMAD-4, Crp-4 and RTD-1 is not associated with antibiotic resistance.
Bactericidal peptide assays were performed in triplicate as described (Fig 2, Materials and Methods). Strains EB151 (A), EB123 (B) and EB135 (C) are clinical isolates from patients whose infections were cleared by vancomycin administration, in contrast to strains JJ593 (D), EB77 (E) and EB378 (F) which were isolated from persistent infections after failure of treatment with vancomycin. All MRSA strains were sensitive to the defensins tested at low micromolar concentrations (Table 1). Each condition was performed in triplicate with error bars denoting standard deviation from the mean. Symbols: Crp-4 (-●-), RMAD-4 (-○-), and RTD-1 (-▼-)
Sensitivity of ciprofloxacin-resistant PA to defensins
PA develops resistance to ciprofloxacin by mutations in DNA gyrase, topoisomerase IV or both and via efflux pumps that remove the drug and prevent its accumulation. Because the α-defensins in this study are membrane disruptive microbicides and peptide accumulation within target cells is not required for activity, we tested whether CipR PA are sensitive to Crp-4 and RMAD-4. Bactericidal assays against ciprofloxacin-sensitive (CipS) and CipR PA isolated from patient sputum showed that most PA sputum isolates were sensitive to both Crp-4 and RMAD-4 at low micromolar peptide levels, regardless of antibiotic resistance (Table 2, Fig. 4, See Supplementary Figure S1). In contrast to the variability of the relative activities of Crp-4 and RMAD-4 against MRSA (Table 1, Fig. 3), Crp-4 consistently was more active than RMAD-4 against most PA strains isolated from sputum (Table 2, Fig. 4). Also, sputum isolates displayed more variability than MRSA strains regarding susceptibility to Crp-4 and RMAD-4. Consistent with the defensin sensitivities of vancomycin-heteroresistant MRSA strains, Crp-4 and RMAD-4 were bactericidal against PA sputum isolates irrespective of strain ciprofloxacin resistance, there being no association between PA isolate sensitivities to Crp-4 or RMAD-4 and antibiotic resistance.
Table 2.
α-Defensin MBC Values against CipR and CipS strains of Pseudomonas aeruginosa
| P. aeruginosa strain | MBC Value (µM) | |
|---|---|---|
| Crp-4 | RMAD-4 | |
| 06397 (CipS)† | 1.5 | 3 |
| ML43 (CipS) | 1.5 | 1.5 |
| 06324 (CipS) | 1.5 | 1.5 |
| 05080 (CipR)§ | 1.5 | 1.5 |
| 06161 (CipR) | 1.5 | 1.5 |
| ML172 (CipR) | 1.5 | 1.5 |
CipS denotes ciprofloxacin-resistant strains;
CipR denotes ciprofloxacin-resistant strains.
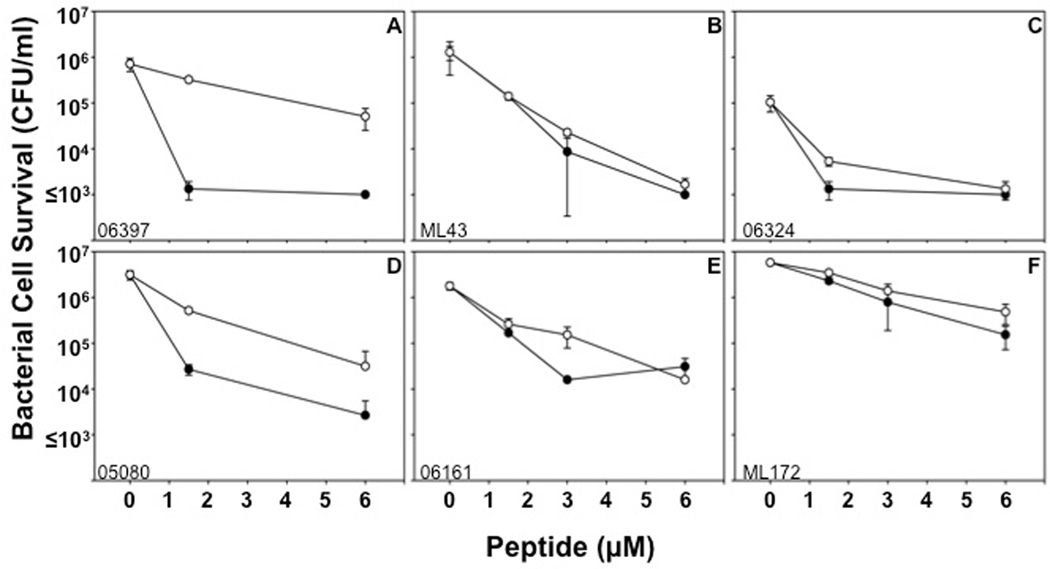
Figure 4. Activities of Crp-4 and RMAD-4 against CipR and CipS strains of PA.
Bactericidal peptide assays were performed in triplicate against clinical isolates from patient sputum. Isolates 06397 (A), ML43 (B), and 06324 (C) are CipR-PA, and isolates 05080 (D), 06161 (E), and ML172 (F) are CipS-PA. No correlation exists between antibiotic resistance and α-defensin activity (Table 2). Bars denote standard deviations from the mean. Symbols: Crp-4 (-●-) and RMAD-4 (-○-).
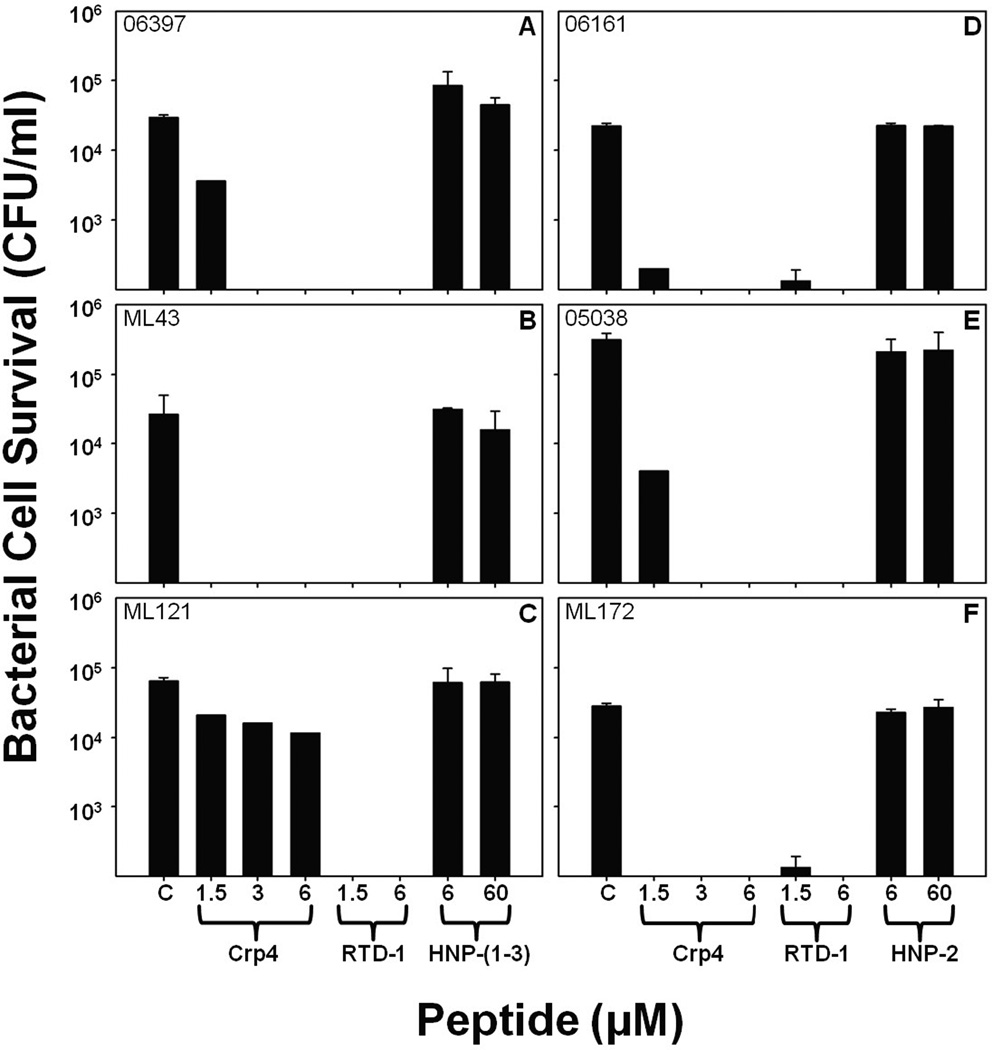
The activities of defensins against PA sputum isolates were tested further by determining the in vitro bactericidal effects of RTD-1, a mixture of natural HNPs 1–3, and purified HNP-2 against sputum PA isolates. RTD-1 at <1.5–6 µM reduced survival of the majority of sputum PA isolates to ≤ 103 CFU/ml, below the limit of detection for these assays (Fig. 5). In contrast, HNP 1–3 (Fig. 4A–C) and HNP-2 (Fig. 4D–F) did not affect survival of PA sputum isolates, even at peptide concentrations 10–fold greater than levels at which Crp-4 and RTD-1 are highly microbicidal. Thus, HNPs, endogenous α-defensins that occur at elevated serum concentrations during septicemia and certain infections in humans53, lacked bactericidal activity against CipR and CipS PA strains under these in vitro conditions. Although HNPs lacked activity against PA strains, they have been shown to have alternative innate host defense roles, including neutralization of anthrax lethal toxin and as chemoattractants54, 55. Because PA isolates were sensitive to Crp-4 and RMAD-4 irrespective of antibiotic resistance, we focused on characterizing the activities of those molecules rather than optimizing the activities of HNPs. Perhaps, the non-human peptides, Crp-4, RMAD-4, and RTD-1, may offer promise as adjunct therapies for treatment of bacterial infections that fail to resolve with conventional treatment.
Figure 5. Attenuated activities of HNPs against PA clinical isolates.
Survival of CipS P. aeruginosa strains 06397 (A), ML43 (B), and ML121 (C) and CipR–PA strains 06161 (D), 05038 (E), and ML172 (F) isolated from patient sputum was measured after exposure to Crp-4, RTD-1, and HNPs. In contrast to Crp-4 and RTD-1, 60 µM HNPs had little bactericidal effects against P. aeruginosa, irrespective of CipR. RTD-1 and HNPs assays were performed in triplicate, Crp-4 activity, assayed once at each peptide concentration, was consistent with Fig. 3 Error bars denote standard deviations from the mean.
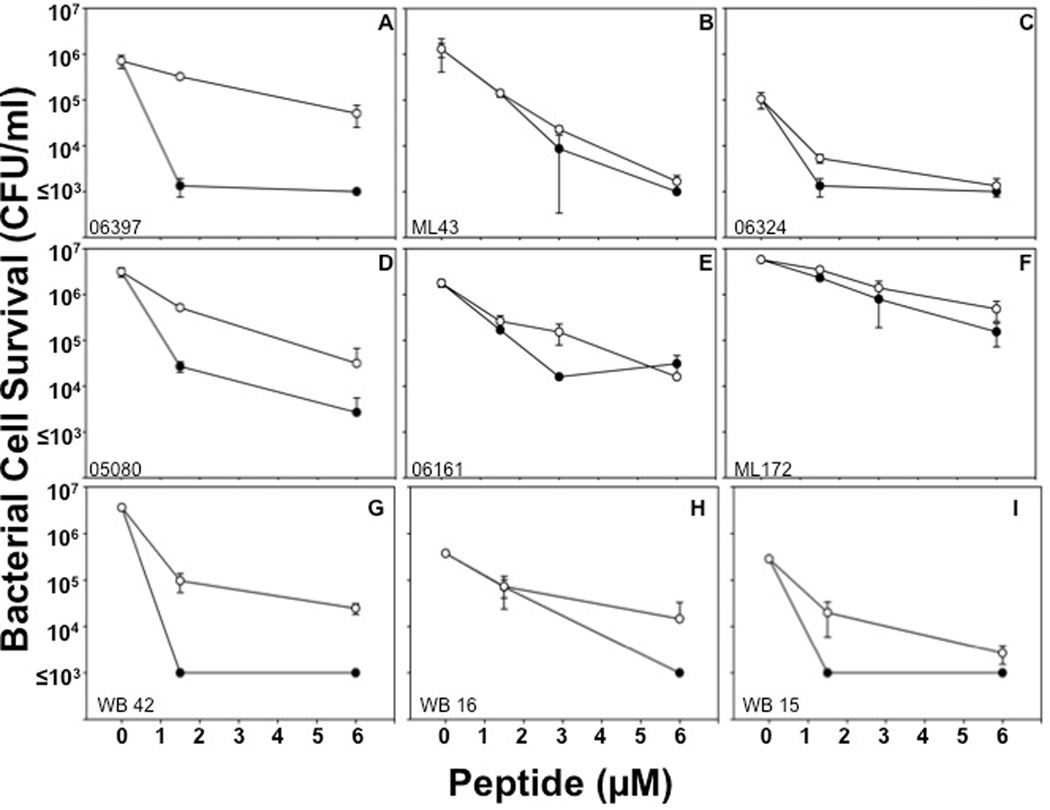
PA sensitivity to α-defensins is independent of the site of isolation
Major sites of PA infections include the urinary tract, lungs of cystic fibrosis patients, and burn wounds, and PA strains may differ phenotypically, depending on the site of infection56. Accordingly, bactericidal peptide assays were performed against PA clinical isolates from patient urine (Fig. 6A, B, C, See Supplementary Figure S2), burn wounds (Fig. 6D, E, F, See Supplementary Figure S3), and blood (Fig. 6G, H, I) to test their sensitivities to Crp-4 and RMAD-4. Crp-4 was highly bactericidal against almost every PA isolate, irrespective of the site of strain isolation. When exposed to ≥ 6 µM Crp4, no PA survivors were detected on plates, i.e., ≤ 103 CFU/ml survived peptide exposure (Figure 6). RMAD-4 had similar activities against PA wound isolates, but PA strains from blood and urine were more variable in RMAD-4 susceptibility (Table 3). Therefore, little relation exists between the site of PA strain isolation and resistance to these α-defensins.
Figure 6. α-Defensin sensitivity is independent of the site of PA isolation.
P. aeruginosa strains WB 25 (A), WB 36 (B), WB 34 (C) isolated from the urine of patients, strains WB 12 (D), WB 35 (E), and WB 44 (F) isolated from patient wounds, and WB 42 (G), WB 16 (H), and WB 15 (I) isolated from patient blood were exposed to 1.5–6 µM peptides (Table 3). Assays were performed in triplicate; error bars denote standard deviation from the mean. Symbols: Crp-4 (-●-) and RMAD-4 (-○-).
Table 3.
α-Defensin MBC Values against P. aeruginosa Blood, Urine, and Wound Isolates
| P. aeruginosa strain | MBC Value (µM) | |
|---|---|---|
| Crp-4 | RMAD-4 | |
| WB 25 (U)† | 3 | 6 |
| WB 36 (U) | 1.5 | 6 |
| WB 34 (U) | 1.5 | >6 |
| WB 12 (W)‡ | 1.5 | 3 |
| WB 35 (W) | 1.5 | 3 |
| WB 44 (W) | 1.5 | 3 |
| WB 42 (B) | 1.5 | 1.5 |
| WB 16 (B) | 1.5 | 1.5 |
| WB 15 (B) | 1.5 | 1.5 |
Symbols denote anatomic source of the clinical isolates:
Urine,
Wounds,
Blood.
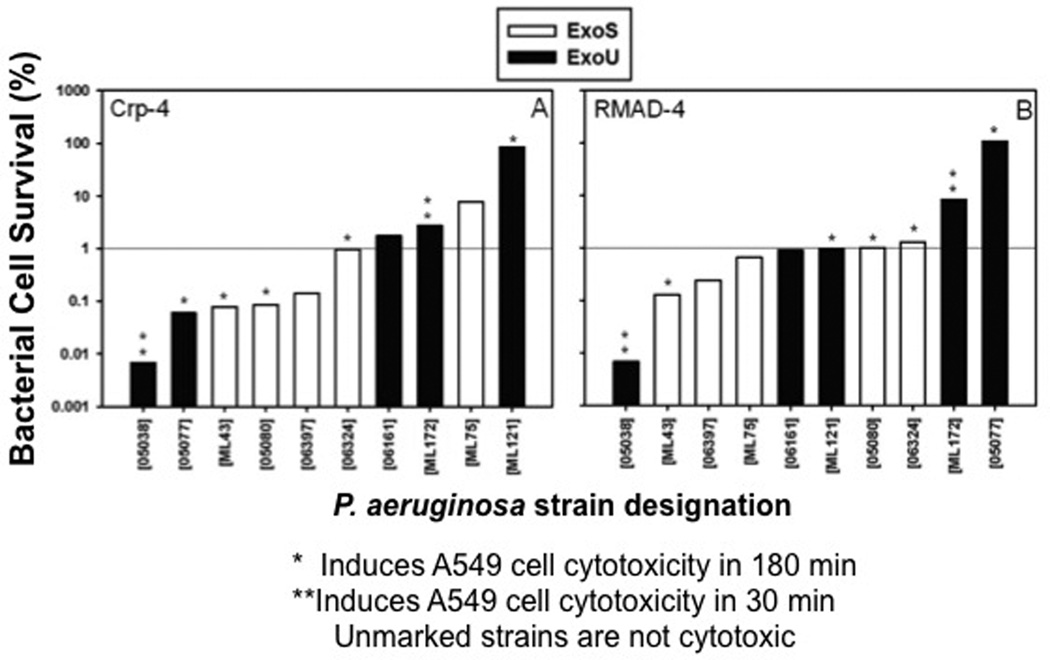
ExoU or ExoS PA genotype and cytotoxicity do not induce α-defensin resistance
Most wild-type PA strains harbor the exoS gene encoding the TTSS effector protein, and CipR PA populations are enriched for the exoU gene that codes for a TTSS effector protein, which contributes to virulence in acute infection50, 57. Notably, ExoU expressing PA are more virulent than ExoS strains in a murine model of acute pneumonia, and they cause more severe disease in humans11, 58. To test whether these PA virulence geno-types have differential α-defensin sensitivities, we performed bactericidal assays and determined whether ExoU or ExoS genotype and α-defensin resistance are related a significantly by constructing contingency tables and performing a Fisher’s exact test. We also grouped the PA strains on the basis of the rate and extent of their cytotoxic effects on exposed A549 lung epithelial cells, and we compared strain α-defensin susceptibility to cytotoxic potential. For this purpose, PA sputum isolates were categorized as α-defensin resistant if less than 99% killing occurred when exposed to 6 µM peptide. A subset of ExoU PA strains were resistant to Crp-4 and RMAD-4 by this definition (Fig. 7), but no significant association existed between genotype and α-defensin resistance (p>>0.1). Reduced sensitivity to Crp-4 or RMAD-4, therefore, was not determined by TTSS effector genotype. Similarly, Crp4 and RMAD-4 bactericidal activities against sputum isolates were independent of PA cytotoxic potential, because highly cytotoxic strains (05038 and ML172) or non-cytotoxic strains (06161 and ML75) were equally peptide sensitive (Fig. 7). Collectively, these studies show that the bactericidal activities of Crp-4 and RMAD-4 against PA clinical isolates are independent of CipR status, site of isolation, TTSS effector genotype, and cytotoxic potential.
Figure 7. Crp-4 or RMAD-4 bactericidal activity against PA are independent of ExoU or ExoS genotype and cytotoxic potential.
PA sputum isolates, characterized as ExoU (Black) or ExoS (white) are arranged in order of increasing bacterial survival following exposure to 6 µM of Crp-4 or RMAD-4. Cytotoxic potentials of each strain are also indicated by asterisks. PA strains were considered resistant if > 1% of bacteria survived α-defensin peptide exposure. TTSS effector genotypes and defensin resistance are not associated (p>0.1), and bactericidal activities were independent of PA cytotoxic potential.
DISCUSSION
Antimicrobial peptides have been proposed as templates for developing new therapeutics to combat multidrug resistant pathogens59, 60, yet the efficacy of α-defensins against antibiotic resistant clinical isolates has not been investigated extensively. Because general mechanisms of α-defensin microbicidal action differ from most antibiotics, they may be useful in combating infections by synergizing in combination with current antibiotic therapies. Because Crp-4, RMAD-4, and RTD-1 resist proteolysis and are non-cytotoxic, potent microbicides in vitro, they may provide new peptide-based platforms for antibiotic development39, 61. To test the hypothesis that Crp-4, RMAD-4, and RTD-1 may be efficacious against clinically important bacteria that also are antibiotic resistant, we assayed the bactericidal activities of these defensins against isolates of MRSA with varying resistance to vancomycin and CipR-PA. Under the in vitro assay conditions, Crp-4, RMAD-4, and RTD-1 were highly active against MRSA and exhibited differential activities against PA, irrespective of antibiotic resistance.
The continuing emergence of MRSA strains with vancomycin resistance, the treatment standard, underscores the need for new therapeutics. Against all strains of MRSA tested, Crp-4, RMAD-4, and RTD-1 were highly active with MBC values of 1.5–3 µM peptide, regardless of vancomycin resistance. In addition, RMAD-4 had a low MBC of 1.5 µM peptide, the lowest concentration assayed, against all but one MRSA strain. The α- defensins investigated have microbicidal mechanisms of action that that disrupt cellular integrity62. For example, certain defensins kill bacteria by sequestering lipid II and inhibiting bacterial cell wall biosynthesis37, 63, 64, HNPs 1–3 mediate non-oxidative microbial cell killing by sequential permeabilization of the outer and inner membranes and formation of stable pores65, and Crp-4, RMAD-4, and RTD-1 disrupt membranes by forming transient pores by induction of saddle-splay curvature66. At concentrations approximately 10-fold greater than the MBC values reported here, Crp4 induces rapid efflux of potassium ions from bacterial cells, a sensitive indicator of membrane disruption and cell death67–69,34, 70. We do not discount inhibition of cell wall biosynthesis as contributing to the defensin mechanism of action in certain settings. On the other hand, over the one-hour course of peptide exposure in the experiments we have presented, membrane disruption is the most likely mechanism of action36, 71, a mechanism that differs from beta lactam inhibitors and vancomycin. Thus, in combination with antibiotics, these defensin peptides could synergize with current antibiotics to improve treatment of MRSA-related infections.
Gram-negative bacteria account for more than 30% of US hospital infections and are the predominant ICU infections72–74. For example, PA is a leading cause of pneumonia, urinary tract, and bloodstream infections72, and the incidence of multidrug resistant PA is rising75. Crp-4, RMAD-4, RTD-1, and HNP 1–3 were tested against PA strains characterized by their ciprofloxacin resistance, site of isolation, TTSS effector genotype and cytotoxic potential to determine whether these traits correlated with defensin resistance. Under the conditions of the in vitro assays, PA strains exhibited variable sensitivity to the defensin peptides tested (Tables 2, 3), but defensin sensitivity was not associated with either of the phenotypic characteristics. Cationic charge contributes to defensin bactericidal activity, but even though Crp-4 and RMAD-4 are equally electropositive, Crp-4 was more active against PA strains but RMAD-4 was more potent against MRSA (Tables 1–3). Perhaps, the differential bactericidal effects are due to differences in the surface charge or hydrophobicity distribution of the two α-defensin peptides51. Although PA isolates from lung, blood, and urine differ phenotypically56, the strains tested were equally sensitive to Crp-4 and RMAD-4, irrespective of their site of isolation.
The α-defensin and θ-defensin peptides tested hold promise toward the eventual development of new therapeutic agents against the growing number of antibiotic resistant pathogens. Although Crp-4, RMAD-4, and RTD-1 are highly active against many of the PA strains tested in vitro, activity against PA strains may be diminished in the setting of chronic lung infections where biofilm formation is induced76, 77, in that biofilm production enhances resistance to bactericidal peptides. Whether biofilms diminish the activities of peptides tested here is unknown78. Also, the ionic strength of assay media inhibits bactericidal activities of Crp4 and RMAD-4, and binding of Crp-4, RMAD-4, and RTD-1 peptides to plasma proteins has not been studied and may limit efficacy in vivo. Against bacteria that are sensitive to these peptides, e.g., MBC ≤1.5 µM, bactericidal effects of Crp-4 and RMAD-4 are partially inhibited by 50 mM NaCl and completely inhibited at 100 mM NaCl. Thus, the inherent salt sensitivity of these native molecules may limit therapeutic application of native, full-length α-defensins, but reiterative selection of salt-insensitive variants of these peptides could lead to drug development based on these peptide scaffolds. In contrast to the α-defensins, however, cyclized RTD-1 is as active in 150 mM NaCl as in low salt media28, 48, 52, an indication that the θ-defensins may provide a more robust platform for therapeutic development. Also, the disulfide array protects the Crp-4 and RMAD-4 from proteolytic degradation, and α-defensins can be recovered from the protease- rich environment of the mouse small and large bowel61, 79. Biophysical studies of Crp-4, RMAD-4, and RTD-1 have established amino acid criteria necessary for their selective membrane disruptive activities29, and Crp-4 and RMAD-4 are non-cytotoxic and nonhemolytic at peptide concentrations as high as 100 µg/ml (data not shown)52.
In addition to their broad spectrum in vitro microbicidal activities, rhesus θ- defensins 1–5 (RTDs 1–5), RTD-1 in particular, reduced the levels of TNF-α, IL-1α, IL- 1β, IL-6, and IL-8 released by mixed blood leukocytes and THP-1 monocytes stimulated by bacteria or LPS, respectively80. Also, systemic administration of RTD-1 to BALB/c mice was non-toxic and stable in serum and plasma, and intravenous delivery of 5 mg/kg RTD-1 significantly improved survival of BALB/c mice with E. coli peritonitis and cecal ligation-and-puncture-induced polymicrobial sepsis80. In contrast to HNPs, which are proinflammatory81–85, Crp4 and RMAD4 also inhibit cytokine and TNF-α release in the same in vitro assays, although both are less inhibitory than RTD-1 (Kamdar et al., submitted for publication). Thus, new antimicrobials that are developed to optimize both defensin-based bactericidal and immunomodulatory properties may prove to be efficacious in combination with current antibiotic therapies against antibiotic resistant bacteria while promoting survival from systemic infections.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grants DK044632, AI059346 (A.J.O.), AI022931, AI058129, DE021341, Southern California Clinical Translational Science Institute UL1RR031986 (M.E.S.), National Institute of Allergy and Infectious Diseases grant AI073467 (A.W.-B.). Also supported in part by the USC Norris Cancer Center Support Grant P30CA014089 from the National Cancer Institute; the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
REFERENCES
- 1.Maragakis LL, Perencevich EN, Cosgrove SE. Clinical and economic burden of antimicrobial resistance. Expert Rev Anti Infect Ther. 2008;6:751–763. doi: 10.1586/14787210.6.5.751. [DOI] [PubMed] [Google Scholar]
- 2.Jarvis WR. Controlling healthcare-associated infections: the role of infection control and antimicrobial use practices. Semin Pediatr Infect Dis. 2004;15:30–40. doi: 10.1053/j.spid.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Klevens RM, Edwards JR, Richards CL, Jr, et al. Estimating health care-associated infections and deaths in U.S. hospitals 2002. Public Health Rep. 2007;122:160–166. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kallen AJ, Mu Y, Bulens S, et al. Health care-associated invasive MRSA infections, 2005–2008. JAMA. 2010;304:641–648. doi: 10.1001/jama.2010.1115. [DOI] [PubMed] [Google Scholar]
- 5.Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century--a clinical super-challenge. N Engl J Med. 2009;360:439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 6.Strateva T, Yordanov D. Pseudomonas aeruginosa - a phenomenon of bacterial resistance. J Med Microbiol. 2009;58:1133–1148. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 7.Neuhauser MM, Weinstein RA, Rydman R, Danziger LH, Karam G and Quinn JP. Antibiotic resistance among gram-negative bacilli in US intensive care units: implications for fluoroquinolone use. JAMA. 2003;289:885–888. doi: 10.1001/jama.289.7.885. [DOI] [PubMed] [Google Scholar]
- 8.Trkanjec Z, Demarin V. Presynaptic vesicles, exocytosis, membrane fusion and basic physical forces. Med Hypotheses. 2001;56:540–546. doi: 10.1054/mehy.2000.1260. [DOI] [PubMed] [Google Scholar]
- 9.Alp S, Skrygan M, Schlottmann R, et al. Expression of beta-defensin 1 and 2 in nasal epithelial cells and alveolar macrophages from HIV-infected patients. Eur J Med Res. 2005;10:1–6. [PubMed] [Google Scholar]
- 10.Georges B, Conil JM, Dubouix A, et al. Risk of emergence of Pseudomonas aeruginosa resistance to beta-lactam antibiotics in intensive care units. Crit Care Med. 2006;34:1636–1641. doi: 10.1097/01.CCM.0000215517.51187.CA. [DOI] [PubMed] [Google Scholar]
- 11.Shaver CM, Hauser AR. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infection and immunity. 2004;72:6969–6977. doi: 10.1128/IAI.72.12.6969-6977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.[New research results give hope. Causal therapy of Crohn disease in sight] MMW Fortschr Med. 2004;146:42–43. [PubMed] [Google Scholar]
- 13.Schito GC. The importance of the development of antibiotic resistance in Staphylococcus aureus. Clin Microbiol Infect. 2006;12(Suppl 1):3–8. doi: 10.1111/j.1469-0691.2006.01343.x. [DOI] [PubMed] [Google Scholar]
- 14.Capitano B, Leshem OA, Nightingale CH, Nicolau DP. Cost effect of managing methicillin-resistant Staphylococcus aureus in a long-term care facility. J Am Geriatr Soc. 2003;51:10–16. doi: 10.1034/j.1601-5215.2002.51003.x. [DOI] [PubMed] [Google Scholar]
- 15.Dye D, Croize J, Brambilla C. [Mechanism of antibiotic resistance in bacteria responsible for respiratory infections] Rev Mal Respir. 1995;12:415–427. [PubMed] [Google Scholar]
- 16.Therrien C, Levesque RC. Molecular basis of antibiotic resistance and beta-lactamase inhibition by mechanism-based inactivators: perspectives and future directions. FEMS Microbiol Rev. 2000;24:251–262. doi: 10.1111/j.1574-6976.2000.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 17.Pechere JC, Michea-Hamzhepour M, Kohler T. [Antibiotic efflux, a mechanism of multiple resistance in Pseudomonas aeruginosa] Bull Acad Natl Med. 1998;182:599–612. discussion 3–5. [PubMed] [Google Scholar]
- 18.Drlica K, Hiasa H, Kerns R, Malik M, Mustaev A, Zhao X. Quinolones: action and resistance updated. Curr Top Med Chem. 2009;9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds PE. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis. 1989;8:943–950. doi: 10.1007/BF01967563. [DOI] [PubMed] [Google Scholar]
- 20.Jovetic S, Zhu Y, Marcone GL, Marinelli F, Tramper J. beta-Lactam and glycopeptide antibiotics: first and last line of defense? Trends Biotechnol. 2010;28:596–604. doi: 10.1016/j.tibtech.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Wang FD, Chen YY, Chen TL, Liu CY. Risk factors and mortality in patients with nosocomial Staphylococcus aureus bacteremia. Am J Infect Control. 2008;36:118–122. doi: 10.1016/j.ajic.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Maor Y, Hagin M, Belausov N, Keller N, Ben-David D, Rahav G. Clinical features of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia versus those of methicillin-resistant S. aureus bacteremia. J Infect Dis. 2009;199:619–624. doi: 10.1086/596629. [DOI] [PubMed] [Google Scholar]
- 23.Ganz T. Defensins: antimicrobial peptides of vertebrates. C R Biol. 2004;327:539–549. doi: 10.1016/j.crvi.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 25.Selsted ME, Tang YQ, Morris WL, et al. Purification, primary structures, and antibacterial activities of beta-defensins, a new family of antimicrobial peptides from bovine neutrophils. The Journal of biological chemistry. 1993;268:6641–6648. [PubMed] [Google Scholar]
- 26.Ouellette AJ. Paneth cell alpha-defensins in enteric innate immunity. Cell Mol Life Sci. 2011;68:2215–2229. doi: 10.1007/s00018-011-0714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rehaume LM, Hancock RE. Neutrophil-derived defensins as modulators of innate immune function. Crit Rev Immunol. 2008;28:185–200. doi: 10.1615/critrevimmunol.v28.i3.10. [DOI] [PubMed] [Google Scholar]
- 28.Tang YQ, Yuan J, Osapay G, et al. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science. 1999;286:498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 29.Januschowski K, Mueller S, Spitzer MS, et al. Investigating the biocompatibility of two new heavy intraocular dyes for vitreoretinal surgery with an isolated perfused vertebrate retina organ culture model and a retinal ganglion cell line. Graefes Arch Clin Exp Ophthalmol. 2012;250:533–45. doi: 10.1007/s00417-011-1895-2. [DOI] [PubMed] [Google Scholar]
- 30.Wimley WC, Selsted ME, White SH. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Protein Sci. 1994;3:1362–1373. doi: 10.1002/pro.5560030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hristova K, Selsted ME, White SH. Interactions of monomeric rabbit neutrophil defensins with bilayers: comparison with dimeric human defensin HNP-2. Biochemistry. 1996;35:11888–11894. doi: 10.1021/bi961100d. [DOI] [PubMed] [Google Scholar]
- 32.Satchell DP, Sheynis T, Shirafuji Y, Kolusheva S, Ouellette AJ, Jelinek R. Interactions of mouse Paneth cell alpha-defensins and alpha-defensin precursors with membranes: Prosegment inhibition of peptide association with biomimetic membranes. J Biol Chem. 2003;278:13838–13846. doi: 10.1074/jbc.M212115200. [DOI] [PubMed] [Google Scholar]
- 33.Satchell DP, Sheynis T, Kolusheva S, et al. Quantitative interactions between cryptdin-4 amino terminal variants and membranes. Peptides. 2003;24:1793–1803. doi: 10.1016/j.peptides.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Hadjicharalambous C, Sheynis T, Jelinek R, Shanahan MT, Ouellette AJ, Gizeli E. Mechanisms of alpha-defensin bactericidal action: comparative membrane disruption by Cryptdin-4 and its disulfide-null analogue. Biochemistry. 2008;47:12626–12634. doi: 10.1021/bi800335e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehrer RI, Barton A and Ganz T. Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry. J Immunol Methods. 1988;108:153–158. doi: 10.1016/0022-1759(88)90414-0. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt NW, Mishra A, Lai GH, et al. Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. J Am Chem Soc. 2011;133:6720–6727. doi: 10.1021/ja200079a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider T, Kruse T, Wimmer R, et al. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science. 2010;328:1168–1172. doi: 10.1126/science.1185723. [DOI] [PubMed] [Google Scholar]
- 38.de Leeuw E, Li C, Zeng P, et al. Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS Lett. 2010;584:1543–1548. doi: 10.1016/j.febslet.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Figueredo S, Mastroianni JR, Tai KP, Ouellette AJ. Expression and purification of recombinant alpha-defensins and alpha-defensin precursors in Escherichia coli. Methods Mol Biol. 2010;618:47–60. doi: 10.1007/978-1-60761-594-1_4. [DOI] [PubMed] [Google Scholar]
- 40.Tanabe H, Qu X, Weeks CS, et al. Structure-activity determinants in Paneth cell alpha-defensins: loss-of-function in mouse cryptdin-4 by charge-reversal at arginine residue positions. J Biol Chem. 2004;279:11976–11983. doi: 10.1074/jbc.M310251200. [DOI] [PubMed] [Google Scholar]
- 41.Satchell DP, Sheynis T, Kolusheva S, et al. Quantitative interactions between cryptdin-4 amino terminal variants and membranes. Peptides. 2003;24:1795–1805. doi: 10.1016/j.peptides.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 42.Rosengren KJ, Daly NL, Fornander LM, et al. Structural and functional characterization of the conserved salt bridge in mammalian Paneth cell alpha-defensins: solution structures of mouse cryptdin-4 and (E15D)-cryptdin-4. J Biol Chem. 2006;281:28068–28078. doi: 10.1074/jbc.M604992200. [DOI] [PubMed] [Google Scholar]
- 43.Shirafuji Y, Tanabe H, Satchell DP, Henschen-Edman A, Wilson CL, Ouellette AJ. Structural determinants of procryptdin recognition and cleavage by matrix metalloproteinase-7. J Biol Chem. 2003;278:7910–7919. doi: 10.1074/jbc.M210600200. [DOI] [PubMed] [Google Scholar]
- 44.Jing W, Hunter HN, Tanabe H, Ouellette AJ, Vogel HJ. Solution Structure of Cryptdin-4, a Mouse Paneth Cell alpha-Defensin. Biochemistry. 2004;43:15759–15766. doi: 10.1021/bi048645p. [DOI] [PubMed] [Google Scholar]
- 45.Rosengren KJ, Daly NL, Fornander LM, et al. Structural and functional characterization of the conserved salt bridge in mammalian Paneth cell alpha-defensins: solution structures of mouse cryptdin-4 and (E15D)-cryptdin-4. The Journal of biological chemistry. 2006;281:28068–28078. doi: 10.1074/jbc.M604992200. [DOI] [PubMed] [Google Scholar]
- 46.Ganz T, Selsted ME, Szklarek D, et al. Defensins. Natural peptide antibiotics of human neutrophils. The Journal of clinical investigation. 1985;76:1427–1435. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selsted ME. HPLC methods for purification of antimicrobial peptides. Methods Mol Biol. 1997;78:17–33. doi: 10.1385/0-89603-408-9:17. [DOI] [PubMed] [Google Scholar]
- 48.Tran D, Tran PA, Tang YQ, Yuan J, Cole T, Selsted ME. Homodimeric thetadefensins from rhesus macaque leukocytes: isolation, synthesis, antimicrobial activities, and bacterial binding properties of the cyclic peptides. J Biol Chem. 2002;277:3079–3084. doi: 10.1074/jbc.M109117200. [DOI] [PubMed] [Google Scholar]
- 49.Leonard SN, Rossi KL, Newton KL, Rybak MJ. Evaluation of the Etest GRD for the detection of Staphylococcus aureus with reduced susceptibility to glycopeptides. J Antimicrob Chemother. 2009;63:489–492. doi: 10.1093/jac/dkn520. [DOI] [PubMed] [Google Scholar]
- 50.Wong-Beringer A, Wiener-Kronish J, Lynch S, Flanagan J. Comparison of type III secretion system virulence among fluoroquinolone-susceptible and -resistant clinical isolates of Pseudomonas aeruginosa. Clin Microbiol Infect. 2008;14:330–336. doi: 10.1111/j.1469-0691.2007.01939.x. [DOI] [PubMed] [Google Scholar]
- 51.Llenado RA, Weeks CS, Cocco MJ, Ouellette AJ. Electropositive charge in alpha-defensin bactericidal activity: functional effects of Lys-for-Arg substitutions vary with the peptide primary structure. Infect Immun. 2009;77:5035–5043. doi: 10.1128/IAI.00695-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tran D, Tran P, Roberts K, et al. Microbicidal properties and cytocidal selectivity of rhesus macaque theta defensins. Antimicrob Agents Chemother. 2008;52:944–953. doi: 10.1128/AAC.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kokryakov VN, Harwig SS, Panyutich EA, et al. Protegrins: leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS letters. 1993;327:231–236. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- 54.Yang D, Chen Q, Chertov O, Oppenheim JJ. Human neutrophil defensins selectively chemoattract naive T and immature dendritic cells. J Leukoc Biol. 2000;68:9–14. [PubMed] [Google Scholar]
- 55.Kim C, Gajendran N, Mittrucker HW, et al. Human alpha-defensins neutralize anthrax lethal toxin and protect against its fatal consequences. Proc Natl Acad Sci U S A. 2005;102:4830–4835. doi: 10.1073/pnas.0500508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woods DE, Schaffer MS, Rabin HR, Campbell GD, Sokol PA. Phenotypic comparison of Pseudomonas aeruginosa strains isolated from a variety of clinical sites. Journal of clinical microbiology. 1986;24:260–264. doi: 10.1128/jcm.24.2.260-264.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sato H, Feix JB, Hillard CJ, Frank DW. Characterization of phospholipase activity of the Pseudomonas aeruginosa type III cytotoxin, ExoU. Journal of bacteriology. 2005;187:1192–1195. doi: 10.1128/JB.187.3.1192-1195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veesenmeyer JL, Howell H, Halavaty AS, Ahrens S, Anderson WF, Hauser AR. Role of the membrane localization domain of the Pseudomonas aeruginosa effector protein ExoU in cytotoxicity. Infection and immunity. 2010;78:3346–3357. doi: 10.1128/IAI.00223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hancock RE. Cationic antimicrobial peptides: towards clinical applications. Expert Opin Investig Drugs. 2000;9:1723–1729. doi: 10.1517/13543784.9.8.1723. [DOI] [PubMed] [Google Scholar]
- 60.Yeung AT, Gellatly SL, Hancock RE. Multifunctional cationic host defence peptides and their clinical applications. Cell Mol Life Sci. 2011;68:2161–2176. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia AE, Tai KP, Puttamadappa SS, Shekhtman A, Ouellette AJ, Camarero JA. Biosynthesis and antimicrobial evaluation of backbone-cyclized alpha-defensins. Biochemistry. 2011;50:10508–10519. doi: 10.1021/bi201430f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White SH, Wimley WC, Selsted ME. Structure, function, and membrane integration of defensins. Curr Opin Struct Biol. 1995;5:521–527. doi: 10.1016/0959-440x(95)80038-7. [DOI] [PubMed] [Google Scholar]
- 63.Schmitt P, Wilmes M, Pugniere M, et al. Insight into invertebrate defensin mechanism of action: oyster defensins inhibit peptidoglycan biosynthesis by binding to lipid II. J Biol Chem. 2010;285:29208–29216. doi: 10.1074/jbc.M110.143388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sass V, Schneider T, Wilmes M, et al. Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci. Infect Immun. 2010;78:2793–2800. doi: 10.1128/IAI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, Selsted ME. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J Clin Invest. 1989;84:553–561. doi: 10.1172/JCI114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt NW, Mishra A, Lai GH, et al. Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. J Am Chem Soc. 2011;133:6720–6727. doi: 10.1021/ja200079a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lambert PA, Hammond SM. Potassium fluxes, first indications of membrane damage in micro-organisms. Biochem Biophys Res Commun. 1973;54:796–799. doi: 10.1016/0006-291x(73)91494-0. [DOI] [PubMed] [Google Scholar]
- 68.Orlov DS, Nguyen T, Lehrer RI. Potassium release, a useful tool for studying antimicrobial peptides. J Microbiol Methods. 2002;49:325–328. doi: 10.1016/s0167-7012(01)00383-9. [DOI] [PubMed] [Google Scholar]
- 69.Tincu JA, Menzel LP, Azimov R, et al. Plicatamide, an antimicrobial octapeptide from Styela plicata hemocytes. J Biol Chem. 2003;278:13546–13553. doi: 10.1074/jbc.M211332200. [DOI] [PubMed] [Google Scholar]
- 70.Shanahan MT, Vidrich A, Shirafuji Y, et al. Elevated expression of Paneth cell CRS4C in ileitis-prone SAMP1/YitFc mice: regional distribution, subcellular localization, and mechanism of action. J Biol Chem. 2010;285:7493–7504. doi: 10.1074/jbc.M109.083220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shanahan MT, Vidrich A, Shirafuji Y, et al. Elevated expression of Paneth cell CRS4C in ileitis-prone SAMP1/YitFc mice: regional distribution, subcellular localization, and mechanism of action. The Journal of biological chemistry. 2010;285:7493–7504. doi: 10.1074/jbc.M109.083220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hidron AI, Edwards JR, Patel J, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infection control and hospital epidemiology : the official journal of the Society of Hospital Epidemiologists of America. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 74.Gaynes R, Edwards JR. Overview of nosocomial infections caused by gramnegative bacilli. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2005;41:848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 75.Kunz AN, Brook I. Emerging resistant Gram-negative aerobic bacilli in hospital-acquired infections. Chemotherapy. 2010;56:492–500. doi: 10.1159/000321018. [DOI] [PubMed] [Google Scholar]
- 76.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 77.Kyd JM, McGrath J, Krishnamurthy A. Mechanisms of bacterial resistance to antibiotics in infections of COPD patients. Curr Drug Targets. 2011;12:521–530. doi: 10.2174/138945011794751519. [DOI] [PubMed] [Google Scholar]
- 78.Otto M. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr Top Microbiol Immunol. 2006;306:251–258. doi: 10.1007/3-540-29916-5_10. [DOI] [PubMed] [Google Scholar]
- 79.Mastroianni JR, Ouellette AJ. Alpha-defensins in enteric innate immunity: functional Paneth cell alpha-defensins in mouse colonic lumen. J Biol Chem. 2009;284:27848–27856. doi: 10.1074/jbc.M109.050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schaal JB, Tran D, Tran P, Osapay G, Trinh K, Roberts KD, Brasky KM, Tongaonkar P, Ouellette AJ, Selsted ME. Rhesus macaque theta-defensins suppress inflammatory cytokines and enhance survival in mouse models of bacteremic sepsis. PLoS One. 2012;7:e51337. doi: 10.1371/journal.pone.0051337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lillard JW, Jr, Boyaka PN, Chertov O, Oppenheim JJ, McGhee JR. Mechanisms for induction of acquired host immunity by neutrophil peptide defensins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:651–656. doi: 10.1073/pnas.96.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaly YV, Paleolog EM, Kolesnikova TS, Tikhonov II, Petratchenko EV, Voitenok NN. Neutrophil alpha-defensin human neutrophil peptide modulates cytokine production in human monocytes and adhesion molecule expression in endothelial cells. Eur Cytokine Netw. 2000;11:257–266. [PubMed] [Google Scholar]
- 83.Presicce P, Giannelli S, Taddeo A, Villa ML, Della Bella S. Human defensins activate monocyte-derived dendritic cells, promote the production of proinflammatory cytokines, and up-regulate the surface expression of CD91. Journal of leukocyte biology. 2009 doi: 10.1189/jlb.0708412. [DOI] [PubMed] [Google Scholar]
- 84.Van Wetering S, Mannesse-Lazeroms SP, Van Sterkenburg MA, Daha MR, Dijkman JH, Hiemstra PS. Effect of defensins on interleukin-8 synthesis in airway epithelial cells. The American journal of physiology. 1997;272:L888–L896. doi: 10.1152/ajplung.1997.272.5.L888. [DOI] [PubMed] [Google Scholar]
- 85.Khine AA, Del Sorbo L, Vaschetto R, et al. Human neutrophil peptides induce interleukin-8 production through the P2Y6 signaling pathway. Blood. 2006;107:2936–2942. doi: 10.1182/blood-2005-06-2314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.