Abstract
Gene silencing in the process of RNA interference is mediated by a ribonucleoprotein complex referred to as RNA-induced silencing complex (RISC). Here we describe the molecular mechanism of target RNA cleavage using affinity-purified minimal RISC from human cells. Cleavage proceeds via hydrolysis and the release of a 3′-hydroxyl and a 5′-phosphate terminus. Substitution of the 2′-hydroxyl group at the cleavage site by 2′-deoxy had no significant effect, suggesting that product release and/or a conformational transition rather than a chemical step is rate-limiting. Substitution by 2′-O-methyl at the cleavage site substantially reduced cleavage, which is presumably due to steric interference. Mutational analysis of the target RNA revealed that mismatches across from the 5′ or the 3′ end of the siRNA had little effect and that substrate RNAs as short as 15 nucleotides were cleaved by RISC.
Keywords: RNAi, endonuclease, 2′ modification
RNAi is a conserved posttranscriptional gene silencing mechanism triggered by the appearance of double-stranded RNA (dsRNA) in most eukaryotic cells (Cerutti 2003; Denli and Hannon 2003; Dykxhoorn et al. 2003). The dsRNA is first processed by Dicer into short duplexes of 21-23-nt in length with characteristic 2-nt 3′-hydroxyl overhanging ends and 5′-phosphate termini, referred to as siRNA duplexes (Hammond et al. 2000; Zamore et al. 2000; Bernstein et al. 2001; Elbashir et al. 2001a). Subsequently, one strand of the siRNA duplex is incorporated into the RNA-induced silencing complex (RISC) to guide the sequence-specific cleavage of complementary target or substrate mRNAs (Elbashir et al. 2001a; Nykänen et al. 2001; Hutvágner and Zamore 2002; Martinez et al. 2002; Liu et al. 2003; Schwarz et al. 2003). The substrate RNA cleavage site has been mapped 10 nt upstream of the nucleotide opposed to the 5′-most residue of the guide siRNA (Elbashir et al. 2001b).
RISC has been partially purified from Drosophila melanogaster Schneider cells as a 500-kD ribonucleoprotein complex that catalyses the complete degradation of a complementary RNA (Hammond et al. 2000, 2001). The complex contains Argonaute 2 (Ago2; Hammond et al. 2001), the Vasa Intronic Gene product (VIG; Caudy et al. 2002), the Drosophila homolog of the Fragile X Mental Retardation Protein (FMRP), dFXR (Caudy et al. 2002), and the Tudor Staphylococcal Nuclease, Tudor SN (Caudy et al. 2003). Tudor SN has been regarded as a potential candidate for the endonuclease component of RISC, because it contains five repeats of a staphylococcal/micrococcal nuclease domain. Chromatographic size fractionation using D. melanogaster embryonic extracts suggested a smaller, <230 kD, size for RISC (Nykänen et al. 2001). Human RISC, which has been affinity-purified from HeLa cell extracts, only showed an apparent molecular mass of 160 kD, containing a single-stranded siRNA, 100 kD eIF2C1 and/or eIF2C2, both members of the Argonaute protein family, and a yet to be identified endonuclease (Martinez et al. 2002).
Sequence and chemical variants of siRNAs have been previously used to study the specificity and structural requirements of the guide RNA embedded in RISC. The results are not readily summarized, but dependent on the number and positions of modifications, differences in gene silencing efficiency were observed. Substitution of 2′-hydroxyl by 2′-fluoro groups (Capodici et al. 2002; Braasch et al. 2003; Chiu and Rana 2003; Harborth et al. 2003) appears to be extremely well tolerated, whereas the introduction of 2′-deoxy residues or the bulkier 2′-O-methyl and 2′-O-allyl group could be more disruptive (Amarzguioui et al. 2003; Chiu and Rana 2003; Czauderna et al. 2003; Holen et al. 2003). These studies also show that the sense siRNA strand is frequently more tolerant to chemical modifications than the guide antisense siRNA strand, indicating different requirements for assembly of RISC and targeting by RISC.
Here we describe the mechanism by which the substrate RNA is recognized and cleaved by the minimal RISC that was prepared by stringent affinity-purification using biotinylated siRNAs and HeLa cell cytoplasmic extract. We determined kinetic parameters and sequence and structural constraints, showing that substrates as short as 15 nt are cleaved by RISC, and that cleavage releases a 5′-phosphomonoester and a 3′-hydroxyl group.
Results and Discussion
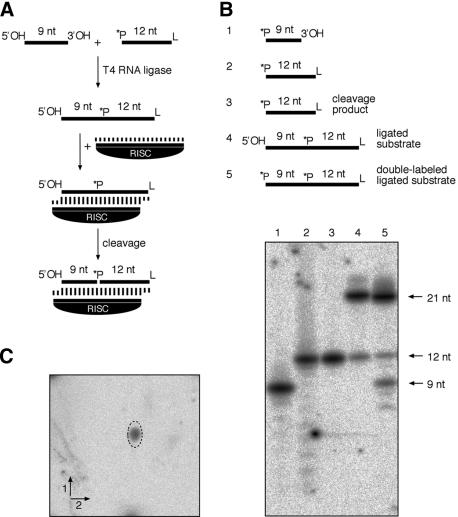
Human RISC was assembled by incubation of a 3′-biotinylated siRNA duplex in HeLa cell cytoplasmic extract, followed by biotin-streptavidin affinity chromatography (Martinez et al. 2002). We noticed that the potassium chloride concentration in the wash steps of the affinity column purification could be increased up to 2.5 M without loss of RISC activity. We refer to this surprisingly stable and highly purified complex as minimal RISC. The high-stringency wash removed a significant amount of unspecific cellular nucleases, and possibly some RISC-specific but partly dispensable proteins, which allowed us to use conventionally radiolabeled target RNAs that would otherwise be unstable in less purified RISC fractions. To determine the minimal length for target RNAs to be cleaved by RISC, we first tested a 5′ 32P-labeled 21-nt RNA (S21) that was identical in sequence to the sense strand of the siRNA duplex used for formation of RISC. Encouraged by the observation that this substrate was effectively cleaved, we prepared a 21-nt substrate site-specifically labeled at the scissile bond located 10 nt upstream of the nucleotide paired up with the 5′-most nucleotide of the guide siRNA (Elbashir et al. 2001a). The preparation of this 21-nt substrate and the RISC cleavage assay are schematized in Figure 1A.
Figure 1.
Target RNA is cleaved endonucleolytically producing 5′-phosphate and 3′-hydroxyl termini. (A) Preparation of site-specifically labeled substrates and cleavage assay. 5′-32P-labeled and 3′ aminolinker (L) protected 12-nt oligoribonucleotide was ligated to nonphosphorylated 9-nt oligoribonucleotide using T4 RNA ligase. An aliquot of the ligation product was further 5′-32P-labeled using T4 polynucleotide kinase. The purified substrates were incubated with affinity-purified RISC programmed with single-stranded guide siRNA. (B) PhosphorImaging of cleavage reactions incubated for 2 h at 30°C, and resolved on a 20% denaturing polyacrylamide gel. 5′-32P-labeled 9- and 12-nt oligoribonucleotides were loaded as marker in lanes 1 and 2, respectively. The cleavage reactions with single- and double-labeled 21-nt substrate are loaded in lanes 4 and 5, respectively. Lane 3 contains the 12-nt cleavage product isolated from a prior cleavage reaction. (C) Two-dimensional thin layer chromatography analysis of the ribonuclease T2-digested RISC-cleavage product. The oval depicts the unlabeled pAp as detected by UV shadowing. The radioactive signal comigrates with the 5′ 32pAp released by ribonuclease T2 digestion from the gel-purified 12-nt cleavage product.
The cleavage reaction of S21 is expected to yield a 9-nt 5′ cleavage product and a 12-nt 3′ cleavage product, but depending on the mechanism of target RNA cleavage, the radiolabeled phosphate might be found at either one of the two products. Comigration analysis indicated that the radiolabeled phosphate was present as a 5′-phosphate at the 12-nt 3′ cleavage product (Fig. 1B, lanes 2-4), indicating that the reaction proceeded via hydrolysis releasing cleavage products carrying 3′-hydroxyl and 5′-phosphate termini.
Thin-layer chromatography of the RNase T2-treated 12-nt cleavage product confirmed that the radiolabeled terminal nucleotide contained a 5′-phosphate based on comigration with synthetic adenosine 3′,5′-diphosphate (Fig. 1C).
In order to visualize both of the cleavage products simultaneously, we also 5′-32P-labeled the internally labeled substrate. As expected, the radiolabeled 9-nt 5′ cleavage product comigrated precisely with the independently 5′-32P-labeled 9-nt oligoribonucleotide (Fig. 1B, lanes 1,5), conclusively demonstrating that minimal RISC cleaves substrate RNA only at a single site. Release of the cleavage products in a more cellular environment would expose the cleavage fragments to further degradation by unspecific nucleases, including 3′ to 5′ exonucleases that can degrade RNAs with free 3′-hydroxyl but not 3′-phosphate termini (Schoenberg and Chernokalskaya 1997).
To determine the kinetic parameters of target RNA cleavage, initial velocities of S21 cleavage were determined as a function of substrate concentrations varied in the low nanomolar range (Supplementary Fig. S1). Fitting the initial rates to the Michaelis-Menten equation, a maximum velocity Vmax of 0.017 ± 0.002 nM/min and a Michaelis constant KM of 1.1 ± 0.3 nM were obtained (Supplementary Fig. S1A). Examination of a second independent preparation of RISC yielded a Vmax of 0.43 ± 0.02 nM/min, and a Michaelis constant KM of 2.3 ± 0.3 nM (Supplementary Fig. S1C). The differences in Vmax are due to differences in the concentration of RISC in the different preparations. Using the more active preparation of RISC, the fit of initial rates at substrate concentrations above 5 nM intersected with the ordinate at a concentration of ∼0.4 nM, which represents the concentration of active enzyme. The presence of this burst is consistent with product release being rate-limiting under our experimental conditions. It was not possible to measure the burst for the low-activity RISC preparation because of its more than 10-fold lower activity.
It is important to note that the cleavage reactions did not proceed to completion, not even at the lowest substrate concentration tested (0.02 nM). This could be explained by the presence of substrate-RNA-binding complexes that are not associated with a nuclease, or free single-stranded siRNA. Presumably, these copurified complexes compete irreversibly during the time course of the experiment for binding of the RNA substrate to active RISC. The IC50 concentration, at which a 2′-O-methyl oligoribonucleotide irreversibly blocked our low- and high-activity RISC was determined to be 0.4 and 0.7 nM, respectively (Hutvágner et al. 2004; Meister et al. 2004). The IC50 concentrations are higher than the estimated concentration of RISC by burst analysis, suggesting competitive binding of the substrate by a non-catalytic affinity-copurified single-stranded antisense siRNA or siRNA-protein complex.
For subsequent comparative analysis of modified substrates, we used the low-activity RISC and 1 nM constant substrate concentrations, and we stopped the reaction before plateau levels of substrate conversion were reached. Changes in the cleavage fraction therefore should reflect maximum changes in the cleavage rate.
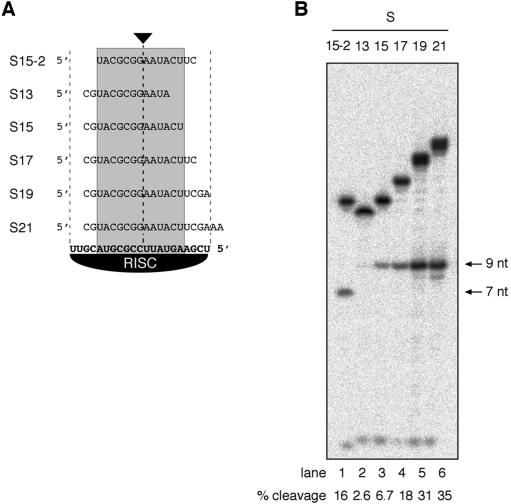
To determine the minimal length of substrate structurally required for cleavage by RISC, we prepared truncated substrates removing 2-nt segments from either the 3′ or the 5′ end (Fig. 2A). Deletion of the 3′-most residues, which are unpaired in the enzyme-substrate complex, reduced cleavage by a marginal 1.1-fold in S19 compared with S21 (Fig. 2B). Subsequent truncations from the 3′ end reduced cleavage by 2.0-fold for S17, 5.2-fold for S15, and 13-fold for S13. Removal of 2 nt from the 5′ end of S17 reduced the reaction rate 2.2-fold compared with S21 and 1.1-fold compared with S17. These data suggest that pairing of the central 13 nt of the guide siRNA of RISC is required for constituting the active site, or for promoting a structural change to constitute the active site. SiRNA residues comprising segments of up to 4 nt at either the 5′ or 3′ terminus can remain unpaired without causing a complete loss of cleavage activity. This observation is reminiscent of studies questioning the specificity of siRNA-guided target RNA degradation, reporting off-target cleavage activity for some substrates that contained only 11-nt segments of complementarity (Jackson et al. 2003).
Figure 2.
Substrates as short as 15-nt are cleaved by RISC. (A) Sequences of the truncated substrates. The 13-nt core sequence required for RISC-guided cleavage is shaded. (B) PhosphorImaging of cleavage reactions incubated for 1 h at 30°C and resolved on a 15% denaturing polyacrylamide gel. Substrate RNAs were radiolabeled only at the 5′ terminus. Arrows indicate the 7- and 9-nt 5′ cleavage product. The fraction of cleaved material is indicated at the bottom of the gel.
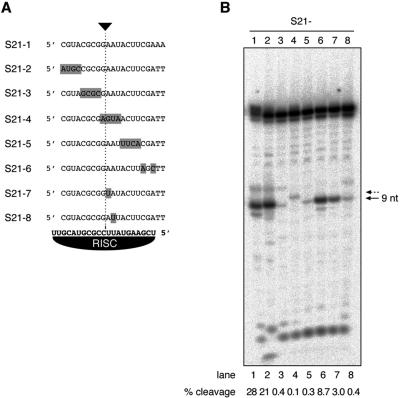
We therefore evaluated the importance of mismatches introduced into the S21 substrate (Fig. 3A). Mismatches were introduced by inverting segments of 3 or 4 nt, or by introducing point mutations. Four mismatches introduced at the 5′ end of the substrate (S21-2) reduced cleavage 1.3-fold, and two mismatches introduced at the 3′ end (S21-6) reduced cleavage 3.2-fold, compared to S21 (Fig. 3B, lanes 2,6). Mutation of the 4-nt segments closer to or overlapping the cleavage site (S21-3, 4, and 5) reduced cleavage at least 70-fold relative to S21 (Fig. 3B, lanes 3-5). The substrate S21-4, which is mismatched across the cleavage site, appears to be miscleaved by 1 nt (Fig. 3B, lane 4, see dotted arrow). This is an interesting observation, as it suggests that naturally expressed microRNAs, which pair imperfectly to their natural target mRNAs, may act partly by cleaving their target mRNAs and not only mediate translational regulation. Single-point mutations introduced proximal to the cleavage site reduced cleavage 9.3-fold for S21-7 and 70-fold for S21-8 relative to S21 (Fig. 3B, lanes 7,8). It is surprising to find that a mutation more proximal to the cleavage site (S21-7) had a less significant effect on cleavage than the mutation in S21-8 that was located 2 nt away from the cleavage site. Mutation and truncation analyses converge to a similar picture defining the minimal requirements for substrate targeting by RISC, and emphasize the need for devoting special attention to the identification of unique 13-nt sequence segments positioned at the center segment of the targeting siRNA in siRNA gene-targeting experiments.
Figure 3.
Specificity of target RNA cleavage. (A) Sequences of modified substrate RNAs. (B) PhosphorImaging of 1 h at 30°C cleavage reactions resolved on a 15% denaturing polyacrylamide gel. Substrate RNAs were radiolabeled at the 5′ terminus. The arrow indicates the 9-nt cleavage product, and the arrow with the dotted line indicates miscleavage guided by substrate S4 that contains a bulge over the cleavage site. The fraction of cleaved material is indicated at the bottom of the gel.
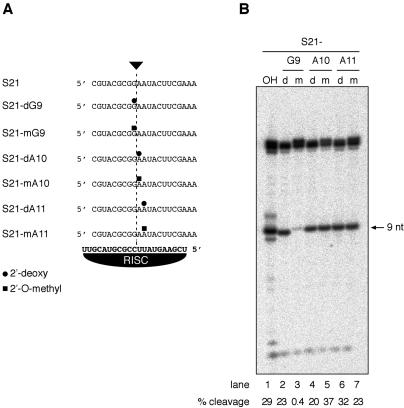
Chemical synthesis of short RNA substrates offers unique advantages for site-specific introduction of single-atom changes, such as the replacement of a single 2′-hydroxyl group by 2′-hydrogen or 2′-O-methyl. We prepared several substrates by substituting the 2′-ribose residue of S21 at G9, A10, or A11 by 2′-deoxy or 2′-O-methyl ribose (Fig. 4A). The cleaved phosphodiester bond is located between G9 and A10. Substitutions by 2′-deoxyribose caused a 1.3-fold reduction in cleavage for S21-dG9, 1.5-fold for S21-dA10, and a 1.1-fold increase for S21-dA11, relative to S21 (Fig. 4B, lanes 2,4,6). Substitutions by 2′-O-methylribose, however, caused a 73-fold reduction in cleavage for S21-meG9, but only a 1.3-fold increase for S21-meA10, and a 1.3-fold reduction for S21-meA11, relative to S21 (Fig. 4B, lanes 3,5,7). Consistent with the conversion of substrate observed in the single time point analysis (Fig. 4B), we did not observe any significant differences in Vmax and KM relative to unmodified S21, when we determined these parameters for S21-dG9 (Vmax = 0.012 ± 0.001 nM min-1, KM = 0.9 ± 0.1 nM, Supplementary Fig. S1B) or S21-dA10 (Vmax = 0.0088 ± 0.0002 nM min-1, KM = 0.9 ± 0.1 nM). Comparable relative rates were obtained using the high-activity RISC. Although individual 2′-deoxynucleotide substitutions in the substrate showed no significant perturbation of cleavage by minimal RISC, a substrate entirely composed of 2′-deoxynucleotide was not cleaved at all (Supplementary Fig. S2).
Figure 4.
Cleavage analysis of 2′-modified substrate RNAs. (A) Sequence and position of 2′-deoxy and 2′-O-methyl modified substrates. (B) PhosphorImaging of cleavage reactions incubated for 1 h at 30°C and resolved on a 15% denaturing polyacrylamide gel. Substrate RNAs were radiolabeled at the 5′ terminus. The arrow indicates the 9-nt cleavage product. The fraction of cleaved material is indicated at the bottom of the gel.
Modification of the ribose 2′ position affects the pKa of the 3′-hydroxyl, and therefore should also affect the rate of hydrolysis of the phosphodiester bond (Izatt et al. 1965; Herschlag et al. 1993; Smith and Pace 1993; Yoshida et al. 2000); the pKa values reported for ribose and 2′-deoxyribose are 12.4 and 15, respectively. The less acidic 3′-hydroxyl of 2′-deoxyribose is expected to be a worse leaving group than the 3′-hydroxyl of regular ribose. However, no significant change in the rate of hydrolysis occurred upon 2′-deoxy modification at the cleavage site, indicating that product release and/or a conformational change rather than the rate of chemical cleavage is limiting for catalysis by minimal RISC. It is conceivable that RNA helicases or RNP remodeling activities (Will and Lührmann 2001), which presumably were removed by our purification protocol, participate in such a transition and promote RISC activity in a more natural environment. In contrast to the 2′-deoxy modification, introduction of the bulky, but chemically inert 2′-O-methyl group at the cleavage site significantly reduced the substrate cleavage, presumably due to steric interference with either a conformational transition or positioning catalytic residues or metal ions at the active site.
The cleavage mechanism of RISC shares molecular features characteristic of dsDNA restriction enzymes (Pingoud and Jeltsch 2001) and RNase III enzymes (Dunn 1982), and is unrelated to conventional RNase A-type endonucleases that cleave via 2′, 3′-cyclic phosphate intermediates and leave a hydrolyzed 3′-phosphomonoester and a free 5′-hydroxyl (Usher et al. 1970). Although these findings might suggest that Dicer RNase III, which is involved in generating siRNAs from dsRNA precursors, might be a good candidate nuclease for RISC, this is unlikely, because cytoplasmic extracts effectively depleted of Dicer were still able to reconstitute RISC (Martinez et al. 2002). In addition, the 220-kD molecular mass of human Dicer (Provost et al. 2002; Zhang et al. 2002) appears too large to be possibly present in our RISC preparation of an estimated apparent molecular mass of 160 kD, that also has to account for a 100-kD Argonaute protein and a 21-nt single-stranded RNA.
A nuclease related to micrococcal nuclease, Tudor SN, was recently reported in association with RISC, and the bacterial recombinantly expressed protein was shown to cleave DNA and RNA nonspecifically (Caudy et al. 2003). Micrococcal nucleases degrade DNA and RNA, however, by generating 3′-phosphonucleotides and 3′-phosphodinucleotides (Reddi 1959; Alexander et al. 1961). The initial cleavage by micrococcal nuclease is endonucleolytic, and is followed by exonucleolytic degradation of the resulting oligonucleotides (Alexander et al. 1961; Williams et al. 1961; Sulkowski and Laskowski 1962). This argues against Tudor SN being the core endonucleolytic component of RISC. This specific example illustrates the importance of defining the mechanism of RISC-endonucleolytic cleavage, and how mechanistic studies provide constraints for the critical evaluation of potential candidate nucleases that may soon emerge from genetic or biochemical studies aimed at uncovering this key RNAi component. The described biochemical assays will also guide future investigation to uncover the role of RISC and RISC-associated proteins.
Materials and methods
Oligonucleotide synthesis
Oligoribonucleotides and modified oligoribonucleotides were chemically synthesized using 5′-silyl, 2′-ACE phosphoramidites, 5′-silyl, 2′-deoxy phosphoramidites, and 5′-silyl, 2′-O-methyl phosphoramidites (Dharmacon Research) on 0.2 μmol synthesis columns using a modified ABI 394 synthesizer (Scaringe 2001). The aminolinker-modified 12-nt RNA was synthesized using a 3′ C6-aminolinker synthesis column (Dharmacon). The phosphate methyl group was removed by flushing the column with 2 mL of 0.2 M 2-carbamoyl-2-cyanoethylene-1,1-dithiolate trihydrate in DMF/water (98:2 v/v) for 30 min at room temperature. The reagent was removed and the column rinsed with 10 mL water followed by 10 mL acetonitrile. Oligonucleotides were cleaved and eluted from the solid support by flushing with 1.9 mL of 40% aqueous methylamine over 2 min, collected in a screw-cap vial, and incubated for 10 min at 55°C. Subsequently, base-treated oligonucleotides were dried down in an Eppendorf concentrator to remove methylamine and water. The residues were dissolved in sterile 2′-deprotection buffer (400 μL of 100 mM acetate-TEMED at pH 3.8 for a 0.2 μmole scale synthesis) and incubated for 30 min at room temperature and another 30 min at 60°C to remove the 2′ ACE group. Oligonucleotides were precipitated from the acetate-TEMED solution by adding 24 μL 5 M NaCl and 1.2 mL of absolute ethanol. Pellets were dissolved in 500 μL of water. DNA oligonucleotides were prepared using 0.2 μmole scale synthesis and standard DNA synthesis reagents (Proligo). Synthesis of 3′ C7-aminolinker-containing siRNAs and biotin conjugation were performed as described (Martinez et al. 2002). Annealing of siRNA duplexes was performed as described (Elbashir et al. 2002).
Labeling of oligoribonucleotide substrates
5′ labeling reactions contained 10 pmol oligonucleotide, 5 pmol γ-32PATP (Amersham, 3000 Ci/mmol), 1 unit T4 polynucleotide kinase (New England Biolabs), and 10 mM MgCl2/5 mM dithiothreitol/70 mM Tris-HCl (pH 7.6) in a final volume of 10 μL. The reaction mixture was incubated for 20 min at 37°C, followed by the addition of 1 μL 100 mM ATP and incubation for another 3 min at 37°C. The reaction was stopped by the addition of 1 volume urea stop solution (8 M urea, 0.05% w/v Bromophenol Blue, 50 mM EDTA at pH 8.0) and 1 μg carrier tRNA. The samples were heat-treated for 2 min at 90°C and separated on a 20% denaturing polyacrylamide gel. The radioactive product bands were excised and eluted overnight at 4°C into 300 μL 0.3 M NaCl containing 1 μg carrier tRNA. To optimize the recovery of the 9- and 12-nt oligoribonucleotides, gel elution was performed using 1 M Na acetate (pH 6.0) containing 1 μg of carrier tRNA. The oligonucleotides were precipitated by the addition of 3 volumes absolute ethanol, collected by centrifugation, and dissolved in water.
RNA ligation
Site-specific internally labeled substrate RNA was prepared by incubating 2 pmol 5′-32P-phosphorylated and linker-modified 12-nt RNA with 20 pmol 9-nt RNA, and 40 units of T4 RNA ligase (Amersham) in 10 mM MgCl2/10 mM 2-mercaptoethanol/50 mM Tris-HCl (pH 7.6)/0.1 mg/mL acetylated bovine serum albumin (Ac-BS, Sigma)/0.2 mM ATP/15% DMSO in a final volume of 20 μL. The reaction mixture was incubated for 3 h at 37°C. The reaction was stopped by the addition of 1 volume urea stop solution and 1 μg of carrier tRNA. The samples were heat-treated for 2 min at 90°C and separated on a 20% denaturing polyacrylamide gel. The ligation product was recovered from the gel as described above, and eluted in the presence of 50 ng of carrier tRNA.
Affinity purification of RISC
RISC was affinity-purified as described using the siRNA duplex containing biotin at both of its 3′ ends (Martinez et al. 2002). The concentration of siRNA duplex was 2 nM during RISC assembly in HeLa cell cytoplasmic extract. The streptavidin resin was washed with increasing concentrations of KCl (0.1-2.5 M) after binding of RISC. The complex was cleaved from the resin by irradiating at 302 nm and recovered in 0.1 M KCl. The affinity-purified RISC only targets sense substrates for degradation, not antisense substrates, indicating completely asymmetric assembly of RISC on the antisense siRNA.
Target RNA cleavage assay
Cleavage assays were performed as described (Martinez et al. 2002), but adjusting the concentration of MgCl2 to 5 mM and omitting phosphocreatine and creatine phosphokinase. Cleavage by RISC is magnesium ion-dependent and plateaus between 2 and 10 mM MgCl2. The concentration of carrier tRNA in the target RNA substrates was adjusted to 10 ng/μL. Cleavage assays were performed using 1 nM concentration of substrate, except for measuring kinetic constants, where the substrate concentration was then varied from 0.02 to 20 nM. Substrate RNA dilutions were heat-treated for 40 sec at 90°C and shortly chilled on ice prior to setting up the reactions. Aliquots were withdrawn at the indicated time points, and the reaction was stopped by the addition of 1 volume of urea-stop solution. To improve the gel resolution of the RNA cleavage products shown in Figure 1, a 100-fold excess of 12-nt RNA relative to substrate was added to the gel loading solution to act as competitor for the radiolabeled product. The gel loading solution was incubated for 2 min at 90°C and then loaded on the gel.
Ribonuclease T2 digestion and two-dimensional thin layer chromatography
The radioactive 12-nt cleavage product shown in Figure 1A was excised from a 20% denaturing polyacrylamide gel, eluted and redissolved in 3.5 μL of water, and digested with 2 units of ribonuclease T2 (Invitrogen) in 50 mM sodium acetate (pH 4.5) for 1 h at 37°C. Samples was cospotted with 1 μg of nonradioactive adenosine 3′,5′-diphosphate (Sigma, catalog no. A-5763) on cellulose HPTLC plates (Merck, catalog no. 1.05787) and separated in the first dimension in isobutyric acid/ammonia 32%/water (577:38:385) for ∼8 h. The TLC plate was dried overnight at room temperature, and the separation in the second dimension was performed using tert-butanol/concentrated hydrochloric acid (37.6%)/water (14:3:3) for ∼8 h. The position of migration of the nonradioactive pAp was detected by UV-shadowing, and the position of the T2-digested radioactive product was determined by PhosphorImaging.
Note added in proof
While this manuscript was under revision, a study by Schwarz et al. (2004) also reported on the endonuclease mechanism of RISC using D. melanogaster lysates.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Acknowledgments
We thank P.Y. Chen and J. Meyer for oligonucleotide synthesis and technical assistance; R. Lührmann (Max-Planck-Institute for Biophysical Chemistry, Göttingen, Germany) for providing HeLa cytoplasmic extracts; M. Konarska for advice, and H. Dormann, W. Fischle, and all members of our laboratory for discussions and comments on the manuscript. This work was supported by NIH grant R01 GM068476-01.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1187904.
Supplemental material is available at http://www.genesdev.org.
References
- Alexander M., Heppel, L.A., and Hurwitz, J. 1961. The purification and properties of micrococcal nuclease. J. Biol. Chem. 236: 3014-3019. [PubMed] [Google Scholar]
- Amarzguioui M., Holen, T., Babaie, E., and Prydz, H. 2003. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 31: 589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. [DOI] [PubMed] [Google Scholar]
- Braasch D.A., Jensen, S., Liu, Y., Kaur, K., Arar, K., White, M.A., and Corey, D.R. 2003. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 42: 7967-7975. [DOI] [PubMed] [Google Scholar]
- Capodici J., Kariko, K., and Weissman, D. 2002. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 169: 5196-5201. [DOI] [PubMed] [Google Scholar]
- Caudy A.A., Myers, M., Hannon, G.J., and Hammond, S.M. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 16: 2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy A.A., Ketting, R.F., Hammond, S.M., Denli, A.M., Bathoorn, A.M., Tops, B.B., Silva, J.M., Myers, M.M., Hannon, G.J., and Plasterk, R.H. 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411-414. [DOI] [PubMed] [Google Scholar]
- Cerutti H. 2003. RNA interference: Traveling in the cell and gaining functions? Trends Genet. 19: 39-46. [DOI] [PubMed] [Google Scholar]
- Chiu Y.L. and Rana, T.M. 2003. siRNA function in RNAi: A chemical modification analysis. RNA 9: 1034-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna F., Fechtner, M., Dames, S., Aygun, H., Klippel, A., Pronk, G.J., Giese, K., and Kaufmann, J. 2003. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 31: 2705-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli A.M. and Hannon, G.J. 2003. RNAi: An ever-growing puzzle. Trends Biochem. Sci. 28: 196-201. [DOI] [PubMed] [Google Scholar]
- Dunn J.J. 1982. Ribonuclease III. In The enzymes (ed. P.D. Boyer), Vol. 15, part B, pp. 485-499. Academic Press, New York. [Google Scholar]
- Dykxhoorn D.M., Novina, C.D., and Sharp, P.A. 2003. Killing the messenger: Short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 4: 457-467. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel, W., and Tuschl, T. 2001a. RNA interference is mediated by 21 and 22 nt RNAs. Genes & Dev. 15: 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Martinez, J., Patkaniowska, A., Lendeckel, W., and Tuschl, T. 2001b. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20: 6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth, J., Weber, K., and Tuschl, T. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26: 199-213. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293-296. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146-1150. [DOI] [PubMed] [Google Scholar]
- Harborth J., Elbashir, S.M., Vandenburgh, K., Manninga, H., Scaringe, S.A., Weber, K., and Tuschl, T. 2003. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 13. [DOI] [PubMed]
- Herschlag D., Eckstein, F., and Cech, T.R. 1993. The importance of being ribose at the cleavage site in the Tetrahymena ribozyme reaction. Biochemistry 32: 8312-8321. [DOI] [PubMed] [Google Scholar]
- Holen T., Amarzguioui, M., Babaie, E., and Prydz, H. 2003. Similar behaviour of single-strand and double-strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 31: 2401-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056-2060. [DOI] [PubMed] [Google Scholar]
- Hutvágner G., Simard, M.J., Mello, C.C., and Zamore, P.D. 2004. Sequence-specific inhibition of small RNA function. PLoS Biol. 2: E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izatt R.M., Hansen, L.D., Rytting, J.H., and Christensen, J.J. 1965. Proton ionization from adenosine. J. Am. Chem. Soc. 87: 2760-2761. [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G., and Linsley, P.S. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotech. 21: 635-637. [DOI] [PubMed] [Google Scholar]
- Liu Q., Rand, T.A., Kalidas, S., Du, F., Kim, H.E., Smith, D.P., and Wang, X. 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921-1925. [DOI] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska, A., Urlaub, H., Lührmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563-574. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler, M., Dorsett, Y., and Tuschl, T. 2004. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10: 544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykänen A., Haley, B., and Zamore, P.D. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309-321. [DOI] [PubMed] [Google Scholar]
- Pingoud A. and Jeltsch, A. 2001. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 29: 3705-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P., Dishart, D., Doucet, J., Frendewey, D., Samuelsson, B., and Radmark, O. 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21: 5864-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi K.K. 1959. Degradation of tobacco mosaic virus nucleic acid with micrococcal phosphodiesterase. Biochim. Biophys. Acta 36: 132-142. [DOI] [PubMed] [Google Scholar]
- Scaringe S.A. 2001. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods 23: 206-217. [DOI] [PubMed] [Google Scholar]
- Schoenberg D.R. and Chernokalskaya, E. 1997. Ribonucleases involved in eukaryotic messenger RNA turnover. In mRNA metabolism and post-transcriptional gene regulation (eds. J.B. Harford and D.R. Morris), pp. 217-240. Wiley, New York.
- Schwarz D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P.D. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199-208. [DOI] [PubMed] [Google Scholar]
- Schwarz D.S., Tomari, Y., and Zamore, P.D. 2004. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr. Biol. (in press). Immediate Early Publication published online March 11, 2004, 10.1016 [DOI] [PubMed]
- Smith D. and Pace, N.R. 1993. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry 32: 5273-5281. [DOI] [PubMed] [Google Scholar]
- Sulkowski E. and Laskowski Sr., M. 1962. Mechanism of action of micrococcal nuclease on deoxyribonucleic acid. J. Biol. Chem. 237: 2620-2625. [PubMed] [Google Scholar]
- Usher D.A., Richardson Jr., D.I., and Eckstein, F. 1970. Absolute stereo chemistry of the second step of ribonuclease action. Nature 228: 663-665. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Lührmann, R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13: 290-301. [DOI] [PubMed] [Google Scholar]
- Williams E.J., Sung, S.C., and Laskowski Sr., M. 1961. Action of venom phosphodiesterase on deoxyribonucleic acid. J. Biol. Chem. 236: 1130-1134. [PubMed] [Google Scholar]
- Yoshida A., Shan, S., Herschlag, D., and Piccirilli, J.A. 2000. The role of the cleavage site 2′-hydroxyl in the Tetrahymena group I ribozyme reaction. Chem. Biol. 7: 85-96. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25-33. [DOI] [PubMed] [Google Scholar]
- Zhang H., Kolb, F.A., Brondani, V., Billy, E., and Filipowicz, W. 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 21: 5875-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]