Abstract
Although antipsychotic drugs can reduce psychotic behavior within a few hours, full efficacy is not achieved for several weeks, implying that there may be rapid, short-term changes in neuronal function, which are consolidated into long-lasting changes. Here, we showed that the antipsychotic drug haloperidol, a dopamine receptor type 2 (D2R) antagonist, stimulated the kinase Akt to activate the mRNA translation pathway mediated by the mammalian target of rapamycin complex 1 (mTORC1). In primary striatal D2R-positive neurons, haloperidol-mediated activation of mTORC1 resulted in increased phosphorylation of ribosomal protein S6 (S6) and eukaryotic translation initiation factor 4E-binding protein (4E-BP). Proteomic mass spectrometry revealed marked changes in the pattern of protein synthesis after acute exposure of cultured striatal neurons to haloperidol, including increased abundance of cytoskeletal proteins and proteins associated with translation machinery. These proteomic changes coincided with increased morphological complexity of neurons that was diminished by inhibition of downstream effectors of mTORC1, suggesting that mTORC1-dependent translation enhances neuronal complexity in response to haloperidol. In vivo, we observed rapid morphological changes with a concomitant increase in the abundance of cytoskeletal proteins in cortical neurons of haloperidol-injected mice. These results suggest a mechanism for both the acute and long-term actions of antipsychotics.
Introduction
Antipsychotics were developed in the late 1950s and currently are used to treat psychosis associated with schizophrenia and refractory depression. First-generation typical antipsychotics, such as haloperidol, primarily antagonize dopamine receptor type 2 (D2R) in the brain. Although many second-generation antipsychotics that antagonize both D2R-like and the serotonin 5HT2-like receptors have been introduced, the initial reports of improved efficacy of these drugs have been questioned (1). Antipsychotics of both groups mainly target positive symptoms of schizophrenia, such as hallucinations and delusions, and their ameliorative effect on these psychotic behaviors can begin as early as two hours after treatment, with distinct improvement seen at 24 hours (2,3). However, full efficacy in patients is not achieved until after three or more weeks of treatment (4), implying that there may be rapid, short-term changes in neuronal function, which are then consolidated into enduring modifications over time (5).
One of the signaling effects of antipsychotics is increased phosphorylation of the kinase Akt [also known as protein kinase B (PKB)], an indication of increased kinase activity (6,7). Increased Akt phosphorylation was measured within two hours of injection of haloperidol (6). Most studies that investigated the effects of D2R antagonists have focused on the phosphorylation and inhibition of glycogen synthase kinase 3β (GSK3β) by Akt, because this signaling pathway is implicated in other behavioral disorders, such as bipolar disorder (8). However, phosphorylation of GSK3β is enhanced by lithium treatment, which has limited efficacy for schizophrenia (9), suggesting that GSK3β phosphorylation may not entirely explain the mechanism of the antipsychotic action of haloperidol.
Long-lasting changes in synaptic function are tightly regulated by transsynaptic signaling and dynamic changes in dendritic protein synthesis (10). One well-described regulator of protein synthesis, including of synaptic proteins involved in synaptic signaling, is the Akt-mTORC1 (mammalian or mechanistic target of rapamycin complex 1) pathway. mTORC1-dependent translation has been implicated in synaptic plasticity, memory consolidation, and autism (11–14). Akt activation relieves inhibition of mTORC1 activity, which in turn promotes cap-dependent translation by phosphorylating and inhibiting 4E-BP. p70 S6 kinase 1 (S6K1), another downstream effector of mTORC1, phosphorylates ribosomal protein S6, which also is associated with increased translation (15,16). Here, we asked whether the Akt-mTORC1 pathway was involved in the neuronal response to antipsychotics.
We analyzed the acute effects of haloperidol on Akt signaling and on the mTORC1 effectors S6 and 4E-BP and subsequent proteomic changes in cultured striatal neurons. We identified proteins synthesized within the first 48 hours of exposure to haloperidol and found that proteins associated with the cytoskeleton and components of the protein synthesis machinery were increased. We also observed increased morphological complexity; in particular increased neuronal projection morphological complexity, which was dependent on mTORC1 effectors. In addition to increases in morphological complexity, we observed an increase in the number of spines in vitro in striatal neurons and in spine formation in vivo in layer 5 cortical pyramidal neurons 24 hours after haloperidol administration. Thus, activation of the Akt-mTORC1 pathway by haloperidol leads to discrete changes in protein synthesis associated with overall increased morphological complexity, representing a previously uncharacterized mechanism for the action of antipsychotics.
Results
Haloperidol increases Akt-mTORC1 signaling
Typical antipsychotics antagonize D2R,, which results in an increase in the phosphorylation of Akt (7,17). Therefore, we asked whether the antipsychotic haloperidol increased Akt phosphorylation in primary striatal neuron cultures at time points shorter than two hours. We focused on striatal neurons because the D2R is abundant in these neurons (18). Haloperidol was selected as a model drug because its binding affinity and clinical profile with regard to D2R are well established (19). We chose a concentration of 20 nM based on its binding affinity and clinical potency profile in human patients (20), and confirmed that this concentration was effective at stimulating phosphorylation of Akt in mouse primary striatal neurons (fig. S1A–B). A 20-minute treatment of 7-days-in-vitro (DIV7) primary striatal neurons with 20 nM haloperidol significantly increased the phosphorylation of Akt at Ser473 (Fig. 1A), which was abrogated by the membrane-permeable Akt PH domain inhibitor, Akti (21).
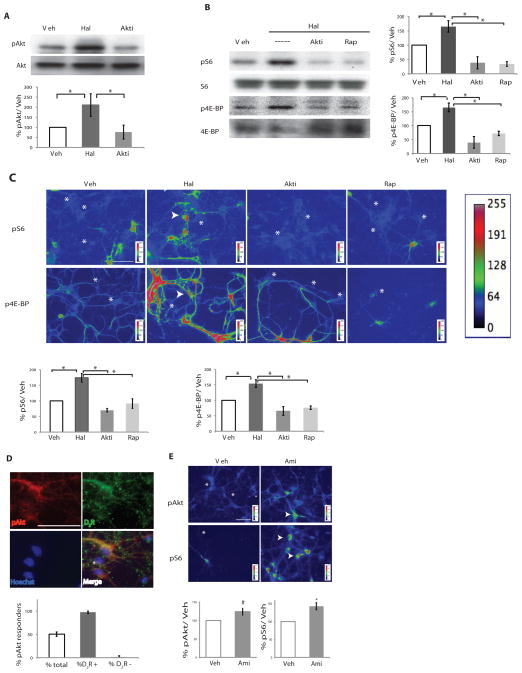
Figure 1. Acute antipsychotic treatment increases Akt-mTORC1 pathway signaling in D2R-positive neurons.
Primary DIV7 mouse striatal neurons were treated with haloperidol (Hal) or DMSO (Veh), in the presence of an Akt inhibitor (Akti) or rapamycin (Rap) for 20 minutes. (A) Western blot analysis of phosphorylated and total Akt in lysates exposed to the indicated conditions (left, representative blot; right quantification of n =4, One Way ANOVA p < 0.0001, post hoc analysis Veh vs Hal p <0.01, Hal vs Akti, p < 0.001). (B) Western blot analysis of phosphorylated and total S6 and 4E-BP in lysates exposed to the indicated conditions [left, representative blot; right quantification of S6 (n=3, One Way ANOVA p < 0.0008, Veh vs Hal p < 0.05, Hal vs Akti and Hal vs Rap p < 0.01) and 4E-BP (n = 3, One Way ANOVA p < 0.0003, Veh vs Hal p <0.05, Hal vs Akti p < 0.001, Hal vs Rap p <0.01). All graphs shown are average ± SEM. (C) Changes in the abundance of the indicated phosphorylated proteins in striatal neurons exposed to the indicated treatments as detected by immunofluorescence. Arrows indicate responding neurons, stars indicate cells that are not responding. Quantification of pS6 (n= 4, One Way ANOVA p < 0.05, all indicated single comparisons p < 0.05) and p4E-BP (n=4, One Way ANOVA p < 0.0001, Veh vs Hal p < 0.01, Hal vs Akti and Hal vs Rap p < 0.001). (D) D2Rs are detected in nearly all cells with increased pAkt (arrow). A nonresponding cell is indicated by asterisk. Quantification of responding cells and D2R-positive (D2R+) and D2R-negative (D2R−) cells (n=3, 109 cells counted). D2R image underwent Iterative deconvolution in ImageJ to increase clarity of signal. (E) The abundance of phosphorylated Akt and S6 in striatal neurons exposed to amisulpride (1 μM) or vehicle for 20 minutes (pAkt, n = 3, p < 0.06; pS6, n = 3, p < 0.03). Scale bars indicate 50 μm.
In conjunction with Akt activation, haloperidol increased the phosphorylation of S6 and 4E-BP, in DIV7 striatal neurons as shown by Western blot (Fig. 1B) and by immunofluorescence (Fig. 1C). Pretreatment of the striatal neurons with either Akti or the mTORC1 inhibitor rapamycin attenuated the number of neurons and the intensity of the haloperidol-induced increase in the phosphorylation of S6 and 4E-BP (Figs. 1B and 1C). Only a subset of cells (40%) responded to haloperidol treatment (Fig. 1D), and these were identified as the D2R-positive striatal neurons (Fig. 1D). That only D2R-positive neurons responded to haloperidol with phosphorylation of Akt also was confirmed in striatal cells derived from a D2R-EGFP transgenic mouse (95% overlap, fig. S1C).
To test whether the observed Akt activation was specific to haloperidol, we tested 1 μM amisulpride (Fig. 1E), an atypical antipsychotic drug with a different affinity profile, and found that this concentration of amisulpride also increased phosphorylated Akt and phosphorylated S6, indicating activation of the mTORC1 pathway. Thus, haloperidol and amisulpride activated the Akt-mTORC1 pathway and its downstream effectors of translation within 20 minutes, suggesting a role for the mTORC1 pathway in the acute mechanism of action of typical and atypical antipsychotics. Furthermore, the response was specifically enriched in a subset of D2R-positive striatal neurons, indicating that this receptor likely mediates the effect of these drugs on protein synthesis.
Haloperidol enhances protein synthesis in cultured striatal neurons
To test whether the increases in the phosphorylation of ribosomal S6 and 4E-BP were coincident with enhanced protein synthesis (15), we utilized SUnSET, a nonradioactive method of monitoring global protein synthesis in cultured cells that uses puromycin to tag nascent proteins (13, 22–23). Puromycin is covalently integrated into a nascent polypeptide chain in the place of a tRNA, thereby “tagging” newly synthesized proteins, which can then be detected with a monoclonal antibody against puromycin. To determine the optimal concentration of puromycin for these experiments, we assayed puromycin incorporation in the presence and absence of haloperidol and found that 1 μg/ml produced the best signal with a greater proportion of large molecular weight proteins labeled, indicating minimal protein truncation and degradation (fig. S2A). Time-course experiments showed that puromycin incorporation was significantly increased in haloperidol-treated samples within 30 minutes and was still increased at one hour, before returning to match vehicle-treated samples by four hours (Fig. 2A). Thus, haloperidol transiently increased protein synthesis in striatal neurons.
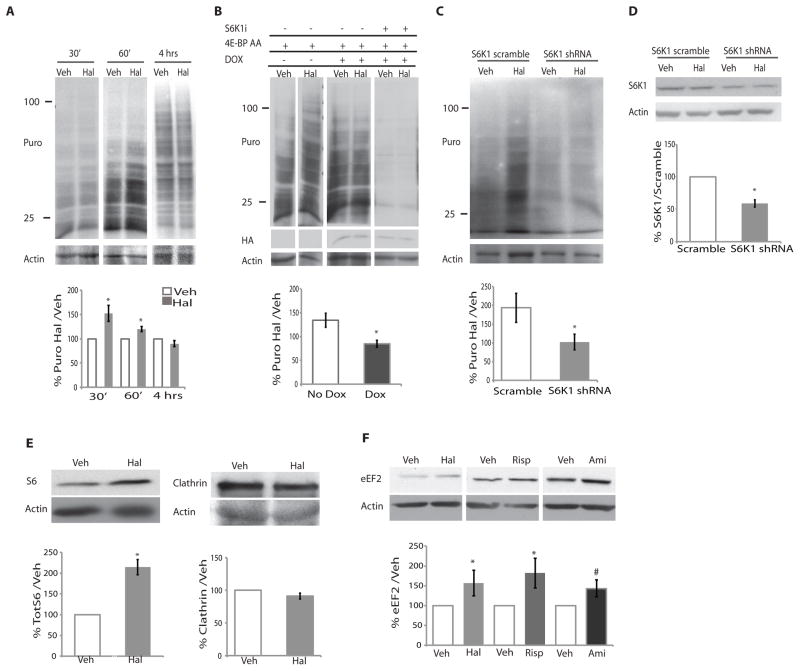
Figure 2. Acute haloperidol treatment leads to transient increases in protein synthesis that are mTORC1 dependent.
(A) Primary DIV7 striatal neurons were treated with haloperidol (Hal) or DMSO (Veh) for 30, 60 minutes, or 4 hours and 1 μg/ml puromycin. Puromycin was detected by Western blotting. A representative blot is shown. The 4-hr blot is a lighter exposure due to the intensity of the signal. Data were quantified as the percent of puromycin signal in the haloperidol-treated samples / puromycin signal in vehicle–treated samples normalized to actin and shown as the average (30 min; n=8, p =0.007; 60 min, n=7, p=0.005; 4 hours, n=4, not significant). Actin is a loading control. (B) The effect of S6K1 shRNA or the doxycycline-inducible (DOX) dominant-negative 4E-BP AA on haloperidol-mediated stimulation of translation. Data were quantified as in panel A for the 4E-BP AA “on” (Dox; n=4) and “off” (No Dox; n=3) conditions. * p=0.02). Doxycycline with and without S6K1 inhibitor (S6K1i), n = 3, not quantified due to comparatively light signal. HA indicates the presence of 4E-BP AA. Actin is a loading control. (C) The effect of S6K1 knockdown (S6K1 shRNA) or control (S6K1 scramble) on the haloperidol-induced increase in puromocyin incorporation. Data are quantified as in panel A. (n=4, p=0.03,). (D) Effectiveness of S6K1 knockdown was quantified for each experiment, knockdown average was 41% (n= 4 p < 0.0003). (E) Western blot analysis of S6 and clathrin abundance in striatal neurons exposed to vehicle or haloperidol under the same conditions used for the puromycin analysis (% Haloperidol/Vehicle normalized to actin loading control). (S6, n=3, p=0.003) (clathrin, n=4, p=not signficant). (F) Western blot analysis of eEF2 abundance in striatal neurons exposed to vehicle or haloperidol (20 μM, n=7, p=0.027), risperidone (Risperidone 100 nM, n=,5, p < 0.06), or amisulpride (Ami, 1 μM, n = 5, p = 0.08) for 4 hours. All graphs shown are average ± SEM and analyzed by Student’s t test. P < 0.05 was considered significant and is marked with asterisks; P< 0.1 is marked with #.
To probe whether the haloperidol-induced increase in translation depended on mTORC1 signaling, we blocked two well-characterized effectors of mTORC1: eukaryotic initiation factor 4E (eIF4E), which is activated when 4E-BP is phosphorylated by mTORC1, and S6K1, a direct mTORC1 substrate. By expressing a doxycycline-inducible phosphorylation-deficient mutant of 4E-BP1 (4E-BP AA) that acts as a constitutively active eIF4E repressor (24) in striatal neurons, we found that the haloperidol-mediated increase in translation was reduced by expression of 4E-BP AA after doxycycline treatment (Fig. 2B). In contrast, in the absence of doxycycline, there was no inhibition of haloperidol-mediated puromycin incorporation (fig. S2B). In both the vehicle and haloperidol conditions, protein synthesis was further reduced upon treatment of neurons expressing 4E-BP AA with an S6K1 inhibitor PF-4708671 (Fig. 2B) (25). To independently examine the role of S6K1 in the haloperidol-induced increase in translation, we utilized a shRNA targeting S6K1 (24). S6K1 knockdown also reduced the haloperidol-induced increase in protein synthesis in primary striatal neuron cultures (Fig. 2C; Fig. 2D).
We investigated how the increase in mTORC1 activation contributed to the mechanism of haloperidol action in affecting neuronal function. One possibility is that mTORC1 activation and the subsequent increase in translation lead to synthesis of specific proteins, rather than just a global increase in translation. Indeed, in mouse embryonic fibroblasts, stimulation of mTORC1 for two hours is sufficient to alter the distribution of mRNA on ribosomes, and thus potentially influence translation of specific transcripts (26). Because proteomic changes also occur following long-term antipsychotic treatment in vivo (27–28), we predicted that early changes in relative protein abundance might accumulate over time to result in large-scale proteomic changes similar to those described after chronic antipsychotic treatment (27–28). To determine whether haloperidol stimulated the production of specific proteins, we combined two techniques. We used stable isotope labeling with amino acids in cell culture (SILAC) to measure relative protein amounts (29) combined with SUnSET to enrich for newly synthesized proteins (23), which enabled the labeling, isolation, and quantification of nascent proteins produced immediately following haloperidol treatment. The specificity of the anti-puromycin antibody for immunoprecipitation was validated by stimulating neurons with an endogenous neurotrophic ligand, brain-derived neurotrophic factor (to stimulate protein synthesis), in the presence and absence of puromycin and showing that the signal was limited to the cells exposed to puromycin (fig. S2C). Additionally, the antibody immunoprecipitated proteins after the cells had been exposed to puromycin for 4 hours (fig. S2D) and the signal was sufficiently strong to enable detection of individual newly synthesized proteins, such as S6, in the immunoprecipitates after a 4-hour exposure to puromycin (fig. S2E).
Protein synthesis increased in cultured striatal neurons after incubation for 5 hours with puromcyin and SILAC labeling and immunoprecipitation with the puromycin-targeted antibody enriched the amount of SILAC-labeled newly synthesized proteins (fig. S3A). Haloperidol treatment produced a detectable shift in the synthesis of a subset of specific proteins (fig. S3A). We identified 269 puromycin-labeled (newly synthesized) proteins in cells that increased in response to a 5-hour exposure to haloperidol and 139 proteins that decreased in abundance (table S1). In addition, after haloperidol treatment and subsequent puromycin immunoprecipitation, we detected an increase in the amount of S6, a protein identified as increased in the 5-hour screen (fig. S2E), suggesting that this technique could be used to evaluate changes in nascent protein synthesis. Because a single dose of antipsychotics can produce changes in behavior within two hours and these changes can become more pronounced after 24 hours (2,3), we also performed analysis of SILAC-labeled and puromycin-enriched proteins from striatal neurons exposed to haloperidol or vehicle for 48 hours (fig. S3B). We identified 3209 proteins measured at least once in the 48-hour study (table S2). We compared our proteomic results from the 5-hour and 48-hour study with mTORC1-specific mRNAs identified in a previous study (26) and noted substantial similarities between proteins that were upregulated with haloperidol treatment in our study and those mRNAs that have decreased ribosomal binding in the presence of a mTORC1 inhibitor, suggesting positive regulation by mTORC1 (table S3). Taken together, these data suggest that haloperidol can increase the abundance of specific mTORC1-regulated proteins.
To validate the results of the mass spectrometry analysis of neurons exposed to haloperidol, we exposed cultured striatal neurons to haloperidol for 4 hours and monitored changes in S6 and clathrin (Fig. 2E), and in eEF2 (Fig. 2F) by Western blot. Consistent with the mass spectrometry results (table S1), we found increased amounts of S6 (Fig. 2E) and eEF2 (Fig. 2F) with haloperidol, but no increases in clathrin (Fig. 2E). To address whether the increase in eEF2 was specific to haloperidol, DIV7 striatal neurons were treated for 4 hours with two atypical antipsychotics, amisulpride or risperidone. Both drug treatments resulted in an increase in eEF2 (Fig. 2E). Taken together, these findings indicated that haloperidol, as well as amisulpride and risperidone, increased the synthesis of components of the translational machinery in cultured striatal neurons.
Mass spectrometry analysis reveals that proteins associated with translation and the cytoskeleton are produced in response to haloperidol in vitro and in vivo
The proteomic analysis of the neurons exposed to haloperidol for 48 hours was performed with three independent replicates. We used three different stringencies to identify proteins that increased in response to 48-hour haloperidol treatment (table S4). Consistent with the 5-hour screen, which identified proteins associated with translation, the 48-hour mass spectrometry study (Fig. 3A) also revealed changes protein abundance and identified several changes in proteins involved in the cytoskeleton (Fig. 3B). Comparing the 5-hour screen results with the 48-hour study data showed that at the later time there were fewer large differences between haloperidol and control samples than were observed in the samples from the 5-hour haloperidol exposure, suggesting that the vehicle and haloperidol-treated neurons had more similar translation profiles at 48 hours (compare Fig. 3A with fig. S3A).Proteins were described as “candidates” if the normalized ratio of the protein (fold change) was at least 1.2 (amount of protein measured with haloperidol treatment/amount of protein measured with vehicle treatment) [criteria based on (29–31)]. We refer to these proteins that increased at 1.2-fold under the least stringent conditions as candidate proteins, because they responded consistently to a change in protein synthesis induced by antipsychotics even though they did not meet the most stringent criteria. Candidate proteins that were classified as translation, cytoskeleton, mRNA processing, membrane, or release-related are listed in table S4 (see Materials and Methods for explanation of the categories). Using the most stringent criteria of a 1.2-fold increase in at least 2 experiments, we found that 17 of the 44 proteins that increased were associated with the cytoskeleton and 10 were associated with translation (Table 1). The candidate proteins appeared to be specific because not all cytoskeletal or translational proteins that changed in response to haloperidol were increased (Fig. 3C).
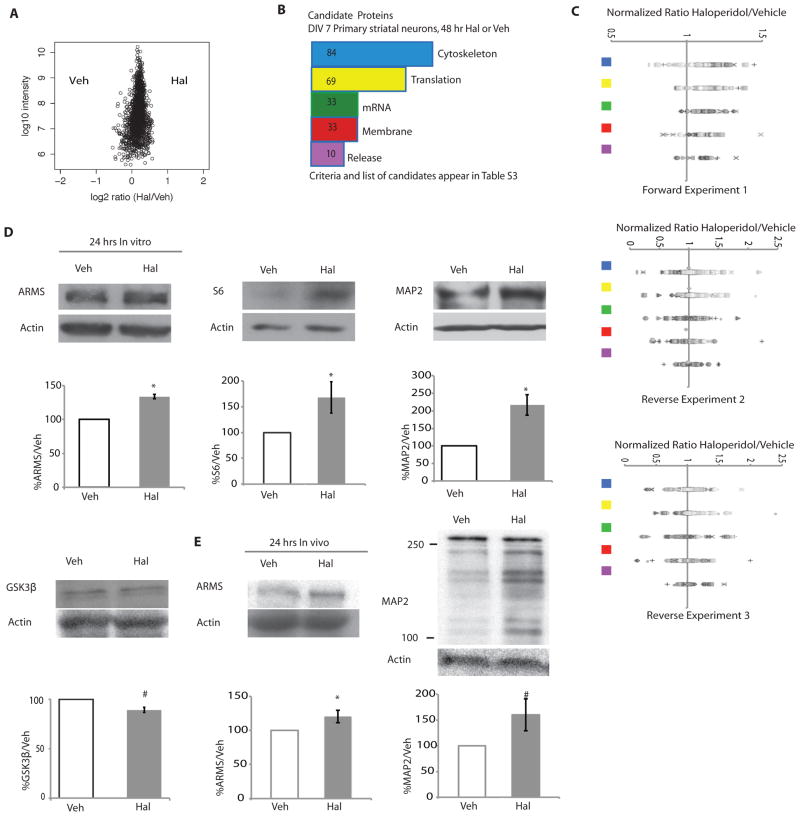
Figure 3. Haloperidol treatment leads to specific early proteomic changes in proteins involved in protein synthesis and associated with the cytoskeleton.
(A) A representative plot of all normalized proteins measured by mass spectrometry after 48 hours of haloperidol or vehicle treatment and SILAC labeling. (B) Bioinformatic analysis of the candidate proteins that increased with haloperidol treatment in the 48-hour mass spectrometry assays. The plot shows the frequency of proteins abundance changes of the top five categories from UniProtKB (see Materials and Methods, table S4). Translation= proteins related to protein translation; mRNA= proteins related to synthesis and trafficking of mRNA; cytoskeletal= proteins related to or part of the cytoskeleton; membrane= proteins integral to the plasma membrane; release= proteins related to synaptic vesicle release or synaptic exocytosis. (C) Normalized fold changes for all proteins in top 5 candidate protein classes. The symbols do not denote any special meaning. The colored squares correspond to the classes as colored in B. (D) Western blot analysis of lysates of DIV7 striatal neurons exposed to haloperidol or vehicle for 24 hours. The candidate proteins S6 (n=4, p =0.06), ARMS (n=3, p <0.0001), and MAP2 (n=3, p= 0.02) were analyzed, as well as GSK3β, a protein that was not a candidate (n=4, p=0.006). (E) Western blot analysis of ARMS and MAP2 in striatal lysates from adult mice treated with either 0.25 mg/kg haloperidol or vehicle for 24 hours. ARMS (n=6/group, p = 0.048); MAP2 (n=6/group, p= 0.08). All graphs shown are average ± SEM and analyzed by Student’s t test. P < 0.05 was considered significant and is marked with asterisks; p < 0.1 is considered trend level and is marked with #.
Table 1. Candidate proteins that met the highest stringency of increasing at least 1.2 fold in at least two experiments.
Proteins are grouped by function as defined by UniProt KB. Protein names or abbreviations and their International Protein Index (IPI) identifiers are provided. A list of proteins that met lower stringencies can be found in table S4.
| Cytoskeleton and trafficking-related proteins |
| Actc1, IPI00194087 |
| Protein linking IAP with cytoskeleton 1,IPI00203373 |
| Gamma-adducin, IPI00209216 |
| Coronin-1A, IPI00210071 |
| Hematopoietic system regulatory peptide, IPI00230925 |
| Developmentally-regulated brain protein, IPI00231407 |
| Ttll12, IPI00361443 |
| plexin A2, IPI00363440 (could also be listed as a membrane protein) |
| Tbc1d17, IPI00364330 |
| Tpm1, IPI00391997 |
| Intersectin-1, IPI00554196 |
| Astrocytic phosphoprotein PEA-15, IPI00555213 |
| Protein-L-isoaspartate(D-aspartate) O-methyltransferase, IPI00555303 |
| IPI00561918 |
| heat shock protein 1, IPI00566286 |
| Tyrosine-protein phosphatase non-receptor type 23, IPI00782007 |
| catenin, alpha 2, IPI00870129 |
| Snx4, IPI00373092 (could also be considered endocytosis protein) |
| Translation, amino acid biosynthesis, and protein folding proteins |
| Ppig, IPI00194996 |
| eif1b, IPI00196316 |
| 60S acidic ribosomal protein P1, IPI00196316 |
| IPI00365169 |
| ER membrane protein complex subunit 2, IPI00366247 |
| Eukaryotic translation initiation factor 4H; IPI00370315 |
| Rrbp1 ribosome binding protein 1, IPI00769110 |
| IPI00870393 |
| IPI00876578 |
| Psat1, IPI00951316 |
| Proteins at the synaptic terminal, involved in synaptic release, or secre proteins |
| Synaptogyrin-1, IPI00209284 |
| Synapsin-3, IPI00390215 |
| Granulins, IPI00213847 |
| Proenkephalin-A, IPI00324459 |
| Pleiotrophin, IPI00204373 |
| complexin 2, IPI00190397 |
| mRNA-related proteins |
| Calcium-regulated heat stable protein 1, IPI00201548 |
| LSM5 homolog, IPI00210564 |
| NOP58 ribonucleoprotein, IPI00214536 |
| Srpk2 SRSF, IPI00372013 |
| CELF1, IPI00373642 |
| U2surp, IPI00560940 |
| Rbmxl1, IPI00763272 |
| Snrpf, IPI00768974 |
| Plasma membrane-associated proteins |
| Nlgn2, IPI00209308 |
| Ephrin-B1, IPI00209416 |
To validate these changes in protein abundance and determine if they also were present at 24 hours, a time where clinical improvement is noted in human patients (2,3), we exposed primary striatal neurons for 24 hours to haloperidol and analyzed protein abundance by Western blot. We chose to investigate the abundance of proteins that were at least low stringency “candidates” in protein classes of interest: proteins associated with translation (ribosomal protein S6), the cytoskeleton [microtubule-associated protein 2 (MAP2)], or morphological complexity [ankyrin repeat-rich membrane-spanning protein (ARMS), also known as kinase D-interacting substrate of 220 kD (Kidins220)]. The ribosomal protein S6 was increased in abundance at 4 hours, and this increase was present in samples exposed to haloperidol for 24 and 48 hours (Fig. 3D, Table 2), indicating a prolonged role for increases in the protein synthesis machinery in the mechanism of action of antipsychotics. MAP2 is a somato-dendritic protein associated with microtubule stability and morphological complexity in neurons (32). Consistent with the proteomic data (Table 2), 24 hours of haloperidol exposure resulted in increased abundance of MAP2 (Fig. 3D). ARMS is associated with branching and spines (33–35) and was increased in abundance after 24 and 48 hours of haloperidol exposure to cultured striatal neurons (Fig. 3D, Table 2). We also examined the abundance of GSK3β, which did not exhibit an increase after 24 hours of haloperidol treatment (Fig. 3D), consistent with the mass spectrometry results where it was not identified as a candidate (table S2).
Table 2.
Summary of proteins analyzed by Western blot and mass spectrometry. Candidate stringency means that the abundance was increased by 1.2 fold (normalized ratio of treated/untreated protein abundance) in one experiment and increased in at least one other independent experiment (complete list in table S4). Increased or decreased by Western blot denotes changes that are statistically significant or trend level as shown in Figures 2 and 3. Complete table of all proteins measured in the mass spectrometry (MS) 5-hour screen and 48-hour mass spectrometry study appear in tables S1 and S2, respectively.
| Protein | Haloperidol 5 hours in culture MS Screen |
Haloperidol 4 hours in culture Western blot |
Haloperidol 48 hours in culture MS |
Haloperidol 24 hours in culture Western blot |
Haloperidol 24 hours in vivo Western blot |
|---|---|---|---|---|---|
| RpS6 Ribosomal Protein S6 |
Increased | Increased | Increased (Candidate stringency) | Increased | Not tested |
| eEF2 eukaryotic Elongation Factor 2 |
Increased | Increased | Variable | Not tested | Not tested |
| ARMS (Kidins220) Ankyrin Repeat-Rich Membrane-Spanning Protein |
Not detected | Not tested | Increased (Candidate stringency) | Increased | Increased |
| MAP2 Microtubule-associated protein 2 |
Increased | Not tested | Increased (Candidate stringency) | Increased | Increased |
| GSK3β Glycogen synthase kinase 3 beta |
Not detected | Not tested | Unchanged | Decreased | Not tested |
| Clathrin | Unchanged (Clathrin heavy chain 1 | Unchanged | Increased (Clathrin heavy chain 1 (unchanged to increased) Clathrin light chain (increased, candidate stringency) |
Not tested | Not tested |
To investigate whether the changes in cytoskeletal proteins also occurred in vivo, adult male mice were injected intraperitoneally with 0.25 mg/kg haloperidol and 24 hours later lysates from the striatum were examined by Western blot. We noted an increase in the abundance of ARMS and MAP2 (Fig. 3E), consistent with both the proteomic analysis and Western blot data from the cultured neurons (Table 2). We also noted that 24-hour haloperidol treatment induced an increase in an immunoreactive band of MAP2 at 270 kD in rat striatal neurons in culture (Fig. 3D and fig. S4A). Haloperidol also stimulated an increase in the abundance of several MAP2-immunoreactive bands between 100 and 250 kD both in striatal lysates from mice (Fig. 3E) and in striatal neurons in culture in vitro (fig. S4B). These additional protein species were not induced by haloperidol when striatal neurons in vitro were pretreated with MAP2 shRNA (fig. S4B). These findings are consistent with the higher number of reported MAP2 isoforms in mice compared with those in rats (36). In summary, haloperidol induced specific changes in the synthesis of proteins that are involved in cytoskeletal architecture and organization, suggesting that haloperidol might alter neuronal morphology.
Haloperidol increases neuronal complexity
Because 24-hour exposure to haloperidol increased the synthesis of MAP2 and ARMS, two key proteins that modulate neuronal arborization, we posited that antipsychotic treatment could lead to changes in morphological complexity. To enhance the ability of cultured neurons to develop a physiologically-relevant morphology (37), we co-cultured rat striatal neurons with cortical neurons. Although there are some D2R-positive cortical neurons, they comprise a low percentage of the total neuronal population. Therefore, we identified striatal neurons by staining for the striatal marker, DARPP32 (38). After DIV14 (Fig. 4), we treated the neurons with haloperidol for 24 hours and quantified neuronal complexity by Sholl analysis of the DARPP32-positive neurons (39).
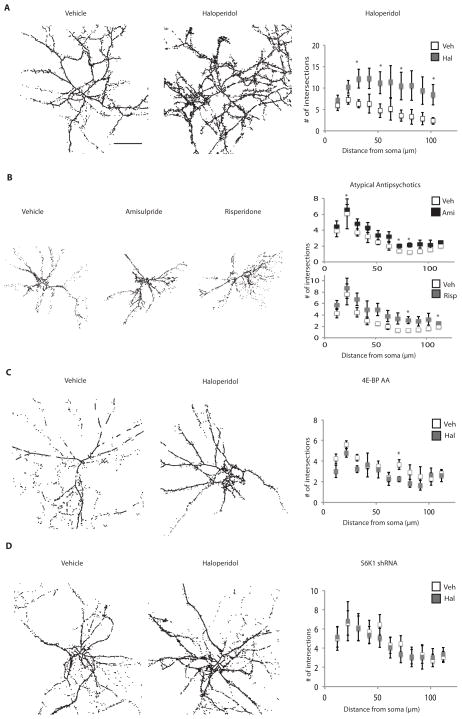
Figure 4. Haloperidol treatment leads to increased morphological complexity in striatal DARPP32-positive neurons.
Primary striatal neurons were cocultured with wild-type cortical neurons (2:3 ratio), grown to DIV14, and exposed for 24 hours to either vehicle, haloperidol, amisulpride, or risperidone. Neurons were stained with an antibody against DARPP32 and morphological complexity was quantified by Sholl analysis. Images shown are line enhanced for visualization. The number of intersections within 100 μm of the soma was quantified on images without line enhancement. (A) The effect of haloperidol on morphological complexity of wild-type striatal neurons. (n=4, p<0.000001, cells analyzed = 19 for vehicle treated, 20 for haloperidol treated) (B) The effect of the indicated atypical antipsychotics on morphological complexity of wild-type striatal neurons. Risperidone (n=3, p=0<0.002, cells analyzed = 24 for vehicle treated, 23 for drug treated); amisulpride (n =3, p<0.01, cells analyzed = 24 for vehicle treated, 21 for drug treated). (C) The effect of haloperidol on striatal neurons expressing 4E-BP AA. 4E-BP AA-expressing striatal neurons were cocultured with wild-type cortical neurons as described above, then induced with doxycycline overnight and exposed to haloperidol for 24 hours. Scholl analysis was performed on cells that were stained positive for both DARPP32 and the HA tag on the 4E-BP AA. (n=3, p = 0.05 decrease, cells analyzed = 12 for vehicle treated, 13 for haloperidol treated). (D) The effect of haloperidol on striatal neurons in which S6K1 is knocked down. S6K1 shRNA-expressing striatal neurons were cocultured with wild-type cortical neurons as described above and exposed to haloperidol for 24 hours. Scholl analysis was performed on cells that were stained positive for both DARPP32 and GFP that marks the shRNA-expressing cells. (n=3, p=not significant). Scale bar indicates 50 μm. All graphs shown are average ± SEM. p ≤ 0.06 is indicated by the asterisk.
Haloperidol triggered an increase in branching (Fig. 4A) after 24 hours, which corresponded to the time that the increase in cytoskeletal proteins was noted by mass spectrometry and Western blot (Table 2). This increase in morphological complexity also occurred in striatal neurons exposed to the atypical antipsychotics amisulpride and risperidone, although it was not as robust as the haloperidol response (Fig. 4B). The increase in morphological complexity induced by haloperidol was attenuated by either the expression of the dominant-negative 4E-BP AA (Fig. 4C) or by treatment of the cultures with S6K1-targeted shRNA (Fig. 4D), consistent with the morphological changes involving mTORC1 signaling. Thus, acute exposure (24 hour) to haloperidol profoundly affected dendritic process branching and growth through a translational mechanism likely mediated by increased mTORC1 signaling, which could alter signal integration and the output of the neurons.
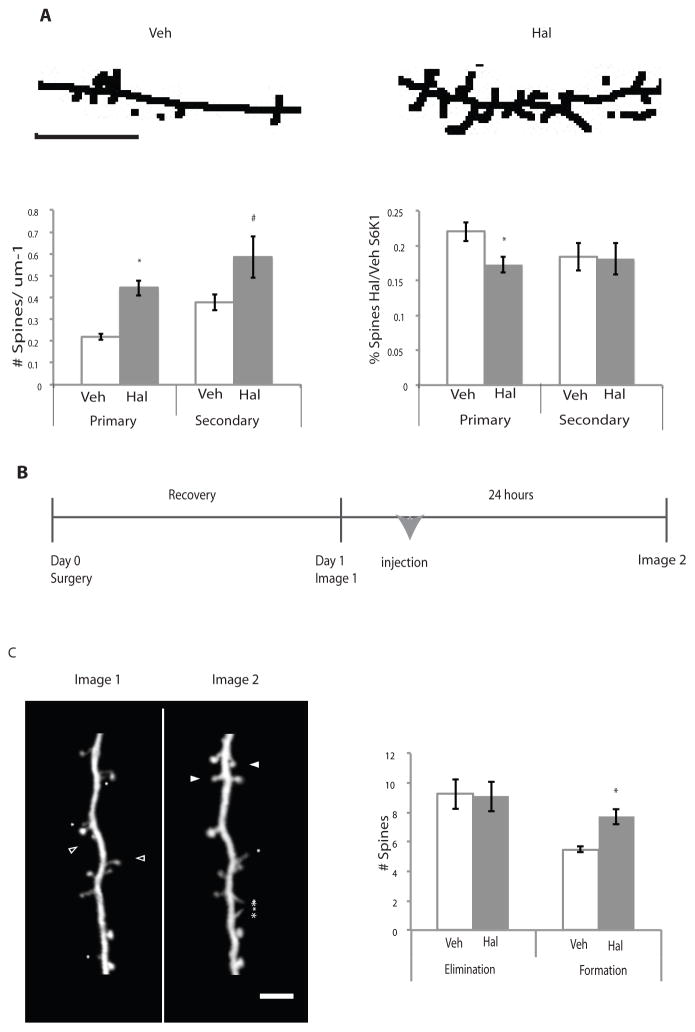
Because ARMS has been implicated in spine stability and maintenance (34), we examined the appearance of spines in DIV14 cultured striatal neurons using a DARPP32 antibody after 24 hours exposure to haloperidol (fig. S5). We noted a significant increase in spines on both primary projections from the soma, and secondary projections, which branch directly off primary projections, in the haloperidol-treated group, both of which were attenuated in neurons expressing S6K1 shRNA (Fig. 5A). These data suggest that haloperidol increases the number of spines in an mTORC1-dependent fashion.
Figure 5. Haloperidol treatment leads to increased spines in striatal neurons in vitro and cortical Layer 5 pyramidal neurons in vivo.
(A) Primary striatal cultures were co-plated with cortical neurons, grown to DIV14, and treated for 24 hours with either haloperidol or vehicle. Images were processed using ImageJ and spines were quantified on primary and secondary projections where detectable. Skeletal representative images are shown and are line enhanced for visualization. Spines that met specified criteria (see Materials and Methods) were quantified (n=4, p<0.0001). Primary indicates spines counted on primary projections from the soma; secondary indicates spines counted on secondary projections that branch from primary projections. The scale bar indicates 5 μm. S6K1 shRNA treated neurons (B) Experimental procedure for in vivo study. One-month-old Thy-1 YFP mice underwent surgery to place head-fixation bars prior to initial imaging session (Day 0), were imaged, and then either injected with haloperidol or vehicle and imaged again 24 hours later. (C) Processed representative images of cortical Thy-1 YFP positive neurons are shown for clarity. Asterisks indicate filopodia, closed arrowheads indicate spines that formed, and open arrowheads indicate spines that were eliminated. The number of spines eliminated or formed is plotted. (Veh=4, Hal=6, p = 0.006). All graphs shown are average ± SEM and analyzed by Student’s t-test for statistical differences. p ≤ 0.05 is indicated by the asterisk.
Haloperidol induces an increase in neuronal morphological complexity and spine number in striatal neurons, a brain region that has a relatively high abundance of D2R compared to other brain regions, such as the cortex. We investigated if haloperidol induced morphological changes in other areas with less abundant D2R, and if we could observe these changes in vivo. To visualize the neuronal morphology in animals treated with haloperidol, we used two-photon microscopy to visualize spines of layer 5 pyramidal neurons in the upper layers of the frontal association cortex in 1-month-old Thy1-YFP mice, which display isolated expression of YFP+ neurons (40). We administered a single intraperitoneal injection of 0.25 mg/kg haloperidol to Thy1-YFP mice and examined spine elimination and formation 24 hours later using a thinned skull preparation (Fig. 5B) (40). This brain region was selected because it is positive for D2R in cortical layers 1/2 and 5 and contains projections that terminate in the striatum (41–42) and has been implicated in cognitive processing performance in primates (43). We noted a significant increase in formation, but not elimination, of spines in apical dendrites of Layer 5 pyramidal neurons after 24 hours of haloperidol treatment (Fig. 5C). Thus, haloperidol altered spine formation within 24 hours in the mouse frontal association cortex in vivo. Together with the increased expression of cytoskeletal-related proteins observed in the striatum both in vitro and in vivo (Table 2), these results suggested that haloperidol induces morphological changes in the striatum and the cortex in an acute manner.
Discussion
Here, we presented a role for the Akt-mTORC1 signaling pathway in response to haloperidol, resulting in the induction of de novo protein synthesis and an increase in morphological complexity in striatal neurons. We also provided evidence that these changes in de novo protein synthesis and morphological complexity can be induced by amisulpride and risperidone, second-generation atypical antipsychotics. Although we cannot rule out the contribution of other cellular processes to the efficacy of antipsychotics, the data support the notion that Akt-mTORC1-induced changes in de novo protein synthesis play a role in the acute mechanism of the action of antipsychotics.
Akt activity is involved in the response of many antipsychotic drugs (7); however, a suggested downstream target, GSK3β, may not fully explain the mechanism of action of antipsychotics. Here, we demonstrated activation of the Akt-mTORC1 pathway and a subsequent increase in protein synthesis that has not been previously described in the action of antipsychotics.
An increase in Akt-mTORC1 signaling in response to haloperidol was observed that was attenuated by an Akt inhibitor and rapamycin. The finding of increased Akt activity in response to antipsychotics is consistent with previous reports (6,7); however, the increase in mTORC1 signaling and its potential role in antipsychotic action have not been fully investigated. Valjent et al. examined S6 phosphorylation in response to haloperidol in vivo and reported that S6 was not affected (44). One explanation for the differences in S6 responsiveness could be due to the different doses and modes of administration of haloperidol. We also found that 4E-BP, another mTORC1 target, was similarly regulated by haloperidol. In addition, the changes in protein synthesis induced by haloperidol were significantly attenuated by knockdown of the mTORC1 effector S6K1, providing evidence that mTORC1 was mediating the response to haloperidol. These data are consistent with Bonito-Oliva et al. (45) who showed that haloperidol induces S6 phosphorylation in an mTORC1-dependent manner in D2R-positive striatal neurons in vivo. Thus, we conclude that the Akt-mTORC1 pathway is activated by haloperidol and induces specific changes in protein synthesis.
Inhibition of either eIF4E with a dominant-negative 4E-BP (4E-BP AA) or S6K1 with a shRNA reduced haloperidol-induced increases in protein synthesis and morphological complexity. These data are contrary to previous reports that 4E-BP is the primary mTORC1 effector that contributes to mTORC1-dependent translation in dividing cells (26). We speculate that the increased protein synthesis and subsequent morphological changes induced by haloperidol may require both translation initiation mediated by phosphorylation of 4E-BP and translation elongation mediated by the phosphorylation and activation S6K1 (43). This may represent a key difference in translational control between dividing cells and post-mitotic neurons.
In addition to demonstrating that haloperidol induced Akt-mTORC1–dependent changes in protein synthesis, the proteomic and cellular biochemistry indicated that proteins involved in translation control were among those which are newly synthesized and increased in abundance. The increase in the abundance of proteins associated with translation persisted despite a return to the basal rate of protein synthesis after a few hours of haloperidol exposure, suggesting that haloperidol may prime the neurons to enable rapid protein synthesis and alteration in the proteomic profile in response to subsequent stimuli.
Because previous studies have indicated that chronic antipsychotic treatment of at least 3 weeks can change protein abundance in both human patient and rodent brains (26,27), we compared our proteomic analysis from 48 hours of treatment to determine if any of the same proteins were altered under both long-term and shorter-term treatment. MAP2, one of the validated proteins meeting the candidate criteria in our proteomic study (Fig. 3), was increased at the mRNA transcript level (47) and its phosphorylation state increased (48), following chronic antipsychotic treatment, further suggesting its role in the antipsychotic response. Comparison of our proteomic data from cultured striatal neurons exposed for 48 hours to haloperidol with 48 proteins that showed altered abundance in the cortex of rats treated with haloperidol for 21 days (27) revealed that 27 of 35 proteins measured in common shifted in the same direction. Of these, 12 of 27 met the criteria we set for candidate proteins (table S4). When compared to a proteomic study of human postmortem cortex in patients treated with antipsychotics (28), our proteomic data also correlated with antipsychotic-induced changes observed previously. Of 34 proteins measured by Chan et al. (27), 13 of 19 proteins measured in common were altered in the same direction, of which 6 met candidate criteria (table S4). Thus, these animal and human results suggest a convergent mechanism by which antipsychotics regulate the abundance of proteins relevant to neuronal function due to increased mTORC1-dependent translation.
We measured a large number of proteins and obtained a unique translational profile in striatal neurons that was altered with antipsychotic treatment. Notably, our study includes validated proteins that are involved with translation and in cytoskeletal function. The commonality between the data set obtained here and in studies in both animals and humans with chronic antipsychotic treatment provide further confirmation that the changes in newly synthesized protein abundance observed in this study were not just the result of short-term exposure to the drug or that were unique to the in vitro system. In addition, the overlap could suggest that the changes may not be limited to striatal neurons, but rather could be indicative of changes in other D2R and D2R-like circuits throughout the brain.
In addition to the activation of mTORC1 in response to haloperidol, the translation of several mRNAs with 5′-terminal oligopyrimidine (TOP) and TOP-like sequences that are linked to mTORC1 signaling (26) were identified. Many of the proteins encoded by these mRNAs were increased by haloperidol at both early and later time points (table S3). The relative abundance of these known mTORC1 targets further supports a specific role of mTORC1 signaling in the acute action of antipsychotics.
Although most of the data presented here were gathered from an in vitro culture system, there are multiple lines of evidence to suggest its relevance to patient and in vivo rodent data. We have provided evidence of an increase in branching and spine number with haloperidol treatment that is mTORC1-dependent and present in the first 24 hours. This finding also supports previous clinical MRI studies showing an increase in striatal volume, which could potentially be explained by increased branching, following chronic treatment with typical antipsychotics and less consistently with atypical antipsychotics [reviewed in (49)]. Although we noted an increase in morphological complexity in striatal neurons when exposed to atypical antipsychotics, this change was much less pronounced than when exposed to haloperidol, which appears consistent with human patient data.
In addition to the increased spine formation that we observed in the cortex, neurite length also increases in hippocampal neurons exposed to atypical antipsychotics (50), which suggests that subtle changes may also occur in non-striatal brain regions where D2R is less abundant. These increases in morphological complexity are consistent with reports in human patients, especially in the early stages of antipsychotic treatment and in rodents treated with antipsychotics (49, 51, 52). In contrast, in rodents treated with antipsychotics and human schizophrenia patients reduced brain volume was reported, and decreased dendritic branching and spine density in the cortex has been observed in postmortem studies of schizophrenia patients (53–56), suggesting a possible decrease in connectivity at later stages of schizophrenia. The volumetric decrease at later stages of treatment is at odds with the increased striatal volume observed in patients and rodents treated with antipsychotics that has been positively correlated with treatment response (49, 55–58) and with the rapid morphological changes that we detected herein. It is unclear if this difference is the result of either the treatment regiment or a failure of antipsychotics to prevent neuronal pruning over time. Although questions about the functional consequences of these volumetric changes remain, the identification of a mechanism for some of these changes represents an important step in exploring the actions of antipsychotics, which will inform future studies.
In summary, here we have presented evidence that could link several observed actions of antipsychotics to a mechanism involving induction of the protein synthesis machinery. We linked the antipsychotic-stimulated increased Akt signaling to mTORC1-dependent enhancement of protein synthesis that in turn increases the abundance of proteins that are components of the translation machinery. Furthermore, antipsychotics increased the production of cytoskeletal-associated proteins that was correlated with increased morphological complexity, which is consistent with previously reported rodent and patient data.
Materials and Methods
Plasmids
The doxycycline-inducible HA-tagged T37A/T46A 4E-BP1 double mutant (4E-BP AA), empty vector, wild-type 4E-BP, scramble shRNA, and S6K1 shRNA 3159 were generated as previously described (24).
Striatal Cell Culture
Primary neurons from E17-18 C57Bl/6 mouse or E18-19 Sprague Dawley rat embryos were prepared as previously described (59) and cultured on plates coated with 0.1 μg/ml Poly-d-lysine (for experiments involving Western blotting of DIV7 neurons) or coverslips coated with 0.1 μg/ml Poly-d-ornithine (for immunofluorescence and Sholl analysis of DIV14 -22 neurons). Striatal neurons were grown in Neurobasal with B27 (Invitrogen, Grand Island, NY). Doxycycline-inducible constitutively active 4E-BP AA, WT 4E-BP, empty vector, S6K1 shRNA, or scramble shRNA was introduced into striatal neurons by electroporation using the AMAXA nucleofection system (Lonza, Allendale, NJ) according to the manufacturer’s instructions at DIV0 before plating. Scramble constructs, S6K1 shRNA and MAP2 shRNA (Open Biosystems, Pittsburgh, PA) were cotransfected with GFP (Lonza, Allendale, NJ). 4E-BP-AA was induced with 0.5 μg/ml doxycycline overnight before treatment with haloperidol. S6K1 inhibitor was used at 10 μM, for 20 minutes before haloperidol treatment. For drug treatments less than 24 hours, neurons were serum starved and treated with haloperidol, amisulpride, or risperidone (Sigma Aldrich) as described. Neurons were pre-incubated with 5 μM of an Akt -VIII inhibitor (Akti) or 125 μM rapamycin (Sigma Aldrich) and then treated with haloperidol. For Western blotting, neurons were cultured at 1 × 106/well in 6-well plates. For immunofluorescence, striatal cells were cultured at 3.33 × 105/well on coverslips in 12-well plates. For co-cultures, striatal neurons were plated with cortical neurons at a 3:4 ratio and grown for 14 days.
Western Blot for Primary Cell Cultures
After treatment, cells were lysed and prepared according to the previously described protocol (59). The majority of Western blots were performed on 10% or 12% acrylamide gels. Antibodies for 4E-BP, phosphorylated 4E-BP, phosphorylated Akt, total Akt, phosphorylated S6 and S6 were from Cell Signaling (Danvers, MA). The antibody recognizing ARMS (892) was previously described (34). Antibodies recognizing GSK3β and clathrin were from Transduction Laboratories/BD Biosciences (San Jose, CA) and the antibody recognizing MAP2 was from Chemicon/Millipore (Billerica, MA).
Immunofluorescence
Cells for immunofluorescence were plated on coverslips, grown for 7 days, and fixed in 4% paraformaldehyde/20% sucrose and stained as previously described (59). The primary antibodies recognized pAkt or pS6 (same as above), DARPP32 [gift from Dr. Hugh Hemmings, Jr. (60)], HA (3F10, Roche Diagnostics, Indianapolis, IN), or GFP (abcam). Some images were taken with an Eclipse E800 microscope equipped with a 60X phase 3, NA 1.40 oil-immersion objective and DigiSight monochrome camera (all Nikon) driven by NIS Elements F 3.0 software (Melville, NY). Image J (National Institutes of Health) was used for intensity quantification by selecting a region of interest of the soma on raw image files, excluding the nucleus, and measuring intensity for every visible cell per view, with views averaged per coverslip, background subtracted, then grouped by treatment, divided by the vehicle baseline and compared to other treatment groups. Confocal imaging was done using a LSM 510 laser-scanning confocal microscope equipped with a 40x Plan Neofluor [numerical aperture (NA) 1.3] DIC oil-immersion objective (Carl Zeiss Microimaging). For Sholl analysis, images were taken, contrast was enhanced, background was removed manually around DARPP32-positive cells, and the cells were skeletonized and examined. Images were processed using ImageJ and the Advanced Sholl Analysis plugin (37). Lines were dilated for ease of demonstration only (Fig. 4D), analysis was performed with thinner, skeletonized lines. Because DARPP32 is present in spines (61), spines were quantified with a DARPP32 stain using ImageJ. Spines were analyzed in the first 100 μm from the soma and identified by the following criteria, they must contain a bright spot with a neck near a projection at least 1 μm × 1 μm in size and present in at least two, 1 μm thick z-stack frames.
SUnSET/SILAC
Striatal cultures were grown at 3 × 107 cells/treatment group in Neurobasal and B27 (Invitrogen). Neurons used in the 48-hour study were changed to SILAC media with haloperidol or vehicle and incubated for 48 hours before lysing. For the 5-hour study, cells were serum starved and incubated in SILAC media and 1 μg/ ml puromycin for 5 hours in the presence and absence of 20 nM haloperidol. After treatment, cells were lysed, centrifuged to remove DNA and debris, and all nascent proteins were immunoprecipitated with an antibody recognizing puromycin (23).
Because the weight of the SILAC amino acid label was associated with the treatment group (vehicle or haloperidol), samples could be combined and immunoprecipitated together to avoid bias and variability between treatment groups. The buffer for immunoprecipitation was 50 mM Tris-HCl (pH 8.0), 140 mM NaCl, 1% NP-40, and 10% glycerol. The puromycin – SILAC initial screen (5-hour study) was performed once with haloperidol labeled with the medium tag and vehicle screen with the heavy tag. For the 48-hour study, 3 independent biological replicates were conducted: the forward experiment in which haloperidol had a heavy tag and the vehicle the medium tag, and 2 reverse experiments in which the tags were reversed. For the 48-hour proteomic study. 3 × 107 striatal cells/treatment group were grown to DIV7 and treated in the presence of 20 nM haloperidol or vehicle for 48 hours in SILAC media. Sample preparation and LC/MS analysis were performed as previously described (29). Briefly, immunoprecipitated proteins were fractionated by SDS-PAGE, digested in-gel overnight with trypsin. Gel bands were cut into small pieces and destained in 25 mM NH4HCO3/50% acetonitrile, dehydrated with acetonitrile and dried. The gel pieces were rehydrated with 12.5 ng/μL trypsin solution in 25 mM NH4HCO3 and incubated overnight at 37C. Peptides were extracted twice with 5% formic acid/50% acetonitrile followed by a final extraction with acetonitrile. Samples were concentrated by vacuum centrifugation to dryness and redissolved with 2% acetonitrile in 0.1% formic acid before further analysis.
The resulting peptides were analyzed by nano-flow RPLC/MS using an LTQ-Orbitrap mass spectrometer equipped with a nanoelectrospray ionization source (Jamie Hill Instrument Services) and prepared for mass spectrometry using published methods (29). For the 48 hour Forward 1 and 5 hour screen samples, an EksigentnanoLC system (Eksigent Technologies) equipped with a self-packed 75-μm × 12-cm reverse phase column (Reprosil C18, 3 μm, Dr. Maisch GmbH, Germany) was coupled directly to the mass spectrometer. Peptides were eluted by a gradient of 3–40% acetonitrile in 0.1% formic acid over 110 min. Mass spectra were acquired in data-dependent mode with one 60 000 resolution MS survey scan by the Orbitrap and up to eight MS/MS scans concurrent with the Orbitrap survey scan in the LTQ for the most intense peaks selected from each survey scan. The automatic gain control target value was set to 500 000 for Orbitrap survey scans and 10 000 for LTQ MS/MS scans. Survey scans were acquired in profile mode and MS/MS scans were acquired in centroid mode.
For analysis of the 48-hour reverse SILAC samples, a Thermo Scientific EASY-nLC 1000 coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific) was used. A self packed 75-μm × 50-cm reverse phase column (Reprosil C18, 1.9 μm, Dr. Maisch GmbH, Germany) was used for peptide separation. Peptides were eluted by a gradient of 3–30% acetonitrile in 0.1% formic acid over 240 min at a flow rate of 250 nL/min. The Q Exactive was operated in the data-dependent mode with survey scans acquired at a resolution of 50,000 at m/z 400 (transient time = 256 ms). Up to the top 10 most abundant precursors from the survey scan were selected with an isolation window of 1.6 Thomsons and fragmented by higher energy collisional dissociation with normalized collision energies of 27. The maximum ion injection times for the survey scan and the MS/MS scans were 60 ms, respectively, and the ion target value for both scan modes were set to 1,000,000.
Protein Identification and Quantitation
The raw files were processed using the MaxQuant computational proteomics platform version 1.2.0.18. The fragmentation spectra were searched against the IPI rat protein database (version 3.68) allowing up to two missed tryptic cleavages. Carbamidomethylation of cysteine was set as a fixed modification. Methionine oxidation, protein N-terminal acetylation, D4-lysine, 13C6-arginine, 13C6-15N2-lysine, and 13C6-15N4-arginine were used as variable modifications for database searching. For LTQ-Orbitrap data, the precursor and fragment mass tolerances were set to 7 ppm and 0.5 Da, respectively. For Q Exactive data, the precursor and fragment mass tolerances were set to 7 ppm and 20 ppm, respectively. The estimated peptide and protein false positive rates based on the decoy database search were 1%.
Proteomic Data Analysis
Identified proteins were manually classified according to their descriptions on UniProtKB into (i) translational-related proteins (chaperones, folding proteins, ribosomal proteins, and tRNA synthetases), (ii) cytoskeletal-related proteins (actin, microtubule, cytoskeletal binding proteins, cytoskeletal modifiers, and motor proteins), (iii) membrane-bound proteins, (iv) membrane signaling proteins (proteins that interact with membrane and have a direct signaling cascade), (v) transcription and nuclear proteins, (vi) mRNA-processing proteins (pre-mRNA to mRNA transport and splicing up to actual ribosomal interaction), (vii) transport proteins (endocytosis and organelle processing), (viii) proteins associated with degradation (for example, those involved in proteasome-mediated degradation, lysosomal proteins, or any other protein breakdown process.), (ix) proteins associated with synaptic release (actively involved in synaptic exocytosis), (x) proteins involved in posttranslational modifications and that not part of a known signaling complex, (xi) mitochondrial proteins, (xii) proteins associated with fat metabolism, and (xiii) other, unclassified. Proteins with inconsistent changes in all three experiments or those that were only measured in one experiment-- were not included in the analysis. A cutoff of 1.2-fold increase (ratio protein amounts of haloperidol/vehicle was 1.2 (to one significant figure) or greater) was selected because reported changes in this range could be reliably detected by Western blot and have been used previously in mass spectrometry analysis of neurons in the literature (30). The criteria that candidates must be measured and increased at least two times out of three replicates is also based on previous publications (29,31). Those proteins that changed 1.2 fold once with at least one other change in the same direction were declared low stringency candidates. Those that changed 1.2 fold once but changed all three times in the same direction where medium stringency. Those with at least two changes in the same direction of greater than 1.2 fold were declared high stringency. Three low stringency candidate proteins were then selected for validation from the pool of candidates. Significance of these candidates was calculated using unnormalized peptide ratios and subjected to t-test analysis for each mass spectrometry experiment.
In Vivo Experiments: Injection of mice with haloperidol
Adult male C57/Bl6 mice were injected i.p. with either 0.25 mg/kg haloperidol or vehicle (DMSO) dissolved in 0.9% saline and returned to their cage for 24 hours. The animals were sacrificed by cervical dislocation, the striatum was microdissected on ice, and the tissue was flash frozen. The striatal lysates were sonicated on ice in the presence of phosphatase and protease inhibitors and analyzed by Western blot. Lysis buffer was as described in (13).
Two-photon imaging
1-month-old Thy1-YFP-positive mice underwent bar implantation surgery for head fixation and skull thinning to 20 μm at the region to be imaged: a 0.2 × 0.2 mm area centered at +2.8 mm Bregma, +1.2 mm midline [frontal association cortex (Day 0)]. Animals were allowed to recover for 24 hours, then they were imaged (40), after which, they were injected with 0.25 mg/kg haloperidol or vehicle dissolved in 0.9% saline and returned to their cage. 24 hours later the same dendrites were re-imaged. ImageJ software was used to analyze image stacks. The same dendritic segments were identified from three-dimensional stacks taken from different time points with high image quality (ratio of signal to background noise >4:1). The number and location of dendritic protrusions (protrusion length was more than one-third the dendritic shaft diameter) were identified in each view without previous knowledge of the animal’s treatment. Filopodia were identified as long, thin structures (generally larger than twice the average spine length, ratio of head diameter to neck diameter<1.2:1 and ratio of length to neck diameter>3:1). The remaining protrusions were classified as spines. No subtypes of spines were separated. Three-dimensional stacks were used to ensure that tissue movements and rotation between imaging intervals did not influence spine identification. Spines or filopodia were considered the same between views if their positions remained the same distance from relative adjacent landmarks. Spines were considered different if they were more than 0.7 mm away from their expected positions based on the first view.
Statistical Analysis
Statistical analyses were performed in GraphPad Prism, InStat Software (GraphPad Softwares, CA, USA), or Microsoft Office Excel. For comparing two groups, a Student’s Two-tailed t-test was used. For comparison of multiple groups, One Way Single Factor ANOVAs were employed. For statistical analysis of Sholl analysis, a Kolmogorov–Smirnov test was used, and the population was found to be normal, so a point-by-point student’s t-test was employed. Significance was set at p<0.05 unless otherwise stated and trend level was set at p<0.1.
Supplementary Material
Supplementary Figure 1: Characterization of haloperidol-mediated pAkt response.
Supplementary Figure 2: Puromycin dosing and control experiments for genetic manipulations.
Supplementary Figure 3: Proteomic analysis of measured proteins
Supplementary Figure 4: Haloperidol-induced proteins that react with a MAP2 antibody in mouse striatal neurons or rat striatal neurons in vitro.
Supplementary Figure 5: DARPP32 can label spines in DIV14 cultured striatal neurons
Supplementary Table 1: Complete results of the 5-hour proteomic screen
Supplementary Table 2: Complete results of the 48-hour proteomic screens
Supplementary Table 3: Haloperidol-induced increases in proteins linked to mTORC1 signaling
Supplementary Table 4: Candidate proteins from cytoskeleton, translation, mRNA, membrane, and release protein groups
Supplementary Table 5: Statistical significance for 3 biological replicates for selected proteins of interest
Acknowledgments
We gratefully acknowledge Hugh Hemmings, Jr. and Philippe Pierre for their generous gift of antibodies (DARPP32 and puromycin respectively) and to Ian Mohr’s lab for the SK61 shRNA, the dominant negative 4E-BP AA, WT and plko.3 scramble. We thank Freddy Jeanneteau, Emanuela Santini, Hanoch Kaphzan and the Klann and Chao labs for their support and discussion; Jeffrey Savas for discussion of proteomic analysis. In addition, we would like to acknowledge the contribution of Robert Schneider and Donald Goff for providing input on the manuscript.
Funding: This work was supported by the NIH (Training Grant T32 MH019524-20 to HB; NS034007 and NS047384 to EK; NS21072 and HD23315 to MVC; NS050276 and RR017990 to TAN).
Footnotes
Author contributions: H.B, G.Z, A.B. and L.M.P performed and analyzed the data. H.B., C.A.H. T.A.N, K.D., W-B G., E.K. and M.V.C. designed the experiments and wrote the paper.
Competing interests: none
Data and materials availability: CHORUS
48h treatment forward experiment: https://chorusproject.org/anonymous/download/experiment/822322165961813368
48h treatment reverse experiment 1: https://chorusproject.org/anonymous/download/experiment/1578932294544382348
48h treatment reverse experiment 2: https://chorusproject.org/anonymous/download/experiment/1788353921704300355
5h treatment:https://chorusproject.org/anonymous/download/experiment/-7618473989342117265
References
- 1.Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO, Keefe RS, Davis SM, Davis CE, Lebowitz BD, Severe J, Hsaio JK. Clinical Antipsychotic Trials of Intervention Effectiveness (CATIE) Investigators. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. New England Journal of Medicine. 2005;353:1209–1223. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- 2.Kapur S, Arenovich T, Agid O, Zipursky R, Lindborg S, Jones B. Evidence for onset of antipsychotic effects within the first 24 hours of treatment. American Journal of Psychiatry. 2005;162:939–946. doi: 10.1176/appi.ajp.162.5.939. [DOI] [PubMed] [Google Scholar]
- 3.Wright P, Lindborg SR, Birkett M, Meehan K, Jones B, Alaka K, Ferchland-Howe I, Pickard A, Taylor CC, Roth J, Battaglia J, Bitter I, Chouinard G, Morris PL, Breier A. Intramuscular olanzapine and intramuscular haloperidol in acute schizophrenia: antipsychotic efficacy and extrapyramidal safety during the first 24 hours of treatment. Canadian Journal of Psychiatry. 2003;48(11):716–21. doi: 10.1177/070674370304801102. [DOI] [PubMed] [Google Scholar]
- 4.Volavka J, Cooper TB, Czobor P, Meisner M. Plasma haloperidol levels and clinical effects in schizophrenia and schizoaffective disorder. Archives of General Psychiatry. 1995;52(10):837–45. doi: 10.1001/archpsyc.1995.03950220047010. [DOI] [PubMed] [Google Scholar]
- 5.Bunney BS. Antipsychotic drug effects on the electrical activity of dopaminergic neurons. Trends Neuroscience. 1984;7:212–5. [Google Scholar]
- 6.Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3β signaling in schizophrenia. Nature Genetics. 2004;36(2):131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- 7.Del’Guidice T, Lemasson M, Beaulieu JM. Role of beta-arrestin 2 downstream of dopamine receptors in the basal ganglia. Frontiers in Neuroanatomy. 2011;5(58) doi: 10.3389/fnana.2011.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jope RS, Roh MS. Glycogen Synthase Kinase-3 (GSK3) in Psychiatric Diseases and Therapeutic Interventions. Current Drug Targets. 2006;7(11):1421–1434. doi: 10.2174/1389450110607011421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leucht S, Kissling W, McGrath J. Lithium for schizophrenia. Cochrane Database of Systematic Reviews. 2003;(3):CD003834. doi: 10.1002/14651858.CD003834. [DOI] [PubMed] [Google Scholar]
- 10.Holt CE, Schuman EM. The Central Dogma Decentralized: New Perspectives on RNA Function and Local Translation in Neurons. Neuron. 2013;80(3):648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antion MD, Merhav M, Hoeffer CA, Reis G, Kozma SC, Thomas G, Schuman EM, Rosenblum K, Klann E. Removal of S6K1 and S6K2 leads to divergent alterations in learning, memory, and synaptic plasticity. Learning and Memory. 2008;15(1):29–38. doi: 10.1101/lm.661908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoeffer CA, Cowansage KK, Arnold EC, Banko JL, Moerke NJ, Rodriguez R, Schmidt EK, Lloyd RE, Pierre P, Wagner G, LeDoux JE, Klann E. Inhibition of the Interaction Between Translation Factors eIF4E and eIF4G Impairs Long-term Associative Memory Consolidation but not Reconsolidation. Proceedings of the National Academy of Science. 2011;108(8):3383–8. doi: 10.1073/pnas.1013063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharya A, Kaphzan H, Alvarez-Dieppa AC, Murphy JP, Pierre P, Klann E. Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron. 2012;76(2):325–37. doi: 10.1016/j.neuron.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santini E, Klann E. Dysregulated mTORC1-dependent translational control: from brain disorders to psychoactive drugs. Frontiers in Behavioral Neuroscience. 2011;5:76. doi: 10.3389/fnbeh.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyuhas O. Physiological roles of ribosomal protein S6: one of its kind. International Review of Cellular and Molecular Biology. 2008;268:1–37. doi: 10.1016/S1937-6448(08)00801-0. [DOI] [PubMed] [Google Scholar]
- 16.Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends in Biochemical Science. 2006;31(6):342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Beaulieu JM, Tirotta E, Sotnikova TD, Masri B, Salahpour A, Gainetdinov RR, Borrelli E, Caron MG. Regulation of Akt signaling by D2 and D3 dopamine receptors in vivo. Journal of Neuroscience. 2007;27(4):881–5. doi: 10.1523/JNEUROSCI.5074-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertran-Gonzalez J, Bosch C, Maroteaux M, Matamales M, Herve D, Valjent E, Girault JA. Opposing patterns of signaling activation in dopamine D1 and D2 receptor-expressing striatal neurons in response to cocaine and haloperidol. The Journal of Neuroscience. 2008;28(22):5671–5685. doi: 10.1523/JNEUROSCI.1039-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donnelly L, Waraich PS, Adams CE, Hamill KM, Marti J, Roque I, Figuls M, Rathbone J. Haloperidol dose for the acute phase of schizophrenia. Cochrane Database of Systematic Reviews. 2010:CD001951. doi: 10.1002/14651858.CD001951. [DOI] [PubMed] [Google Scholar]
- 20.Seeman P, Lee T. Antipsychotic drugs: direct correlation between clinical potency and presynaptic action on dopamine neurons. Science. 1975;188(4194):1217–9. doi: 10.1126/science.1145194. [DOI] [PubMed] [Google Scholar]
- 21.Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, Hoffman R, Shokat KM. Inhibitor hijacking of Akt activation. Nature Chemical Biology. 2009;5(7):484–93. doi: 10.1038/nchembio.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santini E, Huynh TN, MacAskill AF, Carter AG, Pierre P, Ruggero D, Kaphzan H, Klann E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493(7432):411–5. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt EK, Clavarino G, Ceppi M, Pierre PP. SUnSET, a nonradioactive method to monitor protein synthesis. Nature Methods. 2009;6(4):275–7. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi M, Wilson AC, Chao MV, Mohr I. Control of viral latency in neurons by axonal mTOR signaling and the 4E-BP translation repressor. Genes and Development. 2012;26:1527–1532. doi: 10.1101/gad.190157.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearce LR, Alton GR, Richter DT, Kath JC, Lingardo L, Chapman J, Hwang C, Alessi DR. Characterization of PF-4708671, a novel and highly specific inhibitor of p70 ribosomal S6 kinase (S6K1) The Biochemical Journal. 2010;432(2):245–255. doi: 10.1042/BJ20101024. [DOI] [PubMed] [Google Scholar]
- 26.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–13. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma D, Chan MK, Lockstone HE, Pietsch SR, Jones DN, Cilia J, Hill MD, Robbins MJ, Benzel IM, Umrania Y, Guest PC, Levin Y, Maycox PR, Bahn S. Antipsychotic treatment alters protein expression associated with presynaptic function and nervous system development in rat frontal cortex. Journal of Proteomic Research. 2009;8(7):3284–97. doi: 10.1021/pr800983p. [DOI] [PubMed] [Google Scholar]
- 28.Chan MK, Tsang TM, Harris LW, Guest PC, Holmes E, Bahn S. Evidence for disease and antipsychotic medication effects in post-mortem brain from schizophrenia patients. Molecular Psychiatry. 2011;16(12):1189–202. doi: 10.1038/mp.2010.100. [DOI] [PubMed] [Google Scholar]
- 29.Spellman DS, Deinhardt K, Darie CC, Chao MV, Neubert TA. Stable isotopic labeling by amino acids in cultured primary neurons: application to brain-derived neurotrophic factor-dependent phosphotyrosine-associated signaling. Molecular and Cellular Proteomics. 2008;7(6):1067–76. doi: 10.1074/mcp.M700387-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butko MT, Savas JN, Friedman B, Delahunty C, Ebner F, Yates JR, III, Tsien RY. In vivo quantitative proteomics of somatosensory cortical synapses shows which protein levels are modulated by sensory deprivation. Proceedings of the National Academy of Sciences. 2013;110(8):E726–35. doi: 10.1073/pnas.1300424110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodas JJ, Nehring A, Höche N, Sweredoski MJ, Pielot R, Hess S, Tirrell DA, Dieterich DC, Schuman EM. Dopaminergic modulation of the hippocampal neuropil proteome identified by bioorthogonal noncanonical amino acid tagging (BONCAT) Proteomics. 2012;12(15–16):2464–76. doi: 10.1002/pmic.201200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harada A, Teng J, Takei Y, Oguchi K, Hirokawa N. MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. Journal of Cell Biology. 2002;158(3):541–549. doi: 10.1083/jcb.200110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Fu WY, Ip JP, Ye T, Fu AK, Chao MV, Ip NY. Ankyrin repeat-rich membrane spanning protein (kidins220) is required for neurotrophin and ephrin receptor-dependent dendrite development. Journal of Neuroscience. 2012;2(24):8263–8269. doi: 10.1523/JNEUROSCI.1264-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu SH, Arevalo JC, Sarti F, Tessarollo L, Gan WB, Chao MV. Ankyrin Repeat-rich Membrane Spanning/Kidins220 protein regulates dendritic branching and spine stability in vivo. Developmental Neurobiology. 2009;69(9):547–57. doi: 10.1002/dneu.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neubrand VE, Cesca F, Benfenati F, Schiavo G. Kidins220/ARMS as a functional mediator of multiple receptor signalling pathways. Journal of Cell Science. 2012;125(8):1845–1854. doi: 10.1242/jcs.102764. [DOI] [PubMed] [Google Scholar]
- 36.E!Ensembl.org
- 37.Penrod RD, Kourrich S, Kearney E, Thomas MJ, Lanier LM. An embryonic culture system for the investigation of striatal medium spiny neuron dendritic spine development and plasticity. Journal of Neuroscience Methods. 2011;200(1):1–13. doi: 10.1016/j.jneumeth.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walaas SI, Greengard P. DARPP-32, a dopamine- and cyclic AM P-regulated neuronal phosphoprotein. Primary structure and homology with protein phosphatase inhibitor-1. Journal of Biological Chemistry. 1984;61:1890–1903. [PubMed] [Google Scholar]
- 39.Ferreira TA, Iacono LL, Gross CT. Serotonin receptor 1A modulates actin dynamics and restricts dendritic growth in hippocampal neurons. European Journal of Neuroscience. 2010;32(1):18–26. doi: 10.1111/j.1460-9568.2010.07283.x. [DOI] [PubMed] [Google Scholar]
- 40.Yang G, Pan F, Chang PC, Gooden F, Gan WB. Transcranial two-photon imaging of synaptic structures in the cortex of awake head-restrained mice. Methods in Molecular Biology. 2013;1010:35–43. doi: 10.1007/978-1-62703-411-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allen Brain Atlas
- 42.Bouthenet ML, Martres MP, Sales N, Schwartz JC. A detailed mapping of dopamine D-2 receptors in rat central nervous system by autoradiography with [125I]iodosulpride. Neuroscience. 1987;20(1):117–155. doi: 10.1016/0306-4522(87)90008-x. [DOI] [PubMed] [Google Scholar]
- 43.Goldman-Rakic PS. TOPOGRAPHY OF COGNITION: Parallel Distributed Networks in Primate Association Cortex. Annual Reviews in Neuroscience. 1988;11:137–56. doi: 10.1146/annurev.ne.11.030188.001033. [DOI] [PubMed] [Google Scholar]
- 44.Valjent E, Bertran-Gonzalez J, Bowling H, Lopez S, Santini E, Matamales M, Bonito-Oliva A, Herve D, Hoeffer C, Klann E, Girault JA, Fisone G. Haloperidol Regulates the State of Phosphorylation of Ribosomal Protein S6 via Activation of PKA and Phosphorylation of DARPP-32. Neuropsychopharmacology. 2011;36:2561–2570. doi: 10.1038/npp.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonito-Oliva A, Pallottino S, Bertran-Gonzalez J, Girault JA, Valjent E, Fisone G. Haloperidol promotes mTORC1-dependent phosphorylation of ribosomal protein S6 via dopamine- and cAMP-regulated phosphoprotein of 32 kDa and inhibition of protein phosphatase-1. Neuropharmacology. 2013;72:197–203. doi: 10.1016/j.neuropharm.2013.04.043. [DOI] [PubMed] [Google Scholar]
- 46.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nature Reviews Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 47.Law AJ, Hutchinson LJ, Burnet PW, Harrison PJ. Antipsychotics increase microtubule-associated protein 2 mRNA but not spinophilin mRNA in rat hippocampus and cortex. Journal of Neuroscience Research. 2004;76(3):376–82. doi: 10.1002/jnr.20092. [DOI] [PubMed] [Google Scholar]
- 48.Lidow MS, Song ZM, Castner SA, Allen PB, Greengard P, Goldman-Rakic PS. Antipsychotic treatment induces alterations in dendrite- and spine-associated proteins in dopamine-rich areas of the primate cerebral cortex. Biological Psychiatry. 2001;49(1):1–12. doi: 10.1016/s0006-3223(00)01058-1. [DOI] [PubMed] [Google Scholar]
- 49.Navari S, Dazzan P. Do antipsychotic drugs affect brain structure? A systematic and critical review of MRI findings. Psychological Medicine. 2009;39(11):1763–77. doi: 10.1017/S0033291709005315. [DOI] [PubMed] [Google Scholar]
- 50.Park SW, Lee CH, Cho HY, Seo MK, Lee JG, Lee BJ, Seol W, Kee BS, Kim YH. Effects of antipsychotic drugs on the expression of synaptic proteins and dendritic outgrowth in hippocampal neuronal cultures. Synapse. 2013;67(5):224–34. doi: 10.1002/syn.21634. [DOI] [PubMed] [Google Scholar]
- 51.Benes FM, Paskevich PA, Domesick VB. Haloperidol-induced plasticity of axon terminals in rat substantia nigra. Science. 1983;221(4614):969–71. doi: 10.1126/science.6879197. [DOI] [PubMed] [Google Scholar]
- 52.Meredith GE, De Souza IE, Hyde TM, Tipper G, Wong ML, Egan MF. Persistent alterations in dendrites, spines, and dynorphinergic synapses in the nucleus accumbens shell of rats with neuroleptic-induced dyskinesias. Journal of Neuroscience. 2000;20(20):7798–806. doi: 10.1523/JNEUROSCI.20-20-07798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Archives of Geneneral Psychiatry. 2000;57(1):65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 54.Kreczmanski P, Heinsen H, Mantua V, Woltersdorf F, Masson T, Ulfig N, Schmidt-Kastne R, Korr H, Steinbusch HW, Hof PR, Schmitz C. Volume, neuron density and total neuron number in five subcortical regions in schizophrenia. Brain. 2007;130(Pt 3):678–92. doi: 10.1093/brain/awl386. [DOI] [PubMed] [Google Scholar]
- 55.Ho BC, Andreasen NC, Ziebell S, Pierson R, Magnotta V. Long-term antipsychotic treatment and brain volumes: a longitudinal study of first-episode schizophrenia. Archives of General Psychiatry. 2011;68(2):128–37. doi: 10.1001/archgenpsychiatry.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vernon AC, Natesan S, Modo M, Kapur S. Effect of chronic antipsychotic treatment on brain structure: a serial magnetic resonance imaging study with ex vivo and postmortem confirmation. Biological Psychiatry. 2011;69(10):936–44. doi: 10.1016/j.biopsych.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 57.Hutcheson NL, Clark DG, Bolding MS, White DM. Basal ganglia volume in unmedicated patients with schizophrenia is associated with treatment response to antipsychotic medication. Psychiatry Research. 2013 doi: 10.1016/j.pscychresns.2013.10.002. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okugawa G, Nobuhara K, Takase K, Saito Y, Yoshimura M, Kinoshita T. Olanzapine increases grey and white matter volumes in the caudate nucleus of patients with schizophrenia. Neuropsychobiology. 2007;55(1):43–6. doi: 10.1159/000103575. [DOI] [PubMed] [Google Scholar]
- 59.Deinhardt K, Kim T, Spellman DS, Mains RE, Eipper BA, Neubert TA, Chao MV, Hempstead BL. Neuronal growth cone retraction relies on proneurotrophin receptor signaling through Rac. Science Signaling. 2011;4(202):ra82. doi: 10.1126/scisignal.2002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bálint E, Kitka T, Zachar G, Adám A, Hemmings HC, Jr, Csillag A. Abundance and location of DARPP-32 in striato-tegmental circuits of domestic chicks. Journal of Chemical Neuroanatomy. 2004;28(1–2):27–36. doi: 10.1016/j.jchemneu.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 61.Ouimet CC, Greengard P. Distribution of DARPP-32 in the basal ganglia: an electron microscopic study. Journal of Neurocytology. 1990;19(1):39–52. doi: 10.1007/BF01188438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Characterization of haloperidol-mediated pAkt response.
Supplementary Figure 2: Puromycin dosing and control experiments for genetic manipulations.
Supplementary Figure 3: Proteomic analysis of measured proteins
Supplementary Figure 4: Haloperidol-induced proteins that react with a MAP2 antibody in mouse striatal neurons or rat striatal neurons in vitro.
Supplementary Figure 5: DARPP32 can label spines in DIV14 cultured striatal neurons
Supplementary Table 1: Complete results of the 5-hour proteomic screen
Supplementary Table 2: Complete results of the 48-hour proteomic screens
Supplementary Table 3: Haloperidol-induced increases in proteins linked to mTORC1 signaling
Supplementary Table 4: Candidate proteins from cytoskeleton, translation, mRNA, membrane, and release protein groups
Supplementary Table 5: Statistical significance for 3 biological replicates for selected proteins of interest