Abstract
The inorganic polyphosphate (poly-P) is a key regulator of stress responses and virulence in many bacterial pathogens including Campylobacter jejuni. The role of exopolyphosphatases/guanosine pentaphosphate (pppGpp) phosphohydrolases (PPX/GPPA) in poly-P homeostasis and C. jejuni pathobiology remains unexplored. Here, we analyzed deletion mutants (∆ppx1, ∆ppx2) and the double knockout mutant (dkppx), all ∆ppx mutants exhibited increased capacity to accumulate poly-P; however only ∆ppx1 and dkppx mutants showed decreased accumulation of ppGpp, an alarmone molecule that regulates stringent response in bacteria, suggesting potential dual role for PPX1/GPPA. Nutrient survival defect of ∆ppx mutants was rescued by the supplementation of specific amino acids implying that survival defect may be associated with decreased ppGpp and/ or increased poly-P in ∆ppx mutants. The ppk1 and spoT were upregulated in both ∆ppx1 and ∆ppx2 suggesting a compensatory role for SpoT and Ppk1 in poly-P and ppGpp homeostasis. The lack of ppx genes resulted in defects in motility, biofilm formation, nutrient stress survival, invasion and intracellular survival indicating that maintaining a certain level of poly-P is critical for ppx genes in C. jejuni pathophysiology. Both ppx1 and ppx2 mutants were resistant to human complement-mediated killing; however, the dkppx mutant was sensitive. The serum susceptibility did not occur in the presence of MgCl2 and EGTA suggesting an involvement of the classical or lectin pathway of complement mediated killing. Interestingly, the chicken serum did not have any effect on the ∆ppx mutants’ survival. The observed serum susceptibility was not related to C. jejuni surface capsule and lipooligosaccharide structures. Our study underscores the importance of PPX/GPPA proteins in poly-P and ppGpp homeostasis, two critical molecules that modulate environmental stress responses and virulence in C. jejuni.
Keywords: Campylobacter jejuni, ppGpp, PPX, GPPA, poly-P, stress response
Introduction
Campylobacter jejuni is a microaerophilic gram-negative curved bacterium that persistently colonizes the intestine of various livestock especially, poultry.1 However, C. jejuni can infect humans and its infection is considered to be a prevalent cause of bacteria- mediated diarrheal disease worldwide.2 Campylobacteriosis is often a self-limiting disease characterized by fever, abdominal pain, vomiting, and sometimes bloody diarrhea. In addition, fatal post-infectious complications like Guillian–Barre Syndrome, Miller Fisher Syndrome, and reactive arthritis can also occur.3 Efforts to reduce campylobacteriosis in humans are directly linked to a better understanding of the pathobiology of C. jejuni; however, the molecular basis of such virulence mechanisms are not fully elucidated yet.
Inorganic polyphosphate (poly-P), a polymer of ten to hundreds of phosphate residues, linked by high-energy phosphoanhydride bonds, is conserved in every cell in nature.4 It can serve as a source of energy for synthesis of sugars, nucleosides, and proteins as well as an activating precursor for fatty acids, phospholipids, polypeptides, and nucleic acids. Poly-P is also a key regulator of bacterial survival, stress responses, host colonization, and virulence in many pathogenic bacteria including C. jejuni.5-7 Poly-P metabolism is governed by several specialized enzymes; Polyphosphate kinase-1 (PPK1) is responsible for the synthesis of long chain poly-P from ATP. Although the reaction is reversible, the synthesis of poly-P is favored.5,8 Polyphosphate kinase-2 (PPK2) utilizes poly-P to generate GTP at a rate 75-fold greater than that of the poly-P synthesis from GTP.9-11 Exopolyphosphatases (PPXs) degrade poly-P into a smaller branch of inorganic phosphate. PPXs have been classified into two kinds; the classical PPX and the dual function PPX/GPPA. Classical PPX enzymes are primarily monofunctional and their essential role is in poly-P homeostasis.12-15 Classical PPXs have been shown to be involved in motility, biofilm formation, sporulation and providing resistance to complement-mediated killing in different bacteria.16,17 On the other hand, PPX/GPPA enzymes have both exopolyphosphatase and guanosine pentaphosphate (pppGpp) phosphorhydrolase activities to generate guanosine tetraphosphate (ppGpp), an alarmone that regulates stringent response in bacteria.7,14,18,19 In particular, PPX/GPPA enzymes play a supplementary role in ppGpp metabolism related to the bacterial starvation induced stringent response.18,20-23
C. jejuni 81–176 encodes two potential dual function exopolyphosphatases ppx1/gppa (CJJ81176_0377) and ppx2/gppa (CJJ81176_1251). To date, no studies describe the role of these enzymes in poly-P hydrolysis and/or ppGpp synthesis as well as in C. jejuni pathophysiology. Since poly-P homeostasis is critical, and PPX/GPPA enzymes are involved in poly-P metabolism and possibly in ppGpp synthesis, we hypothesize that PPX/GPPA enzymes may have an important role in C. jejuni pathobiology and persistence in different environmental conditions. To test our hypothesis, we generated deletion mutants of ppx1 and ppx2 (∆ppx1 and ∆ppx2) as well as the double knockout for both genes (dkppx) and the analyses of these mutants suggest that ppx1/gppa contributes to both poly-P and ppGpp pools in C. jejuni and ppx/gppa genes are important for the C. jejuni stress responses and virulence related traits.
Results
The PPX/GPPA proteins of C. jejuni
The phylogenetic analysis using MEGA-5 (Center for Evolutionary Medicine and Informatics) showed that the C. jejuni PPX1/GPPA clustered with Helicobacter and Acrobacter whereas PPX2/GPPA with E. coli and Mycobacterium tuberculosis species (Fig. S1). Further ClustalW2, structure based sequence alignment using reference sequence of Aquifex aeolicus also showed that the C. jejuni PPX1/GPPA (CJJ81176_0377) has both the catalytic residue E116 required for PPX activity and arginine residues (R16 and R258) necessary for GPPA activity (Fig. S2A), suggesting that this enzyme might mediate both poly P and (p)ppGpp hydrolysis. Conversely, C. jejuni PPX2/GPPA (CJJ81176_1251) possesses the catalytic residues E103 and R12 but not R254 (Fig. S2A), indicating this enzyme is less likely a mediator of (p)ppGpp hydrolysis. Moreover, the predicted three dimensional (3D) structure of C. jejuni PPX1/GPPA showed closer alignment with the reference sequence from A. aeolicus PPX/GPPA than the PPX2/GPPA further suggesting a role for (Fig. S2B and C) PPX1/GPPA in pppGpp hydrolysis.
The PPX1/GPPA and PPX2/GPPA proteins are conserved across the different strains of C. jejuni with amino acid sequence identity ranging from 97% to 100%. In addition, PPX/GPPA enzymes are also conserved among other sequenced Campylobacter spp. (PPX1/GPPA sequence identity 35% to 97%; PPX2/GPPA sequence identity 46% to 99%). However, the PPX1/GPPA and PPX2/GPPA share 30% identity and 49% similarity in amino acid content as also noted in Corynebacterium glutamicum (PPX1; PPX2 share only 25% identity; 45% similarity) and M. tuberculosis (Rv0496-PPX/GPPA; Rv1026-PPX/GPPA share only 27% identity; 42% similarity).15,24
The ppx/gppa genes contribute to exopolyphosphatase function
In order to verify that the deletion of ppx/gppa genes did not affect the overall growth properties of the mutants, we assessed the ∆ppx mutants growth by determining CFUs at different time interval in MH broth. Our result confirmed that there was no significant difference between CFU forming ability of ∆ppx mutants (∆ppx1, ∆ppx2, dkppx) and wild type (Fig. S3).
To investigate ppx genes role in exopolyphosphatase activity, we measured the intracellular poly-P levels in ∆ppx mutants (∆ppx1, ∆ppx2, and dkppx). Our results indicated that the deletion of ppx genes alters the intracellular poly-P levels when compared with wild type. The ∆ppx1 and dkppx mutants showed significant (P ≤ 0.05, P ≤ 0.001) increase in the poly-P levels (45.5 nM and 48.5 nM/mg of total cellular protein [TCP], respectively) when compared with wild type (35 nM/mg TCP). The ∆ppx2 mutant also exhibited an increased poly-P levels (39.5 nM/mg TCP) but it was not significant (P = 0.079) (Fig. 1). The complemented strains (Cppx1 and Cppx2) displayed levels of poly-P (37 nM/mg TCP) similar to wild type. Our findings confirm that the ppx1/gppa and ppx2/gppa mutants have elevated poly-P and thus may encode exopolyphosphatase activity. Furthermore, these results indicate that ppx1/gppa gene has a major role in poly-P degradation and both genes may have additive effect in poly-P hydrolysis as observed with dkppx mutant.
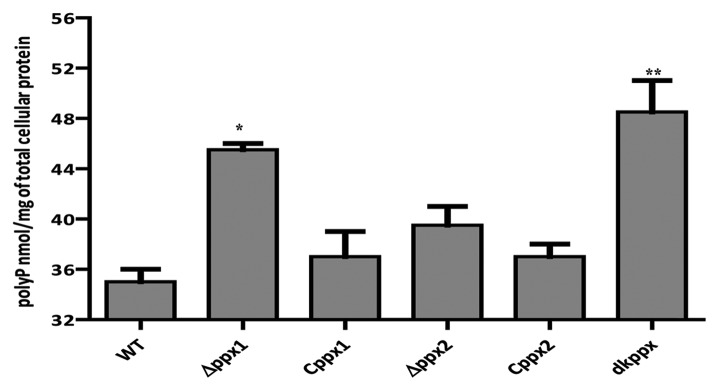
Figure 1. Intracellular poly-P levels in C. jejuni ∆ppx mutants. Poly-P was extracted using glass milk and quantified by toluidine blue O method. Each data point represents the mean ± SE of 3 independent experiments with duplicate samples. *P ≤ 0.05 and **P ≤ 0.001.
The ppx1/gppa gene contributes to ppGpp pool
It has been previously reported that metabolism of ppGpp in C. jejuni is primarily governed by SpoT;25 however, PPX/GPPA proteins in C. jejuni are structurally similar and possess conserved amino acid residues required for the bifunctional activity as seen in A. aeolicus, suggesting that these proteins may have a subsidiary role in ppGpp metabolism. To address this, we determined the levels of ppGpp in the wild type and ∆ppx mutants starved in MOPS for up to 24 h. Our results showed that there was no difference in ppGpp accumulation at 3 h but differences were observed at 6 h and 24 h. At 24 h, the ∆ppx1, and dkppx mutants displayed significantly decreased levels of ppGpp 6% (P < 0.05) and 12% (P < 0.001) respectively, compared with wild type. Conversely, the level of ppGpp in the ∆ppx2 mutant was significantly higher (30%) than wild type (Fig. 2). However, complemented strains (Cppx1 and Cppx2) did not show significant change in the intracellular ppGpp pool (data not shown). The lack of complementation is likely due to chloramphenicol used to grow these strains, since chloramphenicol has been shown to inhibit ppGpp synthesis.26 Further, the pppGpp levels were increased in both ppx1 (4.5%) and dkppx (5.76%) mutants compared with the wild type suggesting a dual role for ppx1. These results suggest that ppx1/gppa contribute to both poly-P and ppGpp pools in C. jejuni. This is consistent with the predicted 3D structure of C. jejuni PPX/GPPA proteins (Fig. S2B and C). Protein sequence and predicted 3D structure of C. jejuni PPX1/GPPA resembles the bi-functional PPX/GPPA protein of A. aeolicus. Only C. jejuni PPX1/GPPA possesses two conserved arginine residues (R16 and R258 corresponding to R22 and R267 of A. aeolicus) necessary for GPPA activity suggesting that the PPX1/GPPA of C. jejuni may be associated with both PPX and GPPA function as shown in our study.
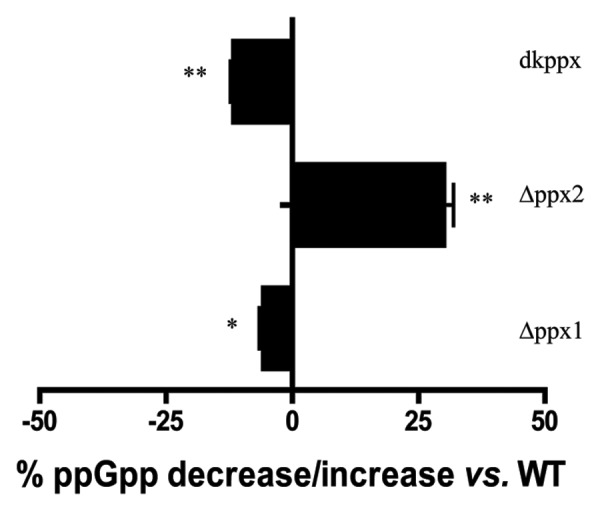
Figure 2. Intracellular ppGpp levels in the C. jejuni ∆ppx mutants. The amount of ppGpp accumulation was assessed in MOPS using early log phase culture labeled with 32P. The nucleotides were resolved by TLC and quantified using densitometry. The graph shows the percentage decrease/increase in the levels of ppGpp in mutants at 24 h. The complemented strains (Cppx1 and Cppx2) did not show a significant change in the intracellular ppGpp pool compared with wild type as chloramphenicol has been shown to inhibit ppGpp synthesis (data not shown). Each bar represents the average from 2 independent experiments performed using duplicate samples in each experiment.
C. jejuni ppx/gppa genes are essential for survival in basal media
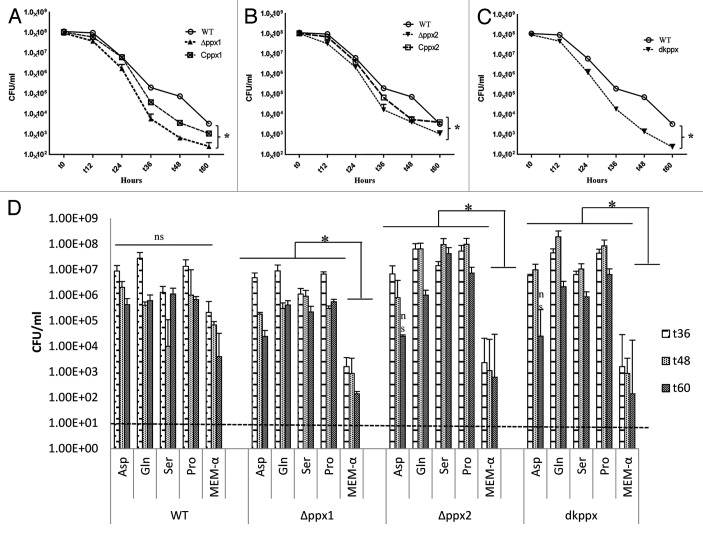
Previous studies have shown that poly-P is essential for C. jejuni to survive under low nutrient stress.6,27 Our results indicate that C. jejuni ∆ppx mutants have altered poly-P homeostasis. Thus, we further studied the mutants’ ability to withstand nutrient limitation stress by monitoring the ∆ppx mutants’ nutrient stress survival in MEM. Results indicate that ∆ppx1, ∆ppx2, and dkppx mutants were significantly (P ≤ 0.05) defective in survival after 36 h. Trans- complementation of ∆ppx1 and ∆ppx2 mutants partially restored the nutrient stress tolerance ability of mutants comparable to wild type (Fig. 3A–C).
Figure 3. Nutrient stress survival of ∆ppx mutants in minimum essential media: (A–C) Sensitivity of C. jejuni ∆ppx mutants to nutrient starvation was assessed by monitoring their survival in chemically defined media (MEM without glutamine) at different time points. All the ∆ppx mutants showed stress survival defect at t 36 h onwards and complemention partially rescued the defect. (D) Nutrient stress survival of ∆ppx mutants in stringent basal media (MEM-α) in presence or absence of specific amino acids. The specific amino acids (l-aspartate, l-glutamate, l-serine and l-proline) were added to basal media at the concentration of 20 mM. CFU/mL value represents mean ± SD of two independent experiments with duplicate samples in each experiment. The dotted line across the bar graph indicates the limit of detection (10 CFU/mL). *P ≤ 0.05. n.s denotes not significant.
Growth of C. jejuni depends upon availability of free amino acids.28 Further studies have shown that the preferential utilization of serine, aspartate, glutamate and proline in vitro by C. jejuni.29 It is also known that perturbation in ppGpp levels is associated with amino acid starvation in E. coli,30 we hypothesize that specific amino acids supplementation to ∆ppx mutants may rescue ppGpp-induced growth defect in basal media (MEM-α). Supplementation of amino acids serine, proline, glutamate, and aspartate at 20 mM in basal media significantly rescued the survival defect of all ∆ppx mutants at t36, t48, and t60 h except for aspartate in ∆ppx2 and dkppx at 60 h (Fig. 3D). Growth of the ∆ppx2 mutant, even though not defective in intracellular ppGpp accumulation, was also rescued by specific amino acids supplementation. However, although the amino acid supplementation in wild-type C. jejuni enhanced its survival, it was not significant. These results suggest that altered poly-P and/ or ppGpp levels in ppx mutants may contribute to C. jejuni nutrient starvation stress survival.
The ppx/gppa genes are essential for motility and biofilm formation in C. jejuni
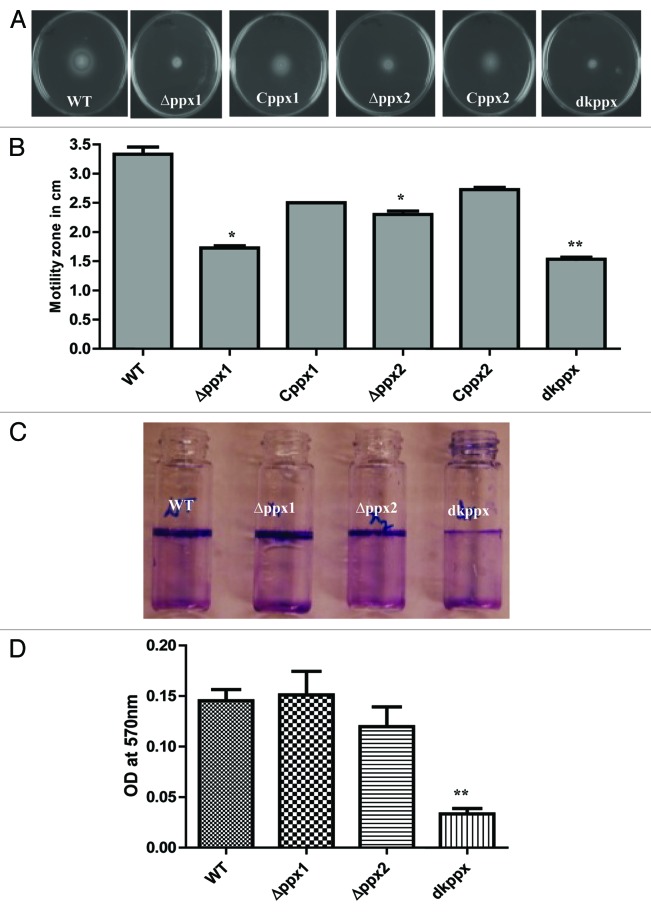
Our results showed that C. jejuni ∆ppx mutants displayed significant (P ≤ 0.05, P ≤ 0.001) defect in motility. The motility zone for ∆ppx1, ∆ppx2, and dkppx were 1.7 cm, 2.3 cm, and 1.5 cm respectively, compared with wild type (3.3 cm). The motility defects were partially restored in complemented strains Cppx1 (2.5 cm) and Cppx2 (2.7 cm) (Fig. 4A and B).
Figure 4. Motility and biofilm formation of C. jejuni ∆ppx mutants. (A) Motility was assessed on 0.4% soft agar at 42 °C for 24 h. (B) Motility was quantified by measuring the swarming zone (diameter in cm) surrounding the stabbed area. Each bar represents the average from 3 independent experiments with duplicate samples. *P ≤ 0.05 **P ≤ 0.001. (C) The biofilm formation was visualized by staining with 1% crystal violet for 15 min. (D) The amount of biofilm was quantified by measuring the absorbance at 570 nm after dissolving in 3 mL DMSO for 48 h. Each bar represents the mean ± SE of 3 independent experiments with triplicate samples in each experiment. **P ≤ 0.001.
We next investigated the role of ppx/gppa genes in C. jejuni biofilm formation. Results in Figure 4C and D indicate that ∆ppx1 and ∆ppx2 mutants showed no impairment in biofilm formation where the amount of biofilm formation for the ∆ppx1 (OD560 = 0.154) and for ∆ppx2 (OD560 = 0.119) was similar to the wild type (OD560 = 0.145). However, the deletion of both genes (dkppx mutant) significantly (P ≤ 0.001) affected the amount of biofilm formation (OD570 = 0.033) (Fig. 4C and D). Altogether, these findings indicate that ppx/gppa genes play a role in transmission related phenotypes such as motility and biofilm formation in C. jejuni.
Deletion of ppx/gppa genes affects C. jejuni survival during osmotic stress but not oxidative stress
The ability of C. jejuni ppx mutants to resist osmotic stress was determined in the presence of NaCl as an osmotic stressor. Our results indicate that there was no significant difference in the osmotic stress tolerance response when tested on MH agar containing 0.17 M NaCl in both ∆ppx1 and ∆ppx2 mutants compared with wild type. Conversely, the dkppx mutant did exhibit a significant (P ≤ 0.05) sensitivity to osmotic stress in comparison to wild type strain (Fig. 5A and B). Furthermore, our CFU data indicated that the survival of the dkppx mutant under liquid osmotic stress (MH broth containing 0.25 M NaCl) was also significantly (P ≤ 0.05) affected at 48 and 60 h. Since, ∆ppx1 and ∆ppx2 mutants did not show defect in osmotic stress tolerance on solid media, we did not test these strains in liquid culture. These results indicate that the ppx/gppa genes may play a role in the C. jejuni osmotic stress tolerance.
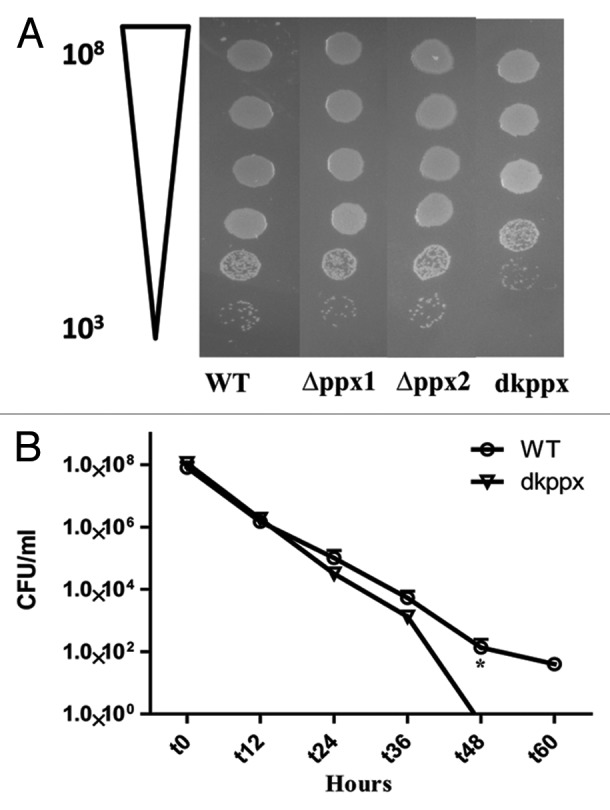
Figure 5. Osmotic stress responses of C. jejuni ∆ppx mutants. The dkppx mutant shows decreased osmotic stress tolerance on solid media (A) as well as liquid media (B). In liquid media osmotic stress was determined by monitoring cells survival at different time points. Each bar represents the mean ± SE from 3 independent experiments with duplicate samples in each experiment. *P ≤ 0.05.
The ∆ppx mutants showed no significant difference between zones of sensitivity to oxidative stressors (0.3% H2O2 or 20 mM Paraquat) compared with wild type (Fig. S4A and B).
The ∆ppx mutants exhibit defect for invasion and intracellular survival
To assay virulence associated phenotypes of invasion and intracellular survival, C. jejuni ∆ppx mutants and wild-type strains were allowed to infect INT 407 human intestinal epithelial cells at different MOIs. Our results indicated that the invasion profile of the ∆ppx mutants and wild type were similar at higher MOI; however, at low MOI the mutants were consistently less invasive. The intracellular survival was also assessed using a MOI of 100:1, although both ∆ppx2 and dkppx mutant were defective in intracellular survival, only the dkppx mutant was significantly (P ≤ 0.001) defective in comparison to wild type (Fig. 6A and B). These data suggest that both PPX/GPPA enzymes are important for the invasion and intracellular survival.
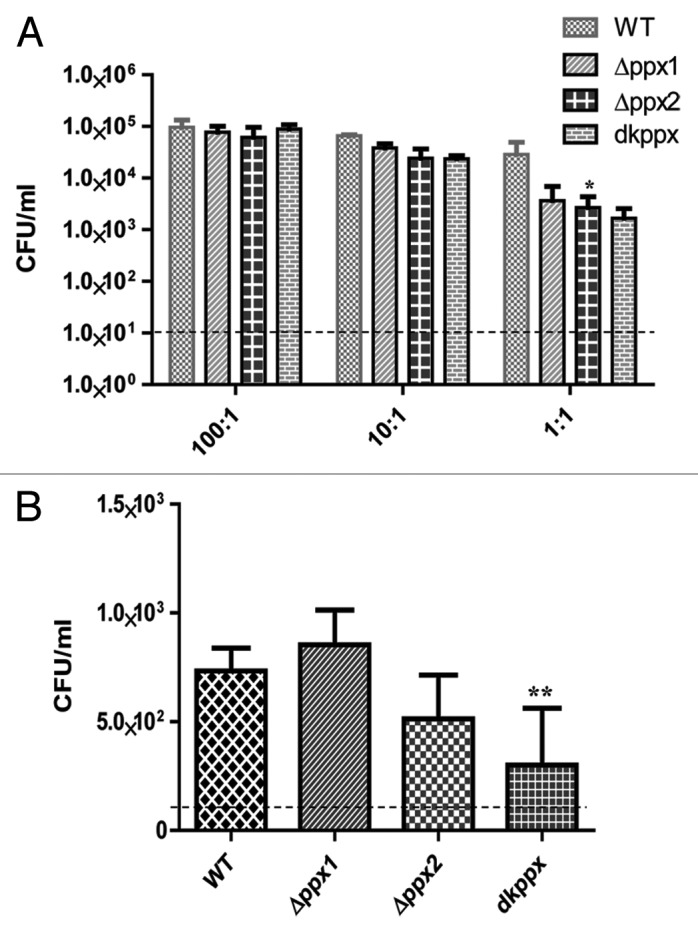
Figure 6. The ppx mutants display defect in invasion and intracellular survival in INT 407 human intestinal epithelial cells. (A) Invasion assay and (B) Intracellular survival assay. The dotted line across the bar graph indicates the limit of detection (10 CFU/mL). Each bar represents the mean ± SE from 2 independent experiments with duplicate samples in each experiment. *P ≤ 0.05 **P ≤ 0.001.
The ppx1 and ppx2 mutants display resistance to complement-mediated killing
The bactericidal activity of complement factors present in serum is an innate defense mechanism against intruding pathogenic bacteria. Since the lack of PPX in Neisseria meningitidis increased its resistance to complement-mediated killing,17 we next asked whether PPX/GPPA may have similar role in C. jejuni. To assess the complement mediated killing we compared the growth and survival of mutant and wild type strains in pooled normal (N) or heat inactivated (HI) human and chicken serum (NHS/NCS), respectively. The ∆ppx1 and ∆ppx2 mutants showed significant (P ≤ 0.05) resistance to killing by human serum complement compared with wild type; surprisingly, the deletion of both the genes (dkppx) significantly (P ≤ 0.05) increased the sensitivity to complement mediated killing. Trans-complementation of ppx1 and ppx2 genes restored the complement mediated killing to levels similar to wild type (Fig. 7A). However, surface structures such as LOS and CPS could not be attributed to complement mediated killing (resistant/sensitivity) observed for the mutants (Fig. S5A and B). This was further confirmed by qRT-PCR analysis for the expression of genes involved in LOS and CPS (kpsM, cstII, waaF, and lgtF) biosynthesis,31 which was similar in all mutants compared with wild type (Fig. S6).
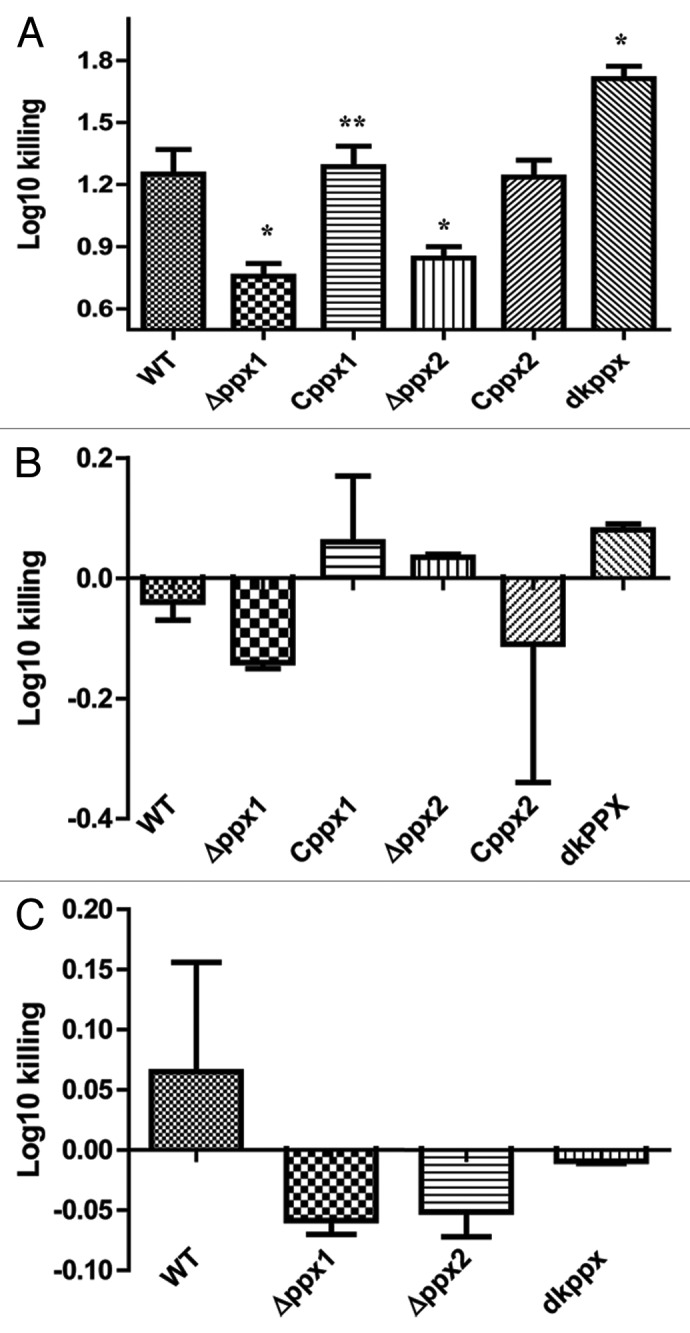
Figure 7. Complement dependent killing of C. jejuni ∆ppx mutants. (A) ∆ppx mutants killing by human serum. (B) ∆ppx mutants killing by human serum in the presence of MgCl2 and EGTA (C) ∆ppx mutants killing by chicken serum. Log10 killing was determined by subtracting the difference between number of CFU determined in heat inactivated human or chicken serum (HINHS or HINCS) to normal serum (NHS or NCS). Bars represent the mean ± SE of 3 independent experiments performed in duplicates each time. *P ≤ 0.05.
To further investigate the complement pathways involved, the activity of the classical and lectin pathways was inhibited in normal sera by adding 10 mM MgCl2 and 10 mM EGTA.17,32 Addition of MgCl2 and EGTA inhibited the observed relative serum resistance in both ∆ppx1 and ∆ppx2 and the sensitivity of dkppx (Fig. 7B) suggesting that serum resistance or sensitivity is likely mediated by the classical and/or the lectin pathway. Further, complement-mediated killing in human serum at 37 °C and 42 °C was similar; however, the killing efficiency was slightly pronounced at 37 °C (data not shown). In contrast to human serum, chicken serum had no effect on the killing of the ∆ppx mutants (Fig. 7C).
spoT and ppk1 are upregulated in ∆ppx mutants
Next, in order to better understand the mechanistic aspects of poly-P homeostasis in C. jejuni ∆ppx mutants, we performed qRT-PCR targeting known key genes involved in poly-P and ppGpp metabolism (spoT, ppk1, and ppk2).6Our results show that spoT expression was upregulated 2-fold or more in all three mutants and was significant (P ≤ 0.05) in ∆ppx1 mutant (>6-fold) (Fig. 8) compared with wild type. Similarly, ppk1 expression was also upregulated more than 2-fold in the ppx1 and dkppx mutants; whereas the expression of ppk2 was similar in all mutants compared with wild type. These data suggest that the upregulation of ppk1 and spoT might be important in regulating poly-P and ppGpp levels thus avoiding excessive accumulation of poly-P in the absence of ppx genes.
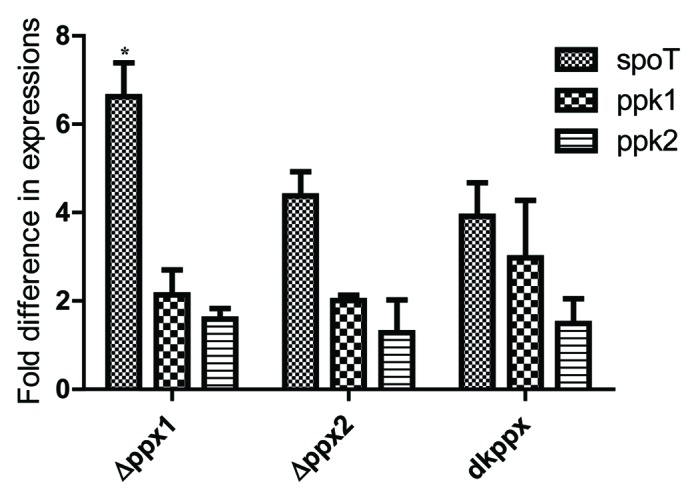
Figure 8. qRT-PCR analysis of C. jejuni ∆ppx mutants for genes involved in poly-P and ppGpp homeostasis. Fold difference in transcript levels was assessed by ∆∆CT method. The expression of target genes was normalized to the 16s-rRNA expression levels from the same strain and then the relative expression of target genes were determined by comparing to expression in wild type strain. Each bar represents the mean ± SE of the relative fold change in expression from 4 independent experiments performed in duplicates each time. *P ≤ 0.05.
Discussion
In contrast to other well studied enteric pathogens such as Salmonella, Shigella, and E. coli, C. jejuni lacks most of the classical virulence factors. Sequencing of several C. jejuni strains, including one known to be highly virulent and invasive strain (C. jejuni 81–176), did not uncover any of the classical virulence factors.33-35 Moreover, relatively small size genome (1.64 Mb) of C. jejuni demands to exploit most of its genes efficiently to meet great challenges to survive and transmit between hosts. Hence, a better understanding of the host-microbe interactions that affect the C. jejuni pathogenesis is of immense importance.
Previous studies have shown that the poly-P plays a critical role in virulence related phenotypic traits in pathogenic bacteria including C. jejuni.6,36,37 In this work, we focused to expand our understanding of poly-P homeostasis in C. jejuni by investigating the role of PPX enzymes. In M. tuberculosis, deficiency of PPX leads to increased accumulation of poly-P and restricted the growth in auxenic cultures and in human macrophages.38 In A. aeolicus, PPX/GPPA enzymes, in addition to poly-P degradation, also breaks down pppGpp to ppGpp,18,23 playing an important role in the starvation induced stringent response. As shown in the present study, C. jejuni has two exopolyphosphatases which individually or additively act in maintaining the poly-P pool (Fig. 1) but are not redundant. In C. glutamicum, deletion of ppx1 or ppx2 resulted in increased poly-P accumulation.15 Determination of exopolyphosphatase activity and intracellular poly-P concentration in C. glutamicum revealed that PPX2 is the major exopolyphosphatase effective on short chain poly-P. C. glutamicum PPX1 and PPX2 share only 25% amino acids identity and it was suggested that non-overlapping substrate preference may be the reason for occurrence of two or more exopolyphosphatase within one species.15 Likewise, PPX1 and PPX2 of C. jejuni share only 30% amino acid identity with the PPX1/GPPA likely serving as a major exopolyphosphatase; however, further analysis to detect poly-P molecules and their sizes are needed to determine C. jejuni PPX1 and PPX2 substrate specificity. The occurrence of two or more exopolyphosphatases within a species is not restricted to C. jejuni as is also seen in E. coli, Vibrio cholera, M. tuberculosis, and C. glutamicum.15,24 If we take into account that poly-P confers improved fitness under environmental stress,36 then the selective maintenance of ppx genes may be strongly associated with changing environments, while an immovable environment, such as a host cell, could cause gene/function loss, as suggested by the lack of ppx genes in Mycoplasma spp.39
Previous studies of PPX/GPPA proteins in A. aeolicus and E. coli have shown that these enzymes possess bifunctional activity.18 However, in M. tuberculosis two PPX-GPPA homologs were shown to have distinct biochemical activities.24 To our knowledge, this is the first study in C. jejuni depicting its PPX/GPPA possible dual role and its significance in numerous transmission and virulence related phenotypes. Our densitometry data for ppGpp production indicated that both ∆ppx1 and dkppx mutants have decreased ppGpp (Fig. 2), while ppx2 mutant has increased ppGpp levels when compared with wild type. This suggests that the ppGpp metabolism in C. jejuni is also regulated by factors other than PPXs. In this context Gaynor et al. showed that C. jejuni stringent response is mediated by SpoT which mediates both ppGpp synthesis and pppGpp hydrolysis and lack of this protein impairs C. jejuni resistance to rifampicin.25,26 Our qRT-PCR data support the concept of SpoT upregulation as a compensatory mechanism in poly-P and ppGpp homeostasis (Fig. 8). However, sensitivity to rifampicin was not affected in the ppx mutants (data not shown) which suggest that ppGpp levels are not below the threshold levels to have any effect on rifampicin sensitivity.25 Further studies are needed to understand the complex mechanisms behind regulation of (p)ppGpp by the PPX-GPPA proteins in C. jejuni.
C. jejuni’s ability to withstand the nutrient limited conditions in environment is critical for waterborne transmission. The biofilm formation and viable but not culturable (VBNC) state are known defense mechanism that a bacterium can exploit.40 The poly-P homeostasis in C. jejuni is shown to be important for biofilm formation, nutrient stress survival, osmotic stress tolerance, and for the maintenance of the VBNC state.6,27 Furthermore, C. jejuni flagellar motility has also been implicated in biofilm formation.41-44 As shown in B. cereus,16 our data indicate that PPX/GPPA enzymes are required for the motility and biofilm formation. Although the motility was affected in both mutants, a pronounced defect was seen in the dkppx mutant, suggesting that C. jejuni ppx1/gppa and ppx2/gppa genes additively contribute to motility. Conversely biofilm formation was only affected in the dkppx mutant (Fig. 4A and B) indicating a complex interconnected network of genes regulating this process in C. jejuni.
Complement-mediated bacterial killing is a key component of the humoral arm of the innate immunity necessary to control systemic infections from mucosal surfaces. Thus encapsulated bacteria counteract the antibacterial effect of complement by preventing deposition of the membrane attack complex on bacterial membranes.45 In N. meningitidis, lack of PPX increases its resistance to complement-mediated killing independently of changes in surface structures such as CPS, LPS, and the factor-H-binding protein. Instead N. meningitidis lack of PPX function modifies the interaction with the components of alternative pathway of complement activation.17 Our findings are in agreement with this previous report, as the deletion of individual ppx genes conferred resistance to complement-mediated killing. Consistent with this finding, in a previous study it was reported that poly-P by itself can inhibit complement-mediated killing.17 Strikingly, our result also showed that in C. jejuni deletion of both genes (dkppx) results in sensitivity to complement-mediated killing (Fig. 7A) suggesting a pleotropic effect of the double knockout contributing to its sensitivity even in the presence of higher levels of poly-P.
Analyses of the CPS and LOS profiles of C. jejuni ppx mutants as well as qRT-PCR analysis of selected LOS/CPS biosynthetic genes suggested that these surface structures may not play a role in complement-mediated killing. Additionally, our results revealed that either classical or lectin mediated complement activation pathway are likely to play a role in C. jejuni ppx-mediated resistance or sensitivity to human serum (Fig. 7B). Perhaps the increased resistance of ppx1 and ppx2 mutants may suggest that the C. jejuni uses it genes optimally to suite its lifestyle in humans and thus facilitating further spread to new host. However, additional studies are required to resolve the mechanism(s) behind observed susceptibility to the complement mediated killing. Interestingly, there was no effect of chicken complement factors on ppx mutants’ killing (Fig. 7C). This could be explained if the functionality of PPX/GPPA proteins depends on several factors such as temperature, host restricted expression of ppx genes, and/or the host immune system tolerance, among others. To our knowledge, this is first study showing differential contribution of ppx genes to complement mediated killing.
In our study in general the significant phenotypic changes (motility, biofilm formation, osmotic stress survival, and serum sensitivity) were observed in the double mutant (dppx) compared with the single mutants. However at present, we do not know what molecules are involved in these phenotypic differences. We believe that it is either due to additive effect of ppx1 and ppx2 deletion leading to higher poly-P levels or decreased ppGpp in the double knockout compared with single mutant; for example, motility was affected more in the double mutant compared with the single mutants. However, we observed osmotic stress defect, biofilm formation defect, and serum sensitivity only in the double mutant, we believe that these results observed in the double mutant is most likely a pleotropic effect of increased poly-P or decreased ppGpp to a threshold levels that is not seen in single mutants. It is also likely that poly-P and/ or ppGpp molecules act separately or in combination toward specific phenotypes. In this regard, a recent study has demonstrated that ppGpp and poly-P signaling molecules uniquely and/or commonly regulated developmental regulators with cell type specific activities in Caulobacter crescentus.46
In conclusion, we report that C. jejuni ppx/gppa genes are important for maintaining critical levels of poly-P and ppGpp in the cell. Besides their role in stress related phenotypic traits, they have a role in complement mediated killing. Our study expands the multi-factorial regulation of poly-P metabolism in C. jejuni.
Materials and Methods
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. All studies were performed with the highly invasive C. jejuni strain 81–176 originally isolated from a diarrheic patient.47 C. jejuni cultures were routinely grown on Mueller–Hinton (MH; Becton Dickinson and Company) agar under microaerobic condition (85% N2, 10% CO2, and 5% O2) in a DG250 Microaerophilic Workstation (Microbiology International). For the growth curve and stress survival assays, C. jejuni strains were cultured in MH broth with or without specific antibiotics at 42 °C with shaking at 200 rpm. All the bacterial strains were enumerated (CFU/mL) on MH agar by plating 10-fold serial dilutions. For the purpose of cloning and plasmid propagation, Escherichia coli DH5α cells were used and routinely cultured on Luria-Bertani (LB; Becton Dickinson and Company) medium at 37 °C. Growth media were supplemented with specific antibiotics: chloramphenicol (20 µg/mL for E. coli, 10 µg/mL for C. jejuni), zeocin (50 µg/mL), and kanamycin (30 µg/mL) wherever required.
Table 1. Bacterial strains and plasmids used in this study.
| Strain/plasmid | Relevant description | Source |
|---|---|---|
| Strains | ||
| C. jejuni 81–176 | Used as wild type strain of C. jejuni | Dr Qijing Zhang |
| E. coli DH5α | E. coli strain used for cloning | Invitrogen |
| ∆ppx1 | C. jejuni 81–176 derivative with deletion in ppx1/gppa gene; ppx1/gppa::Kan | This study |
| ∆ppx2 | C. jejuni 81–176 derivative with deletion in ppx2/gppa gene; ppx2/gppa::Kan | This study |
| dkppx | C. jejuni 81–176 derivative with deletion of ppx1/gppa and ppx2/gppa; ppx1/gppa::Kan, ppx2/gppa::Cm | This study |
| Cppx1 | ∆ppx1 with pRY111- containing ppx1coding region and the upstream promoter sequence for complementation; Cm | This study |
| Cppx2 | ∆ppx2 with pRY111- containing ppx2 coding region and the upstream promoter sequence for complementation; Cm | This study |
| Plasmids | ||
| pRY111 | E. coli-Campylobacter shuttle vector for complementation | Yao et al.50 |
| pRK2013 | Helper plasmid for conjugation, Kan | Akiba et al.58 |
| pZErO-1 | Cloning vector for making suicide vector; Zeo | Invitrogen |
| pUC4K | Source for kanamycin gene, Kan | Amersham |
| pZErO-1-ppx1 | pZErO-1 containing ppx1/gppa region plus 1 kb upstream and downstream sequences from C. jejuni 81–176; Zeo | This study |
| ppx1-suicide vector | pZErO-1-ppx1 with ppx1/gppa replaced by kanamycin resistance gene from pUC4K; Zeo, Kan | This study |
| pZErO-1-ppx2 | pZErO-1 containing ppx2/gppa region plus 1 kb upstream and downstream sequences from C. jejuni 81–176; Zeo | This study |
| ppx2-suicide vector | pZErO-1-ppx2 with ppx2/gppa replaced by kanamycin resistance gene from pUC4K; Zeo, Kan | This study |
| ppx2-suicide vector-Cm | ppx2-suicide vector with kanamycin resistance marker replaced by chloramphenicol resistance gene from pRY111; Zeo, Cm | This study |
| pRY111-ppx1 | pRY111 containing ppx1 coding and upstream sequence for complementation, Cm | This study |
| pRY111-ppx2 | pRY111 containing ppx2 coding and upstream sequence for complementation, Cm | This study |
Kan, kanamycin; Zeo, zeocin; Cm, chloramphenicol
Cloning techniques
The primers for PCR and qRT-PCR were designed using Vector NTI® software (Invitrogen) or Integrated DNA Technologies scientific tool (IDT) and were commercially synthesized by IDT. All the primers used in the present study are listed in the Table S1. QIAquick®PCR purification and a spin miniprep kits for plasmid isolation were purchased from Qiagen. Masterpure® DNA purification and Fast-Link DNA ligation kits were purchased from Epicenter. Restriction enzymes were purchased from Promega. pZEro-1, the zero background cloning vector and E. coli DH5α competent cells were purchased from Invitrogen. Cloning and other molecular biology techniques were performed as previously described.48
Construction of C. jejuni 81–176 ppx deletion mutants and complemented strains
C. jejuni 81–176 genome possesses two exopolyphosphatase genes, ppx1/gppa (CJJ81176_0377) and ppx2/gppa (CJJ81176_1251) encoding 486 and 324 amino acids, respectively (Fig. S7). Further, BLAST search using ppx/gppa gene from Aquifex aeolicus as a reference sequence confirmed that these two genes likely represent exopolyphosphatases. The deletion mutants (∆ppx1, ∆ppx2, and dkppx) were generated by double crossover homologous recombination as we described previously.41 Briefly, to construct the mutant, the target gene, along with its upstream and downstream sequences (approximately 1 kb), was amplified from C. jejuni 81–176 genomic DNA with specific primers. The PCR product was cloned into pZErO-1 plasmid, the resulting construct was amplified by inverse PCR ensuring that most of the ppx1 and ppx2 coding sequences were deleted. The inverse PCR product was ligated to a kanamycin gene from pUC4K and the final suicide vector was introduced into C. jejuni 81–176 by electroporation as described.49 To generate a double knockout (dkppx) mutant, ppx2 suicide vector, in which the kanamycin resistance gene was replaced with chloramphenicol gene, was electroporated into ∆ppx1 mutant. Colonies grown on MH agar containing kanamycin and chloramphenicol were selected as the double knockout mutant and deletion of target genes were confirmed by PCR using primers specific to upstream and downstream sequences of the target gene.
For complementation, target genes along with the potential promoter regions were amplified from genomic DNA with specific set of primers; products were cloned into pRY111, an E. coli–Campylobacter shuttle vector.50 Resulting constructs were introduced into the respective mutants by triparental conjugation. The complementation strain for the dkppx mutant was not generated due to a limited availability of replicating plasmids as well as antibiotic markers of choice for use in C. jejuni. Complemented strains (Cppx1 and Cppx2) were also confirmed by PCR.
Growth curve assay
The growth curve assay was performed as described previously.9 Cultures were grown to mid-log phase on MH agar, washed, suspended in MH broth, adjusted to OD600 0.05, and incubated microaerobically at 42 °C for 60 h with shaking at 200 rpm. For assessing the growth, CFUs were determined at different time points by plating 10-fold serially diluted cultures on MH agar.
Extraction and quantification of poly-P
The extraction of poly-P from C. jejuni strains were performed using the glassmilk method as previously described.6,9 Briefly, C. jejuni cultures were grown in MH broth to mid-stationary phase to reach OD600 of 0.3, cells were harvested followed by lysis with buffer (guanidine isothiocynate-GITC 4 mM, 500 mM TRIS-HCl pH 7.0). Total protein concentration in the lysate was estimated by Bradford analysis (BCA kit, Pierce Scientific) and poly-P content was analyzed using toluidine blue O dye. Briefly, for poly-P quantification, lysates were washed with buffer (5 mM TRIS-HCl [pH-7.5], 50 mM NaCl, 5 mM EDTA, 50% ethanol) and pellets were resuspended in a buffer (50 mM TRIS-HCl [pH 7.4], 10 mM MgCl2, and 20 µg each of DNase and RNase per mL) and the poly-P was eluted by brief centrifugation at 9500 × g with elution buffer (50 mM TRIS-HCl pH 8.0).
The poly-P extracted from C. jejuni lysates were incubated with toluidine blue O dye (TBO 6 mg/L in 40 mM acetic acid) at room temperature followed by assessing absorbance at 630 nm and 530 nm, where the ratio of 530 nm/630 nm provides the amount of poly-P in a given sample. Levels of poly-P in C. jejuni lysates were determined by direct comparison with a phosphorus standard curve and were expressed as nmol/mg of total cellular protein.
ppGpp isolation and detection
Isolation and detection of ppGpp was assayed as described previously.9,25 Briefly, bacterial strains were grown on MH agar for 16–18 h. Cultures were diluted to adjust OD600 of 0.25 in MH broth; cells were pelleted, washed twice in MOPS-MGS at 9500 × g (50 mM MOPS, 55 mM mannitol, 1 mM MgSO4, 0.25 mM CaCl2, 19 mM glutamic acid, and 0.004 mM biotin) and suspended in 250 μL MOPS-MGS.32,51 32P at 100 μCi per mL (3.7 × 1012 Bq) was added to cells and incubated microaerobically for 3, 6, and 24 h at 37 °C. Previously we have observed that the prolonged exposure (starvation stress) in MOPS results in increased accumulation of ppGpp that can be readily detected.9 Labeled bacteria were harvested at the above specified times, washed and treated with lysozyme in 10 mM Tris (pH 8.0) for 20 min. The ppGpp was extracted with equal volume of 2 M formic acid and placed on ice for 15 min. Samples were spun for 5 min at 9500 × g, and 3 μL of supernatant was spotted directly onto cellulose TLC plates, dried, and developed in 1.5 M KH2PO4 and visualized by autoradiography. For TLC separation, loading concentrations were normalized across all strains and approximately equal number of cells were loaded on the TLC plate.
Motility assay
The motility assay was performed as described previously.41 Briefly, C. jejuni grown overnight on MH agar microaerobically at 42 °C was harvested, washed with MH broth and OD600 was adjusted to 0.05. Two microliter culture (~5 × 107 cells) was stabbed onto 0.4% semi-solid agar and incubated microaerobically at 42 °C. After 24 h of incubation, the swarming zone (diameter) around the stabbed area was measured in centimeters.
Biofilm assay
The static biofilm formation was assayed as previously described.6 Briefly, cells were grown microaerobically overnight on MH plate at 42 °C, harvested in MH broth and OD600 was adjusted to 0.05. Then the culture (100 μL) was inoculated to 2 mL of MH broth in borosilicate tubes and incubated microaerobically at 42 °C for 48 h without shaking. Static biofilm formation was visualized by staining with 1% (w/v) crystal violet for 15 min. To quantify amount of biofilms, 3 mL of dimethyl sulfoxide (DMSO, Sigma) was added and incubated at room temperature for 24 h. The absorbance was measured at 570 nm.
Growth in chemically defined medium
Briefly, C. jejuni cultures were grown microaerobically on MH agar for 16–18 h at 42 °C. The cultures were washed twice by centrifuging at 9500 × g for 2 min with minimum essential media (MEM without glutamine) or stringent basal media (MEM-α) with or without supplementation of amino acids l-serine, l-asperatate, l-glutamate, and l-proline at 20 mM concentration. Bacteria were suspended in the defined media to adjust OD600 of 0.05 (approximately 1 × 108 CFU/mL) and incubated microaerobically at 42 °C with shaking at 200 rpm. CFUs were determined by plating on MH agar at different time intervals.
Osmotic stress assay
Osmotic stress was determined as described previously.6 Briefly, mid log-phase grown bacterial cultures were suspended in MH broth and OD600 adjusted to 0.05. Cultures were then serially diluted (10-fold) and a 10 μL samples were spotted on MH agar containing 0.17 M NaCl and the growth was assessed after 48 h of incubation microaerobically at 42 °C. To test survivability of ppx/gppa mutants to osmotic stress in liquid media, cultures were adjusted to OD600 0.05 in MH broth containing 0.25 M NaCl and incubated microaerobically for 60 h at 42 °C with shaking at 200 rpm. CFUs were assessed at different time points by plating on MH agar.
Oxidative stress assay
The ability of C. jejuni ppx mutants to survive under oxidative stress was assessed as previously described.52 Briefly, bacterial strains (~5 × 107 cells) were spread for confluent growth on MH agar. At the center of MH agar, 6 mm diameter hole was made and well bottom was sealed with 50 µL of molten MH agar. Wells were filled with 30 µL 0.3% H2O2 or 20 mM paraquat and plates were incubated microaerobically for 24 h at 42 °C, clear zones of growth inhibition surrounding the wells were measured in centimeter.
Rifampicin sensitivity assay
Fresh overnight cultures of C. jejuni ppx strains were streaked onto MH plate containing different rifampicin concentration (50, 100, 150, 200, 250, and 300 µg/mL) as previously described.25 The plates were incubated at 37 °C, microaerobically for 24–48 h to determine the sensitivity.
Invasion and intracellular survival in INT407 cells
The invasion and intracellular survival in INT 407 cells were performed as previously described.25,53 INT 407 cells (~1.4 × 105) in MEM with 10% (v/v) fetal bovine serum (FBS) were seeded to 24-well tissue culture plate and incubated for 18 h at 37 °C with 5% CO2. MH broth grown mid-log phase C. jejuni cultures were pelleted by centrifuging at 9500 × g for 10 min and washed twice with MEM containing 1% (v/v) FBS, adjusted to OD600 of 0.02, and subsequently used to infect INT 407 cells. Different multiplicities of infection (MOI) were used to infect the cells in duplicate wells. For invasion 1:1, 10:1, 100:1 MOIs were used, and 100:1 MOI was used for intracellular survival assay. To assess invasion, INT 407 cells were incubated with bacteria for 3 h and treated with gentamicin (150 µg/mL), and incubated for additional 2 h. Infected cells were washed with MEM twice, lysed with 0.1% (v/v) Triton-X 100, and 100 µL of an aliquot from each well was 10-fold serially diluted in MEM and plated on MH agar in duplicate to determine CFUs.
In order to determine intracellular survival, following 2 h of gentamicin treatment as above, infected cells were washed twice with MEM and fresh MEM containing gentamicin (10 µg/mL) was added and incubated for additional 24 h. Following incubation, infected cells were processed to assess CFUs as described above for the invasion assay.
Serum bactericidal assay
Serum bactericidal assay was performed as described previously.54,55 Commercially available pooled normal human serum (NHS) and pooled normal chicken serum (NCS) (Innovative Research) were used. Briefly, C. jejuni strains were harvested after 16–18 h of growth on MH plates, washed and suspended to 106 cells/mL in Medium 199 with Hank’s balanced salt solution containing 0.01% glutamine. From each culture, 150 µL aliquots were transferred in duplicate to a 96-well microtiter plate. To each well, 50 µL of either 100% NHS/NCS in Medium 199, 100% heat-inactivated (56 °C for 30 min) HI-NHS/NCS or Medium 199 alone was added to obtain 25% final concentration of NHS/NCS or HI-NHS/NCS and the plate was incubated for 60 min microaerobically at 37 °C for human serum, and at 42 °C for chicken serum. Aliquots (100 μL) from each well were 10-fold serially diluted and plated to assess CFUs. Log10 killing was determined by subtracting the number of CFUs between HI-NHS/NCS to NHS/NCS-treated samples.
Detection of capsular polysaccharide (CPS) and lipooligosaccharide (LOS)
CPS and LOS were analyzed as described previously.55,56 For CPS analysis, C. jejuni wild type and mutant cells were harvested after 48 h of growth, washed in PBS, standardized to an OD600 of 2.0, then solubilized in 200 µL of sample buffer (2% SDS, 4% 2-mercaptoethanol, 10% glycerol, 1 M TRIS-HCl, pH 6.8, and 0.02% bromophenol blue), for 10 min at 100 °C, and then subsequently incubated with 60 µg of proteinase K for 1 h at 60 °C. Whole cell lysates were analyzed by 15% SDS-PAGE at 80 mA for 4 h. Pre-stained protein marker (HyperPAGE, Bioline) was used to determine the molecular mass and bands were resolved with periodic acid silver nitrate (PAS) stain. The presence of LOS (normalized by protein content) was analyzed by 1 dimensional silica gel thin layer chromatography (TLC) using the solvent system n-propanol-water-25% NH4OH (60:30:10 v/v/v), developed with 10% (v/v) sulfuric acid in ethanol, and visualized by heating the TLC at 120 °C for few min as previously described.57
Quantitative RT-PCR
qRT-PCR was performed targeting key genes involved in poly-P homeostasis, CPS, and LOS synthesis. Mid stationary phase grown bacterial cultures were used for total RNA extraction using RNeasy Mini Kit (Qiagen). RNA concentration and purity were determined using NanoDrop ND-2000c spectrophotometer (Thermo Scientific) and agarose gel electrophoresis. DNase treated RNA (200 ng) was used for cDNA synthesis using SuperScript® III First-Strand Synthesis SuperMix kit (Invitrogen). Following cDNA synthesis, cDNA concentration was normalized to 200 ng and gene specific primers were used to amplify the genes involved in poly-P homeostasis (spoT, ppk1, and ppk2), CPS and LOS synthesis (kpsM, cstII, waaF, and lgtF) along with 16S-rRNA as internal control. qRT-PCR was performed using SensiMixPlus® SYBR RT-PCR Kit (Quantace) in a realplex2 mastercycler (Eppendorf). The expression of genes were normalized using 16S-rRNA expression of the same strain and the relative difference in expression of genes was calculated using the comparative threshold cycle (∆∆Ct) method to yield fold-difference in transcript levels compared with wild type.
Statistical analyses
Statistical significance of data was determined using one-way analysis of variance (ANOVA) followed by the Tukey multiple comparison post-test or Student t test (paired 2-tailed). A P value of P ≤ 0.05 (α level) was considered statistically significant.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Mrs Zhe Liu for the technical assistance with qRT-PCR studies. Dr Rajashekara’s laboratory is supported by the Ohio Agricultural Research and Development Center (OARDC), The Ohio State University, and the Agriculture and Food Research Initiative (AFRI) 2012-68003-19679, U. S. Department of Agriculture.
Glossary
Abbreviations:
- PPX
exopolyphosphatase
- GPPA
guanosine pentaphosphate phosphohydrolase
- poly-P
inorganic polyphosphate
- (p)ppGpp
guanosine tetra/pentaphosphate
- CPS
capsular polysaccharide
- LOS
lipooligosaccharide
- MEM
minimum essential media
References
- 1.Beery JT, Hugdahl MB, Doyle MP. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl Environ Microbiol. 1988;54:2365–70. doi: 10.1128/aem.54.10.2365-2370.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adak GK, Meakins SM, Yip H, Lopman BA, O’Brien SJ. Disease risks from foods, England and Wales, 1996-2000. Emerg Infect Dis. 2005;11:365–72. doi: 10.3201/eid1103.040191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Koningsveld R, Rico R, Gerstenbluth I, Schmitz PI, Ang CW, Merkies IS, Jacobs BC, Halabi Y, Endtz HP, van der Meché FG, et al. Gastroenteritis-associated Guillain-Barré syndrome on the Caribbean island Curaçao. Neurology. 2001;56:1467–72. doi: 10.1212/WNL.56.11.1467. [DOI] [PubMed] [Google Scholar]
- 4.Kulaev IS, Vagabov VM. Polyphosphate metabolism in micro-organisms. Adv Microb Physiol. 1983;24:83–171. doi: 10.1016/S0065-2911(08)60385-9. [DOI] [PubMed] [Google Scholar]
- 5.Kornberg A, Rao NN, Ault-Riché D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 6.Candon HL, Allan BJ, Fraley CD, Gaynor EC. Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. J Bacteriol. 2007;189:8099–108. doi: 10.1128/JB.01037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao NN, Gómez-García MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem. 2009;78:605–47. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 8.Tan S, Fraley CD, Zhang M, Dailidiene D, Kornberg A, Berg DE. Diverse phenotypes resulting from polyphosphate kinase gene (ppk1) inactivation in different strains of Helicobacter pylori. J Bacteriol. 2005;187:7687–95. doi: 10.1128/JB.187.22.7687-7695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gangaiah D, Liu Z, Arcos J, Kassem II, Sanad Y, Torrelles JB, Rajashekara G. Polyphosphate kinase 2: a novel determinant of stress responses and pathogenesis in Campylobacter jejuni. PLoS One. 2010;5:e12142. doi: 10.1371/journal.pone.0012142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishige K, Zhang H, Kornberg A. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc Natl Acad Sci U S A. 2002;99:16684–8. doi: 10.1073/pnas.262655299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sureka K, Sanyal S, Basu J, Kundu M. Polyphosphate kinase 2: a modulator of nucleoside diphosphate kinase activity in mycobacteria. Mol Microbiol. 2009;74:1187–97. doi: 10.1111/j.1365-2958.2009.06925.x. [DOI] [PubMed] [Google Scholar]
- 12.Akiyama M, Crooke E, Kornberg A. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J Biol Chem. 1993;268:633–9. [PubMed] [Google Scholar]
- 13.Bolesch DG, Keasling JD. Polyphosphate binding and chain length recognition of Escherichia coli exopolyphosphatase. J Biol Chem. 2000;275:33814–9. doi: 10.1074/jbc.M002039200. [DOI] [PubMed] [Google Scholar]
- 14.Rangarajan ES, Nadeau G, Li Y, Wagner J, Hung MN, Schrag JD, Cygler M, Matte A. The structure of the exopolyphosphatase (PPX) from Escherichia coli O157:H7 suggests a binding mode for long polyphosphate chains. J Mol Biol. 2006;359:1249–60. doi: 10.1016/j.jmb.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 15.Lindner SN, Knebel S, Wesseling H, Schoberth SM, Wendisch VF. Exopolyphosphatases PPX1 and PPX2 from Corynebacterium glutamicum. Appl Environ Microbiol. 2009;75:3161–70. doi: 10.1128/AEM.02705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi X, Rao NN, Kornberg A. Inorganic polyphosphate in Bacillus cereus: motility, biofilm formation, and sporulation. Proc Natl Acad Sci U S A. 2004;101:17061–5. doi: 10.1073/pnas.0407787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q, Li Y, Tang CM. The role of the exopolyphosphatase PPX in avoidance by Neisseria meningitidis of complement-mediated killing. J Biol Chem. 2010;285:34259–68. doi: 10.1074/jbc.M110.154393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristensen O, Laurberg M, Liljas A, Kastrup JS, Gajhede M. Structural characterization of the stringent response related exopolyphosphatase/guanosine pentaphosphate phosphohydrolase protein family. Biochemistry. 2004;43:8894–900. doi: 10.1021/bi049083c. [DOI] [PubMed] [Google Scholar]
- 19.Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–99. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keasling JD, Bertsch L, Kornberg A. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc Natl Acad Sci U S A. 1993;90:7029–33. doi: 10.1073/pnas.90.15.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reizer J, Reizer A, Saier MH, Jr., Bork P, Sander C. Exopolyphosphate phosphatase and guanosine pentaphosphate phosphatase belong to the sugar kinase/actin/hsp 70 superfamily. Trends Biochem Sci. 1993;18:247–8. doi: 10.1016/0968-0004(93)90172-J. [DOI] [PubMed] [Google Scholar]
- 22.Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–3. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 23.Kristensen O, Ross B, Gajhede M. Structure of the PPX/GPPA phosphatase from Aquifex aeolicus in complex with the alarmone ppGpp. J Mol Biol. 2008;375:1469–76. doi: 10.1016/j.jmb.2007.11.073. [DOI] [PubMed] [Google Scholar]
- 24.Choi MY, Wang Y, Wong LL, Lu BT, Chen WY, Huang JD, Tanner JA, Watt RM. The two PPX-GppA homologues from Mycobacterium tuberculosis have distinct biochemical activities. PLoS One. 2012;7:e42561. doi: 10.1371/journal.pone.0042561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaynor EC, Wells DH, MacKichan JK, Falkow S. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol Microbiol. 2005;56:8–27. doi: 10.1111/j.1365-2958.2005.04525.x. [DOI] [PubMed] [Google Scholar]
- 26.Rodionov DG, Ishiguro EE. Direct correlation between overproduction of guanosine 3′,5′-bispyrophosphate (ppGpp) and penicillin tolerance in Escherichia coli. J Bacteriol. 1995;177:4224–9. doi: 10.1128/jb.177.15.4224-4229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gangaiah D, Kassem II, Liu Z, Rajashekara G. Importance of polyphosphate kinase 1 for Campylobacter jejuni viable-but-nonculturable cell formation, natural transformation, and antimicrobial resistance. Appl Environ Microbiol. 2009;75:7838–49. doi: 10.1128/AEM.01603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MD, Newell DG. Campylobacter in poultry: filling an ecological niche. Avian Dis. 2006;50:1–9. doi: 10.1637/7474-111605R.1. [DOI] [PubMed] [Google Scholar]
- 29.Hofreuter D, Novik V, Galán JE. Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe. 2008;4:425–33. doi: 10.1016/j.chom.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–48. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naito M, Frirdich E, Fields JA, Pryjma M, Li J, Cameron A, Gilbert M, Thompson SA, Gaynor EC. Effects of sequential Campylobacter jejuni 81-176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J Bacteriol. 2010;192:2182–92. doi: 10.1128/JB.01222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drogari-Apiranthitou M, Kuijper EJ, Dekker N, Dankert J. Complement activation and formation of the membrane attack complex on serogroup B Neisseria meningitidis in the presence or absence of serum bactericidal activity. Infect Immun. 2002;70:3752–8. doi: 10.1128/IAI.70.7.3752-3758.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, DeBoy RT, Parker CT, Daugherty SC, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, Benitez M, Clark C, Perbost C, Jarvie T, Du L, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun. 2006;74:4694–707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson BM, Gaskin DJ, Segers RP, Wells JM, Nuijten PJ, van Vliet AH. The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828) J Bacteriol. 2007;189:8402–3. doi: 10.1128/JB.01404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown MRW, Kornberg A. Inorganic polyphosphate in the origin and survival of species. Proc Natl Acad Sci U S A. 2004;101:16085–7. doi: 10.1073/pnas.0406909101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao NN, Kornberg A. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J Bacteriol. 1996;178:1394–400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDermott W. Inapparent infection: relation of latent and dormant infections to microbial persistence. Public Health Rep. 1959;74:485–99. doi: 10.2307/4590490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cardona ST, Chávez FP, Jerez CA. The exopolyphosphatase gene from sulfolobus solfataricus: characterization of the first gene found to be involved in polyphosphate metabolism in archaea. Appl Environ Microbiol. 2002;68:4812–9. doi: 10.1128/AEM.68.10.4812-4819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarro Llorens JM, Tormo A, Martínez-García E. Stationary phase in gram-negative bacteria. FEMS Microbiol Rev. 2010;34:476–95. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 41.Rajashekara G, Drozd M, Gangaiah D, Jeon B, Liu Z, Zhang Q. Functional characterization of the twin-arginine translocation system in Campylobacter jejuni. Foodborne Pathog Dis. 2009;6:935–45. doi: 10.1089/fpd.2009.0298. [DOI] [PubMed] [Google Scholar]
- 42.Ferrero RL, Lee A. Motility of Campylobacter jejuni in a viscous environment: comparison with conventional rod-shaped bacteria. J Gen Microbiol. 1988;134:53–9. doi: 10.1099/00221287-134-1-53. [DOI] [PubMed] [Google Scholar]
- 43.Hanning I, Donoghue DJ, Jarquin R, Kumar GS, Aguiar VF, Metcalf JH, Reyes-Herrera I, Slavik M. Campylobacter biofilm phenotype exhibits reduced colonization potential in young chickens and altered in vitro virulence. Poult Sci. 2009;88:1102–7. doi: 10.3382/ps.2008-00307. [DOI] [PubMed] [Google Scholar]
- 44.Reuter M, Mallett A, Pearson BM, van Vliet AH. Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl Environ Microbiol. 2010;76:2122–8. doi: 10.1128/AEM.01878-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moffitt MC, Frank MM. Complement resistance in microbes. Springer Semin Immunopathol. 1994;15:327–44. doi: 10.1007/BF01837364. [DOI] [PubMed] [Google Scholar]
- 46.Boutte CC, Henry JT, Crosson S. ppGpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus. J Bacteriol. 2012;194:28–35. doi: 10.1128/JB.05932-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect Dis. 1985;152:592–6. doi: 10.1093/infdis/152.3.592. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.Wilson DL, Bell JA, Young VB, Wilder SR, Mansfield LS, Linz JE. Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology. 2003;149:3603–15. doi: 10.1099/mic.0.26531-0. [DOI] [PubMed] [Google Scholar]
- 50.Yao R, Alm RA, Trust TJ, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–30. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- 51.Mendrygal KE, González JE. Environmental regulation of exopolysaccharide production in Sinorhizobium meliloti. J Bacteriol. 2000;182:599–606. doi: 10.1128/JB.182.3.599-606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tunpiboonsak S, Mongkolrob R, Kitudomsub K, Thanwatanaying P, Kiettipirodom W, Tungboontina Y, Tungpradabkul S. Role of a Burkholderia pseudomallei polyphosphate kinase in an oxidative stress response, motilities, and biofilm formation. J Microbiol. 2010;48:63–70. doi: 10.1007/s12275-010-9138-5. [DOI] [PubMed] [Google Scholar]
- 53.Svensson SL, Davis LM, MacKichan JK, Allan BJ, Pajaniappan M, Thompson SA, Gaynor EC. The CprS sensor kinase of the zoonotic pathogen Campylobacter jejuni influences biofilm formation and is required for optimal chick colonization. Mol Microbiol. 2009;71:253–72. doi: 10.1111/j.1365-2958.2008.06534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blaser MJ, Duncan DJ. Human serum antibody response to Campylobacter jejuni infection as measured in an enzyme-linked immunosorbent assay. Infect Immun. 1984;44:292–8. doi: 10.1128/iai.44.2.292-298.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keo T, Collins J, Kunwar P, Blaser MJ, Iovine NM. Campylobacter capsule and lipooligosaccharide confer resistance to serum and cationic antimicrobials. Virulence. 2011;2:30–40. doi: 10.4161/viru.2.1.14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karlyshev AV, Wren BW. Detection and initial characterization of novel capsular polysaccharide among diverse Campylobacter jejuni strains using alcian blue dye. J Clin Microbiol. 2001;39:279–84. doi: 10.1128/JCM.39.1.279-284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Besra GS. Preparation of cell-wall fractions from mycobacteria. Methods Mol Biol. 1998;101:91–107. doi: 10.1385/0-89603-471-2:91. [DOI] [PubMed] [Google Scholar]
- 58.Akiba M, Lin J, Barton YW, Zhang Q. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother. 2006;57:52–60. doi: 10.1093/jac/dki419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.