Abstract
Glioblastoma (GBM) contains a self-renewing, tumorigenic cancer stem cell (CSC) population which contributes to tumor propagation and therapeutic resistance. While the tumor microenvironment is essential to CSC self-renewal, the mechanisms by which CSCs sense and respond to microenvironmental conditions are poorly understood. Scavenger receptors are a broad class of membrane receptors that are well characterized on immune cells and instrumental in sensing apoptotic cellular debris and modified lipids. Here we provide evidence that CSCs selectively utilize the scavenger receptor CD36 to promote their maintenance using patient-derived CSCs and in vivo xenograft models. We detected CD36 expression in GBM cells in addition to previously described cell types including endothelial cells, macrophages and microglia. CD36 was enriched in CSCs and was able to functionally distinguish self-renewing cells. CD36 was co-expressed with integrin alpha 6 and CD133, previously described CSC markers, and CD36 reduction resulted in concomitant loss of integrin alpha 6 expression, self-renewal and tumor initiation capacity. We confirmed that oxidized phospholipids, ligands of CD36, were present in GBM and found that the proliferation of CSCs, but not non-CSCs, increased with exposure to oxidized low-density lipoprotein. CD36 was an informative biomarker of malignancy and negatively correlated to patient prognosis. These results provide a paradigm for CSCs to thrive by the selective enhanced expression of scavenger receptors, providing survival and metabolic advantages.
Keywords: cancer stem cells, glioma, stem cell-microenvironment interactions, self-renewal
Introduction
Survival rates for glioblastoma (GBM) patients have not seen major improvement over the last 30 years, with median survival remaining between 12–18 months following diagnosis [1, 2]. Current treatment regimens are palliative in nature, involving surgical resection, radiation, and chemotherapy. Despite these aggressive measures tumor recurrence is frequently observed. GBM is characterized by a high degree of cellular heterogeneity and an increased propensity for invasion, which are barriers to the effective treatment of these tumors. GBMs contain a self-renewing cancer stem cell (CSC) population that drives tumor progression and contributes to therapeutic resistance [3–8]. CSCs have been described in multiple advanced cancers such as leukemia [9], breast [10], colon [11], and prostate [12], and efforts are underway to determine the molecular mechanisms of CSC regulation and their role in both tumor progression and therapeutic resistance.
Normal and neoplastic stem populations rely on interactions with their surrounding microenvironment or niche to control the balance between self-renewal and differentiation, [13, 14]. Niche interactions such as cell-soluble ligand, cell-cell, and cell-extracellular matrix (ECM) communication have been demonstrated to promote CSC maintenance and tumor progression [15]. In contrast to the normal brain, GBM contains regions of increased necrosis and apoptosis leading to the release of cellular debris [16] and activation of numerous inflammatory pathways [17]. The consequence of this environment on CSC function has yet to be fully elucidated. Recent work has demonstrated the regulation of normal neural progenitor cell (NPC) populations by apoptotic cellular debris, in which the phagocytic activity of these cells played a central role [18].
Scavenger receptors are a key mechanism by which cells recognize, phagocytose and clear damage and debris through broad pattern recognition [19]. These receptors are well characterized on immune cells and play a role in a variety of pathological conditions including atherosclerosis, thrombosis, and Alzheimer’s disease [20]. Scavenger receptors are also expressed by non-immune cells and play a role in lipid metabolism. CD36 is a scavenger receptor expressed on multiple cell types in the brain including microglia, endothelial cells, astrocytes, and neurons [21]. CD36 is responsible for immune activation in this setting as well as debris removal [22]. CD36 inhibits vascular growth in GBM via interaction with vasculostatin, an extracellular cleavage product of G protein-coupled receptor brain angiogenesis inhibitor I, leading to induction of endothelial cell apoptosis [23, 24]. These anti-angiogenic and pro-apoptotic effects in endothelial cells were also observed with thrombospondin-1, an additional ligand of CD36 [25, 26]. These studies demonstrate that scavenger receptors play a role in multiple cell types in the brain and recognize a variety of ligands present within the tumor microenvironment. However, the function of scavenger receptors such as CD36 on CSCs has yet to be elucidated. Given GBM is characterized by increased regions of cellular debris release, we considered whether CSCs possessed scavenger receptor function and evaluated surface expression and functional importance of CD36 in this context.
Materials and Methods
GBM tissue
Human GBM and normal brain tissues were obtained following written informed consent and Institutional Review Board (IRB) approval from the Cleveland Clinic and University Hospitals Seidman Cancer Center at Case Western Reserve University. Previously established GBM xenografts were obtained from Duke University and the Mayo Clinic. Tissues were minced and digested with papain (Worthington) as previously described [27] and dissociated cells allowed to recover overnight prior to use. GBM cells were maintained using subcutaneous xenografts as previously described [8, 27–30]. Relevant details for each specimen used are provided in Supplemental Table 1.
Animals
All experiments utilizing mice were approved by the Institutional Animal Care and Use Committee (IACUC) of the Cleveland Clinic Foundation (protocol 2012-0752). Mice were housed in a fully AALAC accredited facility in accordance with all federal and local regulations. Female immunocompromised Nu/Nu mice, aged 4 weeks, were purchased from Charles River Laboratories. Following quarantine, mice were utilized for experiments at 6–8 weeks of age and maintained on sulfamethoxazole/trimethoprim supplemented water. CD36 null and wild type mice generated as previously described [31].
CSC isolation and culture
Following overnight recovery from papain digestion, dissociated xenograft GBM cells were magnetically sorted based on CD133 expression using magnetic beads (Miltenyi). This approach has previously been validated to show difference in tumorigenic potential between CD133 positive and negative fractions [8, 27–30]. After CD133 magnetic bead enrichment, CSC marker expression was evaluated by immunoblotting with CSC markers (Sox 2, Olig 2). CD133 positive CSCs were maintained in Neurobasal media (Life technologies) supplemented with penicillin/streptomycin (50 U/ml final concentration), L-glutamine (2 mM), B27 (Life Technologies), sodium pyruvate (1 mM), epidermal growth factor (EGF 20 ng/ml, R&D Systems) and fibroblast growth factor (FGF 20 ng/ml, R&D Systems). CD133 negative non-CSCs were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (50 U/ml). Cells were maintained at 37°C in 5% CO2. For the induction of differentiation, CSCs were cultured in DMEM medium containing 10% FBS.
Flow cytometry and limiting dilution analysis
Cell sorting and analysis were performed using the BD FACS ARIA II Flow Cytometer. Primarily conjugated antibodies were utilized at recommended dilutions; CD36 (Beckman Coulter), CD105 (BD Biosciences), Iba1 (Abcam), and integrin α6 (BD Biosciences). Limiting dilution analysis was carried out, following 14 days incubation, in a 96 well format with 24 wells of each dilution; 1, 5, 10 and 20 cells/well. Limiting dilution plots and stem cell frequencies were calculated using an online tool available through the Walter and Eliza Hall Institute of Medical Research (http://bioinf.wehi.edu.au/software/elda/index.html).
Immunofluorescence analysis
Human GBM and normal brain tissue samples were snap frozen. Intracranial xenografts were fixed in 4% paraformaldehyde, incubated overnight in 30% sucrose and embedded in OCT. Samples were sectioned at a thickness of 10µm. Blood vessels were visualized by Von Willebrand Factor (Abcam) staining. Sections were additionally stained for CD36 (Novus), integrin α6 (Millipore), CD133 (Miltenyi), and/or Iba1 (Wako). Age-matched wild-type and CD36 knockout mouse brains were prepared in a similar manner as described above and stained with antibodies against proliferating cell nuclear antigen (Abcam) and phospho histone H3 (Millipore). Analysis between wild-type and CD36 knockout mice was done based on 3 different anatomical regions from 3 separate mice. For all immunofluorescence analysis, nuclei were counterstained using 4',6-diamidino-2-phenylindole (Dapi) and images were taken using a Leica SP-5 confocal microscope as previously described [27].
Immunoblotting analysis
Whole cell lysates were collected using a 1% Brij 010, 5mM MgCl2, 25mM Hepes, 150mM NaCl 2.5mM Iodoacetamide, pH 7.0 lysis buffer supplemented with protease and phosphatase inhibitor cocktails (Sigma). CSCs and non-CSCs were analyzed by immunoblotting analysis for CD36 (Novus), Sox2 (R&D Systems), total JNK (Cell Signaling), phospho-JNK (Thr183/Tyr185, Cell Signaling), total Akt (Cell Signaling), phospho-Akt (Ser473, Cell Signaling), total Stat3 (Cell Signaling), and phospho-Stat3 (Ser727, Cell Signaling). Actin (Santa Cruz) was used as a loading control. For differentiation assay, CSCs were differentiated over 7 days and cell lysates collected at 0, 1, 3, 5, and 7 days. Differentiated samples were analyzed by immunoblotting analysis for CD36, Olig2 (Millipore), GFAP (Invitrogen) and Actin.
CD36 knockdown and inhibition
CD36 inhibition was accomplished by siRNA in CSCs. Pooled CD36 (Santa Cruz) and control (Santa Cruz) siRNA knockdown (KD) constructs were applied to 200,000 cells in 500 µl antibiotic free complete neurobasal media. Per well, 100 pmol of siRNA construct was pre-incubated with lipofectamine 2000 in 100µl optimem media without serum. Following pre-incubation, siRNA-lipofectamine solutions were added to the appropriate wells and incubated for 4 hours. Following incubations cells were washed and cultured for 48 hours in complete neurobasal media. Knockdown was confirmed by immunoblotting blot analysis, with additional evaluation of integrin α6 expression. CD36 knockdown was also achieved by shRNA using the Mission shRNA system (Sigma) as previously described [27]. CD36 constructs were generated against different parts of the gene (KD 1 – TRCN0000056998; KD 3 - TRCN0000057000; KD 4 - TRCN0000057001) and applied in a similar fashion to the siRNA treatment. Limiting dilution analysis and tumor implantation were performed as previously described [27] and compared to a non-targeting control.
Lipoprotein preparation
LDL and oxLDL were prepared as previously described [32] and added to cells at a concentration of 50 µg/ml. Cell growth was evaluated using the Cell Titer Glo assay (Promega) and normalized to a time 0 loading control.
2-methylthio-1,4-naphthoquinone (MTN) synthesis and validation
MTN was synthesized from 1,4-naphthoquinone as previously described [33]. The crude material was purified by flash chromatography [34] eluting with 20% v/v ethyl acetate in hexane, and crystallized out of the column fractions. Upon drying under high vacuum this crystalline material gave 1H NMR and MS data in agreement with those published in the literature [35, 36]. To validate MTN inhibition of CD36, macrophages from C57BL/6 wild-type and CD36-null mice were isolated four days after IP injection with 4% thioglycollate and cultured in RPMI/10% FBS. Cells were plated onto coverslips in 12-well dishes, 500,000 cells/well. Two days later, cells were washed with warmed PBS and stained for 20 minutes at 37°C with 2 µM CellTracker Green CMFDA (Invitrogen). After washing with warm PBS, coverslips were moved to wells containing warm RPMI/10% FBS +/− vehicle (DMSO) or MTN at the indicated concentrations for 30 minutes at 37° C. Following pretreatment, coverslips were then moved to wells with RPMI/10%FBS +/− vehicle or MTN, along with DiI-oxLDL at 25 µg/ml. After allowing 2 hours for uptake at 37°C, coverslips were washed thoroughly, fixed with 3% paraformaldehyde (Electron Microscopy Sciences) for 20 minutes and stained with 300 nM DAPI (Invitrogen) for 3 minutes, before mounting Aqua Poly Mount (Polysciences, Inc.). Cells were viewed using a Leica SP-5 confocal microscope (Leica Microsystems). For each assay, at least three fields per coverslip were imaged and used for quantifying the effect of MTN at each concentration. Data from three independent assays were combined to determine the amount of uptake for each condition. Co-localization of the red fluorescent oxLDL particles within the CellTracker Green-stained cytoplasms allowed internalized particles to be distinguished from those adhering to the cell surface. Using Image Pro software, the mean DiI fluorescent intensity for each cell was determined and averaged with all other cells in that field and expressed as a percent of the mean DiI fluorescent of vehicle (DMSO)-treated WT control samples. Self-renewal was assessed by limiting dilution analysis and CSC signaling was evaluated by immunoblotting with CSC signaling pathway antibodies (Sox 2, total and phospho-Akt, total and phospho-Stat 3). Cell death was evaluated using the Caspase 3/7 activity assay (Promega) and normalized to a loading control measured using the Cell Titer Glo assay.
Tumor implantation
Intracranial tumor implantation was performed as previously described [27]. Briefly, 100 luciferase bearing CSCs were implanted into the brain of 4–6 week old Nu/Nu mice in 20 µl of media devoid of growth factors. CSCs were pretreated with control or CD36 siRNA. Mice were monitored daily for the development of neurological signs, at which point they were sacrificed. Tumor formation was monitored by intraperitoneal injection of 3 mg luciferin in 100 ul sterile phosphate buffered saline (PBS) followed by in vivo IVIS imaging.
Bioinformatics analysis
Human expression of CD36 was determined using the human protein atlas (www.proteinatlas.org). The correlation of CD36 with brain tumor expression versus other tumor types, normal versus GBM, NPCs versus GBM, and relationship of CD36 to glioma and GBM patient survival were analyzed using the Oncomine microarray database (Oncomine, Compendia Bioscience, Ann Arbor, MI (http://www.oncomine.org) [37]. The dataset utilized is indicated in the text, figure legends, and directly on the figures. For the evaluation of lipid metabolism, the Murat dataset was used for 5-year survival. High and low groups were determined by patients with expression level above or below 1 standard deviation.
Tissue microarray analysis
Immunohistochemical staining was done for CD36 (Novus) expression on archival deidentified formalin-fixed, paraffin embedded (FFPE) GBM tissue microarrays (TMAs) constructed at the University of Kentucky Markey Biospecimen and Tissue Procurement Shared Resource Facility. Survival data was obtained on each case from the Kentucky Cancer Registry. University of Kentucky institutional review board approval was obtained prior to tissue collection. A total of 44 GBMs were included in the TMAs; each case was sampled by three 2 mm cores apiece, with each core located on a separate TMA block. Slides were digitized by an Aperio ScanScope XT slide scanner and analyzed using Aperio Spectrum Version 11.2.0.780 software (Aperio, Vista, CA). CD36 expression was analyzed using a positive pixel algorithm that analyzed a region of interest and evaluated each pixel within a cell, providing a staining intensity score of 0 to 3+, which were then summed up and expressed as a percent of positive cells over total cells. Visual light microscopic examination was then done to corroborate the results. Only cells that showed strong expression for CD36 were included in subsequent analyses. The cut-off for CD36 high versus low was set by evaluating the staining intensity across all specimens. Data from the 3 cores per case were averaged together to produce a final result for that case.
Statistical analysis
Reported values are mean values +/− standard deviation from studies done using least three replicates. Unless otherwise stated, one-way ANOVA was used to calculate statistical significance; p values are detailed in the text and figure legends. In vivo survival analysis was calculated by log-rank analysis.
Results
CD36 is expressed by GBM cells
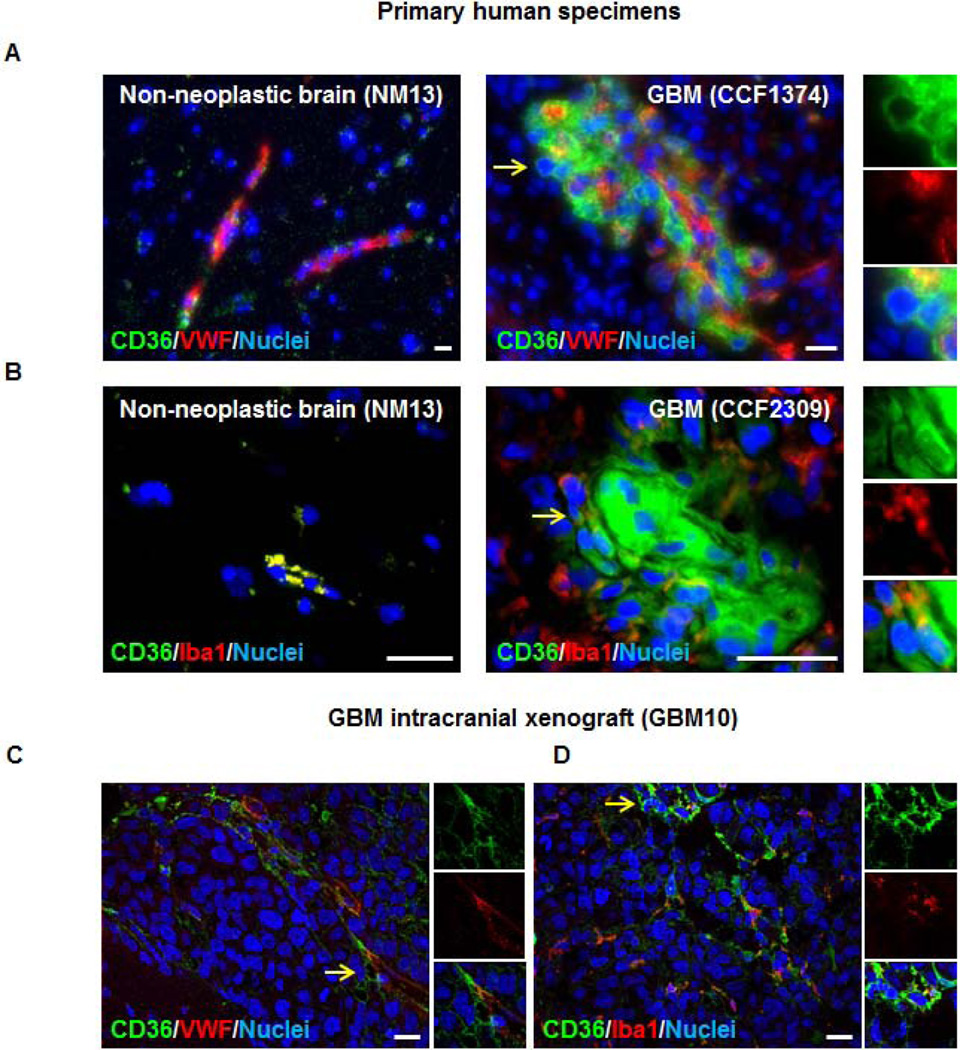
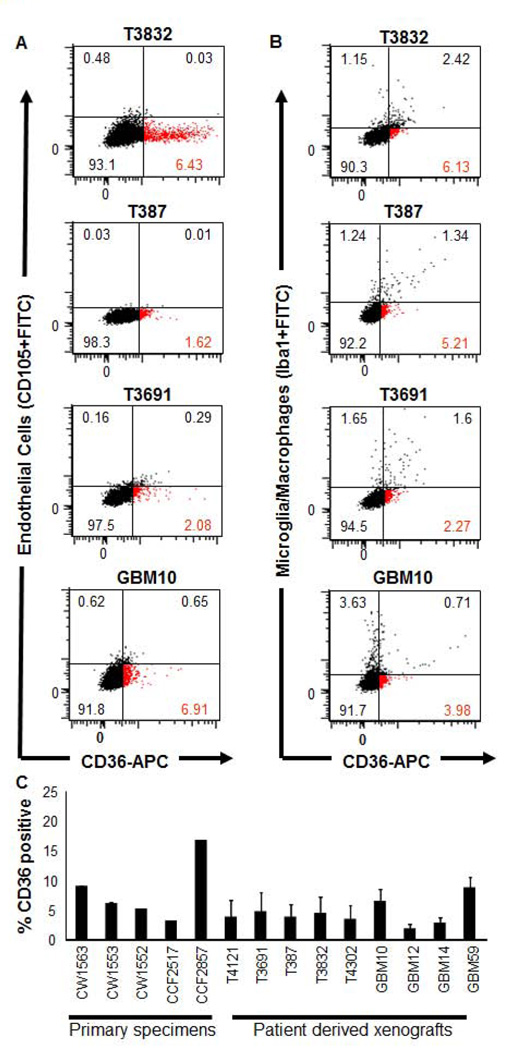
CD36 has previously been described in the regulation of endothelial cell function in multiple cancer types including breast [38], colorectal [39], melanoma [40], and GBM [23, 24]. CD36 has also been characterized on microglia [41, 42]. To evaluate if CD36 expression extended beyond these cell types, we evaluated patient-derived tissues from non-neoplastic (derived from epileptic brain, NM13) and GBM (CCF1374, CCF2309) specimens. Using immunofluorescence, we were able to detect CD36 protein (green) in each condition (Fig. 1A, B). CD36 protein was expressed at higher levels in GBM and we observed expression in the perivascular compartment (as indicated by co-localization with Von Willebrand Factor (VWF) staining on endothelial cells in red, Fig. 1A). We were also able to detect a population of GBM cells that expressed CD36 but were absent for VWF staining (Fig. 1A, yellow arrows and inset). We evaluated CD36 co-expression in microglia/macrophages using the microglia/macrophage specific marker Iba1 (red) [43]; and found while CD36 was co-expressed on a fraction of microglia/macrophages, there were also CD36 positive cells negative for microglia/macrophage marker expression (Fig. 1B, yellow arrows and inset). These expression patterns were also confirmed in GBM intracranial xenograft models where CD36 positive cells were detected in the absence of markers for endothelial cells (Fig. 1C) and microglia/macrophages (Fig. 1D). To confirm the non-endothelial and non-microglial/macrophage cell identity of these GBM cells, we evaluated CD36 expression in patient-derived xenograft tumors (T3832, T387, T3691, GBM10) by flow cytometry and found that an individual population of CD36 expressing GBM cells were absent for both endothelial cell (CD105, Fig. 2A) and microglial/macrophage specific (Iba1, Fig. 2B) marker expression (red population). We expanded our analysis to 5 primary patient-derived tumors (CW1563, CW1553, CW1552, CCF2517, CCF2857) and 9 patient-derived GBM xenografts (T4121, T3691, T387, T3832, T4302, GBM10, GBM12, GBM14, GBM59) and observed consistent CD36 protein expression, detected in 2.3–16.4% of total cells across tumors (Fig. 2C). Taken together, these data demonstrate that CD36 is expressed by a fraction of GBM cells that lack phenotypic surface markers indicative of endothelial cells or microglia/macrophages.
Figure 1. CD36 is expressed in glioblastoma (GBM).
Immunofluorescence images of normal human brain and GBM tissues demonstrates that CD36 (green) is expressed at higher levels in GBM and by vascular (von Willebrand Factor, red, A) and non-vascular cells. CD36 is also expressed by macrophage/microglia (Iba1, red, B) and non-macrophage/microglia in GBM. CD36 expression (green) was confirmed in GBM intracranial xenograft (GBM10) and double labeling with vascular (von Willebrand Factor, red, C) and macrophage/microglia (Iba1, red, D) markers indicates that single positive CD36 cells were present in xenografts. Nuclei counterstained with 4’6-diamidino-2-phenylindole (dapi, blue), yellow arrows indicate magnified insets, and scale bars represent 20 µm. Staining was performed on at least 3 different anatomical regions from 1 control and 4 GBM tissues as well as 2 separate GBM xenografts.
Figure 2. CD36 is expressed by a population of GBM cells negative for endothelial and macrophage/microglia markers.
Flow cytometry analysis of multiple patient-derived GBM xenografts (T3832, T387, T3691, GBM10) demonstrate that a fraction of CD36 positive cells are negative for endothelial cell (CD105, A) and macrophage/microglia (Iba1, B) markers. Red population indicates single CD36 positive population. Summary graph (C) indicates relative CD36 expression levels in primary GBM specimens and patient-derived GBM xenografts. Positive gates set relative to isotype control. Double labeling of CD36 and CD105 or Iba1 was performed in triplicate. CD36 expression analysis was performed once for primary specimens and at least in triplicate for patient-derived xenografts. Summary graph indicates mean +/− standard deviation.
CD36 expression is elevated in CSCs
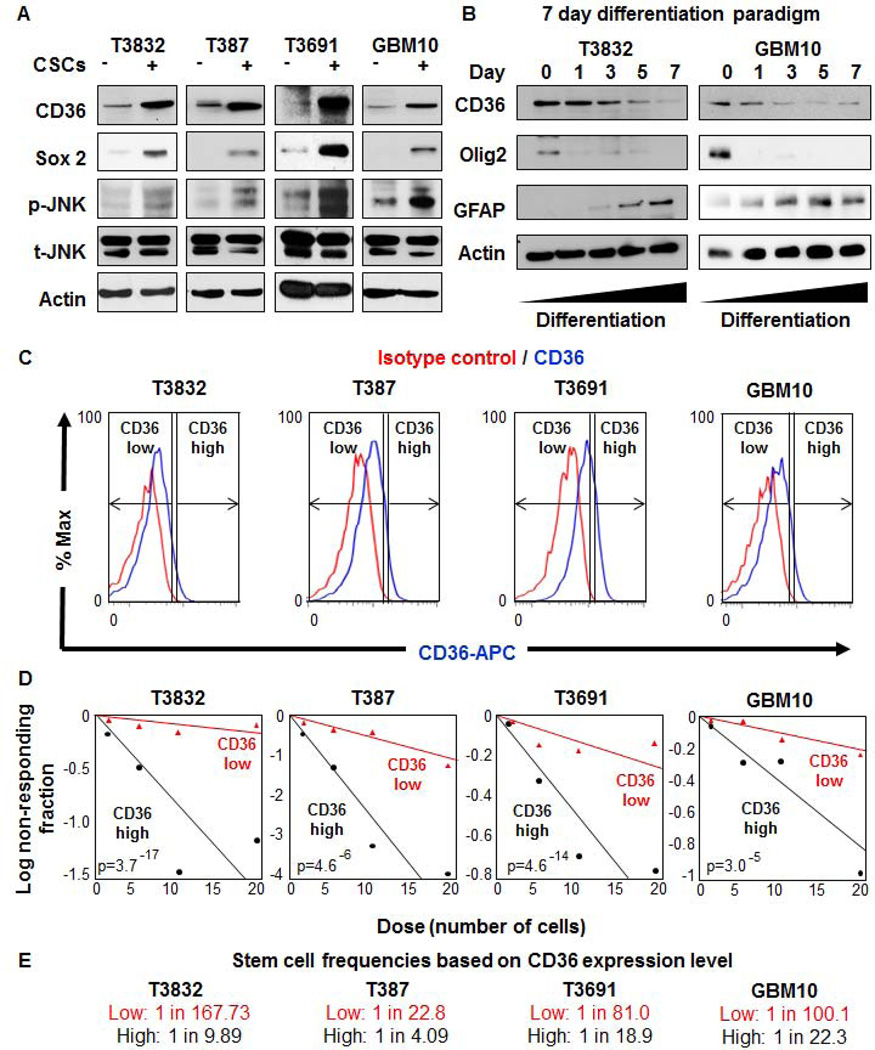
Our staining data indicated that CD36 was present in the perivascular compartment, a known CSC niche [44]. To more clearly define the identity of CD36 positive GBM cells, we compared protein levels of CD36 between enriched CSCs and non-CSCs. Immunoblotting analysis across multiple patient-derived xenografts (T3832, T387, T3691, GBM10) revealed that CSCs expressed more CD36 as compared with non-CSCs from the same tumor (Fig. 3A). CD36 was observed at its predicted molecular weight of 56 kDa, which indicates a deglycosylated form in GBM cells. CD36 has been observed at 88 kDa due to heavy glycosylation in multiple cell types [45], we confirmed our antibody could detect the 56 kDa form following deglycosylation of adipose tissue lysates (data not shown). The degree of CSC enrichment was confirmed by the CSC marker Sox2. Differences in JNK pathway, a key downstream CD36 signaling node [46], were also observed by elevated JNK phosphorylation in CSCs as compared to non-CSCs (Fig. 3A). With elevated expression of CD36 in CSCs, we next wanted to examine if expression was associated with cellular differentiation status. CSCs were enriched from multiple patient-derived xenografts (T3832, GBM10) and subjected to a 7 day differentiation paradigm using media containing 10% serum. To confirm that CSC expression decreased and differentiation marker expression increased over the differentiation time course, Olig2 and GFAP were used as markers of CSCs and astrocytic differentiation, respectively (Fig. 3B). In this paradigm, we observed a decrease in CD36 expression with increased differentiation (Fig. 3B). These data confirm the increased protein level of CD36 in CSCs and support our in vivo observation of CD36 enrichment in the perivascular niche.
Figure 3. CD36 is elevated in cancer stem cells (CSCs), is reduced upon differentiation, and enriches for self-renewal.
Immunoblotting analysis (A) of enriched CSCs (+) and non-CSCs (−) from GBM patient-derived xenografts (T3832, T387, T3691, GBM10) indicates that CD36 expression and its downstream signaling pathway, as indicated by p-JNK, were elevated in CSCs. Sox2 was used as a CSC enrichment control and indicates difference between CSC and non-CSC populations. Total JNK and actin were used as loading controls. Immunoblotting analysis (B) of CSCs which have undergone a serum induced 7 day differentiation time course demonstrate a loss of CD36 expression. Concomitant reduction in Olig2, a CSC marker, and an increase in GFAP, a mature astrocyte marker, were also observed through the time course. Actin was used as a loading control and representative molecular weights are provided on the right hand side of the immunoblots. Comparisons between CSCs and non-CSCs were repeated in triplicate and differentiation studies were repeated in duplicate. A similar trend for differentiation was observed with patient-derived GBM xenograft T387 (data not shown). Representative histograms (C) for patient-derived GBM xenografts (T3832, T387, T3691, GBM10) shows CD36 high and low populations based on CD36 (blue) and isotype control (red) antibodies. Limiting dilution analysis (D) indicates increased sphere formation capacity in CD36 high (black) versus low (red) populations. Stem cell frequencies (E) were calculated by limiting dilution analysis and were increased in CD36 high versus low populations. All limiting dilution analyses were performed in triplicate and p-values are indicated on each graph.
To determine if CD36 expression was elevated in CSCs or shared by normal neural progenitor cells (NPCs), we evaluated CD36 expression in human and mouse tissues. Using the human protein atlas to determine the expression of CD36 in the human brain, we did not observe any positive staining in adult neurogenic niches, subventricular zone (SVZ) or subgranular zone (SGZ) in the hippocampus (Supplemental Fig. 1A). To determine if CD36 plays a major role in adult neurogenesis, we evaluated the proliferation in the mouse SVZ and SGZ using wild-type and age matched CD36 null mice. Using proliferating cell nuclear antigen and phospho-histone H3 analysis (Supplemental Fig. 1B), we observed no difference in proliferation in either the SVZ (Supplemental Fig. 1C) or SGZ (Supplemental Fig. 1D). Taken together, these data suggest that elevated CD36 expression is limited to CSCs.
Cell sorting for CD36 expression can be used to enrich for CSCs
Expanding on our observation of elevated CD36 in CSCs, we next asked whether cell sorting for CD36 could enrich for functional CSCs based on in vitro self-renewal assays. A hallmark of CSCs is their ability to self-renew, which is commonly determined by the ability to form spheres in culture, a surrogate of self-renewal, proliferation, and survival. To determine if CD36 could enrich for sphere formation, we sorted CD36 high and low populations from multiple acutely dissociated xenografts (T3832, T387, T3691, GBM10) using flow cytometry (Fig. 3C). We performed limiting dilution analyses and found that CD36 high cells had a significantly elevated capacity to form spheres across multiple GBM specimens (Fig. 3D). Stem cell frequencies were calculated using a limiting dilution algorithm, and were consistently elevated in CD36 high versus CD36 low populations (Fig. 3E).
As CD36 was shown to be increased in CSCs, we explored the possibility that CD36 may interact with previously described CSC receptors, such as integrin α6 [27], which is a co-receptor for CD36 in platelets [47]. We evaluated CD36 in a patient-derived xenograft (GBM10) using immunofluorescence and observed co-localization between CD36 (green) and integrin α6 (red, Supplemental Fig. 2A). We also evaluated CD36 co-expression with CD133 [6], another well-established CSC marker and observed similar co-localization (Supplemental Fig. 2B). We confirmed this co-expression using flow cytometry from multiple patient-derived xenografts (T3832, T387, T3691, GBM10) and observed a high degree of overlap between CD36 and integrin α6 ranging from 71.6% – 98.1% (Supplemental Fig. 2C) as well as with CD36 and CD133, with overlap ranging from 50.9% – 86.2% (Supplemental Fig. 2D). These results demonstrate CD36 is co-expressed with CSC markers integrin α6 and CD133.
CD36 knockdown in CSCs decreases CSC maintenance signaling, self-renewal, and tumor initiation
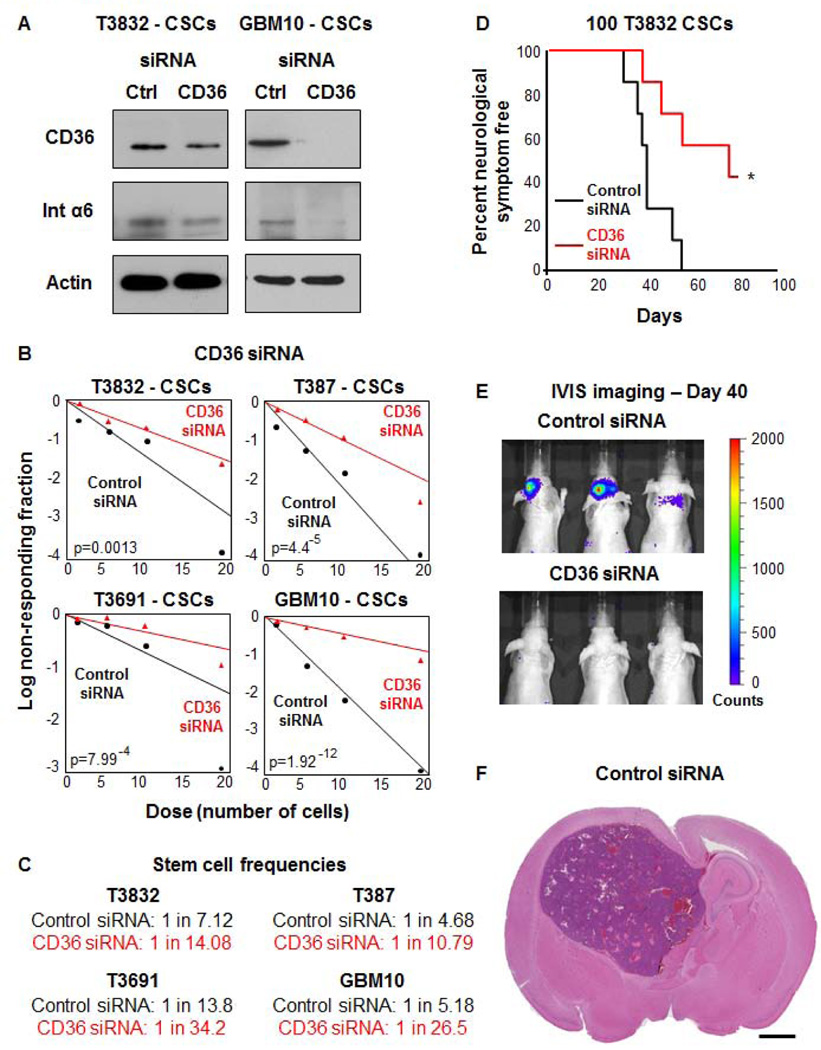
Given the observation that CD36 expression was associated with self-renewal and known CSC markers, we next asked whether inhibition of CD36 would impact CSC status, namely self-renewal and tumorigenic potential. Knockdown of CD36 was reliably accomplished by siRNA (Supplemental Fig. 3A) across multiple xenograft specimens (T3832, T387, T3691, GBM10). Immunoblotting analysis was used to confirm CD36 knockdown (Fig. 4A), with an accompanying reduction in integrin α6 expression observed upon treatment with CD36 siRNA. Evaluation of self-renewal revealed that CD36 knockdown attenuated sphere formation and decreased stem cell frequency (Fig. 4B, C). Tumor initiation capacity was also abrogated upon CD36 siRNA inhibition, evidenced by increased survival and reduced tumor burden in the CD36 siRNA group as compared to the control siRNA group (Fig. 4D–F). We confirmed the specificity of the CD36 siRNA knockdown using a shRNA approach (Supplemental Fig. 3A) and observed a decrease in key CSC maintenance signaling pathways (Sox 2 [48], phospho-Akt [49], and phospho-Stat 3 [50]) as compared to the non-targeting control (Supplemental Fig. 3B). Additionally, CD36 shRNA knockdown also resulted in decreased self-renewal as assessed by sphere formation (Supplemental Fig. 3C).
Figure 4. Inhibition of CD36 expression results in reduced self-renewal, tumor initiation, and integrin α6 expression.
Immunoblotting analysis (A) of control and CD36 siRNA transfected CSCs from patient-derived GBM xenografts (T3832, GBM10) indicates that CD36 siRNA effectively reduces CD36 and integrin α6 expression. Actin was used as a loading control. Limiting dilution analysis (B) indicates reduced sphere formation capacity in CD36 siRNA treatment (red) as compared to control siRNA treatment (black) group. Stem cell frequencies (C) were calculated by limiting dilution analysis and were decreased in CD36 versus control siRNA groups. Kaplan-Meier survival curve (D) of tumor initiated from 100 CSCs demonstrates an increase in tumor latency in CD36 siRNA (red) versus control siRNA (black) treated CSCs. IVIS bioluminescence imaging (E) at day 40 demonstrates detectable tumors in the control siRNA CSC group as compared to the CD36 siRNA CSC tumors. Representative H&E image (F) indicate that resulting tumor contained histological hallmarks of GBM, scale bar represents 1 mm. Immunoblotting to confirm CD36 reduction and limiting dilution analysis were performed in duplicate and p-values indicated on each graph. In vivo tumor initiation studies was performed with 7 mice per group and p-value was calculated using a log-rank analysis.
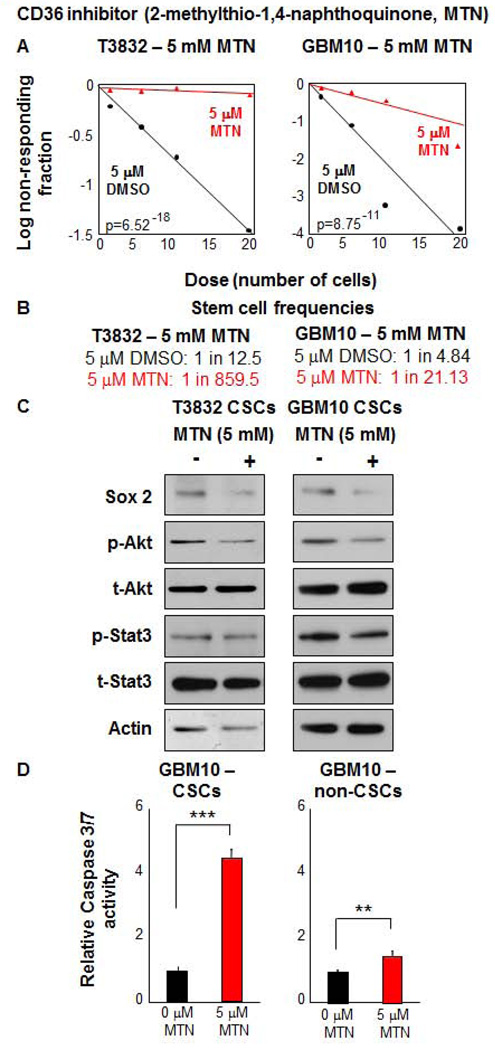
To further evaluate CD36 inhibition of self-renewal, we utilized a specific small molecule CD36 inhibitor, 2-methylthio-1,4-napthoquinone (MTN) [51] that was validated by inhibition of oxidized low density lipoprotein (oxLDL) uptake in wild-type and CD36 deficient macrophages (Supplemental Fig. 4). Using this inhibitor, we found significantly reduced sphere formation capacity and stem cell frequency (Fig. 5A, B). MTN treatment over an 18 hour time course attenuated key CSC signaling pathways such as Sox 2, phospho-Akt, and phospho-Stat 3 (Fig. 5C). MTN treatment over a 3 day time course also induced apoptosis preferentially in CSC populations compared to non-CSCs, as evidenced by increased Caspase 3/7 activity (Fig. 5C). Taken together, these results demonstrate that CD36 is essential for CSC function both in vitro and in vivo.
Figure 5. Alternative inhibition of CD36, by MTN, results in decreased CSC self-renewal.
MTN treatment (red) of CSCs reduced self-renewal capacity (A) and stem cell frequencies (B) compared to DMSO treated controls (black). Effects of MTN treatment were specific to CSCs vs non-CSCs. Immunoblotting analysis (C) CSCs from patient-derived GBM xenografts (T3832, GBM10) treated with MTN for 18 hours indicates that CSC self-renewal pathways (Sox2, p-Akt, p-Stat 3) were decreased. Actin was used as a loading control. MTN treatment (red) specifically induced apoptosis as compared to the DMSO control (black), measured by Caspase 3/7 activity (D) in CSCs after 3 days of treatment. All limiting dilution analyses were performed in triplicate and p-value indicated on each graph. Caspase measurements are represented as a mean +/− standard deviation and conducted in duplicate.
CSCs respond to oxidized lipids
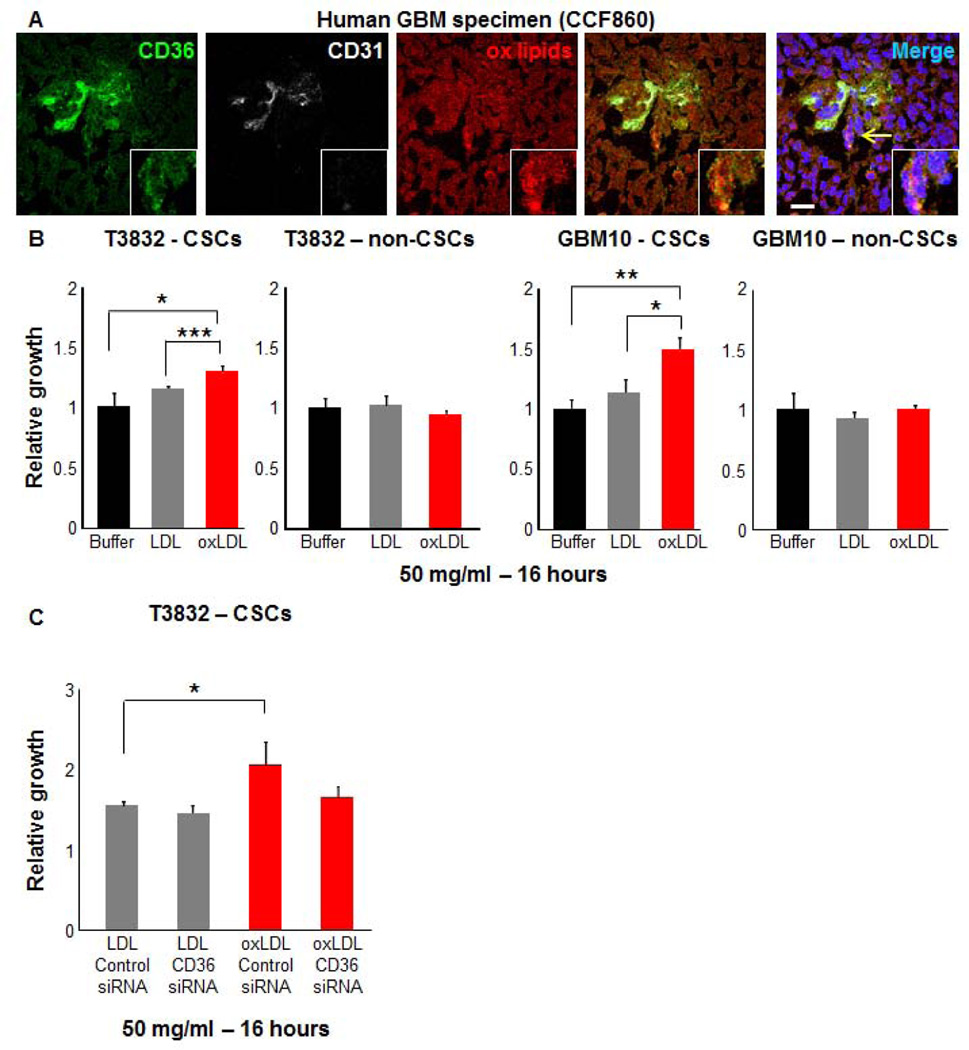
To further characterize the role of CD36 in the regulation of the CSC state, we sought to interrogate natural ligands of CD36. It is well established that oxidized phospholipid moieties, such as oxLDL, are CD36 ligands [32] and that these lipoproteins influence cellular phenotypes in a variety of normal and pathological states. To determine if oxidized phosphatidylcholine species were present in GBMs, we examined human GBM specimens using a monoclonal antibody, E06, that has previously been shown to recognize oxidized phosphatidylcholine containing epitopes that show cross reactivity with multiple structurally specific oxidized phosphatidylcholine species that serve as ligands for scavenger receptor CD36 [52]. Strong staining was observed throughout the tissue including CD36 positive regions (Fig. 6A). To determine whether oxLDL could increase cell growth, we compared its effect on CSCs and non-CSCs from multiple patient-derived xenografts (T3832, GBM10). We found that oxLDL significantly increased CSC proliferation but not non-CSC proliferation in comparison to both control and LDL conditions (Fig. 6B). To determine if this increase in proliferation was due, in part, to CD36, we evaluated control and CD36 knockdown in CSCs using the previously validated siRNA approach (Fig. 6C). There was no significant difference in LDL induced growth between the two groups (p=0.2). A decrease in proliferation in response to oxLDL was observed with CD36 siRNA treatment as compared to the control siRNA condition (p=0.066). When compared to LDL, there was not a significant increase in oxLDL mediated growth in the CD36 siRNA conditions as was observed in the control siRNA conditions (p<0.05). Taken together, these data indicate that oxLDL is present in GBM and drives CSC growth.
Figure 6. Oxidized lipid uptake is elevated in CSCs.
Immunofluorescence images (A) of human GBM tissue (CCF860) demonstrates detectable oxidized lipid species (ox lipids, red) adjacent to CD36 positive cells (green) in perivascular regions (CD31, white). Cell growth analysis (B) of CSCs and non-CSCs derived from patient-derived GBM xenografts (T3832, GBM10) demonstrates that CSC growth is increased by administration of 50 µg/ml for 16 hours in oxidized low density lipoprotein (oxLDL, red) conditions as compared to low density lipoprotein (LDL, grey) and buffer (black) conditions. CD36 siRNA did not reduce the growth in the oxLDL as compared to the LDL condition in CSCs, which was observed in control siRNA treatment (C). Immunofluorescence staining performed in triplicate and observed in tissue from an additional GBM patient. Nuclei in micrographs were counterstained with dapi (blue), yellow arrow indicates inset area, and scale bar represents 20 µm. Growth assays performed in at least duplicate and represent mean +/− standard deviation. P-values are indicated on graphs where significant: * p<0.05, ** p<0.01, *** p<0.001.
CD36 expression is indicative of GBM patient survival
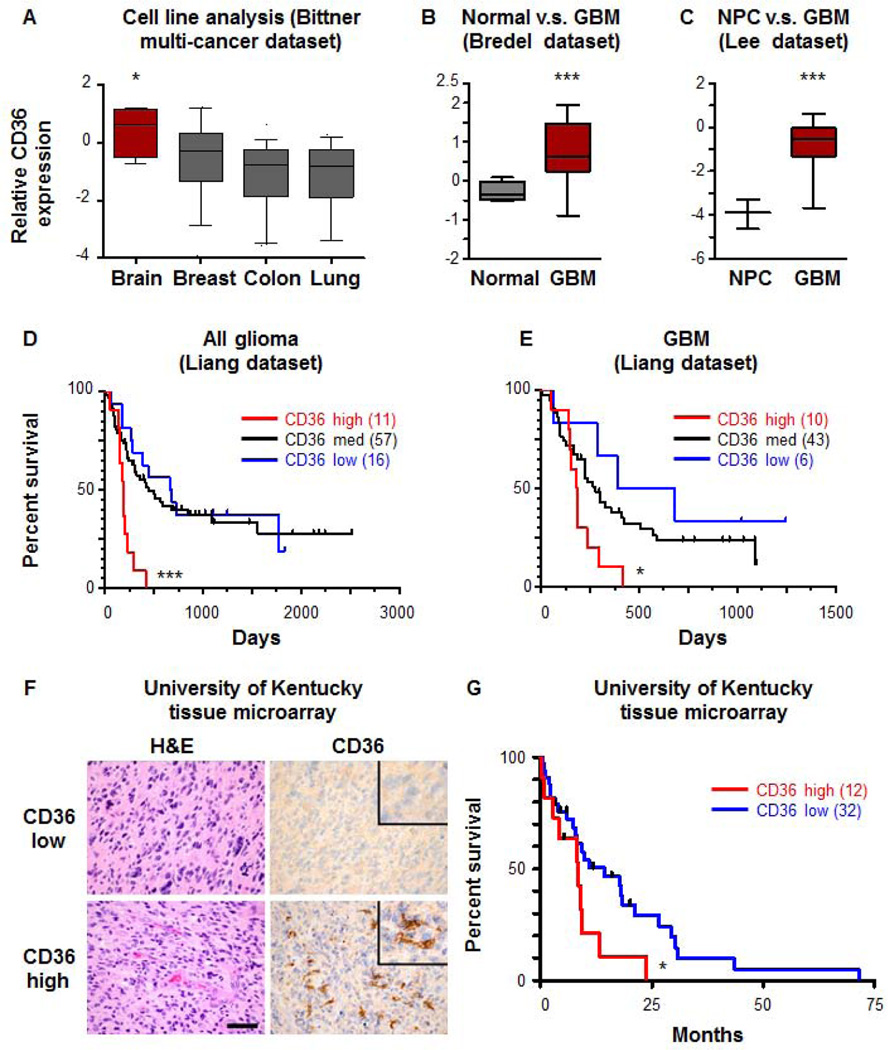
Based on our observations that CD36 is expressed on CSCs and important for CSC maintenance, we wanted to determine whether CD36 expression was a biomarker of malignancy. Using datasets available in Oncomine, we first looked at a comparative analysis of well-established cell lines from advanced cancers (inducing brain, breast, colon, and lung) in the Bittner dataset [37] and found that CD36 expression was significantly elevated in brain tumor cell lines (Fig. 7A). It is worth noting that cell lines were used in this analysis, so these levels are independent of any endothelial cell expression which may be detected in vivo. Using a similar approach, we also confirmed our observation that CD36 was elevated in GBM versus non-neoplastic brain (Fig. 7B) and normal neural progenitor cells (NPCs, Fig. 7C) in the Bredel [53] and Lee [54] datasets. To determine if CD36 expression levels correlated with glioma and GBM patient survival, we interrogated the Liang dataset [55] and found that high CD36 expression correlated with poorer patient prognosis in both glioma (Fig. 7D) and GBM (Fig 7E). To confirm these results at the protein level, we utilized a well annotated tissue microarray from the University of Kentucky and found GBM tumors could be separated into CD36 high and low groups, with CD36 high tumors having a significantly poorer prognosis (Fig. 7F, G). Taken together, these clinical data suggest that CD36 expression is a prognostic biomarker for patient survival.
Figure 7. CD36 expression is elevated in GBM and correlates with patient survival.
CD36 expression from the Bittner dataset in Oncomine (A) indicates that brain tumor cell lines have a higher expression level as compared with other advanced cancer cell lines (breast, colon, lung). Expression from the Bredel dataset in Oncomine (B) indicates higher CD36 expression in GBM versus normal brain and a comparison between normal neural progenitor cells (NPCs) and GBM from the Lee dataset in Oncomine (C) indicated higher CD36 expression in GBM as compared to NPCs. Kaplan-Meier curves demonstrate that high CD36 expression (red) in the Liang dataset in Oncomine correlates with decreased survival in all glioma (D) and GBM (E) as compared with CD36 medium (black) and CD36 low (blue) groups. Representative histoligcal analysis (F) and Kaplan-Meier curve (G) demonstrates that high CD36 expression (red) in the University of Kentucky tissue microarray correlates with decreased survival as compared to the CD36 low expression group (blue). P-values indicated where significant: * p<0.05, *** p<0.001, and scale bar represents 50 µm.
Discussion
The tumor microenvironment contains key extrinsic factors essential to cell survival as well as numerous non-neoplastic cell types including astrocytes, pericytes, microvascular endothelial cells, and microglia that contribute to the pro-tumorigenic environment [56]. While current attention has been focused on identifying self-renewal mechanisms utilized by tumor cells, limited information is available as to how cells communicate and sense changes in the tumor microenvironment. Modulating tumor associated microglia and macrophages, key components of the tumor microenvironment, has been recently proposed as an anti-GBM therapy [57]. These cells have elevated expression of scavenger receptors, key mediators of phagocytosis. Our data suggest that scavenger receptors and their associated ligands may be an additional point of intervention. This presents an interesting hypothesis, that the expression of scavenger receptors is an adaptive mechanism by which CSCs may sense and respond to environments unfavorable to other cell types. Similar adaptive mechanisms have been described for CSCs in conditions of limited oxygen [30], glucose [58], and in response to acidic stress [59]. Based on previous work showing genetic deletion of CD36 resulted in reduced inflammation and deficit in a stroke model [22], additional mechanistic studies are required to further define the cell type specific response to anti-CD36 therapeutic approaches.
Our current work extends previous findings on an anti-angiogenic, pro-apoptotic role of CD36 in endothelial cells in GBM [23–26] and in contrast to these previous findings, demonstrates that CD36 plays a pro-tumorigenic role in CSCs. There are likely multiple reasons for cell-type specific roles of CD36 including differential responses based on different ligands, for which CD36 has several including modified lipoproteins, long chain fatty acids, thrombospondins, β-amyloid, and vasculostatin [20]. While we demonstrate a role for CD36 in CSC maintenance, it remains unclear if CD36 is serving as a driver or passenger in GBM. Our observation that CD36 targeting decreases CSC maintenance suggests an active role, however future studies are required to confirm that CD36 can facilitate tumor progression. In the context of GBM, our findings on the CSC response to oxLDL are of particular interest as oxidized lipoproteins have been found to be toxic to neurons via increased oxidative stress, which also impact astrocytes and microglia [60, 61]. Interestingly, the deleterious effects of oxLDL can be inhibited by A20, in macrophages [62], which is a GBM CSC maintenance factor [63] and may be an additional mechanism by which CSCs are able to respond positively to oxLDL. Additionally, these lipoproteins and other pro-apoptotic ligands are likely to be enriched in necrotic regions of the tumor. Selective utilization may be a key CSC survival mechanism. These findings are also interesting in the context of a recent study where high density lipoproteins were found to have anti-cancer effects [64]. Differences in low and high density lipoproteins with respect to cardioprotection may indeed be mirrored in a pro- and anti-cancer function. With differences in metabolism reported between CSCs and non-CSC [65], the function of CD36 may be to serve as an entry point to fuel key metabolic process such as lipogenesis, which is critical for GBM cell growth [66, 67].
A fundamental challenge to develop novel GBM therapies has been the identification of specific, targetable pathways which are essential for CSCs while dispensible for normal cells such as NPCs. Some signaling pathways, such as Notch, are essential to CSC maintenance in a variety of advanced cancers including GBM [68], but are also crucial to NPC function [69] and therefore do not represent an easily translatable therapeutic paradigm without a way to selectively ablate CSCs. While strategies disrupting interactions in the CSC microenvironment remain a viable therapeutic approach and warrant further investigation, they require the identification of CSC-specific pathways. This issue is timely given that the first generation of CSC targeted therapies are under clinical investigation [70–73]. Our identification of scavenger receptor function in CSCs represents a new paradigm for CSC maintenance and provides an exciting therapeutic target for follow up studies. Additionally, the broad interest in regulating CD36 in a variety of disease states may accelerate the translation of anti-CD36 therapies for GBM.
Supplementary Material
Acknowledgements
We thank Dr. Petra Hamerlik (Danish Cancer Center), Dr. J. Mark Brown (Dept. of Cellular and Molecular Medicine, Cleveland Clinic), and the members of the Lathia laboratory for constructive comments on the experimental design and manuscript. We thank Cathy Shemo, Moneen Morgan, Patrick Barrett, and Sage O’Bryant for flow cytometry assistance and the Cleveland Clinic Foundation Tissue Procurement Service and Dr. Susan Staugaitis, Dr. Robert Weil, and Mary McGraw. We also thank Dana Napier for her excellent work in constructing the TMAs and performing CD36 immunohistochemistry. These studies were supported by a NIH K99/R00 Pathway to Independence Award (CA157948) to JDL. This work was also supported by National Institutes of Health grants NS083629 (JDL), CA112958 (JNR), CA154130 (JNR), CA169117 (JNR), K08 CA155764 (CH), RR020171(CH), P01 HL076491 (SLH), CA152883 (CLG), CA175120 (CLG), CA137443 (AES), NS063971 (AES), CA128269 (AES), CA101954 (AES), and CA116257 (AES). Work in the Lathia lab is also supported by the Lerner Research Institute, Voices Against Brain Cancer, the Ohio Cancer Research Associates, V Scholar Award from the V Foundation for Cancer Research, Grant IRG-91-022-18 to the Case Comprehensive Cancer Center from the American Cancer Society, and the Cleveland Clinic Product Development Fund. CH was also supported by the University of Kentucky College of Medicine Physician Scientist Program. AES was also supported by funding from the Cancer Genome Atlas (TCGA) Project, the Ben & Catherine Ivy Foundation, the Kimble Foundation, the Peter B. Cristal Endowment, and the Ohio Department of Development Technology (09-071).
Footnotes
Author contributions: Conception and design (JSH, JDL), Financial Support (JDL), Provision of study material (BR, AES, RLS, TM, SLH), Collection and/or assembly of data (JSH, BO, MS, AGA, MH, KS, QW, WF, BL, MLJ, DS, JMN, PH), Data analysis and interpretation (JSH, CEG, JAD, JMB, TM, SLH, CH, JNR, JDL), Manuscript writing (JSH, JDL), Final approval of manuscript (all authors)
Disclosure of potential conflicts of interest
None
References
- 1.Lawrence YR, Mishra MV, Werner-Wasik M, et al. Improving prognosis of glioblastoma in the 21st century: who has benefited most? Cancer. 2012;118:4228–4234. doi: 10.1002/cncr.26685. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The lancet oncology. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ignatova TN, Kukekov VG, Laywell ED, et al. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 5.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer research. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 6.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 7.Yuan X, Curtin J, Xiong Y, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 8.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 9.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature medicine. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 10.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 12.Collins AT, Berry PA, Hyde C, et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer research. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 13.Lathia JD, Heddleston JM, Venere M, et al. Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell stem cell. 2011;8:482–485. doi: 10.1016/j.stem.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- 15.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell stem cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Brat DJ, Castellano-Sanchez AA, Hunter SB, et al. Pseudopalisades in glioblastoma are hypoxic, express extracellular matrix proteases, and are formed by an actively migrating cell population. Cancer research. 2004;64:920–927. doi: 10.1158/0008-5472.can-03-2073. [DOI] [PubMed] [Google Scholar]
- 17.Mantovani A, Allavena P, Sica A, et al. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 18.Lu Z, Elliott MR, Chen Y, et al. Phagocytic activity of neuronal progenitors regulates adult neurogenesis. Nature cell biology. 2011;13:1076–1083. doi: 10.1038/ncb2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg ME, Li XM, Gugiu BG, et al. The lipid whisker model of the structure of oxidized cell membranes. The Journal of biological chemistry. 2008;283:2385–2396. doi: 10.1074/jbc.M707348200. [DOI] [PubMed] [Google Scholar]
- 20.Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Science signaling. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Husemann J, Loike JD, Anankov R, et al. Scavenger receptors in neurobiology and neuropathology: their role on microglia and other cells of the nervous system. Glia. 2002;40:195–205. doi: 10.1002/glia.10148. [DOI] [PubMed] [Google Scholar]
- 22.Woo MS, Wang X, Faustino JV, et al. Genetic deletion of CD36 enhances injury after acute neonatal stroke. Annals of neurology. 2012;72:961–970. doi: 10.1002/ana.23727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaur B, Cork SM, Sandberg EM, et al. Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer research. 2009;69:1212–1220. doi: 10.1158/0008-5472.CAN-08-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klenotic PA, Huang P, Palomo J, et al. Histidine-rich glycoprotein modulates the anti-angiogenic effects of vasculostatin. The American journal of pathology. 2010;176:2039–2050. doi: 10.2353/ajpath.2010.090782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rege TA, Stewart J, Jr, Dranka B, et al. Thrombospondin-1-induced apoptosis of brain microvascular endothelial cells can be mediated by TNF-R1. Journal of cellular physiology. 2009;218:94–103. doi: 10.1002/jcp.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson JC, Grammer JR, Wang W, et al. ABT-510, a modified type 1 repeat peptide of thrombospondin, inhibits malignant glioma growth in vivo by inhibiting angiogenesis. Cancer biology & therapy. 2007;6:454–462. doi: 10.4161/cbt.6.3.3630. [DOI] [PubMed] [Google Scholar]
- 27.Lathia JD, Gallagher J, Heddleston JM, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell stem cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eyler CE, Wu Q, Yan K, et al. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 2011;146:53–66. doi: 10.1016/j.cell.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guryanova OA, Wu Q, Cheng L, et al. Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer cell. 2011;19:498–511. doi: 10.1016/j.ccr.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Bao S, Wu Q, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Febbraio M, Abumrad NA, Hajjar DP, et al. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. The Journal of biological chemistry. 1999;274:19055–19062. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- 32.Podrez EA, Febbraio M, Sheibani N, et al. Macrophage scavenger receptor CD36 is the major receptor for LDL modified by monocyte-generated reactive nitrogen species. The Journal of clinical investigation. 2000;105:1095–1108. doi: 10.1172/JCI8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fieser LRaB. R. H. Synthesis of naphthoquinones for studies of the inhibition of enzyme systems. J Am Chem Soc. 1949;71:3609–3614. [Google Scholar]
- 34.Still WC, Kahn M, Mitra A. Rapid chromatographic technique for preparative separations with moderate resolution. J Org Chem. 1978;43:2923–2925. [Google Scholar]
- 35.Kametani T, Nemoto H, Takeuchi M, et al. Studies on the syntheses of heterocyclic compounds. Part 681. A novel alkylation in the 4-position of isoquinoline derivatives. Journal of the Chemical Society Perkin transactions 1. 1977;4:386–390. doi: 10.1039/p19770000386. [DOI] [PubMed] [Google Scholar]
- 36.Coll G, Morey J, Costa A, Saa JM. Direct lithiation of hydroxyaromatics. J Org Chem. 1988;53:5345–5348. [Google Scholar]
- 37.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeFilippis RA, Chang H, Dumont N, et al. CD36 repression activates a multicellular stromal program shared by high mammographic density and tumor tissues. Cancer discovery. 2012;2:826–839. doi: 10.1158/2159-8290.CD-12-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuchida T, Kijima H, Tokunaga T, et al. Expression of the thrombospondin 1 receptor CD36 is correlated with decreased stromal vascularisation in colon cancer. International journal of oncology. 1999;14:47–51. [PubMed] [Google Scholar]
- 40.Hale JS, Li M, Sinyuk M, et al. Context dependent role of the CD36--thrombospondin--histidine-rich glycoprotein axis in tumor angiogenesis and growth. PloS one. 2012;7:e40033. doi: 10.1371/journal.pone.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coraci IS, Husemann J, Berman JW, et al. CD36, a class B scavenger receptor, is expressed on microglia in Alzheimer's disease brains and can mediate production of reactive oxygen species in response to beta-amyloid fibrils. The American journal of pathology. 2002;160:101–112. doi: 10.1016/s0002-9440(10)64354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El Khoury JB, Moore KJ, Means TK, et al. CD36 mediates the innate host response to beta-amyloid. The Journal of experimental medicine. 2003;197:1657–1666. doi: 10.1084/jem.20021546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito D, Imai Y, Ohsawa K, et al. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain research Molecular brain research. 1998;57:1–9. doi: 10.1016/s0169-328x(98)00040-0. [DOI] [PubMed] [Google Scholar]
- 44.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 45.Qiao L, Zou C, Shao P, et al. Transcriptional regulation of fatty acid translocase/CD36 expression by CCAAT/enhancer-binding protein alpha. The Journal of biological chemistry. 2008;283:8788–8795. doi: 10.1074/jbc.M800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rahaman SO, Lennon DJ, Febbraio M, et al. A CD36-dependent signaling cascade is necessary for macrophage foam cell formation. Cell metabolism. 2006;4:211–221. doi: 10.1016/j.cmet.2006.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miao WM, Vasile E, Lane WS, et al. CD36 associates with CD9 and integrins on human blood platelets. Blood. 2001;97:1689–1696. doi: 10.1182/blood.v97.6.1689. [DOI] [PubMed] [Google Scholar]
- 48.Gangemi RM, Griffero F, Marubbi D, et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 49.Eyler CE, Foo WC, LaFiura KM, et al. Brain cancer stem cells display preferential sensitivity to Akt inhibition. Stem cells. 2008;26:3027–3036. doi: 10.1634/stemcells.2007-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sherry MM, Reeves A, Wu JK, et al. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller WE, Thakur NL, Ushijima H, et al. Matrix-mediated canal formation in primmorphs from the sponge Suberites domuncula involves the expression of a CD36 receptor-ligand system. Journal of cell science. 2004;117:2579–2590. doi: 10.1242/jcs.01083. [DOI] [PubMed] [Google Scholar]
- 52.Boullier A, Gillotte KL, Horkko S, et al. The binding of oxidized low density lipoprotein to mouse CD36 is mediated in part by oxidized phospholipids that are associated with both the lipid and protein moieties of the lipoprotein. The Journal of biological chemistry. 2000;275:9163–9169. doi: 10.1074/jbc.275.13.9163. [DOI] [PubMed] [Google Scholar]
- 53.Bredel M, Bredel C, Juric D, et al. High-resolution genome-wide mapping of genetic alterations in human glial brain tumors. Cancer research. 2005;65:4088–4096. doi: 10.1158/0008-5472.CAN-04-4229. [DOI] [PubMed] [Google Scholar]
- 54.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 55.Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Charles NA, Holland EC, Gilbertson R, et al. The brain tumor microenvironment. Glia. 2012;60:502–514. doi: 10.1002/glia.21264. [DOI] [PubMed] [Google Scholar]
- 57.Pyonteck SM, Akkari L, Schuhmacher AJ, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nature medicine. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flavahan WA, Wu Q, Hitomi M, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nature neuroscience. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hjelmeland AB, Wu Q, Heddleston JM, et al. Acidic stress promotes a glioma stem cell phenotype. Cell death and differentiation. 2011;18:829–840. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keller JN, Hanni KB, Kindy MS. Oxidized high-density lipoprotein induces neuron death. Experimental neurology. 2000;161:621–630. doi: 10.1006/exnr.1999.7276. [DOI] [PubMed] [Google Scholar]
- 61.Sugawa M, Ikeda S, Kushima Y, et al. Oxidized low density lipoprotein caused CNS neuron cell death. Brain research. 1997;761:165–172. doi: 10.1016/s0006-8993(97)00468-x. [DOI] [PubMed] [Google Scholar]
- 62.Li HL, Wang AB, Zhang R, et al. A20 inhibits oxidized low-density lipoprotein-induced apoptosis through negative Fas/Fas ligand-dependent activation of caspase-8 and mitochondrial pathways in murine RAW264.7 macrophages. Journal of cellular physiology. 2006;208:307–318. doi: 10.1002/jcp.20665. [DOI] [PubMed] [Google Scholar]
- 63.Hjelmeland AB, Wu Q, Wickman S, et al. Targeting A20 decreases glioma stem cell survival and tumor growth. PLoS biology. 2010;8:e1000319. doi: 10.1371/journal.pbio.1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zamanian-Daryoush M, Lindner D, Tallant TC, et al. The cardioprotective protein apolipoprotein A1 promotes potent anti-tumorigenic effects. The Journal of biological chemistry. 2013;288:21237–21252. doi: 10.1074/jbc.M113.468967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vlashi E, Lagadec C, Vergnes L, et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16062–16067. doi: 10.1073/pnas.1106704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo D, Prins RM, Dang J, et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Science signaling. 2009;2:ra82. doi: 10.1126/scisignal.2000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo D, Hildebrandt IJ, Prins RM, et al. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12932–12937. doi: 10.1073/pnas.0906606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fan X, Khaki L, Zhu TS, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aguirre A, Rubio ME, Gallo V. Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature. 2010;467:323–327. doi: 10.1038/nature09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rudin CM, Hann CL, Laterra J, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. The New England journal of medicine. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Von Hoff DD, LoRusso PM, Rudin CM, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. The New England journal of medicine. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 72.Sheridan C. Genentech obtains proof of concept for hedgehog inhibition. Nature biotechnology. 2009;27:968–969. doi: 10.1038/nbt1109-968. [DOI] [PubMed] [Google Scholar]
- 73.Singh BN, Fu J, Srivastava RK, et al. Hedgehog signaling antagonist GDC-0449 (Vismodegib) inhibits pancreatic cancer stem cell characteristics: molecular mechanisms. PloS one. 2011;6:e27306. doi: 10.1371/journal.pone.0027306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.