Abstract
Coat protein complex I (COPI) and COPII are required for bidirectional membrane trafficking between the endoplasmic reticulum (ER) and the Golgi. While these core coat machineries and other transport factors are highly conserved across species, high-resolution imaging studies indicate that the organization of the ER–Golgi interface is varied in eukaryotic cells. Regulation of COPII assembly, in some cases to manage distinct cellular cargo, is emerging as one important component in determining this structure. Comparison of the ER–Golgi interface across different systems, particularly mammalian and plant cells, reveals fundamental elements and distinct organization of this interface. A better understanding of how these interfaces are regulated to meet varying cellular secretory demands should provide key insights into the mechanisms that control efficient trafficking of proteins and lipids through the secretory pathway.
The secretory pathway in eukaryotic cells is responsible for biogenesis and proper intracellular distribution of a wide range of proteins, complex carbohydrates and lipids. Trafficking in the secretory pathway is highly dynamic and responsive to specific cellular functional demands. Forward transport (also known as anterograde transport) of newly synthesized proteins and lipids is initiated at the endoplasmic reticulum (ER) and, as such, ER-to-Golgi transport represents a vital gateway to the endomembrane system.
Membrane traffic between the ER and the Golgi is bidirectional and occurs via similar mechanisms. In both cases, a carrier forms on the donor organelle and then tethers to and fuses with the target organelle (FIG. 1). Distinct machineries facilitate the formation of carriers for anterograde and retrograde transport, which are thought to ensure fidelity and directionality of trafficking: coat protein complex II (COPII) operates in the anterograde pathway from the ER, and COPI functions in the retrograde route from the Golgi. Although in vitro studies suggest that the formation of COPI- and COPII-coated carriers is mechanistically similar and can occur via the action of minimal components (including heteromeric coat complexes, ADP-ribosylation factor (ARF) and SAR1 (secretion-associated RAS-related 1) GTPases and their regulatory guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (BOX 1; BOX 2), other elements are necessary to coordinate the two trafficking routes in vivo and thereby maintain organelle structure and function. This is supported by evidence demonstrating that chemical disruption of the COPI route with the ARF GEF inhibitor brefeldin A leads to the collapse of ER export1,2, which may be due to depletion of proteins required for the formation of COPII carriers that occurs because Golgi-to-ER retrieval is blocked. Similarly, dominantnegative versions of SAR1 that block COPII assembly rapidly disrupt Golgi structure and function1. While the cellular functions of COPI and COPII as well as interdependence of the anterograde and retrograde trafficking routes are generally conserved across eukaryotes, the organization of the ER–Golgi interface varies greatly in different species. For example, plants and some yeast species have a compact organization of the ER and Golgi, whereas in animal cells the two organelles are separated by a pleomorphic intermediate compartment. The distribution of COPII budding sites on the ER, the nature of the transport vesicles themselves and the dependence on cytoskeletal components also vary. Furthermore, there seems to be enormous plasticity in the ER–Golgi interface, which can adapt to accommodate variations in COPII carrier size and number commensurate with cargo dimensions and quantity.
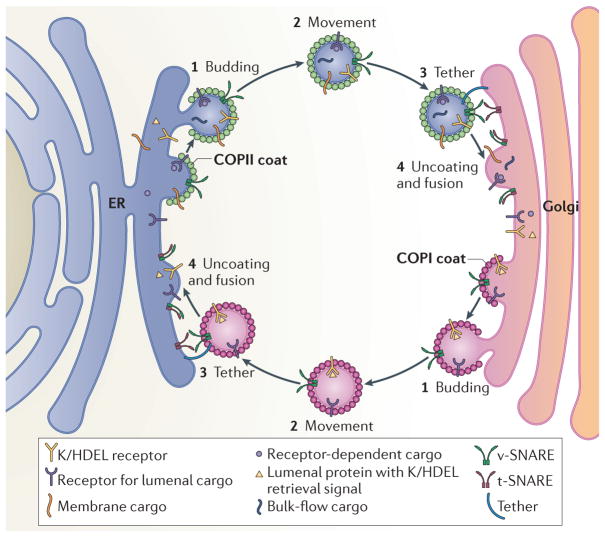
Figure 1. Bidirectional transport between the ER and the Golgi is mediated by COPI and COPII carriers.
Bidirectional transport of secretory cargo between the endoplasmic reticulum (ER) and the Golgi requires budding, movement, tethering, as well as uncoating and fusion of coat protein complex II (COPII) and COPI carriers with their respective compartments. These include bulk-flow cargo, membrane cargo and receptor-dependent luminal cargo. COPII carriers facilitate selective and bulk-flow cargo export towards the Golgi. One important function of COPI is to facilitate retrieval of escaped luminal proteins containing K/HDEL retrieval signals that are recognized by the K/HDEL receptor as well as other machinery required for optimal anterograde transport. Carrier fusion is mediated by vesicular SNARE proteins (v-SNAREs) and target-SNAREs (t-SNAREs) upon anchoring of the carriers to their target compartment via tethers.
Box 1. The COPII coat complex and anterograde transport.

The coat protein complex II (COPII) machinery consists of the SAR1 (secretion-associated RAS-related 1) GTPase and the two subcomplexes SEC23–SEC24 and SEC13–SEC31 (REF. 34) (see the figure, left panel). Activation of SAR1 is coordinated by the endoplasmic reticulum (ER) membrane-anchored guanine nucleotide exchange factor (GEF) SEC12 (top right panel), which produces the GTP-bound form of SAR1. Active SAR1 binds the ER membrane through an amino-terminal α-helix112. SAR1 recruits SEC23–SEC24 heterodimers through interaction with the SEC23 subunit113,114, which functions as a GTPase-activating protein (GAP) for SAR1 (REF. 113). Although diffusion or ‘bulk-flow’ of cargo into COPII carriers occurs115, it has been shown that COPII subunits can recognize specific ER export signals on membrane proteins for selective uptake116,117, and that SEC24 is the main COPII adaptor that recognizes specific sorting sequences in cargo118,119. The COPII coat is completed when SEC13–SEC31 heterodimers polymerize on the underlying SAR1–SEC23–SEC24–cargo complexes. SEC31 interacts directly with SAR1 and SEC23 (REF. 120), and polymerized SEC13–SEC31 subcomplexes provide a scaffold that imposes curvature to the nascent vesicle as it buds from the ER by membrane fission37,121. COPII vesicle biogenesis and uncoating of the transport carrier are regulated by SAR1 GTPase activity and recruitment of the outer coat layer120. ER-derived carriers retain coat subunits until they reach their target membrane, and coat phosphorylation and dephosphorylation are needed for vesicle fusion and budding, respectively102. COPII vesicle budding seems to be independent of dynamin-like GTPases, as purified COPII components are sufficient to generate vesicles (60–90 nm) from synthetic liposomes122,123. SEC12 and the components of the COPII coat are highly conserved with a larger number of paralogues in Arabidopsis thaliana compared with humans89. COPII paralogues display distinct functional complexity as exemplified by the specificity in recognition of cargo export signals among human SEC24 paralogues118, as well as in specific human diseases caused by mutations in the SAR1B, SEC23A and SEC24B isoforms124. Similarly, A. thaliana SEC24A, in contrast to A. thaliana SEC24B and SEC24C, is essential, and partial loss of function of SEC24A causes unique ER morphology defects94.
Box 2. The COPI coat complex and retrograde transport.
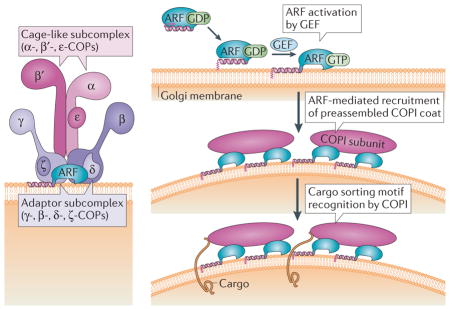
The coat protein complex I (COPI) consists of a heptameric (α, β, β′, γ, δ, ε, ζ) complex, also called coatomer125, with two main subcomplexes: the γ-COP–δ-COP–ζ-COP–β-COP tetrameric complex, which constitutes the inner layer core; and the α-COP–β′-COP–ε-COP trimeric complex, which forms the outer layer of the COPI coat126 (see the figure, left panel). Once activated by ADP-ribosylation factor (ARF) guanine nucleotide exchange factors (GEFs) containing a conserved SEC7 domain, myristoylated membrane-anchored ARF GTPases recruit COPI to Golgi membranes (right panel). The coatomer subunits α-COP, β′-COP, γ-COP and δ-COP recognize sorting motifs on the cytosolic domain of membrane cargo and mediate cargo incorporation into nascent COPI vesicles127. ARF GTPase-activating proteins (GAPs) bind cytoplasmic signals on cargo proteins, γ-COP and β′-COP subunits as well as active ARFs (not shown). Stimulation of the GTPase activity of ARFs by ARF GAPs leads to the release of ARF from the complex and ARF GAP and coat dissociation127,128 (not shown). At the endoplasmic reticulum (ER)–Golgi interface, COPI facilitates retrograde transport from the Golgi and ER–Golgi intermediate compartment (ERGIC). A retrograde Golgi–ER pathway has also been suggested to recycle lipids129 and ER resident proteins130. However, its occurrence in vivo was demonstrated through experiments using brefeldin A (BFA) in which the addition of the drug allowed Golgi-specific oligosaccharide modification of ER-retained vesicular stomatitis virus G-protein131. The results were interpreted as a consequence of a redistribution of resident Golgi proteins to the ER. In accordance with this hypothesis, reversible BFA-induced disruption of the Golgi has been observed in microscopy studies132,133. It is now established that primary targets of BFA are ARF GEFs, such as GBF1 in mammalian cells134,135. BFA stabilizes the interaction of inactive ARFs with their GEFs136 as well as ARF GEF binding to Golgi membranes134, which in turn inactivates ARF. These results underscore that the integrity of the anterograde traffic route depends tightly on the homeostasis of the retrograde pathway, which ensures not only the retrieval of resident proteins that escape the ER but also facilitates the recycling of lipids and trafficking machinery necessary for ER export and carrier fusion to the Golgi. Therefore, inhibition of the retrograde trafficking route leads to the collapse of anterograde trafficking.
In this Review, we discuss how the ER–Golgi interface and COPII-mediated trafficking varies in different species. We focus on the plant and mammalian systems as exquisite examples of how divergent adaptation of common components of the trafficking system has allowed the unique needs of differentiated eukaryotic cell types to be met. Ultimately, a more complete understanding of these observed morphologies in molecular terms should provide crucial insights into regulatory mechanisms that control protein and lipid trafficking in the early secretory pathway.
The ER and Golgi: partners in secretion
The ER is responsible for initiating the synthesis, folding and quality control, as well as priming the glycosylation, of a large part of the cellular proteome. It has a unique architecture that is characterized by a network of interconnected membrane tubules and sheets that form closed polygons. Most proteins that have been synthesized in the ER are transported to the Golgi during their biogenesis. The Golgi has several fundamental cellular roles. First, it functions as a central platform for connecting anterograde and retrograde protein flow within the secretory pathway3. Second, it serves as a complex carbohydrate ‘factory’ that supplies the material needed to build the plant cell wall and the glycoprotein matrix surrounding animal cells, as well as the donor groups for glycosylation of many proteins and lipids4–10. Last, the Golgi also provides a membrane scaffold for the dynamic binding of various signalling and sorting proteins11,12. In most eukaryotes, the membranes of the Golgi assume a characteristic stacked morphology with cisternae that differ in enzymatic content and activity13,14. This highly polarized organization defines cis-, medial- and transcisternae, with the cis-most cisternae facing the ER15. The trans-most cisternae face the trans-Golgi network (TGN), a tubular vesicular cluster that executes final sorting steps to post-Golgi destinations, exchanges material with the endocytic pathway and can, at least in plants, exist as an independent organelle from the Golgi16,17.
Basic elements of the ER–Golgi interface
With a few notable exceptions, including cytoplasmic, nuclear and signal peptide-containing proteins18, secreted proteins follow a conventional ER–Golgi secretory route. To exit the ER, fully folded soluble and membrane cargos are packaged into COPII-coated carriers at specialized, long-lived subdomains of the ER, termed ER exit sites (ERES)19,20. Although the number, size and dynamics of ERES vary across cell types and species, most eukaryotic cells examined so far display these organized export zones on ER membranes. ERES are enriched in COPII and appear as discrete puncta during fluorescence imaging of COPII coat proteins21–24. In addition to the core COPII components, large multidomain SEC16 proteins localize to ERES and are required for export site assembly and function25,26. Indeed, current models suggest that SEC16 establishes an ERES scaffold that recruits COPII to export zones through multiple interactions with coat subunits27. In all cell types studied, the COPII machinery seems to conduct at least two critical functions: first, the inner layer SAR1 and SEC23–SEC24 COPII subunits bind to and select specific cargo for packaging into ER-derived transport vesicles; second, polymerization of the outer layer SEC13–SEC31 COPII complex into a cage structure deforms ER membranes to drive transport vesicle formation (BOX 1).
ER-derived transport intermediates fuse with acceptor membranes through a series of targeting and fusion events that also rely on a highly conserved machinery28. In general, membrane targeting depends on RAB GTPases that function in concert with extended coiled-coil domain proteins, such as p115 (known as Uso1 in yeast), as well as the multisubunit TRAPPI (transport protein particle I) complex29–31. Targeted COPII vesicles proceed to fusion through regulated assembly of ternary SNARE complexes between donor vesicle and acceptor membrane compartments (BOX 3). In the return Golgi-to-ER route, COPI subunits recognize retrograde sorting signals in recycled cargos, and this mediates the incorporation of selected cargos into COPI-coated transport intermediates (BOX 2). Targeting of COPI vesicles to the ER requires the multisubunit DSL1 tethering complex to direct SNARE-mediated membrane fusion of COPI vesicles with ER membranes32.
Box 3. Tethering and SNARE-mediated fusion of COPII vesicles.
The directionality and fidelity of coat protein complex II (COPII) vesicle fusion is mediated by concerted action of RAB GTPases, tethering factors and integral membrane SNARE proteins. In mammalian cells, the vesicle targeting stage depends on RAB1 (REF. 30) and the extended coiled-coil domain tethering factors p115, GM130 (cis-Golgi matrix of 130 kDa) and GRASP65 (Golgi reassembly-stacking protein of 65 kDa)137,138 as well as the 170 kDa multisubunit TRAPPI (transport protein particle I) tethering complex (comprising BET3 (blocked early in transport 3), BET5, TRS20 (TRAPP subunit 20 (also known as Sedlin), TRS23, TRS31 and TRS33) that exerts guanine nucleotide exchange factor (GEF) activity towards RAB1 (REFS 139,140). RAB1·GTP recruits p115 and tethers vesicles to acceptor membranes. Therefore, the activation of RAB1 on acceptor membranes by TRAPPI may generate a localized signal to tether COPII vesicles. The BET3 subunit of TRAPPI also binds directly to the SEC23 subunit of COPII and can tether COPII vesicles at a close distance31. Fusion of COPII-tethered vesicles depends on a set of four tail-anchored integral membrane SNARE proteins named syntaxin 5, membrin (also known as GOSR2), BET1 and SEC22B141–143. SNARE proteins contain a conserved membrane-proximal heptad repeat sequence known as the SNARE motif, and trans-assembly of cognate sets of SNARE proteins from donor and acceptor membranes into four-helix bundles drives bilayer fusion144,145. Therefore, trans-assembly of syntaxin 5, membrin, BET1 and SEC22B between tethered COPII vesicles and Golgi acceptor membranes catalyses fusion. The conserved syntaxin 5-binding protein SLY1 is also required for this vesicle fusion step146 and may serve to coordinate the vesicle tethering and fusion stages. Regulation at these multiple stages in COPII vesicle tethering and fusion may have crucial roles in determining ER–Golgi morphology and levels of coated transport intermediates, but is relatively unexplored.
Mechanisms of interface control
While most cell types examined use these basic conserved ER–Golgi trafficking components, studies are revealing additional layers of control that influence the morphology of the ER–Golgi interface and transport carriers. In some cases, the COPII export machinery seems to have adapted to manage diverse secretory cargos. For example, certain mammalian cell types must export large secretory cargos from the ER such as 300–400 nm procollagen fibres33 or 100–500 nm lipoprotein particles, which are larger than the reported size of a standard 60–90 nm COPII-coated vesicle19,34. Several lines of evidence indicate that the COPII cage is flexible35,36, and this would allow the formation of larger COPII-coated intermediates and thereby the transport of large cargo. A combination of structural approaches has revealed that SEC13–SEC31 subunits can self-assemble into polyhedral cages of distinct orders through moderate variation in geometries of subunit interactions37–39 (FIG. 2). Such flexibility in the COPII cage has also been observed in cells40 and in reconstitution studies using COPII proteins and giant unilamellar vesicles41. These studies show that the COPII coat can form extended tubular structures and constricted tubules resembling ‘beads on a string’ that are of sufficient dimensions to accommodate procollagen fibres.
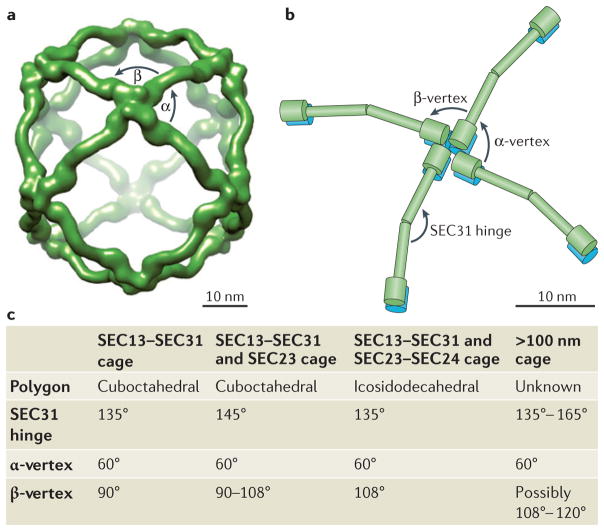
Figure 2. The architecture of the COPII cage facilitates transport of diverse cargo.
Structural analyses have demonstrated that the coat protein complex II (COPII) cage, which consists of the SEC13–SEC31 and SEC23–SEC24 subcomplexes, has a flexible architecture. a | Electron microscope reconstruction model of the SEC13–SEC31 cuboctahedral cage that is formed with purified mammalian SEC13–SEC31 (REF. 37) (Electron Microscopy Data Bank (EMDataBank) accession number: EMDB1232). b | Schematic representation of SEC31–SEC13 (shown in green and blue, respectively) heterotetramers arranged at a vertex point, indicating variable angles in the observed geometry of COPII cages. c | The structures for the SEC13–SEC31 assembly unit38 and the SEC13–SEC31 cage37 in the presence of SEC23 (REF. 147) and SEC23–SEC24 (REF. 35) have been reported. Variations in the SEC31 hinge (135°–165°) as well as in the β-vertex angle (90°–108°) of the SEC13–SEC31 cage have been documented and are listed in the table. The α-vertex angle is constant at 60° in each of the conditions. Under conditions in which SEC23–SEC24 is added to SEC13–SEC31, cuboctahedral (a 24 edged polygon) and icosidodecaheral (a 60 edged polygon) cage geometries were observed with each edge consisting of a SEC13–SEC31 heterotetramer. As the β-angle approaches 120° in the case of very large vesicles or tubules, the COPII cage may produce near-planar lattices35. Distinct arrangements in the COPII cage are thought to allow for the range of COPII vesicle carriers necessary to accommodate varying cargo sizes.
Consistent with the idea that transport carriers can adapt to accommodate particular cargo, genetic studies have revealed that isoform-specific mutations in SEC23A cause developmental defects due to deficiencies in collagen secretion, whereas smaller secretory cargos undergo normal trafficking42–44. Similarly, human chylomicron retention disease has been linked to mutations in the SAR1B isoform, which cause fat malabsorption due to deficiency in the secretion of large lipoproteins from enterocytes45,46. Interestingly, the interaction of SAR1 isoforms with distinct coat subunits and appropriate regulation of SAR1 GTPase activity seem to be crucial for the export of large procollagen and chylomicron cargos in COPII carriers. Recent studies have also revealed that monoubiquitylation of the SEC31 subunit is required for ER export of procollagen in developing mouse embryonic stem cells47. This post-translational modification could alter SEC31 structure and flexibility or recruit a cofactor that influences COPII assembly to generate large vesicles. Collectively, these findings support a model wherein distinct subunits of COPII can be assembled in different geometries and modulated by SAR1 GTPase activity to produce different sized transport carriers in various cell types.
The composition, size and number of ERES also vary across cell types and are influenced by the cargo that is being secreted. Cells that export large procollagen from the ER depend on the transmembrane TANGO1–cTAGE5 (transport and Golgi organization protein 1–cutaneous T cell lymphoma-associated antigen 5) complex that localizes to ERES and has a crucial role in ER export of selected cargo48,49. The TANGO1–cTAGE5 complex interacts with SEC23–SEC24, but is not itself packaged into COPII carriers. Instead, it is thought to coordinate procollagen capture into large vesicles at ERES through binding to lumenal procollagen and regulation of cytoplasmic SEC23–SEC24 (REF. 49). Interestingly, orthologues of TANGO1 and cTAGE5 have only been identified in vertebrates, suggesting that this complex has evolved to manage large cargo in specialized cell types. Indeed, in plant cells, large cellulose fibrils are polymerized directly at the plasma membrane rather than being assembled before their secretion50. In addition to the effects of cargo dimension on ERES composition, secretory cargo load influences ERES size and number. For example, in plant cells the number of ERES and the recruitment of SEC24 to ERES increases in cells transiently expressing ER export-competent membrane cargo. However, this occurs only if the cargo contained an ER export motif51. In mammalian cells, acute increases in cargo load cause ERES to fuse, producing larger but fewer exit sites. Conversely, a chronic rise in secretory cargo increases both the size and number of ERES52 as well as the expression of SEC24 and SEC16 in part through the activation of an ER stress-sensing unfolded protein response (UPR) pathway53. These findings demonstrate that cargo type, size and volume influence ERES structure and abundance, and that the cargo can influence the organization of the ER–Golgi interface. More generally, the UPR is likely to have a prominent role in controlling ER–Golgi membrane structure and function during the development of many cell types.
After secretory cargos have exited the ER in COPII carriers, these intermediates fuse with target membranes using a conserved core machinery. However, the distances that these ER-derived carriers travel and the requirement for cytoskeletal filaments differ depending on the cell type. The target membrane for COPII carriers also varies in addition to the set of tethering factors that are engaged. For example, animal cells rely on microtubules to guide COPII vesicles towards an ER–Golgi intermediate compartment (ERGIC) en route to cis-Golgi compartments54. Indeed, multiple types of microtubule-based motors localize to this intermediate compartment and are thought to generate the pleomorphic membrane structures that are characteristic of the ERGIC55. By contrast, in other species, including yeast that have a more compact ER–Golgi organization, COPII vesicles seem to fuse directly to cis-Golgi compartments56.
In the following sections, we examine more closely the organization of the ER–Golgi interface across species with a focus on plant and mammalian systems as two well-studied examples of how the ER–Golgi interface has been adapted to meet cellular needs. Specifically, we compare current models for COPII carrier-mediated trafficking from the ER in mammalian cells (in which the ER and Golgi compartments are separated and depend on an intermediate compartment for efficient transport) with trafficking in plant cells (in which Golgi stacks are closely associated with the ER and are highly mobile).
Variations in the ER–Golgi interface
Models comparing the ER–Golgi interface in mammalian and plant cells are shown in FIG. 3 and FIG. 4. In cultured mammalian cells, the Golgi stacks are clustered and connected by inter-cisternal tubules to form a Golgi ribbon57. This configuration is fundamentally different from plants, in which the Golgi is composed of individual stacks of flattened cisternae58,59, termed Golgi bodies or stacks. This morphology is seen in the insect Drosophila melanogaster 60 and the yeast Pichia pastoris22 as well as in the unicellular parasites Trypanosoma brucei61 and Toxoplasma gondii62. The yeast Saccharomyces cerevisiae may represent an extreme example, in which cis-, medial- and trans-Golgi cisternae seem to be completely unstacked22. Notably, the plant Golgi is distinct from these other species in that it shows rapid motility (up to 4 μm per sec)58, and this requires the activity of actomyosin motors59,63–65 and close association with tubular ER strands. This latter feature is underscored by the absence in plants of the ERGIC, which is found in mammalian cells66–69.
Figure 3. The ER–Golgi interface and ERES have a distinct organization in mammals and plants.
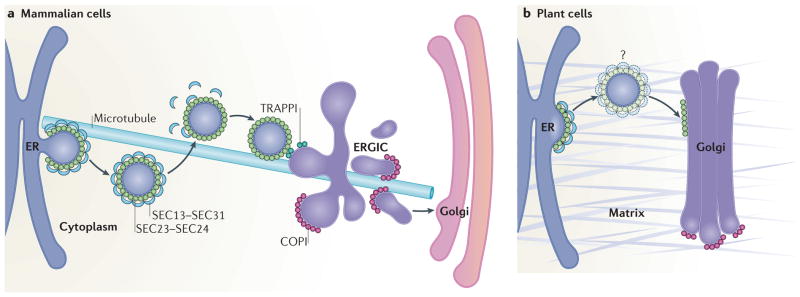
a | In mammalian cells, ER exit sites (ERES) are orientated towards a juxtaposed endoplasmic reticulum (ER)–Golgi intermediate compartment (ERGIC). Coat protein complex II (COPII)-coated vesicles originate within cup-shaped ER subdomains, which are associated with the plus end of microtubules. Upon fission of vesicles from the ERES, the SEC13–SEC31 cage is depolymerized, but the SEC23–SEC24 coat is partially retained. Vesicles reach the ERGIC in a microtubule-independent manner where they are tethered through the interaction between SEC23 and the TRAPPI (transport protein particle I) tethering complex. COPI mediates forward protein transport from the ERGIC towards the Golgi as well as recycling back to the ER membrane (the latter is not shown). b | In plant cells, ERES and Golgi are closely associated, possibly through a matrix (indicated in grey) that holds the ER and the Golgi together. The existence of COPII vesicles in plants is still debated (denoted by the question mark); vesicle-like structures have been seen89,97,99,106 rarely in electron microscopy analyses of high-pressure frozen Arabidopsis thaliana tissues, although it was unclear whether they were undergoing budding or fusion. Unlike mammalian cells, plant cell ER–Golgi transport does not rely on the microtubule cytoskeleton.
Figure 4. Cellular architecture contributes to the ER–Golgi organization and positioning of ERES in mammalian and plant cells.
a,b | Side-view schematic (a) or direct view (b) of a cultured mammalian cell, showing the relative positioning of the nucleus, endoplasmic reticulum (ER) network and the Golgi. The inset shows the distribution of ER exit sites (ERES), which are dispersed over the cytoplasmic ER network and concentrated in regions near the Golgi. c | Confocal image of hepatocarcinoma cells (HHU7 cells) co-transfected with SEC24–mCherry and VSVG–YFP, which label ERES and the ER, respectively. d | Side-view diagram of a plant cell showing that the ER is sandwiched between the tonoplast vacuole and the plasma membrane. e | Diagram illustrating the distribution of the ER network at the cortical region of a plant cell. The inset depicts the organization of the ER, ERES and closely associated Golgi complexes; the mobility of Golgi stacks is indicated by the dashed arrows. f | Confocal images of a tobacco leaf epidermal cell co-expressing the ER and Golgi marker ERD2–GFP (shown in green, right panel) and YFP–SEC24A (shown in red, middle panel). The merged image reveals a large overlap of fluorescent signals at punctae that represent Golgi and ERES. The image in panel c courtesy of K. Hirschberg, Tel Aviv University, Israel. The images in panel f courtesy of L. Renna, Michigan State University, USA.
The ERGIC as a distinct intermediate organelle
Implicit in the ER–Golgi organization of eukaryotic species are different strategies to efficiently transport cargo from the ER to the Golgi. In S. cerevisiae and in plant cells, ER–Golgi transport fits a simple model whereby COPII carriers shuttle cargo from the ER to the Golgi by fusing directly with the cis-Golgi70 or by generating new cisternae by homotypic fusion. In mammalian cells, the situation is more complex as the distances between ERES and the cis-Golgi compartment are longer than in yeast and plant cells. Indeed, the ERGIC may be necessary to facilitate this long-distance transport in mammalian cells. Previous models described the ERGIC as a specialized domain of the ER71 or the cis-Golgi72, although it may be more appropriate to consider the ERGIC as a distinct organelle73,74 that functions in COPI-dependent sorting of retrograde cargo, in concentration of anterograde biosynthetic cargo75 and possibly in post-ER protein quality control76. Early live-cell imaging analyses suggested that ERGIC structures may move from ERES to the Golgi by tracking on microtubules54,77. These results seeded the transport complex model19,78 whereby newly uncoated COPII vesicles would fuse to form transient pleiomorphic vesicular tubular clusters, which would then be transported to the Golgi via long-range microtubule tracks and dynein motors. The tubular clusters would either homotypically fuse to form new cis-Golgi cisternae, or they would fuse with the first cisternae of the cis-Golgi. COPI-coated vesicles formed from tubular clusters and cis-Golgi membranes would then recycle components back to the ER (FIG. 3a).
An alternative model for ER–Golgi protein transport in mammalian cells, termed the stable compartment model, views the ERGIC as a more static structure. This model proposes that positioning of the ERGIC between ERES and the Golgi facilitates a two-step transport process, which involves short-range microtubule-independent transport of COPII vesicles from ERES to the ERGIC followed by long-range microtubule-dependent transport of anterograde cargo-rich domains from the ERGIC to the cis-Golgi. Microtubule-associated proteins such as the microtubule plus enddirected motor kinesin and the minus end-directed motor dynein have been found in association with the ERGIC. Moreover, functional analyses indicate that kinesin operates in recycling of membranes to the ER, whereas dynein is needed for anterograde transport54,79,80. In the stable compartment model, kinesin and dynein would mediate steady-state positioning of the ERGIC but also have an active role in transport: kinesin would facilitate microtubule plus end-directed retrograde trafficking to the ER and dynein–dynactin would facilitate the transport from the ERGIC to the cis-Golgi74. Elements of the actin cytoskeleton such as actin-binding proteins (spectrin)11,81 and regulatory GTPases (CDC42)82 have also been invoked as dynactin-recruiting factors during this process.
Currently, it is not clear whether the stable compartment or the transport complex model better explain ERGIC function in trafficking. Imaging studies have shown that small soluble cargo is sorted into Golgi-directed carriers from long-lived ERGIC structures83, although the machinery responsible for this event has not been defined. It is also possible that both mechanisms operate to transport cargos of varying size that move through the ERGIC. For example, smaller cargo molecules could move through more mobile transport carriers, whereas larger cargo present in mammalian cells may be trafficked in ERGIC transport complexes. Dual-label imaging experiments that simultaneously monitor ERGIC markers and a broad range of secretory cargo may help resolve this issue.
COPII carrier dynamics in plant cells
Analyses in plant cells indicating that ER export can occur while Golgi stacks are moving in close association with ER tubules84 raise a fundamental question of how proteins and membranes are efficiently transported from the ER to mobile Golgi stacks (FIG. 3b). One model has postulated that an active ERES produces a localized signal that uncouples a Golgi stack from actin-associated motors and causes it to pause near this site. Completion of membrane and cargo transfer to and from the ER would be followed by inactivation of this ‘pausing’ cue, allowing the Golgi stack to resume its movement58. This model assumes that the association of ERES with Golgi stacks is not continuous and that COPII foci may be dispersed over the ER, awaiting interaction with a suitable Golgi stack. However, analyses of Arabidopsis thaliana SEC13 expressed in A. thaliana and in tobacco leaf epidermal cells as well as in BY2 cells showed that SEC13 is present in foci that have continuous association with Golgi stacks85 rather than being dispersed over the ER. A continuous association of foci with Golgi stacks has also been validated in several reports in which other COPII proteins were labelled, including SAR1, SEC23 and SEC24, and that investigated both heterologous as well as endogenous plant expression systems51,85–95 (FIG. 4f). Together, this supports the model that ERES are in continuous association with Golgi stacks24,96.
The presence of COPII vesicles in plants is still debated. Vesicle-like structures have been rarely detected89,97–99 in electron microscopy analyses of high-pressure frozen A. thaliana tissues; moreover, the directionality of such structures (that is, whether these are budding or fusing intermediates) could not be established. Nonetheless, in studies of high-pressure freeze-substituted A. thaliana root tip cells, immunogold labelling analyses with antibodies specific for A. thaliana SAR1 and SEC23 showed labelling of some vesicle-like structures67,99. The difficulty to visualize budding COPII vesicles in plants raises the possibility that these entities may be very quickly consumed at the ER surface. Through optical manipulation of individual Golgi stacks with laser tweezers100 in highly vacuolated cells, it has been shown that lateral displacement of individual Golgi stacks is followed by continued link-age and rapid growth of the attached ER tubule. This finding indicates that in these plant cells the Golgi and ER are firmly attached, possibly through a tethering matrix at the Golgi–ER interface that physically links the two organelles together84.
The association of ERES with individual Golgi stacks in plant cells is conceptually feasible in the context of a matrix-mediated attachment of the ER with Golgi stacks. For example, ERES could facilitate export to a Golgi stack at an ER–Golgi interface that is relatively static. Such an organization would be more energy efficient than a system that stochastically produces ERES. This may be analogous to Caenorhabditis elegans in which ERES are closely juxtaposed to the Golgi and proteins such as the SEC-16-interacting protein TFG-1 direct movement of COPII carriers away from the ER101. It is also possible that in addition to ERES-associated Golgi, new ERES may be formed on the ER to initiate de novo assembly of Golgi stacks. Fluorescent protein localization analysis has shown that the YFP–SEC24 fusion protein is enriched at the Golgi surface97, and this was proposed to represent the accumulation of COPII carriers before fusion97. This possibility is consistent with findings in yeast and mammalian cells showing that COPII may be partially retained on ER-derived carriers until they reach the Golgi102. As mentioned above, putative COPII vesicles have been detected in close association with cis-cisternal Golgi elements in electron microscopy analyses of A. thaliana cells, and the formation of these initial cis-cisterna may be functionally analogous to the mammalian ERGIC67. Disassembly of COPII coats is initiated when SAR1 hydrolyses its bound GTP (BOX 1), although coat subunits seem to be slowly released after carrier formation. It has also been shown that fluorescent protein fusions of two plant SAR1 isoforms distribute along the ER facing the Golgi87. Together, these findings suggest that ER export in plant cells relies on a unique organization in which a specialized subdomain of the ER and an associated Golgi stack act as a secretory unit with COPII carriers being formed and quickly released at the ER subdomain and partially coated COPII carriers then associating with the Golgi. Additional validation of this model could be achieved by analysing the subcellular distribution of SEC16, which would be predicted to associate with the ER-facing Golgi stacks. A scenario in which COPII vesicles form and are transported from the ER at a rate that is faster than COPII uncoating at the Golgi is consistent with the observation that fluorescently tagged versions of COPII components can be readily visualized in live plant cells, although COPII vesicles have rarely been seen at an ultrastructural level in plants103–106.
In contrast to mammalian cells, plant ER–Golgi transport does not depend on cytoskeletal components. In highly vacuolated plant cells, the Golgi undergoes actomyosin-dependent movement, but chemical inhibition of this movement does not disrupt the transport between the ER and Golgi. This has been demonstrated by fluorescence recovery after photobleaching (FRAP), in which markers were tracked in immobilized Golgi after treating cells with actin-disrupting chemicals2. Microtubules also do not seem to be involved in ER–Golgi protein transport in plants; for example, rates of ER–Golgi protein traffic are similar in cells with depolymerized actin and in cells depleted of both actin and microtubules2. Fluorescence recovery of cycling proteins has also been observed upon photobleaching of Golgi in untreated cells24, again supporting the model that ER export can occur toward motile Golgi. However, it has not yet been established whether there are differences in ER–Golgi transport kinetics between cells with disrupted versus intact actin, or between fast or slow moving Golgi stacks.
Different forms for unique functions
A conserved core machinery transports biosynthetic cargo forward in the early secretory pathway and is balanced with retrieval routes that maintain the ER and Golgi compartments. In eukaryotic cells, COPII assembly produces transport intermediates from the ER, which then rely on RAB GTPase-dependent tethering factors and the SNARE-dependent membrane fusion machinery for delivery of cargo to the Golgi. Similarly, retrograde transport depends on the COPI machinery to produce retrograde-directed vesicles that also require conserved tethering factors and SNARE proteins for fusion with ER membranes. In spite of this conservation, diversity in eukaryotic cell types and cellular functions generate tremendous variety in the organization of this ER–Golgi interface. Each trafficking step between ER and Golgi compartments provides multiple opportunities for regulation that could influence vesicle and organelle size and shape. Indeed, there are now clear examples in which the expression levels of these core components and their covalent modification (for example, phosphorylation or ubiquitylation) alter the morphology of COPII carriers. Moreover, the additional control provided by accessory factors such as TANGO1 and cTAGE5 and cytoskeletal components in certain cell types also affects ER–Golgi organization. Finally, inherent flexibility in both the COPII35 and COPI107 coats that allows cargos of different sizes to be accommodated can influence the morphology of ER–Golgi transport intermediates in different cell types.
The influence of cargo
The striking differences in the organization of ER–Golgi interfaces across species that are revealed by in vivo morphological analyses probably reflect cellular specialization related to the types of cargo that must be transported as well as the overall cell structure and function. If we consider the possibility that distinct morphologies observed reflect functional adaptation of the core transport machinery, a molecular understanding of these differences should provide important insights into how trafficking and compartmental organization are integrated within the early secretory pathway. For example, the existence of an ERGIC in certain cell types and not in others remains mysterious. As discussed above, P. pastoris yeast cells56 and vacuolated plant cells106 lack an ERGIC but instead contain compact ER–Golgi units (≤300 nm distance between the two organelles) that are thought to be firmly connected through a tethering matrix. However, such a compact arrangement may not provide the space that would be needed for the assembly of large cargo into dissociating transport carriers without generating membrane connections between the ER and Golgi compartments. Indeed, there are no known large secretory cargos (≥300nm) in P. pastoris or A. thaliana. By contrast, active secretory cells in vertebrates generally contain an ERGIC, and these cell types often transport large cargo molecules such as procollagen.
Cellular architecture as a driving force for interface organization
Although the cargo that is being transported may be one driver of ER–Golgi organization, an alternative structural demand for the ERGIC in vertebrate cells may be to bridge the large distance between the perinuclear ER and the plasma membrane. In this context, the ERGIC and a microtubule network may be needed to facilitate efficient transport and limit diffusion of intermediates. By contrast, the plant ER and Golgi are ‘sandwiched’ within a thin layer of cytoplasm between the tonoplast and the plasma membrane due to the presence of a central vacuole that occupies most of the cell volume (FIG. 4d). Cytoskeletal elements may therefore not be needed to facilitate bidirectional ER–Golgi traffic or to restrict diffusion of transport intermediates. Finally, a more extensive set of ER–Golgi tethering factors is present in cells that contain an ERGIC. In plants, there are no apparent homologues of mammalian GRASP65 (Golgi reassembly-stacking protein of 65 kDa), GM130 (cis-Golgi matrix of 130 kDa) and giantin vesicle tethering factors108. However, plants do require a p115 homologue for ER–Golgi transport and express other Golgi-localized coiled-coil domain proteins with sequence identify to tethering factors109. Although the function of these proteins has yet to be determined, this simplified set of tethering factors may underlie matrix-mediated attachment of ER–Golgi units in plants. Nevertheless, questions regarding how the ERGIC is generated and maintained in mammalian cells remain unanswered. Interestingly, loss of GM130 leads to the disruption of the mammalian Golgi ribbon110. The absence of GM130 homologues in plants may suggest that the acquisition or loss of specialized tethering factors may have been a contributing factor for the unique organization of the plant Golgi during evolution from the last common eukaryotic ancestor.
Notably, variations in the ER–Golgi interface can also exist within the same species. For example, most characterizations of ERES in plant cells have been acquired in highly vacuolated cells. In specialized tissues such as root meristem cells and columella cells that are comparatively less vacuolated, a large percentage of Golgi stacks (≤70%) have been reported to not closely associate with the ER106. How ER export occurs towards Golgi stacks in these cells is unknown. Although most cells are highly vacuolated in plants, these findings underscore that the organization of the ER and Golgi can vary and suggest that specific ER export mechanisms are in place to suit the organization of specific cell types, even within the same organism.
Conclusions and perspectives
A useful guide for the biologist’s view of the cell has been that structure reflects function. This perspective has been powerfully applied to the study of membrane traffic with an understanding that the molecular basis for observed membrane structures and dynamics is tightly coupled with function. In this Review, we focus on individual stages in membrane transport that influence organization of the ER–Golgi interface. Through a comparative analysis of model systems, we begin to see how this core structural machinery is adapted to meet cell-specific requirements. However, future studies will need to further integrate cellular regulatory networks that control biosynthetic rates and flux through trafficking routes with this core machinery. For example, changing developmental and environmental conditions that alter the profile of protein and lipid cargos must be integrated in a manner that maintains the organization of the ER–Golgi interface. Global regulation through the UPR clearly has a key homeostatic role in coordinating protein and lipid biosynthetic rates in response to environmental conditions. But studying how the UPR orchestrates such adaptation may be challenging because there are multiple branches of the UPR in animal and plant cells53,111, and these are likely to work in concert with several other signalling networks that regulate membrane traffic. Advancing such ‘systems-level’ questions will depend on the identification of all required factors, in vivo microscopy and cell-free assays to accurately measure cargo biosynthetic and transport rates, as well as the elucidation of the sensor networks that are in place for overall coordination of distinct trafficking steps. Continued comparative analyses across a range of model organisms should ultimately allow us to further define the mechanisms that underlie the regulation of ER–Golgi transport in the context of cellular function.
Acknowledgments
F.B is supported by grants from the US National Institutes of Health (R01 GM101038), Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. DOE (DE-FG02-91ER20021), NASA (NNX12AN71G) and the National Science Foundation (MCB 0948584; 1243792). C.B. acknowledges support from the US National Institutes of Health (R01 GM52549).
Glossary
- Anterograde transport
Membrane traffic pathway in which a linear assembly of membrane-bound compartments facilitates cargo movement towards the cell surface
- Retrograde transport
Membrane traffic pathway in which a linear assembly of membrane-bound compartments facilitates cargo movement towards the ER
- ER exit sites (ERES)
Specialized regions on the surface of endoplasmic reticulum (ER) membranes where coat protein complex II (COPII) coat subunits are recruited and assembled into COPII carrier vesicles that transport secretory cargo from the ER
- Unfolded protein response (UPR)
A system of ancestral signalling pathways that are activated upon increased secretory protein load in the endoplasmic reticulum to ensure maintenance of cellular homeostasis
- ER–Golgi intermediate compartment (ERGIC)
An organelle that is visible in most mammalian cells at the interface between the endoplasmic reticulum (ER) and the Golgi and that is implicated in cargo concentration and sorting
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Federica Brandizzi’s hompage:
http://www.prl.msu.edu/faculty/brandizzi_federica
Charles Barlowe’s homepage:
http://geiselmed.dartmouth.edu/barlowe
Electron Microscopy Data Bank (EMDataBank):
EMDB1232
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol. 2001;155:557–570. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C. Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell. 2002;14:1293–1309. doi: 10.1105/tpc.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altan-Bonnet N, Sougrat R, Lippincott-Schwartz J. Molecular basis for Golgi maintenance and biogenesis. Curr Opin Cell Biol. 2004;16:364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Farquhar MG, Palade GE. The Golgi apparatus: 100 years of progress and controversy. Trends Cell Biol. 1998;8:2–10. doi: 10.1016/S0962-8924(97)01187-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Driouich A, Staehelin LA. In: The Golgi Apparatus. Berger EG, Roth J, editors. Birkhäuser Verlag; 1997. pp. 275–301. [Google Scholar]
- 6.Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 7.Cavalier DM, et al. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell. 2008;20:1519–1537. doi: 10.1105/tpc.108.059873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dirnberger D, Bencur P, Mach L, Steinkellner H. The Golgi localization of Arabidopsis thaliana β1,2-xylosyltransferase in plant cells is dependent on its cytoplasmic and transmembrane sequences. Plant Mol Biol. 2002;50:273–281. doi: 10.1023/a:1016061815748. [DOI] [PubMed] [Google Scholar]
- 9.Fitchette AC, et al. Biosynthesis and immunolocalization of Lewis a-containing N-glycans in the plant cell. Plant Physiol. 1999;121:333–344. doi: 10.1104/pp.121.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keegstra K, Raikhel N. Plant glycosyltransferases. Curr Opin Plant Biol. 2001;4:219–224. doi: 10.1016/s1369-5266(00)00164-3. [DOI] [PubMed] [Google Scholar]
- 11.De Matteis MA, Morrow JS. Spectrin tethers and mesh in the biosynthetic pathway. J Cell Sci. 2000;113:2331–2343. doi: 10.1242/jcs.113.13.2331. [DOI] [PubMed] [Google Scholar]
- 12.Kondoh K, Torii S, Nishida E. Control of MAP kinase signaling to the nucleus. Chromosoma. 2005;114:86–91. doi: 10.1007/s00412-005-0341-9. [DOI] [PubMed] [Google Scholar]
- 13.Saint-Jore-Dupas C, et al. Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell. 2006;18:3182–3200. doi: 10.1105/tpc.105.036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabouille C, et al. Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides. J Cell Sci. 1995;108:1617–1627. doi: 10.1242/jcs.108.4.1617. [DOI] [PubMed] [Google Scholar]
- 15.Klumperman J. The growing Golgi: in search of its independence. Nature Cell Biol. 2000;2:E217–E219. doi: 10.1038/35046635. [DOI] [PubMed] [Google Scholar]
- 16.van Meel E, Klumperman J. Imaging and imagination: understanding the endo-lysosomal system. Histochem Cell Biol. 2008;129:253–266. doi: 10.1007/s00418-008-0384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. Arabidopsis vacuolar H-ATPase subunit E isoform 1 is required for Golgi organization and vacuole function in embryogenesis. Plant Cell. 2006;18:715–730. doi: 10.1111/j.1365-313X.2004.02283.x. [DOI] [PubMed] [Google Scholar]
- 18.Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nature Rev Mol Cell Biol. 2009;10:148–155. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- 19.Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol. 1996;135:19–35. doi: 10.1083/jcb.135.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond AT, Glick BS. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol Biol Cell. 2000;11:3013–3030. doi: 10.1091/mbc.11.9.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaywitz DA, Orci L, Ravazzola M, Swaroop A, Kaiser CA. Human SEC13Rp functions in yeast and is located on transport vesicles budding from the endoplasmic reticulum. J Cell Biol. 1995;128:769–777. doi: 10.1083/jcb.128.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossanese OW, et al. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. Demonstrates that P. pastoris and S. cerevisiae have distinct ERES and Golgi organizations and proposes a model in which Golgi organization correlates with ERES organization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.daSilva LL, et al. Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell. 2004;16:1753–1771. doi: 10.1105/tpc.022673. Shows that ERES and motile Golgi stacks form a secretory unit system that is unique to eukaryotic cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connerly PL, et al. Sec16 is a determinant of transitional ER organization. Curr Biol. 2005;15:1439–1447. doi: 10.1016/j.cub.2005.06.065. [DOI] [PubMed] [Google Scholar]
- 26.Watson P, Townley AK, Koka P, Palmer KJ, Stephens DJ. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic. 2006;7:1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller EA, Barlowe C. Regulation of coat assembly — sorting things out at the ER. Curr Opin Cell Biol. 2010;22:447–453. doi: 10.1016/j.ceb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorente-Rodriguez A, Barlowe C. Entry and exit mechanisms at the cis-face of the Golgi complex. Cold Spring Harb Perspect Biol. 2011 Apr 11; doi: 10.1101/cshperspect.a005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- 31.Cai H, et al. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–944. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- 32.Zink S, Wenzel D, Wurm CA, Schmitt HD. A link between ER tethering and COP-I vesicle uncoating. Dev Cell. 2009;17:403–416. doi: 10.1016/j.devcel.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Bonfanti L, et al. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell. 1998;95:993–1003. doi: 10.1016/s0092-8674(00)81723-7. [DOI] [PubMed] [Google Scholar]
- 34.Barlowe C, et al. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. Reports, for the first time, that COPII-coated transport intermediates exist and reconstitutes COPII carriers in vitro. [DOI] [PubMed] [Google Scholar]
- 35.Stagg SM, et al. Structural basis for cargo regulation of COPII coat assembly. Cell. 2008;134:474–484. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Donnell J, Maddox K, Stagg S. The structure of a COPII tubule. J Struct Biol. 2011;173:358–364. doi: 10.1016/j.jsb.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Stagg SM, et al. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–238. doi: 10.1038/nature04339. Reveals the three-dimensional reconstruction of SEC13–SEC31 cages at 30 Å resolution that defined a flexible cuboctahedron geometry of the COPII outer coat. [DOI] [PubMed] [Google Scholar]
- 38.Fath S, Mancias JD, Bi X, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharya N, JOD, Stagg SM. The structure of the Sec13/31 COPII cage bound to Sec23. J Mol Biol. 2012;420:324–334. doi: 10.1016/j.jmb.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeuschner D, et al. Immuno-electron tomography of ER exit sites reveals the existence of free COPII-coated transport carriers. Nature Cell Biol. 2006;8:377–383. doi: 10.1038/ncb1371. [DOI] [PubMed] [Google Scholar]
- 41.Bacia K, et al. Multibudded tubules formed by COPII on artificial liposomes. Sci Rep. 2011;1:17. doi: 10.1038/srep00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyadjiev SA, et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nature Genet. 2006;38:1192–1197. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- 43.Lang MR, Lapierre LA, Frotscher M, Goldenring JR, Knapik EW. Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nature Genet. 2006;38:1198–1203. doi: 10.1038/ng1880. [DOI] [PubMed] [Google Scholar]
- 44.Fromme JC, et al. The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev Cell. 2007;13:623–634. doi: 10.1016/j.devcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones B, et al. Mutations in a Sar1 GTPase of COPII vesicles are associated with lipid absorption disorders. Nature Genet. 2003;34:29–31. doi: 10.1038/ng1145. [DOI] [PubMed] [Google Scholar]
- 46.Silvain M, et al. Anderson’s disease (chylomicron retention disease): a new mutation in the SARA2 gene associated with muscular and cardiac abnormalities. Clin Genet. 2008;74:546–552. doi: 10.1111/j.1399-0004.2008.01069.x. [DOI] [PubMed] [Google Scholar]
- 47.Jin L, et al. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482:495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saito K, et al. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell. 2009;136:891–902. doi: 10.1016/j.cell.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 49.Saito K, et al. cTAGE5 mediates collagen secretion through interaction with TANGO1 at endoplasmic reticulum exit sites. Mol Biol Cell. 2011;22:2301–2308. doi: 10.1091/mbc.E11-02-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keegstra K. Plant cell walls. Plant Physiol. 2010;154:483–486. doi: 10.1104/pp.110.161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanton SL, Chatre L, Renna L, Matheson LA, Brandizzi F. De novo formation of plant endoplasmic reticulum export sites is membrane cargo induced and signal mediated. Plant Physiol. 2007;143:1640–1650. doi: 10.1104/pp.106.094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP. Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J. 2008;27:2043–2054. doi: 10.1038/emboj.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 54.Presley JF, et al. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. A pioneering study that uses fluorescent protein technology to visualize ER export of secretory cargo along microtubules in live mammalian cells. [DOI] [PubMed] [Google Scholar]
- 55.Stephens DJ. Functional coupling of microtubules to membranes — implications for membrane structure and dynamics. J Cell Sci. 2012;125:2795–2804. doi: 10.1242/jcs.097675. [DOI] [PubMed] [Google Scholar]
- 56.Mogelsvang S, Gomez-Ospina N, Soderholm J, Glick BS, Staehelin LA. Tomographic evidence for continuous turnover of Golgi cisternae in Pichia pastoris. Mol Biol Cell. 2003;14:2277–2291. doi: 10.1091/mbc.E02-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward TH, Brandizzi F. Dynamics of proteins in Golgi membranes: comparisons between mammalian and plant cells highlighted by photobleaching techniques. Cell Mol Life Sci. 2004;61:172–185. doi: 10.1007/s00018-003-3355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nebenfuhr A, et al. Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 1999;121:1127–1142. doi: 10.1104/pp.121.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boevink P, et al. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998;15:441–447. doi: 10.1046/j.1365-313x.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- 60.Bard F, et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- 61.He CY, et al. Golgi duplication in Trypanosoma brucei. J Cell Biol. 2004;165:313–321. doi: 10.1083/jcb.200311076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hager KM, Striepen B, Tilney LG, Roos DS. The nuclear envelope serves as an intermediary between the ER and Golgi complex in the intracellular parasite Toxoplasma gondii. J Cell Sci. 1999;112:2631–2638. doi: 10.1242/jcs.112.16.2631. [DOI] [PubMed] [Google Scholar]
- 63.Avisar D, Prokhnevsky AI, Makarova KS, Koonin EV, Dolja VV. Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 2008;146:1098–1108. doi: 10.1104/pp.107.113647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peremyslov VV, Prokhnevsky AI, Avisar D, Dolja VV. Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 2008;146:1109–1116. doi: 10.1104/pp.107.113654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sparkes IA, Teanby NA, Hawes C. Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. J Exp Bot. 2008;59:2499–2512. doi: 10.1093/jxb/ern114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hawes C, Satiat-Jeunemaitre B. The plant Golgi apparatus — going with the flow. Biochim Biophys Acta. 2005;1744:93–107. doi: 10.1016/j.bbamcr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 67.Donohoe BS, et al. Cis-Golgi cisternal assembly and biosynthetic activation occur sequentially in plants and algae. Traffic. 2013;14:551–567. doi: 10.1111/tra.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bentley M, et al. SNARE status regulates tether recruitment and function in homotypic COPII vesicle fusion. J Biol Chem. 2006;281:38825–38833. doi: 10.1074/jbc.M606044200. [DOI] [PubMed] [Google Scholar]
- 69.Xu D, Hay JC. Reconstitution of COPII vesicle fusion to generate a pre-Golgi intermediate compartment. J Cell Biol. 2004;167:997–1003. doi: 10.1083/jcb.200408135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 71.Sitia R, Meldolesi J. Endoplasmic reticulum: a dynamic patchwork of specialized subregions. Mol Biol Cell. 1992;3:1067–1072. doi: 10.1091/mbc.3.10.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mellman I, Simons K. The Golgi complex: in vitro veritas? Cell. 1992;68:829–840. doi: 10.1016/0092-8674(92)90027-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klumperman J, et al. The recycling pathway of protein ERGIC-53 and dynamics of the ER–Golgi intermediate compartment. J Cell Sci. 1998;111:3411–3425. doi: 10.1242/jcs.111.22.3411. [DOI] [PubMed] [Google Scholar]
- 74.Appenzeller-Herzog C, Hauri HP. The ER–Golgi intermediate compartment (ERGIC): in search of its identity and function. J Cell Sci. 2006;119:2173–2183. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- 75.Martinez-Menarguez JA, Geuze HJ, Slot JW, Klumperman J. Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell. 1999;98:81–90. doi: 10.1016/S0092-8674(00)80608-X. [DOI] [PubMed] [Google Scholar]
- 76.Breuza L, et al. Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J Biol Chem. 2004;279:47242–47253. doi: 10.1074/jbc.M406644200. [DOI] [PubMed] [Google Scholar]
- 77.Scales SJ, Pepperkok R, Kreis TE. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. Demonstrates in vivo a temporal uncoupling of the roles of the COPI and COPII machineries in bidirectional membrane traffic at the ER–Golgi interface in mammalian cells. [DOI] [PubMed] [Google Scholar]
- 78.Stephens DJ, Pepperkok R. Illuminating the secretory pathway: when do we need vesicles? J Cell Sci. 2001;114:1053–1059. doi: 10.1242/jcs.114.6.1053. [DOI] [PubMed] [Google Scholar]
- 79.Lippincott-Schwartz J, Cole NB, Marotta A, Conrad PA, Bloom GS. Kinesin is the motor for microtubule-mediated Golgi-to-ER membrane traffic. J Cell Biol. 1995;128:293–306. doi: 10.1083/jcb.128.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roghi C, Allan VJ. Dynamic association of cytoplasmic dynein heavy chain 1a with the Golgi apparatus and intermediate compartment. J Cell Sci. 1999;112:4673–4685. doi: 10.1242/jcs.112.24.4673. [DOI] [PubMed] [Google Scholar]
- 81.Beck KA. Spectrins and the Golgi. Biochim Biophys Acta. 2005;1744:374–382. doi: 10.1016/j.bbamcr.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 82.Chen JL, et al. Coatomer-bound Cdc42 regulates dynein recruitment to COPI vesicles. J Cell Biol. 2005;169:383–389. doi: 10.1083/jcb.200501157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ben-Tekaya H, Miura K, Pepperkok R, Hauri HP. Live imaging of bidirectional traffic from the ERGIC. J Cell Sci. 2005;118:357–367. doi: 10.1242/jcs.01615. [DOI] [PubMed] [Google Scholar]
- 84.Sparkes IA, Ketelaar T, Ruijter NC, Hawes C. Grab a Golgi: laser trapping of Golgi bodies reveals in vivo interactions with the endoplasmic reticulum. Traffic. 2009;10:567–571. doi: 10.1111/j.1600-0854.2009.00891.x. Uses laser-trap technology in live cells and demonstrates that the ER and the Golgi are physically attached in highly vacuolated plant cells. [DOI] [PubMed] [Google Scholar]
- 85.Hanton SL, Matheson LA, Chatre L, Brandizzi F. Dynamic organization of COPII coat proteins at endoplasmic reticulum export sites in plant cells. Plant J. 2009;57:963–974. doi: 10.1111/j.1365-313X.2008.03740.x. [DOI] [PubMed] [Google Scholar]
- 86.Stefano G, et al. In tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum and of ER export sites depends on active COPI machinery. Plant J. 2006;46:95–110. doi: 10.1111/j.1365-313X.2006.02675.x. [DOI] [PubMed] [Google Scholar]
- 87.Hanton SL, et al. Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant Mol Biol. 2008;67:283–294. doi: 10.1007/s11103-008-9317-5. [DOI] [PubMed] [Google Scholar]
- 88.Wei T, Wang A. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner. J Virol. 2008;82:12252–12264. doi: 10.1128/JVI.01329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robinson DG, Herranz MC, Bubeck J, Pepperkok R, Ritzenthaler C. Membrane dynamics in the early secretory pathway. Crit Rev Plant Sci. 2007;26:199–225. [Google Scholar]
- 90.Sieben C, Mikosch M, Brandizzi F, Homann U. Interaction of the K+-channel KAT1 with the coat protein complex II coat component Sec24 depends on a di-acidic endoplasmic reticulum export motif. Plant J. 2008;56:997–1006. doi: 10.1111/j.1365-313X.2008.03658.x. [DOI] [PubMed] [Google Scholar]
- 91.Osterrieder A, Hummel E, Carvalho CM, Hawes C. Golgi membrane dynamics after induction of a dominant-negative mutant Sar1 GTPase in tobacco. J Exp Bot. 2009;61:405–422. doi: 10.1093/jxb/erp315. [DOI] [PubMed] [Google Scholar]
- 92.Hawes C, Osterrieder A, Hummel E, Sparkes I. The plant ER–Golgi interface. Traffic. 2008;9:1571–1580. doi: 10.1111/j.1600-0854.2008.00773.x. [DOI] [PubMed] [Google Scholar]
- 93.Schoberer J, et al. Arginine/lysine residues in the cytoplasmic tail promote ER export of plant glycosylation enzymes. Traffic. 2009;10:101–115. doi: 10.1111/j.1600-0854.2008.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Faso C, et al. A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. Plant Cell. 2009;21:3655–3671. doi: 10.1105/tpc.109.068262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ito Y, et al. cis-Golgi proteins accumulate near the ER exit sites and act as the scaffold for Golgi regeneration after brefeldin A treatment in tobacco BY-2 cells. Mol Biol Cell. 2012;23:3203–3214. doi: 10.1091/mbc.E12-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lerich A, et al. ER import sites and their relationship to ER exit sites: a new model for bidirectional ER–Golgi transport in higher plants. Front Plant Sci. 2012;3:143. doi: 10.3389/fpls.2012.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Langhans M, Meckel T, Kress A, Lerich A, Robinson DG. ERES (ER exit sites) and the ‘secretory unit concept’. J Microsc. 2012;247:48–59. doi: 10.1111/j.1365-2818.2011.03597.x. [DOI] [PubMed] [Google Scholar]
- 98.Hawes C. The ER/Golgi interface — is there anything in-between? Front Plant Sci. 2012;3:73. doi: 10.3389/fpls.2012.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Donohoe BS, Kang BH, Staehelin LA. Identification and characterization of COPIa- and COPIb-type vesicle classes associated with plant and algal Golgi. Proc Natl Acad Sci. 2007;104:163–168. doi: 10.1073/pnas.0609818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wright GD, Arlt J, Poon WC, Read ND. Optical tweezer micromanipulation of filamentous fungi. Fungal Genet Biol. 2007;44:1–13. doi: 10.1016/j.fgb.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 101.Witte K, et al. TFG-1 function in protein secretion and oncogenesis. Nature Cell Biol. 2011;13:550–558. doi: 10.1038/ncb2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lord C, et al. Sequential interactions with Sec23 control the direction of vesicle traffic. Nature. 2011;473:181–186. doi: 10.1038/nature09969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Craig S, Staehelin LA. High pressure freezing of intact plant tissues. Evaluation and characterization of novel features of the endoplasmic reticulum and associated membrane systems. Eur J Cell Biol. 1988;46:81–93. [PubMed] [Google Scholar]
- 104.Ritzenthaler C, et al. Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell. 2002;14:237–261. doi: 10.1105/tpc.010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Staehelin LA. The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 1997;11:1151–1165. doi: 10.1046/j.1365-313x.1997.11061151.x. [DOI] [PubMed] [Google Scholar]
- 106.Kang BH, Staehelin LA. ER-to-Golgi transport by COPII vesicles in Arabidopsis involves a ribosomeexcluding scaffold that is transferred with the vesicles to the Golgi matrix. Protoplasma. 2008;234:51–64. doi: 10.1007/s00709-008-0015-6. [DOI] [PubMed] [Google Scholar]
- 107.Faini M, et al. The structures of COPI-coated vesicles reveal alternate coatomer conformations and interactions. Science. 2012;336:1451–1454. doi: 10.1126/science.1221443. [DOI] [PubMed] [Google Scholar]
- 108.Latijnhouwers M, Hawes C, Carvalho C. Holding it all together? Candidate proteins for the plant Golgi matrix. Curr Opin Plant Biol. 2005;8:632–639. doi: 10.1016/j.pbi.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 109.Osterrieder A. Tales of tethers and tentacles: golgins in plants. J Microsc. 2012;247:68–77. doi: 10.1111/j.1365-2818.2012.03620.x. [DOI] [PubMed] [Google Scholar]
- 110.Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi–enzyme distribution. Nature Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- 111.Iwata Y, Koizumi N. Plant transducers of the endoplasmic reticulum unfolded protein response. Trends Plant Sci. 2012;17:720–727. doi: 10.1016/j.tplants.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 112.Bielli A, et al. Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. J Cell Biol. 2005;171:919–924. doi: 10.1083/jcb.200509095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yoshihisa T, Barlowe C, Schekman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- 114.Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- 115.Thor F, Gautschi M, Geiger R, Helenius A. Bulk flow revisited: transport of a soluble protein in the secretory pathway. Traffic. 2009;10:1819–1830. doi: 10.1111/j.1600-0854.2009.00989.x. [DOI] [PubMed] [Google Scholar]
- 116.Kuehn MJ, Herrmann JM, Schekman R. COPII–cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- 117.Giraudo CG, Maccioni HJ. Endoplasmic reticulum export of glycosyltransferases depends on interaction of a cytoplasmic dibasic motif with Sar1. Mol Biol Cell. 2003;14:3753–3766. doi: 10.1091/mbc.E03-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wendeler MW, Paccaud JP, Hauri HP. Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 2007;8:258–264. doi: 10.1038/sj.embor.7400893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miller EA, et al. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. Identifies multiple independent cargo-binding sites in the COPII coat subunit SEC24 and describes the plasticity of this adaptor complex in selecting multiple cargo molecules with distinct sorting signals. [DOI] [PubMed] [Google Scholar]
- 120.Bi X, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of Sec23. Sar1 complexed with the active fragment of Sec31. Dev Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Copic A, Latham CF, Horlbeck MA, D’Arcangelo JG, Miller EA. ER cargo properties specify a requirement for COPII coat rigidity mediated by Sec13p. Science. 2012;335:1359–1362. doi: 10.1126/science.1215909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Matsuoka K, et al. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 123.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 124.Routledge KE, Gupta V, Balch WE. Emergent properties of proteostasis–COPII coupled systems in human health and disease. Mol Membr Biol. 2010;27:385–397. doi: 10.3109/09687688.2010.524894. [DOI] [PubMed] [Google Scholar]
- 125.Waters MG, Griff IC, Rothman JE. Proteins involved in vesicular transport and membrane fusion. Curr Opin Cell Biol. 1991;3:615–620. doi: 10.1016/0955-0674(91)90031-s. [DOI] [PubMed] [Google Scholar]
- 126.Eugster A, Frigerio G, Dale M, Duden R. COP I domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 2000;19:3905–3917. doi: 10.1093/emboj/19.15.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Szul T, Sztul E. COPII and COPI traffic at the ER–Golgi interface. Physiol (Bethesda) 2011;26:348–364. doi: 10.1152/physiol.00017.2011. [DOI] [PubMed] [Google Scholar]
- 128.Shiba Y, Randazzo PA. ArfGAP1 function in COPI mediated membrane traffic: currently debated models and comparison to other coat-binding ArfGAPs. Histol Histopathol. 2012;27:1143–1153. doi: 10.14670/HH-27.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wieland FT, Gleason ML, Serafini TA, Rothman JE. The rate of bulk flow from the endoplasmic reticulum to the cell surface. Cell. 1987;50:289–300. doi: 10.1016/0092-8674(87)90224-8. [DOI] [PubMed] [Google Scholar]
- 130.Pelham HR. Evidence that luminal ER proteins are sorted from secreted proteins in a post-ER compartment. EMBO J. 1988;7:913–918. doi: 10.1002/j.1460-2075.1988.tb02896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Doms RW, Russ G, Yewdell JW. Brefeldin A redistributes resident and itinerant Golgi proteins to the endoplasmic reticulum. J Cell Biol. 1989;109:61–72. doi: 10.1083/jcb.109.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. The first demonstration that the Golgi is not a stable entity but that its integrity depends on constant membrane cycling with the ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sciaky N, et al. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J Cell Biol. 1997;139:1137–1155. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Niu TK, Pfeifer AC, Lippincott-Schwartz J, Jackson CL. Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol Biol Cell. 2005;16:1213–1222. doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Richter S, et al. Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature. 2007;448:488–492. doi: 10.1038/nature05967. [DOI] [PubMed] [Google Scholar]
- 136.Peyroche A, et al. Brefeldin A acts to stabilize an abortive ARF-GDP–Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- 137.Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- 138.Shorter J, Beard MB, Seemann J, Dirac-Svejstrup AB, Warren G. Sequential tethering of Golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J Cell Biol. 2002;157:45–62. doi: 10.1083/jcb.200112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sacher M, et al. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol Cell. 2001;7:433–442. doi: 10.1016/s1097-2765(01)00190-3. [DOI] [PubMed] [Google Scholar]
- 140.Cai Y, et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hay JC, Chao DS, Kuo CS, Scheller RH. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- 142.Rowe T, Dascher C, Bannykh S, Plutner H, Balch WE. Role of vesicle-associated syntaxin 5 in the assembly of pre-Golgi intermediates. Science. 1998;279:696–700. doi: 10.1126/science.279.5351.696. [DOI] [PubMed] [Google Scholar]
- 143.Xu D, Joglekar AP, Williams AL, Hay JC. Subunit structure of a mammalian ER/Golgi SNARE complex. J Biol Chem. 2000;275:39631–39639. doi: 10.1074/jbc.M007684200. [DOI] [PubMed] [Google Scholar]
- 144.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 145.Parlati F, et al. Topological restriction of SNAREdependent membrane fusion. Nature. 2000;407:194–198. doi: 10.1038/35025076. [DOI] [PubMed] [Google Scholar]
- 146.Yamaguchi T, et al. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev Cell. 2002;2:295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
- 147.Bhattacharya N, O’Donnell J, Stagg SM. The structure of the Sec13/31 COPII cage bound to Sec23. J Mol Biol. 2012;420:324–334. doi: 10.1016/j.jmb.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]