Summary
Subunit rotation is the mechanochemical intermediate for the catalytic activity of the membrane enzyme FoF1-ATP synthase. Single-molecule studies based on Förster resonance energy transfer (smFRET) have provided insights on the steps sizes of the F1 and Fo motors, internal transient elastic energy storage and controls of the motors. To develop and interpret smFRET experiments, atomic structural information is required. The recent F1 structure of the E. coli enzyme with the ε subunit in an inhibitory conformation initiated a study for real-time monitoring of εȉs conformational changes. This minireview summarizes smFRET rotation experiments and previews new smFRET data on the conformational changes of the C-terminal domain of ε in the E. coli enzyme.
Keywords: FoF1-ATP synthase, rotary motor, ε-subunit, conformational changes, single-molecule FRET
1 Introducing the rotary motors of FoF1-ATP synthase
The rotary engine FoF1-ATP synthase is the molecular protein machine[1] making most of the adenosine-5′-triphosphate (ATP) in living cells. The ubiquitous multi-subunit enzyme is located in the plasma membrane of bacteria, the thylakoid membrane in photosynthetic cells and in the inner mitochondrial membrane of eukaryotes. The enzyme operates as a mechanochemical energy transducer comprising two motors with different step sizes[2]. The current assignment of rotor and stator subunits is shown in Fig. 1A. The F1 part of the enzyme catalyzes the reaction of ADP plus Pi to ATP (ATP synthesis) and the reverse reaction (ATP hydrolysis) at three nucleotide binding sites, and comprises the stator subunits α3β3δ and the rotary subunits γ and ε (Eschericia coli nomenclature is used for subunit names and residue numbers). The membrane-embedded Fo part translocates protons (or Na+ in some organisms[3]) associated with a rotation of the ring of 10 c-subunits in E. coli with respect to the stator complex of a- and b2-subunits. According to this model, a full rotation of the proton-driven c-ring in Fo is subdivided in 10 steps, but the attached γε rotor of F1 induces three sequential open-and-close movements of the nucleotide binding sites in a three-fold symmetry of α3β3, i.e. in 120° steps. The intrinsic mismatch in symmetry and step angles is accommodated by transient elastic deformations[2] and reversible twisting of rotor subunits[4]. The stator connection between the F1 and Fo motors (the b2δ subunits of E. coli FoF1), seen in electron micrographs as a peripheral stalk[5, 6], is much more stiff, as determined from X-ray crystallography[7, 8] and bead-rotation assays[4]. In bacterial enzymes this could be due to the unusual right-handed coiled-coil structure of the b2 dimer[8].
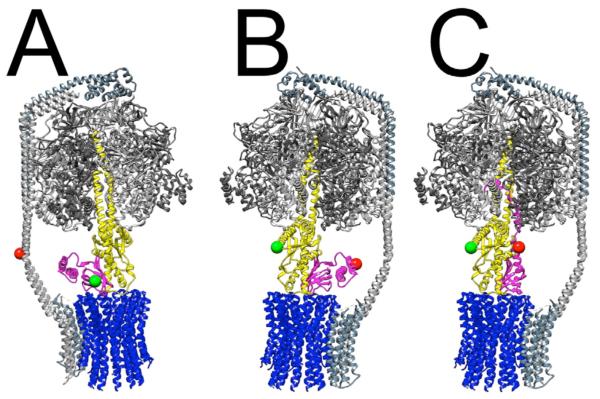
Figure 1. E. coli FoF1-ATP synthase architecture, and cysteine positions for smFRET to monitor rotary subunit movements and ε conformational changes.
Stator subunits are shown in shades of gray (α3β3-δ in F1, ab2 in Fo), and rotor subunits are colored blue (c-ring of Fo), yellow (γ) and magenta (ε). Colored balls mark the locations of engineered cysteines used for labeling with donor (green) or acceptor (red) dyes for smFRET experiments. A: Donor site is ε56, acceptor is b64. During ATP-driven or proton-driven rotation, the labeled ε subunit (i.e. the green ball) stopped at rotary angles in 120° steps so that three distinct distances to the reference position on the b subunits (red ball) were found[42]. B and C: View is rotated 180 °; donor site is γ108, acceptor is ε99. The overall FoF1 architecture shown is from a homology-modeled assembly[60]. In all panels, the α3β3γ complex is from the crystallographic structure[44]. The compact conformation (‘down’) of ε is shown in A and B (structure of isolated ε[61]), and the extended, inhibitory conformation (‘up’) of ε is shown in C, as observed in E. coli F1 [44].
Subunit rotation within the enzyme was predicted by P. Boyer about 30 years ago, based on subunit asymmetry and the cooperative behavior of alternating catalytic sites[1]. Since then, structural studies (and biophysical methods) have supported subunit rotation, beginning with the ‘mother of all F1 structures’ published by J. Walker and collegues[9] in 1994. Many subsequent mitochondrial F1 structures revealed atomic details of the catalytic process in the nucletide binding pocket and further supported the motor view of γ-subunit rotation.
The mode of c-ring rotation in Fo was inferred[10-13] from structural information using chemical crosslink data between introduced cysteines in the a- and c-subunits and NMR structures of isolated c-subunits[14]. Recently, after successful crystallization of c-rings from different organisms, consisting of 8 to 15 subunits[15, 16], atomic simulations of conformational dynamics supported the proposed essential elements of the Fo motor, i.e. electrostatic forces at the interface of the a-subunit and adjacent c-ring and a rotational, swivelling motion of the proton binding and releasing transmembrane helix of the c-subunit (reviewed in[16]).
Biochemical evidence for subunit rotation was first provided by using hybrid F1 complexes and reversible intersubunit crosslinking to show different orientations of F1’s γ rotor with respect to the stator during catalysis in vitro[17, 18]. An advantage to the approach was that it could be applied to membrane-embedded FoF1 to demonstrate changes in rotor orientation during ATP synthesis or hydrolysis. This was later applied to demonstrate that subunit ε also moves as part of the rotor[19]. A similar crosslinking approach provided the first evidence for energy-driven rotation between the c-ring and the a-subunit of FoF1 in E. coli membranes[20]. The disadvantages of these approaches were that they could not measure rotation kinetics or directionality.
The real-time kinetics of γ-subunit rotation were assessed in a spectroscopic experiment[21]. Photoselection by polarized excitation was used for reversible photobleaching of a subset of surface-immobilized F1 parts, and and γ-orientation dependent fluorescence of covalently attached eosin molecules served as the marker of rotation. ATPase-driven changes revealed the rotary movement in milliseconds. However, the direct demonstration of γ-subunit rotation by videomicroscopy[22] in 1997 paved the way for high-resolution biophysical measurements of single F1 motors (reviewed in[23]). The movement of the attached μm-long actin filament magnified the nanometer changes for light microscopy with its diffraction-limited resolution of about 200 nm.
To monitor γ-rotation, the α3β3γ subcomplex was prepared separately and immobilized on a glass surface. Therefore, this approach cannot be used to analyze subunit rotation during ATP synthesis which is driven by proton motive force (PMF) across the lipid bilayer. Very small markers are needed to observe rotation in FoF1-ATP synthase in the physiological membrane environment of living cells. Because of the inherent structural asymmetry caused by the peripheral stalk of FoF1, synchronizing rotor subunit orientations is impossible in vivo. The promising biophysical method for obtaining information about ATP synthesis in vitro and in vivo is the real-time measurement of distance changes within a single enyzme, which requires two different small fluorophore molecules to be attached specifically to one rotor and one stator subunit. During movement of the rotor, the fluorophore distances can be followed in single enzymes based on Förster resonance energy transfer, FRET (translated in 2012[24]). Results of analyzing time trajectories of subunit rotation by single-molecule FRET (smFRET), which are complementary to structural snapshots, are summarized here. This minireview on our current understanding of the motors and controls of single E. coli FoF1-ATP synthase ends with a brief preview of new smFRET evidence for the mechanism of blocking functional rotation by ε’s C-terminal domain (CTD; see conformations in Fig. 1B,C).
2 Single-molecule FRET for subunit rotation in FoF1 ATP synthase
The use of smFRET to measure conformational changes in proteins and nucleic acid complexes has become an increasingly popular and powerful microscopy method since its first proof-of-principle demonstration by T. J. Ha and coworkers published in 1996[25]. With smFRET one can measure fluorophore distances between 2 and 8 nm precisely with 1 Å resolution (but broadened to about 5 Å resolution by stochastic movements of the FRET fluorophores along their linkers[26]) and with sub-millisecond time resolution[27]. We were interested in time trajectories of subunit rotation in single liposome-reconstituted FoF1-ATP synthase. These proteoliposomes allowed creation of a PMF for ATP synthesis conditions using the established buffer mixing approach of the P. Gräber laboratory[28]. For the first successful smFRET rotation experiment with FoF1-ATP synthase shown in 2001[29], the FRET donor fluorophore tetramethylrhodamine (TMR) was placed on the rotating γ-subunit to an introduced cysteine, which was considered to be located far away from the axis of rotation. The FRET acceptor fluorophore Cy5 was attached to one of the peripheral b2 subunits. In the presence of 1 mM ATP and Mg2+, subunit rotation was inferred from stepwise FRET distance changes in sequential order for a single FoF1-ATP synthase in the laser focus[30]. For subsequent smFRET of the F1 and the Fo motor, different positions on the rotor subunits with respect to distinct positions on stator subunits were used[31-35].
Fig. 2 summarizes the actual confocal smFRET measurement and analysis methods using freely diffusing proteoliposomes in buffer solution. Two laser foci are aligned to the same location for alternating excitation of the FRET fluorophores and, as an independent control[36], for the FRET acceptor only. When a FRET-labeled enzyme in a liposome traverses these excitation volumes due to Brownian motion, FRET donor excitation results in a burst of photons from FRET donor and acceptor (“blue laser focus” in Fig. 2A). Nanoseconds later, the FRET acceptor is excited by the second laser (“red laser focus”) to test whether this fluorophore is bound to the same enzyme and in order to exclude photophysical artifacts like spectral fluctuations of the FRET donor fluorophore. For each data point in the photon burst, the fluorophore distance rDA can be calculated from the FRET efficiency according to EFRET = IA/(IA+I) =R06(R06 + rDA6), using ID and IA, intensities of FRET donor and acceptor fluorophores (corrected for background, spectral cross-talk to the other detection channel, detection efficiencies and fluorescence quantum yields), and R0, Förster radius of the given fluorophore pair for 50% energy transfer. Within a photon burst, stepwise changes in EFRET indicating conformational changes or rotary movements of a subunit, respectively, have to be found either be manual inspection[37] or computationally, for example by hidden Markov models[38-40] or change point algorithms[41]. Then, the following information about the motors of FoF1-ATP synthase is obtained.
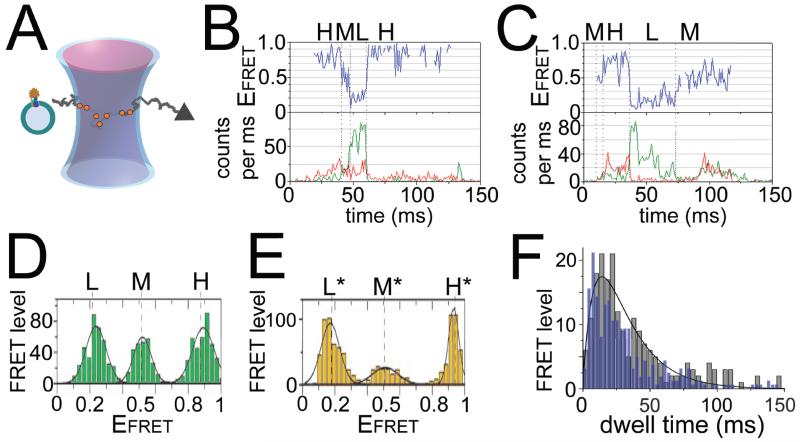
Figure 2. Single-molecule FRET of ε rotation in FoF1-ATP synthase.
A: Alternating laser excitation scheme for confocal smFRET of freely diffusing FoF1-ATP synthase in a liposome. B, C: Photon bursts of single FRET-labeled FoF1, with FRET donor intensities as green traces (donor attached to ε56) and FRET acceptor as red traces (acceptor attached to b64, see Fig. 1A) in the lower panels, and FRET efficiency trajectories as blue traces in upper panels, for ATP hydrolysis (B) or ATP synthesis (C) conditions. H, M, L denote FRET level (see text). D, E: FRET level histograms in the presence of 1 mM Mg2+ATP (D) or 1 mM Mg2+ADP plus 3 mM Pi without PMF (E). H, L, M are the same FRET levels as shown in (B), but H*, L* and M* are different FRET levels. For a visual scheme of these positions in FoF1 see Fig. 7 in[35]. F: Dwell time distribution of ε rotation during ATP hydrolysis as in (B) (blue bars, normalized, 3 ms bins), and in the presence of 20 μm aurovertin (grey bars, 5 ms bins, with fit as black curve)[51]. Figures are reproduced with permissions (B–E from[35], F from[51]).
2.1 Opposite direction of motor rotation during ATP synthesis and hydrolysis
Stepwise changes of FRET efficiencies have been observed for smFRET measurements between the rotary ε-subunit of F1 [35, 42] and the stator b2 of Fo (shown in Figs. 2B,C). Three FRET level called ‘L’ (low EFRET), ‘M’ (medium EFRET) and ‘H’ (high EFRET) with a sequential order of →H→M→L→H→ during ATP hydrolysis but in reverse order →H→L→M→H→ for ATP synthesis indicated the opposite direction of rotation for the distinct catalytic processes, as reported first for γ-subunit rotation in [32] FoF1-ATP synthase in 2004. Each FRET level was consistent and transitions occured within about 200 μs[42].
2.2 Different rotary stopping angles during catalysis
Given the geometrical constraints for the rotary motion of ε or γ in F1, i.e. a 120° stepping at high [ATP] for ATP hydrolysis or high PMF for ATP synthesis, the three stopping positions of the rotary subunits with respect to b2 were very similar for the two catalytic modes as well as in the presence of AMPPNP[35] (Fig. 2D). However, in the presence of ADP and Pi but without PMF, three distinct stopping positions L*, M* and H* were found (Fig. 2E). This correlated with a cryo-EM study of E. coli F1 with a nanogold label on ε’s N-terminal domain: only with ADP and Pi present, ε showed a distinct position relative to α and β subunits[43]. The recently-determined crystal structure of ε-inhibited E. coli F1 [44] is also consistent with the distinct stopping positions of ε seen by smFRET. That is, whereas the main rotary pause should be at the catalytic dwell angle during catalytic turnover with excess substrate, ε-inhibited F1 appears to be paused at a position corresponding to the ATP-binding dwell. This is supported by a structure of mitochondrial F1 [45], thought to be poised at the ATP-binding dwell, that shows a rotary position nearly identical to ε-inhibited E. coli F1 [44, 46]. Finally, recent biochemical studies of E. coli F1 confirmed that the ε-inhibited state is stabilized by MgADP and Pi but reversed by MgAMPPNP[46], consistent with the smFRET L*/M*/H* positions observed only with MgADP and Pi. Several bead-rotational studies with F1 from E. coli and other bacteria showed that ε inhibition pauses rotation for extended times but concluded that ε pauses F1 at the catalytic dwell angle[47-49]. This contrast with the smFRET and bead-rotation results remains to be resolved.
2.3 Smaller step sizes of the rotary Fo motor
Driven by PMF during ATP synthesis, the step sizes of the c-ring with respect to the static a-subunit were smaller and revealed a one-proton-after-another mode of rotation in Fo according to smFRET[34]. Using the geometric constraints of c-ring size and label positions, a 36° step size was most likely for about half of the assigned FRET level changes. Similarly, 10-stepped c-ring rotation was reported during ATP hydrolysis using immobilized FoF1 reconstituted in lipid nanodiscs with a gold nanorod as the marker of c-ring rotation[50].
2.4 Dwell times and rotational speed
The smaller step sizes in c-ring rotation during ATP synthesis were associated with shorter dwell times of the stopping positions[34]. Measuring small dwell time differences with smFRET is possible: for example, the three slightly different calatytic dwell times for the ε-subunit indicated an asymmetry in rotation, eventually related to the asymmetric peripheral stalk affecting the conformational dynamics of the nearby nucleotide binding site[33, 35]. However, large changes of the dwell times were observed after addition of the non-competitive inhibitor aurovertin A, for the F1 as well as the Fo motor[34, 51]. The inhibitor prolonged the dwell time during ATP hydrolysis and also resulted in a double-exponential decay with a rise and a decay components (see Fig. 2F). Dwell time analysis has become an important control using inhibitors to discriminate conformational protein dynamics from single-molecule photophysical artifacts. However, time resolution limits for smFRET apply, by the binning of 1 ms for time trajectories and the difficulties to assign dwell times shorter than 5 to 10 ms from EFRET changes in noisy data.
2.5 Twisting and elastic energy storage with the rotor
SmFRET was also applied to detect a reversible, elastic twisting mode within the rotor subunits ε and c of FoF1-ATP synthase during ATP hydrolysis and synthesis[52, 53]. Transient elastic energy storage had been postulated to address the symmetry mismatch of the F1 and Fo motor step sizes and to ensure maximum efficiency of motor operation (experimental details are summarized in[2, 54]). Using three different specifically attached fluorophores on a single FoF1-ATP synthase (EGFP-a fusion on the stator, Alexa532-ε and Cy5-c on rotor), we could show that the distances between markers on residues ε56 and c2 fluctuated during rotor movement, indicating a twisting up to three single steps of c or 108°, respectively[52].
3 Single-molecule FRET of the C-terminal domain of ε
Here we present our preliminary development of smFRET to monitor conformational changes of ε’s C-terminal domain (CTD) in E. coli F1. Based on the E. coli F1 [44] X-ray structure , we chose ε99 on the first C-terminal α-helix of ε, which does not insert into a β-γ cleft in the ‘up’-conformation (see Fig. 1B,C). The second marker position is γ108, yielding FRET distances of about 3 nm (Fig. 1C) and 6 nm (Fig. 1B) including 0.5 nm for linkers to the fluorophores to ε99 in the ‘up’ or ‘down’ conformations, respectively. These labeling positions were also chosen to avoid perturbing any interactions of ε CTD (either conformation) with the ε NTD or with other subunits. This is in contrast to smFRET experiments of R. Iino and coworkers for the thermophilic enzyme TF1 from Bacillus PS3, in which both γ- and ε-labeling sites would be buried inside F1’s central cavity with ε in the ‘up’ state[55].
Our initial tests with smFRET probes were on freely diffusing F1 under different ligand conditions. Subunit ε was expressed separately with a 6xHis, N-terminal affinity tag and was purified as before [46]. A unique cysteine was included, and ε99C was labeled with Atto647N as FRET acceptor. Maleimide-labeling efficiency was 30%, unbound dye was removed by dialysis. F1(γ108C), depleted of δ and ε[46], was labeled with Atto488 as FRET donor (maleimide-labeling efficiency 55%, unbound dye removed by centrifuge column). Mixing F1 (3 μM) with ε (4 μM) for 30 min yielded FRET-labeled F1, due to ε’s high binding affinity (KD ~0.3 nM[46]). Dilution to less than 1 nM F1-ε immediately before starting smFRET measurements resulted in standard single-molecule detection conditions in solution for our confocal microscope, i.e. one F1-ε molecule at a time. Using alternating laser excitation with 488 nm for FRET between γ and ε, and 635 nm to probe the bound Atto647N-labeled ε to F1 allowed selection of the FRET-labeled enzymes, rejecting any protein aggregates or single-labeled proteins in subsequent analysis.
Diffusion of F1-ε (~10 nm diameter) was fast, i.e. about 3 ms on average through the confocal detection volume (vs. ~300 μs for a free fluorophore). These short observation times allowed us to determine only an average FRET distance for each enzyme, but not time-dependent distance changes or conformational changes between γ and the CTD of ε within a single photon burst. We obtained several hundred burst events with high photon count rates for each biochemical condition using the following thresholds to identify a single FRET-labeled F1-ε: a mean diffusion time longer than 10 ms, maximum peak intensity for the FRET donor fluorophore (to exclude aggregates with multiple dyes), fluorescence intensity thresholds for the FRET acceptor (at least a mean of 4 counts per ms for FRET excitation and 8 counts per ms for direct excitation in the same photon burst) and limited FRET efficiency fluctuations of less than 0.18 (standard deviation within a burst). Figs. 3A, B show two photon bursts of FRET-labeled F1-ε in the presence of 1 mM MgAMPPNP. The FRET efficiencies (“blue traces”) show different average values, about 0.6 and 0.3, indicating different distances between the markers on γ108 and ε99, and corresponding to different conformations of the CTD of ε with respect to γ.
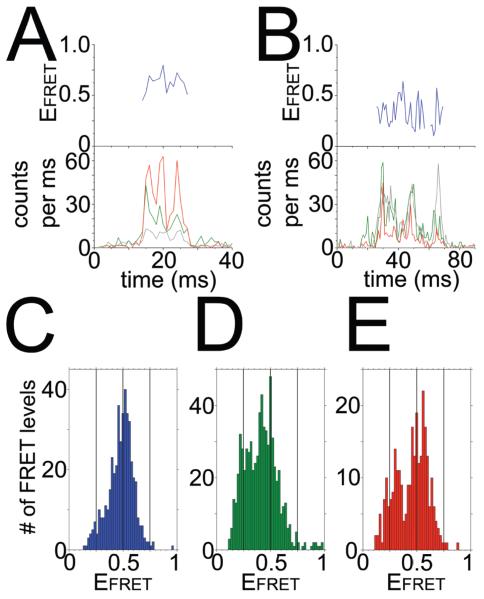
Figure 3. Single-molecule FRET of ε conformational changes in F1.
A, B: Photon bursts of FRET-labeled F1, with donor Atto488 attached to γ108 (green traces in lower panels,) and acceptor Atto647N attached to ε99 (red traces in lower panels, labeling efficiency 30%). Grey traces are Atto647N intensities upon direct excitation with 635 nm. C–E: FRET efficiency histograms for FRET-labeled F1, in the presence of 1 mM Mg2+ADP plus 3 mM Pi (C), 1 mM Mg2+AMPPNP (D), or 1 mM Mg2+ATP (E), respectively. See Fig. 1 for label positions. Reference lines are shown at EFRET 0.25, 0.5 and 0.75.
Addition of different ligand combinations in the presence of Mg2+ resulted in distinct EFRET distributions (Figs. 3C-E). The total number of FRET level in the three histograms depended on the photon burst criteria used to identify a single FRET-labeled F1-ε and, therefore, cannot be compared directly. Biochemical data showed that MgADP and Pi stabilize the ε-inhibited state[46]. In Fig. 3C, additon of ADP/Pi resulted in a dominant population with EFRET about 0.6, similar to the EFRET histogram obtained without adding nucleotides (data not shown). Given a Förster radius of 5.1 nm (Attotec) for EFRET=0.5, this corresponds to a 4.8 nm FRET distance and should represent the ε-inhibited, ‘up’ state, as in the E. coli F1 structure[44] and in Fig. 1C. Adding AMPPNP or ATP resulted in an additional population of EFRET about 0.25. This low EFRET value corresponded to a 6.1 nm distance between the FRET fluorophores and, therefore, should be the ‘down’ conformation of the CTD. However, the majority of F1-ε complexes were still found at EFRET ~0.6. This likely correlates with the strong inhibition of isolated F1 by ε[46]. The distance changes as calculated from the maxima of the two EFRET populations agreed with the changes seen in the structural models in Figs. 1B and C, but the absolute distances were larger than expected, which could be explained by possible photophysical effects of the local protein environment of the fluorophores, like decreased quantum yields or spectral shifts. However, additional smFRET measurements are required to assign unequivocally the different FRET distances with ε’s CTD conformations and its inhibitory role.
4 Outlook
Single-molecule FRET is a complementary approach to measure subunit rotation of the two motors in reconstituted single FoF1-ATP synthase. With a time resolution of 1 ms, dwell times of a few ms for the stopping positions are accessible, and the angular resolution for the rotary movement can be inferred using known structural constraints of the enzyme. In addition, domain movements like the conformational change of the regulatory CTD of ε can be monitored in real time.
Here we reported the nucleotide dependent shifts in the population of the CTD between ‘up’ and ‘down’ states by smFRET of F1-ε in solution. Accordingly more than 50% of F1 on average remained in an inhibited ‘up’ conformation of ε in the presence of Mg2+ATP or AMPPNP, which is in agreement with videomicroscopy results of beads attached to immobilized F1 as a marker for rotation[56] and the role of PMF to activate the enzyme for ATP hydrolysis[57]. We now need to reconstitute FRET-labeled F1 with Fo in liposomes to study dynamics of the ε CTD conformations in the intact ATP synthase.
To improve smFRET-based analysis of the ε CTD, we have to increase the observation time for single enzymes in solution, using either a three dimensional trap (for example the ‘Anti-Brownian electrokinetic trap’, ABELtrap, invented by A. E. Cohen and W. E. Moerner[58]) to hold the FoF1-liposome in place during smFRET recording, or integrating the FRET-labeled enzyme into a ‘black lipid membrane’ (BLM) with access to single-molecule detection. The BLM approach allows to control and change the PMF during the measurement[59]. Furthermore, a three-fluorophore smFRET experiment will be important to correlate rotor movement and the conformation of the CTD of ε, and to minimize photophysical artifacts.
Interpretation of smFRET data requires structural information. More X-ray structures with atomic resolution will be important to advance our understanding of how the rotary motors and their controls operate in this enzyme. These data are also the basis for MD simulations of motors and controls, that provide independent atomic views with high time resolution, but short “observation” times in nanoseconds due to computational limitations. Structural information might elucidate the role of nucleotide (ATP) binding as possible part of the conformational dynamics of ε, and are essential to interpret the nucleotide dependent binding constants of ε to F1. Our ongoing work on ε inhibition is now focussing on the complete enzyme reconstituted into liposomes, and will proceed to probe the regulatory conformational changes of ε and the rotary motors in the native environment of the E. coli enzyme, that is, the plasma membrane of living cells.
Acknowledgements
We want to thank our coworkers M. Renz (Jena) and M. Hutcheon (Syracuse) for their excellent technical assistance for the smFRET measurements of the CTD of ε. This work was funded by a collaborative NIH grant (R01GM083088-03S1 to T.M.D.) and in part by the DFG (grant BO 1891/10-2 to M.B.)
References
- [1].Boyer PD. The ATP synthase--a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- [2].Junge W, Sielaff H, Engelbrecht S. Torque generation and elastic power transmission in the rotary FOF1-ATPase. Nature. 2009;459:364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- [3].von Ballmoos C, Cook GM, Dimroth P. Unique rotary ATP synthase and its biological diversity. Annu Rev Biophys. 2008;37:43–64. doi: 10.1146/annurev.biophys.37.032807.130018. [DOI] [PubMed] [Google Scholar]
- [4].Wachter A, Bi Y, Dunn SD, Cain BD, Sielaff H, Wintermann F, Engelbrecht S, Junge W. Two rotary motors in F-ATP synthase are elastically coupled by a flexible rotor and a stiff stator stalk. Proc Natl Acad Sci U S A. 2011;108:3924–3929. doi: 10.1073/pnas.1011581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wilkens S, Capaldi RA. ATP synthase’s second stalk comes into focus. Nature. 1998;393:29. doi: 10.1038/29908. [DOI] [PubMed] [Google Scholar]
- [6].Bottcher B, Bertsche I, Reuter R, Graber P. Direct visualisation of conformational changes in EF(0)F(1) by electron microscopy. J Mol Biol. 2000;296:449–457. doi: 10.1006/jmbi.1999.3435. [DOI] [PubMed] [Google Scholar]
- [7].Rees DM, Leslie AG, Walker JE. The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci U S A. 2009;106:21597–21601. doi: 10.1073/pnas.0910365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Del Rizzo PA, Bi Y, Dunn SD, Shilton BH. The “second stalk” of Escherichia coli ATP synthase: structure of the isolated dimerization domain. Biochemistry. 2002;41:6875–6884. doi: 10.1021/bi025736i. [DOI] [PubMed] [Google Scholar]
- [9].Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- [10].Junge W, Lill H, Engelbrecht S. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem Sci. 1997;22:420–423. doi: 10.1016/s0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- [11].Vik SB, Antonio BJ. A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J Biol Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- [12].Elston T, Wang H, Oster G. Energy transduction in ATP synthase. Nature. 1998;391:510–513. doi: 10.1038/35185. [DOI] [PubMed] [Google Scholar]
- [13].Dmitriev OY, Jones PC, Fillingame RH. Structure of the subunit c oligomer in the F1Fo ATP synthase: model derived from solution structure of the monomer and cross-linking in the native enzyme. Proc Natl Acad Sci U S A. 1999;96:7785–7790. doi: 10.1073/pnas.96.14.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Girvin ME, Rastogi VK, Abildgaard F, Markley JL, Fillingame RH. Solution structure of the transmembrane H+-transporting subunit c of the F1F0 ATP synthase. Biochemistry. 1998;37:8817–8824. doi: 10.1021/bi980511m. [DOI] [PubMed] [Google Scholar]
- [15].Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci U S A. 2010;107:16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Meier T, Faraldo-Gomez J, Börsch M. In: Molecular Machines in Biology. Frank J, editor. Cambridge University Press; New York: 2012. pp. 208–238. [Google Scholar]
- [17].Duncan TM, Bulygin VV, Zhou Y, Hutcheon ML, Cross RL. Rotation of subunits during catalysis by Escherichia coli F1-ATPase. Proc Natl Acad Sci U S A. 1995;92:10964–10968. doi: 10.1073/pnas.92.24.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou Y, Duncan TM, Cross RL. Subunit rotation in Escherichia coli FoF1-ATP synthase during oxidative phosphorylation. Proc Natl Acad Sci U S A. 1997;94:10583–10587. doi: 10.1073/pnas.94.20.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bulygin VV, Duncan TM, Cross RL. Rotation of the epsilon subunit during catalysis by Escherichia coli FOF1-ATP synthase. J Biol Chem. 1998;273:31765–31769. doi: 10.1074/jbc.273.48.31765. [DOI] [PubMed] [Google Scholar]
- [20].Hutcheon ML, Duncan TM, Ngai H, Cross RL. Energy-driven subunit rotation at the interface between subunit a and the c oligomer in the F(O) sector of Escherichia coli ATP synthase. Proc Natl Acad Sci U S A. 2001;98:8519–8524. doi: 10.1073/pnas.151236798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sabbert D, Engelbrecht S, Junge W. Intersubunit rotation in active F-ATPase. Nature. 1996;381:623–625. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- [22].Noji H, Yasuda R, Yoshida M, Kinosita K., Jr. Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- [23].Adachi K, Oiwa K, Nishizaka T, Furuike S, Noji H, Itoh H, Yoshida M, Kinosita K., Jr. Coupling of rotation and catalysis in F(1)-ATPase revealed by single-molecule imaging and manipulation. Cell. 2007;130:309–321. doi: 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- [24].Förster T. Energy migration and fluorescence. Journal of Biomedical Optics. 2012;17:011002. doi: 10.1117/1.JBO.17.1.011002. [DOI] [PubMed] [Google Scholar]
- [25].Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci U S A. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Antonik M, Felekyan S, Gaiduk A, Seidel CA. Separating structural heterogeneities from stochastic variations in fluorescence resonance energy transfer distributions via photon distribution analysis. J Phys Chem B. 2006;110:6970–6978. doi: 10.1021/jp057257+. [DOI] [PubMed] [Google Scholar]
- [27].Margittai M, Widengren J, Schweinberger E, Schroder GF, Felekyan S, Haustein E, Konig M, Fasshauer D, Grubmuller H, Jahn R, Seidel CAM. Single-molecule fluorescence resonance energy transfer reveals a dynamic equilibrium between closed and open conformations of syntaxin 1. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15516–15521. doi: 10.1073/pnas.2331232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fischer S, Etzold C, Turina P, Deckers-Hebestreit G, Altendorf K, Graber P. ATP synthesis catalyzed by the ATP synthase of Escherichia coli reconstituted into liposomes. Eur J Biochem. 1994;225:167–172. doi: 10.1111/j.1432-1033.1994.00167.x. [DOI] [PubMed] [Google Scholar]
- [29].Borsch M, Diez M, Zimmermann B, Reuter R, Graber P. In: Fluorescence spectroscopy, Imaging and Probes. New Tools in Chemical, Physical and Life Sciences. Kraayenhof R, Visser AJW, Gerritsen HC, editors. Springer-Verlag; Berlin: 2002. pp. 197–207. [Google Scholar]
- [30].Borsch M, Diez M, Zimmermann B, Reuter R, Graber P. Stepwise rotation of the gamma-subunit of EF(0)F(1)-ATP synthase observed by intramolecular single-molecule fluorescence resonance energy transfer. FEBS Lett. 2002;527:147–152. doi: 10.1016/s0014-5793(02)03198-8. [DOI] [PubMed] [Google Scholar]
- [31].Borsch M, Diez M, Zimmermann B, Trost M, Steigmiller S, Graber P. Stepwise rotation of the gamma-subunit of EFoF1-ATP synthase during ATP synthesis: a single-molecule FRET approach. Proc. SPIE. 2003;4962:11–21. [Google Scholar]
- [32].Diez M, Zimmermann B, Borsch M, Konig M, Schweinberger E, Steigmiller S, Reuter R, Felekyan S, Kudryavtsev V, Seidel CA, Graber P. Proton-powered subunit rotation in single membrane-bound FoF1-ATP synthase. Nat Struct Mol Biol. 2004;11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- [33].Duser MG, Bi Y, Zarrabi N, Dunn SD, Borsch M. The proton-translocating a subunit of F0F1-ATP synthase is allocated asymmetrically to the peripheral stalk. J Biol Chem. 2008;283:33602–33610. doi: 10.1074/jbc.M805170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Duser MG, Zarrabi N, Cipriano DJ, Ernst S, Glick GD, Dunn SD, Borsch M. 36 degrees step size of proton-driven c-ring rotation in FoF1-ATP synthase. Embo J. 2009;28:2689–2696. doi: 10.1038/emboj.2009.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zimmermann B, Diez M, Zarrabi N, Graber P, Borsch M. Movements of the epsilon-subunit during catalysis and activation in single membrane-bound H(+)-ATP synthase. Embo J. 2005;24:2053–2063. doi: 10.1038/sj.emboj.7600682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Borsch M, Graber P. Subunit movement in individual H+-ATP synthases during ATP synthesis and hydrolysis revealed by fluorescence resonance energy transfer. Biochemical Society Transactions. 2005;33:878–882. doi: 10.1042/BST0330878. [DOI] [PubMed] [Google Scholar]
- [38].McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zarrabi N, Duser MG, Reuter R, Dunn SD, Wrachtrup J, Borsch M. Detecting substeps in the rotary motors of FoF1-ATP synthase by Hidden Markov Models. Proc. SPIE. 2007;6444:64440E. [Google Scholar]
- [40].Bronson JE, Fei J, Hofman JM, Gonzalez RL, Jr, Wiggins CH. Learning Rates and States from Biophysical Time Series: A Bayesian Approach to Model Selection and Single-Molecule FRET Data. Biophysical Journal. 2009;97:3196–3205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Watkins LP, Yang H. Detection of Intensity Change Points in Time-Resolved Single-Molecule Measurements. The Journal of Physical Chemistry B. 2004;109:617–628. doi: 10.1021/jp0467548. [DOI] [PubMed] [Google Scholar]
- [42].Zimmermann B, Diez M, Borsch M, Graber P. Subunit movements in membrane-integrated EF0F1 during ATP synthesis detected by single-molecule spectroscopy. Biochim Biophys Acta. 2006;1757:311–319. doi: 10.1016/j.bbabio.2006.03.020. [DOI] [PubMed] [Google Scholar]
- [43].Wilkens S, Capaldi RA. Asymmetry and structural changes in ECF1 examined by cryoelectronmicroscopy. Biol Chem Hoppe Seyler. 1994;375:43–51. doi: 10.1515/bchm3.1994.375.1.43. [DOI] [PubMed] [Google Scholar]
- [44].Cingolani G, Duncan TM. Structure of the ATP synthase catalytic complex (F(1)) from Escherichia coli in an autoinhibited conformation. Nat Struct Mol Biol. 2011;18:701–707. doi: 10.1038/nsmb.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rees DM, Montgomery MG, Leslie AG, Walker JE. Structural evidence of a new catalytic intermediate in the pathway of ATP hydrolysis by F1-ATPase from bovine heart mitochondria. Proc Natl Acad Sci U S A. 2012;109:11139–11143. doi: 10.1073/pnas.1207587109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shah NB, Hutcheon ML, Haarer BK, Duncan TM. F1-ATPase of Escherichia coli: the epsilon-inhibited state forms after ATP hydrolysis, is distinct from the ADP-inhibited state, and responds dynamically to catalytic site ligands. J Biol Chem. 2013;288:9383–9395. doi: 10.1074/jbc.M113.451583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Konno H, Murakami-Fuse T, Fujii F, Koyama F, Ueoka-Nakanishi H, Pack CG, Kinjo M, Hisabori T. The regulator of the F1 motor: inhibition of rotation of cyanobacterial F1-ATPase by the epsilon subunit. Embo J. 2006;25:4596–4604. doi: 10.1038/sj.emboj.7601348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tsumuraya M, Furuike S, Adachi K, Kinosita K, Jr., Yoshida M. Effect of epsilon subunit on the rotation of thermophilic Bacillus F1-ATPase. FEBS Lett. 2009;583:1121–1126. doi: 10.1016/j.febslet.2009.02.038. [DOI] [PubMed] [Google Scholar]
- [49].Sekiya M, Hosokawa H, Nakanishi-Matsui M, Al-Shawi MK, Nakamoto RK, Futai M. Single molecule behavior of inhibited and active states of Escherichia coli ATP synthase F1 rotation. J Biol Chem. 2010;285:42058–42067. doi: 10.1074/jbc.M110.176701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ishmukhametov R, Hornung T, Spetzler D, Frasch WD. Direct observation of stepped proteolipid ring rotation in E. coli FF-ATP synthase. Embo J. 2010;29:3911–3923. doi: 10.1038/emboj.2010.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Johnson KM, Swenson L, Opipari AW, Jr., Reuter R, Zarrabi N, Fierke CA, Borsch M, Glick GD. Mechanistic basis for differential inhibition of the F(1)F(o)-ATPase by aurovertin. Biopolymers. 2009;91:830–840. doi: 10.1002/bip.21262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ernst S, Duser MG, Zarrabi N, Borsch M. Three-color Förster resonance energy transfer within single FoF1-ATP synthases: monitoring elastic deformations of the rotary double motor in real time. J Biomed Opt. 2012;17:011004. doi: 10.1117/1.JBO.17.1.011004. [DOI] [PubMed] [Google Scholar]
- [53].Ernst S, Duser MG, Zarrabi N, Dunn SD, Borsch M. Elastic deformations of the rotary double motor of single FoF1-ATP synthases detected in real time by Förster resonance energy transfer. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2012;1817:1722–1731. doi: 10.1016/j.bbabio.2012.03.034. [DOI] [PubMed] [Google Scholar]
- [54].Sielaff H, Borsch M. Twisting and subunit rotation in single FOF1-ATP synthase. Phil Trans R Soc B. 2013;368:20120024. doi: 10.1098/rstb.2012.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Saita E, Iino R, Suzuki T, Feniouk BA, Kinosita K, Jr., Yoshida M. Activation and stiffness of the inhibited states of F1-ATPase probed by single-molecule manipulation. J Biol Chem. 2010;285:11411–11417. doi: 10.1074/jbc.M109.099143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bilyard T, Nakanishi-Matsui M, Steel BC, Pilizota T, Nord AL, Hosokawa H, Futai M, Berry RM. High-resolution single-molecule characterization of the enzymatic states in Escherichia coli F1-ATPase. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368:20120023. doi: 10.1098/rstb.2012.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Fischer S, Graber P, Turina P. The activity of the ATP synthase from Escherichia coli is regulated by the transmembrane proton motive force. J Biol Chem. 2000;275:30157–30162. doi: 10.1074/jbc.275.39.30157. [DOI] [PubMed] [Google Scholar]
- [58].Cohen AE, Moerner WE. The anti-Brownian electrophoretic trap (ABEL trap): fabrication and software. Proc. SPIE. 2005;5699:296–305. [Google Scholar]
- [59].Tabata KV, Sato K, Ide T, Nishizaka T, Nakano A, Noji H. Visualization of cargo concentration by COPII minimal machinery in a planar lipid membrane. Embo J. 2009;28:3279–3289. doi: 10.1038/emboj.2009.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sielaff H, Rennekamp H, Engelbrecht S, Junge W. Functional halt positions of rotary FOF1-ATPase correlated with crystal structures. Biophys J. 2008;95:4979–4987. doi: 10.1529/biophysj.108.139782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wilkens S, Capaldi RA. Solution structure of the epsilon subunit of the F1-ATPase from Escherichia coli and interactions of this subunit with beta subunits in the complex. J Biol Chem. 1998;273:26645–26651. doi: 10.1074/jbc.273.41.26645. [DOI] [PubMed] [Google Scholar]