Abstract
DNA replication origins (ORIs) map close to promoter regions in many organisms, including mammals. However, the relationship between initiation of replication and transcription is not well understood. To address this issue, we have analyzed replication timing and activity of several CpG island-associated ORIs on the transcriptionally active and silent X chromosomes. We find equivalent ORI usage and efficiency of both alleles at sites that are replicated late on the inactive X chromosome. Thus, in contrast to its repressive effect on transcription, heterochromatin does not influence ORI activity. These findings suggest that the relationship between sites of transcription and replication initiation at CpG island regions is restricted to early development, and that subsequent gene silencing and heterochromatin formation influence only the timing of ORI activation.
In the yeast Saccharomyces cerevisiae, replication origins (ORIs) occur at specific sites that are genetically defined. ORIs also occur at specific sites in mammals, but to date there is no evidence for a consensus sequence that marks their location (1, 2). To explain this apparent paradox, it has been proposed that ORIs are epigenetically determined in higher eukaryotes by the origin recognition complex nucleating a specific chromatin conformation (3).
Mammalian ORIs are frequently located in the vicinity of promoter regions (4–12). In particular, cloning of short nascent strands (10), or chromatin immunoprecipitation with α-Orc2 antibodies (12), has shown on a genome-wide basis that ORIs are highly enriched in a class of promoters associated with CpG islands. CpG islands encompass the promoter regions of genes and are found in nearly all housekeeping genes and in >50% of tissue-restricted genes (13). Whilst they are normally unmethylated, regardless of expression status, well documented exceptions include genes on the inactive X chromosome and those subjected to parental imprinting. In these cases, CpG island methylation is a developmentally regulated event that occurs in the early embryo or in germ cells, and is necessary to maintain repression in differentiated cells in the adult (14, 15).
At present, little is known concerning the relationship between sites of initiation of replication and transcription in mammalian cells. To investigate this relationship, we have analyzed several CpG island-associated ORIs on the active and inactive X chromosome (Xa and Xi, respectively). X inactivation occurs early in development in female embryos and ensures transcriptional silencing of one of the two X chromosomes (16). Once established, the silenced state is clonally inherited through subsequent cell division. The Xi is heterochromatinized, replicates late in the S phase, and the CpG islands of most housekeeping genes are methylated (17). Analyzing this system allows us to directly compare ORI activity of CpG island regions of active (unmethylated/euchromatic) and silent (methylated/heterochromatic) alleles. Because X inactivation is developmentally regulated, we also can explore whether changes in transcriptional activity and ORI specification are temporally coregulated through development. In a recent study, Cohen et al. (18) reported equivalent ORI usage for two human CpG island ORIs in hamster cells containing either the active or the inactive human X chromosome. Similarly, analyzing several CpG island-associated ORIs in normal XX cells, we find that X inactivation does not affect ORI usage. Moreover, we find that efficiency of ORI firing on the inactive X is indistinguishable from that on the active X allele. Thus, the specification of ORIs at CpG islands resists gene silencing and DNA methylation occurring in the course of development. Heterochromatin formation does, however, delay replication timing at these sites, suggesting that ORI firing, or replication fork elongation, is dictated by chromatin structure.
Materials and Methods
Mouse Strains and Cell Lines. Mus musculus castaneus and T(X;16)H (T16H) strains were bred inhouse. Fibroblast cell lines were derived from lungs or ear tissues from embryonic or adult mice and grown in DMEM (Invitrogen) supplemented with 10% serum and 50 μM 2-mercaptoethanol. The 129/1 XY embryonic stem (ES) cell line was maintained as described (19). [T16H × Mus castaneus] F1 lymphocytes were isolated from adult spleens and cultured for 3 days in Iscove's modified Dulbecco's medium (Invitrogen) supplemented as above on culture plates coated with 10 μg/ml anti-TCRβ (H57–597; BD Biosciences Pharmingen) and 2 μg/ml anti-CD28 (BD Biosciences Pharmingen) with IL-2 (10 units/ml).
PCR Primers and Conditions. Hprt and Xist primers and PCR and electrophoresis conditions for allele-specific RT-PCR were as described (20). Primer sequences and PCR conditions used for mapping replication initiation by conventional or competitive PCR were verified empirically. Competitor molecules were constructed by recombinant PCR as described (21). Full details are available on request. Single-nucleotide primer extension (SNuPE) primers and annealing conditions are provided in Table 1. Primer extension reactions were performed with Taq polymerase (GIBCO) by using a single PCR cycle of 1 min denaturation at 94°C, 1 min annealing at the indicated temperature, and 2 min extension at 72°C.
Table 1. Single-base polymorphisms mapped at the CpG island regions.
| CpG island | Polymorphism 129/cast | SNuPE primer | Annealing temperature, °C |
|---|---|---|---|
| Ags1 | A/C | 5′-TAAACAACAGAGTGAGACCC | 56 |
| Mecp2 | T/G | 5′-TGGTGAACTACTCAGCAGGG | 63 |
| Mtm1 | T/C | 5′-AACAGGGTTTCCCAGCAGCG | 69 |
| Mtm1r | A/C | 5′-GCTCAGGCTGGGCTGGTTG | 72 |
| Xist | A/T | 5′-TTGATGTACACGGTGTGAGA | 50 |
Tsix was not tested for ORI usage because no polymorphism was found in the 1-kb genomic region surrounding the ORI.
Nascent Strand Fractionation and Immunoprecipitation with Anti-BrdUrd Antibodies. DNA nascent strands were isolated by alkaline sucrose centrifugation essentially as described (10). Exponentially growing cells were pulsed with 50 μM BrdUrd for 30 min. Total DNA was purified, denatured in NaOH 0.2 N, size-fractionated in a seven-step sucrose gradient (5–20% in steps of 2.5%) made up in 0.1 M NaOH, 0.9 M NaCl, and 50 mM EDTA, and centrifuged at 24,000 rpm for 20 h at 4°C in a Beckman SW-40 rotor. One-milliliter fractions were collected after centrifugation, dialyzed against TE buffer (10 mM Tris·HCl/1 mM EDTA, pH 8.0), ethanolprecipitated and resuspended in TE buffer (10 mM Tris·HCl/1mM EDTA, pH 8.0). Equal volumes of each sample were electrophoresed in an alkaline agarose gel to monitor correct fractionation, and an equal dilution of each fraction was used as input for PCR. PCR conditions were 35 cycles at 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min. PCR products were electrophoresed, blotted, and hybridized with the cognate PCR fragment. Nascent strands were purified from potential nonreplicating contaminating genomic DNA by two rounds of immunoprecipitation with anti-BrdUrd antibodies as described (10).
Cell Cycle Fractionation and Cytometry. [T16H × M. castaneus] F1 lymphocytes or fibroblasts were labeled with 50 μM BrdUrd for 60 min, washed in PBS, and stained for 30 min before cell sorting in staining buffer (40 mM Tris·HCl, pH 7.4/0.8% sodium chloride/21 mM magnesium chloride/0.05% Nonidet P-40/propidium iodide (50 μg/ml), and RNase A (1 μg/ml). Cell sorting was performed on a FACS Vantage Instrument (Becton Dickinson). Equal number of cells (between 15,000 and 30,000) were collected for each of the six fractions (G1, S1, S2, S3, S4 and G2), and replicated DNA was purified after immunoprecipitation with BrdUrd antibodies.
Results
ORI Activity at X-Linked CpG Islands. ORI activity was analyzed at several X-linked CpG island regions. Ags1, Mecp2, Mtm1, and Mtm1r were selected as examples of genes that are subject to X inactivation with associated CpG island methylation. We also analyzed the Xist gene, which is expressed exclusively from the inactive X chromosome in XX somatic cells and has a methylated CpG island region specifically on the silent Xa allele (22); and the Tsix gene, which is not expressed in XX somatic cells and shows heterogeneous low level CpG island methylation, both in males and females (23).
To determine ORI activity, we analyzed the representation of CpG islands and flanking regions in size-fractionated nascent strand preparations (10). Exponentially growing XX fibroblasts were pulse labeled with BrdUrd, and nascent strands were purified by size fractionation on alkaline sucrose gradients, followed by immunoprecipitation with anti-BrdUrd antibodies. The closer a region is to an ORI, the smaller the size of nascent strands in which it will be contained. In all cases, we found that sequences within the CpG islands were amplified from fraction 1 (containing nascent strands up to 1 kb in length), whereas flanking regions were detected only in fractions containing longer nascent strands (Fig. 1A). Thus, initiation of bidirectional replication occurs within all of these CpG island regions, which is consistent with previous studies (4, 6, 7, 10–12, 24, 25).
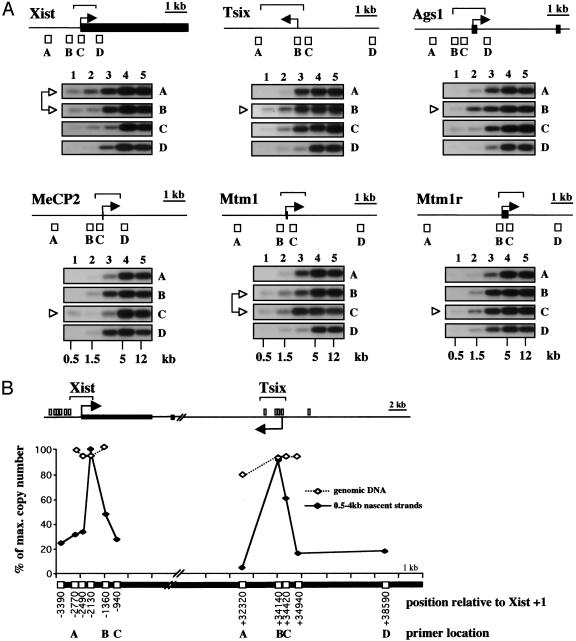
Fig. 1.
Initiation of DNA replication at CpG island regions of X-linked genes. (A) A schematic representation of the genomic regions of the genes Xist (X-inactive specific transcript), Tsix (antisense of Xist), Ags1 (α-galactosidase 1), MeCP2 (methyl CpG-binding protein 2), Mtm1 (myotubularin 1), and Mtm1r (myotubularin-related gene 1). Black rectangles represent exons and arrows indicate the initiation site and direction of transcription. The position of the CpG islands 5′ of each gene is shown with a bracket, and white squares represent the fragments amplified by PCR. Newly replicated nascent strands were purified by size fractionation in alkaline sucrose gradients, immunoprecipitated with anti-BrdUrd antibodies, and used as input for PCR (results from five fractions, named from the top to the bottom of the gradient, are shown). Amplified products were blotted onto filters and were hybridized with the cognate PCR products. Appropriate controls were set up to ascertain that reactions took place under nonsaturating conditions (data not shown). The size of the nascent strands is indicated. A white arrowhead marks the primer pairs located closer to the site of replication initiation. Data shown are representative of three independent nascent strand preparations. (B) Measurement of nascent strand abundance in somatic XX cells at Xist and Tsix CpG island regions by using competitive PCR. Map symbols are as above. Nascent strands (0.5–4 kb in size) were used as input for the PCR analysis (♦). Data on genomic DNA are included as a control to demonstrate that competitor molecules were accurately calibrated (⋄). Positions of the PCR fragments amplified are indicated in base pairs from the center of the PCR product relative to Xist transcription initiation site (+1). The location of the Xist and Tsix primers used in A are marked in B.
High-resolution mapping for the Xist and Tsix CpG island regions by competitive PCR (21) was carried out to verify ORI activity. Competitor molecules spanning the CpG island regions were used to quantify their distribution on nascent strands of 0.5–4 kb size derived from asynchronous somatic XX cells (Fig. 5, which is published as supporting information on the PNAS web site). A site with markedly elevated levels of nascent strands relative to background was detected in both CpG island regions (Fig. 1B), confirming that Xist and Tsix CpG islands are ORIs in XX somatic cells.
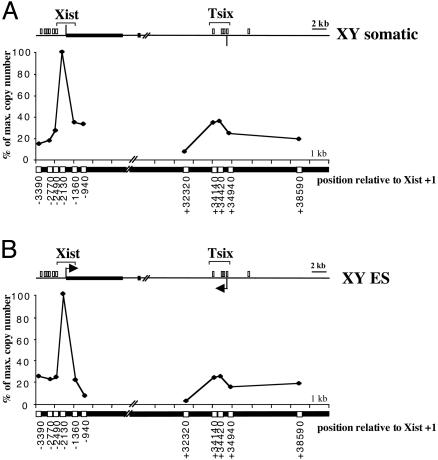
Transcription Is Neither Necessary Nor Sufficient for ORI Activity. Although the Tsix promoter is silent on both the Xa and Xi in XX somatic cells (26), ORI activity was clearly detectable (Fig. 1B, graph). Indeed, the relative abundance of nascent molecules was very similar at the Xist and Tsix CpG island regions (100% vs. 92%), indicating that both ORIs fire with similar efficiency in XX cells. Thus, transcription is not required for activity at the Tsix ORI. To investigate whether the Xist ORI also can function on the silent (Xa) allele, we determined the distribution of nascent strands in XY cells. We first analyzed XY somatic cells where the X chromosome is transcriptionally active and Xist and Tsix genes are silent (26). ORI activity was clearly detectable at the CpG island region of the silent Xist gene (Fig. 2A), demonstrating that for Xist as well as Tsix, transcription is not required for ORI activity. Interestingly, the number of molecules detected along the Tsix CpG island region was similar to background, indicating that this region is not an ORI in XY cells, or that is used in a much lower proportion of cells in the population. The same result was obtained with pluripotent XY ES cells (Fig. 2B), where Tsix is active and there is a low level of Xist expression (26–29). Thus, Tsix transcription is neither necessary nor sufficient to determine ORI activity. Whereas it is not clear why the Tsix ORI is inactive in XY cells, an interesting possibility is that it relates to parental imprinting of this locus (see Discussion).
Fig. 2.
Transcription is neither necessary nor sufficient for ORI activity. Competitive PCR for Xist and Tsix CpG islands, as described for Fig. 1B, was applied to nascent strands isolated from XY somatic cells (A) and XY ES cells (B). The vertical bar and the arrows in Xist and Tsix CpG island maps represent the transcriptional status of the promoters in the cells studied. Neither promoter is active in XY somatic cells (no arrow), and the Xist CpG island is methylated. Both genes are expressed in XY ES cells (black arrows). Graphs in Fig. 1B and here are representative of two independent quantitations and show data expressed as percent of maximum copy number in a given cell type for direct comparison. The position where the maximum abundance of nascent strands was detected at the Xist CpG island was the same in the three cell lines: XX and XY somatic and XY ES cells.
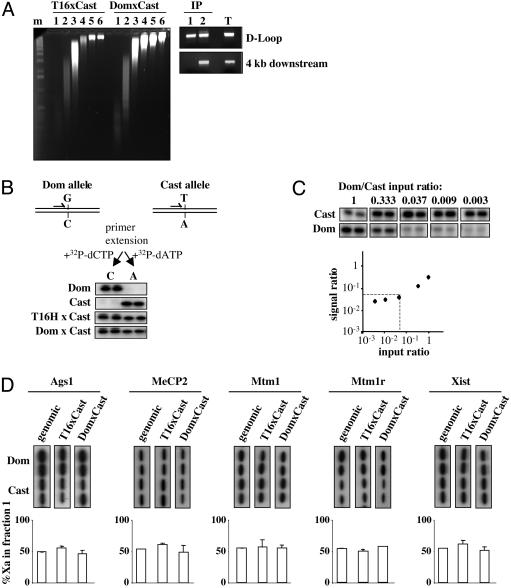
Equivalent ORI Usage on Xa and Xi. The finding that the Xist CpG island is an ORI, even when the island is methylated (Fig. 2A), raised the possibility that this could also be the case for X-linked CpG islands that become methylated during the process of X inactivation. To test this hypothesis, we directly compared ORI usage on the active and inactive X chromosome in XX somatic cells. Interspecific XX fibroblast cell lines carrying either a Mus domesticus (DomxCast) or M. castaneus (T16xCast) inactive X chromosome were established (Fig. 6, which is published as supporting information on the PNAS web site), and single-base polymorphisms were identified in all CpG island regions with the exception of Tsix (Table 1).
Newly replicated DNA from exponentially growing cells was purified by alkaline sucrose fractionation, followed by immunoprecipitation as described above. Enrichment for ORI sequences in fraction 1 (up to 1 kb) was verified by the detection of the mitochondrial replication origin (D-Loop; Fig. 3A). Conversely, a region 4 kb downstream of this unidirectional ORI was only detectable from fraction 2, which contains nascent strands up to 4 kb in length.
Fig. 3.
Equivalent origin usage on active and inactive X-chromosomes. (A) Alkaline sucrose gradient fractionation of DNA derived from exponentially growing T16HxCast and DomxCast XX fibroblasts. m, 1-kb ladder. BrdUrd-immunoprecipitated nascent strands from fractions 1 (IP F1) and 2 (IP F2) were used as input to PCR the mitochondrial replication origin (D-loop) and 4-kb down-stream regions. The result for one of these gradients is shown. T, control PCR by using total DNA as input. (B) Genomic regions surrounding single-base polymorphisms were PCR-amplified, purified products were aliquoted in four identical tubes, and were subjected to primer extension reactions in duplicate with the appropriate 32P-labeled nucleotides triphosphate. The autoradiogram below shows SNuPE results for Ags1 CpG island polymorphism (Table 1) on genomic DNA from XX M. domesticus (Dom), M. castaneus (Cast), and the T16HxCast and DomxCast XX cell lines. (C) One nanogram of nascent DNA from XY M. castaneus cells was mixed in the ratio shown (input) with decreasing amounts of nascent DNA from XY M. domesticus and was subjected to PCR. Amplified products corresponding to Mtm1 CpG island were analyzed by SNuPE. The output ratio between alleles was quantified on a PhosphorImager, normalized to a 1:1 ratio to correct differences in nucleotide incorporation and plotted against the input ratio on a logarithmic scale. (D) ORI-enriched IP F1 from T16xCast and DomxCast cell lines were used as input for SNuPE reactions to analyze the allelic representation of Ags1, MeCP2, Mtm1, Mtm1r, and Xist CpG islands. (Upper) The ratio between alleles found in the IP F1 was quantified on a PhosphorImager and is represented as percent of Xa for each cell line after normalization with genomic DNA from F1 females to correct for differences in the specific activity of the nucleotides. (Lower) The mean values and SE from two independent experiments.
To determine the allelic contribution of CpG island associated ORIs, quantitative SNuPE assays (ref. 30 and Fig. 3B), were carried out on PCR fragments amplified from fraction 1. We first performed control experiments to ensure that the SNuPE assay was sensitive enough to assess allelic levels using small amounts of purified nascent DNA. Known ratios of fraction 1 nascent strands from M. castaneus and M. domesticus XY primary fibroblasts were mixed and used as input for PCR and SNuPE (Fig. 3C). The results show that allelic differences of up to 20-fold can be quantified in the linear range.
We went on to analyze the allelic contribution of the X-linked CpG islands by using ORI-enriched small nascent strands purified from T16xCast and DomxCast XX cell lines. Equal representation of both alleles was found for all of the genes subject to X inactivation; Ags1, MeCp2, Mtm1, and Mtm1r. This finding was true in both cell lines, i.e., regardless of which chromosome is active/inactive (Fig. 3D). The result was entirely reproducible by using the same conditions (Fig. 3D Lower), and also when nascent strands were purified either by λ exonuclease digestion (31), or cesium chloride density centrifugation (ref. 32 and data not shown). We also detected equivalent amounts of nascent strands derived from both alleles at the Xist CpG island region, which is methylated on the Xa (Fig. 3D, far right panel). These findings clearly demonstrate that in the examples analyzed, ORI usage and firing efficiency are equivalent between the transcriptionally active and the silenced X chromosomes.
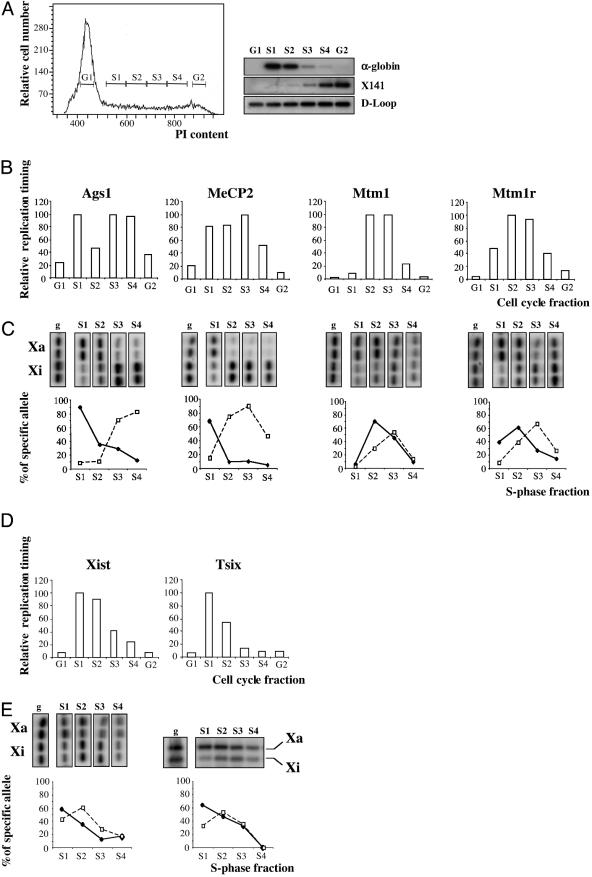
Allele-Specific Replication Timing Analysis. Given that Xi is generally late replicating (33–35), equivalent ORI usage implies that heterochromatin delays ORI firing or fork progression. To confirm that the ORI regions analyzed show differential replication timing on Xa and Xi alleles, we modified a method based on fractionation of S-phase cells by fluorescence-activated cell sorter, combined with quantitative allelic discrimination by SNuPE (36). Exponentially growing lymphocytes or fibroblasts were isolated from [T(X;16)H X M. castaneus] F1 mice. The T(X;16)H translocation gives rise to complete nonrandom X inactivation of the normal (M. castaneus) X chromosome. Cells were pulse-labeled with BrdUrd and were then cell-cycle fractionated according to DNA content by cell sorting (ref. 37 and Fig. 4A Left). Newly replicated DNA was purified from an equal number of cells from each of the six sorted fractions (G1, S1–4, and G2) by immunoprecipitation with anti-BrdUrd antibodies. The fractionation was assessed by determining the relative abundance of markers known to be replicated either early or late in S phase (Fig. 4A Right). The α-globin locus, which replicates in early S phase (38), was most abundant in fractions S1-S2. Conversely, a fragment derived from the DXSmh141 low-repeat sequence (X141), known to replicate in late S phase (39) peaked at S4-G2 phase. As a control for uniform recovery of BrdUrd-labeled DNA, we analyzed the mitochondrial D-loop region, which replicates throughout the cell cycle (40). Similar levels were detected in all fractions (Fig. 4A Bottom Right).
Fig. 4.
Late replication of CpG islands on the inactive X chromosome. (A) Representative profile of propidium iodide content obtained with lymphocytes derived from [T16H × M. castaneus] F1 females, and analysis of early replicating (α-globin) and late-replicating (X141) controls. BrdUrd-labeled cells were separated by cell sorting into six cell-cycle fractions according to their DNA content (G1, S1, S2, S3, S4, and G2). Replication timing was determined by hybridization of cognate probes to PCR products obtained from each fraction after immunoprecipitation with anti-BrdUrd antibodies. Mitochondrial D-loop shows uniform recovery of BrdUrd-labeled DNA. (B) Immunoprecipitated DNA derived from equal number of cells from each cell-cycle fraction were used as input to PCR Ags1, MeCP2, Mtm1, and Mtm1r CpG island regions. The resulting replication profiles were quantified on a PhosphorImager, after hybridization with the cognate PCR product, and represented as a percent of the maximum signal for each region. (C) The resulting PCR products from B were subjected to a second round of PCR with nested primers and the allelic contribution in each S-phase fraction was analyzed by SNuPE. SNuPE reactions were performed in duplicate. The ratio between alleles found in each fraction was quantified on a PhosphorImager, normalized to nucleotide incorporation rates using genomic DNA from F1 females (g), and represented as percent of the active (solid line) or inactive allele (broken line), normalized relative to the replication timing profile shown on B. (D) Relative replication timing of Xist and Tsix CpG islands calculated as in B. (E) Allelic replication timing for Xist CpG island by SNuPE (Left), and for a MnlI restriction site polymorphism (26) located 2,180 bp down-stream of the Tsix ORI mapped in Fig. 2B (Right). The percent specific for Xa and Xi alleles was calculated for each S-phase fraction, as in C. Data shown are representative of three independent cell sorting experiments.
Analysis of the X-linked CpG islands in the different cell cycle fractions revealed a range of patterns from a biphasic distribution, which was consistent with separate replication of Xa and Xi alleles, in the case of Ags-1, to a single peak in mid S phase, in the case of Mtm1 and Mtm1r (Fig. 4B). Both Xist and Tsix CpG islands replicated in early S phase (Fig. 4D). We went on to analyze allelic replication timing in each S-phase fraction by using SNuPE (Fig. 4 C and E). Because the M. castaneus X chromosome is always inactive, we were able to unequivocally determine replication timing profiles for Xa and Xi. As predicted for the Ags1 CpG island, the early and late peaks identified by cell-cycle fractionation were found to correspond to Xa and Xi alleles, respectively. Similarly, Xa and Xi alleles of MeCp2, Mtm1, and Mtm1r genes could all be resolved into early and late replicating peaks (Fig. 4C). The temporal separation for Mtm1 and Mtm1r alleles (which map 200 kb apart on the genome) is relatively small, illustrating that replication timing is affected by local chromosome environment as well as chromosome activity status. Thus, delayed replication of the Xi allele is seen for all of the CpG island regions associated with genes subject to X inactivation. In combination with our data on ORI usage at these sites, we conclude that heterochromatin on the inactive X chromosome delays ORI activation, but has no effect on ORI use or firing efficiency.
In the case of Xist and Tsix CpG islands, allelic discrimination by SNuPE (for Xist) or restriction-site polymorphism [for Tsix (41)] confirmed that the locus replicates early on both chromosomes, although the silent Xist allele (Xa) was consistently seen to replicate slightly earlier (Fig. 4E). This finding is in agreement with early replication timing of the active human XIST locus reported (42). Because the Xist CpG island is methylated on the silent (Xa) allele, we conclude that DNA methylation alone is not sufficient to delay ORI activation.
Discussion
It is well established that the Xi replicates late in S phase relative to Xa, but the molecular basis for this occurrence is unknown. Two main possibilities could be considered. First, a distinct set of ORIs that are independently regulated may be used on the Xi, and second, the same ORIs could be used on both chromosomes with delayed ORI activation on the heterochromatic Xi. Our results support the second model, showing that for several loci the same ORIs are used on Xa and Xi, and moreover, that the efficiency of ORI firing is unaffected by heterochromatin formation.
The ORIs analyzed in this study are associated with CpG islands, and as CpG islands represent more than half of mammalian replication origins (12), we suggest that our observations can be extrapolated to the whole X chromosome. In support of this hypothesis, if a significant number of Xi ORIs were distinct, the relative contribution of shared ORIs to Xi replication should be reduced. We observed indistinguishable levels of Xa and Xi alleles by using accurate quantitative methods at all of the loci analyzed. Thus, although we cannot completely rule out the possibility that additional ORIs could be activated on the Xi, our results are most consistent with equivalent ORI usage occurring chromosome wide. In agreement with this conclusion, a recent report (18) has shown similar ORI usage at the human HPRT and G6PD CpG islands, albeit by using indirect comparison in CHO hybrid cell lines carrying either an active or inactive human X chromosome.
Our observations indicate that DNA methylation and remodeled chromatin structure that accompany transcriptional silencing at CpG island regions (43–46) does not affect ORI function, despite the fact that promoters and ORIs often map close to each other. This observation could also apply to other regions of facultative heterochromatin, for example, at loci that replicate at different times in S phase, depending on their expression status. Indeed, studies on the human β-globin locus have shown that an ORI used in expressing cells (euchromatic/early replicating), is also used in nonexpressing cells (heterochromatic/late replicating; refs. 47 and 48).
Our data also demonstrate that transcription per se is not required for DNA replication initiation from CpG island-associated ORIs in mammalian cells. This finding is best illustrated by the fact that we could detect ORI activity at the Xist CpG island region in somatic XY cells, where the gene is not expressed. The reverse seems also to be true, because the active Tsix promoter is not sufficient to trigger replication initiation in totipotent cells. A similar conclusion was reached in a study on Schizosaccharomyces pombe, where ORI activity was not affected by mutations that abolish transcription from an adjacent promoter (49). Thus, despite colocalization of ORIs with promoter regions in many organisms (5–12, 24, 50, 51), and the fact that transcription factors stimulate replication in several systems (52–57), our data rule out a strict link between initiation of replication and transcription. A possible explanation for this conundrum is that both processes are mechanistically linked only early in development (58), but that ORIs are then stably maintained independent of subsequent changes in transcriptional activity.
Our data indicate that although heterochromatin does not prevent ORI activity, it does delay the assembly or progression of the replication machinery. This conclusion is similar to recent findings in S. cerevisiae that show that heterochromatinization of an ORI, induced by targeting the binding of Sir4p, does not prevent initiation of replication but delays the time of firing of the ORI (59). Conversely, targeting of the major S. cerevisiae acetyltransferase Gcn5p to late replicating origins demonstrates that histone acetylation directly affects the timing of DNA replication initiation and the association of Cdc45p with ORIs (60).
We found that the Xist ORI replicates early on the Xa, where the associated CpG island is methylated, suggesting that DNA methylation of CpG islands is not sufficient to impose late replication on the neighboring ORI. This finding is in agreement with incomplete advancement of replication timing observed at hypomethylated CpG islands in XX cells derived from patients with immunodeficiency, centromeric region instability, and facial anomalies (61). Also, it has been shown recently that loss of DNA methylation at imprinted regions does not alter the asynchronous replication timing observed at imprinted genes (62). Our high-resolution analysis of replication timing on both Xa and Xi indicates that different chromatin features are likely to contribute to the delay in replication timing associated with heterochromatin. Consistent with this hypothesis, a detailed study across the chicken β-globin locus found several early replicating ORIs associated with different patterns of histone modifications (63). This conclusion may be important in interpreting recent findings demonstrating that there is no strict correlation between gene silencing and late replication timing (38, 39, 47, 64).
Based on our observations, we suggest a model in which mammalian ORIs, once specified early in development, are stably maintained in the soma, regardless of transcriptional activity and the methylation status of the associated CpG island. While it is currently unknown how mammalian ORIs are specified, an interesting insight may come from our finding that the Tsix CpG island is an ORI in XX but not in XY cells. This finding raises the possibility that ORI activity could depend on parent of origin as Tsix is subject to parental imprinting (41, 65). Analysis of ORI usage at other imprinted loci will be of considerable interest in terms of understanding ORI specification in a developmental context.
Supplementary Material
Acknowledgments
We thank F. Antequera and members of the X inactivation group for advice and discussion; R. Appanah for assistance with genetic crosses; T. Nesterova for assistance in deriving cell lines; V. Azuara for advice and assistance in activating ex vivo lymphocytes and cell sorting; and F. Antequera, M. Merkenschlager, V. Azuara, and T. Nesterova for helpful comments on the manuscript. Cell sorting analysis was performed with the assistance of G. Warnes at the Cancer Research UK Fluorescence-Activated Cell Sorter Laboratory (Charity no. 1089464). M.G. was supported by Marie Curie IHP-MCIF-99-1 and Human Frontiers Sciences Program LT001721/2000-M Postdoctoral Fellowships.
Abbreviations: ORI, replication origin; SNuPE, single-nucleotide primer extension; ES, embryonic stem.
References
- 1.DePamphilis, M. L. (1999) BioEssays 21, 5-16. [DOI] [PubMed] [Google Scholar]
- 2.Todorovic, V., Falaschi, A. & Giacca, M. (1999) Front. Biosci. 4, D859-D868. [DOI] [PubMed] [Google Scholar]
- 3.Mechali, M. (2001) Nat. Rev. Genet. 2, 640-645. [DOI] [PubMed] [Google Scholar]
- 4.Vassilev, L. & Johnson, E. M. (1990) Mol. Cell. Biol. 10, 4899-4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitsberg, D., Selig, S., Keshet, I. & Cedar, H. (1993) Nature 366, 588-590. [DOI] [PubMed] [Google Scholar]
- 6.Giacca, M., Zentilin, L., Norio, P., Diviacco, S., Dimitrova, D., Contreas, G., Biamonti, G., Perini, G., Weighardt, F., Riva, S., et al. (1994) Proc. Natl. Acad. Sci. USA 91, 7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taira, T., Iguchi-Ariga, S. M. & Ariga, H. (1994) Mol. Cell. Biol. 14, 6386-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao, Y., Tsutsumi, R., Yamaki, M., Nagatsuka, Y., Ejiri, S. & Tsutsumi, K. (1994) Nucleic Acids Res. 22, 5385-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aladjem, M. I., Groudine, M., Brody, L. L., Dieken, E. S., Fournier, R. E., Wahl, G. M. & Epner, E. M. (1995) Science 270, 815-819. [DOI] [PubMed] [Google Scholar]
- 10.Delgado, S., Gomez, M., Bird, A. & Antequera, F. (1998) EMBO J. 17, 2426-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller, C., Ladenburger, E. M., Kremer, M. & Knippers, R. (2002) J. Biol. Chem. 277, 31430-31440. [DOI] [PubMed] [Google Scholar]
- 12.Ladenburger, E. M., Keller, C. & Knippers, R. (2002) Mol. Cell. Biol. 22, 1036-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antequera, F. & Bird, A. (1993) Proc. Natl. Acad. Sci. USA 90, 11995-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, E., Beard, C., Jaenisch, R., Forster, A. C. & Bestor, T. H. (1993) Nature 366, 362-365. [DOI] [PubMed] [Google Scholar]
- 15.Panning, B. & Jaenisch, R. (1996) Genes Dev. 10, 1991-2002. [DOI] [PubMed] [Google Scholar]
- 16.Lyon, M. F. (1961) Nature 190, 372-373. [DOI] [PubMed] [Google Scholar]
- 17.Plath, K., Mlynarczyk-Evans, S., Nusinow, D. A. & Panning, B. (2002) Annu. Rev. Genet. 36, 233-278. [DOI] [PubMed] [Google Scholar]
- 18.Cohen, S. M., Brylawski, B. P., Cordeiro-Stone, M. & Kaufman, D. G. (2003) J. Cell. Biochem. 88, 923-931. [DOI] [PubMed] [Google Scholar]
- 19.Kay, G. F., Penny, G. D., Patel, D., Ashworth, A., Brockdorff, N. & Rastan, S. (1993) Cell 72, 171-182. [DOI] [PubMed] [Google Scholar]
- 20.Carrel, L., Hunt, P. A. & Willard, H. F. (1996) Hum. Mol. Genet. 5, 1361-1366. [DOI] [PubMed] [Google Scholar]
- 21.Giacca, M., Pelizon, C. & Falaschi, A. (1997) Methods 13, 301-312. [DOI] [PubMed] [Google Scholar]
- 22.Norris, D. P., Patel, D., Kay, G. F., Penny, G. D., Brockdorff, N., Sheardown, S. A. & Rastan, S. (1994) Cell 77, 41-51. [DOI] [PubMed] [Google Scholar]
- 23.Prissette, M., El-Maarri, O., Arnaud, D., Walter, J. & Avner, P. (2001) Hum. Mol. Genet. 10, 31-38. [DOI] [PubMed] [Google Scholar]
- 24.Cohen, S. M., Brylawski, B. P., Cordeiro-Stone, M. & Kaufman, D. G. (2002) J. Cell. Biochem. 85, 346-356. [DOI] [PubMed] [Google Scholar]
- 25.Phi-van, L. & Stratling, W. H. (1999) Nucleic Acids Res. 27, 3009-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J. T., Davidow, L. S. & Warshawsky, D. (1999) Nat. Genet. 21, 400-404. [DOI] [PubMed] [Google Scholar]
- 27.Brockdorff, N., Ashworth, A., Kay, G. F., Cooper, P., Smith, S., McCabe, V. M., Norris, D. P., Penny, G. D., Patel, D. & Rastan, S. (1991) Nature 351, 329-331. [DOI] [PubMed] [Google Scholar]
- 28.Panning, B., Dausman, J. & Jaenisch, R. (1997) Cell 90, 907-916. [DOI] [PubMed] [Google Scholar]
- 29.Sheardown, S. A., Duthie, S. M., Johnston, C. M., Newall, A. E., Formstone, E. J., Arkell, R. M., Nesterova, T. B., Alghisi, G. C., Rastan, S. & Brockdorff, N. (1997) Cell 91, 99-107. [DOI] [PubMed] [Google Scholar]
- 30.Singer-Sam, J., LeBon, J. M., Dai, A. & Riggs, A. D. (1992) PCR Methods Appl. 1, 160-163. [DOI] [PubMed] [Google Scholar]
- 31.Gerbi, S. A. & Bielinsky, A. K. (1997) Methods 13, 271-280. [DOI] [PubMed] [Google Scholar]
- 32.Epner, E., Rifkind, R. A. & Marks, P. A. (1981) Proc. Natl. Acad. Sci. USA 78, 3058-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt, M. & Migeon, B. R. (1990) Proc. Natl. Acad. Sci. USA 87, 3685-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen, R. S., Canfield, T. K. & Gartler, S. M. (1995) Hum. Mol. Genet. 4, 813-820. [DOI] [PubMed] [Google Scholar]
- 35.Kawame, H., Gartler, S. M. & Hansen, R. S. (1995) Hum. Mol. Genet. 4, 2287-2293. [DOI] [PubMed] [Google Scholar]
- 36.Xiong, Z., Tsark, W., Singer-Sam, J. & Riggs, A. D. (1998) Nucleic Acids Res. 26, 684-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen, R. S., Canfield, T. K., Lamb, M. M., Gartler, S. M. & Laird, C. D. (1993) Cell 73, 1403-1409. [DOI] [PubMed] [Google Scholar]
- 38.Smith, Z. E. & Higgs, D. R. (1999) Hum. Mol. Genet. 8, 1373-1386. [DOI] [PubMed] [Google Scholar]
- 39.Azuara, V., Brown, K. E., Williams, R. R., Webb, N., Dillon, N., Festenstein, R., Buckle, V., Merkenschlager, M. & Fisher, A. G. (2003) Nat. Cell Biol. 5, 668-674. [DOI] [PubMed] [Google Scholar]
- 40.Clayton, D. A. (1982) Cell 28, 693-705. [DOI] [PubMed] [Google Scholar]
- 41.Lee, J. T. (2000) Cell 103, 17-27. [DOI] [PubMed] [Google Scholar]
- 42.Gartler, S. M., Goldstein, L., Tyler-Freer, S. E. & Hansen, R. S. (1999) Hum. Mol. Genet. 8, 1085-1089. [DOI] [PubMed] [Google Scholar]
- 43.Pfeifer, G. P. & Riggs, A. D. (1991) Genes Dev. 5, 1102-1113. [DOI] [PubMed] [Google Scholar]
- 44.Gilbert, S. L. & Sharp, P. A. (1999) Proc. Natl. Acad. Sci. USA 96, 13825-13830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heard, E., Rougeulle, C., Arnaud, D., Avner, P., Allis, C. D. & Spector, D. L. (2001) Cell 107, 727-738. [DOI] [PubMed] [Google Scholar]
- 46.Boggs, B. A., Cheung, P., Heard, E., Spector, D. L., Chinault, A. C. & Allis, C. D. (2002) Nat. Genet. 30, 73-76. [DOI] [PubMed] [Google Scholar]
- 47.Cimbora, D. M., Schubeler, D., Reik, A., Hamilton, J., Francastel, C., Epner, E. M. & Groudine, M. (2000) Mol. Cell. Biol. 20, 5581-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon, I., Tenzen, T., Mostoslavsky, R., Fibach, E., Lande, L., Milot, E., Gribnau, J., Grosveld, F., Fraser, P. & Cedar, H. (2001) EMBO J. 20, 6150-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gomez, M. & Antequera, F. (1999) EMBO J. 18, 5683-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierron, G., Pallotta, D. & Benard, M. (1999) Mol. Cell. Biol. 19, 3506-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki, T., Sawado, T., Yamaguchi, M. & Shinomiya, T. (1999) Mol. Cell. Biol. 19, 547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marahrens, Y. & Stillman, B. (1992) Science 255, 817-823. [DOI] [PubMed] [Google Scholar]
- 53.Rhode, P. R., Elsasser, S. & Campbell, J. L. (1992) Mol. Cell. Biol. 12, 1064-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiltshire, S., Raychaudhuri, S. & Eisenberg, S. (1997) Nucleic Acids Res. 25, 4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li, R., Yu, D. S., Tanaka, M., Zheng, L., Berger, S. L. & Stillman, B. (1998) Mol. Cell. Biol. 18, 1296-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bosco, G., Du, W. & Orr-Weaver, T. L. (2001) Nat. Cell Biol. 3, 289-295. [DOI] [PubMed] [Google Scholar]
- 57.Bodmer-Glavas, M., Edler, K. & Barberis, A. (2001) Nucleic Acids Res. 29, 4570-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antequera, F. & Bird, A. (1999) Curr. Biol. 9, R661-R667. [DOI] [PubMed] [Google Scholar]
- 59.Zappulla, D. C., Sternglanz, R. & Leatherwood, J. (2002) Curr. Biol. 12, 869-875. [DOI] [PubMed] [Google Scholar]
- 60.Vogelauer, M., Rubbi, L., Lucas, I., Brewer, B. J. & Grunstein, M. (2002) Mol. Cell 10, 1223-1233. [DOI] [PubMed] [Google Scholar]
- 61.Hansen, R. S., Stoger, R., Wijmenga, C., Stanek, A. M., Canfield, T. K., Luo, P., Matarazzo, M. R., D'Esposito, M., Feil, R., Gimelli, G., et al. (2000) Hum. Mol. Genet. 9, 2575-2587. [DOI] [PubMed] [Google Scholar]
- 62.Gribnau, J., Hochedlinger, K., Hata, K., Li, E. & Jaenisch, R. (2003) Genes Dev. 17, 759-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prioleau, M. N., Gendron, M. C. & Hyrien, O. (2003) Mol. Cell. Biol. 23, 3536-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hansen, R. S., Canfield, T. K., Fjeld, A. D., Mumm, S., Laird, C. D. & Gartler, S. M. (1997) Proc. Natl. Acad. Sci. USA 94, 4587-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sado, T., Wang, Z., Sasaki, H. & Li, E. (2001) Development (Cambridge, U.K.) 128, 1275-1286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.