Abstract
The TATA-binding protein (TBP) is a critical general transcription factor that associates with the core promoter and acts as a nexus for gene regulation through its interactions with other factors. A large number of proteins recognize the relatively small yet highly conserved C-terminal domain of TBP. One subset of these proteins (general transcription factors) interacts with the TBP·TATA complex and RNA polymerase II to create the preinitiation complex. To study TBP functions in preinitiation complex and other complexes, we generated a set of RNA aptamers with high affinity to yeast TBP. These aptamers act on TBP in different ways: all of them bind TBP competitively with DNA bearing the TATA element, and some can actively disrupt the TBP·TATA interaction in preformed, higher-order complexes containing the additional general transcription factors TFIIB and TFIIA. In crude cell extracts, the aptamers inhibit transcription in ways that reveal the dynamic nature of TBP interactions during initiation and reinitiation.
Initiation of transcription by RNA polymerase II (Pol II) requires the assembly of a preinitiation complex (PIC) at the core promoter. The first, and often rate-limiting, step in this process is the binding of the TATA-binding protein (TBP) to the TATA element on DNA (1). The core domain of TBP has a saddle-shaped structure with a concave surface that binds and bends DNA severely (2). The PIC is largely built on this TBP·TATA foundation by an interlaced network of polypeptides that interact with each other and with the TBP·DNA complex (3). In vitro experiments suggest that the general transcription factor TFIIB binds to the promoter after TBP, providing a platform for the entry of Pol II/TFIIF and playing a role in determining the transcription start sites (4, 5). TFIIA associates with the PIC through a distinct interaction surface on TBP and stimulates basal as well as activated transcription, presumably by counteracting the inhibitory effects of TBP-binding factors such as NC2 or Mot1 (reviewed in ref. 6). These inhibitory factors are thought to repress transcription by interfering with the TBP·TATA interaction and higher order complex formation (7, 8).
The critical role played by the TBP·TATA interaction in transcription makes it a frequently used target of regulation by numerous proteins that physically interact with TBP. Extensive investigations have been conducted to define the functions of discrete sites on the surface of TBP. Many genetic selections and screens have identified functionally distinct TBP mutants. Most mutants are defective in their interactions with the TATA element, TFIIA, or TFIIB, whereas others disrupt interactions with additional factors, including transcription activators and repressors (reviewed in ref. 9). Particularly informative was a systematic analysis, using alanine scanning mutagenesis of the surface of human TBP, which defined clusters of residues critical for its interactions with individual factors (10). Also, a particular surface of TBP has been shown to bind several transcription factors and TATA DNA competitively (reviewed in ref. 11). As a result, it is difficult to specifically study and control the TBP·TATA interaction in physiologically relevant systems. Moreover, past studies provide only a limited view of the dynamics of TBP interactions during PIC formation and their regulation.
One strategy to probe such dynamic processes is to rapidly disrupt particular interactions and study the immediate consequences. Temperature-sensitive mutants provide a rapid disruption of a protein–protein interaction, but the shift to nonpermissive temperature usually results in the conformational disruption of the entire protein and its degradation along with its partners. Specific ligand-based perturbation of transcription initiation may provide more precise, targeted control. Through binding to the minor groove of DNA, the antibiotic distamycin A and the alkylating benzoyl mustard derivative tallimustine can prevent the recognition of the TATA element by TBP, thereby providing information about TBP·TATA complexes (12). However, these compounds must be used at very high concentration (micromolar) to show their effect, and they are not specific to the TBP·TATA interaction. More specific inhibitors of TBP in the form of small organic compounds have not been isolated.
Alternatives to small-molecular-weight “drugs” are RNA aptamers, which are selected in vitro from a combinatorial sequence pool for their affinity to a target molecule (13, 14). The chemical and biological properties of RNAs that allow efficient production and regeneration have made such aptamers versatile molecular probes. They possess tremendous potential relative to small organic compounds in experimental and therapeutic manipulations. When expressed under the control of specific promoters, they are able to modulate or perturb molecular interactions with high temporal and spatial precision in tissue culture cells or animals (ref. 15; see ref. 16 for a review).
In this study, we describe the selection and properties of a set of RNA aptamers to yeast TBP. These aptamers not only have high affinity and specificity but also act in distinct ways on TBP. Using these aptamers, we performed detailed analyses of the TBP·TATA interaction in the context of the TFIIA·TFIIB·TBP·TATA complex and its subcomplexes. Some of the aptamers were shown to be able to disrupt preformed complexes, and the various complexes responded distinctly to different aptamers. The aptamers were also effective inhibitors of transcription in an in vitro assay, and their different modes of action allowed us to draw more penetrating conclusions when they were used collectively in a study of the dynamics of TBP interactions in a crude cell extract. Furthermore, the efficacy of the inhibition by these aptamers in this complex milieu promises their utility under more physiological conditions.
Materials and Methods
Protein Preparation. Yeast TBP was expressed and purified from bacteria as described (17). Yeast TFIIA was purified by using a protocol obtained from S. Hahn (Fred Hutchinson Cancer Research Center, Seattle), in which subunits Toa1 and Toa2 were expressed separately in Escherichia coli, denatured in 8 M urea, combined and renatured by dialyzing out the urea. Renatured TFIIA was further purified in an AKTA system (Pharmacia) by using a mono Q column. Yeast TFIIB was prepared by using the Intein system (New England Biolabs). It was further purified on an Uno-S column (Bio-Rad).
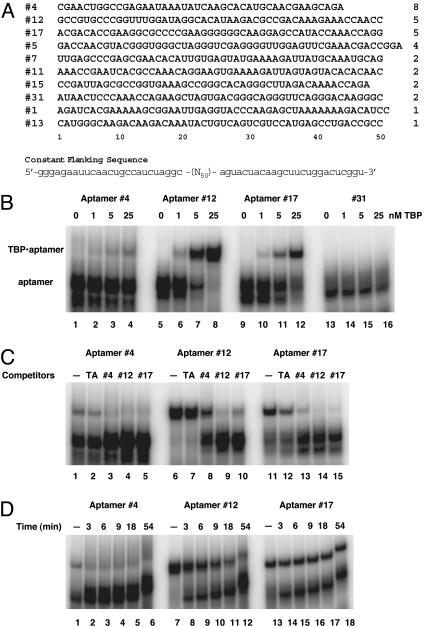
In Vitro Selection. The sequences of the template-primer system have been described (18). The template contains a 50-nt randomized region in the middle. The initial pool contained ≈2.5 × 1015 different sequences. Eleven cycles of selection and amplification were performed against His-tagged yeast TBP. Selection was carried out in 1× binding buffer (12 mM Hepes, pH 7.9/60 mM potassium chloride/5 mM magnesium chloride/1 mM EDTA), followed by partitioning on nitrocellulose filters, except for cycle 8, which was partitioned by using Biacore sensor chip with His-tagged TBP covalently coupled, and cycle 10, which was partitioned by electrophoretic mobility shift. A negative selection step against the filter (i.e., collecting candidates unbound to the filter) was included in every cycle after the fourth. The progress was monitored by electrophoretic mobility-shift assay (EMSA). The final selected pool in the form of DNA was cloned, and 32 individuals were sequenced. Some members of a clone contained minor sequence variations. For example, position 8 in the variable region of no. 4 could be either a C or a G; position 35 of no. 12 could be eitheraGoranA (the version listed in Fig. 1A was used in subsequent studies).
Fig. 1.
Selection and characterization of TBP-aptamers. (A) RNA sequences of the variable region of clones isolated from the 11th generation. The name given each clone is shown to the left of its sequence. The number of isolated individuals is indicated to the right for each clone. The sequence of the flanking constant regions is shown at the bottom. (B) Affinity of TBP-aptamers to yeast TBP. Shown is an EMSA result of 20-μl binding reaction mixture with 0.1 nM radioactive RNA probes and increasing amount of purified His-yTBP. Untagged yTBP was also tested and yielded similar results. (C) TBP-aptamers competing with each other and with TATA-DNA for binding to TBP. Cold competitors (50 nM) were added together with the radioactive probes to reaction mixture containing 25 nM TBP. (D) Stability of the aptamer-TBP complexes. Binding reactions containing 0.1 nM radioactive RNA probes and 25 nM TBP were incubated for 30 min before adding unlabeled RNA to 1 μM final concentration. Aliquots of the mixture were taken out at times indicated and loaded instantly on to a running gel. Lanes labeled with a “minus” sign have no competitor added.
EMSA. EMSA was performed according to the protocol described in ref. 8, with some modifications. All binding reactions were incubated at room temperature for 30 min in 20-μl volume in 1× binding buffer with 100 μg/ml BSA, 25 μg/ml yeast tRNA, 10% glycerol, and 0.1% Nonidet P-40. The RNA probes were internally labeled with [α-32P]UTP by using the T7-MAXIscript in vitro transcription kit (Ambion, Austin, TX) and had a concentration of 0.1 nM in the binding reaction. The TATA-DNA used in binding assays was produced by annealing deoxyoligonucleotides bearing a 30-bp DNA segment derived from the adenovirus major late promoter with the following sequence: 5′-GGGAATTCGGGCTATAAAAGGGGGATCCGG-3′. When used in assays to visualize TATA-containing complexes, DNA was end-labeled by T4 polynucleotide kinase. A typical reaction for complex formation or disruption contained 10 nM TBP. To form higher-order complexes, 2.5 nM TFIIA and/or 100 nM TFIIB were used with 2.5 nM TBP. In competition experiments, the aptamer was mixed with the TATA-DNA, then incubated with the proteins for 30 min. In disruption experiments, the TATA-DNA was incubated with the proteins for 30 min before addition of aptamer. Unless otherwise indicated, the mixture was loaded onto the gel after another 30-min incubation. All complexes were resolved on a 6% polyacrylamide gel run in 0.5× TBE buffer (45 mM Tris/45 mM boric acid/1 mM EDTA, pH 8.3). To stabilize the TBP·TATA complex, 5 mM magnesium acetate was included in both the gel and the buffer whereas 2 mM magnesium acetate was used for EMSA of higher-order complexes. For reactions containing TFIIB·TBP·TATA complex, 60 ng poly(dI)·poly(dC) was added before loading.
In Vitro Transcription. Yeast Strain BJ1991 (prb1 pep4 gal2 leu2 trp1 ura3) was grown in yeast extract/peptone/dextrose (YEPD) to an A600 of 1.5–2.0. Cells were harvested, and whole cell extracts were prepared by using a mortar and pestle (19). Protein concentration was determined by Bradford assay. In vitro transcription was performed based on a protocol previously described (20) at room temperature in a 25-μl final volume using a plasmid (200 ng) that bears the adenovirus major late promoter upstream of a 390-nt G-less cassette. Yeast whole cell extract (100 μg) was incubated for 2 min in transcription buffer (20 mM Hepes, pH 7.6/100 mM potassium glutamate/10 mM magnesium acetate/5 mM EGTA/2.5 mM DTT/10 μM zinc sulfate/10% glycerol/20 units of RNase Inhibitor/SUPERase-In) (Ambion) plus an ATP regeneration system (3 mM ATP/30 mM creatine phosphate/150 ng of creatine kinase). Aptamers were added to the extract mixture at the concentrations indicated, together with, or 30 min after, the addition of DNA template. Transcription was initiated with NTPs [10 μCi (1 Ci = 37 GBq) of [α-32P]UTP, 50 μM UTP, 250 μM CTP and ATP, final concentrations] and terminated with stop solution (20 mM EDTA/0.2 M sodium chloride/10 mM Tris·HCl, pH 7.6/1 μg of glycogen/20 units of RNase T1). The samples were incubated at 37°C for 20 min, digested with proteinase K in the presence of SDS (2%) for 20 min before being phenol/chloroform extracted and ethanol precipitated. RNA products were separated on 6% acrylamide sequencing gels. In control experiments, α-amanitin was added to 20 μg/ml just before NTPs to ensure that the transcription observed was Pol II-dependent, and yeast tRNA was added (up to 1 μg) to demonstrate that nonspecific RNAs did not elicit any effect on transcript levels. Where indicated, sarkosyl was added to 0.25% final concentration 30 s after NTPs to maintain a single round of transcription (21).
Results
Aptamer Selection and Characterization. We performed an in vitro selection experiment with yeast TBP as target and isolated a set of aptamers from a pool of 2.5 × 1015 different RNA sequences. After 11 cycles of selection and amplification, we cloned the final selected pool and sequenced 32 individuals. These individuals belong to 10 clones, each descended from a different sequence in the original pool. As shown in Fig. 1 A, most of these clones have multiple isolates. Other than an 11-nt stretch shared by no. 7 and no. 11, there is no shared consensus sequence among these clones, nor an obvious common secondary structure. Therefore, the different TBP-binding clones are likely to recognize at least partially distinct features of TBP.
Eight of these clones showed specific binding to yeast TBP in an EMSA. The two remaining clones (no. 5 and no. 31) contain sequences defined as multi-G motifs that bind to the nitrocellulose filters used as the partitioning matrix (22). Three aptamer clones were chosen for further characterization. One of them, represented by aptamer no. 4, was the most abundant and was isolated 8 times; the other two, represented by aptamer no. 12 and no. 17, were tied for second in abundance, each represented by 5 individual isolates.
An EMSA with the three representative individuals, no. 4, no. 12, and no. 17, showed that these most abundant clones all have high affinity for TBP (Fig. 1B). Aptamer no. 12 showed highest affinity, with a dissociation constant (Kd) measured by this assay of ≈3 nM. Aptamer no. 17 ranked second whereas aptamer no. 4 appeared to have the lowest affinity. An independent and more direct measurement by nitrocellulose filter-binding showed that no. 4 has a Kd of ≈10 nM whereas no. 12 has a Kd of ≈3 nM. Clone no. 31 showed no detectable RNA·TBP complex even at the highest TBP concentration tested; therefore, it was chosen for use as a negative control.
As shown in Fig. 1C, the three most abundant aptamers competed with each other for binding to TBP (other less abundant aptamers listed in Fig. 1 A also competed with each other; data not shown), suggesting that all of the aptamers interact with overlapping binding sites on TBP. Their relative strength in competition correlated well with their relative affinities. In addition, double-stranded oligodeoxynucleotides bearing the TATA element (TATA-DNA) competed with the aptamers for TBP-binding, suggesting that the DNA-binding site on TBP is affected by the binding of these aptamers. However, all three aptamers exhibited a much higher affinity for TBP than did TATA-DNA (lanes 2, 7, and 12).
We further investigated the stability of the TBP·aptamer complex. After equilibrium had been reached between radio-labeled RNA probes and TBP, a large amount of unlabeled RNA aptamer was added to the reaction, and aliquots of the mixture were removed at the given time points and loaded onto a running native gel. The half-lives of the TBP·aptamer complexes were derived from the results shown in Fig. 1D. The aptamer no. 17·TBP complex had the longest half-life (>1 h) whereas the complex containing the aptamer no. 4 was the least stable (half-life <3 min), in agreement with the lower levels of TBP·aptamer complexes observed in EMSA (Fig. 1B).
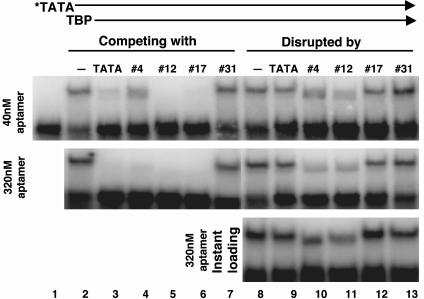
Probing TBP-Containing Complex Formation and Disassembly Using Aptamers. The association of TBP with the TATA element and/or general transcription factors is critical for establishing a PIC. With the TBP aptamers as inhibitory probes, we studied these interactions in the context of multiprotein complexes by examining the perturbation of four different complexes: TBP·TATA, TFIIA·TBP·TATA, TFIIB·TBP·TATA, and TFIIA·TFIIB·TBP·TATA. We started by investigating the ability of the aptamers to prevent the binding of TBP to the TATA element or to disrupt the preformed TBP·TATA complex. Interestingly, the three aptamers acted distinctly on TBP complexes, as shown in Fig. 2. Whereas all three were able to prevent TBP binding to the TATA-DNA (see the “competing with” lanes), the preformed TBP·TATA complex withstood challenge only by no. 17 (see the “disrupted by” lanes). The three aptamers differed in their strength in preventing complex formation, with no. 4 significantly weaker than the other two (compare lane 4 with lanes 5 and 6 in Top). With a preformed TBP·TATA complex, which is very stable under our assay conditions, aptamer no. 17 had a very weak effect, comparable with that caused by unlabeled competitor TATA-DNA (compare lane 12 with lane 9). In contrast, aptamers no. 4 and no. 12 exerted a very strong disruptive effect on the preformed complex, as shown in lanes 11 ands 12. This result indicated an active displacement of TATA-DNA by these aptamers. More interestingly, aptamer no. 4, which had a weaker effect in competing with the TATA-DNA (lane 4), was almost as potent as no. 12 in disrupting the preformed complex.
Fig. 2.
Aptamers acting on TBP·TATA complexes in distinct modes. All reactions contained 0.5 nM radioactive TATA-DNA (indicated by an asterisk) and 10 nM His-yTBP. In competition for complex formation (lanes 2–7), a low (40 nM) and a high (320 nM) concentration of unlabeled RNA or TATA-DNA was added together with the TATA-DNA probe. To examine the effect of aptamers on preformed TBP·TATA complex (lanes 8–13), the same concentrations of unlabeled RNA or TATA-DNA were added after a 30-min incubation of TBP with the radioactive TATA probe. In Top and Middle, the mixtures were incubated for another 30 min before loading onto the gel. In Bottom, the mixtures were loaded right away. The gel contained 5 mM magnesium acetate.
To further investigate the ability of aptamers no. 4 and no. 12 to disrupt a preformed complex, TBP·TATA complexes at equilibrium were challenged with unlabeled aptamers, and the reaction mixture was loaded immediately onto a native gel (Fig. 2 Bottom). Within this short time frame, aptamers no. 4 and no. 12 were able to dissociate the majority of the TBP·TATA complex whereas aptamer no. 17 had little effect. The TBP·TATA complex is normally very stable, with a half-life of 1–2 h (23); thus, the instantaneous disruption of this stable complex further indicated the active role played by these aptamers. We also noticed an increased mobility of the shifted complex remaining in the presence of these aptamers. This new, fast-moving complex was TBP-dependent and was therefore not a TATA·aptamer complex (data not shown). It is likely that this complex contains TBP, TATA-DNA, and aptamer, where the increased mobility of the complex results from the additional negative charge of bound RNA. These results indicate that aptamer no. 17 may simply bind TBP through TBP's concave DNA-binding surface whereas no. 4 and no. 12 seem to be capable of interacting at distinct surfaces to form a TBP·TATA·aptamer complex that then destabilizes the TBP·TATA complex.
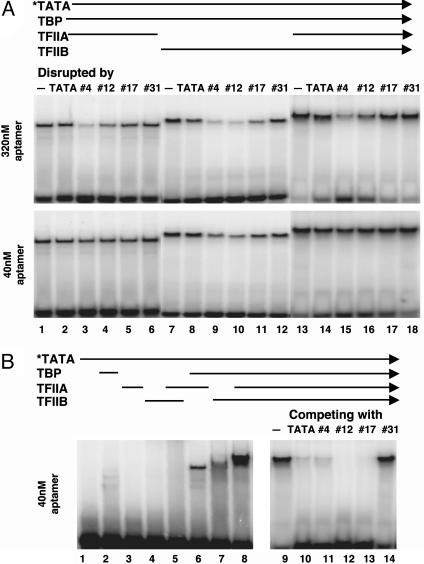
Some higher-order complexes involving the TBP·TATA interaction are significantly more stable than the two-component TBP·TATA complex. For example, the TFIIA·TBP·TATA complex has a half-life of 14 h (24) on a HIS3 promoter whereas the half-life of TBP·TATA complex on the same promoter is 1–2 h (the TFIIB·TBP·TATA complex has a half-life of ≈2 h). If the fast-acting, disruptive effect of aptamers no. 4 and no. 12 is preserved in such higher-order complexes, they could be powerful reagents to study and control the function of TBP both in vitro and in vivo. Moreover, if the higher-order complexes display differential sensitivity to individual aptamers, these results might elucidate the dynamics of TBP-complex formation and disassembly. We probed three higher-order TBP·TATA complexes containing TFIIA, TFIIB, or both. As shown in Fig. 3A, aptamers no. 4 and 12, but not no. 17, were able to disrupt these three-component or four-component complexes. A high concentration of aptamers (320 nM) was required to show significant disruption of TFIIA-containing complexes whereas the TFIIB·TBP·TATA complex was sensitive to a low concentration [40 nM of no. 4 and no. 12 (no. 17 had only weak activity)]. The disruption effect on the TFIIB·TBP·TATA complex was immediate, like that on the TBP·TATA complex, as it could be seen when the “disrupted” reaction mixtures were loaded instantly onto the gel (data not shown). These results further demonstrated that aptamers no. 4 and no. 12, unlike no. 17, were able to gain access to TBP and disrupt a preformed complex when TBP's concave side was occupied by TATA-DNA, even when its second stirrup was occupied by TFIIB. Interestingly, even aptamers no. 4 and no. 12 functioned differently. As shown in Fig. 3A, aptamer no. 4 was more potent than no. 12 in disrupting complexes that contain TFIIA (compare lane 3 with lane 4 and lane 15 with lane 16 in Upper). This is all the more remarkable considering the relatively low levels of stable TBP·aptamer no. 4 complexes observed and the fact that it was weaker than no. 12 and no. 17 in preventing the formation of TFIIA·TFIIB·TBP·TATA complex (compare lane 11 with lanes 12 and 13 in Fig. 3B).
Fig. 3.
Dissecting higher order complexes with aptamers. (A) Different sensitivity of three- and four-component complexes to different TBP-aptamers. All reactions contained 0.5 nM radioactive TATA-DNA (indicated by an asterisk) and 2.5 nM His-yTBP. In addition, 2.5 nM TFIIA (lanes 1–6), 100 nM TFIIB (lanes 7–12), or both (lanes 13–18) were included to form higher order complexes. Two different concentrations of unlabeled RNA or TATA-DNA, as indicated to the left of the gels, were added after a 30-min incubation to disrupt the preformed complexes. The mixtures were incubated for another 30 min before loading onto the gel. (B) Prevention of TFIIA·TFIIB·TBP·TATA complex formation by aptamers. Reactions with same conditions in A except that 40 nM unlabeled RNA or TATA-DNA was added together with the radioactive probe (lanes 9–14). Lanes 1–8 are controls showing the process of TATA-dependent complex formation in the absence of RNA aptamers.
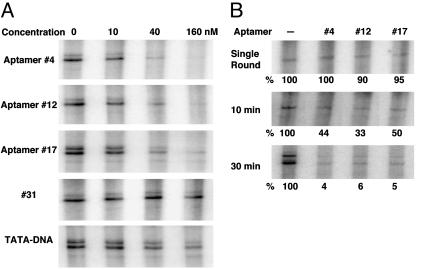
Inhibition of Transcription by Aptamers. An in vitro transcription system, which preserves some key features of the complex milieu of the cell, can be used to test the effect of TBP aptamers on transcription and to evaluate at what step these aptamers act. Because the adenovirus major late promoter has been shown to be TATA-dependent (25), aptamers that are able to bind TBP and prevent TBP binding to the TATA element should block transcription driven by this promoter. We used a yeast transcription system derived from whole cell extracts where total TBP is present at ≈20 nM. Control experiments indicated that the transcription was Pol II-dependent (e.g., sensitive to a low concentration of α-amanitin) and the system was insensitive to high levels of a control RNA (see Materials and Methods). When the RNA aptamers were added to the extract together with the DNA template, we observed inhibition by all three aptamers (no. 4, no. 12, and no. 17) at a concentration as low as 10 nM. As shown in Fig. 4A, with aptamers at 40 nM, which was only twice the approximate TBP concentration in these reactions, the levels of transcription were decreased by 70–85%.
Fig. 4.
Perturbing an in vitro transcription system with TBP-aptamers. A template with a TATA-dependent promoter (2.25 nM) was incubated with yeast whole cell extract (containing ≈20 nM TBP) for 30 min to allow PIC formation. The transcription reaction was started by addition of NTPs and allowed to proceed for 30 min, unless otherwise indicated. (A) Aptamers inhibiting transcription when present during PIC formation. The concentration indicated is that of the aptamers or TATA-DNA included in each reaction. (B) The effect of adding aptamers after PIC formation. Aptamers (160 nM) were added after 30-min incubation of templates with cell extract. To examine single-round transcription (Top), sarkosyl was added to 0.25% final concentration 30 s after transcription initiation. Multiple-round transcription experiments were allowed to proceed for the time indicated. The quantity indicated below each lane represents the percentage of that in the reaction that did not include aptamers.
To pinpoint the step in the transcription process affected by the TBP aptamers, we examined their effects on a single round of transcription after PIC formation. Here, the template was incubated with the cell extract for 30 min to allow PIC formation before adding aptamers. Transcription was initiated 10 min later, with sarkosyl added (to 0.25%) 30 s after the NTPs to allow only a single round of RNA synthesis. As shown in Fig. 4B Top, single-round transcription levels were similar in all reactions, indicating that, once the PIC is formed, the first round of transcription is not affected by the presence of aptamers. In the absence of sarkosyl, transcription reinitiation occurs and continues for at least 30 min in this system (data not shown). Thus, identical reactions performed without sarkosyl treatment allowed us to evaluate the effects of these aptamers on successive rounds of transcription reinitiation.
When aptamers were added after PIC formation and transcription was allowed to proceed for 10 or 30 min, all three aptamers were strong inhibitors and nearly as effective as when added simultaneously with the DNA template (compare 4B with 160 nM reactions in 4A). The transcript level in these inhibited, multiple-round reactions is indistinguishable from that of a single round whereas the uninhibited reactions increased with time. Strikingly, the aptamer that acts as a passive TATA competitor (no. 17) inhibits transcription to similar levels as do aptamers with disruptive activities (no. 4 and no. 12), indicating that TBP's concave (TATA-binding) side was accessible during reinitiation.
Discussion
Most essential cellular functions are actualized by multiprotein complexes. These macromolecular machines undergo multiple steps of assembly and changes in conformation and composition as they carry out their biological functions; and these steps are often regulated by specific interactions with proteins that sense internal or external signals (26). Whereas traditional biochemical and genetic approaches have provided much information about the composition and activities of such complexes, new strategies and methodologies are required to understand the dynamics of these processes at the molecular level and in the context of living cells. In this study, we devised and applied experimental approaches to examine the assembly and function of TBP-containing complexes. What is described here can be easily modified and applied to transcription factors other than TBP or biological processes other than transcription.
Some aptamers we isolated, such as aptamer no. 17, can inhibit TBP binding to TATA element in a manner analogous to some natural TBP inhibitors like NC2 or the NOT complex (reviewed in ref. 6). Others, such as aptamers no. 4 and no. 12, can actively disrupt preformed TBP·TATA complexes, reminiscent of the activity of the protein MOT1 (27). These aptamers can strip TBP off the DNA almost instantaneously. In addition, different TBP·TATA-containing complexes responded differentially to the disruptive activity of aptamers no. 4 and no. 12. One possible explanation for these differential effects is that aptamer no. 4 gains access to the complex at surfaces distinct from the concave portion of TBP and the surfaces of TBP with which TFIIA and TFIIB interact. This initial binding then destabilizes the TBP interaction with TATA-DNA. Aptamer no. 12, in turn, is inhibited in its ability to disrupt complexes containing TFIIA, perhaps because it uses the TFIIA binding surface of TBP as its initial entry point.
In an in vitro functional assay, the TBP aptamers revealed the dynamics of TBP interactions during initiation and reinitiation. This assay system programs proper PIC formation and transcription initiation on the added template DNA, and allows for the partial disassembly and reassembly of the PIC that accompanies each round of Pol II firing and reinitiation. The very effective inhibition seen when aptamers were added before PIC formation indicated that the aptamers could access TBP in the complex environment of a whole cell extract. Moreover, the aptamers were effective inhibitors in multiple round transcription assays, even when added after PIC formation, indicating that the TBP associations are dynamic during reinitiation. The cumulative effect of inhibition during 30 min of transcription was dramatic and was almost the same as seen when aptamers were added to reactions along with template DNA. We propose that the dynamics of the TBP-promoter association allows the TATA-binding surface of TBP to become accessible after the first round of transcription, leading to the inhibition by aptamers observed. Hahn and colleagues have shown that, when Pol II escapes from the promoter, TFIIB and TFIIF are released. In contrast, other components of the PIC, including TBP and TFIIA, as well as upstream activators and the Mediator proteins, remain largely promoter-associated in a structure called the Scaffold, which promotes transcription reinitiation (28). Whereas the TATA-binding surface on TBP becomes accessible during reinitiation (even to our competitively binding aptamer no. 17), TBP may be still associated with the complex through interactions with other proteins, making reinitiation faster than de novo establishment of the PIC (28). Our data provide insight into the dynamics of TBP interactions during transcription reinitiation on a relevant kinetic time scale and suggest that these aptamers could be valuable tools to dissect transcription mechanisms in vivo.
Compared with small-molecule drugs, the size and diversity of macromolecular probes like RNA aptamers may provide a more powerful spectrum of reagents to probe biological processes. Whereas the aptamers described here interfere with a protein–DNA interaction, this approach is not restricted in any way to nucleic acid binding protein complexes because aptamers against many different proteins have been isolated (29). When a nucleic acid binding protein is used as the target of selection, some chemical features of the nucleic acid binding site may be more inviting than other sites for RNA aptamers, resulting in the predominant selection of aptamers homing to this site. To avoid this outcome, the natural nucleic acid ligand, if available, can be used to block this site during selection. We have also developed general methods for targeted destruction of particular sequences in a population that can be used to overcome this problem (22).
As demonstrated previously by us and others, genetically controlled induction of RNA aptamers can provide a means of rapid and/or persistent inactivation of specific proteins, domains, or even discrete surfaces within domains of specific proteins (ref. 15, and reviewed in ref. 16). This result would allow one to assess the primary function of a particular portion of a protein or protein complex in vivo without necessarily disrupting the entire protein or a larger protein complex. When a critical transcription factor or a particular site on a factor is rapidly inactivated, the effects on protein/DNA architecture and function of promoters, regulatory regions, and transcription units can be examined in vivo and in real time. This kind of rationally designed genetic perturbation, when coupled with microarray technologies and dynamic modeling, could be very powerful in the elucidation of regulatory networks in cellular systems (30).
Acknowledgments
We thank Drs. R. Roeder, S. Hahn, and A. Berk for kindly providing strains and plasmids. We thank Drs. J. Fu and M. H. Suh for purified TFIIB, A. Sevilimedu for preparing TFIIA and doing the preliminary tests of aptamer inhibition on TFIIA-containing complexes, and B. Clas for technical assistance. We also thank Dr. Ed Brody and an anonymous reviewer for their critical comments on the manuscript. This work was supported by National Institutes of Health Grant GM40918 (to J.T.L.).
Abbreviations: Pol II, polymerase II; PIC, preinitiation complex; TBP, TATA-binding protein; TF, transcription factor; EMSA, electrophoretic mobility-shift assay.
References
- 1.Buratowski, S., Hahn, S., Guarente, L. & Sharp, P. A. (1989) Cell 56, 549-561. [DOI] [PubMed] [Google Scholar]
- 2.Nikolov, D. B., Hu, S. H., Lin, J., Gasch, A., Hoffmann, A., Horikoshi, M., Chua, N. H., Roeder, R. G. & Burley, S. K. (1992) Nature 360, 40-46. [DOI] [PubMed] [Google Scholar]
- 3.Burley, S. K. & Roeder, R. G. (1996) Annu. Rev. Biochem. 65, 769-799. [DOI] [PubMed] [Google Scholar]
- 4.Parvin, J. D. & Sharp, P. A. (1993) Cell 73, 533-540. [DOI] [PubMed] [Google Scholar]
- 5.Nikolov, D. B., Chen, H., Halay, E. D., Usheva, A. A., Hisatake, K., Lee, D. K., Roeder, R. G. & Burley, S. K. (1995) Nature 377, 119-128. [DOI] [PubMed] [Google Scholar]
- 6.Pugh, B. F. (2000) Gene 255, 1-14. [DOI] [PubMed] [Google Scholar]
- 7.Inostroza, J. A., Mermelstein, F. H., Ha, I., Lane, W. S. & Reinberg, D. (1992) Cell 70, 477-489. [DOI] [PubMed] [Google Scholar]
- 8.Auble, D. T. & Hahn, S. (1993) Genes Dev. 7, 844-856. [DOI] [PubMed] [Google Scholar]
- 9.Hampsey, M. (1998) Microbiol. Mol. Biol. Rev. 62, 465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang, H., Sun, X., Reinberg, D. & Ebright, R. H. (1996) Proc. Natl. Acad. Sci. USA 93, 1119-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burley, S. K. & Roeder, R. G. (1998) Cell 94, 551-553. [DOI] [PubMed] [Google Scholar]
- 12.Bellorini, M., Moncollin, V., D'Incalci, M., Mongelli, N. & Mantovani, R. (1995) Nucleic Acids Res. 23, 1657-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuerk, C. & Gold, L. (1990) Science 249, 505-510. [DOI] [PubMed] [Google Scholar]
- 14.Ellington, A. D. & Szostak, J. W. (1990) Nature 346, 818-822. [DOI] [PubMed] [Google Scholar]
- 15.Shi, H., Hoffman, B. E. & Lis, J. T. (1999) Proc. Natl. Acad. Sci. USA 96, 10033-10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Famulok, M., Blind, M. & Mayer, G. (2001) Chem. Biol. 8, 931-939. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann, A. & Roeder, R. G. (1991) Nucleic Acids Res. 19, 6337-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi, H., Hoffman, B. E. & Lis, J. T. (1997) Mol. Cell. Biol. 17, 1649-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultz, M. C., Choe, S. Y. & Reeder, R. H. (1991) Proc. Natl. Acad. Sci. USA 88, 1004-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woontner, M., Wade, P. A., Bonner, J. & Jaehning, J. A. (1991) Mol. Cell. Biol. 11, 4555-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawley, D. K. & Roeder, R. G. (1987) J. Biol. Chem. 262, 3452-3461. [PubMed] [Google Scholar]
- 22.Shi, H., Fan, X., Ni, Z. & Lis, J. T. (2002) RNA 8, 1461-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoopes, B. C., LeBlanc, J. F. & Hawley, D. K. (1992) J. Biol. Chem. 267, 11539-11547. [PubMed] [Google Scholar]
- 24.Stewart, J. J. & Stargell, L. A. (2001) J. Biol. Chem. 276, 30078-30084. [DOI] [PubMed] [Google Scholar]
- 25.Workman, J. L. & Roeder, R. G. (1987) Cell 51, 613-622. [DOI] [PubMed] [Google Scholar]
- 26.Alberts, B. (1998) Cell 92, 291-294. [DOI] [PubMed] [Google Scholar]
- 27.Auble, D. T., Hansen, K. E., Mueller, C. G., Lane, W. S., Thorner, J. & Hahn, S. (1994) Genes Dev. 8, 1920-1934. [DOI] [PubMed] [Google Scholar]
- 28.Yudkovsky, N., Ranish, J. A. & Hahn, S. (2000) Nature 408, 225-229. [DOI] [PubMed] [Google Scholar]
- 29.Gold, L., Polisky, B., Uhlenbeck, O. & Yarus, M. (1995) Annu. Rev. Biochem. 64, 763-797. [DOI] [PubMed] [Google Scholar]
- 30.Tegner, J., Yeung, M. K., Hasty, J. & Collins, J. J. (2003) Proc. Natl. Acad. Sci. USA 100, 5944-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]