Abstract
The nucleoid-associated protein HU is one of the most abundant proteins in Escherichia coli and has been suggested to play an important role in bacterial nucleoid organization and regulation. Although the regulatory aspects of HU have been firmly established, much less is understood about the role of HU in shaping the bacterial nucleoid. In both functions (local) modulation of DNA architecture seems an essential feature, but information on the mechanical properties of this type of sequence-independent nucleoprotein complex is scarce. In this study we used magnetic tweezers and atomic force microscopy to quantify HU-induced DNA bending and condensation. Both techniques revealed that HU can have two opposing mechanical effects depending on the protein concentration. At concentrations <100 nM, individual HU dimers induce very flexible bends in DNA that are responsible for DNA compaction up to 50%. At higher HU concentrations, a rigid nucleoprotein filament is formed in which HU appears to arrange helically around the DNA without inducing significant condensation.
The nucleoid-associated protein HU is one of the most abundant proteins in Escherichia coli with up to 60,000 copies per cell (1). In E. coli, two types of subunit are found that assemble predominantly into heterodimers. Structural studies have shown that HU, similar to its homologue integration host factor (IHF), consists of a compact body of several intertwined α-helices from which two β-ribbon arms protrude (2, 3) that are inserted into the minor groove of double-stranded (ds) DNA after binding. As a relatively small protein (18 kDa), HU is known to bind sequence-independently to both dsDNA and single-stranded (ss) DNA. Estimates of Kd for undistorted dsDNA vary from 200 to 2,500 nM depending on the ionic strength and experimental method (4, 5). A 10- to 100-fold-higher affinity is found for bent, nicked, or cruciform DNA (4, 6). Despite the vast amount of biochemical data available on the binding properties of HU, its function in vivo is the subject of ongoing debate (7).
Two generic, nonexclusive functions have been proposed for HU. First, its high abundance as a basic protein and its sequence-independent DNA binding suggest a general role in prokaryotic DNA condensation. Indeed, in one of the early articles by Rouviere-Yaniv and Yaniv (8), through the use of electron microscopy the global structure of DNA incubated with HU was found to resemble eukaryotic nucleosomes. Based on this similarity, HU and other small architectural proteins were often referred to as histone-like proteins. Similar to histones, HU is able to induce and constrain DNA supercoiling (9). However, the role of HU in prokaryotic DNA condensation has been questioned recently, and evidence has been put forward that could point at an opposed function (7). Second, HU acts as a regulator in a large number of cellular processes (10–12). In most cases this regulatory function can be attributed to its ability to actively bend DNA or stabilize bent DNA.
The introduction of bends, as demonstrated in ring-closure studies (13, 14), may by itself help to condense DNA in the bacterial nucleoid. However, quantification of this HU-induced DNA bend is difficult, primarily because of the lack of sequence specificity, which is required for bulk, ensemble-averaged measurements. Studies with fluorescence resonance energy transfer (15), chemical nucleases (16), and gel-shift assays (16, 17) have used intrinsically bent DNA substrates, resolving only additional HU-induced DNA bending or specific nicks. More detailed structural information comes from HU–DNA cocrystals (18). X-ray structures of HU–DNA complexes reveal bend angles between 105° and 140°. However, in these structures two extra nonduplexed bases were incorporated, making an independent assessment of HU-induced bending impossible.
In both proposed roles of HU the mechanical properties of the HU–DNA complex are of paramount importance for its function. Single-molecule techniques are not restricted to specific structures in DNA and thus are well suited to address such sequence-independent low-affinity interactions. In this study we use both magnetic tweezers (MT) and atomic force microscopy (AFM) to characterize the mechanical properties of HU–DNA complexes at the single-molecule level.
Materials and Methods
HU Protein. Heteromeric HU from E. coli was purified as described (19). All reactions were done in 60 mM KCl/20 mM Hepes, pH 7.9 (buffer I).
DNA Constructs. For the MT experiments, pSFV1 (Invitrogen) was cleaved with SpeI and BamHI, resulting in a 10-kb linear fragment, which was ligated to two 700-bp PCR fragments, each containing ≈180 biotin- or digoxigenin-modified UTP bases (Roche Diagnostics). Using MT we selected molecules that were not constrained in rotation, i.e., contained a small number of nicks, or were incompletely ligated. Thus, anticipated HU-induced supercoiling does not build up in plectonemes, which can be confused with HU-induced reduction of the DNA tether length. We assume the number of nicks to be small, because a significant fraction of the molecules was nick-free.
For AFM experiments we used a 2,100-bp PCR fragment and commercially available 1,000- and 500-bp EcoRI restriction fragments (Eurogentec, Brussels).
MT. MT were constructed based on the system described by Strick et al. (20). In the optical axis of an inverted microscope (Zeiss Axiovert 200M), a pair of magnets (Goudsmit, Waalre, The Netherlands) was mounted on a translocation stage (Physik Instrumente, Karlsruhe, Germany). Video microscopy was implemented by using a 60-Hz charge-coupled device camera (PULNiX TM-6710, Mannheim, Germany) and a frame grabber (National Instruments, Austin, TX), and real-time image processing was performed by using labview (National Instruments).
The applied force was calculated by quantifying thermal motion of the DNA-tethered bead and substituting it into equipartition theorem (20). By using image processing, 5-nm position accuracy of the bead was obtained in three dimensions. To exclude thermal drift, all positions were measured relative to a reference bead fixed to the bottom of the cell.
Buffers were exchanged in a closed flow cell connected to a syringe pump. The flow cell was constructed from two layers of parafilm with a central cavity sandwiched between cover slides, with two liquid access openings in the top slide. The bottom slide was coated with 1% polystyrene in toluene before incubation for 2 h with 5 × 105 3.2-μm-diameter tosyl beads (Bangs Laboratories, Carmel, IN), 12 h with 100 ng/μl anti-digoxigenin (Roche Diagnostics) in 150 mM KCl/10 mM Tris, pH 7.9/1 mM EDTA (buffer II), and subsequently 12 h with 100 ng/μl sonicated salmon sperm DNA (Sigma–Aldrich) or 100 ng/μl BSA (Sigma–Aldrich).
One microliter of paramagnetic 2.8-μm streptavidin-coated beads (Dynal, Oslo) was washed twice and resuspended in 9 μl of buffer II before incubation with 1 μl of 1 ng/μl DNA construct for 30 min. The reaction mixture was diluted 10 times in 100 ng/μl BSA in buffer II and inserted in the flow cell. After 15 min, the cell was placed in the MT and washed thoroughly with buffer I before the bead–DNA construct was ready for measurements.
AFM. Muscovite mica was cleaved, pretreated with 0.05% polylysine (Sigma–Aldrich), and dried under a flow of nitrogen. After 15 min of incubation with HU, 2.5 μl of 1 ng/μl DNA was deposited onto the polylysine-coated mica. The sample then was rinsed with MilliQ water, quickly dried, and immediately imaged with a Nanoscope IV atomic force microscope (Veeco Instruments, Santa Barbara, CA). Images were acquired in tapping mode in air, with a scan range of 2 μm at 512 × 512 pixels by using silicon tips OMCL-AC160TS-W2 (Olympus, Tokyo). Image processing and analysis were done by using idl (RSI, Boulder, CO). DNA contours were traced semiautomatically (21). End points were assigned to positions at which the DNA trace was reduced to half its height.
Results
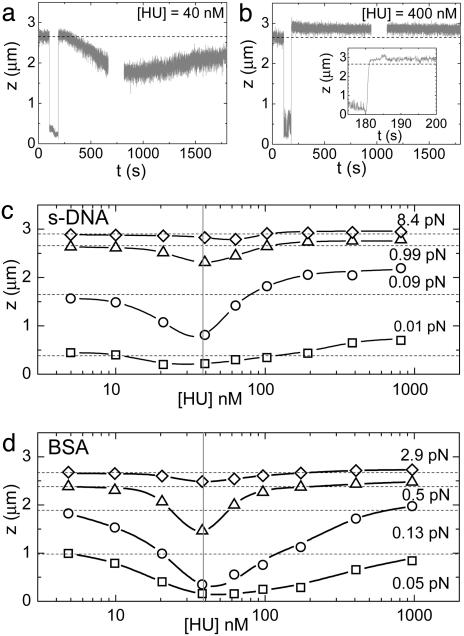
MT Experiments. In the MT setup, a paramagnetic bead was tethered to a glass slide with a single 10-kb DNA molecule. At 1 pN of force, the end-to-end length of the DNA tether was 2.65 μm. After introduction of 40 nM HU, the tether length decreased to 1.75 μm after ≈800 s (Fig. 1a). Subsequently, a slow increase of the end-to-end length was observed, which we attribute to depletion of HU by binding to the walls of the flow cell; successive refreshment of HU solution resulted in saturation of this depletion. The surface passivation of the liquid cell with sonicated salmon sperm DNA would be expected to enhance this HU depletion. However, we observed a very similar HU-concentration dependence of the mechanic properties of the HU–DNA tether for BSA-passivated flow cells. For all measurements we waited until a stable equilibrium was reached, in this case at 2.25 μm after 30 min. Thus, introduction of 40 nM HU resulted in a 15% decrease in tether length.
Fig. 1.
HU-induced compaction and elongation of a single DNA molecule stretched in MT. (a) During exchange of buffer with protein solution (40 nM HU), the bead is displaced by the liquid drag, leading to a low z value between t = 100 and 180 s. Subsequently, a mild compaction of the molecule is observed relative to the length of bare DNA (dashed line) after a long equilibration. In the gap of data points between t = 650 and 800 s no data were recorded. (b) The same experiment as described for a repeated at 400 nM HU. A significant elongation of the DNA tether is observed. (Inset) Directly after the flow is stopped, this elongation is reached. (c) Equilibrium end-to-end length of the nucleoprotein filament as a function of HU concentration and force. The DNA tether length at [HU] = 0 nM is plotted in the dashed lines for all forces. Maximum compaction is observed at 40 nM HU, indicated by the gray vertical line; elongation levels off at 900 nM HU. s-DNA, sonicated salmon sperm DNA. (d) Experiment described in c, repeated with BSA instead of salmon sperm DNA passivation; good agreement was found.
Next to a change in end-to-end length, the addition of 40 nM HU induced a gradual increase in fluctuations of the z position of the bead, as can be seen in Fig. 1a by comparing the noise at t = 0 s with that at t = 1,500 s. In equilibrium, the rms amplitude of the fluctuations increased by 61% relative to bare DNA. Thermal fluctuations in the z direction are proportional to the derivative of the stiffness of the tether (20), and thus this is a direct indication of an increased flexibility of the nucleoprotein complex.
After washing the cell with high-salt buffer, the characteristics of bare DNA were recovered fully. Subsequent addition of 400 nM HU protein, a concentration closer to physiological concentrations, surprisingly resulted in an increase in the end-to-end length by 200 nm or 8%, as shown in Fig. 1b. This elongation was reached immediately after stopping the inflow of HU-containing buffer (Fig. 1b Inset), i.e., within 1 s. The z fluctuations decreased to 80% of the rms amplitude measured before introduction of HU, indicating increased stiffness of the tether.
A gradual transition of the end-to-end length was observed after increasing the HU concentration, as shown in Fig. 1 c and d. Up to 40 nM the end-to-end length decreased as a function of HU concentration. At higher concentrations it increased up to a plateau value near 800 nM. Very good agreement of these trends was found between surface passivation with BSA and with salmon sperm DNA, excluding significant effects of HU depletion by the surface. The end-to-end length modulation was observed over the entire force regime that we studied but was most pronounced at 0.1 pN.
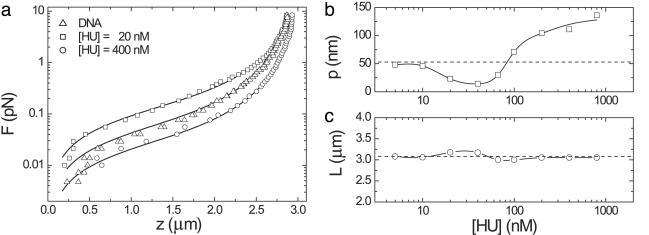
We did not see any indication of the proteins' being driven off the DNA by tension. Based on the slow equilibration time at low HU concentrations, shown in Fig. 1a, we expected to be able to observe a transient response after a rapid decrease in applied force, but this was not observed. In Fig. 2a, force-extension curves are plotted for bare DNA and for low and high HU concentrations. No hysteresis was observed in force-extension measurement cycles. Over the whole force regime, the tether length is reduced at low HU concentration and increased at high HU concentration.
Fig. 2.
The effect of HU on DNA elasticity. (a) Force-extension curves for bare DNA and two representative HU concentrations. Solid lines represent fits to a modified WLC model. (b and c) Shown are the persistence length (b) and contour length (c) resulting from a WLC fit as a function of the HU concentration. Dashed lines represent bare DNA. Elongation and compaction can be attributed to changes in flexibility rather than changes in contour length.
The elastic response of bare DNA can be described by a worm-like chain (WLC) model with two parameters, the persistence length p and the contour length L (22),
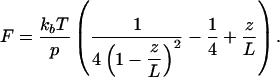 |
[1] |
Bouchiat et al. (23) showed that for intermediate forces, between the entropic and the elastic regimes, a numerical correction improves this fit. Using this corrected model, we obtain p = 55 nm and L = 3.1 μm for bare DNA, in good agreement with its crystallographic length and previously reported values of the persistence length of DNA (24). For the nucleoprotein filament, which is likely not a continuous, homogeneous polymer, it is not a priori clear whether the force-extension curves can be described with a WLC model. However, fitting the same WLC model to the data resulted in good fits (R > 0.995) for all curves. Thus, we obtained an effective persistence length that describes the global stiffness of the filament and can be compared with the persistence length of bare DNA. In Fig. 2 b and c, the fitted parameters are plotted as a function of the HU concentration. Whereas the contour length L of the nucleoprotein filament marginally deviates from the contour length of bare DNA, the effective persistence length p decreases to 15 nm at 40 nM HU to increase to 146 nm at 800 nM HU. Thus, a significant modulation of the tether stiffness rather than a change in contour length is responsible for both the moderate compaction and the small extension observed at low and high concentrations of HU protein, respectively.
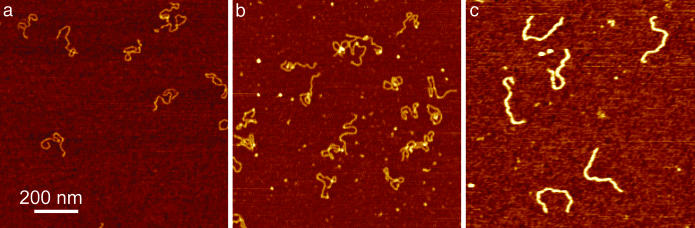
AFM Experiments. Previous AFM work reported rigidification of circular DNA after incubation with HU (7). However, HU molecules on DNA were not resolved, which we attribute to HU dissociation from DNA when deposited on Mg2+-treated mica. We observed reduced DNA binding to mica when incubated with HU. As an alternative approach, we used polylysine-treated mica for sample preparation (25). Based on the strong attractive interaction between DNA and the polylysine-modified substrate, we expected instant immobilization of the nucleoprotein filament, referred to as “kinetic trapping” by Rivetti et al. (26). The strong immobilization by polylysine-coated mica also relieves the requirement to use low-salt MgCl2 buffer for deposition, allowing deposition from the same buffer used in the MT experiments.
By using this alternative immobilization strategy at 18 nM HU, individual HU complexes were visible on DNA (as shown in Fig. 3b) despite the relatively small size of the protein. When the HU concentration is increased to 900 nM (shown in Fig. 3c), the nucleoprotein complexes show entirely different features compared with the low HU concentration complexes shown in Fig. 3b. Filaments with a significantly increased height are formed that appear more elongated compared with bare DNA, reflecting a more rigid structure.
Fig. 3.
AFM imaging of HU–DNA complexes deposited on polylysine-modified mica. (a) Bare 1,000-bp DNA appears more compact than when using MgCl2 for deposition because of the stronger DNA–surface interaction. (b) DNA (1,000 bp) incubated with 18 nM HU (1 dimer per 92 bp) appears somewhat condensed, compared with bare DNA. (c) When incubated with 900 nM HU (1 dimer per 1.8 bp), thick, rigid filaments are formed.
For long polymers, the conformation of the polymer on a substrate resembles 2D projection of the 3D solution structure in the case of kinetic trapping, and thus a somewhat condensed conformation is observed rather than the more elongated, energetically most favorable conformation on a 2D surface. To quantify the flexibility of the nucleoprotein complex we used the relation between the persistence length p, the contour length L, and end-to-end distance R for kinetic trapping derived by Rivetti et al. (26),
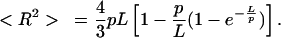 |
[2] |
The coordinates of ≈50 2,100-bp DNA molecules at 0, 18, and 900 nM HU were determined in a semiautomatic fashion. For 900 nM HU (1 dimer per 1.8 bp) the contour length of the nucleoprotein filaments was found to be 720 ± 30 nm, compared with 714 nm for the crystallographic length of bare DNA. For 18 nM HU (1 dimer per 92 bp) most filaments were heavily folded, making it impossible to determine their exact conformation and thus their contour length. For these molecules we assumed the contour length to be the same as the contour length of bare DNA. This assumption was validated by measurement of a few elongated instances of these nucleoprotein complexes and agrees with our MT observations. Substituting the experimentally obtained values for end-to-end length and contour length into Eq. 2, we find that p = 46, 24, and 187 nm for 0, 18, and 900 nM HU, respectively. Thus, the persistence length found by tracing nucleoprotein filament conformations observed with AFM quantitatively matches the values obtained with MT experiments. From the good agreement of these data we conclude that the surface deposition in the AFM experiment does not affect the nucleoprotein filaments and that the conformation and stoichiometry observed with AFM correctly represent the structures formed in bulk.
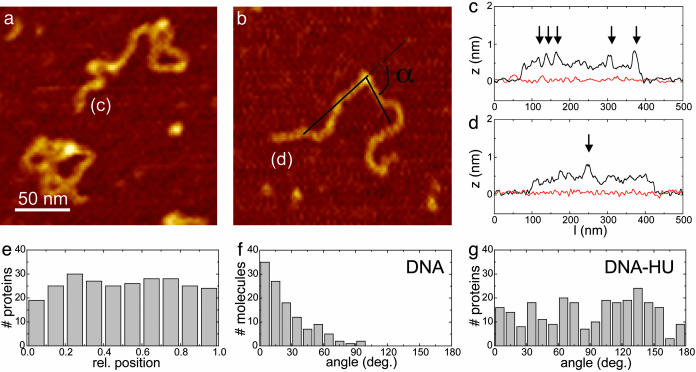
For a more detailed analysis of the AFM data, we used shorter DNA fragments to prevent structures from being obscured by overlap of different parts of the nucleoprotein complex. In Fig. 4 a and b, three 1,000-bp DNA molecules incubated at 18 nM HU (1 dimer per 92 bp) are shown. Two corresponding height traces along the molecules are plotted in Fig. 4 c and d. The small features, which were absent when no HU was added, have a height of 0.75 nm, compared with 0.5 nm of bare DNA. Based on their small and relatively uniform size, we identify these globular features as single HU dimers. The sequence-independent nature of the HU–DNA interaction is reflected in the uniform position distribution of HU proteins along the DNA (shown in Fig. 4e).
Fig. 4.
HU-induced DNA bending. (a and b) Zooms of 1,000-bp dsDNA incubated with 18 nM HU (1 dimer per 92 bp) showing individual proteins. (c and d) Height traces of DNA molecules shown in a and b. HU dimers, indicated by arrows, are 0.75 nm high, compared with 0.5 nm for DNA. A reference trace of the substrate is shown in red. (e) Position distribution of ≈290 HU molecules; no preferential binding is observed. (f) Angle distribution of bare DNA, as shown in b. As expected, no intrinsic curvature is found. (g) HU-induced bending-angle distribution. A large spread in angles is found, indicating high flexibility of the complex.
From the distribution of conformations in the AFM images it is now possible to analyze further the anticipated HU-induced flexibility of DNA. Good agreement for various sequence-specific proteins has been reported between bending-angle measurements obtained with AFM and by other techniques such as crystal structures, gel shifts, etc. (27). Next to the average protein-induced angle, the distribution of angles, representing the local flexibility of the complex, can be quantified uniquely by using this AFM-imaging-based method. To exclude the possibility that pinning of DNA to the polylysine-treated substrate generated the broad distribution of bending angles, we compared both deposition procedures with a control protein, yielding good agreement for the width and position of the observed bending angle (see Fig. 6, which is published as supporting information on the PNAS web site).
In Fig. 4f, the bending angle of bare DNA is plotted, and no intrinsic curvature is observed, as expected. The width of the angle distribution reflects the flexibility, and in the case of DNA it is inversely proportional to the persistence length of DNA. The bending angle of HU–DNA complexes (shown in Fig. 4g) clearly deviates from the distribution found for bare DNA. It exhibits no preferential angle, but a rather broad distribution is found, indicating that the flexibility of these HU–DNA complexes is significantly higher than that of dsDNA.
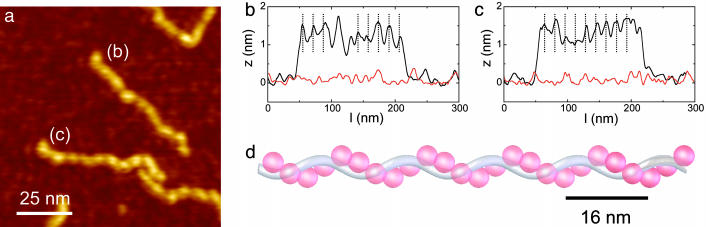
A high-resolution image of rigid filaments that are formed on 500-bp DNA at 1 dimer per 1.8 bp is shown in Fig. 5a. The nucleoprotein filaments appear significantly higher (1.5 nm) than the individual HU dimers on DNA (0.75 nm) that form at low HU concentrations. Within the filaments, a quite regular, 16-nm periodic structure is observed, as indicated by the corresponding height traces in Fig. 5 b and c. The increased height in combination with a negligible change in length of the filaments implies that HU wraps around DNA in a rather regular fashion.
Fig. 5.
Filament formation by HU. (a) Zoom of 500-bp dsDNA incubated with 900 nM HU (1 dimer per 1.8 bp) showing thick, rigid filaments. (b and c) Height traces over the DNA molecules shown in a. Nucleoprotein filaments appear as regular 16-nm-spaced periodic structures, with a height of 1.7 nm. Dotted lines represent 16-nm intervals. A reference trace of the substrate is shown in red. (d) Low-resolution structure of HU–DNA filaments based on 1 HU dimer (red) per 9 bp. The HU-induced increase in supercoiling, as reported in the literature, is realized by introducing an extra helicity with a 16-nm pitch as observed with AFM.
Discussion and Conclusions
In this single-molecule study of the mechanical parameters of HU nucleoprotein complexes we show that HU not only introduces sharp, flexible bends but that it also is capable of forming rigid nucleoprotein filaments at higher HU concentrations. The high flexibility of the HU–DNA complex that we found from analyzing the bending angle in AFM images concurs with suggestions based on the large variation found in three related HU–DNA crystal structures (18), which had bending angles between 105° and 139°. Although almost half of the 240 complexes that we measured fall in this range, we did not observe a clear preferential bending angle. This result may reflect either different stable conformations that contribute to the angle distribution or very flexible hinges formed by the HU–DNA complex, after which thermal fluctuations inflict a high diversity in conformations. The increased thermal fluctuations of the nucleoprotein filament length in the MT that we observed in this concentration regime confirm such a highly flexible hinge. As a consequence, HU not only stabilizes bends but also may affect kinetics, effectively increasing the rate of bend formation.
Ali et al. (28) recently reported force-extension curves of the structural homologue IHF measured with MT. For sequence-independent interactions they observed a 30% compaction of single DNA tethers incubated with IHF, similar to the compaction observed for the low-concentration regime of HU. At high force the compaction decreases for both nucleoprotein filaments, which they attributed to proteins being pulled off the DNA by tension. We did not see indications of such dissociation and propose that the high flexibility of the HU-induced bends can accommodate force-induced elongation of the filament. Indeed, Yan and Marko (29) argued that force-induced dissociation is less likely in flexible protein–DNA complexes than when rigid bends are formed. Our fits to a simple WLC in the low HU concentration regime are consistent with this more advanced model, which explicitly takes protein-induced flexibility into account.
Contrary to our MT experiments, our AFM measurements did not reveal a gradual transition as the HU concentration was increased. Between 600 and 900 nM, the observed structures changed from highly condensed, thin, and flexible DNA–protein complexes to thick, rigid filaments. The formation of these nucleoprotein filaments seemed highly cooperative, because partially coated or bare DNA was never observed in combination with thick filaments under these buffer conditions. However, at low salt concentration (data not shown), we did observe sparsely HU-decorated DNA, partial and complete filaments within one sample. The dependence on the ionic strength indicates an electrostatic origin of the high degree of cooperativity of filament formation.
The apparent inconsistency in the HU-concentration dependence between the AFM and MT data cannot be attributed to HU depletion to the surface of the flow cell, because it does not depend on the chosen passivation layer. The stretching of DNA in the MT, however, may facilitate (partial) filament formation, effectively shifting the equilibrium to filament formation at lower concentrations. Likewise, it has been reported that RecA binds more strongly to stretched DNA rather than unstretched DNA (30). In Fig. 1 c and d it seems that the concentration at which the plateau is reached shifts to higher HU concentration at lower forces, which is consistent with this explanation.
Dame and Goosen (7) argue in a recent review that glutaraldehyde cross-linking, necessary for sample preparation, induced the nucleosome-like structures observed in early electron microscopy experiments (8). In the same work, however, 80% of the nucleoprotein complexes were reported to be thick, rigid filaments (very much like the ones we observed) when no fixation was used. These structures were discarded, because they did not seem consistent with reported HU-induced supercoiling. In the current study, fixation artifacts can be excluded, because we did not use fixation agents. Moreover, we found quantitatively similar mechanical parameters for both air-dried, surface-immobilized nucleoprotein filaments and filaments tethered in an aqueous environment. By using two complementary techniques on the same system we were able to relate mechanical properties sensitively measured with MT with high-resolution visualization of the complex with AFM.
Based on the 16-nm periodicity observed in AFM images, we propose that HU induces a superhelical filament in which HU and DNA spiral around each other as shown in Fig. 5d. Close contact between neighboring HU dimers 9 bp apart (9) can explain the apparent cooperativity that was observed. Stabilizing dimer–dimer interactions also would explain the paradox between extra flexibility observed at low HU concentrations and rigidification observed at higher HU concentrations. Furthermore, a superhelical filament is consistent with reported HU-induced supercoiling (9, 31) while only marginally changing the filament contour length.
Despite their strong homology, HU behaves quite differently from its structural homologue IHF at concentrations >100 nM. Contrary to our results with HU, no extension or rigidification was observed for IHF (28). Large differences between HU–DNA and IHF–DNA complexes have been reported in gelretardation assays (32). Where HU has a binding site of ≈9 bp, only one IHF binds nonspecifically to a 35-bp strand of DNA (32), even at high concentrations. This indicates that IHF may be unable to interact with neighboring IHF proteins and thus may be unable to form filaments as HU can. In fact, at high concentrations, the mechanical properties of HU–DNA complexes rather resemble the increased bending rigidity recently measured on histone-like nucleoid structuring protein (H-NS) (33). However, contrary to HU, H-NS dimers have two DNA-binding sites that have been proposed to stabilize large DNA loops, as observed in various microscopy studies. The physiological significance of the found rigidification effect was questioned, because the single-molecule assay drives the nucleoprotein complex from loop to filament formation because of a high protein/DNA ratio that can occur in single-molecule experiments (34). The good agreement between AFM and MT data confirms that filament formation is a general structure found in HU–DNA complexes.
How does this view on HU–DNA complexes apply to the in vivo situation? Azam et al. (1) measured the HU content per E. coli cell to vary between 15,000 and 60,000 depending on its growth stage, which corresponds to HU concentrations in the micromolar range and thus to rigid filament formation based on our experiments. However, in vivo, only 1 dimer per ≈100 bp is present, ruling out complete coating of the bacterial chromosome. The abundant presence of at least 10 other nucleoid-associated proteins further complicates this issue. Until now, most experimental evidence pointed at a regulatory role of the highly flexible hinges created by individual HU proteins. HU–DNA filaments, which may have an antagonizing effect on DNA condensation, have not been reported before, but the cooperativity of filament formation together with the high HU concentration found in E. coli makes it likely that these structures also act in the bacterial nucleoid. Because the amount of HU is not sufficient to cover the whole length of the bacterial chromosome, filament formation in vivo is anticipated to occur only locally and next to DNA bending. As a consequence on the scale of the nucleoid, the two effects could be balanced such that the net effect of HU on nucleoid volume is limited.
It has been argued that the nonspecific regulatory role of HU in transcription could be related to locally antagonizing the condensatory effects of H-NS (see ref. 7 and references therein). It is not clear a priori whether this would require the bending activity of single HU proteins or the local formation of a filament. It seems likely that the primary antagonistic effect depends on competition for the same type of preferential binding sites (35). We speculate that the subsequent cooperative formation of an HU–DNA filament effectively renders certain regions on the bacterial chromosome (in particular genes sensitive to repression by H-NS) transcriptionally more active. This effect could be at the level of transcription initiation by “opening up” H-NS-condensed promotor regions or during elongation, at which long tracts of extensively H-NS-bridged DNA (36) could form a stronger barrier to transcription by the progressing RNA polymerase instead of an HU filament (37).
In conclusion, we have shown that two structurally and mechanically quite different HU–DNA nucleoprotein filaments can be formed depending on the HU concentration. The insight on the structure and mechanics of HU nucleoprotein filaments will help to gain a better understanding of the role of protein-mediated structuring of the bacterial nucleoid.
Supplementary Material
Acknowledgments
We thank Thijn van der Heijden and Peter Veenhuizen for technical assistance and Keith Firman (School of Biological Sciences, University of Portsmouth, Portsmouth, U.K.) for kindly providing the methyltransferase used in the validation of the bending-angle analysis (see legend for Fig. 6).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IHF, integration host factor; ds, double-stranded; ss, single-stranded; MT, magnetic tweezers; AFM, atomic force microscopy; WLC, worm-like chain; H-NS, histone-like nucleoid structuring protein.
References
- 1.Azam, T. A., Iwata, A., Nishimura, A., Ueda, S. & Ishihama, A. (1999) J. Bacteriol. 181, 6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rice, P. A., Yang, S., Mizuuchi, K. & Nash, H. A. (1996) Cell 87, 1295-1306. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka, I., Appelt, K., Dijk, J., White, S. W. & Wilson, K. S. (1984) Nature 310, 376-381. [DOI] [PubMed] [Google Scholar]
- 4.Pinson, V., Takahashi, M. & Rouviere-Yaniv, J. (1999) J. Mol. Biol. 287, 485-497. [DOI] [PubMed] [Google Scholar]
- 5.Wojtuszewski, K., Hawkins, M. E., Cole, J. L. & Mukerji, I. (2001) Biochemistry 40, 2588-2598. [DOI] [PubMed] [Google Scholar]
- 6.Castaing, B., Zelwer, C., Laval, J. & Boiteux, S. (1995) J. Biol. Chem. 270, 10291-10296. [DOI] [PubMed] [Google Scholar]
- 7.Dame, R. T. & Goosen, N. (2002) FEBS Lett. 529, 151-156. [DOI] [PubMed] [Google Scholar]
- 8.Rouviere-Yaniv, J. & Yaniv, M. (1979) Cell 17, 265-274. [DOI] [PubMed] [Google Scholar]
- 9.Broyles, S. S. & Pettijohn, D. E. (1986) J. Mol. Biol. 187, 47-60. [DOI] [PubMed] [Google Scholar]
- 10.Aki, T., Choy, H. E. & Adhya, S. (1996) Genes Cells 1, 179-188. [DOI] [PubMed] [Google Scholar]
- 11.Haykinson, M. J. & Johnson, R. C. (1993) EMBO J. 12, 2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavoie, B. D. & Chaconas, G. (1993) Genes Dev. 7, 2510-2519. [DOI] [PubMed] [Google Scholar]
- 13.Hodges-Garcia, Y., Hagerman, P. J. & Pettijohn, D. E. (1989) J. Biol. Chem. 264, 14621-14623. [PubMed] [Google Scholar]
- 14.Paull, T. T., Haykinson, M. J. & Johnson, R. C. (1993) Genes Dev. 7, 1521-1534. [DOI] [PubMed] [Google Scholar]
- 15.Wojtuszewski, K. & Mukerji, I. (2003) Biochemistry 42, 3096-3104. [DOI] [PubMed] [Google Scholar]
- 16.Kamashev, D., Balandina, A. & Rouviere-Yaniv, J. (1999) EMBO J. 18, 5434-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider, G. J., Sayre, M. H. & Geiduschek, E. P. (1991) J. Mol. Biol. 221, 777-794. [DOI] [PubMed] [Google Scholar]
- 18.Swinger, K. K., Lemberg, K. M., Zhang, Y. & Rice, P. A. (2003) EMBO J. 22, 3749-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dame, R. T. (2003) Ph.D. thesis (Leiden Univ., Leiden, The Netherlands).
- 20.Strick, T. R., Allemand, J. F., Bensimon, D. & Croquette, V. (1998) Biophys. J. 74, 2016-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Noort, J., Van Der Heijden, T., De Jager, M., Wyman, C., Kanaar, R. & Dekker, C. (2003) Proc. Natl. Acad. Sci. USA 100, 7581-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marko, J. F. & Siggia, E. D. (1995) Macromolecules 28, 8759-8770. [Google Scholar]
- 23.Bouchiat, C., Wang, M. D., Allemand, J. F., Strick, T., Block, S. M. & Croquette, V. (1999) Biophys. J. 76, 409-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bustamante, C., Marko, J. F., Siggia, E. D. & Smith, S. (1994) Science 265, 1599-1600. [DOI] [PubMed] [Google Scholar]
- 25.Brewer, L. R., Friddle, R., Noy, A., Baldwin, E., Martin, S. S., Corzett, M., Balhorn, R. & Baskin, R. J. (2003) Biophys. J. 85, 2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivetti, C., Guthold, M. & Bustamante, C. (1996) J. Mol. Biol. 264, 919-932. [DOI] [PubMed] [Google Scholar]
- 27.Bustamante, C. & Rivetti, C. (1996) Annu. Rev. Biophys. Biomol. Struct. 25, 395-429. [DOI] [PubMed] [Google Scholar]
- 28.Ali, B. M. J., Amit, R., Braslavsky, I., Oppenheim, A. B., Gileadi, O. & Stavans, J. (2001) Proc. Natl. Acad. Sci. USA 98, 10658-10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan, J. & Marko, J. F. (2003) Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Top. 68, 011905. [Google Scholar]
- 30.Leger, J. F., Robert, J., Bourdieu, L., Chatenay, D. & Marko, J. F. (1998) Proc. Natl. Acad. Sci. USA 95, 12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka, H., Yasuzawa, K., Kohno, K., Goshima, N., Kano, Y., Saiki, T. & Imamoto, F. (1995) Mol. Gen. Genet. 248, 518-526, and erratum (1995) 249, 570. [DOI] [PubMed] [Google Scholar]
- 32.Bonnefoy, E. & Rouviere-Yaniv, J. (1991) EMBO J. 10, 687-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amit, R., Oppenheim, A. B. & Stavans, J. (2003) Biophys. J. 84, 2467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dame, R. T. & Wuite, G. J. L. (2003) Biophys. J. 85, 4146-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuber, F., Kotlarz, D., Rimsky, S. & Buc, H. (1994) Mol. Microbiol. 12, 231-240. [DOI] [PubMed] [Google Scholar]
- 36.Dame, R. T., Wyman, C. & Goosen, N. (2000) Nucleic Acids Res. 28, 3504-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morales, P., Rouviere-Yaniv, J. & Dreyfus, M. (2002) J. Bacteriol. 184, 1565-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.