Abstract
We recently reported the development of nonhomologous random recombination (NRR) as a method for nucleic acid diversification and applied NRR to the evolution of DNA aptamers. Here, we describe a modified method, protein NRR, that enables proteins to access diversity previously difficult or impossible to generate. We investigated the structural plasticity of protein folds and the ability of helical motifs to function in different contexts by applying protein NRR and in vivo selection to the evolution of chorismate mutase (CM) enzymes. Functional CM mutants evolved using protein NRR contained many insertions, deletions, and rearrangements. The distribution of these changes was not random but clustered in certain regions of the protein. Topologically rearranged but functional enzymes also emerged from these studies, indicating that multiple connectivities can accommodate a functional CM active site and demonstrating the ability to generate new domain connectivities through protein NRR. Protein NRR was also used to randomly recombine CM and fumarase, an unrelated but also α-helical protein. Whereas the resulting library contained fumarase fragments in many contexts before functional selection, library members surviving selection for CM activity invariably contained a CM core with fumarase sequences found only at the termini or in one loop. These results imply that internal helical fragments cannot be swapped between these proteins without the loss of nearly all CM activity. Our findings suggest that protein NRR will be useful in probing the functional requirements of enzymes and in the creation of new protein topologies.
Directed evolution has been used to alter (1–4) enzyme activity as well as to investigate sequence constraints on protein folding (5–7) and catalysis (8). The sequences and therefore the properties of proteins that emerge from directed evolution depend on the diversification method used to generate the protein library before screening or selection. Theoretical studies (9) suggest that diversification methods, including point mutagenesis, homologous recombination, secondary structure swapping, and nonhomologous recombination differ in their ability to access protein sequences representing new folds and improved functions. Of the methods listed above, nonhomologous recombination has been theorized to be the most effective at enabling new structures and functions to emerge during protein evolution. Additional studies on enzyme superfamilies suggest that the reorganization of structurally similar components can result in altered substrate specificity or function (10). Because these components often lack DNA sequence homology, their combinatorial diversification can in general only be accomplished through nonhomologous recombination.
Methods that enable recombination to take place at defined sites without sequence homology have been recently described (11). For example, it is possible to recombine unrelated protein-encoding genes by using synthetic oligonucleotides to encode each desired crossover (12, 13). Although this strategy can result in a high likelihood of preserving function after diversification, many fewer sites of recombination and therefore fewer novel structures are accessible than if crossover sites are randomly generated. Alternatively, methods allowing a single random nonhomologous crossover of two protein-encoding genes have been developed (14, 15), and additional nonhomologous recombination events can be obtained by fragmenting and homologously recombining the resulting genes (16). Despite efforts to enhance the number of crossovers obtained (17), existing methods for diversifying proteins by nonhomologous recombination have thus far yielded only modest numbers of recombination events (three or fewer per 500 bp) in protein-encoding sequences, with even fewer crossovers (one to two per 500 bp) among sequences encoding active proteins.
We previously described nonhomologous random recombination (NRR), a simple method that enables the random recombination of any number of DNA sequences with no homology or ordering requirements (18). In side-by-side comparisons, NRR enabled the evolution of DNA aptamers with significantly higher target-binding affinity than aptamers obtained by using point mutagenesis alone (18). In the present work, we have modified NRR for protein evolution and applied this method, protein NRR, to the evolution of the monomeric Methanococcus jannaschii chorismate mutase (mMjCM) previously described by Hilvert and coworkers (7). Our results demonstrate the ability of protein NRR to create protein diversity that is difficult or impossible to access otherwise. In addition, our findings dissect functional requirements of CM enzymes in a broad and unbiased manner.
Materials and Methods
Molecular Biology Reagents. Restriction enzymes, Vent DNA polymerase, T4 DNA polymerase, and T4 DNA ligase were purchased from New England Biolabs. PCR reagents were purchased from Promega. SDS/PAGE gels were stained for analysis by using GelCode blue stain (Pierce) and quantitated by densitometry. Chorismic acid for in vitro kinetic assays on purified proteins was purchased from Sigma-Aldrich. Escherichia coli strain KA12 (19) was generously provided by D. Hilvert and P. Kast (Eidgenössische Technische Hochschule, Zürich). E. coli strain BL21(DE3)/pLysS was purchased from Novagen.
Oligonucleotides. The 5′-phosphorylated and PAGE-purified hairpin oligonucleotides PL1 and PL2 were purchased from Sigma-Genosys. PL1 (5′-CATACACGTCATCCGAATTCAGGCCTCCGGGCGCGCCCGGAGGCCTGAATTCGGATGACGTGT ATG-3′) contains an AflIII site (underlined) and PL2 (5′-CATGGTGACCCATCCGAATTCAGGCCTGCCGGCGCGCCGGCAGGCCTGAATTCGGATGGGTCACCATG-3′) contains a BstEII site (underlined) for ligation into the selection plasmid. Both contain a StuI site for removal of hairpin ends (italicized), and both end with NsiI half-sites (ATG/CAT) for digesting hairpin dimers and to provide a start codon for translation. PCR primers PL3 (CCTGAATTCGGATGACGTGTATG) and PL4 (CCTGAATTCGGATGGGTCACCATG) were synthesized by standard phosphoramidite chemistry on an Expedite 8909 DNA synthesizer and purified by reverse-phase HPLC.
Construction of Selection Plasmid pCM. Standard PCR, restriction digestion, and DNA ligation methods were used to assemble selection plasmid pCM, which contains the following key components: (i) the p15A replication origin from pACYC184; (ii) tyrA and pheC genes as in pKIMP-UAUC (20); (iii) the β-lactamase gene from pBR322; (iv) the chloramphenicol acetyl-transferase (CAT) gene from pACYC184 for expression as a C-terminal protein fusion (lacking its natural start codon) located immediately downstream of restriction sites for protein library cloning; and (v) a tac promoter upstream of the library cloning site. The library insertion site was created by using synthetic PCR primers containing AflIII and BstEII sites. The library promoter and insertion sites and the CAT gene were confirmed by DNA sequencing, and the tyrA, pheC, and β-lactamase genes, as well as the P15A origin, were confirmed to be functional by in vivo activities. All plasmid fragments were amplified by using Vent DNA polymerase.
Protein NRR of CM. The mMjCM gene (7), with class II-optimized codons for E. coli, was constructed from overlapping synthetic oligonucleotides, confirmed by sequencing after cloning into a vector, and amplified by PCR using 5′-phosphorylated primers (5′-TTTTTTGTTTTTGTTCTGGGTTTCTTCCAGG-3′ and 5′-ATGATCGAAAAACTGGCAGAAATCCG-3′). A total of 4 μg of the 321-bp product was randomly digested by using 2 μl of DNase I (Sigma, 7.6 μg/μl, 31.3 units/μg, diluted 1,000-fold) in a buffer of 20 mM Tris·HCl (pH 8.0) containing 10 mM MgCl2 at 25°C. Aliquots were analyzed by gel electrophoresis and the digestion was terminated by phenol/chloroform extraction when the fragments reached the desired size range. The fragments were subjected to gel filtration (Princeton Separations) then blunt-ended by using T4 DNA polymerase (T4 DNA polymerase buffer, 50 μg/ml BSA, 200 μM dNTP, 1–3 units of T4 DNA polymerase per μg of DNA for 30 min at 16°C). The reaction was extracted with phenol/chloroform and subjected to gel filtration. The desired size range (e.g., 75–125 bp) of pieces was purified by agarose gel electrophoresis into dialysis membrane (6,000 molecular weight cutoff) to provide ≈1 μg of fragments for NRR assembly. In a typical reaction, 10 pmol of fragments was combined with the desired ratio of hairpins PL1 and PL2 in blunt ligation buffer (T4 DNA ligase buffer with 50 μM ATP/15% polyethylene glycol 6000/18 Weiss units T4 DNA ligase) at 25°C for 16 h. The ligation reaction was digested with StuI and NsiI, then was amplified by PCR using primers PL3 and PL4. The PCR was subjected to gel purification to capture products of desired size (e.g., 300–800 bp), digested with AflIII and BstEII, and gel-purified again before ligation into pCM.
Protein NRR of CM with Fumarase. The E. coli fumarase gene was obtained by PCR from E. coli genomic DNA using 5′-phosphorylated primers 5′-ATGAATACAGTACGCAGCGAAAAAGATTCG-3′ and 5′-ACGCCCGGCTTTCATACTGCCGACC-3′. The 1,401-bp PCR product was gel-purified and digested for NRR as described above. A 3:1 ratio of fumarase:CM fragments was used in the NRR ligation. The resulting library was amplified and cloned as above.
CM Activity Selection. The library in pCM was transformed into 320 μl of electrocompetent DH10B cells and recovered in 8 ml of 2× yeast/tryptone (YT) medium at 37°C for 30 min. Ampicillin (100 μg/ml) and isopropyl β-d-thiogalactoside (1 mM) were added and the cells grown at 30°C for 90 min. A fraction of culture was plated on both 2× YT plus ampicillin (100 μg/ml) and 2× YT plus chloramphenicol (40 μg/ml) to determine the size of the library and the fraction of clones expressing CAT. For in-frame preselection, the culture was diluted into 500 ml of 2× YT plus chloramphenicol (40 μg/ml) and grown at 30°C until saturated before plasmids were isolated and transformed into KA12. Transformed KA12 cells were recovered as above, washed, and plated on agar containing M9c media (21) plus 20 μg/ml phenylalanine plus 100 μg/ml ampicillin plus 1 mM isopropyl β-d-thiogalactoside at 30°C. For growth without preselection, the initial library ligation was transformed directly into KA12 cells and grown as above. After incubation up to 10 days, colonies were picked, regrown on fresh plates to confirm growth, and then grown in liquid M9c medium plus 20 μg/ml phenylalanine at 30°C. Active plasmids were isolated and activity was confirmed by recovery of the putative active insert by PCR, religation into pCM, and retransformation into KA12 cells.
Sequence Analysis. Plasmids were sequenced by using standard protocols on an ABI Prism 3100 DNA Sequencer. Unselected sequences were obtained by isolating individual colonies from the plates used to determine the size of the initial library. Nonhomologous crossovers were located by using vectornti (Invitrogen) and macaw software (22).
Purification and Analysis of Proteins. Representative active clones were subcloned (without the CAT gene) into pET23a (introducing a C-terminal His6 tag) and transformed into BL21(DE3)/pLysS. A 500-ml culture of a transformant was grown at 37°C to OD600 = 0.8 before addition of isopropyl β-d-thiogalactoside to 1 mM. Induced cells were grown at 25°C for 4 h. The cells were harvested by centrifugation and were lysed by sonication and treatment with lysozyme. Mutant CM proteins in PBS were captured with TALON cobalt-agarose resin (BD Biosciences), washed with 40 ml of 5 mM imidazole in PBS, and eluted with 75 mM imidazole plus 2 mM EDTA in PBS. The eluted protein was dialyzed against PBS containing 1 mM 2-mercaptoethanol and 10–30% glycerol. Final protein solutions were quantitated by SDS/PAGE, staining, and densitometry comparing with prequantitated protein standards.
CM activity was assayed as described (23) in 0.1 M potassium phosphate buffer (pH 7.5). Absorbance at 274 and 304 nm was followed by using a Hewlett–Packard 8453 spectrophotometer. Kinetic parameters were extracted by direct fitting of initial rate data to the Michaelis–Menton equation. Contamination by endogenous E. coli CM (EcCM) was ruled out by the absence of activity from control purifications using plasmids lacking a functional mutase gene, and from the inability to saturate the protein for the less active proteins. The oligomeric state of the proteins was analyzed by gel filtration chromatography on an AKTA FPLC system (Pharmacia) with a Superdex 75 (10/300) column in PBS containing 10% glycerol (vol/vol) by using mMjCM and EcCM as standards.
Results
Protein NRR. The application of NRR to the evolution of proteins faces additional challenges compared with nucleic acid evolution by using NRR. Assembled genes from protein NRR must be cloned into an expression vector and transformed into cells. Products of the original NRR method (18), which uses a single hairpin to terminate random intermolecular ligation events, do not clone into expression vectors efficiently due to their identical termini. We therefore used two hairpins, each with a different nonpalindromic restriction endonuclease cleavage site, to terminate random ligation. This approach generates a statistical mixture of products, 50% of which are terminated with two different hairpins. These products were efficient substrates for cloning and expression.
A second challenge of protein NRR is the generation of nonsense mutations through frameshifting or the misorientation of gene fragments. These events reduce the meaningful diversity of protein libraries due to the introduction of internal stop codons that truncate recombined protein products. To minimize the impact of this problem, we designed an expression vector for protein NRR that fuses diversified gene products to CAT. Products of protein NRR that contain internal stop codons when introduced into this vector are mostly unable to propagate in E. coli cells in the presence of chloramphenicol, although internal ribosome-binding sites and start codons could allow chloramphenicol resistance even when following a stop codon. As an added benefit of this preselection step, diversified genes encoding proteins that are unable to be expressed or that are insoluble should also be eliminated. Preselection in a highly competent strain (DH10B) increases the meaningful diversity of the selected library because the selection strain, KA12, is much less efficiently transformed.
Nonhomologous Recombination of mMjCM. Protein NRR (Fig. 1) was used to diversify mMjCM. Blunt-ended DNA gene fragments that ranged from 75 to 125 bp were generated and recombined as described (18) using a 2:1 ratio of fragments to terminator hairpins. In theory, this stoichiometry should result in an average of four fragments recombining before being terminated by a hairpin at each end. Based on the average fragment size, this combination would create recombined genes of approximately the same size as the parental gene (321 nt), while containing an average of three crossovers.
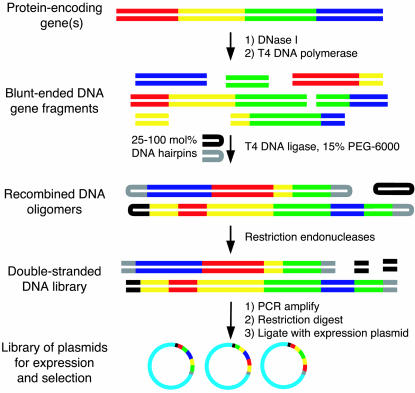
Fig. 1.
Protein NRR. One or more parental genes are digested with DNase I. Fragments are blunt-ended with T4 DNA polymerase, size-selected, and ligated under conditions that favor intermolecular ligation. Two hairpin sequences are added in a defined stoichiometry to the ligation reaction to generate recombined products of the desired average size. The ends of the hairpins are removed by restriction digestion, and the PCR-amplified pool is cloned for protein expression and selection.
The resulting NRR products were digested by using enzymes that cleave the closed end of each hairpin and ligated into selection plasmid pCM (Fig. 7, which is published as supporting information on the PNAS web site). The plasmid library was transformed into highly competent DH10B cells, providing libraries consistently comprising >108 ampicillin-resistant transformants. These transformants were preselected for in-frame and soluble proteins by incubation in liquid media containing chloramphenicol. Approximately 2.5% of the initial library (≈8 × 106 clones) was chloramphenicol-resistant.
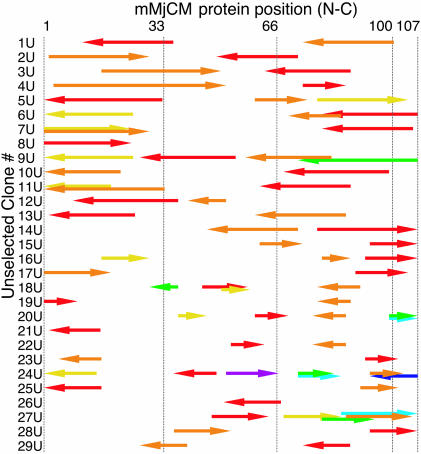
To evaluate the diversity introduced by protein NRR, we sequenced genes encoding library members before selection for CM activity. Fig. 2 depicts a representative set of sequences obtained from two independent NRR libraries with average target fragment sizes of 75–125 bp (clones 1U–14U) or 40–60 bp (clones 15U–29U). The sequences contain one to seven fragments of the mMjCM gene, with each fragment ranging in size from 21 to 210 bp. The size range of recombined fragments was consistent with target fragment sizes, and no apparent bias in the orientation of the fragments was observed. These results indicate that protein NRR is able to diversify proteins by high-resolution NRR events (under these conditions generating crossovers at a density of up to 9 per 500 bp).
Fig. 2.
Sequence diversity created by NRR. Unselected (inactive) sequences were obtained from two libraries. Clones 1U–14U were derived from an average fragment size of 100 bp; clones 15U–29U were derived from an average fragment size of 50 bp. Numbering across the top corresponds to the residue position in the mMjCM protein. Each arrow represents a recombined fragment. The arrow positions indicate the origin of each fragment within the parental mMjCM gene. Arrow colors indicate the order of fragment reassembly (5′-red-orange-yellow-green-teal-blue-violet-3′). The direction of each arrow indicates the sense (Right) or antisense (Left) strand of mMjCM. Overlapping arrows indicate sequence that appears more than once in a clone.
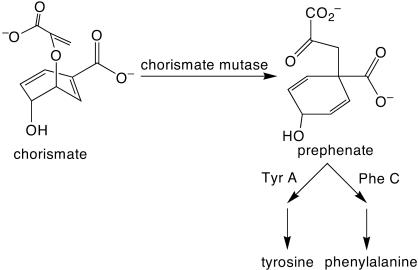
Active CM Sequences Contain Deletions, Repetitions, Appendages, and Rearrangements. CM catalyzes the Claisen rearrangement of chorismate to prephenate, an essential step in the biosynthesis of tyrosine and phenylalanine (Fig. 3). Plasmids containing the preselected NRR-diversified mMjCM library were transformed into the CM-deficient E. coli strain KA12 developed by Kast et al. (19), resulting in 3 × 107 chloramphenicol-resistant clones before selection for CM activity. This complexity is sufficient to ensure representation of the substantial majority of the preselected clones. The transformed KA12 library was selected for CM activity on minimal media lacking tyrosine. Approximately 2,600 active clones were observed, representing a survival rate of one in 11,500 preselected sequences and one in 4.5 × 105 initial library clones.
Fig. 3.
The Claisen rearrangement catalyzed by CM during amino acid biosynthesis. Cells lacking CM activity are unable to grow on media lacking tyrosine.
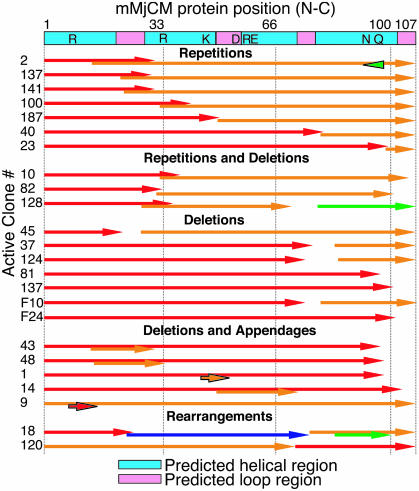
The sequences of active clones reveal many significant modifications to mMjCM. Only 42% of the sequenced active clones (27 of 64) contained full-length mMjCM or mMjCM containing only polymerase-induced point mutations. These clones could arise from undigested starting material, or from fragment reassembly of the full-length sequence. The remaining sequences (22 unique clones and 15 duplicates, likely arising from PCR amplification) each contained at least one recombination event, with up to three crossovers observed per active clone. Several of the selected protein sequences contained a variety of appended, inserted, or deleted amino acids compared with mMjCM (Fig. 4). In contrast with the recombined fragments before selection for CM activity, among which only 11% (4 of 35) were expressed in-frame relative to the start codon, 94% (46 of 49) of the fragments within active clones were in the same frame as the parental gene.
Fig. 4.
Protein sequences of active NRR-diversified mMjCM clones. The labeling scheme is identical to that used in Fig. 2. Arrows outlined in black indicate out-of-frame protein fragments. The colored bar at the top indicates predicted helical (blue) and loop (pink) regions based on homology with EcCM (26). The type of mutation is indicated: overlapping arrows indicate a duplication of one or more residues; gaps indicate a deletion. Predicted active site residues are indicated at the top.
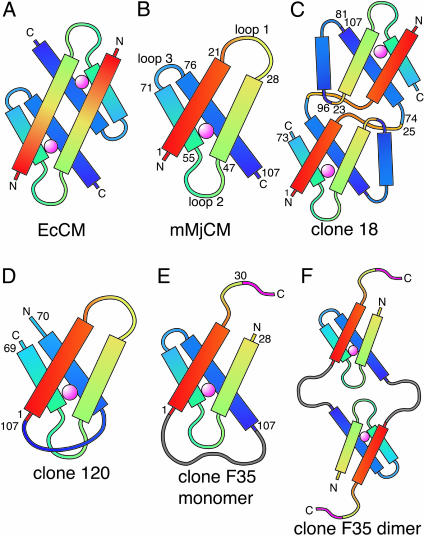
Two active sequences, clones 18 and 120, contain significant rearrangements of α-helix connectivity within the protein. Clone 18 contains four gene fragments (three crossovers) that reorder the four α-helices in the enzyme from 1–2-3–4 to 1–4-4′-2–3 (Figs. 4 and 6B). Clone 120 is a circular permutant that begins with residue 70, continues to the original C-terminal residue 107, and ends with residues 1–69. Taken together, the sequence diversity found among active CM variants highlight regions of low and high structural plasticity within the protein. The implications of specific selected sequences and the distribution of mutations are presented in Discussion.
Fig. 6.
Structural models of CMs. (A) EcCM. Coloring proceeds in spectral order from the N to the C terminus. (B) Structural model of mMjCM based on homology between the MjCM dimer and the EcCM dimer. Numbering indicates the residue at the approximate start and the end of each helix. (C–F) Diagrammatic models of rearranged clones, reflecting the observed oligomeric states, that preserve the active-site region (indicated with the pink sphere). Coloring is maintained from B to illustrate crossovers. Numbers indicate amino acid residues at each nonhomologous crossover based on the numbering in B. Out-of-frame fumarase residues are gray and out-of-frame mMjCM residues are magenta.
Recombination with an Unrelated Protein Results in Active Chimeric Proteins That Preserve CM α-Helices. E. coli fumarase is unrelated to CM in sequence or function but, like CM, is largely α-helical (24). To evaluate in a broad and unbiased manner the ability of foreign protein fragments to substitute regions of CM, fumarase was recombined with mMjCM by using protein NRR. Small fumarase gene fragments (averaging 40 bp each) were used to enhance the resolution of crossovers. A 3:1 molar ratio of fumarase to mMjCM fragments applied significant statistical pressure favoring the incorporation of fumarase sequences. The resulting plasmid library was transformed either into DH10B cells to characterize diversification, or directly into KA12 (107 transformants) for CM activity selection. Fifty colonies survived on minimal media lacking tyrosine and were confirmed by recloning to encode functional CMs. This survival rate of 1 in 2 × 105 was comparable with that of the all-mMjCM library.
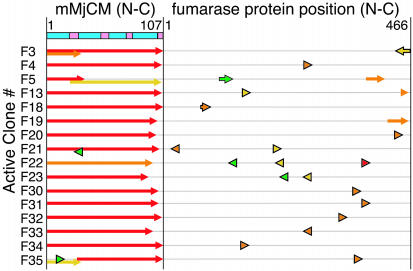
The sequences of clones from the recombined CM-fumarase library confirmed that most library members contained a mixture of sequence fragments encoding fumarase and CM. Among 15 unselected clones, 11 mMjCM and 51 fumarase fragments were found. Fumarase fragments ranged in size from 8 to 77 bp and mMjCM fragments ranged from 17 to 92 bp, which is consistent with the average size and fragment stoichiometry used to create the library. Recombined sequences contained up to 12 fragments (11 crossovers). The composition of the most highly recombined clone, F15U, contained seven fragments from mMjCM and five from fumarase (Table 2, which is published as supporting information on the PNAS web site).
Only two of 18 active clones (11%) that were sequenced lacked any fumarase sequence (F10 and F24); both contain deletions similar to those seen in the all-mMjCM library (Fig. 4). Interestingly, the sequences of 14 of 16 active hybrid clones revealed a nearly full-length mMjCM core preceded and/or followed by appendages of the fumarase gene (Fig. 5). Only two internal insertions of fumarase were found. One of the insert-containing clones, F5, is similar to previously characterized all-mMjCM mutants containing insertions at loop 1 but contains an in-frame 39-aa fragment of fumarase. While this insertion is longer than any of those described above, all residues of mMjCM are present at least once, indicating that the fumarase insertion in F5 need not assume the function of any part of mMjCM. Indeed, deletion of four or eight mMjCM helix 1 residues upstream of this fumarase insertion results in the loss of activity (data not shown). The other internal insertion, clone F35, is a chimeric circular permutant beginning at mMjCM residue 28 that contains an out-of-frame fumarase linker connecting the former termini of the protein (Fig. 5).
Fig. 5.
Protein sequences of active CM-fumarase hybrids. The labeling scheme is identical to that used in Figs. 2 and 4. The amino acid positions of each fumarase fragment are indicated by their position in the gene as indicated at the top.
Selected Clones Diversified by Protein NRR Exhibit CM Activity in Vitro. A subset of proteins surviving selection (from clones 18, 120, 128, and F35) were individually overexpressed (replacing the CAT fusion with a C-terminal His6 tag) and purified. The purified proteins were confirmed to catalyze the conversion of chorismate to prephenate in vitro with 5-fold to 9,000-fold lower kcat/Km values compared with mMjCM (Table 1). Analysis by analytical gel filtration chromatography suggested that proteins from clones 18 and 128 are dimeric, whereas protein from clone 120 is monomeric. The analysis of protein from clone F35 was consistent with a mixture of monomeric and dimeric states.
Table 1. In vitro activities of NRR-diversified CMs.
| Protein | Modification | kcat,s-1 | Km,μM | kcat/Km,μM-1·s-1 | Relative activity |
|---|---|---|---|---|---|
| mMjCM | None | 41.6 ± 2.7 | 222 ± 39 | (1.9 ± 0.4) × 105 | 1 |
| 18 | Rearranged connectivity | 14.9 ± 0.5 | 366 ± 29 | (40.7 ± 3.5) × 104 | 1/5 |
| 120 | Circular permutant | ND | ND | (2.1 ± 0.3) × 101 | 1/9,000 |
| 128 | Insertion plus deletion | ND | ND | (8.1 ± 0.4) × 102 | 1/230 |
| F35 | Chimeric circular permutant | 1.7 ± 0.2 | 146 ± 57 | (1.1 ± 0.5) × 104 | 1/17 |
ND, not determined.
Discussion
Protein NRR is a simple method that diversifies proteins in ways that are difficult to achieve by existing methods. The implementation of protein NRR is straightforward, enabling starting DNA to be converted into a diversified library in ≈1 day. The frequency of nonhomologous recombination events by using protein NRR can be tuned by modulating fragment sizes and fragment:hairpin stoichiometries during intermolecular ligation reactions, inducing in the above examples up to 11 crossovers within a recombined 664-nt CM-fumarase hybrid gene, or up to six crossovers within a 260-nt CM gene. In addition, protein diversification by NRR does not impose any restrictions on the original location of the recombining fragments within parental sequences, enabling dramatic gene rearrangements as observed in the inactive and active CM mutants described above. As expected, the ability to access this unusual degree of protein diversification comes at the expense of a lower frequency of active proteins due to frameshifting and the translation of formerly noncoding fragments; the latter problems can be partially avoided with an in-frame preselection by using highly competent cells.
Although the three-dimensional structures of mMjCM and its natural progenitor, the dimeric mMjCM (25), have not been determined, both are homologous to the structurally characterized dimeric EcCM (26). An alignment of mMjCM with the E. coli protein (G. MacBeath, personal communication) provides a reasonable model for the location of helical, loop, and active-site residues (ref. 7 and Figs. 4 and 6B). The active mutants generated in this study can be interpreted in light of this structural model. Without exception, each of the functional mutants retains the active-site residues present in mMjCM (Figs. 4 and 5). Two-thirds (10 of 15) of the observed insertion and deletion mutations align with predicted loop regions (Fig. 4). All seven of the larger insertions (2–19 amino acids) occur in or within three residues of loop 1, the region previously altered to confer the monomeric state of mMjCM. This loop may be unusually tolerant of insertions, perhaps as a result of these previous mutations. It is also likely that some of these loop 1 insertions revert the resulting protein to a dimeric state.
The observed deletions among active mutants occurred either near loop 3, or at the C terminus of the protein within the last 13 residues of helix 4. It is tempting to speculate that the junction of helices 3 and 4 does not have stringent sequence requirements because residues that are helical in the wild-type protein may play the role of deleted loop residues; previous reports support this hypothesis (27). Indeed, one mutant containing both an insertion in loop 1 and a deletion of all but one residue in the predicted loop 3 (clone 128) maintained significant in vitro activity (230-fold lower kcat/Km than mMjCM; Table 1). These results also indicate that the C-terminal 13 residues in mMjCM are nonessential, a result that is consistent with a similar finding for the EcCM (28). Additionally, our results suggest that loop 2 is highly intolerant of mutations, because the only change in this region observed among active clones was the repetition of a single glycine residue (Fig. 4, clone 187). This finding may be due to the proximity of loop 2 to the active site, with one active site residue (Asp 54) predicted to lie within this loop.
The three rearranged CM enzymes obtained through protein NRR (clones 18, 120, and F35) are of special interest because they each represent secondary structure connectivities previously not known to support catalysis of the Claisen rearrangement of chorismate to prephenate. Based on the structure of EcCM (ref. 26 and Fig. 6A), the homology model of the wild-type mMjCM (Fig. 6B), and gel filtration chromatography analysis, we constructed diagrammatic models for the rearranged mutants (Fig. 6 C–F) that preserve the active-site region of each protein and demonstrate the types of topological diversification that can yield functional CM enzymes. Taken together, the in vivo and in vitro activities (Table 1) of these rearranged mutants, while reduced compared with the starting mMjCM enzyme, establish that multiple secondary structure topologies are capable of providing CM activity.
The two evolved circular permutants provide insight into functional ways of joining the termini of mMjCM. Clone 120 (Fig. 6D) is a perfect circular permutant with no added or deleted residues. As shown above, the last 13 residues of mMjCM are not essential for activity and therefore in principle could exist either as part of a loop or as part of the last α-helix. As a loop (but not as a helix), these residues could connect the former N and C termini of the protein. The other circular permutant, clone F35, uses a long fumarase linker to connect the former C and N termini and exists as both a monomer (Fig. 6E) and a dimer (Fig. 6F). The additional linker could allow dimer formation by avoiding interactions between loop 2 that do not occur in the native dimer; clone 120, which lacks an analogous long linker, exists only as a monomer. A comparison of the activities of these two circular permutants (Table 1) reveals a 550-fold higher kcat/Km for F35 compared with clone 120, suggesting that a longer linker between the C and N terminus minimizes conformational distortions that reduce enzyme activity.
Coupled with an efficient functional selection or screen, protein NRR can serve as a useful tool for determining an enzyme's functional requirements in a broad and unbiased manner. In addition, the ability of protein NRR to combine two unrelated proteins can reveal the degree to which the function of secondary structure elements are protein-specific. Although active-site residues are expected to be intolerant to substitution, in principle, it is possible for secondary structure elements to be exchanged without loss of function when they play similar structural roles in both contexts and do not form precise and crucial interactions with neighboring residues. Consistent with this hypothesis, libraries of sequences matching only the hydrophobic pattern of the wild-type MjCM have been found to result in functional variants (6). However, fumarase substitutions within predicted helical regions of CM genes were prevalent in libraries only before functional selection. The complete disappearance of these substitutions after selection (leaving fumarase fragments only at the termini or in loop 1) suggests that the helical regions of CM, including those not involved in active-site contacts, are involved in unique interactions that cannot easily be replicated by regions of foreign helical proteins.
The simple metabolic selection (20) used in this work was not designed to differentiate mutants of various activities above a low threshold (8) (mutase activity 9,000-fold lower than that of mMjCM was sufficient to confer survival). It is possible, however, that protein NRR may enable proteins of improved activity to be evolved even though the vast majority of genes immediately after NRR diversification encode inactive proteins. In addition, the discovery of new connectivities that maintain CM activity suggests that protein NRR may also be useful to protein engineering efforts that seek an optimal orientation, arrangement, and spacing of structural elements to maximize desired properties. For example, protein NRR may enable the evolution of multifunctional proteins when simple fusion fails to provide the specific and unpredictable contexts necessary for desired function.
Supplementary Material
Acknowledgments
We thank Prof. Gavin MacBeath for the structural model of mMjCM and for helpful discussions; Prof. Donald Hilvert and Dr. Peter Kast for providing strain KA12; and Bryan Wang, Mathias Gruen, and Alex Ruthenberg for assistance. This work was supported by National Science Foundation CAREER Award MCB-0094128 and by National Institutes of Health/National Institute of General Medical Sciences Grant R01 GM65400-01. J.A.B. and J.M.L. acknowledge Howard Hughes Medical Institute and National Science Foundation Graduate Research Fellowships, respectively.
Abbreviations: NRR, nonhomologous random recombination; CM, chorismate mutase; mMjCM, monomeric Methanococcus jannaschii CM; EcCM, E. coli CM; CAT, chloramphenicol acetyltransferase; YT, yeast/tryptone.
References
- 1.Santoro, S. W. & Schultz, P. G. (2002) Proc. Natl. Acad. Sci. USA 99, 4185-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang, L., Brock, A., Herberich, B. & Schultz, P. G. (2001) Science 292, 498-500. [DOI] [PubMed] [Google Scholar]
- 3.Crameri, A., Whitehorn, E. A., Tate, E. & Stemmer, W. P. (1996) Nat. Biotechnol. 14, 315-319. [DOI] [PubMed] [Google Scholar]
- 4.Stemmer, W. P. (1994) Nature 370, 389-391. [DOI] [PubMed] [Google Scholar]
- 5.Lim, W. A. & Sauer, R. T. (1989) Nature 339, 31-36. [DOI] [PubMed] [Google Scholar]
- 6.Taylor, S. V., Walter, K. U., Kast, P. & Hilvert, D. (2001) Proc. Natl. Acad. Sci. USA 98, 10596-10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacBeath, G., Kast, P. & Hilvert, D. (1998) Science 279, 1958-1961. [DOI] [PubMed] [Google Scholar]
- 8.Kast, P., Grisostomi, C., Chen, I. A., Li, S., Krengel, U., Xue, Y. & Hilvert, D. (2000) J. Biol. Chem. 275, 36832-36838. [DOI] [PubMed] [Google Scholar]
- 9.Bogarad, L. D. & Deem, M. W. (1999) Proc. Natl. Acad. Sci. USA 96, 2591-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babbitt, P. C. & Gerlt, J. A. (1997) J. Biol. Chem. 272, 30591-30594. [DOI] [PubMed] [Google Scholar]
- 11.Hiraga, K. & Arnold, F. H. (2003) J. Mol. Biol. 330, 287-296. [DOI] [PubMed] [Google Scholar]
- 12.O'Maille, P. E., Bakhtina, M. & Tsai, M. D. (2002) J. Mol. Biol. 321, 677-691. [DOI] [PubMed] [Google Scholar]
- 13.Tsuji, T., Onimaru, M. & Yanagawa, H. (2001) Nucleic Acids Res. 29, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sieber, V., Martinez, C. A. & Arnold, F. H. (2001) Nat. Biotechnol. 19, 456-460. [DOI] [PubMed] [Google Scholar]
- 15.Ostermeier, M., Shim, J. H. & Benkovic, S. J. (1999) Nat. Biotechnol. 17, 1205-1209. [DOI] [PubMed] [Google Scholar]
- 16.Lutz, S., Ostermeier, M., Moore, G. L., Maranas, C. D. & Benkovic, S. J. (2001) Proc. Natl. Acad. Sci. USA 98, 11248-11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawarasaki, Y., Griswold, K. E., Stevenson, J. D., Selzer, T., Benkovic, S. J., Iverson, B. L. & Georgiou, G. (2003) Nucleic Acids Res. 31, e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bittker, J. A., Le, B. V. & Liu, D. R. (2002) Nat. Biotechnol. 20, 1024-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kast, P., AsifUllah, M. & Hilvert, D. (1996) Tetrahedron Lett. 2691-2694.
- 20.Kast, P., Asif-Ullah, M., Jiang, N. & Hilvert, D. (1996) Proc. Natl. Acad. Sci. USA 93, 5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamper, M., Hilvert, D. & Kast, P. (2000) Biochemistry 39, 14087-14094. [DOI] [PubMed] [Google Scholar]
- 22.Schuler, G. D., Altschul, S. F. & Lipman, D. J. (1991) Proteins 9, 180-190. [DOI] [PubMed] [Google Scholar]
- 23.Cload, S. T., Liu, D. R., Pastor, R. M. & Schultz, P. G. (1996) J. Am. Chem. Soc. 118, 1787-1788. [Google Scholar]
- 24.Weaver, T. & Banaszak, L. (1996) Biochemistry 35, 13955-13965. [DOI] [PubMed] [Google Scholar]
- 25.MacBeath, G., Kast, P. & Hilvert, D. (1998) Biochemistry 37, 10062-10073. [DOI] [PubMed] [Google Scholar]
- 26.Lee, A. Y., Karplus, P. A., Ganem, B. & Clardy, J. (1995) J. Am. Chem. Soc. 117, 3627-3628. [Google Scholar]
- 27.MacBeath, G., Kast, P. & Hilvert, D. (1998) Protein Sci. 7, 325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen, S., Vincent, S., Wilson, D. B. & Ganem, B. (2003) Eur. J. Biochem. 270, 757-763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.