Abstract
Chromosome replication origins were mapped in vivo in the two hyperthermophilic archaea, Sulfolobus acidocaldarius and Sulfolobus solfataricus, by using microarray-based marker frequency analysis. Bidirectional replication was found to be initiated in near synchrony from three separate sites in both organisms. Two of the three replication origins in each species were located in the vicinity of a cdc6/orc1 replication initiation gene, whereas no known replication-associated gene could be identified near the third origin in either organism. In contrast to initiation, replication termination occurred asynchronously, such that certain replication forks continued to progress for >40 min after the others had terminated. In each species, all replication forks advanced at similar DNA polymerization rates; this was found to be an order of magnitude below that displayed by Escherichia coli and thus closer to eukaryotic elongation rates. In S. acidocaldarius, a region containing short regularly spaced repeats was found to hybridize aberrantly, as compared to the rest of the chromosome, raising the possibility of a centromere-like function.
The prokaryotes are divided into two main lineages, the Bacteria and Archaea domains (1). Strikingly, proteins involved in replication, transcription, translation, and recombination in archaea are closely related to the corresponding eukaryotic proteins, whereas the bacterial information machinery is considerably less similar (2).
The mode of chromosome replication is a fundamental distinguishing feature between bacteria and eukaryotes. Whereas bacteria replicate their chromosome(s) bidirectionally from a single replication origin, eukaryotic chromosomes contain multiple start sites for DNA synthesis. Through the use of multiple origins, the chromosome replication stage (S phase) of a eukaryotic cell cycle may be similar in length to the corresponding stage (C period) of a bacterium containing a considerably smaller genome.
Sulfolobus acidocaldarius and Sulfolobus solfataricus belong to the Crenarchaeota phylum within the Archaea domain. The organisms are hyperthermophilic acidophiles that display optimal growth at ≈80°C and pH 3 and were originally isolated from geothermal hot springs (3, 4). We have initiated studies of the cell cycle in the two species, with the main focus on chromosome replication, genome segregation, and cell division (5–9).
We decided to investigate the chromosome replication characteristics of S. acidocaldarius and S. solfataricus by marker frequency (MF) analysis, by using whole-genome DNA microarrays developed in our laboratory. The MF technique is based on the fact that the copy number of a chromosomal DNA marker located close to a replication origin, on average, will be higher than that of a marker located near a terminus in a replicating cell population (see description of the principle of MF analysis in supporting information, which is published on the PNAS web site). The approach has been successfully used in combination with microarrays to study replication characteristics and to map chromosome replication origins in bacteria (Escherichia coli; ref. 10) and eukaryotes (Saccharomyces cerevisiae; ref. 11). We have previously identified two replication origins in S. solfataricus by 2D gel electrophoresis (12). Here, we use MF analysis to demonstrate that the number of replication origins is in fact three, and that this is the case also in S. acidocaldarius. We also present information about the in vivo chromosome replication properties of the organisms, including replication initiation, elongation, and termination characteristics, as well as possible centromere-related functions.
Materials and Methods
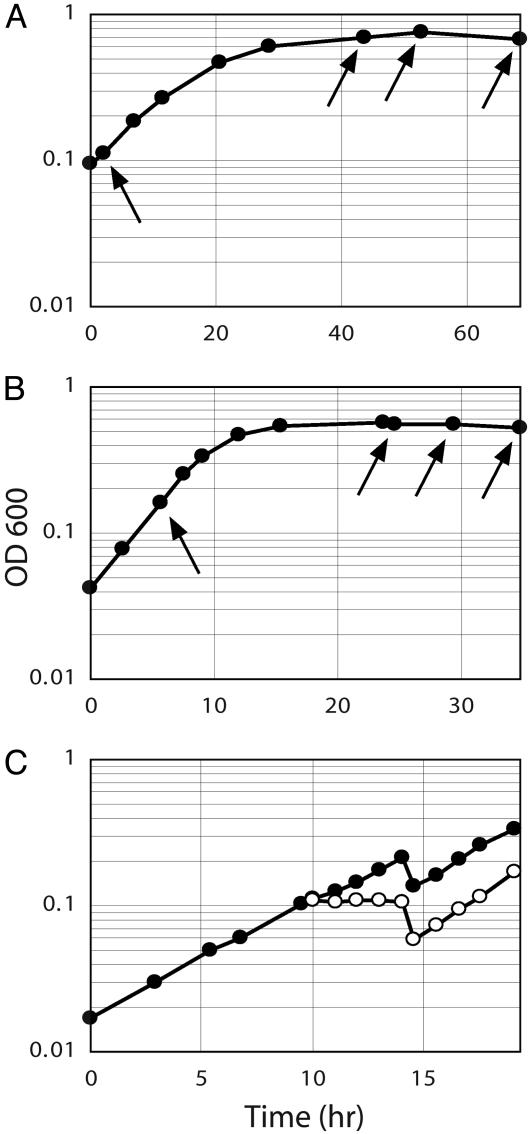
Strains, Growth Conditions, and Sampling. S. acidocaldarius Deutsche Sammlung von Mikroorganismen (DSM) 639 and S. solfataricus DSM 1617 were grown at 79°C in modified Allen mineral base medium (13) containing 0.2% tryptone. Growth was monitored by optical density measurements at 600 nm. Samples for DNA extraction and flow cytometry were collected from exponential growth and stationary phase as indicated in Fig. 1. The samples for DNA extraction were centrifuged at 2,300 × g for 15 min at room temperature, and the pellets were stored at -20°C.
Fig. 1.
Optical density measurements. (A) Batch culture of S. solfataricus grown into stationary phase. Sampling time points from exponential and stationary phase are indicated by arrows. (B) Batch culture of S. acidocaldarius. (C) Synchronization of an exponentially growing S. acidocaldarius batch culture. At an optical density of 0.1, the culture was split into two flasks, one of which was treated with acetic acid. After 4 h, the acetic acid was removed by centrifugation and fresh medium added. Open circles, culture treated with acetic acid; filled circles, untreated control.
Synchronized S. acidocaldarius Cultures. A 450-ml culture was grown in a 2-liter Erlenmeyer flask. At an optical density (OD) of 0.1, 20 ml was transferred to a separate flask (control), whereas acetic acid was added to 400 ml of the remaining culture (final concentration, 3 mM). The culture was then centrifuged at 2,700 × g for 15 min at room temperature. The cells were resuspended in a total of 500 ml of preheated medium, and incubation was continued. Samples for DNA extraction (50 ml) were collected 60, 105, 120, 135, and 150 min after resuspension. The experiment was monitored by OD measurements and flow cytometry at all stages.
DNA Purification. The cell pellets were resuspended in 560 μl of cell suspension mix (14), and 30 μl of cell lysis solution (14) was added. The samples were carefully mixed and incubated for 15 min at 55°C, after which 8 μl of protease mix (14) was added and incubation continued for another 60 min at 55°C. DNA was purified by phenol and chloroform extraction and concentrated by ethanol precipitation by using standard protocols (15). The precipitate was dissolved in 200 μl of H2O, 16 μl of RNase mix (14) was added, and the samples were incubated for 15 min at 37°C. The DNA was repurified with phenol and chloroform and again ethanol precipitated. The pellets were dried, resuspended in H2O, and stored at -20°C.
DNA Labeling. Purified DNA (2 μg) was mixed with 100 pmol of random nonamer oligonucleotides (Qiagen, Chatsworth, CA) in a final volume of 10 μl. The DNA was denatured for 5 min at 95°C, followed by incubation on ice for 2 min. Labeling mix (10 μl) containing 2× REact 2 (Invitrogen); 0.5 mM each of dATP, dCTP, and dGTP; 0.2 mM dTTP; 4 mM potassium phosphate buffer (pH 8.0); 0.1 mM Cy3/Cy5 dUTP (Amersham Pharmacia Biosciences); and 5 units of DNA polymerase I Klenow fragment (Invitrogen) was added, and the samples were incubated for 2 h at 37°C. Samples to be cohybridized were mixed and purified by using a Minelute kit (Qiagen) and eluted in 2 × 10 μl of the elution buffer provided with the kit.
Whole-Genome DNA Microarray Design and Fabrication. DNA microarrays containing 1,914 and 2,488 PCR-amplified gene-specific tags (GSTs) were produced for S. acidocaldarius and S. solfataricus, respectively. Primers were selected by using an in-house-developed software (unpublished work) that ensures high specificity of the GSTs by excluding PCR products that display >70% similarity over 50 base pairs, to a gene other than the intended one. The program utilizes the primer3 software (www.genome.wi.mit.edu/genome_software/other/primer3.html) for the actual primer design.
PCR amplifications were performed in 100-μl reaction volumes. All products were inspected for single bands of expected sizes on agarose gels before purification with MultiScreen SEQ384 filter plates (Millipore). The purified PCR products were dissolved in 60 μl of 50% DMSO.
All gene-specific tags were printed in three to four replicates on Ultra GAPS microarray slides (Corning) by using a QArray (Genetix, New Milton, Hampshire, UK) instrument with SMP2.5 pins (TeleChem International, Sunnyvale, CA).
Microarray Hybridization and Scanning. The microarray slides were prehybridized in 5× SSC/0.1% SDS/10 mg/ml BSA (Sigma) at 42°C, then washed three times in water and once in isopropanol before being dried in a microarray centrifuge (TeleChem International).
The labeled DNA was mixed with 60 μl of hybridization mixture containing 63% formamide, 6× SSC, 0.16% SDS, 10 μg of tRNA (S. cerevisiae; Sigma), and 10 μg of herring sperm DNA (Sigma). The samples were denatured for 2 min at 95°C and then incubated on ice for 1 min. The array was placed in a hybridization cassette (TeleChem International) and a coverslip (number 2 LifterSlip; Erie Scientific Company, Portsmouth, NH) was applied. The hybridization mixture was added, and the chamber was humidified with 20 μl of H2O. The reaction was incubated for 16–20hina42°C water bath in the dark. After hybridization, the coverslip was removed, and the arrays were washed for 5 min in 2× SSC and 0.1% SDS at 42°C, then for 10 min in 0.1× SSC and 0.1% SDS at room temperature, and finally 5 times in 0.1× SSC at room temperature, and dried by centrifugation.
The arrays were scanned at 10-μm resolution by using an Agilent Microarray Scanner model G2565BA (Agilent Technologies, Palo Alto, CA) at 635 and 532 nm, with laser power and photomultiplier tubes set at 100%. The raw data are available as supporting information on the PNAS web site.
Data Processing. Microarray image processing was performed by using genepix pro 5.0 software (Axon Instruments, Foster City, CA). Low-quality spots were excluded by filtering out those where the ratio of medians deviated from the regression ratio by >20% or if, for any channel, <70% of the foreground pixels had intensities exceeding median of background plus 2 SD of background (16). The ratios of the median pixel intensities were log2 transformed. Each slide was normalized such that the mean of the log2 ratios was equal to zero. Log2 ratios from replicate (two to six) slides were averaged after first averaging the spot replicates within the slides. From these average log2 ratios, the MF graphs were calculated. A marker was used only if all spots on all replicate arrays passed filtering. Finally, the obtained ratios for all plots were normalized, such that the baseline of the nonreplicated chromosomal regions or, for exponential vs. stationary phase plots, the terminus with the lowest ratio, was positioned at a value of one.
Microarray-Based MF Analysis. The principle and application of MF analysis are available as supporting information on the PNAS web site.
Theoretical Calculations and Simulations. The formula Rori/Rter = 2/(2-S/T) was used to calculate the length of the S phase, by inserting the maximum origin-to-terminus ratios from the MF distributions. S and T denote the length of the S phase and the generation time, respectively, and R, the marker ratio between two conditions. The prereplicative period was assumed to be negligible, based on flow cytometry data (5). Conversely, the maximal origin-to-terminus ratio was calculated by inserting S period lengths estimated from the DNA content distributions obtained by flow cytometry. When appropriate, values were adjusted for the exponential age distribution (5), i.e., that there are twice as many newborn as dividing cells in an exponentially growing population.
Theoretical MF gradients were modeled for cell populations by using the origins simultaneously or individually (Fig. 2C). In the simultaneous model, the formula (2(1-S/T) + (2-d/D)(S/T))/k was used to calculate the ratio for a marker located at a distance d from the nearest origin. D denotes the longest origin-to-terminus distance, and the constant k was adjusted such that the terminus with the lowest ratio was located at a ratio of one. In the individual model, the formula (2(1-S/T) + ((2-d1/L)/3 + (2-d2/L)/3 + (2-d3/L)/3)S/T)/k was used to calculate the ratio for markers located at distances of d1, d2, and d3 from origin1, origin2, and origin3, respectively. L denotes half the chromosome length, and the same value for the constant k was used as above.
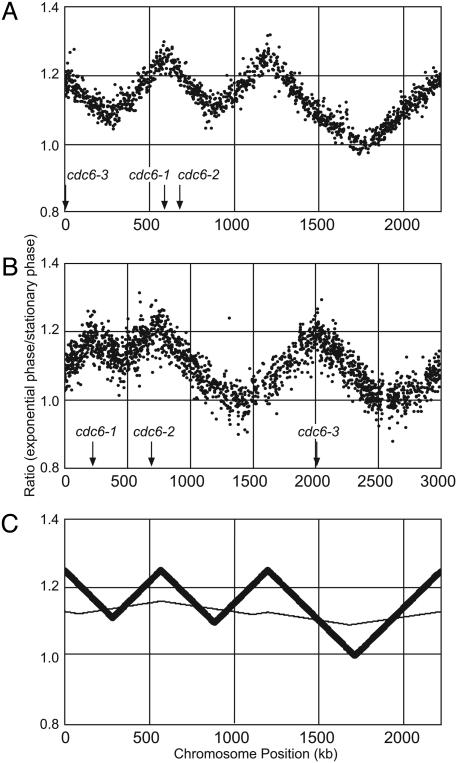
Fig. 2.
MF distributions. (A) Ratio of hybridization signals from exponentially growing vs. stationary phase S. acidocaldarius cells. (B) Exponential growth vs. stationary phase for S. solfataricus.(C) Theoretical simulations (see Materials and Methods) of different replication initiation modes in S. acidocaldarius cell populations. The thick line denotes initiation from all three replication origins in each cell. The thin line shows initiation from a single origin, randomly chosen among the three.
Flow Cytometry. Flow cytometry sampling and analysis were performed as described (5).
Results
Identification of Three Origins in Asynchronously Growing Cultures. Both Sulfolobus species were grown in batch cultures and displayed exponential growth, followed by a gradual decrease in growth rate and subsequent entry into stationary phase (Fig. 1 A and B). Flow cytometry showed that 30–40% of the exponentially growing cells contained between 1 and 2 genome equivalents of DNA (the cell size and DNA content distributions are available as supporting information on the PNAS web site), and that all cells in the resting populations contained two fully replicated chromosomes, in accordance with previous results (5).
Chromosomal DNA was isolated from the exponentially grown and stationary phase cells, and the DNA preparations were differentially labeled, mixed, and hybridized against whole-genome DNA microarrays. The resulting MF ratios displayed three prominent peaks (Fig. 2 A and B), indicating that DNA synthesis was initiated from three separate chromosome locations in both organisms. In both species, the replication origins were unevenly distributed along the chromosome. The distances between the origins were 630, 570, and 1,020 kb in S. acidocaldarius and 520, 1,280, and 1,200 kb in S. solfataricus.
The cdc6-1 and cdc6-3 replication initiation genes coincided with peaks in the MF distributions in both species (Fig. 2). However, the cdc6-2 gene was located between a peak and a valley in S. acidocaldarius and was offset ≈80 kb to the left of the second peak in the S. solfataricus MF distribution. Thus, one origin in each species was devoid of an immediately adjacent cdc6 gene and, in both organisms, it was the cdc6-2 gene that did not colocalize with an origin.
We have discovered that addition of 3 mM acetic acid allows ongoing rounds of replication to continue to termination (replication runout) in S. acidocaldarius, while no new rounds of replication are initiated. After acetic acid treatment, all cells contained two fully replicated chromosomes (Fig. 3, 0-min time point). In hybridizations against chromosomal DNA from stationary-phase cells, flat MF distributions were obtained (data not shown; cf. 60-min time point in Fig. 4), as expected if all markers had become equally abundant in both samples and demonstrating that the peaks were associated with ongoing chromosome replication.
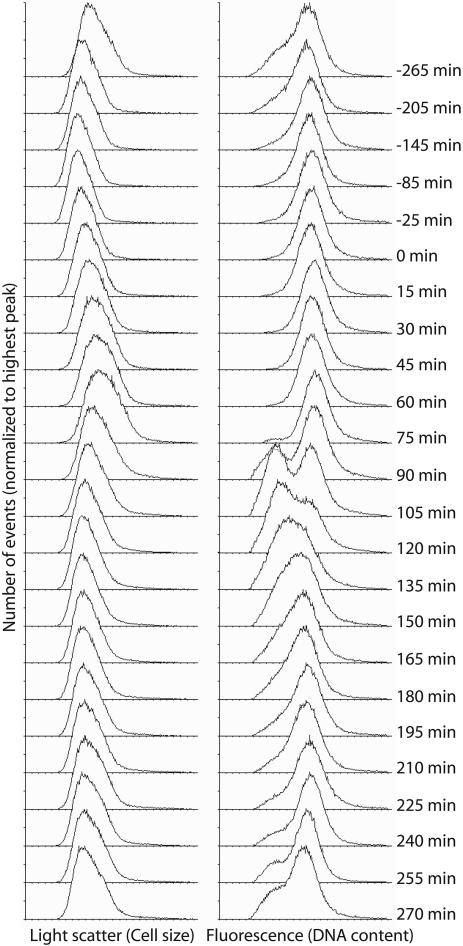
Fig. 3.
Cell size and DNA content distributions from S. acidocaldarius synchronization experiment. Acetic acid was added to the culture at -265 min and removed at 0 min; samples for flow cytometry were removed at the indicated time points. Note the decreased average cell size after the 75-min time point, concomitant with the appearance of cells containing a single chromosome, demonstrating ongoing cell division.
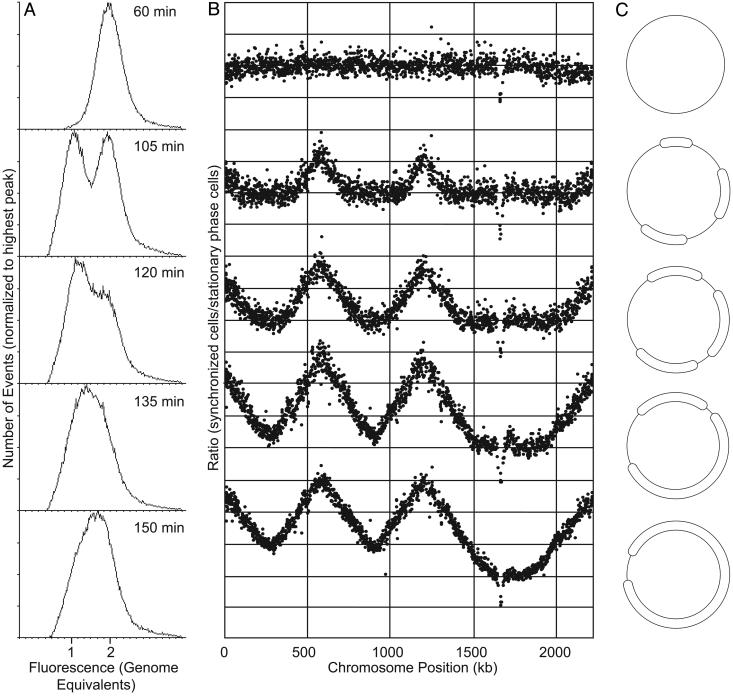
Fig. 4.
(A) Flow cytometry DNA content (fluorescence) distributions from an S. acidocaldarius population progressing through the cell cycle in synchrony. (B) MF distributions from the same time points. (C) Progression of replication forks around the circular S. acidocaldarius chromosome.
Bidirectionality, Replication Termination, and Origin Utilization. The MF gradients were similar in both directions from all origins (Fig. 2), showing that replication was bidirectional in both organisms and demonstrating that all six replication forks progressed at similar rates. The depth of the intervening valleys, corresponding to termination events, was proportional to the distance from the two flanking origins, as expected. Also, the maximum origin to terminus ratios revealed that 33% and 40% of the growing S. solfataricus and S. acidocaldarius cell populations, respectively, were in the S phase of the cell cycle, which correlated well with the flow cytometry data (above). Replication termination was found to occur asynchronously in both organisms, as a consequence of the uneven origin distribution along the chromosomes. In S. acidocaldarius, two termination events first took place during a short time interval, whereas replication continued for about twice as long before the third termination occurred (Fig. 2 A). In contrast, in S. solfataricus, a single termination first took place, followed by two near-synchronous termination events after more than twice the time (Fig. 2B).
Synchronous Replication Initiation in S. acidocaldarius. In eukaryotes, different sets of origin are initiated in succession at different stages during S phase (11), conferring possibilities for temporal control of replication. We wanted to investigate this possibility for the three Sulfolobus origins. We found that removal of the acetic acid from an S. acidocaldarius culture in which replication runout had been induced (above) resulted in that a considerable part of the cell population progressed through the cell cycle in synchrony (Fig. 3).
At 60 min after acetic acid removal, the DNA content distribution obtained by flow cytometry revealed that all cells still contained two chromosomes, showing that no cell division had occurred at this time point (Fig. 4A). No peaks were evident in the MF distribution, demonstrating that chromosome replication had not initiated (Fig. 4B).
At 105 min, approximately half of the cell population contained a single chromosome in the DNA content distribution, showing that cell division had occurred. Furthermore, the one-chromosome peak had begun to trail toward the right, i.e., toward a DNA content of more than one chromosome, indicating that replication had started. This is in accordance with previous data from our laboratory showing that chromosome replication in Sulfolobus species does not begin until after cell division has taken place (7), and that the prereplicative (G1; Fig. 4B) period occupies ≤5% of the cell cycle (5). In parallel, three distinct peaks appeared in the MF distribution, showing that replication had initiated. Unreplicated chromosome sections were evident as flat regions in the MF distribution: replication at this time point had proceeded ≈120–200 kb per replication fork in the cells that first initiated replication.
At the 120-min time point, the one-chromosome peak had become larger than the two-chromosome peak in the flow cytometry analysis, showing that the cell population continued to divide. In the MF distribution, replication had progressed a further 70–100 kb, extending the peak heights and shortening the unreplicated regions.
After another 15 min, at 135 min, the replication forks between the second and third origins had met (in the leading cell population), and the forks between the first and second origins were close to termination. In contrast, an unreplicated region of ≈380 kb remained between the third and first origins.
At the final time point (150 min), the unreplicated region between the third and first origins had become reduced to ≈170 kb.
The similar peak heights in the MF distributions indicated that all three origins were initiated in near synchrony in each cell. However, an alternative possibility would be that each cell selected only one of three origins and then replicated the entire chromosome as a result of such a single initiation event. We simulated the MF distributions that would result from alternative modes of replication initiation in asynchronously growing cultures (Fig. 2C), assuming that the replication forks move at similar rates, and that all origins initiate bidirectional replication. The experimentally obtained MF graph (Fig. 2 A) corresponded to the model in which all origins were initiated simultaneously in each cell, whereas it differed markedly from the single-origin utilization model, which thus could be ruled out.
Replication Rate and S Phase Length. From the successive reduction of the unreplicated chromosome regions (Fig. 4), fork rates of 80–110 bp/s were calculated, or approximately one-third of previous estimates, based on a single replication origin (9). The total length of the S phase in S. acidocaldarius, under these conditions, was determined to 96 min, or 40% of the cell cycle.
An Aberrantly Hybridizing Region in S. acidocaldarius. A short region between the third and first origins consistently displayed considerably lower ratios in the hybridizations between synchronized and stationary phase cultures (Fig. 4). The region was found to be centered on a cluster of short regularly spaced repeats (SRSR, ref. 18; other SRSR regions were not represented on the microarrays).
Discussion
In this report, we demonstrate that in the hyperthermophilic archaea S. acidocaldarius and S. solfataricus, chromosome replication is initiated in synchrony from three separate replication origins. The single cdc6 gene of the euryarchaeon Pyrococcus abyssi is situated immediately adjacent to the single replication origin (18), and one of the cdc6 paralogues in Halobacterium NRC-1 is also located next to an origin (19). Because the number of origins and cdc6 genes correlates in both S. acidocaldarius and S. solfataricus, it might have been anticipated that each origin should be flanked by a cdc6 gene. However, in both species, one origin is devoid of an immediately adjacent cdc6 gene and, in both cases, it is the cdc6-2 gene that does not colocalize. We have fine-mapped two of the origins in S. solfataricus by 2D gel electrophoresis (12), aided by the MF distributions and the assumption that the origins are located immediately adjacent to cdc6 genes. However, because 2D gel analysis can be applied only to short DNA stretches, the exact location of the third origin could not be determined in the absence of a recognizable replication initiation gene to guide the search. We have identified conserved repeated sequence elements in the other two S. solfataricus origins (12) and, because the MF analysis limits the location of the third origin to a well defined 40-kb region in S. acidocaldarius, this region was searched for repeats, as well as the corresponding S. solfataricus region. No additional copies of the previously identified repeats were found, but the TTTTGTTGTTTATTGTC consensus motif is repeated three times in both organisms. The organization is similar to that of those in the other two origins, with one repeat being inverted; the sequence is distantly related to the others; and the repeats are well centered in the MF peaks. However, in S. solfataricus, the repeats are located within a possible coding sequence (SSO0867), and they lack the internal symmetry characteristic of the other repeats. Thus, other locations cannot be ruled out, and the significance of the repeats needs to be experimentally investigated.
We previously suggested that the Cdc6-2 protein may act as negative regulator of replication (12), based on our demonstration that the protein is present during the G2 phase of the cell cycle and in stationary phase, i.e., in nonreplicating cells, whereas it is absent in S phase, i.e., during active replication. Although the negative function may explain the lack of a need for colocalization of the cdc6-2 gene with any particular origin, it still leaves open the question of why a linked positively acting replication initiation gene is not required at only one of the origins. We envisage that synchronous replication initiation is achieved not only by temporal coordination but also by physical origin proximity within the cell, perhaps in the form of a large centrally located replication factory. It is conceivable that one of the three initiation sites might act as the master regulator in such a complex, with the other two origins being subordinate and therefore different in sequence and/or organization. This might explain the apparent lack of a linked replication gene at one of the origins. In addition, the fact that the origins differ in organization and gene context opens up a range of questions regarding the structure of multiple origins also in eukaryotic organisms.
In contrast to initiation, replication in Sulfolobus species is terminated asynchronously, such that either two or four of the replication forks, depending on species, continue to progress for >40 min after the other(s) has terminated. This has also not been described previously and highlights issues about coordination of multiple termination events, and how concatenated chromosomes can be disentangled in a system with multiple terminations in both time and space.
The deviating MF ratio of the SRSR region in S. acidocaldarius is intriguing and could indicate that the replication kinetics differ from those of the rest of the chromosome. Also, SRSR clusters have been proposed to function as centromere-like regions during chromosome segregation (17). Peng et al. (17) also suggested that the SRSR regions may be involved in higher-order structuring of the chromosome DNA, and that the region might be condensed. This could influence the labeling efficiency and hybridization pattern, provided that the condensation level differs between the cell cycle stages, and that it is retained in the purified DNA. A third possibility is that tightly associated proteins may affect the recovery of the region during purification by removal of DNA through association with protein precipitates. In this alternative, the amount of protein bound to SRSR repeats would vary significantly during the cell cycle, presumably being at its highest during genome segregation.
The replication characteristics of Sulfolobus species differ from those of members of the genus Pyrococcus (18), in terms of both replication rates and number of origins. The organisms belong to the Crenarchaeota and Euryarchaeota phyla, respectively, and it will be of considerable interest to study additional species from both lineages to determine the generality of the characteristics and to correlate this to archaeal evolution and diversity.
Because multiple replication origins so far have been exclusive to eukaryotic organisms, it may be assumed that this feature is coupled to the presence of a nucleus; however, the results presented here and in our previous analysis (12) demonstrate that this is not the case. Furthermore, the Sulfolobus replication rate was found to be similar to rates displayed by eukaryotic organisms and an order of magnitude slower than that of E. coli, adding to the list of eukaryotic-like cell cycle features. Thus, in addition to the well-known similarities in terms of the molecular components of the replication machinery (2), the multiple replication origin mode of chromosome replication, as well as the replication rate, unites eukaryotes with Sulfolobus species. The results, therefore, have implications not only for the biology of archaea and extremophiles but also for the origin and evolution of the eukaryotic replication system. Thus, questions are raised about whether the first eukaryotic organisms may have coevolved with, and then branched off from, an archaeal lineage that contained multiple origins, if the multiple origin mode of replication has been horizontally transferred between the lineages, or whether this replication mode may have been invented independently on multiple occasions during evolution.
The results underscore the potential of archaea as simple model systems for complex eukaryotic processes and indicate that Sulfolobus species are particularly well suited for dissection of the mechanism that coordinate initiation at multiple replication origins and for elucidation of how asynchronous termination of replication in different chromosome regions is organized.
Supplementary Material
Acknowledgments
The genome sequencing of S. acidocaldarius by L.C. was performed in the laboratory of Roger Garrett, who is gratefully acknowledged. We thank Jasper Walther for help with pilot experiments, Björn Brindefalk for similarity searches, and Stefan Eriksson for excellent technical assistance. This work was supported by the European Union 5th Framework Program, the Swedish Research Council, and the Swedish Graduate School in Genomics and Bioinformatics.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MF, marker frequency; SRSR, short regularly spaced repeats.
References
- 1.Woese, C. R. & Fox, G. E. (1977) Proc. Natl. Acad. Sci. USA 74, 5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsen, G. J. & Woese, C. R. (1997) Cell 89, 991-994. [DOI] [PubMed] [Google Scholar]
- 3.Brock, T. D., Brock, K. M., Belly, R. T. & Weiss, R. L. (1972) Arch. Mikrobiol. 84, 54-68. [DOI] [PubMed] [Google Scholar]
- 4.Zillig, W., Stetter, K. O., Wunderl, S., Schulz, W., Priess, H. & Scholz, I. (1980) Arch. Microbiol. 125, 259-269. [Google Scholar]
- 5.Bernander, R. & Poplawski, A. (1997) J. Bacteriol. 179, 4963-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poplawski, A. & Bernander, R. (1997) J. Bacteriol. 179, 7625-7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hjort, K. & Bernander, R. (1999) J. Bacteriol. 181, 5669-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernander, R., Poplawski, A. & Grogan, D. W. (2000) Microbiology 146, 749-757. [DOI] [PubMed] [Google Scholar]
- 9.Hjort, K. & Bernander, R. (2001) Mol. Microbiol. 40, 225-234. [DOI] [PubMed] [Google Scholar]
- 10.Khodursky, A. B., Peter, B. J., Schmid, M. B., DeRisi, J., Botstein, D., Brown, P. O. & Cozzarelli, N. R. (2000) Proc. Natl. Acad. Sci. USA 97, 9419-9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raghuraman, M. K., Winzeler, E. A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D. J., Davis, R. W., Brewer, B. J. & Fangman, W. L. (2001) Science 294, 115-121. [DOI] [PubMed] [Google Scholar]
- 12.Robinson, N. P., Dionne, I., Lundgren, M., Marsh, V. L., Bernander, R. & Bell, S. D. (2004) Cell 116, 25-38. [DOI] [PubMed] [Google Scholar]
- 13.Grogan, D. W. (1989) J. Bacteriol. 171, 6710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramakrishnan, V. & Adams, M. W. W. (1995) in Archaea, A Laboratory Manual: Thermophiles, eds. Robb, F. T., Place, A. R., Sowers, K. R., Schreier, H. J., DasSarma, S. & Fleischmann, E. M. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 95-96.
- 15.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 16.Handran, S., Pickett, S. & Verdnik, D. (2002) in DNA Array Image Analysis Nuts and Bolts, eds. Kamberova, G. & Shah, S. (DNA Press, Eagleville, PA), pp. 83-98.
- 18.Peng, X., Brügger, K, Shen, B., Chen, L., She, Q. & Garrett, R. A. (2003) J. Bacteriol. 185, 2410-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myllykallio, H., Lopez, P., López-García, P., Heilig, R., Saurin, W., Zivanovic, Y., Philippe, H. & Forterre, P. (2000) Science 288, 2212-2215. [DOI] [PubMed] [Google Scholar]
- 20.Berquist, B. R. & DasSarma, S. (2003) J. Bacteriol. 185, 5959-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.