Abstract
DNA damage response genes play vital roles in the maintenance of a healthy genome. Defects in cell cycle checkpoint and DNA repair genes, especially mutation or aberrant downregulation, are associated with a wide spectrum of human disease, including a predisposition to the development of neurodegenerative conditions and cancer. On the other hand, upregulation of DNA damage response and repair genes can also cause cancer, as well as increase resistance of cancer cells to DNA damaging therapy. In recent years, it has become evident that many of the genes involved in DNA damage repair have additional roles in tumorigenesis, most prominently by acting as transcriptional (co-) factors. Although defects in these genes are causally connected to tumor initiation, their role in tumor progression is more controversial and it seems to depend on tumor type. In some tumors like melanoma, cell cycle checkpoint/DNA repair gene upregulation is associated with tumor metastasis, whereas in a number of other cancers the opposite has been observed. Several genes that participate in the DNA damage response, such as RAD9, PARP1, BRCA1, ATM and TP53 have been associated with metastasis by a number of in vitro biochemical and cellular assays, by examining human tumor specimens by immunohistochemistry or by DNA genomewide gene expression profiling. Many of these genes act as transcriptional effectors to regulate other genes implicated in the pathogenesis of cancer. Furthermore, they are aberrantly expressed in numerous human tumors and are causally related to tumorigenesis. However, whether the DNA damage repair function of these genes is required to promote metastasis or another activity is responsible (e.g., transcription control) has not been determined. Importantly, despite some compelling in vitro evidence, investigations are still needed to demonstrate the role of cell cycle checkpoint and DNA repair genes in regulating metastatic phenotypes in vivo.
INTRODUCTION
DNA is continuously damaged by genotoxic agents generated either in the environment (e.g., UV light, ionizing radiation, etc.) or intracellularly (e.g., reactive oxygen species as byproducts of routine metabolic processes). In normal cells, the integrity of the genome is ensured by a very efficient DNA damage response signaling network that includes cell cycle checkpoints and DNA repair pathways. On the other hand, cancer cells are thought to arise through the accumulation of numerous genetic alterations that confer growth and survival advantages. Dysregulation (either loss or gain) of DNA repair factors can promote the accumulation of DNA errors and genomic instability, which is implicated in aging, immune deficiencies, neurodegenerative disorders and cancer. Germline mutations in cell cycle checkpoint or DNA repair genes can predispose to hereditary forms of cancer, whereas somatic mutations and epigenetic silencing of DNA damage response genes are common in cancers with no inherent genetic link (1). DNA repair genes involved in nucleotide excision repair (2), mismatch repair (3), non-homologous end joining (4, 5) and homologous recombination (6) can predispose to different types of cancer.
Dysregulation of DNA repair genes affects the response of cells to DNA damaging anti-cancer treatment. Upregulation of DNA repair pathways can cause resistance to chemotherapy and radiotherapy, so inhibitors of these pathways have the potential to sensitize cancer cells to these agents (7, 8). Conversely, cancer cells that have lost a repair pathway and are solely dependent on another, alternative pathway, can be rendered vulnerable by targeting the functional pathway using the principle of synthetic lethality, whereas noncancer cells (with two functional repair pathways) would demonstrate resistance (7, 8).
In recent years, there has been accumulating evidence that DNA damage response genes are involved in additional cellular functions beyond mending damaged DNA and cell cycle checkpoint control, such as transcriptional regulation, chromatin remodeling and apoptosis. In this article, we provide an overview of the evidence that DNA damage response (DDR) genes participate not only in tumor initiation, at least in part when they fail to ensure proper repair of damaged DNA, but in tumor progression and metastasis as well.
DNA REPAIR GENES AND CANCER INITIATION
Genomic instability is characteristic of most human malignancies and it is considered a hallmark of cancer cells. Genomic instability is caused by downregulating DNA damage response pathways, such as those controlled by p53, ataxia telangiectasia mutated (ATM) and AT and Rad3-related (ATR) kinases. Alternatively, genomically unstable tumors can arise from acquired defects in any one of six DNA repair or damage tolerance pathways, base excision repair (BER), nucleotide excision repair (NER), DNA mismatch repair (MMR), homologous recombination repair (HR), non-homologous end joining (NHEJ), and translesion DNA synthesis (TLS). The connection between DNA repair defects and carcinogenesis is highlighted by the fact that inherited defects in DNA repair mechanisms that cause progeroid or accelerated aging syndromes (9), including Ataxia telangiectasia, Nijmegen syndrome, Werner syndrome, Bloom syndrome, Rothmund-Thomson syndrome, Xeroderma pigmentosum or Trichothiodystrophy also carry a higher cancer risk as well (2, 10).
In addition to DNA damage response gene mutations, which can either be inherited or somatically acquired, epigenetic gene silencing may also promote tumorigenesis. Epigenetic inactivation of DNA repair genes in cancer has been reported and has been related to several DNA repair pathways including BER, NER and other DNA damage processing mechanisms (11). In sporadic cancers, one of the most common mechanisms of inactivation of DNA repair pathways is the epigenetic silencing of a critical gene (e.g., FANCF, BRCA1) through methylation of the promoter region. Epigenetic silencing of DNA repair genes, such as MGMT, MLH1, BRCA1, WRN and FANCF, can boost mutation rates and promote genomic instability in cancer cells (12). Below is a brief description of the connection between specific aberrant DNA repair pathways and human cancer.
Direct Repair
The simplest form of DNA repair is the direct reversal of a lesion. MGMT (O6-Methylguanine DNA methyltransferase) participates in this kind of pathway and is often at higher levels in tumors compared with normal cells where it can confer resistance to DNA-alkylating agents. The MGMT enzyme repairs O6-alkylated guanine residues in genomic DNA. O6-methylguanine pairs with thymine and would lead to a G-to-A transition during DNA replication if left unrepaired. MGMT is epigenetically silenced (13) in a variety of tumors, including glioblastoma (14–16), colon cancer (17, 18), non-small cell lung cancer (19, 20), gastric carcinoma (21), and head and neck squamous cell carcinoma (22–24). In the case of colorectal cancer, MGMT promoter methylation is associated with G-to-A mutations in KRAS (25) and in p53 (26).
Base Excision Repair
The base excision repair (BER) pathway is the main mechanism that protects the genome from deleterious effects of exposure to reactive oxygen species. This pathway removes damaged bases from DNA and it can also repair DNA single-strand breaks (27). A number of DNA glycosylases (e.g., OGG1: 8-oxoguanine DNA glycosylase; MUTYH: the human homolog of the E. coli mutY gene), endonucleases (APE1: apurinic/apyrimidinic endonuclease 1; FEN1: flap structure-specific endonuclease 1), XRCC1 (X-ray repair complementing defective repair 1), DNA polymerase β (Polβ), DNA ligase III and PARP-1 participate in this DNA repair pathway.
Inherited mutations in BER genes are rare. However, polymorphisms in genes like OGG1, APE1 and XRCC1 have been genetically linked to cancer (28). MUTYH, (29) is the first BER gene to have been associated with a human cancer syndrome (MUTYH-associated polyposis), as biallelic germline mutations in MUTYH were identified in individuals with a pre-disposition to multiple colorectal adenomas and carcinomas (30, 31). Other DNA glycosylases are also found dysregulated in various cancers. For example, OGG1, which repairs oxidatively damaged guanine bases in DNA, is involved in tumorigenesis (32– 34), whereas expression of the thymine DNA glycosylase gene (TDG) is decreased in several multiple myeloma cell lines compared with normal plasma cells by promoter methylation (35). Production of APE1, an enzyme that follows the action of DNA glycosylases in the BER pathway, is frequently increased in germ cell tumors (36), and higher APE1 protein levels have been associated with increased drug and radiation resistance (36, 37). Further downstream in the BER pathway, Polβ, which is the major DNA polymerase that fills in the nucleotide gap created by APE1, is overexpressed in prostate, ovary, uterus and stomach cancers (38), as well as prostate, breast and colon cancer cell lines (39). In addition, elevated levels of Polβ lead to genomic instability through the accumulation of DNA single- and double-strand breaks (40, 41), and these effects are particularly evident after exposure of Polβ-overexpressing cells to oxidative stress-inducing DNA damaging agents (42).
FEN1, which participates in BER (43), non-homologous end joining (44) and homologous recombination (45), and is important for genomic stability (46), demonstrates increased expression in many tumors. It is highly abundant in testis, lung and brain tumors (47) and in prostate cancer (48), metastatic prostate cancer cells (49), neuroblastomas (50) and pancreatic cancer (51). In breast cancer, this increase is due to the absence of FEN1 promoter methylation (52).
Finally, glioblastoma cells overexpressing EGFRvIII, an oncogenic variant of epidermal growth factor receptor (EGFR), become hyper-dependent on a variety of DNA repair genes (53), including an enrichment of base excision repair genes required for repair of reactive oxygen speciesinduced DNA damage. One example is PARP-1 (53), which generally shows higher abundance in tumors. Besides PARP-1, other BER enzymes (TDG, OGG1) were also upregulated in EGFRvIII-containing cells after radiation exposure. The increased reliance on BER in these cells suggests the presence of elevated reactive oxygen species levels abundance cells with resultant genomic instability.
Nucleotide Excision Repair
The nucleotide excision repair (NER) pathway is a major DNA repair process that safeguards genome integrity by repairing numerous DNA modifications, especially bulky helix-distorting damage (54). However, recent work has revealed that some proteins in NER have activities that go beyond DNA repair and include nucleosome remodeling, histone ubiquitylation, and transcriptional activation of genes involved in nuclear receptor signaling, stem cell reprogramming and post-natal mammalian growth (55).
Tumors with enhanced NER have an intrinsic resistance to radiotherapy and chemotherapy (56), leading to continued growth and metastasis after treatment (57). On the other hand, NER is often disrupted in testicular germ cell tumors due to loss of XPA expression (58). Likewise, XPC, a critical component of global genome NER, is controlled by promoter methylation in bladder cancer (59). In addition, ERCC1 is also inactivated through promoter methylation in glioma tumors (60). Moreover, mutations of XP (xeroderma pigmentosum complementation group) A, B, C, E, F and G have been found in skin and testicular cancer, and variant expression of ERCC1 (excision repair complementation group 1) or XPD was demonstrated in lung cancer (61).
Mismatch Repair
Mismatch repair (MMR) targets incorrectly paired nucleotides introduced accidentally by DNA polymerases or after treatment with base-modifying chemotherapeutic drugs (e.g., alkylating agents) (62). MMR disruption causes microsatellite instability (MSI), a form of genetic instability associated with cancer. Familial cases of colonic tumors with MSI in Lynch syndrome result from germline mutations in mismatch repair genes, primarily MSH2 and MLH1 (63). However, most MSI-high tumors arise from an epigenetic defect in sporadic cases of cancer (64). Methylation in the promoter region of MLH1 correlates with decreased activity in sporadic colon cancer (65, 66). Likewise, MLH1 is also controlled by aberrant methylation in sporadic endometrial carcinoma (67), gastric cancer (68) and many other cancers.
Homologous Recombination
DNA double-strand breaks (DSBs) pose the most serious threat among all genotoxic assaults to the survival of cells and are repaired by either homologous recombination (HR) or non-homologous end joining (NHEJ). HR is disrupted in breast and ovarian cancer (69). BRCA1 and BRCA2, two important players in the HR pathway, are mutated in early onset breast and ovarian cancer (70), prostatic (71) and pancreatic cancer (72). Loss of BRCA1 expression by promoter hypermethylation is also seen in nonhereditary breast and ovarian cancer (73). In addition, the gene that encodes NBS1, which along with MRE11 and RAD50 constitute a heterotrimeric complex that senses DNA damage mainly in the form of double-strand breaks (74), is often mutated in lymphoma (Nijmegen breakage syndrome) (75). However, loss of NBS1 expression is found in prostate cancer (76). Additionally, RAD50 frameshift mutations, which result in a truncated protein, occur in a third of gastrointestinal cancers (77). The Fanconi Anemia/BRCA pathway, which repairs DNA crosslinks (74), is often impaired in a number of hematogenous and solid tumors. Thus, homozygous mutation of numerous FA genes (A, B, C, D1, E, F, G, I, J, L, M and BRCA2) or heterozygous mutation of some FA genes (e.g., FANCA, FANCC, FANCG and BRCA2) has been shown in hereditary breast, ovarian, cervical, prostatic, lung, pancreatic, gastric cancers, as well as melanoma and leukemia (78).
Non-homologous End Joining
DSBs are predominantly repaired by non-homologous end joining (NHEJ). The gene encoding DNA ligase IV, a major mediator of this pathway, is mutated in leukemia (Lig4 syndrome) (5), whereas Artemis, a structure-specific endonuclease, is mutated in lymphoma (Omenn syndrome) (4). Loss of Ku70 expression in cervical, rectal and colon cancer has also been reported (79–81), whereas Ku86 protein abundance is reduced in some rectal cancers (80).
Translesion DNA Synthesis
The translesion synthesis (TLS) machinery bypasses DNA adducts during DNA replication with the help of low stringency DNA polymerases (β, ι, κ). As mentioned above, Polβ is overexpressed in prostate, ovary, uterus and stomach cancers (38), whereas Polι is overexpressed in breast cancer (82) and Polκ is overexpressed in lung cancer (83). Elevation in expression and activity of the error-prone polymerase, Polβ, accounts for the increase in cisplatin resistance and mutagenesis of many cancers (84).
DNA DAMAGE INDUCED CELL CYCLE CHECKPOINT MECHANISMS
Cell cycle checkpoints are activated to arrest transiently cell proliferation, allowing extra time to repair DNA damage. When lesions are repaired, cells resume cell cycle progression (checkpoint recovery). However, when damage is irreparable cells either remain permanently arrested (senescence) or undergo programmed cell death (74). Defects in cell cycle related DNA damage response pathways result in genome instability and lead to carcinogenesis. Central to the DNA damage response are members of the phosphoinositide 3-kinase (PI3K)-related protein kinase family, ATM, ATR (ATM and Rad3-related), and DNA-PKcs (DNA protein kinase catalytic subunit) that sense the damage, and amplify the signal by phosphorylating numerous downstream substrates, including checkpoint kinases 1 and 2 (74). Activation of the upstream kinases require recognition of the damage. For DSBs, this is achieved by the heterotrimeric MRE11-RAD50-NBS1 (MRN) complex that directly binds to the exposed ends of DNA, recruits ATM and initiates its activation (85). Active ATM phosphorylates histone variant H2AX, which serves as a docking site for MDC1 (86) and many other proteins, including 53BP1, RNF8, RNF168 and BRCA1 (87).
Single-stranded DNA lesions generated from stalled replication forks are rapidly coated by replication protein A complexes, which then recruit ATR and its binding partner ATRIP (88), and independently Rad17 that in turn recruits the heterotrimeric RAD9-RAD1-HUS1 (9–1–1) complex to the site of damage. Subsequently, ATR phosphorylates the 9-1-1 complex, then RAD9 binds TopBP1 and RHINO (89, 90), which enhance ATR activity. Active ATR phosphorylates many substrates, including CHK1, an event important for establishing the cell cycle checkpoint (91).
Transition between the different phases of the cell cycle is dependent on cyclin-dependent kinases. Their activity is negatively regulated by WEE1 and MYT1 kinase mediated phosphorylation and, conversely, they become activated by the CDC25 phosphatases that dephosphorylate the inhibitory phosphorylations (92). After DNA damage, CHK1 induces CDC25A degradation by phosphorylating CDC25A and targeting it for proteosomal degradation (93). Alternatively, CHK1 activates WEE1 or NEK11 that further phosphorylates CDC25A, again preparing it for degradation (94). CHK2, although redundant for checkpoint activation in p53-proficient cells, becomes important for IR-induced cell cycle arrest in p53-deficient cells (95). Another mechanism implicated in G1 checkpoint induction is rapid degradation of cyclin D1 and release of cyclin-dependent kinase inhibitor p21waf1/cip1, culminating in cdk2/cyclin E inhibition and blocking G1/S transition (96). Cyclin D1 destruction is induced by GSK3β phosphorylation and SCF-dependent proteosomal degradation (97) or direct phosphorylation of the F-box protein FBXO31 by ATM (98).
CHK1 is crucial for maintaining genomic stability as it is required for monitoring replication fork progression during S phase of the cell cycle, and inhibition of CHK1 leads to stalling of replication forks and irreversible fork collapse (99–101). Furthermore, CHK1 also influences many aspects of mitosis by controlling cyclin B/Cdk1 activation, contributing to spindle checkpoint function, chromosome segregation and cytokinesis (102–104).
Tumor suppressor p53 plays an important role in cell cycle checkpoints. Following genotoxic stress, ATM/ATR/ DNA-PKcs and CHK1/2 phosphorylate as well as stabilize p53. Subsequently, p53 drives a transcription program that includes p21waf1/cip1, which plays a pivotal role to initiate G1 and sustain G2 arrest (105, 106). Furthermore, p53 transcriptionally represses expression of cyclin B, CDC25B and polo-like kinase 1 (Plk1) that are required for mitotic entry (107–109). p53 also participates in DNA repair and it is the main factor that determines the choice between DNA damage repair or the induction of senescence or apoptosis.
CELL CYCLE CHECKPOINT CONTROL AND CANCER
The cell cycle response to DNA damage is a highly efficient barrier against tumorigenesis. Premalignant cells have to overcome this barrier to progress into more malignant states (110, 111). Failure of mechanisms that regulate DNA damage checkpoint control leads to chromosomal aberrations (112) and genomic instability (113), both of which contribute to neoplastic transformation (114). Furthermore, alterations of proteins that control the DNA damage response-signaling pathway and impair the stringency of cell cycle checkpoints create a permissive environment that allows mutations to accumulate. The rapid accumulation of mutations in the genome of a cell leads to the so-called ‘‘mutator phenotype’’, which contributes to tumor progression by creating tumor heterogeneity and subsequent emergence of aggressive types of cancer (115). The importance of an intact checkpoint related DDR mechanism is further underscored by the existence of hereditary cancer predisposition syndromes that are the result of a germ line mutation in a DDR gene, e.g., ataxia telangiectasia mutated (ATM), ataxia telangiectasia-like disorder (MRE11), Nijmegen breakage syndrome (NBS1), hereditary breast/ovarian cancer (BRCA1/2), Fanconi anemia (FA pathway genes) and Li-Fraumeni syndrome (TP53) (116). As mentioned previously, sporadic cancers have at least one defect in the DDR pathway, manifested as an altered cell cycle profile or sensitivity/resistance to genotoxic stress (7, 8), illustrating the importance of the DDR signaling in tumorigenesis.
Haploinsufficiency for a variety of DDR proteins, including ATM, ATR, γ-H2AX and CHK1 (117–121) or knockdown/knockout of NBS1 (122, 123), RPA (124, 125), RAD17 (126), BRCA1 (127) and BRCA2 (128) is associated with genomic instability. Combined ATM and RAD9 (129) and ATM and HUS1 (130) haploinsufficiency led also to increased genomic instability. ATR and CHK1 participate in HR repair (131–133), besides regulating the S and G2 checkpoints and replication initiation and fork stability (134–139), thus facilitating repair of DNA breaks and preserving genomic integrity. CHK2 is an important mediator of cell cycle checkpoints, DNA repair and apoptosis. CHK2 is usually absent or downregulated in non-small cell lung cancer (140). Heterozygous mutations in ATR and CHK1 have been found in a subset of stomach, colon and endometrial cancers (141–143). In animal studies, ATR and CHK1 hypomorphic mutations could contribute to increased risk of tumorigenesis (118, 144).
Other members of the DDR network, such as PLK1 (145), and Aurora kinases (146) are frequently overexpressed in human tumors and promote chromosomal instability through defects in the spindle assembly checkpoint that result in chromosome missegregation and centrosome amplification. In addition, cyclin-dependent kinases (CDK) are overactive in many tumors and a number of proteins that enhance or reduce CDK activation display oncogenic or tumor suppressive traits, respectively (147). CDC25 phosphatase (148), which is required for CDK activation, can act as an oncogene, whereas WEE1 that opposes the activation of CDK1 and enforces G2/M arrest after DNA damage acts as a tumor suppressor (149, 150). Moreover, dysregulation of cyclin E is considered a major factor of tumorigenesis. Elevated abundance of cyclin E is associated with various neoplasias (151) and its prolonged expression induces chromosomal instability (152). A number of CDK-specific inhibitors are currently being tested in clinical trials for the treatment of patients with a variety of cancers, such as multiple myeloma, pancreatic, and lung carcinomas (147).
ROLE OF CELL CYCLE CHECKPOINT CONTROL AND DNA REPAIR GENES IN METASTASIS
Metastasis is the most clinically important attribute of cancer as more than 90% of cancer-related deaths are due to lack of local control. Tumor metastasis consists of a series of complex steps that need to be executed successfully to give rise to detectable tumors at sites distal to the organs where primary tumors initiate (153). Large-scale analyses of gene expression profiles of human cancers have revealed aberrant expression patterns of a number of genes involved in cell adhesion, migration, angiogenesis, kinase activation and other tumor-related functions. Although there is ample evidence that DNA repair genes are associated with the onset of tumorigenesis and DNA repair gene deficiencies cause inherent predisposition to cancer, a direct role of DNA repair or cell cycle checkpoint proteins in the etiology of metastasis has not been shown conclusively. Currently, the role of DNA repair genes in metastasis has only been inferred by DNA gene expression microarray analyses or by in vitro assays that serve as surrogates for in vivo metastasis phenotypes. Some of these genes are downregulated as tumors progress to a more malignant stage, while other genes are actually overexpressed and can affect the metastatic process. Whether these genes control the metastatic process due to their role in DNA repair or cell cycle control and the maintenance of genomic stability or because of some novel functions (e.g., acting as transcription factors or co-factors, or by more direct involvement for example in cell adhesion to matrix, etc.) is currently not known.
A genome-wide screen that compared gene expression in metastatic prostate and primary prostate tumors identified a strong correlation between high proliferation rates in metastatic cancers and overexpression of genes that participate in cell cycle regulation, DNA replication and DNA repair (49). Oncomine analyses showed that numerous human cancers, such as of the prostate, brain, cervix, head and neck, kidney, bladder and pancreas, displayed elevated levels of DNA repair proteins [for review see ref. (154)]. Moreover, in melanoma, there is considerable evidence that DNA repair genes are upregulated in metastases compared with primary tumors (57, 155). Gene expression microarray analyses as well as immunohistochemical examination of human melanoma specimens have shown an increase in expression of genes involved in HR and NER, but not BER (155). In contrast to the progression from melanocytes to primary melanoma, genetic stability appears to be necessary for a melanoma cell to give rise to distant metastasis (156). Therefore, the majority of neoplastic cells, found in primary melanomas poised to metastasize, have overexpressed genes responsible for efficient repair, ultimately resulting in genetically stable cells that are able to metastasize and grow at distant sites (155). Based on these results, it has been hypothesized that genomic instability is beneficial for the early stages of tumor development, whereas advanced and metastatic tumors overexpress an array of DNA repair genes to ensure a minimum of genomic stability (157). This inactivation– activation mode of DNA repair genes is not without precedent in other contexts (e.g., chemoresistance). It has been shown experimentally that a repair pathway may become inactivated early in carcinogenesis resulting in chromosomal instability, whereas consequent secondary mutations confer a selective advantage to the tumor. Subsequently, the repair pathway is reactivated (78). A case in point is the loss of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian cancer and re-gain of the pathway activity after prolonged treatment with cisplatin, which results in resistance to the drug (158).
A number of genes that participate in DNA damage induced checkpoints and DNA repair, and are either upregulated (e.g., RAD9, PARP1) or downregulated/ mutated (e.g., BRCA1/2, ATM and TP53) have been associated with metastasis that was demonstrated using a variety of in vitro assays and by examining human tumor specimens by immunohistochemistry. A review of some of these genes and their potential relationship to metastasis follows.
RAD9 Activities
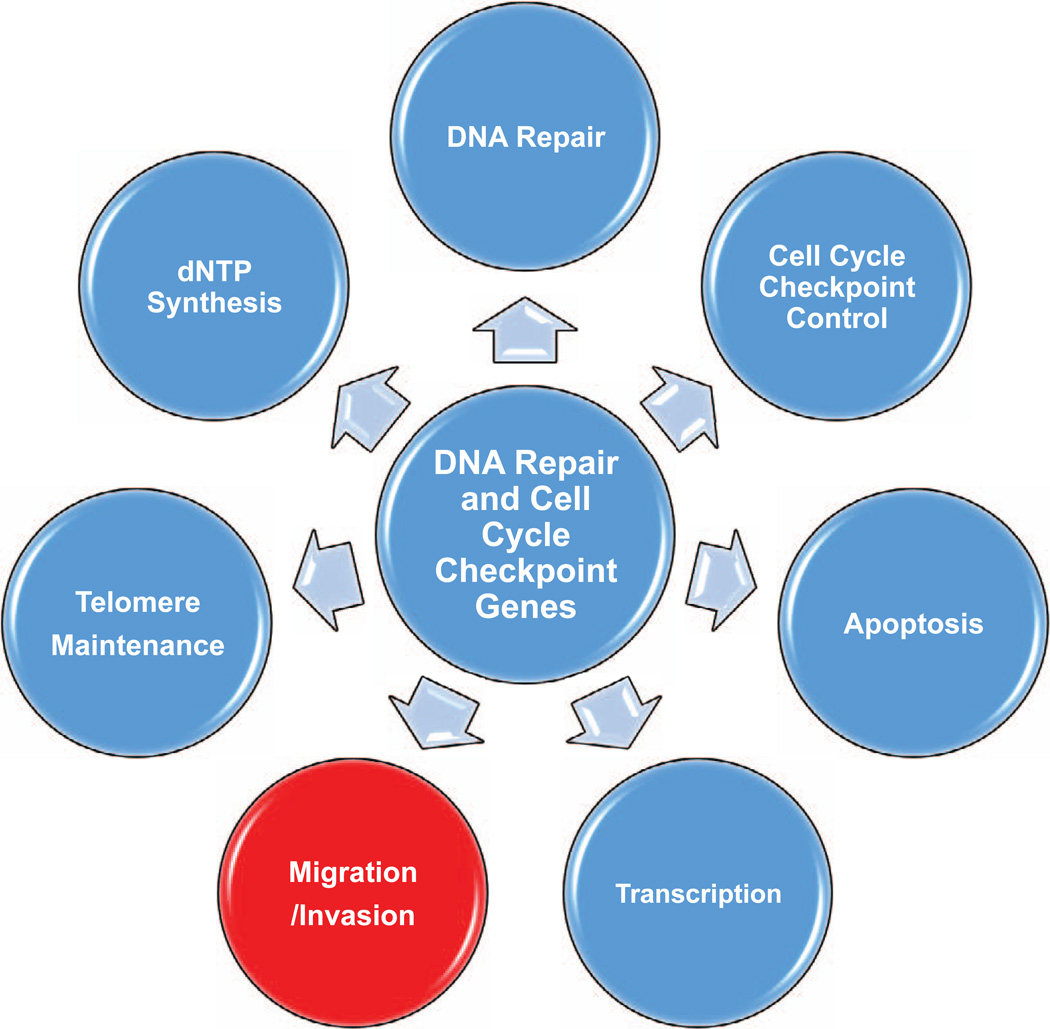
RAD9 can function as part of a heterotrimer with RAD1 and HUS1 (the RAD9-RAD1-HUS1 complex, 9-1-1), which is recruited to DNA damage sites by the RAD17-RFC (replication factor C) complex and is required for the subsequent activation of CHK1 and cell cycle arrest (159). However, RAD9 is a versatile protein that participates in numerous cell functions besides cell cycle checkpoint activation, such as DNA repair, telomere maintenance, dNTP biosynthesis, apoptosis and transcriptional regulation of genes (160). In addition, RAD9 can interact with several other proteins outside the context of the 9-1-1 complex and checkpoint functions (160).
Human RAD9 is involved in almost all aspects of DNA repair, including base excision repair (161, 162), nucleotide excision repair (163), mismatch repair (164) and homologous recombination, but not non-homologous end joining (165). Telomere instability and ionizing radiation sensitivity are linked to defective DNA repair (166) and RAD9 affects both. Moreover, chromosome end-to-end associations have been connected to genomic instability and carcinogenesis (167–169). When RAD9 is inactivated, increases in chromosome end-to-end associations and frequency of telomere loss are observed (165). Studies in murine embryonic stem cells lacking Mrad9 (Mrad9–/– ES cells) demonstrate a marked increase in spontaneous chromosome aberrations (an increase in the frequency of chromosome and chromatid breaks) and HPRT (hypoxanthine phosphorybosyl transferase) mutations even in the absence of exposure to exogenous DNA damaging agents, indicating a role in the maintenance of genomic integrity (170).
RAD9 functions in apoptosis, in addition to its role in cell cycle checkpoint control and DNA damage repair pathways. Mrad9 deficiency causes midgestational embryonic death, accompanied by increased apoptosis and reduced cellular proliferation (170). The lack of Mrad9 in mouse ES cells also causes enhanced spontaneous apoptosis (171). RAD9 can interact and neutralize the action of anti-apoptotic Bcl-XL and Bcl-2 (172), and induce pro-apoptotic Bax activation (173). The pro-apoptotic action of RAD9 is potentiated by c-Abl phosphorylation (174) and protein kinase C delta (175), as well as the p63 transcription factor (176).
Another important and largely unexplored activity of RAD9 is its ability to function as a transcription factor and regulate a number of downstream target genes, most notably p21waf1/Cip1 (177). Human RAD9 has also been identified as a coregulator that can suppress androgen-androgen receptor transactivation in prostate cancer cell lines (178).
The Role of RAD9 in Metastasis
Reduction in RAD9 levels is associated with genomic instability manifested as telomere dysfunction, aberrant chromosomal segregation, high spontaneous levels of mutations, as well as defective DNA repair. Thus, given the roles of RAD9 in maintaining genomic stability, it is reasonable to hypothesize that the protein is important for tumorigenesis. Indeed, studies by a number of laboratories have linked aberrations in RAD9 abundance to a variety of cancers or an impact on phenotypes representing hallmark features characteristic of neoplastic transformation (160). Aberrant RAD9 expression has been associated with breast, lung, skin, thyroid and gastric cancers. RAD9 is frequently overexpressed in human prostate cancer tissue specimens as well as prostate cancer cell lines and, importantly, downregulation of RAD9 in human tumor cell line xenografts impairs growth in nude mice, thus establishing a causative role for RAD9 in prostate cancer (179).
Cancer metastasis is a multi-step process in which tumor cells progressively acquire traits, including detachment from the extracellular matrix, anoikis resistance (defined as resistance to cell death triggered when cells lose adhesion to extracellular matrix), migration and invasion through the basement membrane, intravasation to blood and lymphatic vessels, extravasation from the circulation to distant sites, the ability to stimulate angiogenesis, and, finally, formation of macroscopic secondary malignant growths. At the molecular level, a number of signaling pathways, including those that involve integrins and Akt, contribute to the survival and progression of a tumor. Integrins are heterodimeric αβ transmembrane receptors that connect the extracellular matrix to the cytoskeleton and play important roles in migration, invasion and anoikis resistance. In particular, β1 integrin is known to confer higher survival and metastatic capacity to a number of cancer cells, including those of prostate origin (180, 181). The serine/ threonine protein kinase Akt is a downstream effector of PI3K and an important regulator of various cellular functions, including cell metabolism, transcription, survival and proliferation. Activation of Akt, due to mutations of the phosphatidylinositol 3' kinase (PI3K) p110 catalytic subunit or to loss of the phosphatase and tensin homolog (PTEN) tumor suppressor gene, occurs frequently in human cancers. The cancer cells rely heavily on active Akt to survive after experiencing a number of insults, such as genotoxic stress or growth factor depletion, and to regulate metastasis (182).
The first indication that RAD9 may be related to metastasis came from immunohistochemical analyses of human noncancer and cancer prostate specimens where the protein levels were positively correlated with more advanced stages of the disease (179). Furthermore, a number of in vitro metastasis markers such as cell motility, invasion, anoikis resistance and anchorage-independent growth, as well as activation of tumor promoting signaling pathways, specifically integrin expression and Akt activation were examined (183). Suppression of RAD9 protein abundance, by RNA interference, reduced both migration and invasion of DU145 as well as PC3 human prostate cancer cell lines, whereas ectopically expressing Mrad9, the mouse homolog of human RAD9, restored the phenotype in these cells (183). Likewise, anchorage-independent growth, which reflects most faithfully the in vivo metastatic potential of a cancer cell, was impaired when RAD9 was silenced in DU145 prostate cancer cells. Malignant cells have developed mechanisms to evade anoikis and either proliferate without matrix support or enter quiescence until a more suitable environment is presented. Anoikis resistance is, therefore, a prerequisite of tumor metastasis and is considered a hallmark of cancer. Akt kinase plays a pivotal role in the resistance of malignant cells to anoikis. RAD9 downregulation impaired Akt phosphorylation when prostate cancer DU145 and LNCaP cells were maintained in suspension. Conversely, when Mrad9 was ectopically expressed in DU145 with reduced levels of endogenous RAD9, Akt phosphorylation was restored, and cells became more resistant to anoikis (183). Silencing of RAD9 leads to a marked down-regulation of integrin β1. In addition, ectopic expression of Mrad9 restores integrin β1 levels when endogenous RAD9 expression is knocked down in DU145 cells. Furthermore, reduction of integrin β1 protein levels by a specific siRNA negated the effect of Mrad9 on migration and invasion, suggesting that RAD9 affects these metastasis-related processes through the activity of integrin β1 (183).
In addition to immunochemical data with human prostate specimens and the in vitro metastasis assays, gene expression profiling information for human prostate cancer also provides evidence of a role for RAD9 in tumor progression. Querying publically available datasets (184– 186) revealed that the relative RAD9 mRNA abundance in metastatic prostate tumors is twice as high as in primary prostate tumors (CGB and HBL, unpublished observations).
Given the function of RAD9 in tumorigenesis, it is reasonable to also consider whether other members of the 9-1-1 complex or RAD17 can impact the process. ATM/ATR-mediated phosphorylation of human RAD17, which recruits 9-1-1 to DNA damage sites, is required for claspin recruitment and CHK1 activation in response to genotoxic responses (187). However, a phosphorylation-defective RAD17 can neither recruit the 9-1-1 complex to the damage site, nor induce G2 checkpoint arrest in response to DNA damage (188). The potential association of RAD17 with cancer has been demonstrated in a number of studies. It has been shown that RAD17 acts as a haploinsufficient tumor suppressor that responds to oncogenic stress and loss of RAD17 is associated with poor prognosis in human B-cell lymphoma patients (189). Likewise, RAD17 is downregulated in head and neck squamous cell carcinomas (190). In contrast, elevated levels of RAD17 have been associated with breast (191) and lung carcinomas (192). Furthermore, immunohistochemical analyses have demonstrated that expression of human RAD17 might correlate with more advanced stages of non-small cell lung carcinoma (NSCLC). Abundance of RAD17 mRNA was correlated with lymph node metastasis, whereas RAD17 protein was highly prevalent at the advancing margin of the tumor of lung cancer tissue but not within the normal lung tissue (193).
Downregulation of HUS1 and RAD1 (as well as RAD9) lead to defects in DNA replication and cell cycle checkpoint control (194). HUS1 deficiency, for example, sensitizes mouse embryonic fibroblasts to etoposide-induced apoptosis (195), and HUS1 downregulation sensitizes human lung carcinoma cells to cisplatin (196). Similarly, HUS1 or RAD9 downregulation renders cells susceptible to ionizing radiation (197, 198). However, unlike RAD9, there is little information regarding the role of HUS1 and RAD1 in tumor initiation and/or progression. In one study, HUS1 levels correlated significantly with a number of adverse clinicopathologic factors in ovarian cancer, including stage, p53 and BAX expression, mitotic index, and apoptotic index (199). On the other hand, heterozygous deletion of mouse Mrad1 facilitates the development of experimental skin cancer in response to treatment with the carcinogen 7,12-dimethylbenzanthracene (DMBA) (200).
PARP-1
The PARP-1 protein is an abundant nuclear enzyme that modifies substrates by poly(ADP-ribose)ylation and is involved in the repair of single-strand breaks (116). In response to genotoxic insult, PARP-1 is recruited to sites of damage, where it becomes activated, and mediates the assembly as well as function of the base excision repair machinery (201). Distinct from its role in DNA repair, PARP-1 can act as a transcriptional regulator to control a diverse array of functions, including enhancer binding, association with insulators, modulation of chromatin structure, and/or transcription factor regulation (202). PARP-1 is overexpressed aberrantly in a number of human cancers, including those of breast and prostate (203, 204).
PARP-1 elicits pro-tumorigenic effects in androgen receptor-positive prostate cancer cells, in both the presence and absence of genotoxic drugs. Mechanistically, enzymatically active PARP-1 plays a critical role in the control of androgen receptor (AR) function. Moreover, in models of advanced prostate cancer, PARP-1 enzymatic activity is enhanced and regulates castration-resistant AR activity, further linking PARP-1 to AR activity and disease progression (205).
Recent evidence has uncovered a possible role of PARP-1 in metastasis. Chromosomal rearrangements involving genes encoding ETS transcription factors (ERG, ETV1) are found in 50% of human prostate cancer cases (206, 207). Translocations place the coding region or ERG or ETV1 under control of androgen-responsive promoters, such as TMPRSS2, thereby activating expression in response to androgens. At least in the case of ETS expressing cancers, the role of PARP-1 in metastasis appears to be due to its function as a transcription factor, although a role in DNA repair cannot be excluded (205, 208). ETS gene-mediated transcription and cell invasion require expression and activity of PARP-1 and DNA-PKcs, the kinase involved in NHEJ (204).
PARP-1 is also able to control SNAIL-1 transcription and Snail protein stability (208). Snail is a master regulator of the epithelial-mesenchymal transition (EMT) and has been implicated in key tumor biological processes, such as invasion and metastasis (209). PARP-1 is involved in the activation of SNAIL-1 gene transcription through binding to the integrin-linked kinase (ILK) promoter (210). PARP-1 downregulation has a clear effect on the EMT phenotype, with SNAIL-1 repression and E-cadherin upregulation, decreased cell elongation and invasiveness. Furthermore, PARP-1 and NF-kB together with Snail1 drive expression of the fibronectin gene, which is a typical mesenchymal gene (211). In contrast, PARP-1 has been shown in HaCaT keratinocyte cells to attenuate SMAD-mediated transcription and negatively regulate TGFβ-controlled genes involved in the EMT program, such as fibronectin (FN1) and N-cadherin (CDH2) (212).
BRCA1/2
The breast and ovarian cancer predisposition genes, BRCA1 and BRCA2, encode proteins that are required for efficient homologous recombination repair (213, 214). Germline mutations of BRCA1 predispose women to breast and ovarian cancers (70). Since its discovery, BRCA1 has been reported to be involved in multiple functions, all of which control genomic stability in the nucleus, such as cell cycle regulation and checkpoint activation, DNA repair (specifically HR), centrosome regulation, apoptosis and chromatin remodeling (215). BRCA1 also functions as a transcription regulatory cofactor (216).
Mutations in BRCA1 do not directly result in tumor formation, but instead cause genomic instability, subjecting cells to a high risk of malignant transformation (217). Furthermore, disruption of BRCA1 transcriptional activity can be crucial for tumor formation (218). Recent reports have identified novel roles of BRCA1 in the regulation of caveolin-1 (CAV-1) transcription and the inhibition of cell invasiveness (219), as well as the physical association with plasma membrane proteins ezrin-radixin-moesin, thus controlling cell spreading and motility, which have significant implications for tumor invasion and metastasis (220). Interestingly, the CAV-1 gene is upregulated in cells treated with ionizing radiation and its expression protects cells after exposure through modulating activities of both the HR and NHEJ pathways (221).
BRCA2 is considered a tumor suppressor gene involved in homologous recombination repair of DNA double-stranded breaks (222). In addition to its role in mediating DNA repair and genome stability, BRCA2 plays a role in the stabilization of stalled DNA replication forks, centrosome duplication, cytokinesis, and transcriptional regulation (214, 223–225). BRCA2 can suppress tumor development by inhibiting cancer cell growth (226). Loss of BRCA2 triggers a proliferative response upon prostate cancer cell interaction with basement membrane proteins (227, 228). Besides its role in cell proliferation, BRCA2 functions in tumor progression as it can negatively impact on the metastatic potential of prostate cancer cells by down-regulating metalloprotease MMP-9 production through inhibition of PI3K/Akt, thus hindering cancer cell migration and invasion (229).
There is scant clinical information regarding the connection of BRCA1/2 mutations to the etiology of tumor metastasis. However, it has recently been shown that germline mutations in BRCA1/2 confer a more aggressive prostate cancer phenotype, with a higher probability of nodal involvement and distant metastasis (230).
NBS1
Nijmegen breakage syndrome (NBS) is a chromosomal instability disorder associated with cancer predisposition, radiosensitivity, microcephaly and growth retardation (231). The gene defective in NBS is NBS1 (p95, nibrin) and is a member of the DNA double strand break repair complex that also includes MRE11 and RAD50 (232). NBS1 is a putative tumor suppressor gene as shown by the existence of mutations discovered in different tumors (233, 234). NBS1 is also a prostate cancer susceptibility gene (76). In line with a pro-tumorigenic activity, c-MYC oncogene directly activates NBS1 expression (235), and NBS1 overproduction stimulates PI3K activity and enhances cell transformation (236). The induction of tumorigenicity by NBS1 overexpression may proceed through activation of an oncogenic pathway or the repression of a tumor suppressor, whereas mutations of NBS1 could also contribute to tumorigenesis through deficiency in DNA repair leading to genomic instability (233, 234, 237).
Evidence that NBS1 may be associated with metastasis is: (1) NBS1 overexpression correlates with head and neck squamous cell carcinoma metastasis; (2) NBS1 overexpression induces EMT through the upregulation of Snail1 levels and matrix metalloprotease, MMP-2 and increases invasiveness/metastasis of head and neck cancer cells as observed both in vivo and in vitro (238); and (3) NBS1 upregulates heat shock proteins A4 and A14 with a concomitant increase in the in vitro migration, invasion and soft agar colony formation of the lung adenocarcinoma H1299 cell line (239). In addition, it has recently been demonstrated that although NBS1 haploinsufficiency leads to increased mammary tumor latency in the MMTV-neu mouse model, the tumors that do form are characterized by high metastatic potential (240), further highlighting the role of NBS1 in tumor metastasis.
RAD51
A critical step in the formation of metastases is cell survival in the bloodstream. Normal and most cancer cells undergo programmed cell death (anoikis) when detached from their matrices, and metastatic cells must develop specific molecular strategies to survive or even proliferate in an anchorage-independent fashion before they localize to the metastatic site and extravasate. RAD51 is a DNA repair gene involved in tumorigenesis. Its downregulation has been associated with defects in error-free HR DNA repair. However, overexpression of RAD51, a rather common occurrence in human cancers (241), contributes to carcinogenesis as it is also associated with aberrant recombination between short repetitive elements and homologous sequences (241). Results with prostate cancer cell lines indicate that anchorage-independence sensitizes cells to genotoxic agents: however it also attenuates a faithful component of DNA repair by targeting the stability of RAD51. This temporal attenuation of HR may contribute to the accumulation of new mutations after DNA damage, which confer a selective advantage to the cells for survival under anchorage independent conditions (242).
XRCC3
The DNA repair protein known as X-ray complementing protein 3 (XRCC3), a member of the RAD51 family, participates in HR and is important for the maintenance of chromosome stability as well as DNA damage repair. XRCC3 affects the invasive behavior of MCF-7 and BT20 human breast cancer cell lines. Specifically, stable or transient overexpression of XRCC3 increased invasiveness in vitro (243). Moreover, XRCC3 overexpressing MCF-7 cells also showed a high frequency of tumorigenesis in vivo, and this phenotype was associated with increased activity of the metalloproteinase MMP-9 and the expression of known modulators of cell-cell adhesion and metastasis, such as CD44 (Receptor for hyaluronic acid), ID-1 (inhibitor of DNA binding 1), DDR1 (discoidin domain receptor tyrosine kinase 1), and TFF1 (trefoil factor 1). These findings suggest a role for XRCC3 in breast cancer cell line invasiveness and expression of genes associated with cell adhesion and invasion.
ATM
The ATM gene product is a serine/threonine protein kinase involved in cell cycle control, DNA repair and chromosomal stability. Its importance to cancer is underscored by the fact that inactivating mutations of ATM predispose individuals to the disease Ataxia Telangiectasia (244). Evidence that ATM may promote tumor progression and metastasis has come from recent studies revealing that ATM kinase is hyperactive in late stage breast tumors with lymph node metastasis (245). It was further demonstrated that ATM phosphorylates and stabilizes the epithelialmesenchymal transition transcription factor SNAIL1, with a concomitant increase in cell migration and invasion in vitro and metastasis in vivo, which was reversed by inhibiting SNAIL phosphorylation by ATM (245). Likewise, inhibiting ATM kinase activity not only radiosensitizes human glioma cells, but it also impairs migration and invasion in vitro, possibly by reducing Akt and ERK activation (246). The stimulus that maintains high levels of ATM activation in advanced tumors is not known, but hypoxia or oxidative stress, which is able to promote metastasis and ATM activation, could be examples (247–250).
TP53
The p53 protein is involved in DNA damage repair and cell cycle checkpoint control (251). It plays critical roles in maintaining genetic stability, and TP53 is the most commonly mutated tumor suppressor gene in human cancers (251). Mutations of TP53 are mainly seen in later tumor stages and coincide with more aggressive types of cancer (252–254). Interestingly, the majority of TP53 alterations are missense mutations within the DNA binding domain, thus maintaining a full-length protein that has lost its tumor suppressor function (255). However, in addition to losing this function, many mutant p53 proteins also acquire novel, oncogenic activities (256). These gain-of-function p53 mutants lead to increased genomic instability by interfering with proper ATM activation and DNA repair (257, 258). This causes increased incidence of chromosomal translocations (259) and chymothripsis (260), as well as gene amplifications (261). Aneuploidy is promoted by inhibiting assembly of the mitotic spindle checkpoint, as well as inducing centrosome amplification (256). This can inhibit apoptosis by, among other mechanisms, suppressing the function of p73 that induces p53-independent apoptosis after DNA damage (262). p53 inhibits tumor metastasis by multiple mechanisms. For example, p53 controls the transcription of SMAR1 (scaffold/matrix attachment region binding protein 1), which in turn controls cyclin D1 (CCND1) gene expression and inhibits migration as well as invasion by interfering with TGFβ signaling in breast cancer (263). Not surprisingly, breast tumors in advanced stages show reduced expression of SMAR1 (263). In contrast, mutant p53 protein can promote aspects of the metastatic process (264), such as migration and invasion, epithelial to mesenchymal transition, or through the inhibition of p63 protein, which results in increased trafficking of β1 integrin (265), an integrin that is intimately involved in metastasis in human breast and prostate carcinomas (266–268). The increased rate of recycling of this integrin to the cell plasma membrane correlates positively with cell migration and invasion (269).
GADD45a
The growth arrest and DNA damage gene, GADD45a, plays important roles in the control of cell cycle checkpoints, DNA repair (270) and apoptosis (271). Mouse embryonic fibroblasts derived from GADD45a–null mice exhibit genomic instability, single oncogene-mediated transformation, loss of normal cellular senescence, increased cellular proliferation, centrosome amplification and reduced DNA repair. A high frequency of GADD45a point mutations has been identified in human pancreatic cancer (272), whereas increased gene methylation and decreased protein levels have been shown in breast cancer (273).
A role for GADD45a in metastasis is inferred by the observations that it is involved in the control of cell contact inhibition and cell-cell adhesion by enhancing β-catenin protein stability and translocation to the cell membrane (274). Furthermore, GADD45a inhibits cell migration and invasion by altering expression of various genes encoding extracellular matrix, cell communication, and cell adhesion proteins (275).
NM23
The gene NM23 functions in DNA repair and determining whether metastases will form (276). Unlike the other examples where established DNA repair genes are examined for their role in metastasis, NM23 was first established as an antimetastatic gene, and its significance as a metastasis suppressor has been highlighted in numerous studies (276). Low NM23 expression in primary melanomas is correlated with poor clinical outcome, suggesting relevance of NM23 deficiency to initiation and/or progression in earlier stages of this tumor (277). Subsequent studies revealed that NM23 participates in DNA repair as well. Importantly, the protein’s DNA repair activity is required for its metastasis suppressing function, albeit the exact mechanism remains elusive (278). Recent findings have shown that NM23-H1 (isoform H1) participates in nucleotide excision repair (279), however, it is not known whether this function is required for the antimetastatic role.
Other DDR Proteins
A number of other DDR proteins, such as MCPH1, 14-3-3σ, CDC25A, TIP60 and H2AX, have been associated with tumor metastasis. Evidence is mainly based on immunohistochemical analyses of clinical specimens or in vitro studies.
MCPH1 (microcephalin 1, also known as BRIT1), a repressor of human telomerase reverse transcriptase (hTERT) function and a key regulator in the DDR pathway, ensures genomic stability and acts as a barrier to the development of cancer (280). MCPH1 level is inversely correlated with the likelihood of breast cancer metastasis (281) or prostate cancer (282). Likewise, inactivation of the G2/M checkpoint protein 14-3-3σ correlates with lymph node metastasis in nasopharyngeal carcinoma (283). The axis PLK1-CDC25A that permits cells to enter mitosis is often dysregulated in metastatic hepatocellular carcinoma, and cisplatin treatment of metastatic cells does not lead to CDC25A degradation or PLK1 inactivation as normally happens after DNA damage is incurred. As a result, cells enter mitosis, but without mitotic catastrophe, thus leading to increased genetic instability (284). On the other hand, nonmetastatic cells responded to cisplatin with the degradation of CDC25A and the downregulation of PLK1 activity (284).
The tumor suppressor TIP60 is a protein lysine acetytransferase involved in DNA damage response and repair particularly of double-strand breaks (285), by acetylating and activating, among other substrates, ATM (286). The TIP60 gene is frequently downregulated in colon and lung carcinomas (287). Moreover, downregulation of TIP60 correlates with distant metastasis in colon cancer (288), as well as melanoma (289). In the latter case, ectopic expression of Tip60 in melanoma cells reduced and knockdown increased in vitro cell migration, pointing further to a potential role of Tip60 in metastasis (289).
Histone H2AX is an important effector of the DNA damage response that is responsible for recruiting cell cycle checkpoint and DNA repair factors to sites of double-strand breaks (290). By facilitating the DNA damage response and repair, H2AX functions as a tumor suppressor. H2AX maps to chromosome 11q23, a region that is deleted or mutated in a variety of human malignancies, including leukemia, breast and head and neck cancers (290). However, H2AX can also promote tumor growth and pathologic angiogenesis under conditions of hypoxia (291) and therefore aid the dissemination of tumor cells to metastasize to distant sites. H2AX is needed for endothelial cell proliferation under hypoxic conditions and for hypoxia-driven neovascularization, whereas genetic ablation of H2AX reduces the proliferation of these cells in vitro and in vivo (291). However, it is still not clear whether the DNA repair function of H2AX is required for the regulation of endothelial cell proliferation under hypoxia conditions (292).
CONCLUSIONS
In recent years, it has become apparent that many genes classically thought to operate in DNA repair or checkpoint control also have roles in carcinogenesis and in particular tumor metastasis as well. However, the molecular mechanisms involved are not completely understood. Most published experiments are correlative in nature, and therefore, at present, cause-effect relationships between most of these genes and cancer metastasis cannot be unambiguously assigned. The evidence that DNA repair or checkpoint genes participate in metastasis comes mainly from genome profiling data of primary versus metastatic tumors, or from immunohistochemical analyses of patient tumor specimens. In addition, in vitro data have linked expression of some of these genes to increased migration and invasion, anoikis resistance and anchorage-independent growth. Molecularly, DNA repair or cell cycle checkpoint genes affect pro-tumorigenic and pro-metastatic pathways involving Akt activation, integrin expression, as well as transcriptional control of genes involved in epithelial-to-mesenchymal transition and metalloproteases. It is therefore necessary to prove that the encoded proteins drive metastasis in animal models to demonstrate directly a cause-effect relationship. So far, this has been shown only for NBS1 in head and neck cancer cell lines by an in vivo tail vein metastasis assay (238).
Proteins that participate in DNA repair or cell cycle checkpoint control usually have multiple cellular, biochemical or molecular functions, as shown schematically in Fig. 1 and detailed in Tables I and II. An emerging common theme for many DNA repair proteins is that they can function in gene regulation as transcriptional factors or cofactors. RAD9, for example, can function as a sequence specific transcription factor and regulate a number of genes including p21waf1/Cip1 (177), whereas BRCA1, which lacks sequence specific DNA binding, can be recruited to promoters by sequence specific transcription factors and act as a transcriptional co-activator or co-repressor (216). Likewise, GADD45a alters global transcript abundance, affecting proteins important for cell migration and invasion, such as those impacting on cell–cell adhesion (275).
FIG. 1.
The multiple functions of DNA damage response and repair genes. Many genes discussed in this review have at least a subset of the activities depicted. For more details see Tables I and II.
TABLE I.
Functional Roles of DNA Damage Response and Repair Genes
| Gene | Role in DDR | Other functions | Refs. |
|---|---|---|---|
| 14-3-3σ | Cell cycle checkpoint | — | (283) |
| ATM | DDR | Cell metabolism | (244, 293) |
| BRCA1 | HRR, cell cycle checkpoint | Centrosome regulation, chromatin remodeling, apoptosis, transcriptional regulation | (213–216) |
| BRCA2 | HRR, cell cycle checkpoint | Centrosome duplication, transcriptional regulation | (214, 222–225) |
| CDC25A | Cell cycle checkpoint | — | (284) |
| GADD45a | NER | Transcriptional regulation, apoptosis | (270, 271, 275) |
| H2AX | DDR, DSBR | — | (290) |
| MCPH1/BRIT1 | HRR | — | (280) |
| NBS1 | DDR | — | (232) |
| NM23 | NER | Nucleoside diphosphate kinase | (276, 279) |
| PARP-1 | BER, HRR | Transcriptional regulation, apoptosis | (116, 201, 202) |
| RAD9 | BER, NER, MMR HRR, TLS, cell cycle checkpoint | Telomere maintenance, dNTP synthesis, apoptosis, transcription factor | (160–166, 170–176, 179) |
| RAD17 | DDR, cell cycle checkpoint | — | (187, 188) |
| RAD51 | HRR | — | (241) |
| TIP60 | DDR, DSBR | — | (285, 286) |
| TP53 | DDR | Transcription factor, apoptosis | (251, 256) |
| XRCC3 | NER | — | (243) |
Abbreviations: DDR, DNA damage response; BER, base excision repair; NER, nucleotide excision repair; MMR, mismatch repair; HRR, homologous recombination repair; NHEJ, non-homologous end joining; DSBR, double-strand break repair; TLS, translesion synthesis.
TABLE II.
Role of DNA Damage Response and Repair Genes in Tumor Growth and Metastasis
| Gene | Tumor growth | Metastasis | Refs. |
|---|---|---|---|
| 14-3-3σ | Tumor suppressor | Inactivation associates with metastasis | (283) |
| ATM | Tumor promoter | SNAIL1 phosphorylation and stabilization, correlates with metastasis | (245–250) |
| BRCA1 | Tumor suppressor | Metastasis suppressor, regulation of cav-1 | (70, 217–200, 230) |
| BRCA2 | Tumor suppressor | Downregulation associates with metastasis | (226–230) |
| CDC25A | Tumor promoter | Persistence of CDC25A expression associates with metastasis | (284) |
| GADD45a | Tumor suppressor | Migration, invasion inhibition, β-catenin exclusion from nucleus, correlates with decreased metastasis | (272–275) |
| H2AX | Tumor suppressor | Angiogenesis, correlates with metastasis | (290, 291) |
| MCPH1/BRIT1 | Tumor suppressor | Expression is inversely correlated with metastasis | (281, 282) |
| NBS1 | Tumor suppressor in some cancers, tumor promoter in others | SNAIL1, MMP-2 upregulation, EMT, migration, invasion, soft agar colony formation, elevated in vivo metastasis | (76, 231, 233–240) |
| NM23 | No effect | Metastasis suppressor | (276–278) |
| PARP-1 | Primary tumor growth | SNAIL1 transcription, EMT, invasion; may associate with metastasis | (203–205, 208) |
| RAD9 | Tumor promoter in some cancers, tumor suppressive activity in others | Migration, invasion, anoikis resistance, anchorage-independent growth, ITGB1 expression control, increased levels correlate with metastasis | (160, 179, 183) |
| RAD17 | Tumor promoter in some cancers, tumor suppressor in others | Overexpression correlates with metastasis | (189–193) |
| RAD51 | Tumor promoter | Destabilized under anchorage-independent conditions that may increase mutations and favor metastasis | (241, 242) |
| TIP6 | Tumor suppressor | Downregulation correlates with metastasis | (287–289) |
| TP53 | Tumor suppressor | Metastasis inhibitor, mutant p53 promotes metastasis by modulating ITGB1 stability | (251–254, 263–265) |
| XRCC3 | Tumor promoter | Invasion, MMP-9, CD44, ID-1, DDR1, TFF1 expression, associates with metastasis | (243) |
Notes. A direct link between protein/mRNA expression levels and/or gene mutations and metastasis has been demonstrated only for NBS1 and NM23, whereas in all other cases there is only an indirect correlation with metastasis, based either on gene expression microarray or immunohistochemical analyses of human cancer specimens or in vitro biological characteristics that may predict in vivo metastasis behavior. Abbreviations: EMT, epithelial to mesenchymal transition; MMP, matrix metalloprotease; ITGB1, integrin β1; CD44, CD44 antigen; ID-1, inhibitor of differentiation-1; DDR1, discoidin domain receptor tyrosine kinase 1; TFF1, trefoil factor 1; cav-1, caveolin-1.
Which of the multiple functions of DNA repair or cell cycle checkpoint control proteins is actually required for regulating metastases is not clear. One can speculate that DNA repair activity is required. On the other hand, the transcriptional regulatory activity of numerous DNA repair proteins will certainly be important. It is already known that DNA repair proteins control, at the transcriptional level, the abundance of many other proteins involved in metastatic pathways and certainly more will be discovered as studies progress. Finally, specific protein–protein interactions of DNA repair or checkpoint factors with as yet unidentified proteins will also likely influence metastasis. A very complex picture is emerging from all of these investigations. Nevertheless, from a pragmatic perspective, resolution of the molecular mechanisms involved is important to facilitate the design of therapies that target DNA repair and cell cycle checkpoint proteins as novel anti-cancer agents.
ACKNOWLEDGMENTS
This work was supported, in whole or part, by National Institutes of Health grants R01CA130536, R01GM079107 and P01CA49062 (HBL).
REFERENCES
- 1.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–817. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 2.de Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21:453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 3.Rustgi AK. The genetics of hereditary colon cancer. Genes Dev. 2007;21:2525–2538. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 4.Moshous D, Pannetier C, Chasseval RD, Deist FL, Cavazzana-Calvo M, Romana S, et al. Partial T and B lymphocyte immunodeficiency and predisposition to lymphoma in patients with hypomorphic mutations in Artemis. J Clin Invest. 2003;111:381–387. doi: 10.1172/JCI16774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roddam PL, Rollinson S, O’Driscoll M, Jeggo PA, Jack A, Morgan GJ. Genetic variants of NHEJ DNA ligase IV can affect the risk of developing multiple myeloma, a tumour characterised by aberrant class switch recombination. J Med Genet. 2002;39:900–905. doi: 10.1136/jmg.39.12.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li ML, Greenberg RA. Links between genome integrity and BRCA1 tumor suppression. Trends Biochem Sci. 2012;37:418–424. doi: 10.1016/j.tibs.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 8.Lieberman HB. DNA damage repair and response proteins as targets for cancer therapy. Curr Med Chem. 2008;15:360–367. doi: 10.2174/092986708783497328. [DOI] [PubMed] [Google Scholar]
- 9.Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- 10.Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 11.Lahtz C, Pfeifer GP. Epigenetic changes of DNA repair genes in cancer. J Mol Cell Biol. 2011;3:51–58. doi: 10.1093/jmcb/mjq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toyota M, Suzuki H. Epigenetic drivers of genetic alterations. Adv Genet. 2010;70:309–323. doi: 10.1016/B978-0-12-380866-0.60011-3. [DOI] [PubMed] [Google Scholar]
- 13.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 14.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 15.Mellai M, Caldera V, Annovazzi L, Chio` A, Lanotte M, Cassoni P, et al. MGMT promoter hypermethylation in a series of 104 glioblastomas. Cancer Genomics Proteomics. 2009;6:219–227. [PubMed] [Google Scholar]
- 16.Shamsara J, Sharif S, Afsharnezhad S, Lotfi M, Raziee HR, Ghaffarzadegan K, et al. Association between MGMT promoter hypermethylation and p53 mutation in glioblastoma. Cancer Invest. 2009;27:825–829. doi: 10.1080/07357900902783211. [DOI] [PubMed] [Google Scholar]
- 17.Herfarth KK, Brent TP, Danam RP, Remack JS, Kodner IJ, Wells SA, et al. A specific CpG methylation pattern of the MGMT promoter region associated with reduced MGMT expression in primary colorectal cancers. Mol Carcinog. 1999;24:90–98. doi: 10.1002/(sici)1098-2744(199902)24:2<90::aid-mc3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 18.Ogino S, Hazra A, Tranah GJ, Kirkner GJ, Kawasaki T, Nosho K, et al. MGMT germline polymorphism is associated with somatic MGMT promoter methylation and gene silencing in colorectal cancer. Carcinogenesis. 2007;28:1985–1990. doi: 10.1093/carcin/bgm160. [DOI] [PubMed] [Google Scholar]
- 19.Wolf P, Hu YC, Doffek K, Sidransky D, Ahrendt SA. O(6)-Methylguanine-DNA methyltransferase promoter hypermethyla-tion shifts the p53 mutational spectrum in non-small cell lung cancer. Cancer Res. 2001;61:8113–8117. [PubMed] [Google Scholar]
- 20.Wu JY, Wang J, Lai JC, Cheng YW, Yeh KT, Wu TC, et al. Association of O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation with p53 mutation occurrence in non-small cell lung cancer with different histology, gender, and smoking status. Ann Surg Oncol. 2008;15:3272–3277. doi: 10.1245/s10434-008-0078-9. [DOI] [PubMed] [Google Scholar]
- 21.Oue N, Shigeishi H, Kuniyasu H, Yokozaki H, Kuraoka K, et al. Promoter hypermethylation of MGMT is associated with protein loss in gastric carcinoma. Int J Cancer. 2001;93:805–809. doi: 10.1002/ijc.1403. [DOI] [PubMed] [Google Scholar]
- 22.Goldenberg D, Harden S, Masayesva BG, Ha P, Benoit N, Westra WH, et al. Intraoperative molecular margin analysis in head and neck cancer. Arch Otolaryngol Head Neck Surg. 2004;130:39–44. doi: 10.1001/archotol.130.1.39. [DOI] [PubMed] [Google Scholar]
- 23.Maruya S, Issa JP, Weber RS, Rosenthal DI, Haviland JC, Lotan R, et al. Differential methylation status of tumor-associated genes in head and neck squamous carcinoma: incidence and potential implications. Clin Cancer Res. 2004;10:3825–3830. doi: 10.1158/1078-0432.CCR-03-0370. [DOI] [PubMed] [Google Scholar]
- 24.Steinmann K, Sandner A, Schagdarsurengin U, Dammann RH. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol Rep. 2009;22:1519–1526. doi: 10.3892/or_00000596. [DOI] [PubMed] [Google Scholar]
- 25.Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–2371. [PubMed] [Google Scholar]
- 26.Esteller M, Risques RA, Toyota M, Capella G, Moreno V, Peinado MA, et al. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61:4689–4692. [PubMed] [Google Scholar]
- 27.Dianov GL, Hübscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41:3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson DM3rd, Kim D, Berquist BR, Sigurdson AJ. Variation in base excision repair capacity. Mutat Res. 2011;711:100–112. doi: 10.1016/j.mrfmmm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slupska MM, Baikalov C, Luther WM, Chiang JH, Wei YF, Miller JH. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J Bacteriol. 1996;178:3885–3892. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, et al. Inherited variants of MYH associated with somatic G:C-.T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 31.Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, et al. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C->T:A mutations. Hum Mol Genet. 2002;11:2961–2967. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- 32.Arai K, Morishita K, Shinmura K, Kohno T, Kim SR, Nohmi T, et al. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 33.Chevillard S, Radicella JP, Levalois C, Lebeau J, Poupon MF, Oudard S, et al. Mutations in OGG1, a gene involved in the repair of oxidative DNA damage, are found in human lung and kidney tumours. Oncogene. 1998;16:3083–3086. doi: 10.1038/sj.onc.1202096. [DOI] [PubMed] [Google Scholar]
- 34.Shinmura K, Yokota J. The OGG1 gene encodes a repair enzyme for oxidatively damaged DNA and is involved in human carcinogenesis. Antioxid Redox Signal. 2001;3:597–609. doi: 10.1089/15230860152542952. [DOI] [PubMed] [Google Scholar]
- 35.Peng B, Hurt EM, Hodge DR, Thomas SB, Farrar WL. DNA hypermethylation and partial gene silencing of human thymine-DNA glycosylase in multiple myeloma cell lines. Epigenetics. 2006;1:138–145. doi: 10.4161/epi.1.3.2938. [DOI] [PubMed] [Google Scholar]
- 36.Robertson KA, Bullock HA, Xu Y, Tritt R, Zimmerman E, Ulbright TM, et al. Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res. 2001;61:2220–2225. [PubMed] [Google Scholar]
- 37.Silber JR, Bobola MS, Blank A, Schoeler KD, Haroldson PD, Huynh MB, et al. The apurinic/apyrimidinic endonuclease activity of Ape1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin Cancer Res. 2002;8:3008–3018. [PubMed] [Google Scholar]
- 38.Albertella MR, Lau A, O’Connor MJ. The overexpression of specialized DNA polymerases in cancer. DNA Repair (Amst) 2005;4:583–593. doi: 10.1016/j.dnarep.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Srivastava DK, Husain I, Arteaga CL, Wilson SH. DNA polymerase beta expression differences in selected human tumors and cell lines. Carcinogenesis. 1999;20:1049–1054. doi: 10.1093/carcin/20.6.1049. [DOI] [PubMed] [Google Scholar]
- 40.Canitrot Y, Cazaux C, Fréchet M, Bouayadi K, Lesca C, Salles B, et al. Overexpression of DNA polymerase beta in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc Natl Acad Sci U S A. 1998;95:12586–12590. doi: 10.1073/pnas.95.21.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace SS, Murphy DL, Sweasy JB. Base excision repair and cancer. Cancer Lett. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3:998–1001. [PubMed] [Google Scholar]
- 43.Hiraoka LR, Harrington JJ, Gerhard DS, Lieber MR, Hsieh CL. Sequence of human FEN-1, a structure-specific endonuclease, and chromosomal localization of the gene (FEN1) in mouse and human. Genomics. 25:220–225. doi: 10.1016/0888-7543(95)80129-a. 199. [DOI] [PubMed] [Google Scholar]
- 44.Wu X, Wilson TE, Lieber MR. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc Natl Acad Sci U S A. 1999;96:1303–1308. doi: 10.1073/pnas.96.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikuchi K, Taniguchi Y, Hatanaka A, Sonoda E, Hochegger H, Adachi N, et al. Fen-1 facilitates homologous recombination by removing divergent sequences at DNA break ends. Mol Cell Biol. 2005;25:6948–6955. doi: 10.1128/MCB.25.16.6948-6955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh P, Zheng L, Chavez V, Qiu J, Shen B. Concerted action of exonuclease and Gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n-and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J Biol Chem. 2007;282:3465–3477. doi: 10.1074/jbc.M606582200. [DOI] [PubMed] [Google Scholar]
- 47.Nikolova T, Christmann M, Kaina B. FEN1 is overexpressed in testis, lung and brain tumors. Anticancer Res. 2009;29:2453–2459. [PubMed] [Google Scholar]
- 48.Lam JS, Seligson DB, Yu H, Li A, Eeva M, Pantuck AJ, et al. Flap endonuclease 1 is overexpressed in prostate cancer and is associated with a high Gleason score. BJU Int. 2006;98:445–451. doi: 10.1111/j.1464-410X.2006.06224.x. [DOI] [PubMed] [Google Scholar]
- 49.LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 50.Krause A, Combaret V, Iacono I, Lacroix B, Compagnon C, Bergeron C, et al. Genome-wide analysis of gene expression in neuroblastomas detected by mass screening. Cancer Lett. 2005;225:111–120. doi: 10.1016/j.canlet.2004.10.035. [DOI] [PubMed] [Google Scholar]
- 51.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh P, Yang M, Dai H, Yu D, Huang Q, Tan W, et al. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol Cancer Res. 2008;6:1710–1717. doi: 10.1158/1541-7786.MCR-08-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nitta M, Kozono D, Kennedy R, Stommel J, Ng K, Zinn PO, et al. Targeting EGFR induced oxidative stress by PARP1 inhibition in glioblastoma therapy. PLoS One. 2010;5(5):e10767. doi: 10.1371/journal.pone.0010767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugasawa K. Regulation of damage recognition in mammalian global genomic nucleotide excision repair. Mutat Res. 2010;685:29–37. doi: 10.1016/j.mrfmmm.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Kamileri I, Karakasilioti I, Garinis GA. Nucleotide excision repair: new tricks with old bricks Trends Genet. 2012;28:566–573. doi: 10.1016/j.tig.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 56.Zeng-Rong N, Paterson J, Alpert L, Tsao MS, Viallet J, Alaoui-Jamali MA. Elevated DNA repair capacity is associated with intrinsic resistance of lung cancer to chemotherapy. Cancer Res. 1995;55:4760–4764. [PubMed] [Google Scholar]
- 57.Wei Q, Cheng L, Xie K, Bucana CD, Dong Z. Direct correlation between DNA repair capacity and metastatic potential of K-1735 murine melanoma cells. J Invest Dermatol. 1997;108:3–6. doi: 10.1111/1523-1747.ep12285608. [DOI] [PubMed] [Google Scholar]
- 58.Köberle B, Masters JR, Hartley JA, Wood RD. Defective repair of cisplatin-induced DNA damage caused by reduced XPA protein in testicular germ cell tumours. Curr Biol. 1999;9:273–276. doi: 10.1016/s0960-9822(99)80118-3. [DOI] [PubMed] [Google Scholar]
- 59.Yang J, Xu Z, Li J, Zhang R, Zhang G, Ji H, et al. XPC epigenetic silence coupled with p53 alteration has a significant impact on bladder cancer outcome. J Urol. 2010;184:336–343. doi: 10.1016/j.juro.2010.03.044. [DOI] [PubMed] [Google Scholar]
- 60.Chen HY, Shao CJ, Chen FR, Kwan AL, Chen ZP. Role of ERCC1 promoter hypermethylation in drug resistance to cisplatin in human gliomas. Int J Cancer. 2010;126:1944–1954. doi: 10.1002/ijc.24772. [DOI] [PubMed] [Google Scholar]
- 61.Kiyohara C, Yoshimasu K. Genetic polymorphisms in the nucleotide excision repair pathway and lung cancer risk: a meta-analysis. Int J Med Sci. 2007;4:59–71. doi: 10.7150/ijms.4.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin SA, Lord CJ, Ashworth A. Therapeutic targeting of the DNA mismatch repair pathway. Clin Cancer Res. 2010;16:5107–5113. doi: 10.1158/1078-0432.CCR-10-0821. [DOI] [PubMed] [Google Scholar]
- 63.Pineda M, González S, Lázaro C, Blanco I, Capellá G. Detection of genetic alterations in hereditary colorectal cancer screening. Mutat Res. 2010;693:19–31. doi: 10.1016/j.mrfmmm.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 64.Cunningham JM, Kim CY, Christensen ER, Tester DJ, Parc Y, Burgart LJ, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet. 2001;69:780–790. doi: 10.1086/323658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 66.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Esteller M, Levine R, Baylin SB, Ellenson LH, Herman JG. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–2417. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]
- 68.Fleisher AS, Esteller M, Wang S, Tamura G, Suzuki H, Yin J, et al. Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res. 1999;59:1090–1095. [PubMed] [Google Scholar]
- 69.Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 70.Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 71.Li D, Kumaraswamy E, Harlan-Williams LM, Jensen RA. The role of BRCA1 and BRCA2 in prostate cancer. Front Biosci. 2013;18:1445–1459. doi: 10.2741/4191. [DOI] [PubMed] [Google Scholar]
- 72.Iqbal J, Ragone A, Lubinski J, Lynch HT, Moller P, Ghadirian P, et al. The incidence of pancreatic cancer in BRCA1 and BRCA2 mutation carriers. Br J Cancer. 2012;107:2005–2009. doi: 10.1038/bjc.2012.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 74.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Varon R, Reis A, Henze G, von Einsiedel HG, Sperling K, Seeger K. Mutations in the Nijmegen Breakage Syndrome gene (NBS1) in childhood acute lymphoblastic leukemia (ALL) Cancer Res. 2001;61:3570–3572. [PubMed] [Google Scholar]
- 76.Cybulski C, Górski B, Debniak T, Gliniewicz B, Mierzejewski M, Masojc B, et al. NBS1 is a prostate cancer susceptibility gene. Cancer Res. 2004;64:1215–1219. doi: 10.1158/0008-5472.can-03-2502. [DOI] [PubMed] [Google Scholar]
- 77.Kim NG, Choi YR, Baek MJ, Kim YH, Kang H, Kim NK, et al. Frameshift mutations at coding mononucleotide repeats of the hRAD50 gene in gastrointestinal carcinomas with microsatellite instability. Cancer Res. 2001;61:36–38. doi: 10.1186/bcr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kennedy RD, D’Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24:3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 79.Wilson CR, Davidson SE, Margison GP, Jackson SP, Hendry JH, West CM. Expression of Ku70 correlates with survival in carcinoma of the cervix. Br J Cancer. 2000;83:1702–1706. doi: 10.1054/bjoc.2000.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Komuro Y, Watanabe T, Hosoi Y, Matsumoto Y, Nakagawa K, Tsuno N, et al. The expression pattern of Ku correlates with tumor radiosensitivity and disease free survival in patients with rectal carcinoma. Cancer. 2002;95:1199–1205. doi: 10.1002/cncr.10807. [DOI] [PubMed] [Google Scholar]
- 81.Rigas B, Borgo S, Elhosseiny A, Balatsos V, Manika Z, Shinya H, et al. Decreased expression of DNA-dependent protein kinase, a DNA repair protein, during human colon carcinogenesis. Cancer Res. 2001;61:8381–8384. [PubMed] [Google Scholar]
- 82.Yang J, Chen Z, Liu Y, Hickey RJ, Malkas LH. Altered DNA polymerase iota expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenesis. Cancer Res. 2004;64:5597–5607. doi: 10.1158/0008-5472.CAN-04-0603. [DOI] [PubMed] [Google Scholar]
- 83.O-Wang J, Kawamura K, Tada Y, Ohmori H, Kimura H, Sakiyama S, et al. DNA polymerase kappa, implicated in spontaneous and DNA damage-induced mutagenesis, is overexpressed in lung cancer. Cancer Res. 2001;61:5366–5369. [PubMed] [Google Scholar]
- 84.Boudsocq F, Benaim P, Canitrot Y, Knibiehler M, Ausseil F, Capp JP, et al. Modulation of cellular response to cisplatin by a novel inhibitor of DNA polymerase beta. Mol Pharmacol. 2005;67:1485–1492. doi: 10.1124/mol.104.001776. [DOI] [PubMed] [Google Scholar]
- 85.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 86.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 87.Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst) 2010;9:1219–1228. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 88.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 89.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cotta-Ramusino C, McDonald ER3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–1217. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smits VA, Reaper PM, Jackson SP. Rapid PIKK-dependent release of CHK1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol. 2006;16:150–159. doi: 10.1016/j.cub.2005.11.066. [DOI] [PubMed] [Google Scholar]
- 92.Mailand N, Falck J, Lukas C, Syljua^sen RG, Welcker M, Bartek J, et al. Rapid destruction of human CDC25A in response to DNA damage. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 93.Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, et al. SCFbeta-TRCP links CHK1 signaling to degradation of the CDC25A protein phosphatase. Genes Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Melixetian M, Klein DK, Sørensen CS, Helin K. NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat Cell Biol. 2009;11:1247–1253. doi: 10.1038/ncb1969. [DOI] [PubMed] [Google Scholar]
- 95.Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–1909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Agami R, Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- 97.Takahashi-Yanaga F, Sasaguri T. GSK-3beta regulates cyclin D1 expression: a new target for chemotherapy. Cell Signal. 2008;20:581–589. doi: 10.1016/j.cellsig.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 98.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, et al. Aberrant cell cycle checkpoint function and early embryonic death in CHK1(-/-) mice. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 100.Syljua˚sen RG, Sørensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, et al. Inhibition of human CHK1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA. CHK1 regulates the density of active replication origins during the vertebrate S phase. EMBO J. 2007;26:2719–2731. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Krämer A, Mailand N, Lukas C, Syljua˚sen RG, Wilkinson CJ, Nigg EA, et al. Centrosome-associated CHK1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 103.Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC, et al. CHK1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–260. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peddibhotla S, Lam MH, Gonzalez-Rimbau M, Rosen JM. The DNA-damage effector checkpoint kinase 1 is essential for chromosome segregation and cytokinesis. Proc Natl Acad Sci U S A. 2009;106:5159–5164. doi: 10.1073/pnas.0806671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Iliakis G, Wang Y, Guan J, Wang H. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene. 2003;22:5834–5847. doi: 10.1038/sj.onc.1206682. [DOI] [PubMed] [Google Scholar]
- 106.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 107.Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, et al. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol Cell Biol. 2005;25:3737–3751. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dalvai M, Mondesert O, Bourdon JC, Ducommun B, Dozier C. CDC25B is negatively regulated by p53 through Sp1 and NF-Y transcription factors. Oncogene. 2011;30:2282–2288. doi: 10.1038/onc.2010.588. [DOI] [PubMed] [Google Scholar]
- 109.McKenzie L, King S, Marcar L, Nicol S, Dias SS, Schumm K, et al. p53-dependent repression of polo-like kinase-1 (PLK1) Cell Cycle. 2010;9:4200–4212. doi: 10.4161/cc.9.20.13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 111.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 112.Gollin SM. Mechanisms leading to chromosomal instability. Semin Cancer Biol. 2005;15:33–42. doi: 10.1016/j.semcancer.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 113.Eyfjord JE, Bodvarsdottir SK. Genomic instability and cancer: networks involved in response to DNA damage. Mutat Res. 2005;592:18–28. doi: 10.1016/j.mrfmmm.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 114.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 115.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 116.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 117.Barlow C, Eckhaus MA, Schäffer AA, Wynshaw-Boris A. Atm haploinsufficiency results in increased sensitivity to sublethal doses of ionizing radiation in mice. Nat Genet. 1999;21:359–360. doi: 10.1038/7684. [DOI] [PubMed] [Google Scholar]
- 118.Fang Y, Tsao CC, Goodman BK, Furumai R, Tirado CA, Abraham RT, et al. ATR functions as a gene dosage-dependent tumor suppressor on a mismatch repair-deficient background. EMBO J. 2004;23:3164–3174. doi: 10.1038/sj.emboj.7600315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 121.Lam MH, Liu Q, Elledge SJ, Rosen JM. CHK1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 2004;6:45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 122.Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van-Laethem F, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- 123.Zhang Y, Lim CU, Williams ES, Zhou J, Zhang Q, Fox MH, et al. NBS1 knockdown by small interfering RNA increases ionizing radiation mutagenesis and telomere association in human cells. Cancer Res. 2005;65:5544–5553. doi: 10.1158/0008-5472.CAN-04-4368. [DOI] [PubMed] [Google Scholar]
- 124.Balajee AS, Geard CR. Replication protein A and gamma-H2AX foci assembly is triggered by cellular response to DNA doublestrand breaks. Exp Cell Res. 2004;300:320–334. doi: 10.1016/j.yexcr.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 125.Wang Y, Putnam CD, Kane MF, Zhang W, Edelmann L, Russell R, et al. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat Genet. 2005;37:750–755. doi: 10.1038/ng1587. [DOI] [PubMed] [Google Scholar]
- 126.Budzowska M, Jaspers I, Essers J, de Waard H, van Drunen E, Hanada K, et al. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 2004;23:3548–3558. doi: 10.1038/sj.emboj.7600353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shen SX, Weaver Z, Xu X, Li C, Weinstein M, Chen L, et al. A targeted disruption of the murine Brca1 gene causes gammairradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–3124. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- 128.Tutt A, Ashworth A. The relationship between the roles of BRCA genes in DNA repair and cancer predisposition. Trends Mol Med. 2002;8:571–576. doi: 10.1016/s1471-4914(02)02434-6. [DOI] [PubMed] [Google Scholar]
- 129.Smilenov LB, Lieberman HB, Mitchell SA, Baker RA, Hopkins KM, Hall EJ. Combined haploinsufficiency for ATM and RAD9 as a factor in cell transformation, apoptosis, and DNA lesion repair dynamics. Cancer Res. 2005;65:933–938. [PubMed] [Google Scholar]
- 130.Balmus G, Zhu M, Mukherjee S, Lyndaker AM, Hume KR, Lee J, Riccio ML, Reeves AP, et al. Disease severity in a mouse model of ataxia telangiectasia is modulated by the DNA damage checkpoint gene Hus1. Hum Mol Genet. 2012;21:3408–3420. doi: 10.1093/hmg/dds173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang H, Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004;64:7139–7143. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- 132.Sørensen CS, Hansen LT, Dziegielewski J, Syljuåsen RG, Lundin C, Bartek J, et al. The cell-cycle checkpoint kinase CHK1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 133.Hu B, Wang H, Wang X, Lu HR, Huang C, Powell SN, et al. hit and CHK1 have opposing effects on homologous recombination repair. Cancer Res. 2005;65:8613–8616. doi: 10.1158/0008-5472.CAN-05-1966. [DOI] [PubMed] [Google Scholar]
- 134.Sørensen CS, Syljua˚sen RG, Falck J, Schroeder T, Rönnstrand L, Khanna KK, et al. CHK1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of CDC25A. Cancer Cell. 2003;3:247–258. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 135.Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, et al. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Petermann E, Maya-Mendoza A, Zachos G, Gillespie DA, Jackson DA, Caldecott KW. CHK1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26:3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Petermann E, Woodcock M, Helleday T. CHK1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A. 2010;107:16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, et al. Activation of mammalian CHK1 during DNA replication arrest: a role for CHK1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154:913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 140.Zhang P, Wang J, Gao W, Yuan BZ, Rogers J, Reed E. CHK2 kinase expression is down-regulated due to promoter methylation in non-small cell lung cancer. Mol Cancer. 2004;3:14. doi: 10.1186/1476-4598-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bertoni F, Codegoni AM, Furlan D, Tibiletti MG, Capella C, Broggini M. CHK1 frameshift mutations in genetically unstable colorectal and endometrial cancers. Genes Chromosomes Cancer. 1999;26:176–180. [PubMed] [Google Scholar]
- 142.Lewis KA, Mullany S, Thomas B, Chien J, Loewen R, Shridhar V, et al. Heterozygous ATR mutations in mismatch repair-deficient cancer cells have functional significance. Cancer Res. 2005;65:7091–7095. doi: 10.1158/0008-5472.CAN-05-1019. [DOI] [PubMed] [Google Scholar]
- 143.Menoyo A, Alazzouzi H, Espín E, Armengol M, Yamamoto H, Schwartz S., Jr Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001;61:7727–7730. [PubMed] [Google Scholar]
- 144.Fishler T, Li YY, Wang RH, Kim HS, Sengupta K, Vassilopoulos A, et al. Genetic instability and mammary tumor formation in mice carrying mammary-specific disruption of CHK1 and p53. Oncogene. 2010;29:4007–4017. doi: 10.1038/onc.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.van Vugt MA, Medema RH. Getting in and out of mitosis with Polo-like kinase-1. Oncogene. 2005;24:2844–2859. doi: 10.1038/sj.onc.1208617. [DOI] [PubMed] [Google Scholar]
- 146.Katayama H, Sen S. Aurora kinase inhibitors as anticancer molecules. Biochim Biophys Acta. 2010;1799:829–839. doi: 10.1016/j.bbagrm.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Canavese M, Santo L, Raje N. Cyclin dependent kinases in cancer: potential for therapeutic intervention. Cancer Biol Ther. 2012;13:451–457. doi: 10.4161/cbt.19589. [DOI] [PubMed] [Google Scholar]
- 148.Galaktionov K, Lee AK, Eckstein J, Draetta G, Meckler J, Loda M, et al. CDC25 phosphatases as potential human oncogenes. Science. 1995;269:1575–1577. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 149.Backert S, Gelos M, Kobalz U, Hanski ML, Böhm C, Mann B, et al. Differential gene expression in colon carcinoma cells and tissues detected with a cDNA array. Int J Cancer. 1999;82:868–874. doi: 10.1002/(sici)1097-0215(19990909)82:6<868::aid-ijc16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 150.Yoshida T, Tanaka S, Mogi A, Shitara Y, Kuwano H. The clinical significance of Cyclin B1 and Wee1 expression in non-small-cell lung cancer. Ann Oncol. 2004;15:252–256. doi: 10.1093/annonc/mdh073. [DOI] [PubMed] [Google Scholar]
- 151.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–2786. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 152.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 153.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mathews LA, Cabarcas SM, Farrar WL. DNA repair: the culprit for tumor-initiating cell survival? Cancer Metastasis Rev. 2011;30:185–197. doi: 10.1007/s10555-011-9277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 156.Kauffmann A, Rosselli F, Lazar V, Winnepenninckx V, Mansuet-Lupo A, Dessen P, et al. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27:565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- 157.Sarasin A, Kauffmann A. Overexpression of DNA repair genes is associated with metastasis: a new hypothesis. Mutat Res. 2008;659:49–55. doi: 10.1016/j.mrrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 158.Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 159.Niida H, Nakanishi M. DNA damage checkpoints in mammals. Mutagenesis. 2006;21:3–9. doi: 10.1093/mutage/gei063. [DOI] [PubMed] [Google Scholar]
- 160.Broustas CG, Lieberman HB. Contributions of Rad9 to tumorigenesis. J Cell Biochem. 2012;113:742–751. doi: 10.1002/jcb.23424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Toueille M, El-Andaloussi N, Frouin I, Freire R, Funk D, Shevelev I, et al. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilization efficiency: Implications for DNA repair. Nucleic Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang W, Brandt P, Bambara RA. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc Natl Acad Sci U S A. 2004;101:16762–16767. doi: 10.1073/pnas.0407686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li T, Wang Z, Zhao Y, He W, An L, Liu S, et al. Checkpoint protein Rad9 plays an important role in nucleotide excision repair. DNA Repair (Amst) 2013;12:284–292. doi: 10.1016/j.dnarep.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 164.He W, Zhao Y, Zhang C, An L, Hu Z, Liu Y, et al. Rad9 plays an important role in DNA mismatch repair through physical interaction with MLH1. Nucleic Acids Res. 2008;36:6406–6417. doi: 10.1093/nar/gkn686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pandita RK, Sharma GG, Laszlo A, Hopkins KM, Davey S, Chakhparonian M, et al. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol. 2006;26:1850–1864. doi: 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Günes C, Rudolph KL. The role of telomeres in stem cells and cancer. Cell. 2013;152:390–393. doi: 10.1016/j.cell.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 167.Riboni R, Casati A, Nardo T, Zaccaro E, Ferretti L, Nuzzo F, et al. Telomeric fusions in cultured human fibroblasts as a source of genomic instability. Cancer Genet Cytogenet. 1997;95:130–136. doi: 10.1016/s0165-4608(96)00248-8. [DOI] [PubMed] [Google Scholar]
- 168.Meeker AK, Argani P. Telomere shortening occurs early during breast tumorigenesis: a cause of chromosome destabilization underlying malignant transformation? J Mammary Gland Biol Neoplasia. 2004;9:285–296. doi: 10.1023/B:JOMG.0000048775.04140.92. [DOI] [PubMed] [Google Scholar]
- 169.Stewart SA, Weinberg RA. Telomeres: cancer to human aging. Annu Rev Cell Dev Biol. 2006;22:531–557. doi: 10.1146/annurev.cellbio.22.010305.104518. [DOI] [PubMed] [Google Scholar]
- 170.Hopkins KM, Auerbach W, Wang XY, Hande MP, Hang H, Wolgemuth DJ, et al. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol Cell Biol. 2004;24:7235–7248. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhu A, Zhou H, Leloup C, Marino SA, Geard CR, Hei TK, Lieberman HB. Differential impact of mouse Rad9 deletion on ionizing radiation-induced bystander effects. Radiat Res. 2005;164:655–661. doi: 10.1667/rr3458.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Komatsu K, Miyashita T, Hang H, Hopkins KM, Zheng W, Cuddeback S, et al. Human homologue of S. pombe Rad9 interacts with BCL-2/BCL-xL and promotes apoptosis. Nat Cell Biol. 2000;2:1–6. doi: 10.1038/71316. [DOI] [PubMed] [Google Scholar]
- 173.Ishii H, Inageta T, Mimori K, Saito T, Sasaki H, Isobe M, et al. Frag1, a homolog of alternative replication factor C subunits, links replication stress surveillance with apoptosis. Proc Natl Acad Sci U S A. 2005;102:9655–9660. doi: 10.1073/pnas.0504222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yoshida K, Komatsu K, Wang HG, Kufe D. c-Abl tyrosine kinase regulates the human Rad9 checkpoint protein in response to DNA damage. Mol Cell Biol. 2002;22:3292–3300. doi: 10.1128/MCB.22.10.3292-3300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yoshida K, Wang HG, Miki Y, Kufe D. Protein kinase Cdelta is responsible for constitutive and DNA damage-induced phosphor-ylation of Rad9. EMBO J. 2003;22:1431–1441. doi: 10.1093/emboj/cdg134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gressner O, Schilling T, Lorenz K, Schulze Schleithoff E, Koch A, et al. TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 2005;24:2458–2471. doi: 10.1038/sj.emboj.7600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yin Y, Zhu A, Jin YJ, Liu YX, Zhang X, Hopkins KM, et al. Human RAD9 checkpoint control/proapoptotic protein can activate transcription of p21. Proc Natl Acad Sci U S A. 2004;101:8864–8869. doi: 10.1073/pnas.0403130101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang L, Hsu CL, Ni J, Wang PH, Yeh S, Keng P, et al. Human checkpoint protein hRad9 functions as a negative coregulator to repress androgen receptor transactivation in prostate cancer cells. Mol Cell Biol. 2004;24:2202–2213. doi: 10.1128/MCB.24.5.2202-2213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhu A, Zhang CX, Lieberman HB. Rad9 has a functional role in human prostate carcinogenesis. Cancer Res. 2008;68:1267–1274. doi: 10.1158/0008-5472.CAN-07-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 181.Zeng ZZ, Jia Y, Hahn NJ, Markwart SM, Rockwood KF, Livant DL. Role of focal adhesion kinase and phosphatidylinositol 3'-kinase in integrin fibronectin receptor-mediated, matrix metal-loproteinase-1-dependent invasion by metastatic prostate cancer cells. Cancer Res. 2006;66:8091–8099. doi: 10.1158/0008-5472.CAN-05-4400. [DOI] [PubMed] [Google Scholar]
- 182.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Broustas CG, Zhu A, Lieberman HB. Rad9 protein contributes to prostate tumor progression by promoting cell migration and anoikis resistance. J Biol Chem. 2012;287:41324–41333. doi: 10.1074/jbc.M112.402784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 185.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 186.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang X, Zou L, Lu T, Bao S, Hurov KE, Hittelman WN, et al. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol Cell. 2006;23:331–341. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 188.Bao S, Tibbetts RS, Brumbaugh KM, Fang Y, Richardson DA, Ali A, et al. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature. 2001;411:969–974. doi: 10.1038/35082110. [DOI] [PubMed] [Google Scholar]
- 189.Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhao M, Begum S, Ha PK, Westra W, Califano J. Downregulation of RAD17 in head and neck cancer. Head Neck. 2008;30:35–42. doi: 10.1002/hed.20660. [DOI] [PubMed] [Google Scholar]
- 191.Zhou Z, Jing C, Zhang L, Takeo F, Kim H, Huang Y, et al. Regulation of Rad17 Protein Turnover Unveils an Impact of Rad17-APC Cascade in Breast Carcinogenesis and Treatment. J Biol Chem. 2013;288:18134–18145. doi: 10.1074/jbc.M113.456962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang X, Wang L, Callister MD, Putnam JB, Mao L, Li L. Human Rad17 is phosphorylated upon DNA damage and also overexpressed in primary non-small cell lung cancer tissues. Cancer Res. 2001;61:7417–7421. [PubMed] [Google Scholar]
- 193.Sasaki H, Chen LB, Auclair D, Moriyama S, Kaji M, Fukai I, et al. Overexpression of Hrad17 gene in non-small cell lung cancers correlated with lymph node metastasis. Lung Cancer. 2001;34:47–52. doi: 10.1016/s0169-5002(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 194.Bao S, Lu T, Wang X, Zheng H, Wang LE, Wei Q, et al. Disruption of the Rad9/Rad1/Hus1 (9-1-1) complex leads to checkpoint signaling and replication defects. Oncogene. 2004;23:5586–5593. doi: 10.1038/sj.onc.1207753. [DOI] [PubMed] [Google Scholar]
- 195.Meyerkord CL, Takahashi Y, Araya R, Takada N, Weiss RS, Wang HG. Loss of Hus1 sensitizes cells to etoposide-induced apoptosis by regulating BH3-only proteins. Oncogene. 2008;27:7248–7259. doi: 10.1038/onc.2008.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kinzel B, Hall J, Natt F, Weiler J, Cohen D. Downregulation of Hus1 by antisense oligonucleotides enhances the sensitivity of human lung carcinoma cells to cisplatin. Cancer. 2002;94:1808–1814. doi: 10.1002/cncr.10383. [DOI] [PubMed] [Google Scholar]
- 197.Brandt PD, Helt CE, Keng PC, Bambara RA. The Rad9 protein enhances survival and promotes DNA repair following exposure to ionizing radiation. Biochem Biophys Res Commun. 2006;347:232–237. doi: 10.1016/j.bbrc.2006.06.064. [DOI] [PubMed] [Google Scholar]
- 198.Wang X, Hu B, Weiss RS, Wang Y. The effect of Hus1 on ionizing radiation sensitivity is associated with homologous recombination repair but is independent of nonhomologous end-joining. Oncogene. 2006;25:1980–1983. doi: 10.1038/sj.onc.1209212. [DOI] [PubMed] [Google Scholar]
- 199.de la Torre J, Gil-Moreno A, García A, Rojo F, Xercavins J, Salido E, et al. Expression of DNA damage checkpoint protein Hus1 in epithelial ovarian tumors correlates with prognostic markers. Int J Gynecol Pathol. 2008;27:24–32. doi: 10.1097/pgp.0b013e31812dfaef. [DOI] [PubMed] [Google Scholar]
- 200.Han L, Hu Z, Liu Y, Wang X, Hopkins KM, Lieberman HB, et al. Mouse Rad1 deletion enhances susceptibility for skin tumor development. Mol Cancer. 2010;9:67. doi: 10.1186/1476-4598-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- 202.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gonc¸alves A, Finetti P, Sabatier R, Gilabert M, Adelaide J, Borg JP, et al. Poly(ADP-ribose) polymerase-1 mRNA expression in human breast cancer: a meta-analysis. Breast Cancer Res Treat. 2011;127:273–281. doi: 10.1007/s10549-010-1199-y. [DOI] [PubMed] [Google Scholar]
- 204.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 207.Gopalan A, Leversha MA, Satagopan JM, Zhou Q, Al-Ahmadie HA, Fine SW, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–146. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Rodríguez MI, González-Flores A, Dantzer F, Collard J, de Herreros AG, Oliver FJ. Poly(ADP-ribose)-dependent regulation of Snail1 protein stability. Oncogene. 2011;30:4365–4372. doi: 10.1038/onc.2011.153. [DOI] [PubMed] [Google Scholar]
- 209.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 210.McPhee TR, McDonald PC, Oloumi A, Dedhar S. Integrin-linked kinase regulates E-cadherin expression through PARP-1. Dev Dyn. 2008;237:2737–2747. doi: 10.1002/dvdy.21685. [DOI] [PubMed] [Google Scholar]
- 211.Stanisavljevic J, Porta-de-la-Riva M, Batlle R, de Herreros AG, Baulida J. The p65 subunit of NF-κB and PARP1 assist Snail1 in activating fibronectin transcription. J Cell Sci. 2011;124:4161–4171. doi: 10.1242/jcs.078824. [DOI] [PubMed] [Google Scholar]
- 212.Lönn P, van der Heide LP, Dahl M, Hellman U, Heldin CH, Moustakas A. PARP-1 attenuates Smad-mediated transcription. Mol Cell. 2010;40:521–532. doi: 10.1016/j.molcel.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 213.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4(4):511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 214.Tutt A, Gabriel A, Bertwistle D, Connor F, Paterson H, Peacock J, et al. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9:1107–1110. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 215.Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006;34:1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gorski JJ, Savage KI, Mulligan JM, McDade SS, Blayney JK, Ge Z, et al. Profiling of the BRCA1 transcriptome through micro-array and ChIP-chip analysis. Nucleic Acids Res. 2011;39:9536–9548. doi: 10.1093/nar/gkr679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386(761) doi: 10.1038/386761a0. 763. [DOI] [PubMed] [Google Scholar]
- 218.Monteiro AN. BRCA1: exploring the links to transcription. Trends Biochem Sci. 2000;25:469–474. doi: 10.1016/s0968-0004(00)01632-7. [DOI] [PubMed] [Google Scholar]
- 219.Wang Y, Yu J, Zhan Q. BRCA1 regulates caveolin-1 expression and inhibits cell invasiveness. Biochem Biophys Res Commun. 2008;370:201–206. doi: 10.1016/j.bbrc.2008.03.031. [DOI] [PubMed] [Google Scholar]
- 220.Coene ED, Gadelha C, White N, Malhas A, Thomas B, Shaw M, et al. A novel role for BRCA1 in regulating breast cancer cell spreading and motility. J Cell Biol. 2011;192:497–512. doi: 10.1083/jcb.201004136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhu H, Yue J, Pan Z, Wu H, Cheng Y, Lu H, et al. Involvement of Caveolin-1 in repair of DNA damage through both homologous recombination and non-homologous end joining. PLoS One. 2010;5(8):e12055. doi: 10.1371/journal.pone.0012055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 223.Milner J, Ponder B, Hughes-Davies L, Seltmann M, Kouzarides T. Transcriptional activation functions in BRCA2. Nature. 1997;386:772–773. doi: 10.1038/386772a0. [DOI] [PubMed] [Google Scholar]
- 224.Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–879. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- 225.Costanzo V. Brca2, Rad51 and Mre11: performing balancing acts on replication forks. DNA Repair (Amst) 2011;10:1060–1065. doi: 10.1016/j.dnarep.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 226.Wang SC, Shao R, Pao AY, Zhang S, Hung MC, Su LK. Inhibition of cancer cell growth by BRCA2. Cancer Res. 2002;62:1311–1314. [PubMed] [Google Scholar]
- 227.Moro L, Arbini AA, Marra E, Greco M. Down-regulation of BRCA2 expression by collagen type I promotes prostate cancer cell proliferation. J Biol Chem. 2005;280:22482–22491. doi: 10.1074/jbc.M414091200. [DOI] [PubMed] [Google Scholar]
- 228.Moro L, Arbini AA, Marra E, Greco M. Up-regulation of Skp2 after prostate cancer cell adhesion to basement membranes results in BRCA2 degradation and cell proliferation. J Biol Chem. 2006;281:22100–22107. doi: 10.1074/jbc.M604636200. [DOI] [PubMed] [Google Scholar]
- 229.Moro L, Arbini AA, Yao JL, di Sant’Agnese PA, Marra E, Greco M. Loss of BRCA2 promotes prostate cancer cell invasion through up-regulation of matrix metalloproteinase-9. Cancer Sci. 2008;99:553–563. doi: 10.1111/j.1349-7006.2007.00719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Castro E, Goh C, Olmos D, Saunders E, Leongamornlert D, Tymrakiewicz M, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748–1757. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Demuth I, Digweed M. The clinical manifestation of a defective response to DNA double-strand breaks as exemplified by Nijmegen breakage syndrome. Oncogene. 2007;26:7792–7798. doi: 10.1038/sj.onc.1210876. [DOI] [PubMed] [Google Scholar]
- 232.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Karran P. DNA double strand break repair in mammalian cells. Curr Opin Genet Dev. 2000;10:144–150. doi: 10.1016/s0959-437x(00)00069-1. [DOI] [PubMed] [Google Scholar]
- 234.Petrini JH. The Mre11 complex and ATM: collaborating to navigate S phase. Curr Opin Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 235.Chiang YC, Teng SC, Su YN, Hsieh FJ, Wu KJ. c-Myc directly regulates the transcription of the NBS1 gene involved in DNA double-strand break repair. J Biol Chem. 2003;278:19286–19291. doi: 10.1074/jbc.M212043200. [DOI] [PubMed] [Google Scholar]
- 236.Chen YC, Su YN, Chou PC, Chiang WC, Chang MC, et al. Overexpression of NBS1 contributes to transformation through the activation of phosphatidylinositol 3-kinase/Akt. J Biol Chem. 2005;280:32505–32511. doi: 10.1074/jbc.M501449200. [DOI] [PubMed] [Google Scholar]
- 237.D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 238.Yang MH, Chang SY, Chiou SH, Liu CJ, Chi CW, Chen PM, et al. Overexpression of NBS1 induces epithelial-mesenchymal transition and co-expression of NBS1 and Snail predicts metastasis of head and neck cancer. Oncogene. 2007;26:1459–1467. doi: 10.1038/sj.onc.1209929. [DOI] [PubMed] [Google Scholar]
- 239.Wu CY, Lin CT, Wu MZ, Wu KJ. Induction of HSPA4 and HSPA14 by NBS1 overexpression contributes to NBS1-induced in vitro metastatic and transformation activity. J Biomed Sci. 2011;18:1. doi: 10.1186/1423-0127-18-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wan R, Crowe DL. Haploinsufficiency of the Nijmegen breakage syndrome 1 gene increases mammary tumor latency and metastasis. Int J Oncol. 2012;41:345–352. doi: 10.3892/ijo.2012.1435. [DOI] [PubMed] [Google Scholar]
- 241.Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 2008;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wang JY, Ho T, Trojanek J, Chintapalli J, Grabacka M, Stoklosa T, et al. Impaired homologous recombination DNA repair and enhanced sensitivity to DNA damage in prostate cancer cells exposed to anchorage-independence. Oncogene. 2005;24:3748–3758. doi: 10.1038/sj.onc.1208537. [DOI] [PubMed] [Google Scholar]
- 243.Martinez-Marignac VL, Rodrigue A, Davidson D, Couillard M, Al-Moustafa AE, Abramovitz M, et al. The effect of a DNA repair gene on cellular invasiveness: XRCC3 over-expression in breast cancer cells. PLoS One. 2011;6(1):e16394. doi: 10.1371/journal.pone.0016394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Derheimer FA, Kastan MB. Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS Lett. 2010;584:3675–3681. doi: 10.1016/j.febslet.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Sun M, Guo X, Qian X, Wang H, Yang C, Brinkman KL, et al. Activation of the ATM-Snail pathway promotes breast cancer metastasis. J Mol Cell Biol. 2012;4:304–315. doi: 10.1093/jmcb/mjs048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Golding SE, Rosenberg E, Valerie N, Hussaini I, Frigerio M, Cockcroft XF, et al. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol Cancer Ther. 2009;8:2894–2902. doi: 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Sarsour EH, Kumar MG, Chaudhuri L, Kalen AL, Goswami PC. Redox control of the cell cycle in health and disease. Antioxid Redox Signal. 2009;11:2985–3011. doi: 10.1089/ars.2009.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Pani G, Giannoni E, Galeotti T, Chiarugi P. Redox-based escape mechanism from death: the cancer lesson. Antioxid Redox Signal. 2009;11:2791–2806. doi: 10.1089/ars.2009.2739. [DOI] [PubMed] [Google Scholar]
- 249.Ditch S, Paull TT. The ATM protein kinase and cellular redox signaling: beyond the DNA damage response. Trends Biochem Sci. 2012;37:15–22. doi: 10.1016/j.tibs.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tsai YP, Wu KJ. Hypoxia-regulated target genes implicated in tumor metastasis. J Biomed Sci. 2012;19:102. doi: 10.1186/1423-0127-19-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 252.Baker SJ, Preisinger AC, Jessup JM, Paraskeva C, Markowitz S, Willson JK, et al. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990;50:7717–7722. [PubMed] [Google Scholar]
- 253.Bookstein R, MacGrogan D, Hilsenbeck SG, Sharkey F, Allred DC. p53 is mutated in a subset of advanced-stage prostate cancers. Cancer Res. 1993;53:3369–3373. [PubMed] [Google Scholar]
- 254.Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- 255.Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 256.Hanel W, Moll UM. Links between mutant p53 and genomic instability. J Cell Biochem. 2012;113:433–439. doi: 10.1002/jcb.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 258.Liu DP, Song H, Xu Y. A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene. 2010;29:949–956. doi: 10.1038/onc.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 260.Rausch T, Jones DT, Zapatka M, Stütz AM, Zichner T, Weischenfeldt J, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Livingstone LR, White A, Sprouse J, Livanos E, Jacks T, Tlsty TD. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 262.Li Y, Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- 263.Singh K, Mogare D, Giridharagopalan RO, Gogiraju R, Pande G, Chattopadhyay S. p53 target gene SMAR1 is dysregulated in breast cancer: its role in cancer cell migration and invasion. PLoS One. 2007;2(7):e660. doi: 10.1371/journal.pone.0000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 266.Shibue T, Weinberg RA. Integrin beta1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc Natl Acad Sci U S A. 2009;106:10290–10295. doi: 10.1073/pnas.0904227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Huck L, Pontier SM, Zuo DM, Muller WJ. beta1-integrin is dispensable for the induction of ErbB2 mammary tumors but plays a critical role in the metastatic phase of tumor progression. Proc Natl Acad Sci U S A. 2010;107:15559–15564. doi: 10.1073/pnas.1003034107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Reymond N, Im JH, Garg R, Vega FM, Borda d’Agua B, Riou P, et al. Cdc42 promotes transendothelial migration of cancer cells through b1 integrin. J Cell Biol. 2012;199:653–668. doi: 10.1083/jcb.201205169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Caswell PT, Vadrevu S, Norman JC. Integrins: masters and slaves of endocytic transport. Nat Rev Mol Cell Biol. 2009;10:843–853. doi: 10.1038/nrm2799. [DOI] [PubMed] [Google Scholar]
- 270.Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, et al. Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 271.Hildesheim J, Bulavin DV, Anver MR, Alvord WG, Hollander MC, Vardanian L, et al. Gadd45a protects against UV irradiation-induced skin tumors, and promotes apoptosis and stress signaling via MAPK and p53. Cancer Res. 2002;62:7305–7315. [PubMed] [Google Scholar]
- 272.Yamasawa K, Nio Y, Dong M, Yamaguchi K, Itakura M. Clinicopathological significance of abnormalities in Gadd45 expression and its relationship to p53 in human pancreatic cancer. Clin Cancer Res. 2002;8:2563–2569. [PubMed] [Google Scholar]
- 273.Wang W, Huper G, Guo Y, Murphy SK, Olson JA, Jr, Marks JR. Analysis of methylation-sensitive transcriptome identifies GAD-D45a as a frequently methylated gene in breast cancer. Oncogene. 2005;24:2705–2714. doi: 10.1038/sj.onc.1208464. [DOI] [PubMed] [Google Scholar]
- 274.Ji J, Liu R, Tong T, Song Y, Jin S, Wu M, et al. Gadd45a regulates beta-catenin distribution and maintains cell-cell adhesion/contact. Oncogene. 2007;26:6396–6405. doi: 10.1038/sj.onc.1210469. [DOI] [PubMed] [Google Scholar]
- 275.Shan Z, Li G, Zhan Q, Li D. Gadd45a inhibits cell migration and invasion by altering the global RNA expression. Cancer Biol Ther. 2012;13:1112–1122. doi: 10.4161/cbt.21186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Marino N, Nakayama J, Collins JW, Steeg PS. Insights into the biology and prevention of tumor metastasis provided by the Nm23 metastasis suppressor gene. Cancer Metastasis Rev. 2012;31:593–603. doi: 10.1007/s10555-012-9374-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Flørenes VA, Aamdal S, Myklebost O, Maelandsmo GM, Bruland OS, Fodstad O. Levels of nm23 messenger RNA in metastatic malignant melanomas: inverse correlation to disease progression. Cancer Res. 1992;52:6088–6091. [PubMed] [Google Scholar]
- 278.Zhang Q, McCorkle JR, Novak M, Yang M, Kaetzel DM. Metastasis suppressor function of NM23-H1 requires its 3' −5' exonuclease activity. Int J Cancer. 2011;128:40–50. doi: 10.1002/ijc.25307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Jarrett SG, Novak M, Dabernat S, Daniel JY, Mellon I, Zhang Q, et al. Metastasis suppressor NM23-H1 promotes repair of UV-induced DNA damage and suppresses UV-induced melanoma-genesis. Cancer Res. 2012;72:133–143. doi: 10.1158/0008-5472.CAN-11-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Chaplet M, Rai R, Jackson-Bernitsas D, Li K, Lin SY. BRIT1/ MCPH1: a guardian of genome and an enemy of tumors. Cell Cycle. 2006;5:2579–2583. doi: 10.4161/cc.5.22.3471. [DOI] [PubMed] [Google Scholar]
- 281.van ‘t-Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 282.Rai R, Dai H, Multani AS, Li K, Chin K, Gray J, et al. BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell. 2006;10:145–157. doi: 10.1016/j.ccr.2006.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Yi B, Tan SX, Tang CE, Huang WG, Cheng AL, Li C, et al. Inactivation of 14-3-3 sigma by promoter methylation correlates with metastasis in nasopharyngeal carcinoma. J Cell Biochem. 2009;106:858–866. doi: 10.1002/jcb.22051. [DOI] [PubMed] [Google Scholar]
- 284.Wang XQ, Zhu YQ, Lui KS, Cai Q, Lu P, Poon RT. Aberrant Polo-like kinase 1-CDC25A pathway in metastatic hepatocellular carcinoma. Clin Cancer Res. 2008;14:6813–6820. doi: 10.1158/1078-0432.CCR-08-0626. [DOI] [PubMed] [Google Scholar]
- 285.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 286.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Lleonart ME, Vidal F, Gallardo D, Diaz-Fuertes M, Rojo F, Cuatrecasas M, et al. New p53 related genes in human tumors: significant downregulation in colon and lung carcinomas. Oncol Rep. 2000;16:603–608. doi: 10.3892/or.16.3.603. [DOI] [PubMed] [Google Scholar]
- 288.Sakuraba K, Yasuda T, Sakata M, Kitamura YH, Shirahata A, Goto T, et al. Down-regulation of Tip60 gene as a potential marker for the malignancy of colorectal cancer. Anticancer Res. 2009;29:3953–3955. [PubMed] [Google Scholar]
- 289.Chen G, Cheng Y, Tang Y, Martinka M, Li G. Role of Tip60 in human melanoma cell migration, metastasis, and patient survival. J Invest Dermatol. 2012;132:2632–2641. doi: 10.1038/jid.2012.193. [DOI] [PubMed] [Google Scholar]
- 290.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Economopoulou M, Langer HF, Celeste A, Orlova VV, Choi EY, Ma M, et al. Histone H2AX is integral to hypoxia-driven neovascularization. Nat Med. 2009;15:553–558. doi: 10.1038/nm.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Rankin EB, Giaccia AJ, Hammond EM. Bringing H2AX into the angiogenesis family. Cancer Cell. 2009;15:459–461. doi: 10.1016/j.ccr.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]