Abstract
Mice in which lung epithelial cells can be induced to express an oncogenic KrasG12D develop lung adenocarcinomas in a manner analogous to humans. A myriad of genetic changes accompany lung adenocarcinomas, many of which are poorly understood. To get a comprehensive understanding of both the transcriptional and post-transcriptional changes that accompany lung adenocarcinomas, we took an omics approach in profiling both the coding genes and the non-coding small RNAs in an induced mouse model of lung adenocarcinoma. RNAseq transcriptome analysis of KrasG12D tumors from F1 hybrid mice revealed features specific to tumor samples. This includes the repression of a network of GTPase related genes (Prkg1, Gnao1 and Rgs9) in tumor samples and an enrichment of Apobec1-mediated cytosine to uridine RNA editing. Furthermore, analysis of known SNPs revealed not only a change in expression of Cd22 but also that its expression became allele-specific in tumors. The most salient finding however, came from small RNA sequencing of the tumor samples, which revealed that a cluster of ~53 microRNAs and mRNAs at the Dlk1-Dio3 locus on mouse chromosome 12qF1 was dramatically and consistently increased in tumors. Activation of this locus occurred specifically in sorted tumor-originating cancer cells. Interestingly, the 12qF1 RNAs were repressed in cultured KrasG12D tumor cells but reactivated when transplanted in vivo. These microRNAs have been implicated in stem cell pleuripotency and proteins targeted by these microRNAs are involved in key pathways in cancer as well as embryogenesis. Taken together our results strongly imply that these microRNAs represent key targets in unraveling the mechanism of lung oncogenesis.
Keywords: Lung adenocarcinoma, NSCLC, RNAseq, microRNAs
Introduction
Lung cancer is the leading cause of cancer-related mortality in both men and women worldwide. Lung cancer subtypes include small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), which is subdivided into adenocarcinoma, squamous cell carcinoma and large cell carcinoma. Adenocarcinomas represent approximately 30% of all lung tumors. Each of these subtypes has different prognosis, disease signature and risk factors (e.g. smoking).
The role that KRAS plays during development, organ homeostasis and in the development of lung cancer has been extensively studied, leading to an understanding of its role in uncontrolled tumor growth, angiogenesis and inhibition of apoptosis.1 KRAS is a GTP binding protein that is localized to the inner face of the plasma membrane. Guanine nucleotide exchange factors activate KRAS and enable cell growth and survival through downstream signaling pathways.2, 3 KRAS is inactivated by GTPase activating proteins; however, many point mutations identified in KRAS prevent this GTP hydrolysis and thus maintain a constitutively active KRAS4 KRAS mutations are very frequent in lung adenocarcinoma, occurring in approximately 25% of tumors5, 6 Among KRAS mutations, variants at amino acid 12 represent 90 percent of the cases and a mouse bearing a conditional KrasG12D mutation has been generated to study the effects of this mutation on cancer initiation.5, 7, 8
The decreased cost of sequencing technologies has sparked an interest in identifying the genetic changes that occur during tumor progression. Targeted re-sequencing9 in addition to whole exome and whole transcriptome studies10, 11 of tumor biopsies have provided several new candidate mutations. The analysis of the effect of additional somatic mutations and gene expression changes in mouse models of human tumors have complemented existing mutation data and provide a genetic framework for understanding tumor development. Additionally, genomic sequencing of several mouse strains has revealed several coding SNPs that can be used to identify parent of origin expression from F1 offspring from hybrid mouse strain crosses. Large scale non-coding RNA profiling studies have also identified microRNAs involved in oncogenesis including the miR-17 to miR-92 cluster of six microRNAs that are upregulated in B-cell lymphoma and SCLC12, 13 and the common microRNA, let-7, has been shown to regulate the 3′UTR of Kras.14 These previous studies have been insightful in understanding tumor development in lung adenocarcinoma, despite of focus solely on either coding or noncoding RNA populations.
We employed an integrative omics approach to identify transcriptional changes in a defined mouse model of lung adenocarcinoma. Sequencing was performed of bulk tumors, which reflect a mixed population of both tumor cells and infiltrating stromal cells; notable genetic changes could then be validated in purified sorted tumor cells. This approach revealed dysregulation of a complex stem cell associated microRNA locus in lung adenocarcinoma.
Results
Gene expression analysis indicates that a subset of genes are up and down regulated specifically in lung tumors
We performed a high throughput RNA sequencing analysis of the small and large RNA populations from three wildtype lungs and three KrasG12D-driven lung adenocarcinomas. Two of these sets were derived from the offspring of an F1 between 129S4 and Molf/EiJ parents and one set from 129S4 homozygous parents (Table 1).
Table 1.
Profile of samples subject to RNA sequencing.
| Tumor1 | Tumor2 | Tumor3 | Normal1 | Normal2 | Normal3 | |
|---|---|---|---|---|---|---|
| Maternal strain | MOLF/EiJ | MOLF/EiJ | 129S4 | MOLF/EiJ | MOLF/EiJ | 129S4 |
| Paternal strain | 129S4 | 129S4 | 129S4 | 129S4 | 129S4 | 129S4 |
| Kras | G12D | G12D | G12D | wildtype | G12D* | wildtype |
| Cre-addition | Yes | Yes | Yes | Yes | No | No |
| p53 status | wildtype | wildtype | Inactive (flox/flox) | wildtype | wildtype | wildtype |
| Sequence reads | 165,569,498 | 149,208,283 | 164,380,208 | 171,553,662 | 171,393,687 | 180,671,287 |
| % mapped reads | 87.17 | 84.62 | 86.01 | 80.76 | 84.14 | 81.45 |
Mutation is in inactive state until Cre recombinase is expressed
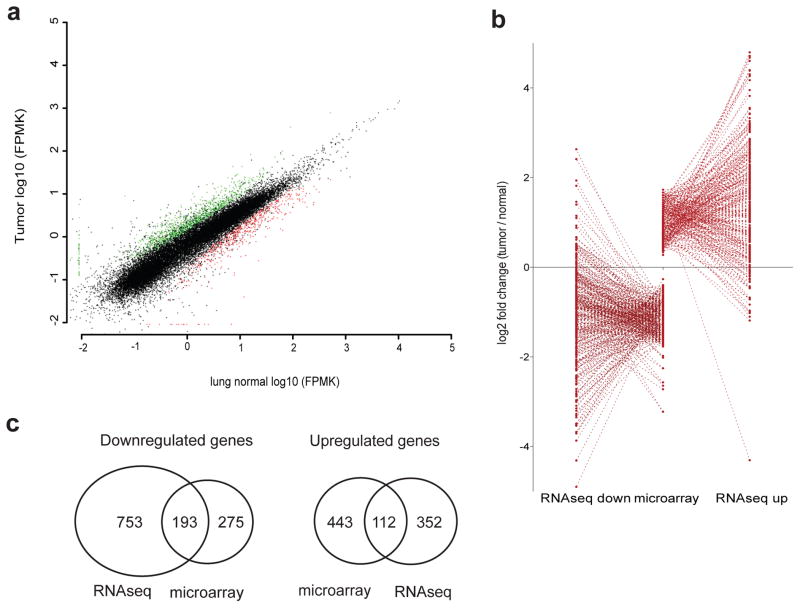
Differential expression analysis yielded ~450 significantly up-regulated and twice as many down-regulated genes in tumor versus normal lung samples (Figure 1a, Supplementary Table 1), distributed more or less evenly across the genome (Supplementary Figure 1). Genes significantly up in tumors include the Ros1 proto-oncogene (Supplementary Figure 2a) and Clec4n, both previously implicated in Kras transcriptomics and lung cancer.15, 16 The top three genes decreased in tumors are the cGMP-dependent protein kinase Prkg1, the guanine nucleotide binding protein alpha (Gnao1) and the regulator of G-protein signaling 9 (Rgs9). These genes and Kras thus potentially are directly related or at least perform similar function as all are G-protein signal proteins. The Kras gene itself had similar levels of expression between tumors (p-value = 0.545; Supplementary Figure 2b).
Figure 1.
Expression profile of aligned RNA sequences defines tumor versus normal differentially expressed sequences. (a) A scatterplot of mean FPKM values from all genes with a minimum FPKM of 0.01 (N = 21110). Genes significantly enriched in tumor samples are in green while those significantly down in tumor are red. Overall correlation between samples had an R2 value of 0.90782. (b) Gene expression profiling is comparable with microarray data from lung adenocarcinoma tumors.17 Significant values from the microarray data set are plotted in the middle column. For genes significantly up in this data set, the corresponding RNAseq log2 fold change was plotted to the rightmost column. For genes significantly down in microarrays, the corresponding RNAseq values were plotted in the left-most column. (c) Venn Diagram of overlapping genes in this data set versus microarray examples.
Ingenuity Pathway Analysis of the most significantly differentially expressed genes provided broad categories of nucleic acid metabolism, embryonic/organ development and cell signaling and cancer that were enriched in differentially regulated genes in the tumor (Supplementary Figure 3a, 3b). In addition, pathway analysis revealed that direct and indirect connections could be established between the Kras gene, the p53 gene and the three most differentially down-regulated G-protein associated genes Prkg1, Rgs9 and Gnao1 (Supplementary Figure. 3c).
RNA expression profiles have been evaluated previously in similar KrasG12D-driven lung tumors by microarray analysis that revealed 657 significantly differentially expressed genes.17 The corresponding fold-changes of these genes in our RNAseq data were quite similar (Pearson r of 0.631; p < 0.001; Figure 1b) and several genes were considered significantly differentially expressed (112 up and 193 down) in each data set (Figure 1c).
Somatic mutations accumulate at similar frequencies in normal lung and tumor samples
The extensive coverage afforded by high throughput sequencing enabled us to identify 18, 34 and 23 nonsynonymous variants in the three tumor samples and 44, 40 and 80 variants in the three control lung samples (Supplementary Table 2). Genes with nonsynonymous variants from lung adenocarcinoma samples were not more frequently present in the Cosmic database18 than the corresponding normal lung sample genes. Thus, in this model, Kras mutations do not appear to act together with multiple commonly mutated genes in lung or all cancers (Table 2).
Table 2.
mRNA variant identification between samples.
| Tumor1 | Tumor2 | Tumor3 | Normal1 | Normal2 | Normal3 | |
|---|---|---|---|---|---|---|
| GATK total variants | 2,498,695 | 2,243,002 | 951,637 | 2,358,541 | 3,639,468 | 1,328,724 |
| Passing filters | 117,167 | 238,903 | 27,091 | 248,036 | 186,095 | 50,882 |
| In exons | 3,300 | 8,443 | 550 | 9,243 | 4,051 | 815 |
| Unique | 41 | 79 | 40 | 105 | 89 | 180 |
| Synonymous | 21 | 45 | 17 | 61 | 48 | 100 |
| Nonynonymous | 18 | 34 | 23 | 44 | 40 | 80 |
| NS/S | 0.86 | 0.76 | 1.35 | 0.72 | 0.83 | 0.80 |
| Nonsyn genes in lung Cosmic mutations | 5 | 0 | 1 | 6 | 5 | 9 |
| Nonsyn genes in all Cosmic entries | 11 | 28 | 10 | 42 | 32 | 73 |
| Indels | 13 | 23 | 6 | 19 | 23 | 14 |
| Fusion transcripts | 51 | 43 | 74 | 23 | 29 | 28 |
Analysis of known SNPs between MOLF and 129S4 mice identifies allele specific expression and potential areas of loss of heterozygosity
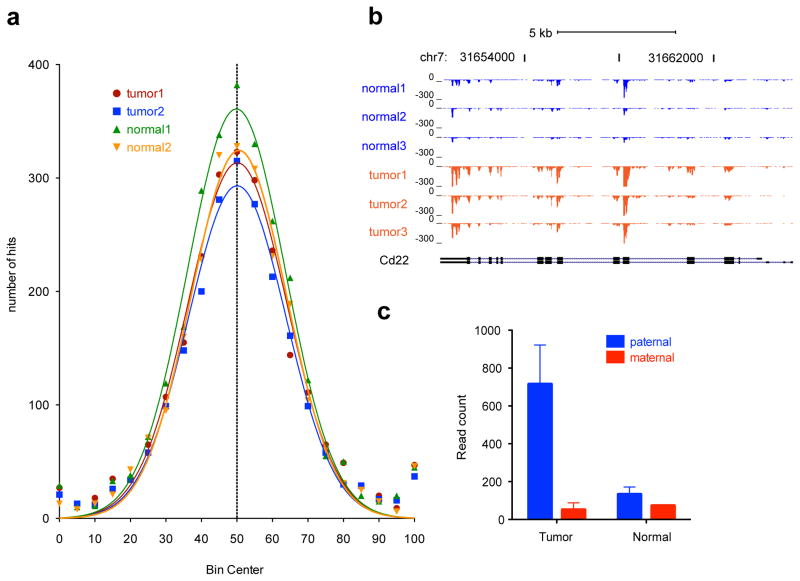
A total of 8065 coding variants in 4234 unique genes differentiate MOLF and 129S4 mice. This enabled analysis of allele specific expression of certain genes in addition to locations of loss of heterozygosity. Binning of SNPs based on their percent maternal expression did not show broad differences of parent-of-origin expression between tumor and normal lung samples (Figure 2a, Supplementary Figure 4). However, we could use the SNP information to identify individual genes with a biased allele specific expression (Supplementary Table 3). Cd22 had high levels in tumor samples (FPKM of 10.21 in tumor and 0.32 in normal lung; p=0.0001, Figure 2b). Surprisingly however, this expression largely or exclusively came from only the paternal allele while wildtype samples had bi-allelic expression (2 way ANOVA p-value of 0.028; Figure 2c). This primarily mono-allelic expression has been observed previously as a mechanism to retain specific antigen activity.19 Sanger-based sequencing revealed no chromosomal amplifications and confirmed an allele-specific expression bias for Cd22 mRNA (Supplementary Figure 5). Allelic expression analysis for Kras revealed that neither the wildtype nor mutant allele was amplified, as can be the case in certain tumors with KRAS mutations.20
Figure 2.
RNAseq analysis of coding variants reveals parent of origin specific expression or amplification of alleles. A total of 8065 coding variants that differ between 129S4/SvJae and MOLF/EiJ mice were interrogated for parent of origin expression. (a) Binning of percent maternal expression reveals that most SNPs follow a binomial distribution surrounding equal (50%) expression. (b) RNAseq read coverage for Cd22. Y-axis values represent read depth at each position adjusted by the total mapped reads for that sample relative to the mean mapped reads for all samples set from 0 to −300. Reads are negative because the gene is transcribed from right to left. (c) An analysis of genes that have the greatest fold difference of paternal to maternal allele expression reveals paternal-specific enhancement of Cd22 mRNA expression while maternal read counts remain similar between tumor and normal lung (2-way ANOVA p-value of 0.028 for tumor status). Error bars represent S.E.M. of the two F1 mice in each condition.
Analysis of RNA editing sites indicate that Apobec mediated C-to-U editing is common in tumors but not in wildtype lung
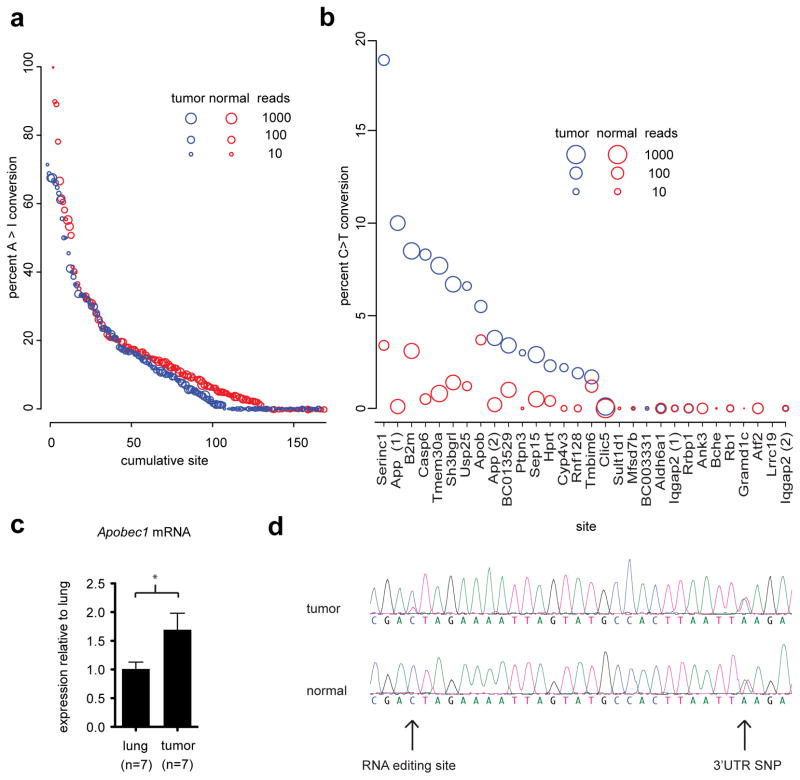
Post-transcriptional modifications, including RNA editing, can also be evaluated from RNAseq data. Several adenosine to inosine RNA editing sites have been identified in mice.21 We found no evidence of differential adenosine to inosine editing in these tumor samples at these known positions (Figure 3a; Wilcoxon signed rank test p=0.19). However, the Apobec enzyme performs an alternate form of RNA editing, namely cytidine deamination leading to a uridine residue. Of the 30 editing sites that were identified in studies of Apobec1−/− mice22 and were expressed in our lung samples, just over half (16) were C-to-U edited in tumor samples with editing ranging from 1.7% to 18.8% and the levels of editing were higher than corresponding rates in controls (Figure 3b). Detection of expression levels of Apobec1 revealed a modest 1.5 fold increase in expression in tumors (p<0.05; Figure 3c). A C-to-U edited site present in the Serinc1 3′UTR was validated by Sanger sequencing (Figure 3d). This data indicate that C-to-U editing is enriched in lung tumorigenesis, though it does not distinguish between whether the editing arises within the tumor during its formation or is the result of editing in immune derived cells in response to the tumor. Indeed, sorting for pure tumor originating cells (described below) revealed low Apobec1 mRNA levels and an absence of C-to-U editing at the Serinc1 site. While this cannot exclude the possibility that C-to-U editing in surrounding cells contributes to tumor progression it suggests that editing is not an inherent property of the tumor-originating cells.
Figure 3.
Lung tumors display an increase in Apobec but not Adar RNA editing. (a) Mean percent editing for 168 previously identified Adar mediated A to I edited sites in the mouse with the size of the point reflecting the number of mapped reads at the given location. (b) Mean percent Apobec editing at 30 previously identified sites for the three samples, plotted as in panel A. (c) Apobec1 mRNA expression levels from the three sequenced mice in each condition plus tumors and normal lungs from four additional mice each, error bars represent SEM; * = p < 0.05 by t-test. (d) Sanger verification of C to T editing in the Serinc1 3′UTR. Primers were designed to amplify the edited site along with a coding SNP that indicated equal expression from both parental alleles.
Small RNA sequencing identifies a cluster of microRNAs upregulated in lung adenocarcinomas
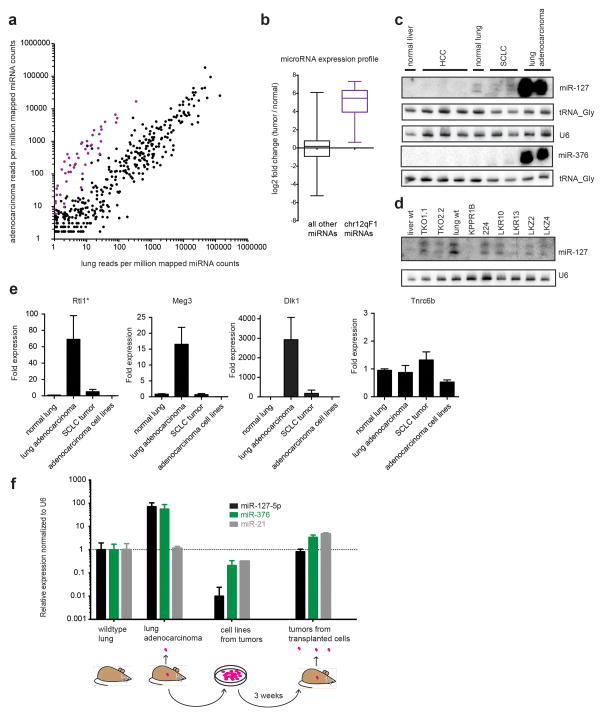
Our most salient finding arose when we complemented RNA sequencing by performing small RNA sequencing on the normal lung and adenocarcinoma samples. Almost all of the most differentially expressed microRNAs aligned to an ~800kb region (nucleotides 110,691,433-111,519,307) on mouse chromosome 12qF1 (Figure 4a; Supplementary Table 4). 53 microRNAs align to this region with a mean fold increase of 31.6 (t-test p=3.8×10−87 relative to all other microRNAs; Figure 4b). This fraction of chr12qF1 microRNAs represents ~9% of all lung adenocarcinoma miRNA reads, compared with 0.1–0.2% of miRNA reads in the normal lung. Outside this locus the two closest microRNAs, miR-345 at ~600kb proximal and miR-203 at ~1.8mb distal, had equivalent expression between tumors and normal lung samples. This level of microRNA induction was substantially higher and more prominent than was observed for large RNAs suggesting that microRNAs may be key mediators of oncogenic drive in this mouse model.
Figure 4.
A cluster of microRNAs on chromosome 12qF1 is upregulated in lung adenocarcinomas. (a) A scatterplot of microRNA counts normalized to one million mapped microRNA reads. Points in purple represent microRNAs that arise on chromosome 12qF1. (b) Boxplot of log2 based fold change for chr12qF1 microRNAs (in purple) and all remaining microRNAs (in black). (c) Small RNA northern blot of two representative microRNAs that align on chromosome 12qF1 (miR-127 and miR-376c), re-probed for tRNA sequences and/or U6. Small cell lung cancer (SCLC) and hepatocellular carcinoma (HCC) tumors were additional controls. (d) Cell lines derived from lung and liver tumor did not show high levels of chr12qF1 microRNA expression. (e) Quantitative RT-PCR analysis indicates that non-coding RNA expression of genes in the chromosome 12qF1 interval are highly expressed in lung adenocarcinomas relative to normal lung, SCLC tumors, and human lung adenocarcinoma tumor-derived cell lines. (* Probes for Rtl1 detect both sense and antisense transcripts) Values are plotted as mean +/− standard error of the mean of at least three samples run in triplicate. Note the differences in the y-axis for each of the plots. (f) microRNA qPCR indicates that an upregulation of the chr12qF1 locus is specific to tumors in vivo. Levels of two microRNAs from this cluster, miR-127-5p and miR-376a were normalized to U6 snRNA and then to wildtype lung levels. Cell lines derived from tumors were grown in culture or transplanted back into mice.
Northern blot analysis validated representative chr12qF1 microRNA expression patterns in lung adenocarcinoma samples from all 12 mice tested (two shown in Figure 4c). However, cell lines derived from the KrasG12D mouse tumors, normal lung and SCLC samples had levels of chr12qF1 microRNAs comparable to wildtype lung (Figure 4c, 4d). Thus, the up-regulated microRNA cluster is a hallmark only of the tumors in vivo and not of the associated derived cell lines.
The mouse chr12qF1 region, also known as the Dlk1-Dio3 locus,23 it is an area that is extensively methylated with a set of genes that are expressed specifically from the maternal or paternal chromosome. The cluster of microRNAs is transcribed from the chromosome inherited from the mother, as are the noncoding RNAs Meg3, Meg8, Rtl1as and Rian. Conversely, paternally expressed genes include Dlk1, Rtl1 and Dio3. SNPs in these genes were used to verify that this established parent of origin expression pattern was maintained in the tumor samples. From the RNAseq dataset, only the Rian gene was significantly up-regulated in the lung adenocarcinoma samples (FPKM of 6.48 for tumors versus 1.58; q-value < 0.005). However, when we examined previous microarray data from lung adenocarcinoma mouse tumors, probes for Meg3 were ranked second and sixth as the most abundantly represented in adenocarcinoma versus control (10.56 fold and 5.33 fold differences respectively) and a probe for Dlk1 ranked ninth on this list (5.18 fold increase).17 We confirmed this up-regulation via quantitative real-time PCR with probes against Rtl1 (sense and antisense), Dlk1 and Meg3 (Figure 4e). Consistent with data from microRNAs in this locus, tumor samples displayed a marked increase in expression of these co-expressed noncoding RNAs (Figure 4e). Differentially methylated regions exist at the Meg3 promoter and intergenic to Meg3 and Dlk1 that are fully methylated in the adult lung. While hypomethylation at the maternal allele is typically associated with microRNA expression,24 we observed no change in methylation in bulk tumors (Supplementary Figure 6) suggesting a different mode of locus activation is involved.
Increase of chr12qF1 microRNA expression is present in tumor-originating cells
Differential expression of genes or small RNAs from bulk tumors could arise from stromal cells that have infiltrated and become mixed with bona fide cancer cells. To reconcile whether the chromosome 12qF1 microRNAs and mRNAs are specific to the cancer cells, we generated tumors in KrasLSL-G12D mice that also carried a Rosa26LSL-tdTomato Cre reporter allele. In this situation, when tumors were initiated with viral Cre, all resulting cancer cells would express red fluorescent protein along with the activated KrasG12D allele. Fluorescence activated cell sorting of Tomato-positive cells from tumors enabled an evaluation of genes that are specific to the cancer cells. Validation of chromosome 12qF1 microRNAs revealed a ~35 fold induction of miR-127 and miR-376a in KrasG12D expressing cells relative to wildtype lung (Figure 4f). Further, the Meg3, Dlk1 and Rtl1 genes were dramatically up-regulated (Supplementary Figure 7a). This demonstrates that the chromosome 12qF1 locus is activated in a manner specific to cells carrying a KrasG12D mutation.
As noted above, cell lines derived from lung adenocarcinomas exhibited low levels of chr12qF1 gene and microRNA activation. However, when the KrasG12D cells were transplanted back into mice, tumors arose within 3 weeks. By evaluating three of the tumors derived from transplanted cells, we were able to demonstrate that the same population of cells could reactivate their chr12qF1 microRNAs (Figure 4f) and mRNAs (Supplementary Figure 7b) when propagated in vivo. Taken together, this implies that the cell lines derived from lung adenocarcinomas do not display some of the more prominent features of solid tumors that are dependent on their in vivo environment.
miRNAs alter select target protein levels without globally influencing mRNA target expression
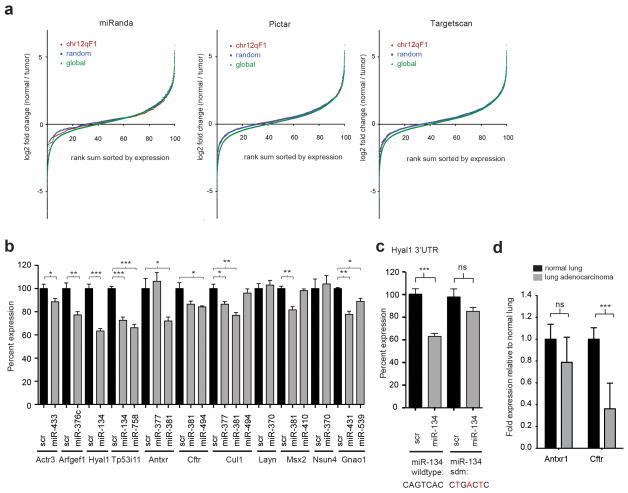
The large scale small RNA and large RNA sequencing datasets enabled global comparisons between predicted microRNA targets and their expression changes. The 53 microRNAs that are up-regulated on chr12qF1 were compared to 51 microRNAs that had a similar expression profile (within 10% of each other) between tumor and normal lung. Target mRNAs had no significant difference in expression for chr12qF1 microRNAs relative to the control microRNA set for Miranda (p=0.18), Pictar (p=0.46) and TargetScan (p=0.39) prediction programs (Figure 5a). This indicates that chr12qF1 microRNAs do not lead to reduction in mRNA levels of predicted targets.
Figure 5.
Chr12qF1 microRNAs can repress protein levels of a subset of genes involved in oncogenesis. (a) The cumulative distribution of mRNA expression is unchanged for mRNAs predicted to be targets of the 53 chr12qF1 microRNAs upregulated in lung adenocarcinoma (in red) versus 51 control unchanged microRNAs (in blue) and overall mRNAs (in green). Three prediction programs were queried, Miranda (395 chr12qF1 targets and 376 control targets), Pictar (1231 and 1117 respective targets) and Targetscan (1002 and 1125 respective targets). (b) Luciferase expression of UTRs of mRNAs predicted to be targets of chr12qF1 microRNAs. Expression is normalized to firefly luciferase within the same construct and to a scrambled control shRNA (scr; black bars), transfected in E10.5 mouse embryonic fibroblasts (MEFs) where chr12qF1 microRNAs are not expressed. Significance was determined by a two-tailed t-test compared with a corresponding control shRNA (* = p < 0.05; ** = p < 0.01; *** = p < 0.001). Values represent the mean +/− SEM of at least two experiments performed in triplicate. (c) Site directed mutagenesis of three nucleotides of the miR-134 minding site in the Hyal1 3′UTR abrogated the repression of miR-134 on this 3′UTR. (d) Quantitative RT-PCR showed a reduction in Cftr but not Antxr1 mRNA levels in lung adenocarcinoma samples (n = 7) relative to normal lung (n = 7).
We utilized a luciferase reporter system to identify whether protein levels were altered by the up-regulation of these microRNAs. Twelve 3′UTRs that were predicted by TargetScan to be strong targets of ten of the most highly expressed chr12qF1 microRNAs. Co-transfection of the luciferase constructs and its corresponding target miRNA led to the down-regulation of 8 of these 11 targets from nine of the ten microRNAs relative to a scrambled control shRNA (Figure 5b). Three point mutations in the miR-134 binding site of the Hyal1 3′UTR caused a loss of miR-134 mediated repression of this 3′UTR (Figure 5c). Quantitative RT-PCR analysis revealed that Cftr but not Antxr1 showed a reduction in mRNA levels (Figure 5d). This is just a select subset of genes that are potential targets, yet it indicates that the microRNAs do have the intended effect of mediating post-transcriptional effects. Given that microRNAs have many more predicted targets, the number of proteins influenced by these 53 microRNAs is exponentially larger and has the potential to dramatically re-shape the living tissue environment.
Dlk1-Dio3 locus activation is characteristic of a subset of human lung adenocarcinoma samples
The small RNA population from 346 human lung adenocarcinoma samples has been subject to high throughput sequencing, available for analysis as part of the cancer genome atlas (TCGA). The percent of chr12qF1 microRNAs were increased 3.31-fold in these TCGA adenocarcinoma samples relative to 40 matched normal lung samples (p=0.014; Supplementary Figure 8a). However, from the tumor samples, a bimodal expression pattern was noted whereby several samples exhibited elevated expression patterns (Supplementary Figure 8a). Parsing of tumor samples into the 34 with high locus expression (~10%) with all other tumor samples revealed a consistent and specific activation of all Dlk1-Dio3 locus microRNAs (Supplementary Figure 8b). Thus, this locus is aberrantly activated in a subset of human lung adenocarcinomas.
Discussion
A central tenet of cancer genetics is that rapidly dividing tissues over time have the potential to accumulate enough mutations and chromosomal alterations in oncogenes and tumor suppressors such that a critical threshold is obtained and tumorigenesis ensues. However, in organs that do not undergo this rapid cell division and turnover – such as the lung – a reversal back to an embryonic state or a proliferation of stem cells (cancer stem cells) with their associated rapid growth and development is one of the mechanisms that is postulated to be involved. This is indicative of the scenario that we observe in this murine lung adenocarcinoma dataset in which a cluster of stem-cell associated microRNAs are up-regulated.
Several lines of evidence point to the role of the chr12qF1 locus in stem cell biology, lung development and oncogenesis. The sustained expression of this locus is essential in the development of induced pluripotent cells with a common loss of chr12qF1 microRNA expression resulting in the low proportion of cells that maintain an iPS state.25 The proper expression of mmu-miR-127 is essential for lung development as its overexpression led to fewer terminal buds indicating impaired lung branching.26 Meanwhile, removal of the maternally-derived (but not paternally-derived) Meg3 allele in mice led to thin-walled lungs with reduced radial alveolar counts and early postnatal lethality.27 In addition, knockout of the Dicer1 gene, critical for RNA interference, had effects that were more specific to the lung,28 while loss of one Dicer1 allele reduced survival in the same KrasLSL-G12D lung adenocarcinoma model.29 Several of the chr12qF1 microRNAs were among the most up-regulated by microarray analysis in a completely distinct mouse model of lung adenocarcinoma with sustained high levels of cyclin E and up-regulation of miR-376a and miR-136 was validated in human lung adenocarcinomas.30 Differential expression of the chr12qF1 locus has also been identified in other cancer subtytpes31 including up-regulation in mouse and human hepatocellular carcinoma samples,32 gastrointestinal stromal tumors,33 acute promyelocytic leukemia34 and associated with epithelial to mesenchymal transition in endrometrial carcinoma.35 Notably, miR-127-3p was significantly up in colorectal cancer associated with KRAS mutations.36
When an entire cluster of genes and miRNAs is up-regulated, it becomes difficult to identify and target a single gene or miRNA that may be responsible for the tumorigenic phenotype. It will be interesting to determine whether one or a few microRNAs are sufficient to recapitulate this oncogenic event, though a more likely scenario is that the cohort of small RNAs and non-coding RNAs act coordinately to regulate a multitude of genes. Several genes that are targets of chr12qF1 by luciferase analysis have implications in oncogenesis and lung development. The p53 interacting protein Tp53i11, repressed by miR-134 and miR-758 is a putative tumor suppressor in liver cancer.37 The actin related protein Actr3 is a major constituent of the Arp2/3 protein complex down-regulated in gastric cancer,38 Arfgef1 in breast cancer,39 and Cul1 in various tissues.40 Relevant to lung biology, the cystic fibrosis transmembrane receptor Cftr which is hypermethylated and down-regulated in lung adenocarcinoma41 and mutated in non-small cell lung cancers,42 is targeted by miR-381 and miR-494. Indeed several oncogenic pathways are implicated upon microRNA activation. How these microRNAs are specifically activated (or how a stem cell like population can continue to proliferate unabated) will be of interest in future studies. While methylation patterns do not appear to change in the tumor samples (contradictory to established methylation patterns at this locus), other epigenetic marks certainly may be involved, particularly histone marks. Alternatively since we examined methylation patterns of tumor samples in bulk, it remains possible that demethylation at a subset of cells is sufficient to activate the chr12qF1 locus. The results of this and other mouse models43 appear to be limited to a subset of human lung tumors, and it will be interesting to determine if KRAS mutations in human samples can induce a similar activation of the chr12qF microRNAs.
By sequencing the transcriptome to a considerable depth we could search for additional mutations and expression changes that could aid in progression of tumorigenicity. An evaluation of both normal and tumorigenic lungs indicated no difference in the number of coding changes. However, if a variant caused nonsense-mediated decay or induces loss of expression, it would be more difficult to detect using this approach and would require whole genome or exome DNA sequencing. By examining allele-specific expression, we identified genes with modifications in expression of maternal versus paternal alleles depending on tumor status. This includes the paternal-specific enhancement of Cd22 mRNA expression specifically in tumor samples. CD22 is a B-cell lymphocyte cell surface marker that incurs mutations and splicing defects in human B-precursor leukemia44 and is the target of Epratuzumab, a humanized monoclonal antibody therapeutic for B cell tumors.45 Interestingly, a group recently independently identified CD22 cell surface expression in A549 cells and solid tumors and have shown that anti-CD22 antibodies can delay tumor progression.46 Quantification of RNAseq read hits enabled a calculation of expression changes across the genome. The three most down-regulated genes (Prkg1, Gnao1 and Rgs9) are implicated in G-protein coupling and which have indirect connections with Kras and p53. Of note, a point mutation in Gnao1 has been recently identified in breast cancer,11 and this mutation appears to function in a manner analogous to the Kras G12D mutation in that it maintains the gene in a constitutively active state.47
The presence of editing in these lung adenocarcinoma samples is quite striking, yet it is difficult to ensure that this effect is specific to tumor progression or infiltration of B cells in tumor samples. Of the 32 Apobec1 edited sites identified previously,22 all but one are present in the 3′UTR of transcripts which can have multiple effects including influencing poly-A usage.48 This can have regulatory consequences if regions such as microRNA binding sites are precluded from the edited RNA transcript. Our work suggests that much of the editing does not occur in tumor-originating cells; nonetheless, Apobec1 mRNA was increased overall in tumors and Apobec transgene expression in the liver has been shown to inadvertently drive HCC.52 Separate from RNA editing, APOBEC mediated DNA mutagenesis from up-regulated APOBEC family proteins was reported to be a property of several human cancers49, 50 after it was noted that cascades of localized C-to-T changes, termed kataegis, were found in breast cancer samples.51
In conclusion, through use of high throughput sequencing technology, we have uncovered several novel genetic abnormalities that exist in the coding and non-coding transcriptome of extracted solid tumors of the lung. By small RNA sequencing we were able to detect consistent up-regulation of a cluster of microRNAs typically associated with a stem cell like state. It is the activation of this locus and the multitude of mRNA targets of the ~53 microRNAs that we believe are crucial for oncogenic drive in this Kras mutant mouse model of lung adenocarcinoma.
Materials and Methods
Mouse breeding
The Stanford Institute of Medicine Animal Care and Use Committee approved all animal studies and procedures. 129S4 males heterozygous for a KrasLSL-G12D allele were mated with MOLF/EiJ females. Littermate controls that were used include a KrasLSL-G12D+ mouse with no Adeno-Cre administered, and a wildtype Kras mouse with Adeno-Cre (to control for adenovirus exposure effects). In addition, tumors and normal lungs were extracted from a distinct mouse lung adenocarcinoma model, namely the Kras LA mouse which does not require Cre delivery and is present on an inbred background.8
KrasLSL-G12D mice were bred as previously described.53, 54 To evaluate allele-specific expression, KrasLSL-G12D mice on the 129S4/SvJae background were bred with MOLF mice. Offspring were genotyped for the presence of the KrasLSL-G12D allele. To activate Kras, an adenovirus bearing Cre-recombinase (AdCre:CaPi co-precipitates, Baylor Vector Development Lab) was intra-nasally administered at 6 weeks of age. Tumors were dissected ~4 months later.
Tumor cell sorting and transplantation
The Rosa-LSL-tdTomato Cre reporter allele was bred into KrasLSL-G12D; p53flox mice. Tumors were initiated by intratracheal infection of mice with a lentiviral vector expressing Cre recombinase.54 Single cell suspensions were generated from individual lung tumors harvested from mice 8–14 months after tumor initiation. Tumors were minced and then digested for 30 min at 37°C in 2ml of HBSS-free containing trypsin, collagenase IV and dispase. Subsequently, 4ml of Quench Solution (L15 media supplemented with FBS and DNase) was added and samples were then pressed through 40mm cell strainers (BD Biosciences). Finally, samples were centrifuged at 1,000 r.p.m. for 5 min and re-suspended in FACS media (PBS, 2% FBS, 2mM EDTA). Sorting for td-Tomato-positive cells was performed at the Stanford Shared FACS Facility.
High throughput large RNA sequencing
The Ribo-Zero ribosomal removal kit (Epicentre) was used to remove ribosomal sequences from total Trizol-extracted RNA. 200ng of purified RNA was subjected to strand-specific RNAseq using the ScriptSeq library preparation kit (Epicentre). 50bp paired end reads were generated on an Illumina HiSeq 2000 machine. Trimmed sequences were mapped to the mm9 genome using TopHat version 1.3.3 under default settings allowing multiple alignments with the following specific parameters: -r 100, --mate-std-dev 100 --segment-length 20–library-type fr-unstranded.55 To accomodate SNPs between the Molf/EiJ and 129S4/SvJae strains, three mismatches were allowed in the mapping process for these samples, while two mismatches were allowed for samples with homozygous parents. FPKM values were calculated using Cufflinks v.1.3.0.56, 57 Transcripts with a q-score of < 0.05 were considered significant and we further required a minimum FPKM of 5 in at least one of the two conditions.
SNP analysis
GATK (v.1.5) was used to calculate non-synonymous variants present in tumor and normal lung samples.58, 59 Candidate variants in tumors were filtered based on a minimum read depth of 10, an absence in control lung samples, and that did not exhibit a bias in end-of-sequence. Raw reads were manually inspected for accuracy of GATK calls.
Allele specific expression
To evaluate allele specific expression, we extracted a list of all coding single nucleotide variants between MOLF and 129S4 mice as computed from the Mouse Genome Informatics dataset (Jackson labs). The Samtools BCFtools program60 was used to call the number of variants in each sequenced sample. This data was filtered to have a minimum of 20 reads per sample. Variants in individual genes were sorted based on combined fold-changes between tumor and normal samples. Genes with the greatest fold-differences were manually analyzed for additional exonic SNPs.
RNA editing analysis
Allele calls at adenosine to inosine21 and cytosine to uridine22 editing sites were identified using BCFtools. The Serinc1 variant at chr10:57,235,791 (mm9) was validated by Sanger sequencing using the primers 5′-ACATTAGGCTCGGGTTAGGCACTA and 5′-AAGGCTGGAACATGAAGATGAACT for both genomic DNA and cDNA.
Small RNA sequencing
3ug of a mirVana (Invitrogen) extracted small RNA fraction was ligated to a 3′linker in ATP-free buffer. Samples were resolved on a 12% polyacrylamide gel and 17–28nt fragments were excised. A barcoded linker was ligated to the 5′ end of the extracted RNA using T4 RNA ligase and these RNAs were reverse transcribed using Superscript II (Invitrogen). 5ul of this product was subject to 21–24 PCR amplification cycles in a total of 50ul volume using Taq polymerase (NEB). The product was resolved on a 4% Nusieve GTG agarose gel (Lonza, Rockland, ME). 20ng of this product was subjected to 36 base pair high throughput sequencing on an Illumina GAII machine. Small RNA reads (17–28bp) were aligned to miRBase (release 15)61 using the Bowtie program release 0.12.762 allowing for two mismatches.
Small RNA northern blot
10ug of Trizol (Invitrogen) extracted RNA was resolved on a 15% acrylamide gel, transferred to a Hybond N+ membrane (Amersham). Membranes were scanned using a phosphoimager.
Quantitative PCR
2ug of total RNA was reverse-transcribed using Superscript II (Invitrogen). Gene-specific probes used included Rtl1 (Mm02392620_s1), Dlk1 (Mm00494477_m1), Meg3 (Mm00522599_m1), Tnrc6b (Mm00523487_m1), Cftr (Mm00445197_m1) and Antxr1 (Mm00712952_m1) (Life Technologies). MicroRNA Taqman analysis was performed on 250ng of Trizol-extracted small RNAs using probes that detect mature miR-21, miR-127-5p, miR-376a and U6 (Life Technologies). Quantitative RT-PCR was performed on a CFX384 Real-Time system (BioRad).
Luciferase analysis
Dual-luciferase assays (Promega) were performed 24hr after transfection according to manufacturer’s protocol and detected by a Modulas Microplate Luminometer (Turner Biosystems). For transfection, 250ng of psi-check reporter plasmids were co-transfected with 250ng of miRNA overexpression plasmids (Sh-constructs) in E10.5 mouse embryonic fibroblasts using TransIT-LT1 (Mirus). Cell seeding was performed at a concentration of 2.5×104 cells per well in a 24-well plate.
For cloning of the psi-check constucts, the entire 3′UTR of each gene was PCR amplified (see Supplementary Table 5 for primer sequences) from mouse genomic DNA and cloned in psi-check-2 vector (Promega) between the XhoI and SpeI sites using the In-fusion HD cloning kit. The quickchange II site directed mutagenesis kit (Agilent) was used to introduce three mutations in the Hyal1 3′UTR using the primer 5′-GGACTTCCTCAAATACTGACTCATGCCCATAAGTC and the reverse complement thereof (mismatches are listed in bold). For generation of the microRNA over-expression constructs, shRNA sequences (Supplementary Table 6) were chemically synthesized; both strands were annealed and inserted between BglII and KpnI sites downstream of the U6 Pol III promoter.
Bisulfite sequencing
Genomic DNA was extracted from tumor and normal lung samples by the DNeasy tissue kit (Qiagen) and converted by bisulfite treatment using an EZ DNA methylation kit (Zymo research). PCR primers were as follows for Meg3: 5′-GTTATAGTAATTTGTTATAGAATTTGGGG (forward) and 5′-AAACTTTCAACCACCAAAACC (reverse), and for an intergenic differentially methylated region: 5′-GGTTTGGTATATATGGATGTATTGTAATATAGG (forward) and 5′-ATAAAACACCAAATCTATACCAAAATATACC (reverse).63 Products were cloned using the Topo TA cloning system (Invitrogen) and at least eight clones were sequenced per sample per locus.
miRNA target prediction
For Miranda, 395 targets were identified for chr12qF1 microRNAs versus 376 in controls using a cutoff mirSVR score of -1.364. The Pictar program predicted 1231 chr12qF1 and 1171 control target mRNAs. Finally, 1002 chr12qF1 and 1125 control mRNAs were identified as TargetScan 61, 65 targets (within the 95th percentile of hits).
Pathway analysis
Transcripts significantly differentially expressed between tumor and normal samples (false discovery rate < 0.001) were analyzed by the ingenuity pathway analysis program using default settings.
Human lung adenocarcinoma samples
Small RNA deep sequencing reads corresponding to human microRNAs were queried from samples collected as part of the cancer genome atlas (TCGA). Data was available from a total of 346 samples from patients with stage I–IV lung adenocarcinomas collected from centers throughout the United States. This was compared with data from 40 matched normal samples.
Data access
Sequences have been deposited in the NCBI Gene Expression Omnibus (accession number GSE43028).
Supplementary Material
Acknowledgments
PNV is a Banting Postdoctoral Fellow supported by the Canadian Institutes of Health Research. MMW is funded by a Baxter Foundation Scholar Award and C-HC is supported by a Stanford Dean’s Fellowship. We would like to thank Yue Zhang for help in variant detection and Julien Sage for small cell lung cancer samples. This work was supported by grant R01 DK078424 (Kay).
Footnotes
Conflict of interest
The authors have no conflicts of interest to report.
References
- 1.Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: ‘it ain’t over ‘til it’s over’. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 2.Simon MA, Dodson GS, Rubin GM. An SH3-SH2-SH3 protein is required for p21Ras1 activation and binds to sevenless and Sos proteins in vitro. Cell. 1993;73:169–177. doi: 10.1016/0092-8674(93)90169-q. [DOI] [PubMed] [Google Scholar]
- 3.Rogge RD, Karlovich CA, Banerjee U. Genetic dissection of a neurodevelopmental pathway: Son of sevenless functions downstream of the sevenless and EGF receptor tyrosine kinases. Cell. 1991;64:39–48. doi: 10.1016/0092-8674(91)90207-f. [DOI] [PubMed] [Google Scholar]
- 4.Al-Mulla F, Milner-White EJ, Going JJ, Birnie GD. Structural differences between valine-12 and aspartate-12 Ras proteins may modify carcinoma aggression. J Pathol. 1999;187:433–438. doi: 10.1002/(SICI)1096-9896(199903)187:4<433::AID-PATH273>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 5.Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, Chitale DA, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santos E, Martin-Zanca D, Reddy EP, Pierotti MA, Della Porta G, Barbacid M. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661–664. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- 7.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 8.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 9.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ju YS, Lee WC, Shin JY, Lee S, Bleazard T, Won JK, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–445. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 12.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 14.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Lee S, Kang J, Cho M, Seo E, Choi H, Kim E, et al. Profiling of transcripts and proteins modulated by K-ras oncogene in the lung tissues of K-ras transgenic mice by omics approaches. Int J Oncol. 2009;34:161–172. [PubMed] [Google Scholar]
- 17.Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 18.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chess A. Mechanisms and consequences of widespread random monoallelic expression. Nat Rev Genet. 2012;13:421–428. doi: 10.1038/nrg3239. [DOI] [PubMed] [Google Scholar]
- 20.Mita H, Toyota M, Aoki F, Akashi H, Maruyama R, Sasaki Y, et al. A novel method, digital genome scanning detects KRAS gene amplification in gastric cancers: involvement of overexpressed wild-type KRAS in downstream signaling and cancer cell growth. BMC Cancer. 2009;9:198. doi: 10.1186/1471-2407-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danecek P, Nellaker C, McIntyre RE, Buendia-Buendia JE, Bumpstead S, Ponting CP, et al. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 2012;13:r26. doi: 10.1186/gb-2012-13-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat Struct Mol Biol. 2011;18:230–236. doi: 10.1038/nsmb.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagan JP, O’Neill BL, Stewart CL, Kozlov SV, Croce CM. At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS One. 2009;4:e4352. doi: 10.1371/journal.pone.0004352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Current opinion in cell biology. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, et al. MicroRNA-127 modulates fetal lung development. Physiol Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahashi N, Okamoto A, Kobayashi R, Shirai M, Obata Y, Ogawa H, et al. Deletion of Gtl2, imprinted non-coding RNA, with its differentially methylated region induces lethal parent-origin-dependent defects in mice. Hum Mol Genet. 2009;18:1879–1888. doi: 10.1093/hmg/ddp108. [DOI] [PubMed] [Google Scholar]
- 28.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298–1309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benetatos L, Voulgaris E, Vartholomatos G. DLK1-MEG3 imprinted domain microRNAs in cancer biology. Crit Rev Eukaryot Gene Expr. 2012;22:1–15. doi: 10.1615/critreveukargeneexpr.v22.i1.10. [DOI] [PubMed] [Google Scholar]
- 32.Luk JM, Burchard J, Zhang C, Liu AM, Wong KF, Shek FH, et al. DLK1-DIO3 genomic imprinted microRNA cluster at 14q32. 2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J Biol Chem. 2011;286:30706–30713. doi: 10.1074/jbc.M111.229831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haller F, von Heydebreck A, Zhang JD, Gunawan B, Langer C, Ramadori G, et al. Localization- and mutation-dependent microRNA (miRNA) expression signatures in gastrointestinal stromal tumours (GISTs), with a cluster of co-expressed miRNAs located at 14q32. 31. J Pathol. 2010;220:71–86. doi: 10.1002/path.2610. [DOI] [PubMed] [Google Scholar]
- 34.Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One. 2008;3:e2141. doi: 10.1371/journal.pone.0002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castilla MA, Moreno-Bueno G, Romero-Perez L, Van De Vijver K, Biscuola M, Lopez-Garcia MA, et al. Micro-RNA signature of the epithelial-mesenchymal transition in endometrial carcinosarcoma. J Pathol. 2011;223:72–80. doi: 10.1002/path.2802. [DOI] [PubMed] [Google Scholar]
- 36.Mosakhani N, Sarhadi VK, Borze I, Karjalainen-Lindsberg ML, Sundstrom J, Ristamaki R, et al. MicroRNA profiling differentiates colorectal cancer according to KRAS status. Genes Chromosomes Cancer. 2012;51:1–9. doi: 10.1002/gcc.20925. [DOI] [PubMed] [Google Scholar]
- 37.Ricketts SL, Carter JC, Coleman WB. Identification of three 11p11. 2 candidate liver tumor suppressors through analysis of known human genes. Mol Carcinog. 2003;36:90–99. doi: 10.1002/mc.10101. [DOI] [PubMed] [Google Scholar]
- 38.Kaneda A, Kaminishi M, Sugimura T, Ushijima T. Decreased expression of the seven ARP2/3 complex genes in human gastric cancers. Cancer Lett. 2004;212:203–210. doi: 10.1016/j.canlet.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 39.Kim JH, Kim TW, Kim SJ. Downregulation of ARFGEF1 and CAMK2B by promoter hypermethylation in breast cancer cells. BMB Rep. 2011;44:523–528. doi: 10.5483/bmbrep.2011.44.8.523. [DOI] [PubMed] [Google Scholar]
- 40.Lee J, Zhou P. Cullins and cancer. Genes Cancer. 2010;1:690–699. doi: 10.1177/1947601910382899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Son JW, Kim YJ, Cho HM, Lee SY, Lee SM, Kang JK, et al. Promoter hypermethylation of the CFTR gene and clinical/pathological features associated with non-small cell lung cancer. Respirology. 2011;16:1203–1209. doi: 10.1111/j.1440-1843.2011.01994.x. [DOI] [PubMed] [Google Scholar]
- 42.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma Y, Fiering S, Black C, Liu X, Yuan Z, Memoli VA, et al. Transgenic cyclin E triggers dysplasia and multiple pulmonary adenocarcinomas. Proc Natl Acad Sci U S A. 2007;104:4089–4094. doi: 10.1073/pnas.0606537104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uckun FM, Goodman P, Ma H, Dibirdik I, Qazi S. CD22 EXON 12 deletion as a pathogenic mechanism of human B-precursor leukemia. Proc Natl Acad Sci U S A. 2010;107:16852–16857. doi: 10.1073/pnas.1007896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leung SO, Goldenberg DM, Dion AS, Pellegrini MC, Shevitz J, Shih LB, et al. Construction and characterization of a humanized, internalizing, B-cell (CD22)-specific, leukemia/lymphoma antibody, LL2. Mol Immunol. 1995;32:1413–1427. doi: 10.1016/0161-5890(95)00080-1. [DOI] [PubMed] [Google Scholar]
- 46.Tuscano JM, Kato J, Pearson D, Xiong C, Newell L, Ma Y, et al. The CD22 Antigen is Broadly Expressed on Lung Cancer Cells and is a Target for Antibody-based Therapy. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-12-0173. [DOI] [PubMed] [Google Scholar]
- 47.Garcia-Marcos M, Ghosh P, Farquhar MG. Molecular basis of a novel oncogenic mutation in GNAO1. Oncogene. 2011;30:2691–2696. doi: 10.1038/onc.2010.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamanaka S, Balestra ME, Ferrell LD, Fan J, Arnold KS, Taylor S, et al. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 62.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sekita Y, Wagatsuma H, Irie M, Kobayashi S, Kohda T, Matsuda J, et al. Aberrant regulation of imprinted gene expression in Gtl2lacZ mice. Cytogenet Genome Res. 2006;113:223–229. doi: 10.1159/000090836. [DOI] [PubMed] [Google Scholar]
- 64.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA. org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.