Abstract
Hepatitis B virus (HBV) is a member of the hepadnavirus family. Hepadnaviruses can be found in both mammals (orthohepadnaviruses) and birds (avihepadnaviruses). The genetic variability of HBV is very high. There are eight genotypes of HBV and three clades of HBV isolates from apes that appear to be additional genotypes of HBV. Most genotypes are now divided into subgenotypes with distinct virological and epidemiological properties. In addition, recombination among HBV genotypes increases the variability of HBV. This review summarises current knowledge of the epidemiology of genetic variability in hepadnaviruses and, due to rapid progress in the field, updates several recent reviews on HBV genotypes and subgenotypes.
Keywords: Orthohepadnavirus; Avihepadnavirus; Hepatitis B virus; Genotype, Subgenotype; Recombi-nation
INTRODUCTION
Hepatitis B virus (HBV) is the prototype member of a steadily growing family of viruses called hepadnaviruses[1]. Hepadnaviruses can be found in both mammals (orthohepadnaviruses) and birds (avihepadnaviruses). HBV, the hepadnavirus infecting humans, is classified into eight genotypes today. HBV genotypes differ by at least 8%[2]. Since the first definition of the genotypes A, B, C and D[2], genotypes E[3], F[4], G[5] and H[6] have been detected. Due to the genetic diversity of HBV, numerous subgenotypes of HBV have been described[7] (Table 1). HBV subgenotypes differ by at least 4%[8].
Table 1.
Orthohepadnaviruses and their host
| Host | Ref. | |
| Hepatitis B Virus | Man | [75] |
| Homo sapiens sapiens | ||
| Chimpanzee Hepatitis B Virus | Chimpanzee | [76] |
| Pan troglodytes | ||
| Gibbon Hepatitis B Virus | White handed gibbon | [77] |
| Hylobates lar | ||
| Orangutan Hepatitis B Virus | Orangutan | [78] |
| Pongo pygmaeus pygmaeus | ||
| Gorilla Hepatitis B Virus | Gorilla | [79] |
| Gorilla gorilla | ||
| Woolly Monkey Hepatitis B Virus | Woolly monkey | [80] |
| Lagothrix lagotricha | ||
| Woodchuck Hepatitis Virus | Woodchuck | [19] |
| Marmota monax | ||
| Ground Squirrel Hepatitis Virus | Ground Squirrel | [20] |
| Spermophilus beecheyi | ||
| Arctic Squirrel Hepatitis Virus | Arctic Squirrel | [81] |
| Spermophylus parryi kennicotti |
HBV genotypes and most subgenotypes show a distinct geographic distribution. In Asia, where there is a high prevalence of HBV carriers, strong evidence suggests that HBV genotypes influence the course of disease. Several recent reviews have summarised knowledge on different aspects of HBV genotypes[7-12] and on hepadnaviruses that infect species other than homo sapiens[13-15]. This review will update recent developments in understanding HBV genotypes and taxonomy.
TAXONOMY
HBV is a partially double stranded virus that uses reverse transcriptase in its replication cycle. Thus, HBV is similar to many retroviruses found in animals and pararetroviruses in plants[16,17].
After cloning and sequencing the HBV genome[18], several related viruses were discovered in woodchucks (Marmota monax)[19], ground squirrels (Spermophilus beecheyi)[20] and pekin duck (Anas domesticus)[21]. Subsequently, numerous new viruses that are similar to HBV were found in mammals and birds and have been cloned (Tables 1 and 2). All these viruses are classified in the family of hepadnaviridae, including the genus orthohepdnavirus (mammals; Figure 1), and the genus avihepadnavirus (birds; Figure 2). In addition to the avihepadnaviruses listed in Table 2, five new hepadnaviruses were cloned from exotic duck and goose species; i.e., the Chiloe wigeon, mandarin duck, puna teal, Orinoco sheldgoose, and ashy-headed sheldgoose. Sequence comparisons revealed that 4 virus isolates were closely related to existing isolates of duck hepatitis B virus (DHBV), while the mandarin duck virus was closely related to Ross goose hepatitis B virus[22].
Table 2.
Avihepadnaviruses and their host
| Host | Ref. | |
| Duck Hepatitis B Virus | Pekin duck | [21] |
| DHBV | Anas domesticus | |
| Grey Teal Hepatitis B Virus | Grey Teal | [82] |
| (GTHBV) | Anas gibberifrons gracilis | |
| Heron Hepatitis B Virus | Heron | [83] |
| (HHBV) | Adrea cinerea | |
| Maned Duck Hepatitis B Virus | Maned Duck | [82] |
| (MDHBV) | Chenonetta jubata | |
| Ross Goose Hepatitis Virus | Ross Goose | [4] |
| (RGHV) | Anser rossi | |
| Snow Goose Hepatitis B Virus | Snow Goose | [84] |
| (SGHBV) | Anser caerulescens | |
| Stork Hepatitis B Virus | White Stork | [85] |
| (STHBV) | Ciconia ciconia | |
| Demoiselle cranes | [86] | |
| Crane Hepatitis B Virus | Anthropoides virgo | |
| (CHBV) | Grey crowned cranes | |
| Balearica regulorum |
Figure 1.
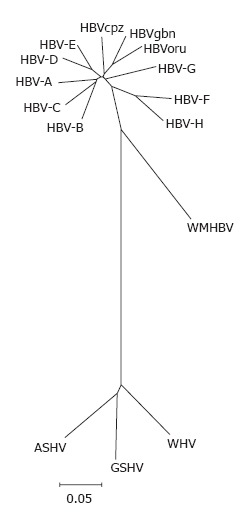
Phylogenetic tree of orthohe-padnaviruses. Complete genomes of HBV genotypes A (X02763), B (D00330), C (M12906), D (V01460), E (X75657), F (X69798), G (AF160501) and H (AY090454); HBVcpz (D00220), HBVoru (NC 002168), and HBVgbn (U46935) were aligned using clustal w with orthohepadnavirus genomes from woolly monkey (AF046996) woodchuck (J02442), ground squirrel (K02715) and the tentative member from arctic squirrel (nc_001719). The alignment was tested with the neighbour-joining method.
Figure 2.
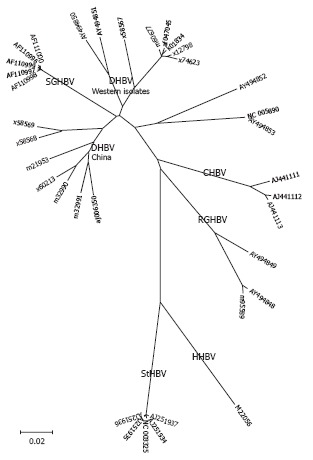
Phylogenetic tree of the genus avihepadnavirus.
In chimpanzees, gorillas, orangutans and gibbons new putative members of hepadnaviridae were discovered and sequenced completely[14]. It is now widely accepted that primate hepadnaviruses are indigenous to their hosts. Because hepadnaviruses isolated from apes are grouped as HBV genotypes in phylogenetic analyses, it has been suggested that isolates from apes should be named following the nomenclature used for immune deficiency viruses[23] (Table 1), e.g. HBV found in chimpanzees should be called HBVcpz. With only 5% divergence from the chimpanzee HBV isolates, the HBV isolate from gorilla is categorized in the HBV genotype (Figure 3, unpublished results). Thus, three HBV genotypes from apes can now be differentiated. The chimpanzee and gorilla isolates from Africa are categorized as one genotype, i.e., HBVcpz. The isolates from the South-East-Asian apes, gibbon and orang-utan, are categorized into two genotypes, i.e., HBVgbn and HBVoru, respectively. These genotypes diverge by 8%. Within the gibbon genotype, distinct strains of HBV circulating in geographically separated populations have been described[24].
Figure 3.
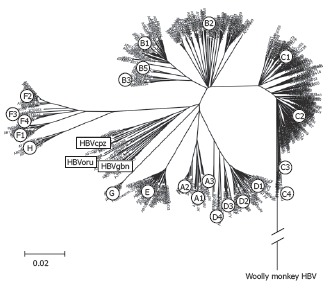
Phylogenetic tree of complete HBV genomes. An alignment of 601 complete HBV sequences was performed with Clustal X in the program DNAstar. The alignment was further analysed by boot-strapping using the Neighbourhood-Joining method contained in MEGA version 3.1[104].
Avihepdnaviruses are the most distant relatives of HBV with a nucleic acid homology of only 40%. WHV and GSHV as mammalian hepadnaviruses are more closely related to HBV and differ by only 17%. Complete WHV and GSHV genomes from GenBank show a high degree of homology and only one genotype is listed[25-27]. However, using degenerate primers, several variant WHV isolates from wild-captured woodchucks were found that showed high divergence with sequencing of small parts of the genome[28]. DHBV has two genotypes, in contrast to WHV and GSHV, which have a narrow host range and geographical distribution[25,26], DHBV is found in different avian species with independent isolates in many countries around the world[29] (Figure 4).
Figure 4.
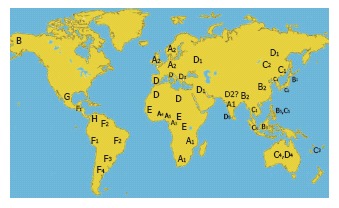
Geographic distribution of HBV genotypes and subgenotypes.
Human HBV can be grouped into eight genotypes (based on more than 8% difference)[7,9-12]. Several attempts have been made to reconstruct the evolution of hepadnaviruses[30-34]. Estimating the rate of synonymous substitutions for HBV to be 4.57 × 10-5 per site per year, DHBV has been proposed to have diverged about 30 000 years ago from a common ancestor while GSHV and WHV should have diverged about 10 000 years ago from HBV and the HBV serotypes would be separated by about 3000 years[31]. However, as long as we are not able to accurately estimate the mutation rate of HBV over centuries or even millennia, it is not possible to calculate a time point for the separation of HBV genotypes or hepadnaviral species.
HBV GENOTYPES AND SUBGENOTYPES
HBV genotypes differ by more than 8%[2,3]. Phylogenetic analyses using alignments of whole genomes have shown that 8 genotypes, called A, B, C, D, E, F, G and H, of HBV can be distinguished[7,11,12,35] (Figure 1). In general, HBV isolates found in apes diverge similarly to HBV genotypes in phylogenetic analyses and have been named HBVcpz, -oru, -gor and -gbn for their host’s, i.e. chimpanzee, orang-utan, gorilla and gibbon, respectively (Table 1)[23]. However, as elucidated above, the isolate from gorillas is always categorized into the chimpanzee clade.
A prototypic HBV genome may have a length of 3215 nt, as found in HBV genotypes B, C, F and H. Due to deletions and insertions (Table 3), the other HBV genotypes differ slightly in length of genome (Table 3). Thus HBV genotype G with 3248 nt. is 66 nt longer than genotype D with 3182 bp.
Table 3.
Fundamental properties of genomes and differences between HBV genotypes
| Genotype | Genome length in bp | ORF-differences |
| A | 3221 | Insertion of aa 153 and 154 in HBc |
| B | 3215 | |
| C | 3215 | |
| D | 3182 | Deletion of aa 1-11 in preS1 |
| E | 3212 | Deletion of aa 11 in preS1 |
| F | 3215 | |
| G | 3248 | Insertion of 12 aa in HBc |
| Deletion of aa 11 in preS1 | ||
| H | 3215 |
Extensive phylogenetic analyses have shown that HBV genotypes can be further subdivided into subgenotypes (Table 4). HBV subgenotypes differ by at least 4%[8]. In genotypes A, B and C, epidemiological data show that the respective subgenotype pairs A1/A2 (formerly termed Aa/Ae)[36], B1/B2 (formerly Bj/Ba)[36] and C1/C2 (formerly Cs/Ce)[37-39] differ substantially in many virological and probably some clinical parameters. Subgenotypes also show distinct geographic distribution (Figure 3). However, this is not true for genotype D with subgenotypes D1, D2 and D3 being described as widespread in the world; e.g. D3 was found in Asia (East India)[40], South Africa[41] and Europe (Serbia) (Stanojevic et al, unpublished results).
Table 4.
HBV subgenotypes and geographic prevalence
| Subgenotype | Synonyms | Geographic origin | Ref. | |
| A | A1 | Aa, A’ | Africa, (Asia, South America) | [41,87] |
| A2 | Ae, A-A' | Europe | ||
| A3 | Ac | Gabon, Cameroon | [88,89] | |
| (A4) | Mali | [59] | ||
| (A5) | Nigeria | [59] | ||
| B | B1 | Bj | Japan | [67,90] |
| B2 | Ba | Asia without Japan | ||
| B3 | Indonesia, Philippines | [7] | ||
| B4 | Vietnam | [7] | ||
| B5 | Philippines | [91,92] | ||
| C | C1 | Cs | South East Asia (Vietnam, Myanmar, Thailand, Southern China) | [37-39] |
| C2 | Ce | Far East (Korea, Japan, Northern China) | ||
| C3 | Micronesia | [7] | ||
| C4 | Australia | [93] | ||
| C5 | Philippines, Vietnam | [92,94] | ||
| D | D1 | Mongolia, Belarus, Europe? | ||
| D2 | India? | |||
| D3 | South Africa, East India, Serbia | [40,41] | ||
| D4 | Australia | [93] | ||
| D5 | East India | [40] | ||
| F | F1 | South and Central America | [95,96] | |
| F2 | South America | [4] | ||
| F3 | Bolivia | [97,98] | ||
| F4 | Argentina | [97,98] |
Except for genotype E and G, all HBV genotypes can be divided into subgenotypes. The absence of subgenotypes in HBV genotype E has been assumed to be the consequence of a recent genesis for genotype E[42-45]. Furthermore, genotype E is not present in Americans of African origin from Venezuela and Brazil[46,47]. The case for HBV genotype G appears to be less clear. Genotype G was originally found in the USA, France[5] and Germany[48]. Later, partial sequencing of HBV genes pointed to a high prevalence of HBV genotype G in Mexico[49]. Nevertheless, the geographic origin of HBV genotype G remains unknown[50]. To date only a limited number of complete HBV genotype G sequences have been deposited in GenBank that are not classified into subgenotypes.
DOUBLE INFECTIONS AND RECOMBI-NANTS
Double infections with two different HBV genotypes have been known since typing was done serologically[51,52]. Subsequently, evidence of super infection with HBV isolates of the same or different genotype was described in chronic HBV patients[53]. Super infection was accompanied by acute exacerbation of the chronic disease. Additional observations came from patients treated with interferon. Before treatment, HBV genotype A was prevalent. After treatment and relapse, a switch of the genotype to HBV genotype D was described[54,55].
Using different methods for genotyping, several reports described high rates of double infection with two different HBV genotypes in all parts of the world. Using these methods double infections have been found in 4.4%[56], 10.9%[57], 12.5%[58], 14.1% (Kirschberg et al, unpublished results), 17.3%[59] and 17.5%[60] of HBV infected patients. Even triple infections with HBV of genotype A, B and C have been described in 0.9% of HBV infected intravenous drug users[60].
Infection with HBV of genotype G seems to be associated very often with an infection of HBV genotype A[61]. This was found in 4 individuals from the USA and in one patient from France[62].
Coinfection with two different HBV genotypes in one patient may lead to an exchange of genetic material between the two strains. However, with current knowledge of HBV replication, the mechanism for this supposed recombination remains enigmatic. No mechanism can be envisioned that would allow an exchange of genetic material between two hepadnaviral genomes at the level of transcription. Nevertheless, numerous authors described changes in the genome of HBV that appear to be the consequences of a recombinatorial event (Figure 5 and Table 5).
Figure 5.
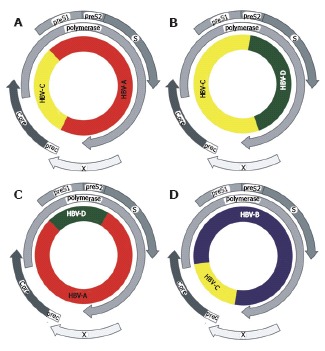
Schematic genome organisation of recombinants between HBV genotypes. HBV recombinants were described from materials sampled in A: Vietnam[99]; B: Tibet[68]; C: Africa[100] and D: Asia[67]. The ORF coding for the HBV proteins are shown as arrows, the inner circle represents the HBV genome.
Table 5.
Examples for recombination events between of HBV genotypes
|
Genotype of |
Recombination Breakpoint |
No. in literature | Ref. | ||
| Backbone | Insert | 5’ | 3’ | ||
| A | C | 1801 | 2865 | 3 | [99] |
| A | D | 2895 | 327 | ||
| 2820 | 386-586 | 3 | [100] | ||
| ? | 670 | ||||
| B | C | 1740-1838 | 2443- 2485 | 41 | [67,73,101,102] |
| B | C | 3120 | 3171 | 1 | [60] |
| 3060 | 3191 | 1 | |||
| 2910 | 2950 | 1 | |||
| C | B | 1731-1838 | 2437- 2479 | 1 | [102] |
| D | A | 129 | 2339 | 3 | [73,101,102] |
| 495 | 780 | ||||
| 822 | 1775 | ||||
| G | C | 1860 | 2460 | 1 | [103] |
| A | E | 882 | 1060 | 1 | [88] |
Two recent works have comprehensively analysed the prevalence of events in the HBV genome that are reminiscent of recombinations[63,64]. About 87% of the putative recombinants were B/C (120) and A/D (29) hybrids. The other recombinants comprised A/B/C, A/C, A/E, A/G, C/D, C/F, C/G, C/U (U for unknown genotype) and B/C/U hybrids. Genotypes A and C showed a higher recombination tendency than did other genotypes. The results also demonstrated region priority and breakpoint hot spots in the intergenotype recombination. Recombination breakpoints were found to be concentrated mainly in the vicinity of the DR1 region (nt 1640-1900), the preS1/S2 region (nt 3150-100), the 3’-end of the Core gene (nt 2330-2450) and the 3’-end of the Surface gene (nt 650-830)[63,64].
Recombination events between human and chim-panzee[65]or gibbon[63] HBV sequences have also been described. Discrepant genotyping results from different parts of the genome are indicative of a recombination between genotype A and F[66]. Even mosaic genomes with sequences derived from three different genotypes have been described[59,64].
Some recombinants among HBV genotypes have become the dominant subgenotype prevalent in certain geographic regions. Recombination between genotypes B and C has led to the generation of two different strains with distinct geographic distribution[67]. Strains of genotype B without recombination are found in Japan (subgenotype B1), whereas strains with recombination between genotype B and C are found throughout Asia (subgenotype B2), sparing Japan[67]. Recombinants between HBV genotypes C and D are the leading HBV subgenotype in Tibet[68-70].
It remains open for discussion whether the observed exchanges are the consequence of direct genetic recombination taking place between two HBV strains or if they are the consequence of fast adaptation of HBV to a certain genetic and immunologic environment in different human populations in the world. The high replication capacity of HBV with a release of up to 1013 viral particles per day[71,72] and the high error rate of the viral polymerase, lead to the production of HBV genomes with all possible single mutations and double mutations of every nucleotide of the HBV genome every day[72]. Thus, a fast adaptation of HBV to a new environment is also a possibility.
A hypothetical mosaicism of the HBV genome has already been proposed by Bowyer and Sim[73]. This work and later works described most HBV genotypes as a modular genome[63] that represents a mixture of small segments coming from many different HBV genotypes. If we expand on this observation, the HBV genome may be made up of a number of allelic modules with different properties; e.g. different binding sites for transcription factors or antigenic epitopes. Thus, a certain combination of these modules would make up an HBV genotype. The findings of Fischer et al are in support of this speculation. The authors described genotype specific activation or repression of HBV enhancer II, preCore-pregenomic promoter by the transcription factor COUP-TF1[74].
CONCLUSION
HBV has been recognised as a prototype member of a family of viruses infecting mammals and birds. Due to its high replication capacity and the high error rate of the viral reverse transcriptase, HBV is able to adapt to the host’s environment. This adaptation has led to the emergence of eight genotypes in humans and three closely related genotypes in apes. The human genotypes have further diverged into at least 24 subgenotypes, with certainly many more to come, and a plethora of recombinants. From the analysis of recombinants there are indications that at least one more genotype remains to be detected.
Footnotes
S- Editor Liu Y L- Editor Lutze M E- Editor Ma WH
References
- 1.Mason WS, Burrell CJ, Casey J, Gerlich WH, Howard CR, Kann M, Newbold J, Schaefer S, Taylor JM, Will H Hepadnaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (editors). Virus Taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Amsterdam: Elsevier; 2005. [Google Scholar]
- 2.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69 (Pt 10):2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 3.Norder H, Couroucé AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 4.Naumann H, Schaefer S, Yoshida CF, Gaspar AM, Repp R, Gerlich WH. Identification of a new hepatitis B virus (HBV) genotype from Brazil that expresses HBV surface antigen subtype adw4. J Gen Virol. 1993;74 (Pt 8):1627–1632. doi: 10.1099/0022-1317-74-8-1627. [DOI] [PubMed] [Google Scholar]
- 5.Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 6.Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059–2073. doi: 10.1099/0022-1317-83-8-2059. [DOI] [PubMed] [Google Scholar]
- 7.Norder H, Couroucé AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 8.Kramvis A, Kew MC. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy. J Viral Hepat. 2005;12:456–464. doi: 10.1111/j.1365-2893.2005.00624.x. [DOI] [PubMed] [Google Scholar]
- 9.Chu CJ, Lok AS. Clinical significance of hepatitis B virus genotypes. Hepatology. 2002;35:1274–1276. doi: 10.1053/jhep.2002.33161. [DOI] [PubMed] [Google Scholar]
- 10.Kramvis A, Kew M, François G. Hepatitis B virus genotypes. Vaccine. 2005;23:2409–2423. doi: 10.1016/j.vaccine.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 11.Miyakawa Y, Mizokami M. Classifying hepatitis B virus genotypes. Intervirology. 2003;46:329–338. doi: 10.1159/000074988. [DOI] [PubMed] [Google Scholar]
- 12.Schaefer S. Hepatitis B virus: significance of genotypes. J Viral Hepat. 2005;12:111–124. doi: 10.1111/j.1365-2893.2005.00584.x. [DOI] [PubMed] [Google Scholar]
- 13.Schaefer S, Tolle T, Lottmann S, Gerlich W. Animal Models and Experimental Systems in Hepatitis B Virus Research. in: Koshy R, Caselmann W (eds). Hepatitis B Virus: Molecular Mechanisms in Disease and Novel Strategies for Therapy. London: Imperial College Press; 1998. pp. 51–74. [Google Scholar]
- 14.Robertson BH, Margolis HS. Primate hepatitis B viruses - genetic diversity, geography and evolution. Rev Med Virol. 2002;12:133–141. doi: 10.1002/rmv.348. [DOI] [PubMed] [Google Scholar]
- 15.Burda MR, Günther S, Dandri M, Will H, Petersen J. Structural and functional heterogeneity of naturally occurring hepatitis B virus variants. Antiviral Res. 2001;52:125–138. doi: 10.1016/s0166-3542(01)00177-2. [DOI] [PubMed] [Google Scholar]
- 16.Abravaya K, Huff J, Marshall R, Merchant B, Mullen C, Schneider G, Robinson J. Molecular beacons as diagnostic tools: technology and applications. Clin Chem Lab Med. 2003;41:468–474. doi: 10.1515/CCLM.2003.070. [DOI] [PubMed] [Google Scholar]
- 17.Li MD, Bronson DL, Lemke TD, Faras AJ. Phylogenetic analyses of 55 retroelements on the basis of the nucleotide and product amino acid sequences of the pol gene. Mol Biol Evol. 1995;12:657–670. doi: 10.1093/oxfordjournals.molbev.a040231. [DOI] [PubMed] [Google Scholar]
- 18.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 19.Summers J, Smolec JM, Snyder R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci USA. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marion PL, Oshiro LS, Regnery DC, Scullard GH, Robinson WS. A virus in Beechey ground squirrels that is related to hepatitis B virus of humans. Proc Natl Acad Sci USA. 1980;77:2941–2945. doi: 10.1073/pnas.77.5.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mason WS, Seal G, Summers J. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol. 1980;36:829–836. doi: 10.1128/jvi.36.3.829-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo H, Mason WS, Aldrich CE, Saputelli JR, Miller DS, Jilbert AR, Newbold JE. Identification and characterization of avihepadnaviruses isolated from exotic anseriformes maintained in captivity. J Virol. 2005;79:2729–2742. doi: 10.1128/JVI.79.5.2729-2742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartholomeusz A, Schaefer S. Hepatitis B virus genotypes: comparison of genotyping methods. Rev Med Virol. 2004;14:3–16. doi: 10.1002/rmv.400. [DOI] [PubMed] [Google Scholar]
- 24.Sall AA, Starkman S, Reynes JM, Lay S, Nhim T, Hunt M, Marx N, Simmonds P. Frequent infection of Hylobates pileatus (pileated gibbon) with species-associated variants of hepatitis B virus in Cambodia. J Gen Virol. 2005;86:333–337. doi: 10.1099/vir.0.80274-0. [DOI] [PubMed] [Google Scholar]
- 25.Marion PL Ground Squirrel Hepatitis Virus. In: MacLachlan A (editor). Molecular Biology of the Hepatitis B Virus. Boca Raton: CRC Press; 1991. pp. 39–51. [Google Scholar]
- 26.Paronetto F, Tennant BC. Woodchuck hepatitis virus infection: a model of human hepatic diseases and hepatocellular carcinoma. Prog Liver Dis. 1990;9:463–483. [PubMed] [Google Scholar]
- 27.Roggendorf M, Tolle TK. The woodchuck: an animal model for hepatitis B virus infection in man. Intervirology. 1995;38:100–112. doi: 10.1159/000150418. [DOI] [PubMed] [Google Scholar]
- 28.Huang Z, Buckwold VE. A TaqMan PCR assay using degenerate primers for the quantitative detection of woodchuck hepatitis virus DNA of multiple genotypes. Mol Cell Probes. 2005;19:282–289. doi: 10.1016/j.mcp.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Schödel F, Weimer T, Fernholz D, Schneider R, Sprengel R, Wildner G, Will H The Biology of Avian Hepatitis B Viruses. In: MacLachlan A (editor). Molecular Biology of the Hepatitis B Virus. Boca Raton: CRC Press; 1991. pp. 53–80. [Google Scholar]
- 30.Bollyky PL, Holmes EC. Reconstructing the complex evolutionary history of hepatitis B virus. J Mol Evol. 1999;49:130–141. doi: 10.1007/pl00006526. [DOI] [PubMed] [Google Scholar]
- 31.Orito E, Mizokami M, Ina Y, Moriyama EN, Kameshima N, Yamamoto M, Gojobori T. Host-independent evolution and a genetic classification of the hepadnavirus family based on nucleotide sequences. Proc Natl Acad Sci USA. 1989;86:7059–7062. doi: 10.1073/pnas.86.18.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simmonds P. The origin and evolution of hepatitis viruses in humans. J Gen Virol. 2001;82:693–712. doi: 10.1099/0022-1317-82-4-693. [DOI] [PubMed] [Google Scholar]
- 33.Simmonds P. Reconstructing the origins of human hepatitis viruses. Philos Trans R Soc Lond B Biol Sci. 2001;356:1013–1026. doi: 10.1098/rstb.2001.0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fares MA, Holmes EC. A revised evolutionary history of hepatitis B virus (HBV) J Mol Evol. 2002;54:807–814. doi: 10.1007/s00239-001-0084-z. [DOI] [PubMed] [Google Scholar]
- 35.Chu CJ, Hussain M, Lok AS. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology. 2002;122:1756–1762. doi: 10.1053/gast.2002.33588. [DOI] [PubMed] [Google Scholar]
- 36.Sugauchi F, Kumada H, Acharya SA, Shrestha SM, Gamutan MT, Khan M, Gish RG, Tanaka Y, Kato T, Orito E, et al. Epidemiological and sequence differences between two subtypes (Ae and Aa) of hepatitis B virus genotype A. J Gen Virol. 2004;85:811–820. doi: 10.1099/vir.0.79811-0. [DOI] [PubMed] [Google Scholar]
- 37.Chan HL, Tsui SK, Tse CH, Ng EY, Au TC, Yuen L, Bartholomeusz A, Leung KS, Lee KH, Locarnini S, et al. Epidemiological and virological characteristics of 2 subgroups of hepatitis B virus genotype C. J Infect Dis. 2005;191:2022–2032. doi: 10.1086/430324. [DOI] [PubMed] [Google Scholar]
- 38.Huy TT, Ushijima H, Quang VX, Win KM, Luengrojanakul P, Kikuchi K, Sata T, Abe K. Genotype C of hepatitis B virus can be classified into at least two subgroups. J Gen Virol. 2004;85:283–292. doi: 10.1099/vir.0.19633-0. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka Y, Orito E, Yuen MF, Mukaide M, Sugauchi F, Ito K, Ozasa A, Sakamoto T, Kurbanov F, Lai CL, et al. Two subtypes (subgenotypes) of hepatitis B virus genotype C: A novel subtyping assay based on restriction fragment length polymorphism. Hepatol Res. 2005;33:216–224. doi: 10.1016/j.hepres.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 40.Banerjee A, Kurbanov F, Datta S, Chandra PK, Tanaka Y, Mizokami M, Chakravarty R. Phylogenetic relatedness and genetic diversity of hepatitis B virus isolates in Eastern India. J Med Virol. 2006;78:1164–1174. doi: 10.1002/jmv.20677. [DOI] [PubMed] [Google Scholar]
- 41.Kimbi GC, Kramvis A, Kew MC. Distinctive sequence characteristics of subgenotype A1 isolates of hepatitis B virus from South Africa. J Gen Virol. 2004;85:1211–1220. doi: 10.1099/vir.0.19749-0. [DOI] [PubMed] [Google Scholar]
- 42.Fujiwara K, Tanaka Y, Orito E, Ohno T, Kato T, Sugihara K, Hasegawa I, Sakurai M, Ito K, Ozasa A, et al. Distribution of HBV genotypes among HBV carriers in Benin: phylogenetic analysis and virological characteristics of HBV genotype E. World J Gastroenterol. 2005;11:6410–6415. doi: 10.3748/wjg.v11.i41.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huy TT, Ishikawa K, Ampofo W, Izumi T, Nakajima A, Ansah J, Tetteh JO, Nii-Trebi N, Aidoo S, Ofori-Adjei D, et al. Characteristics of hepatitis B virus in Ghana: full length genome sequences indicate the endemicity of genotype E in West Africa. J Med Virol. 2006;78:178–184. doi: 10.1002/jmv.20525. [DOI] [PubMed] [Google Scholar]
- 44.Kramvis A, Restorp K, Norder H, Botha JF, Magnius LO, Kew MC. Full genome analysis of hepatitis B virus genotype E strains from South-Western Africa and Madagascar reveals low genetic variability. J Med Virol. 2005;77:47–52. doi: 10.1002/jmv.20412. [DOI] [PubMed] [Google Scholar]
- 45.Mulders MN, Venard V, Njayou M, Edorh AP, Bola Oyefolu AO, Kehinde MO, Muyembe Tamfum JJ, Nebie YK, Maiga I, Ammerlaan W, et al. Low genetic diversity despite hyperendemicity of hepatitis B virus genotype E throughout West Africa. J Infect Dis. 2004;190:400–408. doi: 10.1086/421502. [DOI] [PubMed] [Google Scholar]
- 46.Motta-Castro AR, Martins RM, Yoshida CF, Teles SA, Paniago AM, Lima KM, Gomes SA. Hepatitis B virus infection in isolated Afro-Brazilian communities. J Med Virol. 2005;77:188–193. doi: 10.1002/jmv.20435. [DOI] [PubMed] [Google Scholar]
- 47.Quintero A, Martínez D, Alarcón De Noya B, Costagliola A, Urbina L, González N, Liprandi F, Castro De Guerra D, Pujol FH. Molecular epidemiology of hepatitis B virus in Afro-Venezuelan populations. Arch Virol. 2002;147:1829–1836. doi: 10.1007/s00705-002-0842-2. [DOI] [PubMed] [Google Scholar]
- 48.Vieth S, Manegold C, Drosten C, Nippraschk T, Günther S. Sequence and phylogenetic analysis of hepatitis B virus genotype G isolated in Germany. Virus Genes. 2002;24:153–156. doi: 10.1023/a:1014572600432. [DOI] [PubMed] [Google Scholar]
- 49.Sánchez LV, Maldonado M, Bastidas-Ramírez BE, Norder H, Panduro A. Genotypes and S-gene variability of Mexican hepatitis B virus strains. J Med Virol. 2002;68:24–32. doi: 10.1002/jmv.10166. [DOI] [PubMed] [Google Scholar]
- 50.Lindh M. HBV genotype G-an odd genotype of unknown origin. J Clin Virol. 2005;34:315–316. doi: 10.1016/j.jcv.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Hess G, Arnold W, Koesters W, Biswas R, Hütteroth TH, zum Büschenfelde KH. Simultaneous presence of HBsAg and anti-HBs in the serum of different subtypes (serological and immunofluorescent studies) Z Immunitatsforsch Immunobiol. 1977;153:143–151. [PubMed] [Google Scholar]
- 52.Tabor E, Gerety RJ, Smallwood LA, Barker LF. Coincident hepatitis B surface antigen and antibodies of different subtypes in human serum. J Immunol. 1977;118:369–370. [PubMed] [Google Scholar]
- 53.Kao JH, Chen PJ, Lai MY, Chen DS. Acute exacerbations of chronic hepatitis B are rarely associated with superinfection of hepatitis B virus. Hepatology. 2001;34:817–823. doi: 10.1053/jhep.2001.28188. [DOI] [PubMed] [Google Scholar]
- 54.Gerner PR, Friedt M, Oettinger R, Lausch E, Wirth S. The hepatitis B virus seroconversion to anti-HBe is frequently associated with HBV genotype changes and selection of preS2-defective particles in chronically infected children. Virology. 1998;245:163–172. doi: 10.1006/viro.1998.9126. [DOI] [PubMed] [Google Scholar]
- 55.Hannoun C, Krogsgaard K, Horal P, Lindh M. Genotype mixtures of hepatitis B virus in patients treated with interferon. J Infect Dis. 2002;186:752–759. doi: 10.1086/342599. [DOI] [PubMed] [Google Scholar]
- 56.Ding X, Gu H, Zhong ZH, Zilong X, Tran HT, Iwaki Y, Li TC, Sata T, Abe K. Molecular epidemiology of hepatitis viruses and genotypic distribution of hepatitis B and C viruses in Harbin, China. Jpn J Infect Dis. 2003;56:19–22. [PubMed] [Google Scholar]
- 57.Kato H, Orito E, Sugauchi F, Ueda R, Koshizaka T, Yanaka S, Gish RG, Kurbanov F, Ruzibakiev R, Kramvis A, et al. Frequent coinfection with hepatitis B virus strains of distinct genotypes detected by hybridization with type-specific probes immobilized on a solid-phase support. J Virol Methods. 2003;110:29–35. doi: 10.1016/s0166-0934(03)00095-8. [DOI] [PubMed] [Google Scholar]
- 58.Osiowy C, Giles E. Evaluation of the INNO-LiPA HBV genotyping assay for determination of hepatitis B virus genotype. J Clin Microbiol. 2003;41:5473–5477. doi: 10.1128/JCM.41.12.5473-5477.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olinger CM, Venard V, Njayou M, Oyefolu AO, Maïga I, Kemp AJ, Omilabu SA, le Faou A, Muller CP. Phylogenetic analysis of the precore/core gene of hepatitis B virus genotypes E and A in West Africa: new subtypes, mixed infections and recombinations. J Gen Virol. 2006;87:1163–1173. doi: 10.1099/vir.0.81614-0. [DOI] [PubMed] [Google Scholar]
- 60.Chen BF, Kao JH, Liu CJ, Chen DS, Chen PJ. Genotypic dominance and novel recombinations in HBV genotype B and C co-infected intravenous drug users. J Med Virol. 2004;73:13–22. doi: 10.1002/jmv.20051. [DOI] [PubMed] [Google Scholar]
- 61.Kato H, Orito E, Gish RG, Bzowej N, Newsom M, Sugauchi F, Suzuki S, Ueda R, Miyakawa Y, Mizokami M. Hepatitis B e antigen in sera from individuals infected with hepatitis B virus of genotype G. Hepatology. 2002;35:922–929. doi: 10.1053/jhep.2002.32096. [DOI] [PubMed] [Google Scholar]
- 62.Kremsdorf D, Garreau F, Capel F, Petit MA, Brechot C. In vivo selection of a hepatitis B virus mutant with abnormal viral protein expression. J Gen Virol. 1996;77 (Pt 5):929–939. doi: 10.1099/0022-1317-77-5-929. [DOI] [PubMed] [Google Scholar]
- 63.Simmonds P, Midgley S. Recombination in the genesis and evolution of hepatitis B virus genotypes. J Virol. 2005;79:15467–15476. doi: 10.1128/JVI.79.24.15467-15476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J, Xing K, Deng R, Wang J, Wang X. Identification of Hepatitis B virus putative intergenotype recombinants by using fragment typing. J Gen Virol. 2006;87:2203–2215. doi: 10.1099/vir.0.81752-0. [DOI] [PubMed] [Google Scholar]
- 65.Magiorkinis EN, Magiorkinis GN, Paraskevis DN, Hatzakis AE. Re-analysis of a human hepatitis B virus (HBV) isolate from an East African wild born Pan troglodytes schweinfurthii: evidence for interspecies recombination between HBV infecting chimpanzee and human. Gene. 2005;349:165–171. doi: 10.1016/j.gene.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 66.Gutiérrez C, Devesa M, Loureiro CL, León G, Liprandi F, Pujol FH. Molecular and serological evaluation of surface antigen negative hepatitis B virus infection in blood donors from Venezuela. J Med Virol. 2004;73:200–207. doi: 10.1002/jmv.20076. [DOI] [PubMed] [Google Scholar]
- 67.Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, Ishida T, Chutaputti A, Lai CL, Ueda R, et al. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J Virol. 2002;76:5985–5992. doi: 10.1128/JVI.76.12.5985-5992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cui C, Shi J, Hui L, Xi H, Zhuoma G. The dominant hepatitis B virus genotype identified in Tibet is a C/D hybrid. J Gen Virol. 2002;83:2773–2777. doi: 10.1099/0022-1317-83-11-2773. [DOI] [PubMed] [Google Scholar]
- 69.Wang Z, Liu Z, Zeng G, Wen S, Qi Y, Ma S, Naoumov NV, Hou J. A new intertype recombinant between genotypes C and D of hepatitis B virus identified in China. J Gen Virol. 2005;86:985–990. doi: 10.1099/vir.0.80771-0. [DOI] [PubMed] [Google Scholar]
- 70.Zeng G, Wang Z, Wen S, Jiang J, Wang L, Cheng J, Tan D, Xiao F, Ma S, Li W, et al. Geographic distribution, virologic and clinical characteristics of hepatitis B virus genotypes in China. J Viral Hepat. 2005;12:609–617. doi: 10.1111/j.1365-2893.2005.00657.x. [DOI] [PubMed] [Google Scholar]
- 71.Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, McDade H. Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci USA. 1996;93:4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brechtbuehl K, Whalley SA, Dusheiko GM, Saunders NA. A rapid real-time quantitative polymerase chain reaction for hepatitis B virus. J Virol Methods. 2001;93:105–113. doi: 10.1016/s0166-0934(01)00260-9. [DOI] [PubMed] [Google Scholar]
- 73.Bowyer SM, Sim JG. Relationships within and between genotypes of hepatitis B virus at points across the genome: footprints of recombination in certain isolates. J Gen Virol. 2000;81:379–392. doi: 10.1099/0022-1317-81-2-379. [DOI] [PubMed] [Google Scholar]
- 74.Fischer SF, Schmidt K, Fiedler N, Glebe D, Schüttler C, Sun J, Gerlich WH, Repp R, Schaefer S. Genotype-dependent activation or repression of HBV enhancer II by transcription factor COUP-TF1. World J Gastroenterol. 2006;12:6054–6058. doi: 10.3748/wjg.v12.i37.6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dane DS, Cameron CH, Briggs M. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet. 1970;1:695–698. doi: 10.1016/s0140-6736(70)90926-8. [DOI] [PubMed] [Google Scholar]
- 76.Vaudin M, Wolstenholme AJ, Tsiquaye KN, Zuckerman AJ, Harrison TJ. The complete nucleotide sequence of the genome of a hepatitis B virus isolated from a naturally infected chimpanzee. J Gen Virol. 1988;69 (Pt 6):1383–1389. doi: 10.1099/0022-1317-69-6-1383. [DOI] [PubMed] [Google Scholar]
- 77.Norder H, Ebert JW, Fields HA, Mushahwar IK, Magnius LO. Complete sequencing of a gibbon hepatitis B virus genome reveals a unique genotype distantly related to the chimpanzee hepatitis B virus. Virology. 1996;218:214–223. doi: 10.1006/viro.1996.0181. [DOI] [PubMed] [Google Scholar]
- 78.Warren KS, Heeney JL, Swan RA, Heriyanto EJ. A new group of hepadnaviruses naturally infecting orangutans (Pongo pygmaeus) J Virol. 1999;73:7860–7865. doi: 10.1128/jvi.73.9.7860-7865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grethe S, Heckel JO, Rietschel W, Hufert FT. Molecular epidemiology of hepatitis B virus variants in nonhuman primates. J Virol. 2000;74:5377–5381. doi: 10.1128/jvi.74.11.5377-5381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lanford RE, Chavez D, Brasky KM, Burns RB, Rico-Hesse R. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc Natl Acad Sci USA. 1998;95:5757–5761. doi: 10.1073/pnas.95.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Testut P, Renard CA, Terradillos O, Vitvitski-Trepo L, Tekaia F, Degott C, Blake J, Boyer B, Buendia MA. A new hepadnavirus endemic in arctic ground squirrels in Alaska. J Virol. 1996;70:4210–4219. doi: 10.1128/jvi.70.7.4210-4219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li L, Dixon RJ, Gu X, Newbold JE: Comparison of the sequences of the Grey Teal, Maned Duck and Duck Hepatitis B Viruses. In The molecular biology of Hepatitis B Virus. The United States: University of California San Diego; 1998. p. 13. [Google Scholar]
- 83.Sprengel R, Kaleta EF, Will H. Isolation and characterization of a hepatitis B virus endemic in herons. J Virol. 1988;62:3832–3839. doi: 10.1128/jvi.62.10.3832-3839.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chang SF, Netter HJ, Bruns M, Schneider R, Frölich K, Will H. A new avian hepadnavirus infecting snow geese (Anser caerulescens) produces a significant fraction of virions containing single-stranded DNA. Virology. 1999;262:39–54. doi: 10.1006/viro.1999.9844. [DOI] [PubMed] [Google Scholar]
- 85.Pult I, Netter HJ, Fröhlich K, Kaleta EF, Will H: Identification, structural and functional analysis of a new avian Hepadnavirus from storks (STHBV) In The molecular biology of Hepatitis B Virus. The United States: University of California San Diego; 1998. p. 2. [Google Scholar]
- 86.Prassolov A, Hohenberg H, Kalinina T, Schneider C, Cova L, Krone O, Frölich K, Will H, Sirma H. New hepatitis B virus of cranes that has an unexpected broad host range. J Virol. 2003;77:1964–1976. doi: 10.1128/JVI.77.3.1964-1976.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kramvis A, Weitzmann L, Owiredu WK, Kew MC. Analysis of the complete genome of subgroup A' hepatitis B virus isolates from South Africa. J Gen Virol. 2002;83:835–839. doi: 10.1099/0022-1317-83-4-835. [DOI] [PubMed] [Google Scholar]
- 88.Kurbanov F, Tanaka Y, Fujiwara K, Sugauchi F, Mbanya D, Zekeng L, Ndembi N, Ngansop C, Kaptue L, Miura T, et al. A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in Cameroon. J Gen Virol. 2005;86:2047–2056. doi: 10.1099/vir.0.80922-0. [DOI] [PubMed] [Google Scholar]
- 89.Makuwa M, Souquière S, Telfer P, Apetrei C, Vray M, Bedjabaga I, Mouinga-Ondeme A, Onanga R, Marx PA, Kazanji M, et al. Identification of hepatitis B virus subgenotype A3 in rural Gabon. J Med Virol. 2006;78:1175–1184. doi: 10.1002/jmv.20678. [DOI] [PubMed] [Google Scholar]
- 90.Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, Ishida T, Chutaputti A, Lai CL, Gish RG, et al. Epidemiologic and virologic characteristics of hepatitis B virus genotype B having the recombination with genotype C. Gastroenterology. 2003;124:925–932. doi: 10.1053/gast.2003.50140. [DOI] [PubMed] [Google Scholar]
- 91.Nagasaki F, Niitsuma H, Cervantes JG, Chiba M, Hong S, Ojima T, Ueno Y, Bondoc E, Kobayashi K, Ishii M, et al. Analysis of the entire nucleotide sequence of hepatitis B virus genotype B in the Philippines reveals a new subgenotype of genotype B. J Gen Virol. 2006;87:1175–1180. doi: 10.1099/vir.0.81525-0. [DOI] [PubMed] [Google Scholar]
- 92.Sakamoto T, Tanaka Y, Orito E, Co J, Clavio J, Sugauchi F, Ito K, Ozasa A, Quino A, Ueda R, et al. Novel subtypes (subgenotypes) of hepatitis B virus genotypes B and C among chronic liver disease patients in the Philippines. J Gen Virol. 2006;87:1873–1882. doi: 10.1099/vir.0.81714-0. [DOI] [PubMed] [Google Scholar]
- 93.Sugauchi F, Mizokami M, Orito E, Ohno T, Kato H, Suzuki S, Kimura Y, Ueda R, Butterworth LA, Cooksley WG. A novel variant genotype C of hepatitis B virus identified in isolates from Australian Aborigines: complete genome sequence and phylogenetic relatedness. J Gen Virol. 2001;82:883–892. doi: 10.1099/0022-1317-82-4-883. [DOI] [PubMed] [Google Scholar]
- 94.Cavinta L, Sun J, Zarnekow M, Barzaga N, Schaefer S. New hepatitis B virus subgenotype C5 from the Philippines. submitted [Google Scholar]
- 95.Kato H, Fujiwara K, Gish RG, Sakugawa H, Yoshizawa H, Sugauchi F, Orito E, Ueda R, Tanaka Y, Kato T, et al. Classifying genotype F of hepatitis B virus into F1 and F2 subtypes. World J Gastroenterol. 2005;11:6295–6304. doi: 10.3748/wjg.v11.i40.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Norder H, Arauz-Ruiz P, Blitz L, Pujol FH, Echevarria JM, Magnius LO. The T(1858) variant predisposing to the precore stop mutation correlates with one of two major genotype F hepatitis B virus clades. J Gen Virol. 2003;84:2083–2087. doi: 10.1099/vir.0.19034-0. [DOI] [PubMed] [Google Scholar]
- 97.Devesa M, Rodríguez C, León G, Liprandi F, Pujol FH. Clade analysis and surface antigen polymorphism of hepatitis B virus American genotypes. J Med Virol. 2004;72:377–384. doi: 10.1002/jmv.20015. [DOI] [PubMed] [Google Scholar]
- 98.Huy TT, Ushijima H, Sata T, Abe K. Genomic characterization of HBV genotype F in Bolivia: genotype F subgenotypes correlate with geographic distribution and T(1858) variant. Arch Virol. 2006;151:589–597. doi: 10.1007/s00705-005-0671-1. [DOI] [PubMed] [Google Scholar]
- 99.Hannoun C, Norder H, Lindh M. An aberrant genotype revealed in recombinant hepatitis B virus strains from Vietnam. J Gen Virol. 2000;81:2267–2272. doi: 10.1099/0022-1317-81-9-2267. [DOI] [PubMed] [Google Scholar]
- 100.Owiredu WK, Kramvis A, Kew MC. Hepatitis B virus DNA in serum of healthy black African adults positive for hepatitis B surface antibody alone: possible association with recombination between genotypes A and D. J Med Virol. 2001;64:441–454. doi: 10.1002/jmv.1070. [DOI] [PubMed] [Google Scholar]
- 101.Bollyky PL, Rambaut A, Harvey PH, Holmes EC. Recombination between sequences of hepatitis B virus from different genotypes. J Mol Evol. 1996;42:97–102. doi: 10.1007/BF02198834. [DOI] [PubMed] [Google Scholar]
- 102.Morozov V, Pisareva M, Groudinin M. Homologous recombination between different genotypes of hepatitis B virus. Gene. 2000;260:55–65. doi: 10.1016/s0378-1119(00)00424-8. [DOI] [PubMed] [Google Scholar]
- 103.Suwannakarn K, Tangkijvanich P, Theamboonlers A, Abe K, Poovorawan Y. A novel recombinant of Hepatitis B virus genotypes G and C isolated from a Thai patient with hepatocellular carcinoma. J Gen Virol. 2005;86:3027–3030. doi: 10.1099/vir.0.81241-0. [DOI] [PubMed] [Google Scholar]
- 104.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]


