Abstract
The hepatitis B virus (HBV) particle consists of an envelope containing three related surface proteins and probably lipid and an icosahedral nucleocapsid of approximately 30 nm diameter enclosing the viral DNA genome and DNA polymerase. The capsid is formed in the cytosol of the infected cell during packaging of an RNA pregenome replication complex by multiple copies of a 21-kDa C protein. The capsid gains the ability to bud during synthesis of the viral DNA genome by reverse transcription of the pregenome in the lumen of the particle. The three envelope proteins S, M, and L shape a complex transmembrane fold at the endoplasmic reticulum, and form disulfide-linked homo- and heterodimers. The transmembrane topology of a fraction of the large envelope protein L changes post-translationally, therefore, the N terminal domain of L (preS) finally appears on both sides of the membrane. During budding at an intracellular membrane, a short linear domain in the cytosolic preS region interacts with binding sites on the capsid surface. The virions are subsequently secreted into the blood. In addition, the surface proteins can bud in the absence of capsids and form subviral lipoprotein particles of 20 nm diameter which are also secreted.
Keywords: Hepatitis B virus morphogenesis, HBsAg, Hepatitis B virus capsid, Virus envelopment
INTRODUCTION
In vitro systems for efficient production of hepatitis B virus (HBV) capsids and subviral particles and for experimental examination of their morphogenesis are available. These systems allowed to draw a quite detailed, although still fragmentary, picture of these processes. However, in vitro production of virions by transfection of certain cell lines derived from hepatocellular carcinomas, such as HepG2 or Huh7, with cloned genomic HBV DNA[1,2] or by in vitro infection[3] is still quite inefficient, and this hampers many approaches to study the morphogenesis of the complete virus. In natural HBV infections one single hepatocyte in the liver releases 1 to 10 viruses per day[4]. In vitro the production rate seems to be similar. Therefore, the direct observation of HBV budding by electron microscopy or the characterization of the process by biochemical approaches is difficult to achieve[5]. It seems that the virus production rate in duck hepatitis B virus (DHBV) infection is higher at least in vitro, making these techniques suitable for studying DHBV morphogenesis[6].
THE HBV CAPSID FORMATION AND STRUCTURE
The C protein forming the shell of the HBV capsid consists of 183 or 185 amino acid (aa) residues depending on the genotype. The protein is relatively conserved among HBV isolates[7]. It can be expressed in a broad range of pro- and eukaryotic cell types and self-assembles into capsids. The first step is the formation of homodimers[8] linked by a disulfide bridge between the cystein residue 61[9,10]. Higher oligomers containing chaperons have been described[11] but the pathway leading from dimers to complete capsids has not been elucidated in more detail. In the final capsid, the inter-dimer interactions are rather weak[12]. Two different types of capsids are formed[13]: particles with an icosahedral T = 3 symmetry have a diameter of 30 nm and consist of 90 C dimers, whereas particles with an icosahedral T = 4 symmetry are larger (the diameter is 34 nm) and contain 120 C dimers. Both particle species can also be found in infected human liver[14]. In infectious virions, T = 4 capsids have been found[15].
The primary amino acid (aa) sequence of the C protein can be divided into two parts (Figure 1): the N-terminal 149 or 151 aa (depending on the genotype) form the so called assembly domain because this part of the protein is sufficient to direct the self-assembly of capsids. The C-terminal 34 aa are dispensable for capsid formation, rich in arginine residues, and involved in packaging of the pregenome/reverse transcriptase complex. Deletion of this domain abolishes the encapsidation of nucleic acid[16]. Expression of the C-terminally truncated C protein in E coli produces high amounts of T = 4 capsids and relatively little T = 3 particles[17]. Using this material a model for the folding of the C protein in the capsid first at lower resolution by cryo-electron microscopy[18,19] and finally after crystallization at a resolution of 3.3 Å[20,21] has been proposed. The C protein dimer forms a structure like an upside down “T”. The horizontal bar mediates the inter-dimer contacts with 5 and 6 dimers arranged around the 5-fold and quasi 6-fold symmetry axes, respectively, and the vertical bar forms a spike protruding outwards from the capsid surface (Figure 2). The tip of the spike forms the major epitope of the capsid antigen (HBcAg). The capsid shell contains pores with a diameter between 12 Å and 15 Å. These pores allow the diffusion of nucleotides into and out of the capsid lumen during the synthesis of the viral DNA genome.
Figure 1.
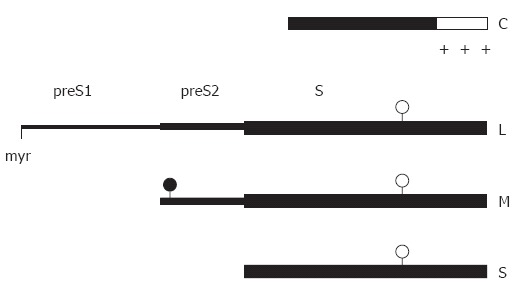
Linear map of the main structural HBV proteins. The C-terminal region (open box) of the capsid protein C is rich in arginine residues (+). The sequence of the small envelope S is also present at the C termini of the M and L protein. The two larger envelope proteins contain the additional N-terminal preS2 and preS2 + preS1 domain, respectively. The L protein is myristylated at glycin 2 (myr), the preS2 domain of M is N-glycosylated (filled circles), and the S domain of all 3 proteins is partially N-glycosylated (open circles).
Figure 2.
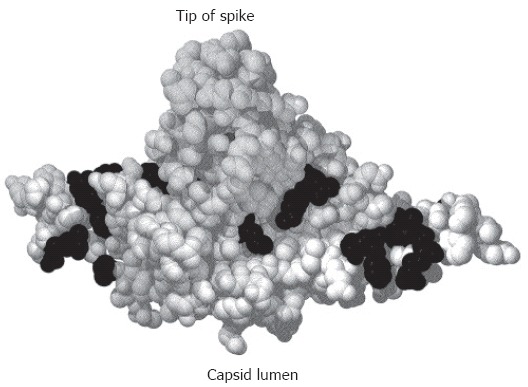
Crystal structure of a C-terminally truncated C protein dimer[21]. The spike protrudes upwards. The lumen of the capsid would be below the figure. Mutational analysis identified aa (shown in black) where the mutation was compatible with capsid formation and viral DNA synthesis in the lumen of the particle but blocked nucleocapsid envelopment[121].
The arginine-rich domain is not present in the capsid crystals but thought to interact with the viral genome in the lumen of the particle[16,22,23]. However, a monoclonal antibody directed against this region binds to intact HBV capsids[24], and trypsin can clip off this domain from approximately half of the C protein chains in recombinant HBV capsids (Daniela Lieder, PhD thesis, Goettingen, 2002). It therefore seems possible that the C-terminal region of one fraction of C proteins reach into the lumen of the particle, while the domains from the other fraction appear on the external surface of the same particle. The peptide at the boundary between the assembly and arginine-rich domains of C forms a mobile array[25] and may allow an extreme mobility of the C terminal domain.
Capsid formation during recombinant expression of the C protein requires a higher concentration of C protein dimers relative to nucleocapsid formation in the context of an infection[26]. During authentic capsid formation, not only the viral pregenomic RNA bound to the viral P protein[27-30] but also cellular factors such as chaperones[31-33] and a protein kinase phosphorylating serine residues in the arginine-rich domain of C[34-37] are encapsidated. Apparently, the threshold concentration of C dimers needed for the initiation of capsid formation is lowered by one or more of these factors. This mechanism assures the efficient encapsidation of replication complexes and prevents that large amounts of empty capsids are formed in the presence of free replication complexes.
C proteins from human and woodchuck HBV can form mixed capsids, while this is not possible between human and duck C proteins[38] which are less homologous. Foreign protein domains can be incorporated into capsids when fused to the N or C terminus or at the tip of the spike[39-44]. The assembly of HBV capsids can be blocked by low molecular weight compounds, possibly offering new options for antiviral treatments in the future[45-48].
THE HBV ENVELOPE PROTEINS
The HBV envelope contains three related viral surface proteins. They are expressed from one open reading frame (ORF) referred to as E containing 389 or 400 codons depending on the genotype and three start sites for translation[49] (Figure 1). Transcription is initiated at a promoter upstream of the ORF and, in addition, at an internal promoter upstream of the second translation initiation site[50]. Translation of the larger mRNA yields the large envelope protein (L) consisting of 389 or 400 aa. Translation of the shorter transcripts gives rise to the middle sized, 281 aa long M protein and, in addition, to the small S protein consisting of 226 aa[51] depending on which translation initiation site is used. The aa sequence present at the C termini of L and M is identical to the S protein and is referred to as the S domain. The 55 aa long additional N-terminal domain of M being central in L is called preS2, and the 108 or 119 aa long N-terminal domain unique to L is named preS1. The E ORF of avian hepadnaviruses contains only 2 start codons, therefore, these viruses possess only two envelope proteins (L and S).
Like typical membrane proteins, the HBV envelope proteins are synthesized at the endoplasmic reticulum (ER). They gain a relatively complex topology (Figure 3).Insertion of the S protein into the ER membrane is initiated by an N-terminal signal sequence (aa 8 to 22) which is, however, not cleaved by the host’s signal peptidase. A second signal (aa 80 to 98) directs the translocation of the peptide chain downstream of this signal through the ER membrane into the ER lumen[52], whereas the region upstream of the signal remains in the cytosol. The signal itself anchors the protein as a transmembrane domain in the lipid bilayer. The C-terminal hydrophobic 57 aa of S are believed to be embedded in the ER membrane. Foreign domains fused to the C terminus of S are oriented towards the ER lumen, suggesting that the C terminus of S is also oriented toward this compartment[52]. This configuration causes the region between residues 23 and 79 to form a loop at the cytosolic side of the ER membrane, whereas the loop between aa 99 and approximately 169 is on the luminal side. The luminal loop carries the major conformational epitope of the HBV surface protein antigen (HBsAg) and is N-glycosylated in approximately half of the S molecules at asparagine (asn) residue 146[53]. After budding the HBsAg epitopes are located at the external surface of viral particles.
Figure 3.
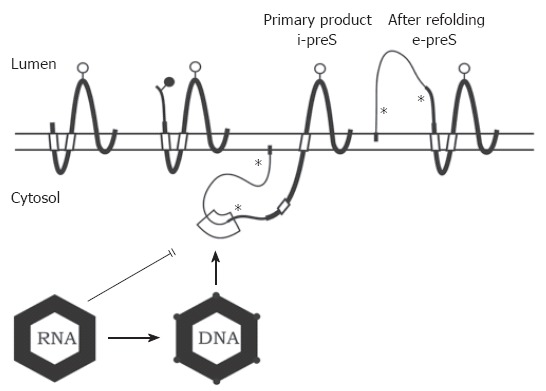
Transmembrane topology of the HBV envelope proteins and model for envelope-capsid interaction. The transmembrane folding of the S protein is determined by an N-terminal and an internal signal shown as open boxes. The C-terminal domain is hydrophobic and probably embedded in the lipid bilayer (horizontal open bar). The C terminus is oriented towards the ER lumen. The folding of the M protein is similar to S. The preS2 domain of M (thinner line) is located in the ER lumen. In the initial folding of the L protein, the preS domains are located in the cytosol (i-preS). Whether the N-terminal myristate group (filled box) is inserted into the membrane as shown here is unknown. After refolding approximately half of the L chains expose the preS domains at the luminal side of the membrane (e-preS). Open and filled circles: see Figure 1. Asterisks indicate potential but unused N-glycosylation sites in preS of L. A domain in i-preS (boxed area) and in the cytosolic loop of S may interact with the capsid during budding. Immature capsids containing pregenomic RNA are not capable to bud. During viral DNA synthesis, the capsid shell changes (indicated by filled circles at the edges) and becomes competent for envelopment.
The transmembrane topology of the M protein is identical to S. The N-terminal preS2 domain (55 aa) is translocated into the ER lumen probably by the action of the first signal in the S domain[54]. The M protein is N-glycosylated at asn 4[55]. In addition, the preS2 domain is O-glycosylated in some but not all HBV genotypes[56].
Glycine residue 2 of the L protein is myristylated[57]. The preS1 and preS2 domains at the N terminus of L initially remain at the cytosolic side of the ER membrane during L translation (i-preS conformation). The central signal in the S domain of L anchors the protein in the ER membrane and causes the translocation of downstream sequences. Therefore, asn 146 in the S domain of L is partially N-glycosylated, while asn 4 of the preS2 as well as a further potential N-glycosylation site in preS1 remain unmodified (Figure 3). These sites are used when the N terminus of L is forced to cotranslationally translocate by the artificial fusion of a signal sequence to the N terminus of preS1[58].
In about half of the L chains, the transmembrane topology changes after translation[59-63]. The preS domains then appear on the luminal side of the ER membrane (e-preS conformation). Probably the N-terminal signal in the S domain crosses the membrane in this conformation similar to the M and S proteins. How the preS domains move post-translationally through the membrane is not known. Cytosolic chaperones like Hsc70 bind to preS1 and deletion of the binding site causes cotranslational translocation of preS[64-66]. Luminal chaperones binding to the e-preS domain[67] could support this process. It might be possible that oligomerized S domains form a channel in the membrane for the preS transport[68]. For HBV, the S and M proteins are not required for the translocation process[69], whereas the S protein is essential for DHBV[70,71]. In DHBV, a C-terminally truncated S protein has been described to fulfil a chaperone function during preS translocation[72]. These facts suggest that the preS translocation mechanism might be different between HBV and DHBV[73]. Both L isoforms have their own function: in the e-preS conformation, the preS1 and preS2 domains of L are exposed on the surface of virions and participate in virus receptor binding[74,75], while in the i-preS conformation, the preS1 and preS2 domains of L are internal in the virion and probably important for contacting the nucleocapsid (see below). In addition, the i-preS domain can activate a variety of promoter elements[76]; however, the significance of this function is not clear.
The S domain but not the preS domain contains multiple cystein residues. Cystein residues in the luminal loop crosslink the envelope proteins with each other by multiple disulfide bridges. Shortly after synthesis, disulfide-linked homo- and heterodimers between S, M, and L proteins can be found[77,78]. The cytosolic loop contains 4 cystein residues. Mutational analysis demonstrated that the exchange of 1 out of 3 of the 4 cysteine residues in this loop by a serine residue blocked subviral particle formation[79]. However, these cysteins are not involved in disulfide bridge formation in subviral particles[77], and a covalent modification of these sites, for example, by fatty acid acylation, has not been found[57]. The DHBV L protein is partially phosphorylated[80,81], mainly at serine 118[82]. However, mutational analyses could not demonstrate an essential role for this modification in the DHBV life cycle[83]. Phosphorylation of the HBV L protein could not be found.
SUBVIRAL PARTICLES
The HBV surface proteins are not only incorporated into virion envelopes. Rather, they also bud very efficiently from intracellular, post-ER pre-Golgi membranes[84,85] without envelopment of capsids, appear as subviral quasi-spherical or filamentous lipoprotein particles in the lumen of the compartment, and are released from the cell by secretion. The quasi-spherical particles have a diameter of 20 nm and an octahedral symmetry[86], the filaments have variable lengths. Subviral particles are highly over-expressed relative to virions and reach a 10 000-fold higher concentration in serum. Subviral particles and virions carry identical surface antigens (HBsAg), although the protein composition is not identical. Spherical subviral particles contain only low amounts of L protein, whereas the relative amount of L is higher in filaments and even higher in the virion envelope[49]. It is assumed that the massive HBsAg overproduction influences the host’s immune system in a way that is advantageous for the virus.
Recombinant expression of the S protein (e.g. in yeast) yields highly immunogenic intracellular 20-nm HBsAg particles which can be used as an active vaccine against hepatitis B[87]. S protein expressed in mammalian cells is efficiently secreted as 20-nm HBsAg particles. How the protein escapes the membrane and mobilizes lipid during subviral particle formation is unclear. Chaperons, such as calnexin[88] and BiP[67], bind to S and support the maturation of the protein. The relative amount of lipid is only 25% by weight in subviral particles[89], suggesting that the lipid is not organized as in a conventional membrane bilayer
The M protein essentially behaves like the S protein with respect to subviral particle formation. However, the L protein can not be secreted from cells when expressed by itself. In fact, the L protein causes a dose-dependant inhibition of particle release when coexpressed with the S protein[90,91] and a storage of subviral particles in the ER lumen[88]. This can cause cell stress and even cell death or cancer[92,93]. The significance of the secretion inhibition function of L for the viral life cycle has remained unclear. This function can be abolished by blocking L myristylation (e.g. by a point mutation of the acceptor glycine residue) or by the deletion of the N-terminal 19 aa of preS1[94-96]. Also, the fusion of a secretion signal to the N terminus of L forcing the protein to exclusively generate the e-preS conformation abrogates secretion inhibition[58]. This, however, is different for the DHBV L protein[97].
Host proteins are efficiently excluded during the morphogenesis of subviral particles. Even the HBV and DHBV S proteins sharing 25% identical aa do not form mixed particles during coexpression[98]. However, this is possible with the more closely related S proteins from HBV and the woodchuck hepatitis B virus (WHV). Apparently, the S protein subunits interact tightly with each other during 20 nm particle formation. However, foreign protein domains can be incorporated into subviral HBsAg particles when they are fused to the S protein[99,100].
CAPSID MATURATION
During HBV nucleocapsid formation, the RNA pregenome is packaged into the particle’s lumen and first converted into single stranded and than into partially double-stranded DNA. While nucleocapsids showing all stages of the viral DNA synthesis can be found within cells, secreted virions contain only rather mature circular, partially double-stranded DNA[101,102]. Therefore, it was proposed that early RNA-containing capsid can not be incorporated into virions and that the viral DNA synthesis is associated with a structural change in the capsid shell that allows only mature capsids to be enveloped[103]. This hypothesis was supported by several genetic experiments. C-terminal truncations of the DHBV core protein blocked viral DNA synthesis and also inhibited capsid envelopment[104]. Missense mutations inhibited the reverse transcriptase activity of HBV and DHBV P protein and locked nucleocapsids in an immature state. The incorporation of these capsids into virions was greatly reduced[105,106]. Using a synchronized DHBV replication system, it was shown that envelopment of capsids happened only late in the replication cycle[107].
The nature of the maturation signal has only been described insufficiently. A comparison of capsids containing RNA and DNA by cryo-electronmicroscopy revealed structural differences[15]. There is also evidence that the phosphorylation state of the arginine-rich domain of the C protein might be part of the signal[108-110]. A validation of this hypothesis by a genetic approach is not suitable because substitutions of phosphorylation sites with alanine or glutamic/aspartic acid also influence pregenome packaging and DNA synthesis[111]. Interestingly, the point mutation of isoleucine 97 to leucine in the C protein caused the envelopment of immature capsids[112]. The side chain of this residue is located in the inner space of the spike and close to a hydrophobic pocket showing structural differences in mature versus immature capsids[15]. Possibly, the I97L mutation induces a conformational change causing a constitutive or early expression of the envelopment signal. An additional point mutation (P130T) in a quite distant area of the core protein restored the wild-type phenotype[113], demonstrating the complex nature of the maturation signal for envelopment.
The exclusion of immature nucleocapsids from envelopment causes only replication-competent capsids to become part of virions. This may be one reason for the high specific infectivity of DHBV which is close to the optimum of 1 infectious particle per virion[114]. Also for HBV the specific infectivity seems to be very high[115].
The disassembly of capsids occurs in the basket of nuclear pores upon nuclear transport of the viral genome[116] either during infection or intracellular amplification of the viral genome copy number. Capsid destabilization can also be induced by a tumor necrosis factor alpha-mediated non-cytopathic pathway and may play a role as an antiviral mechanism in natural infections[117].
CAPSID ENVELOPMENT AND VIRION FORMATION
In contrast to retroviruses or togaviruses, it is difficult to directly observe the envelopment of HBV capsid by electron microscopy probably because budding events are less frequent in the available in vitro expression systems. It has even not been clarified whether the HBV envelope contains lipid, although this seems to be likely due to the composition of subviral HBsAg particles. Nevertheless, based on the molecular characterization of HBV formation (see below), it is assumed that hepatitis B virions are formed by budding in analogy to other enveloped viruses.
Mature hepadnaviral nucleocapsids originate in the cytosol. How the capsids move to post-ER, pre-Golgi membranes where the envelopment by the surface proteins supposedly occurs[84,85] is unknown. For DHBV capsids, there is evidence that mature capsids lacking C protein hyperphosphorylation, like capsids in virions, attach to intracellular membranes independent of viral envelope proteins[118]. Immature capsids are hyperphosphorylated and do not bind. This observation suggests that the discrimination between immature and mature capsids happens during the transport of the particle to budding sites before the contact to envelope proteins is established.
Several enveloped viruses utilize a host cell machinery for budding of vesicles into the lumen of so-called multivesicular bodies for their own virus budding[119,120]. Viral capsid proteins interact with the host factor of this pathway via so-called late domains. The HBV C protein contains the sequence PPAY (aa 129-132) exposed on the capsid surface[21] resembling the late domain motif PPXY. However, mutations at this site either blocked capsid formation or reproduced the wild-type phenotype[121]. Therefore, future experiments have to decide whether this pathway is involved in HBV morphogenesis.
The envelopment of HBV capsids strictly depends on viral envelope proteins[122-124] in contrast to type C retroviruses or lentiviruses where mutants blocked in envelope protein expression still release capsids wrapped with a lipid layer. A natural HBV point mutant unable to express the M protein was isolated from a patient and demonstrated that this protein is not required[125], whereas suppression of L or S expression impeded virion formation[122,123]. An L construct with an N-terminally fused secretion signal generating only the e-preS conformation was secreted as a component of subviral particles[126] but failed to support virion formation[58]. Apparently, the i-preS conformation of L exposing the preS domain at the cytosolic side of the ER was essential for nucleocapsid envelopment. This finding is compatible to a model where this part of the L protein contains regions (matrix domains) mediating a contact to the capsid required for budding.
In DHBV, the L protein influences the fate of cytoplasmic capsids[124,127]. If the L protein is absent, capsids deliver the viral genome to the nucleus like in the initial infection of the cell and amplify the intracellular viral genome copy number, whereas capsids are mainly secreted as enveloped virions when the L protein is present. This function mapped to aa 116 to 137 of the 161 aa long DHBV preS domain[128]. A similarly short linear stretch between aa 103 and 124 (or aa 92 and 113 depending on the genotype) genetically mapping in HBV preS was found to be important for virion formation[129]. The exchange of two adjacent aa by alanine residues in this area also prevented nucleocapsid envelopment.
Therefore, it is hypothesized that this part of L interacts with the capsid during envelopment and serves the function of a matrix domain similar to the cytoplasmic tail of the alpha virus E2 protein[130]. This model is supported by an HBV double point mutant where the I97L C protein mutation causing the envelopment of relatively immature capsids is suppressed by the A119F mutation in the putative matrix domain of L[131]. Also in vitro binding assays, using HBV envelope-derived peptides and liver-derived as well as recombinant capsids favour the model[132]. These results also suggest that the discrimination between immature and mature capsids might not occur on the level of capsid-envelope protein interactions. As aforementioned, this selection may happen during surface protein-independent membrane association of capsids.
The loop between the first and second transmembrane region in the S protein is also located at the cytoplasmic side of intracellular membranes and may establish a contact between envelope and capsid. Indeed, short deletions in the C-terminal half of this loop inhibited virion but not 20-nm particle formation[133]. However, point mutations (substitutions of two adjacent aa by alanine residues) in this area were not sufficient to block envelopment (V. Bruss, umpublished).
Potential binding sites on the capsid for envelope protein domains have also been mapped by mutational analyses. A screening of random insertions and deletions in the C protein[134] identified a few mutations allowing nucleocapsid formation and genome synthesis but blocking nucleocapsid envelopment[135]. A similar phenotype was found for naturally occurring HBV mutants isolated from chronically infected virus carriers[136]. Eleven point mutations generated on the basis of the crystal structure of the HBV capsid also induced a loss of nucleocapsid envelopment[121] (Figure 2). They are clustered around the base of the spike and in the grove between spikes. The minimal distance from the matrix domain in the preS region of L to the transmembrane region in the S domain of L allowing virion formation as mapped by deletion mutagenesis[137] is sufficient to allow the matrix domain to reach these sites on the capsid surface. Mutations at the tip or stem of the spike had no impact on capsid envelopment. However, HBV budding from transfected cells can be suppressed by a peptide binding to the tip of the spike[138,139], possibly by steric hindrance.
As in the case of subviral 20-nm particles, the incorpo-ration of foreign proteins into the virion envelope is strictly suppressed. Host membrane proteins could not be detected in virions and even envelope proteins from avian hepadnaviruses do not mix. However, the L protein from WHV can substitute with low efficiency for the HBV L protein in HBV morphogenesis[98]. The matrix domains of WHV and HBV L protein are highly conserved in contrast to the matrix domains of DHBV and HBV L. Foreign domains can be integrated into the HBV envelope by fusion to the N terminus of the S protein and addition of an N-terminal secretion signal[122]. This configuration results in a transmembrane topology similar to the M protein with the preS2 domain substituted by the foreign sequence. When coexpressed with wild-type virus, the chimeric protein is phenotypically mixed into virions and the foreign domain is exposed on the virus surface.
Footnotes
S- Editor Liu Y L- Editor Kumar M E- Editor Ma WH
References
- 1.Acs G, Sells MA, Purcell RH, Price P, Engle R, Shapiro M, Popper H. Hepatitis B virus produced by transfected Hep G2 cells causes hepatitis in chimpanzees. Proc Natl Acad Sci USA. 1987;84:4641–4644. doi: 10.1073/pnas.84.13.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci USA. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, McDade H. Viral dynamics in hepatitis B virus infection. Proc Natl Acad Sci USA. 1996;93:4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roingeard P, Sureau C. Ultrastructural analysis of hepatitis B virus in HepG2-transfected cells with special emphasis on subviral filament morphogenesis. Hepatology. 1998;28:1128–1133. doi: 10.1002/hep.510280431. [DOI] [PubMed] [Google Scholar]
- 6.Funk A, Hohenberg H, Mhamdi M, Will H, Sirma H. Spread of hepatitis B viruses in vitro requires extracellular progeny and may be codetermined by polarized egress. J Virol. 2004;78:3977–3983. doi: 10.1128/JVI.78.8.3977-3983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chain BM, Myers R. Variability and conservation in hepatitis B virus core protein. BMC Microbiol. 2005;5:33. doi: 10.1186/1471-2180-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou S, Standring DN. Hepatitis B virus capsid particles are assembled from core-protein dimer precursors. Proc Natl Acad Sci USA. 1992;89:10046–10050. doi: 10.1073/pnas.89.21.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nassal M, Rieger A, Steinau O. Topological analysis of the hepatitis B virus core particle by cysteine-cysteine cross-linking. J Mol Biol. 1992;225:1013–1025. doi: 10.1016/0022-2836(92)90101-o. [DOI] [PubMed] [Google Scholar]
- 10.Zheng J, Schödel F, Peterson DL. The structure of hepadnaviral core antigens. Identification of free thiols and determination of the disulfide bonding pattern. J Biol Chem. 1992;267:9422–9429. [PubMed] [Google Scholar]
- 11.Lingappa JR, Martin RL, Wong ML, Ganem D, Welch WJ, Lingappa VR. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J Cell Biol. 1994;125:99–111. doi: 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceres P, Zlotnick A. Weak protein-protein interactions are sufficient to drive assembly of hepatitis B virus capsids. Biochemistry. 2002;41:11525–11531. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- 13.Crowther RA, Kiselev NA, Böttcher B, Berriman JA, Borisova GP, Ose V, Pumpens P. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell. 1994;77:943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 14.Kenney JM, von Bonsdorff CH, Nassal M, Fuller SD. Evolutionary conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure. 1995;3:1009–1019. doi: 10.1016/s0969-2126(01)00237-4. [DOI] [PubMed] [Google Scholar]
- 15.Roseman AM, Berriman JA, Wynne SA, Butler PJ, Crowther RA. A structural model for maturation of the hepatitis B virus core. Proc Natl Acad Sci USA. 2005;102:15821–15826. doi: 10.1073/pnas.0504874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallina A, Bonelli F, Zentilin L, Rindi G, Muttini M, Milanesi G. A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J Virol. 1989;63:4645–4652. doi: 10.1128/jvi.63.11.4645-4652.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zlotnick A, Cheng N, Conway JF, Booy FP, Steven AC, Stahl SJ, Wingfield PT. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
- 18.Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 19.Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 20.Zlotnick A, Palmer I, Kaufman JD, Stahl SJ, Steven AC, Wingfield PT. Separation and crystallization of T = 3 and T = 4 icosahedral complexes of the hepatitis B virus core protein. Acta Crystallogr D Biol Crystallogr. 1999;55:717–720. doi: 10.1107/s090744499801350x. [DOI] [PubMed] [Google Scholar]
- 21.Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 22.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zlotnick A, Cheng N, Stahl SJ, Conway JF, Steven AC, Wingfield PT. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc Natl Acad Sci USA. 1997;94:9556–9561. doi: 10.1073/pnas.94.18.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machida A, Ohnuma H, Tsuda F, Yoshikawa A, Hoshi Y, Tanaka T, Kishimoto S, Akahane Y, Miyakawa Y, Mayumi M. Phosphorylation in the carboxyl-terminal domain of the capsid protein of hepatitis B virus: evaluation with a monoclonal antibody. J Virol. 1991;65:6024–6030. doi: 10.1128/jvi.65.11.6024-6030.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watts NR, Conway JF, Cheng N, Stahl SJ, Belnap DM, Steven AC, Wingfield PT. The morphogenic linker peptide of HBV capsid protein forms a mobile array on the interior surface. EMBO J. 2002;21:876–884. doi: 10.1093/emboj/21.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seifer M, Zhou S, Standring DN. A micromolar pool of antigenically distinct precursors is required to initiate cooperative assembly of hepatitis B virus capsids in Xenopus oocytes. J Virol. 1993;67:249–257. doi: 10.1128/jvi.67.1.249-257.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch RC, Lavine JE, Chang LJ, Varmus HE, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 28.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lott L, Notvall L, Lanford RE. Transcomplementation of core and polymerase functions of the woolly monkey and human hepatitis B viruses. Virology. 2003;308:330–339. doi: 10.1016/s0042-6822(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 30.Lott L, Beames B, Notvall L, Lanford RE. Interaction between hepatitis B virus core protein and reverse transcriptase. J Virol. 2000;74:11479–11489. doi: 10.1128/jvi.74.24.11479-11489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beck J, Nassal M. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J Biol Chem. 2003;278:36128–36138. doi: 10.1074/jbc.M301069200. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Grammatikakis N, Hu J. Role of p50/CDC37 in hepadnavirus assembly and replication. J Biol Chem. 2002;277:24361–24367. doi: 10.1074/jbc.M202198200. [DOI] [PubMed] [Google Scholar]
- 33.Hu J, Flores D, Toft D, Wang X, Nguyen D. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J Virol. 2004;78:13122–13131. doi: 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kann M, Gerlich WH. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J Virol. 1994;68:7993–8000. doi: 10.1128/jvi.68.12.7993-8000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kau JH, Ting LP. Phosphorylation of the core protein of hepatitis B virus by a 46-kilodalton serine kinase. J Virol. 1998;72:3796–3803. doi: 10.1128/jvi.72.5.3796-3803.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daub H, Blencke S, Habenberger P, Kurtenbach A, Dennenmoser J, Wissing J, Ullrich A, Cotten M. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J Virol. 2002;76:8124–8137. doi: 10.1128/JVI.76.16.8124-8137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enomoto M, Sawano Y, Kosuge S, Yamano Y, Kuroki K, Ohtsuki K. High phosphorylation of HBV core protein by two alpha-type CK2-activated cAMP-dependent protein kinases in vitro. FEBS Lett. 2006;580:894–899. doi: 10.1016/j.febslet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Chang C, Zhou S, Ganem D, Standring DN. Phenotypic mixing between different hepadnavirus nucleocapsid proteins reveals C protein dimerization to be cis preferential. J Virol. 1994;68:5225–5231. doi: 10.1128/jvi.68.8.5225-5231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulrich R, Nassal M, Meisel H, Krüger DH. Core particles of hepatitis B virus as carrier for foreign epitopes. Adv Virus Res. 1998;50:141–182. doi: 10.1016/s0065-3527(08)60808-8. [DOI] [PubMed] [Google Scholar]
- 40.Skamel C, Ploss M, Böttcher B, Stehle T, Wallich R, Simon MM, Nassal M. Hepatitis B virus capsid-like particles can display the complete, dimeric outer surface protein C and stimulate production of protective antibody responses against Borrelia burgdorferi infection. J Biol Chem. 2006;281:17474–17481. doi: 10.1074/jbc.M513571200. [DOI] [PubMed] [Google Scholar]
- 41.Nassal M, Skamel C, Kratz PA, Wallich R, Stehle T, Simon MM. A fusion product of the complete Borrelia burgdorferi outer surface protein A (OspA) and the hepatitis B virus capsid protein is highly immunogenic and induces protective immunity similar to that seen with an effective lipidated OspA vaccine formula. Eur J Immunol. 2005;35:655–665. doi: 10.1002/eji.200425449. [DOI] [PubMed] [Google Scholar]
- 42.Geldmacher A, Skrastina D, Borisova G, Petrovskis I, Krüger DH, Pumpens P, Ulrich R. A hantavirus nucleocapsid protein segment exposed on hepatitis B virus core particles is highly immunogenic in mice when applied without adjuvants or in the presence of pre-existing anti-core antibodies. Vaccine. 2005;23:3973–3983. doi: 10.1016/j.vaccine.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 43.Ulrich R, Koletzki D, Lachmann S, Lundkvist A, Zankl A, Kazaks A, Kurth A, Gelderblom HR, Borisova G, Meisel H, et al. New chimaeric hepatitis B virus core particles carrying hantavirus (serotype Puumala) epitopes: immunogenicity and protection against virus challenge. J Biotechnol. 1999;73:141–153. doi: 10.1016/s0168-1656(99)00117-0. [DOI] [PubMed] [Google Scholar]
- 44.Kratz PA, Böttcher B, Nassal M. Native display of complete foreign protein domains on the surface of hepatitis B virus capsids. Proc Natl Acad Sci USA. 1999;96:1915–1920. doi: 10.1073/pnas.96.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deres K, Schröder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Krämer T, Niewöhner U, Pleiss U, Stoltefuss J, et al. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science. 2003;299:893–896. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 46.Stray SJ, Bourne CR, Punna S, Lewis WG, Finn MG, Zlotnick A. A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. Proc Natl Acad Sci USA. 2005;102:8138–8143. doi: 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stray SJ, Johnson JM, Kopek BG, Zlotnick A. An in vitro fluorescence screen to identify antivirals that disrupt hepatitis B virus capsid assembly. Nat Biotechnol. 2006;24:358–362. doi: 10.1038/nbt1187. [DOI] [PubMed] [Google Scholar]
- 48.Asif-Ullah M, Choi KJ, Choi KI, Jeong YJ, Yu YG. Identification of compounds that inhibit the interaction between core and surface protein of hepatitis B virus. Antiviral Res. 2006;70:85–90. doi: 10.1016/j.antiviral.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Heermann KH, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich WH. Large surface proteins of hepatitis B virus containing the pre-s sequence. J Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cattaneo R, Will H, Hernandez N, Schaller H. Signals regulating hepatitis B surface antigen transcription. Nature. 1983;305:336–338. doi: 10.1038/305336a0. [DOI] [PubMed] [Google Scholar]
- 51.Sheu SY, Lo SJ. Preferential ribosomal scanning is involved in the differential synthesis of the hepatitis B viral surface antigens from subgenomic transcripts. Virology. 1992;188:353–357. doi: 10.1016/0042-6822(92)90764-g. [DOI] [PubMed] [Google Scholar]
- 52.Eble BE, MacRae DR, Lingappa VR, Ganem D. Multiple topogenic sequences determine the transmembrane orientation of the hepatitis B surface antigen. Mol Cell Biol. 1987;7:3591–3601. doi: 10.1128/mcb.7.10.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson DL, Nath N, Gavilanes F. Structure of hepatitis B surface antigen. Correlation of subtype with amino acid sequence and location of the carbohydrate moiety. J Biol Chem. 1982;257:10414–10420. [PubMed] [Google Scholar]
- 54.Eble BE, Lingappa VR, Ganem D. The N-terminal (pre-S2) domain of a hepatitis B virus surface glycoprotein is translocated across membranes by downstream signal sequences. J Virol. 1990;64:1414–1419. doi: 10.1128/jvi.64.3.1414-1419.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stibbe W, Gerlich WH. Structural relationships between minor and major proteins of hepatitis B surface antigen. J Virol. 1983;46:626–628. doi: 10.1128/jvi.46.2.626-628.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitt S, Glebe D, Tolle TK, Lochnit G, Linder D, Geyer R, Gerlich WH. Structure of pre-S2 N- and O-linked glycans in surface proteins from different genotypes of hepatitis B virus. J Gen Virol. 2004;85:2045–2053. doi: 10.1099/vir.0.79932-0. [DOI] [PubMed] [Google Scholar]
- 57.Persing DH, Varmus HE, Ganem D. The preS1 protein of hepatitis B virus is acylated at its amino terminus with myristic acid. J Virol. 1987;61:1672–1677. doi: 10.1128/jvi.61.5.1672-1677.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bruss V, Vieluf K. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J Virol. 1995;69:6652–6657. doi: 10.1128/jvi.69.11.6652-6657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bruss V, Lu X, Thomssen R, Gerlich WH. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273–2279. doi: 10.1002/j.1460-2075.1994.tb06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prange R, Streeck RE. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ostapchuk P, Hearing P, Ganem D. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 1994;13:1048–1057. doi: 10.1002/j.1460-2075.1994.tb06353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swameye I, Schaller H. Dual topology of the large envelope protein of duck hepatitis B virus: determinants preventing pre-S translocation and glycosylation. J Virol. 1997;71:9434–9441. doi: 10.1128/jvi.71.12.9434-9441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo JT, Pugh JC. Topology of the large envelope protein of duck hepatitis B virus suggests a mechanism for membrane translocation during particle morphogenesis. J Virol. 1997;71:1107–1114. doi: 10.1128/jvi.71.2.1107-1114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lambert C, Prange R. Chaperone action in the posttranslational topological reorientation of the hepatitis B virus large envelope protein: Implications for translocational regulation. Proc Natl Acad Sci USA. 2003;100:5199–5204. doi: 10.1073/pnas.0930813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Löffler-Mary H, Werr M, Prange R. Sequence-specific repression of cotranslational translocation of the hepatitis B virus envelope proteins coincides with binding of heat shock protein Hsc70. Virology. 1997;235:144–152. doi: 10.1006/viro.1997.8689. [DOI] [PubMed] [Google Scholar]
- 66.Prange R, Werr M, Löffler-Mary H. Chaperones involved in hepatitis B virus morphogenesis. Biol Chem. 1999;380:305–314. doi: 10.1515/BC.1999.042. [DOI] [PubMed] [Google Scholar]
- 67.Cho DY, Yang GH, Ryu CJ, Hong HJ. Molecular chaperone GRP78/BiP interacts with the large surface protein of hepatitis B virus in vitro and in vivo. J Virol. 2003;77:2784–2788. doi: 10.1128/JVI.77.4.2784-2788.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berting A, Hahnen J, Kröger M, Gerlich WH. Computer-aided studies on the spatial structure of the small hepatitis B surface protein. Intervirology. 1995;38:8–15. doi: 10.1159/000150409. [DOI] [PubMed] [Google Scholar]
- 69.Lambert C, Prange R. Dual topology of the hepatitis B virus large envelope protein: determinants influencing post-translational pre-S translocation. J Biol Chem. 2001;276:22265–22272. doi: 10.1074/jbc.M100956200. [DOI] [PubMed] [Google Scholar]
- 70.Grgacic EV. Identification of structural determinants of the first transmembrane domain of the small envelope protein of duck hepatitis B virus essential for particle morphogenesis. J Gen Virol. 2002;83:1635–1644. doi: 10.1099/0022-1317-83-7-1635. [DOI] [PubMed] [Google Scholar]
- 71.Grgacic EV, Kuhn C, Schaller H. Hepadnavirus envelope topology: insertion of a loop region in the membrane and role of S in L protein translocation. J Virol. 2000;74:2455–2458. doi: 10.1128/jvi.74.5.2455-2458.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grgacic EV, Anderson DA. St, a truncated envelope protein derived from the S protein of duck hepatitis B virus, acts as a chaperone for the folding of the large envelope protein. J Virol. 2005;79:5346–5352. doi: 10.1128/JVI.79.9.5346-5352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lambert C, Mann S, Prange R. Assessment of determinants affecting the dual topology of hepadnaviral large envelope proteins. J Gen Virol. 2004;85:1221–1225. doi: 10.1099/vir.0.19737-0. [DOI] [PubMed] [Google Scholar]
- 74.Urban S, Gripon P. Inhibition of duck hepatitis B virus infection by a myristoylated pre-S peptide of the large viral surface protein. J Virol. 2002;76:1986–1990. doi: 10.1128/JVI.76.4.1986-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gripon P, Cannie I, Urban S. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J Virol. 2005;79:1613–1622. doi: 10.1128/JVI.79.3.1613-1622.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hildt E, Saher G, Bruss V, Hofschneider PH. The hepatitis B virus large surface protein (LHBs) is a transcriptional activator. Virology. 1996;225:235–239. doi: 10.1006/viro.1996.0594. [DOI] [PubMed] [Google Scholar]
- 77.Wounderlich G, Bruss V. Characterization of early hepatitis B virus surface protein oligomers. Arch Virol. 1996;141:1191–1205. doi: 10.1007/BF01718824. [DOI] [PubMed] [Google Scholar]
- 78.Mangold CM, Unckell F, Werr M, Streeck RE. Secretion and antigenicity of hepatitis B virus small envelope proteins lacking cysteines in the major antigenic region. Virology. 1995;211:535–543. doi: 10.1006/viro.1995.1435. [DOI] [PubMed] [Google Scholar]
- 79.Mangold CM, Streeck RE. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J Virol. 1993;67:4588–4597. doi: 10.1128/jvi.67.8.4588-4597.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grgacic EV, Anderson DA. The large surface protein of duck hepatitis B virus is phosphorylated in the pre-S domain. J Virol. 1994;68:7344–7350. doi: 10.1128/jvi.68.11.7344-7350.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rothmann K, Schnölzer M, Radziwill G, Hildt E, Moelling K, Schaller H. Host cell-virus cross talk: phosphorylation of a hepatitis B virus envelope protein mediates intracellular signaling. J Virol. 1998;72:10138–10147. doi: 10.1128/jvi.72.12.10138-10147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Borel C, Sunyach C, Hantz O, Trepo C, Kay A. Phosphorylation of DHBV pre-S: identification of the major site of phosphorylation and effects of mutations on the virus life cycle. Virology. 1998;242:90–98. doi: 10.1006/viro.1997.9004. [DOI] [PubMed] [Google Scholar]
- 83.Grgacic EV, Lin B, Gazina EV, Snooks MJ, Anderson DA. Normal phosphorylation of duck hepatitis B virus L protein is dispensable for infectivity. J Gen Virol. 1998;79(Pt 11):2743–2751. doi: 10.1099/0022-1317-79-11-2743. [DOI] [PubMed] [Google Scholar]
- 84.Huovila AP, Eder AM, Fuller SD. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J Cell Biol. 1992;118:1305–1320. doi: 10.1083/jcb.118.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Patzer EJ, Nakamura GR, Simonsen CC, Levinson AD, Brands R. Intracellular assembly and packaging of hepatitis B surface antigen particles occur in the endoplasmic reticulum. J Virol. 1986;58:884–892. doi: 10.1128/jvi.58.3.884-892.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gilbert RJ, Beales L, Blond D, Simon MN, Lin BY, Chisari FV, Stuart DI, Rowlands DJ. Hepatitis B small surface antigen particles are octahedral. Proc Natl Acad Sci USA. 2005;102:14783–14788. doi: 10.1073/pnas.0505062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McAleer WJ, Buynak EB, Maigetter RZ, Wampler DE, Miller WJ, Hilleman MR. Human hepatitis B vaccine from recombinant yeast. Nature. 1984;307:178–180. doi: 10.1038/307178a0. [DOI] [PubMed] [Google Scholar]
- 88.Xu Z, Bruss V, Yen TS. Formation of intracellular particles by hepatitis B virus large surface protein. J Virol. 1997;71:5487–5494. doi: 10.1128/jvi.71.7.5487-5494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gavilanes F, Gonzalez-Ros JM, Peterson DL. Structure of hepatitis B surface antigen. Characterization of the lipid components and their association with the viral proteins. J Biol Chem. 1982;257:7770–7777. [PubMed] [Google Scholar]
- 90.Persing DH, Varmus HE, Ganem D. Inhibition of secretion of hepatitis B surface antigen by a related presurface polypeptide. Science. 1986;234:1388–1391. doi: 10.1126/science.3787251. [DOI] [PubMed] [Google Scholar]
- 91.Ou JH, Rutter WJ. Regulation of secretion of the hepatitis B virus major surface antigen by the preS-1 protein. J Virol. 1987;61:782–786. doi: 10.1128/jvi.61.3.782-786.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu Z, Jensen G, Yen TS. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J Virol. 1997;71:7387–7392. doi: 10.1128/jvi.71.10.7387-7392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 94.Gazina E, Gallina A, Milanesi G. Common localization of retention determinants in hepatitis B virus L protein from different strains. J Gen Virol. 1996;77(Pt 12):3069–3075. doi: 10.1099/0022-1317-77-12-3069. [DOI] [PubMed] [Google Scholar]
- 95.Kuroki K, Russnak R, Ganem D. Novel N-terminal amino acid sequence required for retention of a hepatitis B virus glycoprotein in the endoplasmic reticulum. Mol Cell Biol. 1989;9:4459–4466. doi: 10.1128/mcb.9.10.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Prange R, Clemen A, Streeck RE. Myristylation is involved in intracellular retention of hepatitis B virus envelope proteins. J Virol. 1991;65:3919–3923. doi: 10.1128/jvi.65.7.3919-3923.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gazina EV, Lin B, Gallina A, Milanesi G, Anderson DA. Intracellular retention of duck hepatitis B virus large surface protein is independent of preS topology. Virology. 1998;242:266–278. doi: 10.1006/viro.1997.9015. [DOI] [PubMed] [Google Scholar]
- 98.Gerhardt E, Bruss V. Phenotypic mixing of rodent but not avian hepadnavirus surface proteins into human hepatitis B virus particles. J Virol. 1995;69:1201–1208. doi: 10.1128/jvi.69.2.1201-1208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lambert C, Thomé N, Kluck CJ, Prange R. Functional incorporation of green fluorescent protein into hepatitis B virus envelope particles. Virology. 2004;330:158–167. doi: 10.1016/j.virol.2004.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bruss V, Ganem D. Mutational analysis of hepatitis B surface antigen particle assembly and secretion. J Virol. 1991;65:3813–3820. doi: 10.1128/jvi.65.7.3813-3820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mason WS, Aldrich C, Summers J, Taylor JM. Asymmetric replication of duck hepatitis B virus DNA in liver cells: Free minus-strand DNA. Proc Natl Acad Sci USA. 1982;79:3997–4001. doi: 10.1073/pnas.79.13.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weiser B, Ganem D, Seeger C, Varmus HE. Closed circular viral DNA and asymmetrical heterogeneous forms in livers from animals infected with ground squirrel hepatitis virus. J Virol. 1983;48:1–9. doi: 10.1128/jvi.48.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Summers J, Mason WS. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 104.Yu M, Summers J. A domain of the hepadnavirus capsid protein is specifically required for DNA maturation and virus assembly. J Virol. 1991;65:2511–2517. doi: 10.1128/jvi.65.5.2511-2517.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gerelsaikhan T, Tavis JE, Bruss V. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J Virol. 1996;70:4269–4274. doi: 10.1128/jvi.70.7.4269-4274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wei Y, Tavis JE, Ganem D. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J Virol. 1996;70:6455–6458. doi: 10.1128/jvi.70.9.6455-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perlman D, Hu J. Duck hepatitis B virus virion secretion requires a double-stranded DNA genome. J Virol. 2003;77:2287–2294. doi: 10.1128/JVI.77.3.2287-2294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu M, Summers J. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J Virol. 1994;68:4341–4348. doi: 10.1128/jvi.68.7.4341-4348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu M, Summers J. Phosphorylation of the duck hepatitis B virus capsid protein associated with conformational changes in the C terminus. J Virol. 1994;68:2965–2969. doi: 10.1128/jvi.68.5.2965-2969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Perlman DH, Berg EA, O'connor PB, Costello CE, Hu J. Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc Natl Acad Sci USA. 2005;102:9020–9025. doi: 10.1073/pnas.0502138102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gazina EV, Fielding JE, Lin B, Anderson DA. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J Virol. 2000;74:4721–4728. doi: 10.1128/jvi.74.10.4721-4728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yuan TT, Sahu GK, Whitehead WE, Greenberg R, Shih C. The mechanism of an immature secretion phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human hepatitis B virus core antigen. J Virol. 1999;73:5731–5740. doi: 10.1128/jvi.73.7.5731-5740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yuan TT, Shih C. A frequent, naturally occurring mutation (P130T) of human hepatitis B virus core antigen is compensatory for immature secretion phenotype of another frequent variant (I97L) J Virol. 2000;74:4929–4932. doi: 10.1128/jvi.74.10.4929-4932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jilbert AR, Miller DS, Scougall CA, Turnbull H, Burrell CJ. Kinetics of duck hepatitis B virus infection following low dose virus inoculation: one virus DNA genome is infectious in neonatal ducks. Virology. 1996;226:338–345. doi: 10.1006/viro.1996.0661. [DOI] [PubMed] [Google Scholar]
- 115.Barker LF, Murray R. Relationship of virus dose to incubation time of clinical hepatitis and time of appearance of hepatitis--associated antigen. Am J Med Sci. 1972;263:27–33. doi: 10.1097/00000441-197201000-00005. [DOI] [PubMed] [Google Scholar]
- 116.Rabe B, Vlachou A, Panté N, Helenius A, Kann M. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc Natl Acad Sci USA. 2003;100:9849–9854. doi: 10.1073/pnas.1730940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Biermer M, Puro R, Schneider RJ. Tumor necrosis factor alpha inhibition of hepatitis B virus replication involves disruption of capsid Integrity through activation of NF-kappaB. J Virol. 2003;77:4033–4042. doi: 10.1128/JVI.77.7.4033-4042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mabit H, Schaller H. Intracellular hepadnavirus nucleocapsids are selected for secretion by envelope protein-independent membrane binding. J Virol. 2000;74:11472–11478. doi: 10.1128/jvi.74.24.11472-11478.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.von Schwedler UK, Stuchell M, Müller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 120.Freed EO. Viral late domains. J Virol. 2002;76:4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ponsel D, Bruss V. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J Virol. 2003;77:416–422. doi: 10.1128/JVI.77.1.416-422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ueda K, Tsurimoto T, Matsubara K. Three envelope proteins of hepatitis B virus: large S, middle S, and major S proteins needed for the formation of Dane particles. J Virol. 1991;65:3521–3529. doi: 10.1128/jvi.65.7.3521-3529.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Summers J, Smith PM, Huang MJ, Yu MS. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fernholz D, Stemler M, Brunetto M, Bonino F, Will H. Replicating and virion secreting hepatitis B mutant virus unable to produce preS2 protein. J Hepatol. 1991;13 Suppl 4:S102–S104. doi: 10.1016/0168-8278(91)90036-b. [DOI] [PubMed] [Google Scholar]
- 126.Prange R, Werr M, Birkner M, Hilfrich R, Streeck RE. Properties of modified hepatitis B virus surface antigen particles carrying preS epitopes. J Gen Virol. 1995;76(Pt 9):2131–2140. doi: 10.1099/0022-1317-76-9-2131. [DOI] [PubMed] [Google Scholar]
- 127.Summers J, Smith PM, Horwich AL. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J Virol. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lenhoff RJ, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bruss V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol. 1997;71:9350–9357. doi: 10.1128/jvi.71.12.9350-9357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cheng RH, Kuhn RJ, Olson NH, Rossmann MG, Choi HK, Smith TJ, Baker TS. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Le Pogam S, Shih C. Influence of a putative intermolecular interaction between core and the pre-S1 domain of the large envelope protein on hepatitis B virus secretion. J Virol. 2002;76:6510–6517. doi: 10.1128/JVI.76.13.6510-6517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Poisson F, Severac A, Hourioux C, Goudeau A, Roingeard P. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology. 1997;228:115–120. doi: 10.1006/viro.1996.8367. [DOI] [PubMed] [Google Scholar]
- 133.Löffler-Mary H, Dumortier J, Klentsch-Zimmer C, Prange R. Hepatitis B virus assembly is sensitive to changes in the cytosolic S loop of the envelope proteins. Virology. 2000;270:358–367. doi: 10.1006/viro.2000.0268. [DOI] [PubMed] [Google Scholar]
- 134.Koschel M, Thomssen R, Bruss V. Extensive mutagenesis of the hepatitis B virus core gene and mapping of mutations that allow capsid formation. J Virol. 1999;73:2153–2160. doi: 10.1128/jvi.73.3.2153-2160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Koschel M, Oed D, Gerelsaikhan T, Thomssen R, Bruss V. Hepatitis B virus core gene mutations which block nucleocapsid envelopment. J Virol. 2000;74:1–7. doi: 10.1128/jvi.74.1.1-7.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Le Pogam S, Yuan TT, Sahu GK, Chatterjee S, Shih C. Low-level secretion of human hepatitis B virus virions caused by two independent, naturally occurring mutations (P5T and L60V) in the capsid protein. J Virol. 2000;74:9099–9105. doi: 10.1128/jvi.74.19.9099-9105.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kluge B, Schläger M, Pairan A, Bruss V. Determination of the minimal distance between the matrix and transmembrane domains of the large hepatitis B virus envelope protein. J Virol. 2005;79:7918–7921. doi: 10.1128/JVI.79.12.7918-7921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dyson MR, Murray K. Selection of peptide inhibitors of interactions involved in complex protein assemblies: association of the core and surface antigens of hepatitis B virus. Proc Natl Acad Sci USA. 1995;92:2194–2198. doi: 10.1073/pnas.92.6.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Böttcher B, Tsuji N, Takahashi H, Dyson MR, Zhao S, Crowther RA, Murray K. Peptides that block hepatitis B virus assembly: analysis by cryomicroscopy, mutagenesis and transfection. EMBO J. 1998;17:6839–6845. doi: 10.1093/emboj/17.23.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]


