Abstract
Competing endogenous RNA (ceRNA) interactions form a multilayered network that regulates gene expression in various biological pathways. Recent studies have demonstrated novel roles of ceRNA interactions in tumorigenesis, but the dynamics of the ceRNA network in cancer remain unexplored. Here, we examine ceRNA network dynamics in prostate cancer from the perspective of alternative cleavage and polyadenylation (APA) and reveal the principles of such changes. Analysis of exon array data revealed that both shortened and lengthened 3′UTRs are abundant. Consensus clustering with APA data stratified cancers into groups with differing risks of biochemical relapse and revealed that a ceRNA subnetwork enriched with cancer genes was specifically dysregulated in high-risk cancers. The novel connection between 3′UTR shortening and ceRNA network dysregulation was supported by the unusually high number of microRNA response elements (MREs) shared by the dysregulated ceRNA interactions and the significantly altered 3′UTRs. The dysregulation followed a fundamental principle in that ceRNA interactions connecting genes that show opposite trends in expression change are preferentially dysregulated. This targeted dysregulation is responsible for the majority of the observed expression changes in genes with significant ceRNA dysregulation and represents a novel mechanism underlying aberrant oncogenic expression.
MicroRNAs are approximately 22-nucleotide RNAs that regulate gene expression through complementary binding with microRNA response elements (MREs) in target transcripts1. Although positive influences on target transcripts have been reported2, microRNAs are largely negative regulators of protein production whose effects predominantly involve the destabilization of target mRNAs3. The coherent role of microRNAs in mRNA regulation and their many-to-many interaction paradigm allow microRNAs to function as common resources for which different transcripts compete4,5. Such interactions have been termed the ceRNA network and represent a novel form of regulation. Multiple studies have demonstrated that ceRNA networks regulate essentially all known biological processes, and their dysregulation could represent novel disease mechanisms6,7,8.
Although coding regions and 5′UTRs have been reported to harbor MREs9,10, the vast majority of microRNA-mRNA interactions are mediated by 3′UTRs1. Polymorphism in 3′UTR regions is pervasive throughout the human genome, as about half of all known genes are estimated to undergo polyadenylation (APA)11. Because long 3′UTRs tend to harbor more MREs, changing the sizes of 3′UTRs could influence key biological processes by strengthening or weakening the repressive effects of microRNAs. Sandberg et al. first described the profound role of 3′UTR APA dynamics in T cell proliferation12. Through clever probe-level analysis of microarray data, they showed that proliferating T cells utilized shorter 3′UTRs compared to their resting counterparts. The shortening of 3′UTRs enables key genes to escape microRNA repression, thus leading to higher expression and promoting proliferation. Additionally, 3′UTR shortening has been established as a key mechanism of oncogene activation and has demonstrated promising potential as a prognostic marker13,14,15,16. In addition to 3′UTR shortening, neural-specific lengthening of 3′UTRs has been reported during Drosophila development and in the mammalian brain17,18, suggesting that there is considerable versatility in 3′UTR APA dynamics in diverse biological processes.
Because 3′UTRs are essential building blocks of the ceRNA network (together with microRNAs)5, we hypothesized that the dynamics of 3′UTR APAs modulate the structure and strength of the relevant ceRNA networks. We applied the Bayesian change point (BCP) approach19,20 to analyze tandem 3′UTR APA dynamics using a large prostate cancer dataset21. Our results demonstrated that prostate cancers can be stratified into subsets with differing risks of biochemical relapse according to 3′UTR APA dynamics. A densely connected sub-network enriched with prostate cancer genes was specifically dysregulated in high-risk prostate cancers. The dysregulation is not random; rather, it preferentially targets genes showing opposite expression changes, effectively shifting the total ceRNA balance and driving gene up-/downregulation. These results demonstrated that in addition to direct effects on mRNAs, 3′UTR APA dynamics can exert profound influences on ceRNA networks; furthermore, the results, revealed a fundamental rule governing the dysregulation of the ceRNA network in cancer.
Results
Bayesian change point analysis of exon array data reveals complex 3′UTR APA dynamics in prostate cancer
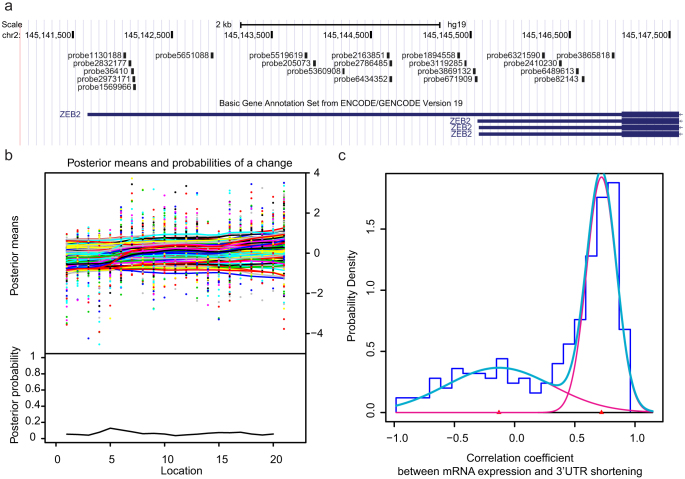
We first established the 3′UTR APA landscape across a large cohort of prostate cancers consisting of 185 exon arrays from normal samples, primary and metastatic cancers and cell lines21. We focused on tandem 3′UTRs because multiple studies have supported their roles in tumorigenesis13,15,22, and several studies have successfully analyzed tandem 3′UTR shortening with exon array data12,23. The exon array probes were first mapped to hg19 using methods similar to PLATA12 and Rmodel23 (Fig. 1a). Instead of applying a modified t-test to individual samples in a manner similar to those used in previous studies, we adopted a multivariate Bayesian approach such that for each tandem 3′UTR, all samples were considered in the same calculation (see Methods). Furthermore, the test estimated the probes' posterior probabilities as the change point and their posterior means in the same Bayesian procedure, thus substantially reducing the number of statistical tests (Fig. 1b). The estimated 3′UTR shortening/lengthening values were then filtered to retain only those with significant change probabilities and median absolute deviations (MADs), resulting in 279 tandem 3′UTRs.
Figure 1. Bayesian change point approach for APA analysis with exon array data.
(a) Mapping of exon array probes to ZEB2 3′UTR. (b) BCP analysis results. The upper panel shows the input probe intensities (dots) and posterior probe mean intensities (solid lines) for all samples. The lower panel shows the posterior change point probabilities for the probes. (c) The correlation coefficient between gene expression and 3′UTR shortening follows a bimodal distribution. The histogram represents the distribution of the correlation coefficient (blue). The estimated densities are shown for individual (magenta) and combined (turquoise) distributions.
The shortening of 3′UTRs has been predominantly associated with elevated gene expression12,13. However, a recent study has shown that 3′UTR sizes have limited effects on mRNA stabilities24. Taking advantage of the large dataset, we demonstrated that the correlation coefficient between 3′UTR shortening and gene expression clearly followed a bimodal distribution (Fig. 1c). As expected, the majority of the genes demonstrated a strong positive correlation. However, a subset of 51 genes displayed a significant negative correlation, indicating that longer 3′UTRs could be associated with higher gene expression (we termed those the “negative gene set”). Those genes included the known oncogene RUNX1 (Supplementary Fig. 1) and prostate cancer-associated genes KLK2 and KLK3. Although functional enrichment analysis did not reveal significant results, most of those tandem 3′UTRs (42 out of 51) did fall within the same cluster when hierarchical clustering was performed using the 3′UTR APA dynamics data (Fig. 2c), suggesting that their 3′UTR lengths are regulated in concert. We further explored possible mechanisms linking longer 3′UTRs with increased gene expression via motif enrichment analysis. Several hexamers representing C-rich elements were significantly enriched in sequence regions that follow the identified APA sites in the negative set genes (Supplementary Table 1). C-rich elements are recognized by poly(C)-binding proteins and have been predominantly designed as stabilizers of mRNAs25. Thus, this bimodal pattern indicated that mRNA stabilities are context dependent, and both microRNAs and RNA binding proteins could be the dominant factors.
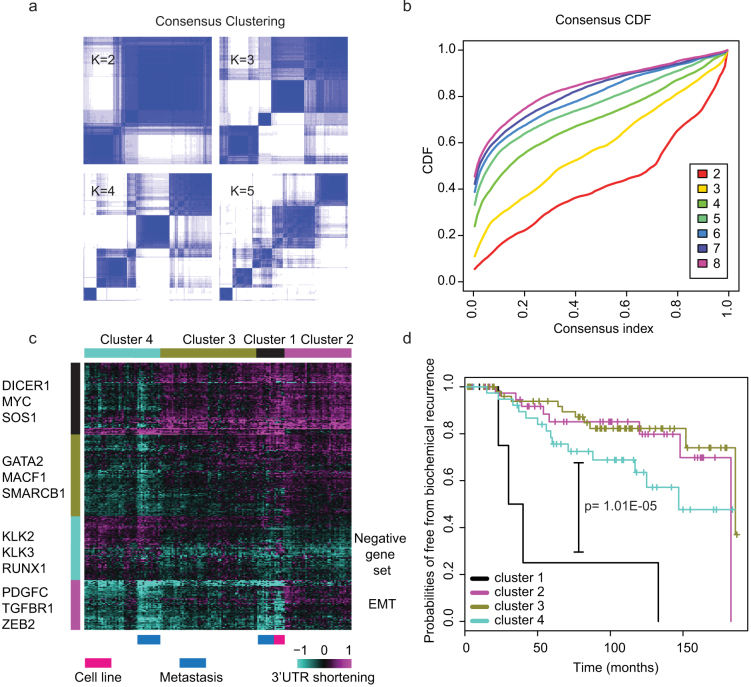
Figure 2. APA dynamics defines stable clusters with differing risks of biochemical relapse.
(a) Consensus clustering matrix of prostate cancer samples for k = 2 to k = 5. (b) Consensus clustering CDF for k = 2 to k = 5. (c) Heatmap of the clustering result. (d) Survival analysis using classifications generated from consensus clustering. Cluster 1 displays a significantly higher probability of relapse.
Consensus clustering of 3′UTR changes defines distinct prostate cancer subtypes with differing risks of relapse
We then investigated whether 3′UTR shortening data can produce biologically relevant stratification of prostate cancers. Consensus clustering generated four stable clusters (Fig. 2a, 2b). There are strong differences in biochemical relapse probabilities among the four subtypes (Fig. 2d). For the cluster with highest relapse probability, the samples were mostly metastatic tumors. The high-risk cluster also displayed the largest absolute change in 3′UTR shortening/lengthening (Fig. 2c). Prostate cancer cell lines also appeared in the same cluster. Moreover, all high-risk samples are from the two copy number variation (CNV) clusters with the largest CNV changes that have been reported previously21, thus confirming their status as advanced tumors. For the majority of the genes (including known oncogenes such as MYC, RUNX1, etc.), the high-risk cluster samples showed the highest expression of those genes (having shorter 3′UTRs, or longer 3′UTRs for the negative set genes). However, a subset of genes showed the opposite trend (Fig. 2c): high-risk cluster samples and metastatic samples displayed lower expression (longer 3′UTRs). This subset contained several EMT-related genes such as PDGFC, TGFBR1 and ZEB2. Because ZEB2 has been designated as a key regulator of EMT via the antagonizing microRNA 200 family members26, we further investigated the 3′UTR lengthening of ZEB2 in metastatic samples and cell lines (Supplementary Fig. 1d). We also analyzed the expression of PDGFD, a known growth factor expressed in normal prostate tissues that could upregulate ZEB227. Those two genes displayed a significant positive correlation, and both genes were expressed at a lower level in cell lines and metastatic tumors (Supplementary Fig. 1e). This finding raised the interesting possibility that reduced expression of the PDGFD-ZEB2 axis may render cells more sensitive to EMT stimuli, thus leading to higher relapse probabilities.
Targeted dysregulation of ceRNA networks in high-risk prostate cancers
We next examined the dysregulation of the ceRNA network in these subtypes via a mutual information- (MI-) based approach. A subnetwork of 5,185 significantly dysregulated ceRNA interactions was identified in the high-risk cluster (Fig. 3a, Table 1). The directions of the changes were homogeneous, in that removing high-risk samples substantially increased MIs for all dysregulated gene pairs (Table 1), indicating that the ceRNA network was uniformly weakened in high-risk cancers. In contrast to the high-risk cluster, minimal numbers of significantly dysregulated ceRNA interactions were identified for the other three groups (Table 1). This observation suggested that substantial dysregulations of the ceRNA network are only evident in samples with large 3′UTR length dynamics, supporting our initial hypothesis. In addition, the medium-risk group was dominated by genes utilizing longer 3′UTRs (Fig. 2c, cluster 4) and was the only group with significantly strengthened ceRNA interactions (Table 1). This pattern further supported the idea that shortening 3′UTRs would release microRNAs and weaken the relevant ceRNA interactions.
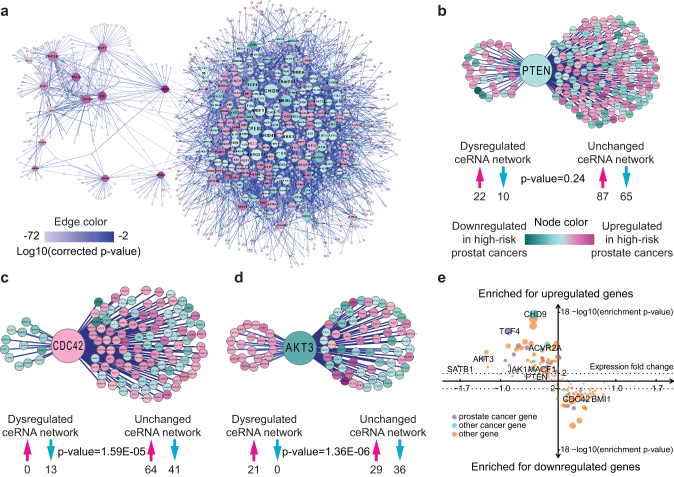
Figure 3. Targeted dysregulation of the ceRNA network.
(a) The dysregulated ceRNA network in high-risk prostate cancers with 5,185 dysregulated ceRNA interactions. Node colors represent expression changes, and edge colors represent the significance of ΔMI. The node sizes of the 182 significantly dysregulated genes are proportional to the number of dysregulated ceRNA interactions. The node sizes for the non-significant genes are set to a small value to allow visual separation between significant and non-significant genes. (b, c and d) Enrichment of genes displaying opposite expression change in dysregulated ceRNA interactions for PTEN (b), CDC42 (c) and AKT3 (d). Only genes with significant expression change between high-risk and low-risk cancer samples were considered (SAM q-value < 0.05). Numbers under arrows indicate the counts for upregulated (upward facing arrows, magenta) and downregulated (downward facing arrows, turquoise) genes. (e) Expression changes of dysregulated genes display a strong correlation with the enrichment of genes with opposite directions of expression change in their dysregulated ceRNA interactions. The size of each dysregulated gene is proportional to its -log10(enrichment p-value). The dotted line represents the cutoff for significant enrichment (p-value = 0.01).
Table 1. Distribution of dysregulated ceRNA network signatures Gain/loss of MI indicates that the removal of a particular cluster from the MI calculation increases/decreases the MI between ceRNA interactions.
| Consensus cluster | Significantly dysregulated genes | Loss of MI | Gain of MI |
|---|---|---|---|
| Cluster 1 (high-risk) | 182 | 0 | 5185 |
| Cluster 2 (low-risk) | 20 | 0 | 162 |
| Cluster 3 (low-risk) | 7 | 0 | 22 |
| Cluster 4 (medium-risk) | 29 | 109 | 4 |
The dysregulated network in high-risk prostate cancers defined 182 genes with unusually high numbers of dysregulated ceRNA interactions (Fig. 3a). These included well-known cancer genes such as PTEN, AKT3 and CDC25A. We first looked for pathways enriched in these 182 dysregulated genes. DAVID28 analysis identified 21 pathways (q-value < 0.05, Supplementary Table 2). Among the top identified were key pathways involved in tumorigenesis, including those in prostate cancer. We next analyzed the number of prostate cancer records in PubMed that were associated with these 182 genes. Compared to simulated random gene sets, the set of 182 genes returned on average 20.47 PubMed records, which was significantly higher than the random gene sets (p-value = 4.19E-11). These results confirmed that the dysregulated ceRNA network in high-risk prostate cancers specifically targeted cancer-related genes and pathways.
Dysregulation of the ceRNA network is associated with 3′UTR APA dynamics
A straightforward explanation for the observed dysregulation is that the mediating microRNAs for those interactions are upregulated. To test this possibility, we first compared the microRNA expression in samples strongly displaying ceRNA network dysregulation (cluster 1) with those showing few ceRNA network changes (clusters 2, 3 and 4) (Supplementary Fig. 2a). Significant Analysis of Microarray29 (SAM) identified 29 differentially expressed microRNAs (q-value < 0.01). Among those microRNAs, only 6 were over-expressed in dysregulated samples. This result indicated that in this particular dataset, differentially expressed microRNAs are not likely to dysregulate the ceRNA network because a reduced (but not depleted) level of microRNAs helps to strengthen ceRNA interactions. Additionally, we compared the set of differentially expressed microRNAs with the microRNAs that had been predicted to mediate the dysregulated ceRNA interactions (Supplementary Fig. 2b). Only approximately 2.3% of the dysregulated ceRNA interactions had more than 20% overlap with the differentially expressed microRNAs. This small overlap further supported the conclusion that differentially expressed microRNAs are not responsible for the observed ceRNA network dysregulation.
We next examined whether the 279 3′UTRs with significant APA dynamics shared any microRNAs with the set of dysregulated ceRNA interactions. The MREs in the extended 3′UTR regions of the 279 genes were collected from computational predictions (TargetScan30, PITA31 and miRanda32). Overlaps between this set of MREs and the MREs predicted to mediate dysregulated ceRNA interactions were computed (Supplementary Fig. 2c, 2d). Compared to randomly selected ceRNA network interactions, the disrupted interactions in high-risk prostate cancers displayed a significantly higher number of total overlaps, as well as a strong bias toward a larger number of overlapping MREs, further strengthening the idea that 3′UTR dynamics could mediate the dysregulation of the ceRNA network.
Dysregulated ceRNA interactions preferentially target genes with opposite directions of expression change
Because the core function of the ceRNA network is to regulate gene expression, we next investigated the principles governing the observed dysregulations. We first applied SAM to identify genes that were differentially expressed between high-risk and low-risk cancers (q-value < 0.05). We then examined the PTEN ceRNA network in detail because it is one of the most well-studied cancer genes. As expected, PTEN was slightly downregulated in high-risk prostate cancers (fold change = −0.40, SAM q-value = 0.12), which is consistent with its known haploinsufficiency33. We then compared differentially expressed genes that are directly connected with PTEN in dysregulated and unchanged ceRNA interactions. While there are larger numbers of downregulated genes in the unchanged ceRNA interactions, there are twice as many genes upregulated in the dysregulated interactions (Fig. 3b). Although the difference is not significant (p-value = 0.24), the trend indicates that dysregulated ceRNA interactions preferentially interrupt the regulatory relationship between PTEN (downregulated) and upregulated genes. The trends are much more obvious for CDC42 (Fig. 3c) and AKT3 (Fig. 3d). Both demonstrated significant enrichment of genes having opposite trends in expression change in dysregulated ceRNA interactions. To gain a systems-level insight, we analyzed all 182 significantly dysregulated genes for such enrichment (Fig. 3e). For 145 of the 182 dysregulated genes, significant (p-value < 0.05) consistent enrichments were observed: if the gene is up/downregulated, the dysregulated ceRNA interactions are enriched for down/upregulated genes. Re-sampling analysis indicated that over 94% of SAM identified genes were consistently observed, confirmed that the observed dysregulation principle is not due to variations in differential expression detection. Twenty-six genes showed a consistent (albeit not significant) trend similar to that of PTEN. Only 11 of the 182 dysregulated genes exhibited the opposite trend (four were significant) of dysregulated ceRNA interactions enriched for genes with the same trend in expression change as the dysregulated gene. However, none of the 11 dysregulated genes displayed a significant change in expression (smallest SAM q-value = 0.17), and the magnitudes of fold change were all close to zero, suggesting that those anomalies are not representative.
We next investigated whether the enrichment of genes with opposite expression change could contribute to the observed expression change of the 182 significantly dysregulated genes. We first examined the contribution by driving CNV to the observed expression changes. Although there was a significant positive correlation between CNV and expression change for the 182 dysregulated genes, the correlation coefficient was smaller and less significant than that of ceRNA dysregulation enrichment (0.49 vs. −0.64, Supplementary Fig. 3). Moreover, multivariate linear regression analysis demonstrated that more than 60% of the explained variance was due to ceRNA dysregulation enrichment (Supplementary Fig. 4). This result clearly showed that for the 182 genes with significant ceRNA dysregulation, their expression changes were mostly due to preferential disruption of ceRNA interactions connecting genes with opposite trends in expression changes. Overall, 84 of the 182 dysregulated genes displayed significant expression changes (SAM q-value < 0.05). The list can be further expanded with haploinsufficient genes such as PTEN, indicating that ceRNA dysregulation could have a significant impact on aberrant gene expression in high-risk prostate cancers.
Discussion
In contrast to the well-studied signal transduction and transcription regulatory networks, the properties and dynamics of the emerging ceRNA network remain elusive. Recent studies have shown that ceRNA network interactions are strongest when the interaction partners are expressed near threshold level34, indicating that the underlying ceRNA network could be considerably dynamic. From the perspective of the ceRNA network, gene expression levels reflect the dynamic balance among the many ceRNA regulatory partners4,34. In contrast to single gene-based disruptions such as upregulation of TGFBR3 by HMGA235, we showed, from a systems perspective, that ceRNA-based aberrant oncogenic expression can also be achieved by specifically removing genes with opposite trends in expression from the ceRNA equation. Despite the presence of thousands of dysregulated ceRNA interactions, we demonstrated that simple rules govern such changes. These findings not only substantially enhance our understanding of ceRNA networks but also could serve as inspirations to reveal the rules governing the dynamics of other types of biological networks.
Because 3′UTRs are the main entities containing MREs and are thus the essential building blocks of the ceRNA network, the theory that changes in 3′UTRs could influence the underlying ceRNA network is logically grounded. However, given the enormous diversity in 3′UTRs and the large number of participating microRNAs, global changes in 3′UTRs must be non-random to induce dysregulation of the ceRNA network. Our analysis of 3′UTR shortening patterns as well as the unusually high number of shared MREs between altered 3′UTRs and dysregulated ceRNA interactions support this hypothesis. Although our analysis was constrained to only a subset of all possible 3′UTRs due to the limited genomic coverage of exon array probes, the presence of dysregulated ceRNA networks in high-risk prostate cancers indicates that 3′UTR dynamics directly influence the ceRNA network by modulating the pool of microRNAs targeting dysregulated ceRNA interactions. The novel connection between alternative splicing and ceRNA network dysregulation further highlights the advantages of approaching tumorigenesis from a systems perspective.
A key observation from our findings with important clinical implications is that significant ceRNA dysregulation is evident only in high-risk prostate cancers; this finding may indicate that remarkable dynamic robustness exists in the ceRNA network. Such robustness is partly due to the scale-free topology36 similar to that in many other biological networks, and it could be further enhanced by the intrinsic redundancy among ceRNA interactions. This observation suggests that constraining ceRNA may be one of the more difficult barriers to overcome during tumorigenesis, and as a late-stage event, its dysregulation may serve as a novel biomarker for prognosis.
Methods
Generation of tandem 3′UTR dataset
To identify the set of transcripts with tandem 3′UTRs that could be analyzed with the Affymetrix Human Exon 1.0 ST Array, we first queried the Ensembl database with the following filters: Transcript count > = 2 and with Affymetrix Microarray huex 1.0 st v2 probeset IDs. We then used the UCSC table browser to retrieve the 3′UTRs of the returned Ensembl Transcript IDs. For each gene, the genomic coordinates of its 3′UTRs were compared, and tandem 3′UTRs were identified (3′UTRs with the same start position and different APA sites).
We used a similar approach, as described previously, to identify probes that could measure 3′UTR APA dynamics12,23. Briefly, the probes of the Affymetrix Human Exon 1.0 ST Array were mapped to the hg19 genome. Probes mapped to multiple genomic locations were discarded. The number of uniquely mapped probes in each tandem 3′UTR was examined. The set of tandem 3′UTRs with at least two probes before and after APAs was kept for 3′UTR expression analysis, resulting in 7,059 tandem 3′UTRs.
Microarray Data
The raw Affymetrix Human Exon 1.0 ST Array data files were obtained from the Gene Expression Omnibus (GEO) accession GSE21034. The processed mRNA and microRNA expression data from the same study were downloaded from the MSKCC Prostate Cancer Genomics Data Portal (http://cbio.mskcc.org/prostate-portal/, Date of access: 10/10/2011). Gene level CNV data were obtained from cBioPortal37.
Exon array data analysis
The CEL files were processed with aroma.affymetrix38 using RMA background correction and quantile normalization. The processed probe intensities were extracted from the intermediate CEL files to allow probe-level analysis. The extracted probe intensities were first log2 transformed. We then applied a similar approach to that outlined in PLATA12 to normalize the probe-level data. Briefly, the probe intensities were first normalized to the median intensity of all probes mapped to the transcript. The gene-level normalized probe intensities were then mean centered to remove probe-specific effects.
Calculation of tandem 3′UTR APA expression changes
The cancer sample probe intensities were first normalized to the median of the normal sample probe intensities to create relative fold changes. We then modeled the tandem 3′UTR APA dynamics problem as a change point problem and utilized the R package BCP20 to perform Bayesian analysis of the change points. Briefly, the approach treated all samples for a tandem 3′UTR as a multivariate series with a common change point, and an n (number of probes for the 3′UTR) by m (number of samples) matrix was provided as algorithm input. The position (probe) with the highest posterior change probability was chosen as the change point. The probes were then partitioned into common and extended groups using the identified change point. For each group, the median of the posterior means of all probes was calculated, and the fold change in the expression between the common and extended regions was calculated as the difference between the two medians. Positive numbers indicated higher expression of common regions (shortening), and negative numbers indicted higher expression of extended regions (lengthening).
Tandem UTR expression data filtering
Several filtering steps were applied to remove unreliably measured tandem 3′UTR expression changes. We first selected tandem 3′UTRs with significant change points. The largest change point posterior probabilities for all tested tandem 3′UTRs were collected. Assuming that the majority of the detected changes were not significant, we estimated the mean and standard deviation by fitting the data to a normal distribution. The z-score for each change point according to the estimated normal distribution was then calculated. We then applied a cutoff of z-score > 2 to select tandem 3′UTRs harboring significant changes. Similarly, tandem 3′UTRs with a MAD z-score ≤ 2 were excluded from further analysis. We then examined the selected tandem 3′UTRs for multiple significant change points. If such change points in a tandem 3′UTR were discontinuous, the tandem 3′UTR was excluded from further analysis because the discontinuity represented complex splicing patterns and did not reflect simple 3′UTR shortening or lengthening.
Clustering analysis
We utilized the Bioconductor package ConsensusClusterPlus39 to identify robust clusters. Hierarchical clustering was performed with Ward's minimum variance method and Euclidean distance. The procedure was run using the 279 filtered tandem 3′UTRs over 1000 iterations and with a sub-sampling ratio of 0.8.
Dysregulation analysis of ceRNA networks
The ceRNA network developed by Sumazin et al.6 was downloaded from the publication website. We used an approach similar to the IDEA algorithm40 developed by Mani et al. to analyze the perturbation of the ceRNA network coupled with 3′UTR APA dynamics. Briefly, MIs between ceRNA gene pairs were first calculated using all prostate cancer samples (MIall). Samples for each subtype identified in the consensus clustering analysis were then removed, and the MIs were recalculated (MIall-k). A positive ΔMI (MIall-k - MIall) indicated that samples of subtype k weakened the underlying ceRNA interaction. We then calculated the significance of ΔMI using the permutation approach outlined in the IEAD method and applied a Bonferroni-corrected p-value of 0.01 to select significantly altered ceRNA interactions. We further filtered the identified interactions and retained only those with positive correlation because they represented modulators with a strong sponge effect. Finally, Fisher's exact test was performed to select genes with a significant number of dysregulated ceRNA interactions using a Bonferroni-corrected p-value of 0.01. The final dysregulated network was filtered to only contain those genes that were directly connected to the significant genes.
Differential expression and driving CNV analysis
We adopted the re-sampling approach outlined in Multiple Survival Screening (MSS)41 to identify the set of genes demonstrating robust differential expression. Briefly, the ratio of high-risk vs. low-risk cancer samples was maintained and 1000 rounds of re-sampling were performed with a sub-sampling ration of 0.8. For driving CNV analysis, we adopted the definition established by Zaman et al.42. Briefly, a driving CNV has a GISTIC score > 0.3, plus among top (for amplification) or bottom (for deletion) 50% of the expressed genes.
Statistical and microRNA analysis of dysregulated ceRNA networks
Network visualizations were produced with Cytoscape43. The pathway enrichment analysis was performed with the NCBI DAVID tool28. For PubMed analysis, PubMed records were retrieved using the keywords “prostate cancer” and the query gene's official gene symbol. For genes whose names matched regular English words (such as REST or SET), the full gene names were used. Significance was assessed with 10,000 simulations using randomly selected genes. To calculate the significance of overlapping microRNAs between dysregulated ceRNA network interactions and the 279 3′UTRs, a Chi-squared test was performed using the average overlapping data from 10,000 random ceRNA networks as a reference distribution. All statistical computations were performed in R.
Author Contributions
L.L., D.W. and M.X. performed the study. X.M. contributed to the array and cluster analysis. Y.L. contributed to the network analysis. P.W. designed the study, contributed to the computational analysis and wrote the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
The authors would like to thank Dr. Jiarui Wu and Dr. Luonan Chen for their critical comments on the manuscript. We would also like to thank members of the Laboratory of Systems Biology and the Center for Stem Cell and NanoMedicine for participating in helpful discussions. This work was supported in part by the National Natural Science Foundation of China (NSFC) grant 31271413, the Science and Technology Commission of Shanghai Municipality grant 12DZ1910800 and a startup fund from the Chinese Academy of Sciences “Hundred Talent Program” to P.W.
References
- Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233, 10.1016/j.cell.2009.01.002 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Tong Y. & Steitz J. A. Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934, 10.1126/science.1149460 (2007). [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N. T., Weissman J. S. & Bartel D. P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840, 10.1038/nature09267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y., Rinn J. & Pandolfi P. P. The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352, 10.1038/nature12986 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., Kats L. & Pandolfi P. P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language.? Cell 146, 353–358, 10.1016/j.cell.2011.07.014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazin P. et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147, 370–381, 10.1016/j.cell.2011.09.041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y. et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147, 344–357, 10.1016/j.cell.2011.09.029 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth F. A. et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147, 382–395, 10.1016/j.cell.2011.09.032 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman J. J., Legesse-Miller A. & Coller H. A. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proceedings of the National Academy of Sciences of the United States of America 105, 14879–14884, 10.1073/pnas.0803230105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle J. R., Yario T. A. & Steitz J. A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proceedings of the National Academy of Sciences of the United States of America 104, 9667–9672, 10.1073/pnas.0703820104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Hu J., Zhang H. & Lutz C. S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic acids research 33, 201–212, 10.1093/nar/gki158 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R., Neilson J. R., Sarma A., Sharp P. A. & Burge C. B. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647, 10.1126/science.1155390 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. & Bartel D. P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684, 10.1016/j.cell.2009.06.016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo A., Di Cunto F. & Provero P. Shortening of 3′UTRs Correlates with Poor Prognosis in Breast and Lung Cancer. PloS one 7, e31129, 10.1371/journal.pone.0031129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P. et al. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer research 69, 9422–9430, 10.1158/0008-5472.CAN-09-2236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapuk A. et al. Exon-Level Microarray Analyses Identify Alternative Splicing Programs in Breast Cancer. Molecular Cancer Research 8, 961–974, 10.1158/1541-7786.mcr-09-0528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P. et al. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell reports 1, 277–289, 10.1016/j.celrep.2012.01.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura P., Shenker S., Andreu-Agullo C., Westholm J. O. & Lai E. C. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome research 23, 812–825, 10.1101/gr.146886.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman C. & Emerson J. W. A fast Bayesian change point analysis for the segmentation of microarray data. Bioinformatics 24, 2143–2148, 10.1093/bioinformatics/btn404 (2008). [DOI] [PubMed] [Google Scholar]
- Erdman C. & Emerson J. W. bcp: An R Package for Performing a Bayesian Analysis of Change Point Problems. Journal of Statistical Software 23, 1–13 (2007). [Google Scholar]
- Taylor B. S. et al. Integrative genomic profiling of human prostate cancer. Cancer cell 18, 11–22, 10.1016/j.ccr.2010.05.026 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. et al. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome research 21, 741–747, 10.1101/gr.115295.110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury J., Hutchison K. W., Wigglesworth K., Eppig J. J. & Graber J. H. Probe-level analysis of expression microarrays characterizes isoform-specific degradation during mouse oocyte maturation. PloS one 4, e7479, 10.1371/journal.pone.0007479 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N., Burge C. B. & Bartel D. P. 3′ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome research 23, 2078–2090, 10.1101/gr.156919.113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M., Wang X. & Liebhaber S. A. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. The EMBO journal 14, 4357–4364 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C. P. et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer research 68, 7846–7854, 10.1158/0008-5472.CAN-08-1942 (2008). [DOI] [PubMed] [Google Scholar]
- Ustach C. V. & Kim H. R. Platelet-derived growth factor D is activated by urokinase plasminogen activator in prostate carcinoma cells. Molecular and cellular biology 25, 6279–6288, 10.1128/MCB.25.14.6279-6288.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4, 44–57, 10.1038/nprot.2008.211 (2009). [DOI] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R. & Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America 98, 5116–5121, 10.1073/pnas.091062498 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A. et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell 27, 91–105, 10.1016/j.molcel.2007.06.017 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M., Iovino N., Unnerstall U., Gaul U. & Segal E. The role of site accessibility in microRNA target recognition. Nature genetics 39, 1278–1284, 10.1038/ng2135 (2007). [DOI] [PubMed] [Google Scholar]
- John B. et al. Human MicroRNA targets. PLoS biology 2, e363, 10.1371/journal.pbio.0020363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwabi-Addo B. et al. Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proceedings of the National Academy of Sciences of the United States of America 98, 11563–11568, 10.1073/pnas.201167798 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ala U. et al. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proceedings of the National Academy of Sciences of the United States of America 110, 7154–7159, 10.1073/pnas.1222509110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M. S. et al. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature 505, 212–217, 10.1038/nature12785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Aldana M. & Cluzel P. A natural class of robust networks. Proceedings of the National Academy of Sciences of the United States of America 100, 8710–8714, 10.1073/pnas.1536783100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2, 401–404, 10.1158/2159-8290.CD-12-0095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson H., Wirapati P. & Speed T. P. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWideSNP 5 & 6. Bioinformatics 25, 2149–2156, 10.1093/bioinformatics/btp371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson M. D. & Hayes D. N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573, 10.1093/bioinformatics/btq170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani K. M. et al. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Molecular systems biology 4, 169, 10.1038/msb.2008.2 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nature communications 1, 34, 10.1038/ncomms1033 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman N. et al. Signaling network assessment of mutations and copy number variations predict breast cancer subtype-specific drug targets. Cell reports 5, 216–223, 10.1016/j.celrep.2013.08.028 (2013). [DOI] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13, 2498–2504, 10.1101/gr.1239303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information