Loss-of-function mutations in hematopoietic transcription factors occur in most cases of B-progenitor acute lymphoblastic leukemia (B-ALL). Here, Liu et al. used transgenic RNAi to reversibly suppress endogenous Pax5 expression in the hematopoietic compartment of mice. Restoring Pax5 expression in established B-ALL triggers immunophenotypic maturation and durable disease remission by engaging a transcriptional program reminiscent of normal B-cell differentiation. Similar findings in human B-ALL cell lines establish that Pax5 hypomorphism promotes B-ALL self-renewal by impairing a differentiation program that can be re-engaged despite the presence of additional oncogenic lesions.
Keywords: PAX5, leukemia, B-ALL, differentiation, transcription factor
Abstract
Loss-of-function mutations in hematopoietic transcription factors including PAX5 occur in most cases of B-progenitor acute lymphoblastic leukemia (B-ALL), a disease characterized by the accumulation of undifferentiated lymphoblasts. Although PAX5 mutation is a critical driver of B-ALL development in mice and humans, it remains unclear how its loss contributes to leukemogenesis and whether ongoing PAX5 deficiency is required for B-ALL maintenance. Here we used transgenic RNAi to reversibly suppress endogenous Pax5 expression in the hematopoietic compartment of mice, which cooperates with activated signal transducer and activator of transcription 5 (STAT5) to induce B-ALL. In this model, restoring endogenous Pax5 expression in established B-ALL triggers immunophenotypic maturation and durable disease remission by engaging a transcriptional program reminiscent of normal B-cell differentiation. Notably, even brief Pax5 restoration in B-ALL cells causes rapid cell cycle exit and disables their leukemia-initiating capacity. These and similar findings in human B-ALL cell lines establish that Pax5 hypomorphism promotes B-ALL self-renewal by impairing a differentiation program that can be re-engaged despite the presence of additional oncogenic lesions. Our results establish a causal relationship between the hallmark genetic and phenotypic features of B-ALL and suggest that engaging the latent differentiation potential of B-ALL cells may provide new therapeutic entry points.
Acute lymphoblastic leukemia (ALL) is the most common pediatric cancer, and despite cure rates approaching 90%, it remains a leading cause of childhood morbidity and mortality in developed countries (Inaba et al. 2013). Most ALL cases involve transformation of B-cell progenitors at the highly proliferative pre-B-cell stage of differentiation (B-ALL). B-ALL subtypes have traditionally been defined by recurrent cytogenetic abnormalities, including chromosome translocations encoding the fusion oncoproteins ETV6-RUNX1 (TEL-AML1) or breakpoint cluster region (BCR)-ABL1. However, in recent years, our understanding of the genetic basis of ALL has been revolutionized by high-resolution genome-wide studies of large ALL cohorts, which have identified recurrent submicroscopic genetic alterations influencing leukemia onset and therapy response (Inaba et al. 2013). Remarkably, loss-of-function mutations or focal deletions in genes encoding transcriptional regulators of lymphoid development, including PAX5, IKZF1, TCF3, and EBF1, occur in approximately two-thirds of B-ALL cases (Mullighan et al. 2007; Inaba et al. 2013). Most common among these is PAX5, with monoallelic deletions, fusion translocations, or point mutations that disrupt PAX5 DNA-binding or transcriptional regulatory functions occurring in approximately one-third of B-ALLs (Kuiper et al. 2007; Mullighan et al. 2007). Somatic PAX5 alterations occur in up to 50% of the high-risk BCR-ABL1-positive and Ph-like ALL subtypes (Mullighan et al. 2008; Roberts et al. 2012) and are also acquired during progression of chronic myeloid leukemia (CML) to lymphoid blast crisis (Mullighan et al. 2008). Germline hypomorphic mutations in PAX5 have recently been associated with B-ALL susceptibility (Shah et al. 2013).
In mice, Pax5 acts downstream from the essential B-lineage transcription factors Tcf3 (E2A) and Ebf1 to commit lymphoid progenitors to a B-cell fate (Cobaleda et al. 2007; Nutt and Kee 2007). B-cell development in Pax5-deficient mice is arrested at the early pro-B-cell stage in the bone marrow (Nutt et al. 1997), and Pax5-null pro-B cells can differentiate into multiple non-B-cell hematopoietic lineages in vitro and in vivo (Nutt et al. 1999; Rolink et al. 1999; Schaniel et al. 2002). Pax5 drives B-lineage commitment by transcriptionally activating B-lineage-specific gene expression networks while repressing alternative lineage genes and remains essential in mature B-lineage cells (Cobaleda et al. 2007).
A conserved tumor suppressor role for Pax5 was recently demonstrated in a transgenic B-ALL mouse model driven by hematopoietic expression of a constitutively active form of signal transducer and activator of transcription 5 (STAT5), which is activated downstream from several human B-ALL oncogenes, including BCR-ABL1, activated JAK kinases, and activated IL7R (Malin et al. 2010). These STAT5b-CA mice normally develop B-ALL with a relatively long latency and low penetrance (Burchill et al. 2003; Nakayama et al. 2008), but this is dramatically accelerated by Pax5 heterozygosity (Heltemes-Harris et al. 2011). Tumors arising in STAT5b-CA;Pax5+/− mice invariably retain the wild-type Pax5 allele (Heltemes-Harris et al. 2011), consistent with mutations in human B-ALL that reduce rather than ablate PAX5 function (Mullighan et al. 2007; Shah et al. 2013).
Although these studies clearly define PAX5 and related transcription factors as B-ALL tumor suppressors, the critical question of how their loss contributes to leukemogenesis remains unexplored. It has been postulated that these transcription factor mutations are involved in the differentiation block characteristic of B-ALL; however, experimental evidence supporting this concept is lacking. Moreover, it remains unclear whether inactivating mutations in transcriptional regulators of B-cell development promote leukemogenesis by simply creating an aberrant progenitor compartment that is susceptible to malignant transformation through accumulation of secondary mutations or whether they retain driver functions in established leukemia. Understanding whether these hallmark mutations are required for B-ALL maintenance provides important rationale for therapeutic strategies targeting their downstream effectors. To directly address these questions, we developed a transgenic RNAi-based B-ALL mouse model allowing inducible suppression and restoration of endogenous Pax5 expression in vivo and used it to define leukemogenic mechanisms and transcriptional programs imposed by hypomorphic Pax5 states in leukemia. We demonstrate that restoration of Pax5 re-engages B-lineage differentiation, leading to progressive tumor clearance and long-term survival.
Results
Stable Pax5 knockdown disrupts B-cell development in vivo
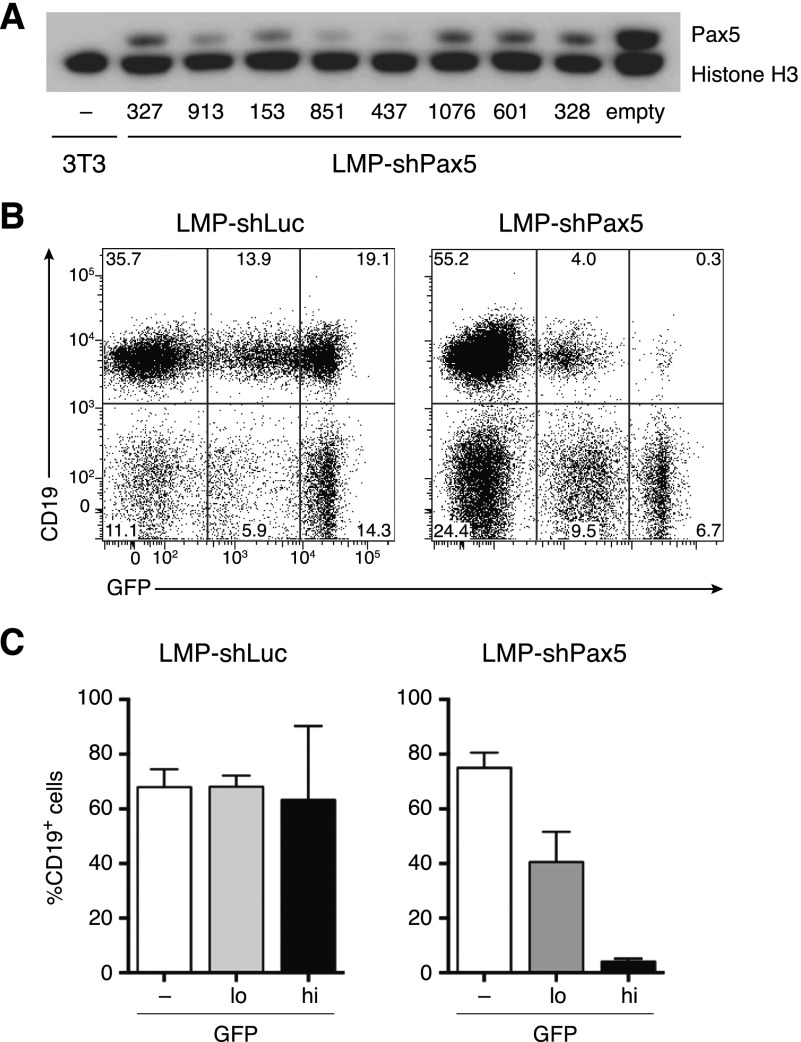
Hypomorphic PAX5 mutations are a common feature of B-ALL (Mullighan et al. 2007; Shah et al. 2013). To model this in mice, we generated several retroviral vectors encoding microRNA-based shRNAs that effectively inhibited Pax5 protein expression in a mouse B-cell line in vitro (Fig. 1A). To examine the effects of stable Pax5 knockdown in vivo, we reconstituted lethally irradiated recipient mice with fetal liver-derived hematopoietic stem and progenitor cells transduced with effective LMP-shPax5 vectors that stably coexpress green fluorescent protein (GFP). Flow cytometry showed normal proportions of CD19+ B-lineage cells in spleens of mice reconstituted with cells transduced with control shRNAs targeting firefly luciferase (shLuc) but a decreased proportion of GFP+ B-lineage cells in shPax5-reconstituted mice (Fig. 1B,C). In this context, GFP intensity reports multiplicity of infection; therefore, an inverse correlation between shPax5 (GFP) expression and CD19 expression suggests that B-lineage development is Pax5 dose-dependent in vivo (Fig. 1B,C). These data demonstrate that shRNA-mediated Pax5 inhibition disrupts normal B-cell development in vivo, in keeping with observations in Pax5-null mice (Nutt et al. 1999).
Figure 1.
Pax5 knockdown inhibits B-cell differentiation in vivo. (A) Western blot of Pax5 expression in WEHI-231 B lymphoma cells transduced with LMP-shPax5 vectors, with numbers indicating different shRNAs. NIH3T3 fibroblasts are included as a Pax5-negative control, and histone H3 is shown as a loading control. (B) Flow cytometric analysis of GFP expression in splenocytes from representative mice reconstituted with fetal liver hematopoietic stem and progenitor cells transduced with LMP-shPax5 and control LMP-shLuc vectors. Mice were analyzed 12 wk following reconstitution. B-lineage cells are identified by surface CD19 expression. (C) Relative proportions of B-lineage cells in the spleen using GFP gating as indicated in B. Mean ± SEM; n = 3 for shLuc; n = 4 for shPax5.
Reversible Pax5 knockdown in transgenic mice
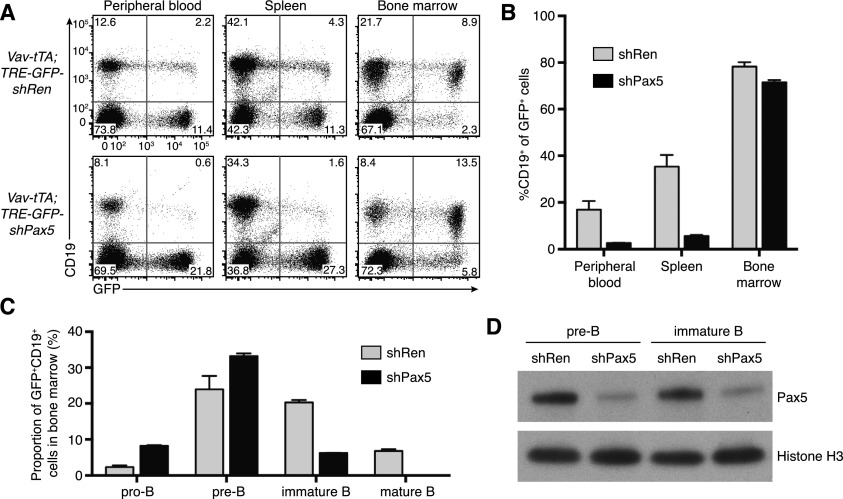
To reversibly manipulate endogenous Pax5 expression in vivo, we generated transgenic mice allowing tetracycline (tet)-regulated Pax5 knockdown. Tet-regulated RNAi comprises three components: a tet-responsive element (TRE) promoter driving shRNA expression, a tet transactivator that conditionally activates the TRE promoter, and doxycycline (Dox), which reversibly controls transactivator function. Dox inhibits the tTA (tet-off) transactivator, whereas the rtTA (tet-on) transactivator is Dox-dependent. Using a recently established strategy (Premsrirut et al. 2011), we produced transgenic mice in which a TRE promoter targeted to the type I collagen (Col1a1) locus controls coexpression of GFP and the effective Pax5.437 shRNA. We crossed TRE-GFP-shPax5 mice with Vav-tTA transgenic mice, which have pan-hematopoietic expression of tTA (Kim et al. 2007; Takiguchi et al. 2013). Consistent with our retroviral Pax5 knockdown experiments, the proportion of B-lineage cells within the GFP+ cell population in the blood, spleen, and bone marrow of Vav-tTA;TRE-GFP-shPax5 bitransgenic mice was reduced relative to control mice expressing an shRNA targeting Renilla luciferase (shRen) (Fig. 2A,B). Analysis of B-lineage development in the bone marrow revealed an increase in the proportion of pro-B cells and a decreased proportion of immature and recirculating B cells within the GFP+ cell population of Vav-tTA;TRE-GFP-shPax5 bitransgenic mice (Fig. 2C), and shPax5-expressing B-cell progenitors had reduced Pax5 protein expression (Fig. 2D). These data verify that transgenic, tet-regulatable Pax5 knockdown inhibits Pax5 expression and blocks B-lineage differentiation in vivo.
Figure 2.
Tet-regulated Pax5 knockdown in transgenic mice in vivo. (A) Flow cytometry analysis of expression of the B-lineage marker CD19 and GFP in the peripheral blood, spleen, and bone marrow of representative Vav-tTA;TRE-GFP-shRen and Vav-tTA;TRE-GFP-shPax5 mice. (B) Decreased proportion of GFP+ B cells in the peripheral blood (P < 0.05, Student’s t-test), spleen (P < 0.01), and bone marrow (P < 0.05) of Vav-tTA;TRE-GFP-shPax5 mice relative to Vav-tTA;TRE-GFP-shRen mice (mean ± SEM; n = 3 for each group). (C) Analysis of GFP+CD19+ B lineage cells in the bone marrow of Vav-tTA;TRE-GFP-shRen and Vav-tTA;TRE-GFP-shPax5 mice (mean ± SEM; n = 3 for each group). Relative to controls, Vav-tTA;TRE-GFP-shPax5 mice have an increased proportion of pro-B cells (Kit+CD25− IgM−; P < 0.001), similar pre-B cells (Kit−CD25+ IgM−), and fewer immature and recirculating mature B cells (IgM+IgD− and IgM+IgD+, respectively; both P < 0.001). (D) Western blot of Pax5 expression in GFP+ pre-B and immature B cells isolated from the bone marrow of representative Vav-tTA;TRE-GFP-shRen and Vav-tTA;TRE-GFP-shPax5 mice, with histone H3 as a loading control.
Pax5 knockdown cooperates with constitutively active STAT5 in leukemogenesis
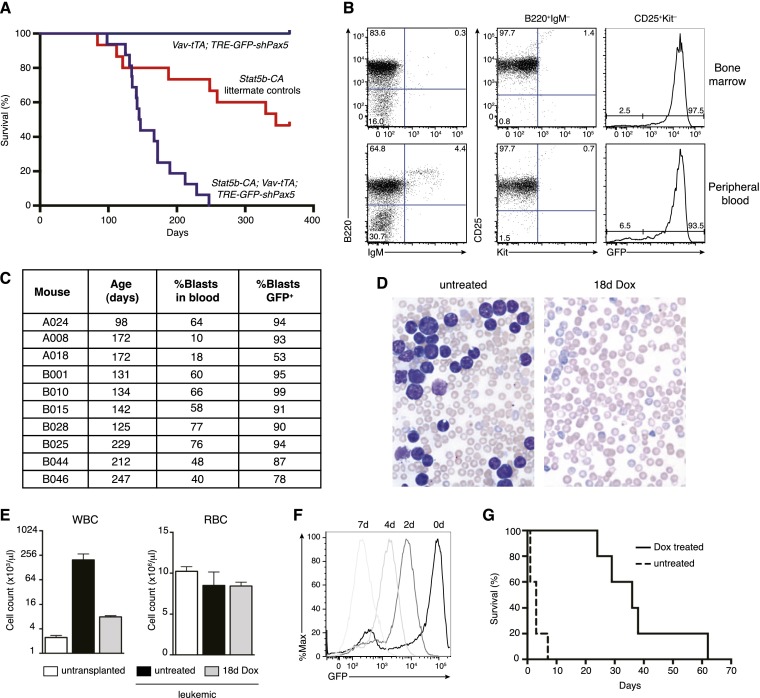
Germline Pax5 heterozygosity accelerates leukemogenesis in a mouse model of B-ALL driven by transgenic expression of constitutively active STAT5 (Heltemes-Harris et al. 2011). To reproduce this genetic interaction using RNAi-based Pax5 inhibition, we generated STAT5b-CA;Vav-tTA;TRE-GFP-shPax5 tritransgenic mice along with littermate STAT5b-CA single- and bitransgenic controls. Tritransgenic mice developed fully penetrant B-ALL with a median latency of 146 d, compared with partially penetrant disease in control mice with a median latency of 345 d (P < 0.0001, log rank test) (Fig. 3A). Consistent with previous findings in STAT5b-CA;Pax5+/− mice (Heltemes-Harris et al. 2011), most tritransgenic leukemias were B220+Kit−CD19+CD25+ and negative for surface IgM, indicating a differentiation block at the pre-B-cell stage, with GFP expression indicating that Pax5 knockdown contributed to leukemogenesis (Fig. 3B,C; Supplemental Table S1). RNA sequencing (RNA-seq) of the leukemia from a representative primary tritransgenic mouse revealed monoclonal rearrangement of the immunoglobulin heavy chain (Igh) locus (Supplemental Fig. S1). These findings suggest that additional cooperating genetic changes likely contribute to leukemogenesis in this model.
Figure 3.
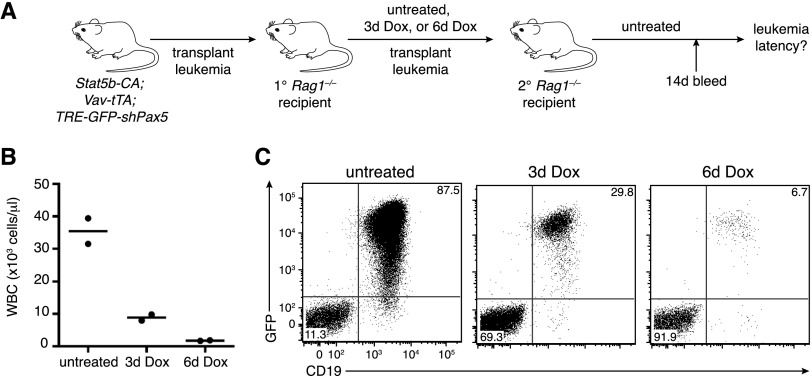
Transgenic Pax5 knockdown promotes reversible B-ALL development. (A) Kaplan-Meier survival curve of STAT5-CA;Vav-tTA;TRE-GFP-shPax5 mice (purple line; n = 16) relative to pooled littermate controls (STAT5-CA mice, STAT5-CA;Vav-tTA mice, and STAT5-CA;TRE-GFP-shPax5 mice; red line; n = 15). Vav-tTA;TRE-GFP-shPax5 control mice (blue line; n = 7) are also shown. (B) Immunophenotype of bone marrow (top panels) and blood (bottom panels) cells from a representative STAT5-CA;Vav-tTA;TRE-GFP-shPax5 leukemic mouse A024 showing B220 and IgM expression (left panels), CD25 expression on B220+IgM− cells (middle panels), and GFP expression in CD25+Kit− cells (right panels). (C) Summary of leukemic tritransgenic mouse phenotypes. (D) Blood smears from representative Rag1−/− mice transplanted with STAT5-CA;Vav-tTA;TRE-GFP-shPax5 leukemia A024 either untreated or with Dox administered at leukemia onset as indicated. (E) Peripheral white (WBC) and red (RBC) blood cell counts from mice as shown in D. Counts from control untransplanted Rag1−/− mice are included. Mean ± SEM; n = 3 for each group; P < 0.05 for WBC (untreated vs. 18d Dox), Student’s t-test. (F) Histogram of GFP expression in CD19+ leukemia cells in the peripheral blood of mice transplanted with STAT5-CA;Vav-tTA;TRE-GFP-shPax5 leukemia cells and Dox-treated as indicated. (G) Kaplan-Meier survival curve for Rag1−/− mice transplanted with STAT5-CA;Vav-tTA;TRE-GFP-shPax5 leukemia. Dox treatment of leukemic mice was initiated at day 0. n = 5 untreated mice and Dox-treated mice; log rank test, P < 0.005.
Pax5 restoration causes B-ALL regression
To restore Pax5 expression in established leukemias in vivo, we initially transplanted leukemia cells from representative tritransgenic mouse A024 into cohorts of Rag1−/− mice. Recipient mice developed aggressive GFP+ leukemia within 2–3 wk, with very high peripheral white blood cell (WBC) counts (Fig. 3D,E), palpable splenomegaly, and enlarged lymph nodes. As anticipated, Dox treatment of mice with a significant peripheral tumor burden caused a progressive decrease in GFP fluorescence intensity of circulating leukemia cells, indicating shutoff of the TRE-GFP-shPax5 transgene (Fig. 3F; see below). Whereas untreated leukemic mice succumbed to disease within 1 wk, the condition of Dox-treated mice markedly improved, and, after 18 d, their WBC counts had almost normalized (Fig. 3D,E). Tumor regression brought about by continuous Dox treatment consistently extended host life span, with mice surviving for many weeks without overt signs of leukemia (Fig. 3G). Tumor growth in mice bearing control STAT5b-CA or STAT5b-CA;Pax5+/− leukemias was unabated upon Dox treatment (data not shown), ruling out nonspecific effects. These results demonstrate that ongoing Pax5 suppression is required for B-ALL maintenance in vivo.
Pax5 restoration releases the pre-B stage differentiation block in B-ALL
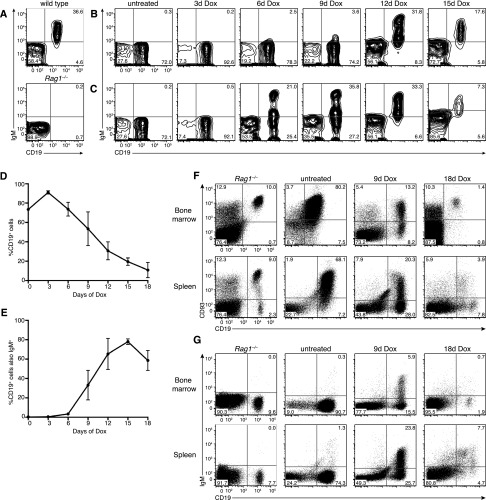
Having observed tumor regression upon sustained Dox treatment, we monitored leukemia clearance kinetics in several diseased mice using flow cytometry of peripheral blood at 3-d intervals following Dox administration. Transplanted leukemias recapitulated the primary CD19+Kit−CD25+IgM− pre-B stage disease; therefore, CD19 flow cytometry readily identified leukemia cells in the blood of lymphocyte-deficient Rag1−/− recipient mice (Fig. 4A–C). Remarkably, the proportion of leukemia cells in the blood of Dox-treated mice decreased incrementally from day 3 onward, falling to ∼10% at day 18, indicating that Pax5 restoration results in gradual leukemia clearance rather than acute regression (Fig. 4B–D).
Figure 4.
Pax5 restoration triggers leukemia differentiation and regression. (A) Flow cytometry of CD19 and IgM expression in peripheral blood of wild-type (top panel) and untransplanted Rag1−/− (bottom panel) control mice. (B) Flow cytometry of CD19 and IgM expression on mononuclear cells from the peripheral blood of a representative Rag1−/− mouse transplanted with STAT5-CA;Vav-tTA;TRE-GFP-shPax5 leukemia and Dox-treated upon leukemia development as indicated. (C) Time course analysis as shown in B for an independent leukemic recipient mouse. (D) Leukemia burden (proportion of CD19+ cells in the blood) upon Dox treatment (mean ± SEM; n = 3 mice). (E) Proportion of CD19+ cells coexpressing IgM upon Dox treatment (mean ± SEM; n = 3 mice). (F) Flow cytometry of CD19 and CD93 expression on bone marrow cells (top panels) and splenocytes (bottom panels) isolated from representative leukemic Rag1−/− mice at various times during Dox treatment as indicated. The left panels are profiles from untransplanted Rag1−/− control mice, indicating a small population of CD19+ pro-B cells in the bone marrow. (G) Flow cytometry of CD19 and IgM expression as shown in F.
During normal B-cell development in the bone marrow, productive Igh gene rearrangement results in pre-B-cell receptor (pre-BCR) assembly and signaling in pre-B cells, triggering their clonal expansion (Rolink et al. 2000). These “large cycling” pre-B cells (also known as large pre-BII cells [Rolink et al. 1994] or fraction C′ cells [Hardy et al. 1991]) then exit the cell cycle to become “small resting” pre-B cells (small pre-BII or fraction D cells), whereupon productive rearrangement of the immunoglobulin light chain genes Igk or Igl allows the surface IgM expression characteristic of immature B cells. Notably, Dox treatment of leukemic mice resulted in expression of IgM in an increasing proportion of circulating CD19+ leukemia cells over time (Fig. 4B,C,E). Indeed, after 15 d, when tumor burden had significantly decreased, ∼80% of cells were IgM+. The kinetics of IgM acquisition varied between individual leukemic mice but occurred in the majority of CD19+ cells within a 3-d period during Dox treatment (Fig. 4B,C). Similar effects were seen in mice transplanted with independent tritransgenic leukemia A008, although this leukemia showed a more striking immunophenotypic maturation after ∼10 d on Dox, with the majority of antecedent leukemia cells coexpressing the mature marker IgD along with IgM (Supplemental Fig. S2A–F). Together, these data suggest that re-engaging endogenous Pax5 function releases the pre-B-cell stage differentiation block in leukemia, promoting Igk or Igl recombination and IgM expression characteristic of mature B cells.
The bone marrow and spleen of mice bearing leukemia A024 was dominated by CD19+ leukemia cells coexpressing the immature marker CD93, which is normally expressed by pro-B, pre-B, and immature B cells in the bone marrow and by immature transitional B cells in the spleen (Fig. 4F). Nine days of Dox treatment led to down-regulation of CD93 expression on a substantial proportion of CD19+ cells in the bone marrow and spleen, indicating maturation of antecedent leukemia cells in these organs (Fig. 4F). This was accompanied by surface IgM expression in a significant proportion of cells (Fig. 4G). Consistent with changes in peripheral leukemia burden, flow cytometry analysis demonstrated near-complete clearance of leukemia cells from the bone marrow and spleen following 18 d of Dox treatment (Fig. 4F,G). Although continuous Dox treatment caused leukemia regression and extended host life span (Fig. 3G), after many weeks, mice became moribund, with splenomegaly and enlarged lymph nodes. Unexpectedly, in all mice examined, these organs were mainly comprised of host-derived cells, potentially expanded in response to antecedent leukemia cells (Supplemental Fig. S3). In contrast, while sustained Pax5 restoration in independent tritransgenic leukemia A008 caused similar acute regression and robust differentiation, in this case, mature leukemia-derived cells accumulated in lymph nodes and spleens of all mice analyzed after 2 wk on Dox (Supplemental Fig. S2G,H). In summary, although long-term Dox responses of individual primary leukemias varied (Supplemental Table S1), potentially due to different genetic changes during leukemogenesis, Pax5 restoration in pre-B-cell leukemias uniformly caused acute tumor clearance and/or differentiation in vivo.
Identification of Pax5-regulated genes in B-ALL
Expression profiling of Pax5-deficient mice has identified Pax5-regulated genes in normal pro-B and mature B cells (Delogu et al. 2006; Schebesta et al. 2007; Pridans et al. 2008; Revilla-I-Domingo et al. 2012); however, Pax5-regulated genes in pre-B-ALL remain poorly defined. To identify transcriptional responses to acute Pax5 restoration in leukemias in vivo, STAT5b-CA;Vav-tTA;TRE-GFP-shPax5 leukemia cells were isolated by FACS from the bone marrow of triplicate, transplanted leukemic mice that were either untreated or Dox-treated for 3 d (GFPhigh and GFPmid cells, respectively) (Fig. 3F). Although the leukemia immunophenotype was stable during these initial stages of Pax5 restoration (Fig. 4), we reasoned that expression profiling at this point would enrich for primary Pax5-dependent gene expression changes. RNA-seq revealed that leukemia cells from untreated mice expressed Pax5 mRNA at levels ∼40% of those in normal bone marrow CD19+Kit−CD25+IgM− pre-B cells, which are mainly small resting pre-B cells (Fig. 5A; Hoffmann et al. 2002). Three days of Dox treatment effectively restored Pax5 mRNA to near wild-type levels, with a corresponding increase in protein expression (Fig. 5A,B). Surprisingly, this was accompanied by significant up-regulation of 3343 genes and down-regulation of 3240 genes (false discovery rate [FDR] 0.05), together comprising more than half of the expressed genes in leukemia cells (Fig. 5C). The majority (52%) of these differentially expressed genes was previously identified as direct Pax5 targets by Bio-ChIP sequencing in pro-B cells (Revilla-I-Domingo et al. 2012), including well-established Pax5 target genes such as Cd19 and Blnk (Fig. 5D; Nutt et al. 1997; Schebesta et al. 2002). Genes insensitive to Pax5 restoration showed proportionally less (45%) Pax5 binding (P < 10−13, Fisher’s exact test). Therefore, despite differences in the stages of B-cell differentiation examined by ChIP-seq (chromatin immunoprecipitation [ChIP] combined with deep sequencing) (pro-B cells) and RNA-seq (pre-B ALL cells), our RNAi-based approach preferentially identified genes under the direct transcriptional control of Pax5.
Figure 5.
Pax5 restoration causes rapid repression of Myc and DNA replication factors. (A) Expression (RNA-seq reads per kilobase per million mapped reads [RPKM]) of Pax5 in STAT5-CA;Vav-tTA;TRE-GFP-shPax5 leukemia cells harvested from untreated and Dox-treated leukemic recipient mice compared with normal pre-B cells (mean ± SEM; n = 3 mice for each group). (B) Western blot of Pax5 and GFP expression in B-ALL cells harvested from duplicate leukemic mice with Dox treatments indicated. Histone H3 is a loading control. (C) Scatter plot of average RNA-seq differential gene expression in B-ALL cells comparing untreated with 3 d of Dox treatment (three mice per condition). Genes with significantly increased (red) or decreased (blue) expression in leukemia cells upon Pax5 restoration are indicated (FDR 0.05). Pax5 and Rag1/2 are highlighted. (D,E) RNA-seq expression of Cd19 and Blnk (D) and Rag1/2 (E) in B-ALL cells as indicated in A. (F) Gene set analysis barcode plot. The RNA-seq differential gene expression data set upon Pax5 restoration in STAT5-CA;Vav-tTA;TRE-GFP-shPax5 leukemia cells in vivo is shown as a shaded rectangle, with genes horizontally ranked by moderated t statistic; genes up-regulated upon Pax5 restoration are shaded pink (t > 1), and down-regulated genes are shaded blue (t < −1). Overlaid are a set of previously described genes induced (red bars) or repressed (blue bars) upon the transition from large cycling pre-B cells to small resting pre-B cells during normal B-cell development in the bone marrow (Hoffmann et al. 2002). Red/blue traces above/below the bar represent relative enrichment. (G) Heat map of average RNA-seq differential gene expression in STAT5-CA;Vav-tTA;TRE-GFP-shPax5 leukemia cells harvested from triplicate untreated and Dox-treated leukemic recipient mice (RNA-seq RPKM is expressed as Z-score, with red and blue indicating up-regulation and down-regulation, respectively). Components of the pre-BCR complex and the DNA prereplication complex are in bold. (H) Myc mRNA expression as described in A. (I) Cell cycle profiles of CD45.1−CD19+ leukemia cells freshly isolated from untreated and Dox-treated leukemic mice. Cells were stained with DAPI and analyzed by flow cytometry. Percentages of G0/G1-phase and S/G2/M-phase cells are indicated.
Acute Pax5 restoration causes transcriptional changes resembling normal B-cell differentiation
Among the genes most sensitive to Pax5 restoration were the V(D)J recombination activation genes Rag1 and Rag2, with 15-fold and 38-fold up-regulation, respectively (Fig. 5C,E; Supplemental Table S2). In normal B-cell development, Rag1/2 are highly expressed during Igh recombination in pro-B cells, silenced during the proliferative expansion of large cycling pre-B cells, and then re-expressed in small resting pre-B cells to promote Igl/Igk rearrangement (Grawunder et al. 1995). Leukemia cells isolated from untreated mice had low Rag1/2 expression, in keeping with their large cycling pre-B stage immunophenotype (Fig. 5E), but marked Rag1/2 induction upon Pax5 restoration suggested transition toward the small resting pre-B-cell stage. Consistent with this, gene set analysis revealed that global expression changes accompanying dynamic Pax5 restoration in B-ALL were specifically and highly correlated (P < 0.0001) with expression changes that occur at the transition from large cycling to small resting pre-B cells during normal cell differentiation in murine bone marrow (Fig. 5F; Supplemental Fig. S4; Hoffmann et al. 2002). This differentiation step is characterized by proliferative exit (Rolink et al. 1994; Hoffmann et al. 2002), and, accordingly, genes significantly down-regulated upon Pax5 restoration were heavily enriched (40 of the top 75) for critical DNA replication and cell cycle genes, including those encoding the catalytic subunits of the major nuclear DNA polymerases α, δ, and ε (Pola1, Pold1, and Pole); DNA prereplication complex components Cdc6, Cdt1, and Mcm2–7; DNA replisome components, including Pcna; and nucleotide synthesis enzymes (Fig. 5G; Supplemental Table S3). Unbiased gene ontology analysis (Supplemental Table S4) confirmed that the proliferative transcriptional signature of B-ALL cells is rapidly suppressed by Pax5 restoration.
Pax5 restoration induces pre-BCR signaling components and represses Myc
Assembly of a pre-BCR complex following productive Igh recombination triggers the clonal expansion of large pre-BII cells. The pre-BCR consists of nascent immunoglobulin heavy chains associated with the surrogate light chain (SLC) components Vpreb1/2 and λ5 (Igll1) and the signal transducing subunits Cd79a (Igα) and Cd79b (Igβ) (Supplemental Fig. S5A; Herzog et al. 2009). Surprisingly, genes encoding all pre-BCR components except Vpreb2, which are among the most highly expressed genes in the ALL transcriptome (Supplemental Table S5), were significantly induced following acute re-establishment of Pax5 expression in B-ALL (Fig. 5G; Supplemental Fig. S5B,C). Furthermore, genes encoding several critical components of the pre-BCR signal transduction cascade were similarly induced, including Blnk (SLP-65), Plcg1 (PLC-γ1), and the tyrosine kinases Syk, Btk, Blk, and Lyn (Fig. 5G; Supplemental Fig. S5D). Notably, Blnk and Btk also suppress B-ALL in mice, suggesting that Pax5 loss may promote B-ALL pathogenesis by attenuating the expression of multiple tumor suppressor genes (see the Discussion).
Particularly notable among down-regulated genes was the proto-oncogene Myc (Fig. 5H), ranked 39th by significance. Myc stimulates proliferation and inhibits differentiation by directly modulating the expression of thousands of genes (Pelengaris et al. 2002). Accordingly, gene set analysis revealed loss of transcriptional signatures associated with Myc activation upon Pax5 restoration (Supplemental Fig. S6). Myc is required for the normal proliferative expansion of pre-B cells, and elevated Myc expression is a hallmark of leukemia and other malignancies (Pelengaris et al. 2002; Habib et al. 2007). Our data indicate that abnormally elevated Myc expression in B-ALL results primarily from a pre-B-cell stage differentiation block, which in turn is imposed by Pax5 loss.
Acute Pax5 restoration disables the leukemia-initiating capacity of B-ALL cells
DNA content analysis indicated that at least one-third of STAT5b-CA;Vav-tTA;TRE-GFP-shPax5 B-ALL cells were cycling in vivo (Fig. 5I). In keeping with coordinated repression of DNA replication and cell cycle factors upon Pax5 restoration, we observed a rapid decrease in the proportion of proliferating ALL cells from primary leukemias A008 and A024 following acute Dox treatment in vivo (Fig. 5I; Supplemental Fig. S2I–K). To test whether the proliferative shutdown caused by brief Pax5 restoration altered the leukemogenic potential of ALL cells, we isolated CD19+ A024 leukemia cells from untreated and Dox-treated primary recipient mice by FACS and transplanted equal cell numbers into secondary recipient mice (Fig. 6A). While leukemia cells isolated from untreated primary mice produced rapid leukemia in secondary recipients, tumorigenesis in recipients of leukemia cells from Dox-treated donors was significantly delayed (Fig. 6B,C). Leukemias eventually arising in these secondary recipients were GFP+, were immunophenotypically indistinguishable from primary leukemias, and regressed upon Dox treatment, suggesting that relapse was driven by renewed Pax5 knockdown in leukemia cells upon transfer into untreated mice. Together, these results indicate that brief Pax5 restoration in B-ALL cells in vivo significantly mitigates their leukemogenic potential.
Figure 6.
Pax5 restoration disables the leukemia-initiating capacity of B-ALL cells. (A) Strategy for testing leukemia initiation properties of B-ALL cells following Pax5 restoration. (B) Peripheral WBC counts from secondary recipient mice 14 d following transplant with 2 million leukemia cells isolated from primary recipient mice that were treated as indicated. n = 2 mice for each time point. Horizontal lines indicate average counts. (C) Flow cytometric analysis of leukemia burden (GFP+CD19+ cells) in the blood of secondary recipient mice 14 d following transplant as described in B.
PAX5 inhibits the self-renewal of human PAX5 mutant B-ALL cell lines
To explore the relevance of our findings in human leukemia, we established a system for tet-on-inducible PAX5 re-expression in three human B-ALL cell lines harboring PAX5 aberrations that reflect the common pathogenic mutation types. REH cells harbor a PAX5 frameshift mutation (p.Arg225fs) encoding a protein lacking the C-terminal transactivation domain (Dorfler and Busslinger 1996), identical to a recurrent mutation in primary B-ALL (Mullighan et al. 2007). 697 cells harbor a similar truncating PAX5 mutation (p.Pro321fs) (Barretina et al. 2012), whereas NALM-6 cells have a focal, heterozygous deletion at the PAX5 promoter (Fig. 7A). As a control, we examined the BCR-ABL1+ lymphoblastic cell line BV173, with normal PAX5 copy number (Fig. 7A) and sequence (Barretina et al. 2012).
Figure 7.
PAX5 re-expression is specifically detrimental to PAX5 mutated human B-ALL cell lines. (A) Copy number heat map showing a focal, heterozygous PAX5 promoter deletion in the NALM-6 cell line (indicated by a horizontal line). PAX5 point mutations are also indicated. (B) Strategy for generating tet-on-competent human B-ALL cell lines with inducible PAX5 expression. (C) Western blot of PAX5 expression in B-ALL cell lines following Dox-induced expression of IRES-GFP or PAX5-IRES-GFP as indicated, with tubulin as a loading control. (D) Time course showing the proportion of GFP+ cells following induction of PAX5-IRES-GFP in the indicated cell lines relative to matched control IRES-GFP cells. All data were initially normalized to percentage GFP 4 d after Dox administration. Data are representative of two independent experiments. (E,F) Flow cytometric analysis of cell size/FSC (E) and the cell surface markers CD19, CD38, and CD10 (F) in GFP− (red line) and GFP+ (green line) cells in B-ALL cell lines 3 d after Dox-dependent induction of PAX5-IRES-GFP. These parameters were unaffected by GFP expression alone (Supplemental Fig. S7C,D).
To facilitate inducible PAX5 re-expression in human B-ALL cells, we first generated “tet-on-competent” cell lines by stably expressing the reverse tet transactivator rtTA3 together with the ecotropic receptor and then introduced retroviral vectors encoding human PAX5 and GFP under the control of the improved TRE promoter (TRE3G) (Fig. 7B). While enforced expression of PAX5 triggered by Dox treatment (Fig. 7C) had no major effect on proliferation of BV173 cells, all three PAX5 mutant cell lines showed a rapid depletion of PAX5-overexpressing cells in competitive proliferation assays (Fig. 7D). Cell cycle analysis indicated a reduction of cells in S phase that was most pronounced in 697 cells (Supplemental Fig. S7A,B). In both frameshift mutant cell lines, PAX5 expression led to a marked reduction in cell size reminiscent of the large-to-small pre-B-cell transition, which was not observed in NALM-6 cells (Fig. 7E; Supplemental Fig. S7C). Remarkably, in all three mutant cell lines, PAX5 overexpression triggered clear shifts in the expression of surface markers associated with B-cell differentiation. Specifically, enforced PAX5 expression led to up-regulation of the well-established B-lineage differentiation marker CD19, while CD38, a highly expressed marker in normal human bone marrow B-cell progenitors that is down-regulated in mature B cells in the blood (Kumagai et al. 1995), was strongly repressed (Fig. 7F; Supplemental Fig. S7D). Intriguingly, PAX5 expression in both frameshift cell lines also triggered robust expression of CD10, also known as common ALL antigen (CALLA) (Fig. 7F). In summary, while the specific phenotype in response to enforced PAX5 expression varies between human cell lines, likely due to different background mutation profiles and culture histories (Barretina et al. 2012), those derived from PAX5 mutant B-ALL remain addicted to PAX5 hypomorphism.
To identify PAX5-regulated genes in REH cells, we performed RNA-seq analysis following 2 d of PAX5 induction to enrich for direct PAX5 targets. Overall, 203 genes were up-regulated more than twofold (Supplemental Fig. S7E), corresponding to 106 genes expressed in our mouse B-ALL RNA-seq data set. Many genes up-regulated following Pax5 restoration in our in vivo mouse model were unaffected in REH cells (Supplemental Fig. S7E), which could reflect technical differences or biases introduced during long-term culture. Despite this, gene set analysis demonstrated significant enrichment (P < 0.0005, roast test) of the 106 REH up-regulated genes among Pax5-activated genes in our mouse B-ALL model (Supplemental Fig. S7F), with 31 of these genes (29%) exceeding twofold induction in both B-ALL models (Supplemental Fig. S7E; Supplemental Table S6). Notably, genes commonly induced in both species included those encoding TNFRSF13C/BAFF-R, a major regulator of peripheral B-cell survival (Mackay et al. 2010); BACH2, a recently described tumor suppressor in B-ALL (Swaminathan et al. 2013); and the immunoglobulin λ constant region (Iglc3/IGLC7), consistent with a conserved differentiation response (Supplemental Fig. S7E).
Discussion
Mutations in transcription factors and chromatin regulators that govern cell fate decisions are the most striking finding of recent oncogenomic profiling (Vogelstein et al. 2013); however, most of these driver events remain functionally unexplored. Importantly, in many cases, it remains unclear whether these aberrations act predominantly during tumor initiation (e.g., by locking cells into progenitor states that are more vulnerable to the acquisition of transforming mutations) or remain essential in full-blown disease. Loss-of-function mutations in PAX5 and related transcription factors occur in the majority of cases of B-ALL, a disease characterized by an accumulation of highly proliferative lymphoblasts that fail to undergo normal differentiation. In this study, we used a purpose-built mouse model to demonstrate that aberrant PAX5 activity specifically acts to enforce a differentiation block in B-ALL, causally linking the hallmark genetic and phenotypic features of this disease for the first time. Moreover, we found that PAX5 loss not only is important for B-ALL pathogenesis but remains critical for the maintenance of established disease. These properties of PAX5 are likely in part shared by the other essential B-lineage transcription factors TCF3 (E2A) and EBF1, which directly induce PAX5 and are also frequently mutationally inactivated in B-ALL (Kuiper et al. 2007; Mullighan et al. 2007; Nutt and Kee 2007). Hence, our results define the biological consequences of the predominant class of tumor suppressor gene mutation in B-ALL.
Restoring Pax5 function in B-ALL driven by its knockdown triggers a rapid and coordinated in vivo differentiation response that closely mimics the transition of large cycling pre-B cells to small resting pre-B cells during normal B lymphopoiesis. Both processes are characterized by proliferative exit and concomitant global down-regulation of genes involved in DNA replication and cell cycle progression and Rag1/2 up-regulation in preparation for Igl/Igk gene recombination. In keeping with these acute molecular changes, sustained Pax5 restoration leads to a striking immunophenotypic maturation of antecedent B-ALL cells, reminiscent of normal B-cell differentiation and durable tumor regression.
Given the molecular and phenotypic similarities between Pax5-induced B-ALL differentiation and normal B-progenitor differentiation, it is likely that partial Pax5 deficiency promotes B-ALL self-renewal by disengaging mechanisms that normally limit the clonal expansion of large pre-BII cells during B-cell development. Signaling from the IL-7 receptor and pre-BCR normally collaborate to drive a burst of large pre-BII-cell proliferation (Herzog et al. 2009), and differentiation to the small resting pre-BII stage is thought to involve shutdown of these pathways. Notably, Pax5-induced B-ALL differentiation in our model proceeds despite enforced STAT5 activity, which partially mimics sustained IL-7 receptor signaling. Similarly, Pax5-induced B-ALL cell cycle exit occurs despite ongoing expression of the critical pre-BCR components λ5 (Igll1) and Vpreb1. Together, these findings are consistent with previous work showing that pre-B-cell expansion can be curtailed regardless of IL-7 signaling and despite enforced SLC expression (Milne et al. 2004; van Loo et al. 2007), suggesting alternative Pax5-dependent mechanisms of proliferative shutdown. Interestingly, we found that proliferative exit of B-ALL cells upon Pax5 restoration is associated with significant induction of genes encoding (1) the core pre-BCR components Vpreb1, λ5, Igα, and Igβ; (2) critical pre-BCR signaling proteins, including Blnk, Syk, and Btk; and (3) known targets of pre-BCR signaling, including Ikzf3 (Aiolos), Irf4, and Irf8 (Thompson et al. 2007). This suggests that full Pax5 expression may re-engage pre-BCR-dependent mechanisms that limit clonal expansion. A tumor-suppressive role for the pre-BCR been previously proposed in murine B-ALL models (Trageser et al. 2009; Duy et al. 2010) and is also supported by recurrent IGLL1 and VPREB1 deletions in human B-ALL (Mullighan et al. 2007; Mangum et al. 2014).
Our results emphasize the importance of Pax5 dosage in leukemia suppression. B-cell differentiation in Pax5-null mice is completely blocked at the early pro-B-cell stage (Urbánek et al. 1994; Nutt et al. 1997). Surprisingly, while Pax5 heterozygous mice have apparently normal B-cell development, they are remarkably susceptible to B-ALL driven by constitutively active STAT5, and leukemias arising in these mice retain the wild-type Pax5 allele (Nutt et al. 1997; Heltemes-Harris et al. 2011). Similarly, genetic alterations in PAX5 in pediatric B-ALL characteristically reduce rather than ablate PAX5 transcriptional activity (Kuiper et al. 2007; Mullighan et al. 2007), and pre-B leukemias arising in our RNAi-based model express Pax5 at approximately half of normal levels. These observations suggest a threshold level of Pax5 activity in B-ALL that is sufficient to sustain the clonal expansion state characteristic of normal large pre-B cells but insufficient to engage differentiation mechanisms that enforce quiescence. It is notable that simply doubling the Pax5 dose in this context drives synchronous leukemia differentiation.
Remarkably, Pax5 hypomorphic B-ALL cells retain the ability to re-engage a B-lineage differentiation program despite ongoing expression of activated STAT5 and the presence of additional oncogenic lesions, providing rationale for the development of therapeutic approaches aimed at restoring or mimicking full PAX5 activity in B-ALL. Approximately 2.5% of B-ALL cases harbor chromosomal rearrangements producing dominant-negative PAX5 fusion proteins that are, in principle, “druggable” (Mullighan et al. 2007; Nebral et al. 2009; Kawamata et al. 2012). However, the majority of PAX5 aberrations in B-ALL generally reduce its expression or DNA binding, and in these cases restoring PAX5 activity is therapeutically intractable. Despite this, our results suggest that elevated Myc expression in B-ALL is a critical, albeit indirect, consequence of the pre-B-cell differentiation arrest enforced by PAX5 loss. Hence, although MYC is required for normal B-cell development (Habib et al. 2007), our data implicate MYC as a rational drug target in PAX5-deficient B-ALL. Notably, recent studies demonstrate that the BET bromodomain inhibitor JQ1, which dramatically reduces MYC transcription, has anti-leukemic activity in primary human B-ALL cell lines and xenografts (Ott et al. 2012). Our results suggest that further interrogation of PAX5-dependent gene expression networks in B-ALL may uncover other novel therapeutic entry points for this disease.
Materials and methods
Additional details are provided in the Supplemental Material.
Fetal liver transduction and reconstitution
Retroviral supernatants were produced by calcium chloride transfection of 293T cells with plasmids encoding packaging components and LMP vectors (Dickins et al. 2005) encoding Pax5 shRNAs. Embryonic day 13.5–14.5 (E13.5–E14.5) fetal liver cell suspensions were immunomagnetically depleted of erythryoid cells and transduced using plates treated with retronectin and viral supernatant. Transduced cells were cultured for 24 h in IMDM supplemented with 10% FCS and cytokines (100 ng/mL SCF, 10 ng/mL IL-6, 50 ng/mL TPO, 5 ng/mL Flt3) and injected intravenously into lethally irradiated (twice at 5.5 Gy) recipient mice.
Transgenic mice
The TREtight-GFP-Pax5.437 and TREtight-GFP-Ren.713 transgenes were targeted to the type I collagen (Col1a1) locus using previously described protocols (Premsrirut et al. 2011). Genotyping details are provided in the Supplemental Material. Dox (Sigma-Aldrich) was administered in the diet at 600 mg per kilogram of food (Specialty Feeds). All mouse experiments were approved by the Walter and Eliza Hall Institute Animal Ethics Committee.
Blood and flow cytometry analysis
Mice were monitored for signs of leukemia, including loss of activity or weight, breathing difficulty, enlarged lymph nodes, and palpable splenomegaly. Blood was collected from the retro-orbital plexus, mandible, or tail vein, and parameters were measured with an Advia 2120 hematological analyzer (Bayer). Single-cell suspensions were prepared from bone marrow, spleen, lymph nodes, or peripheral blood and treated with red cell lysis buffer. FACS antibody details are provided in the Supplemental Material. Stained cells were analyzed or sorted by flow cytometry (LSR II or LSRFortessa, BD Biosciences). FACS data were analyzed with FlowJo software (Tree Star).
Leukemia transplantation and human B-ALL cell line culture
Primary mouse leukemia cells were transplanted intravenously into immunocompromised CD45Ly5.1Rag1−/− recipient mice. Approximately 2 × 106 primary or secondary leukemia cells were transplanted. Transplant recipients generally developed overt leukemia 2–3 wk after transplantation. The human B-ALL cell line culture is described in the Supplemental Material.
DNA content analysis
Following staining with antibodies described above, cells were permeabilized and fixed using the Cytofix/Cytoperm kit (BD Biosciences). Cells were then stained with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (Sigma-Aldrich) and analyzed for DNA content by flow cytometry.
Western blotting
Lysates prepared from either cultured cell lines or ex vivo sorted cells were Western-blotted with anti-Pax5 antibody 1H9, anti-GFP antibody A6455 (Invitrogen), and anti-acetyl-Histone H3 antibody 06-599 (Millipore).
RNA-seq, bioinformatics, and statistical analysis
Total RNA was extracted using the RNeasy Plus minikit (Qiagen) and profiled on Illumina HiSeq 2000 sequencers. Reads were mapped to the mouse or human genomes using subread (Liao et al. 2013), summarized using featureCounts (Liao et al. 2014), and analyzed for differential expression using voom (Law et al. 2014). Enrichment analysis used roast (Wu et al. 2010). Details are described in the Supplemental Material. RNA-seq data are available at the Gene Expression Omnibus (GEO) through accession numbers GSE52868, GSE52870, and GSE57480.
Acknowledgments
We thank M. Salzone, M. Dayton, P. Kennedy, K. Stoev, C. Smith, L. Wilkins, M. Howell, T. Carle, E. Lanera, S. Brown, and other Walter and Eliza Hall Institute Bioservices staff; W. Alexander for embryonic stem cell resources; E. Viney and J. Sarkis at the Australian Phenomics Network Transgenic RNAi service; M. Everest and M. Tinning at the Australian Genome Research Facility; and R. Lane, T. Willson, J. Corbin, and C. Hyland for technical expertise. We thank S. Lowe for vectors, D. Largaespada for Vav-tTA transgenic mice, and M. Jaritz and M. Busslinger for Pax5 ChIP-seq data. We thank S. Chappaz, D. Tarlinton, D. Hilton, M. Busslinger, and S. Lowe for advice and discussions. This work was supported by the National Health and Medical Research Council (NHMRC) of Australia project grants 575535, 1024599, and 1023454; Australian Government NHMRC Independent Research Institute Infrastructure Support Scheme (IRIISS); an Australian Research Council Future Fellowship (to S.L.N.); an NHMRC Research Fellowship (to G.K.S.); National Institutes of Health USA grants R01 CA151845 and CA154998 (to M.A.F.); Austrian Science Fund grant F4710-B20 (to J.Z.); Boehringer Ingelheim Institutional funding (to J.Z. and J.G.J.); the Leukaemia Foundation of Australia (G.J.L., M.T.W., and M.D.M.); a Sylvia and Charles Viertel Charitable Foundation Fellowship (to R.A.D.); Victorian State Government Operational Infrastructure Support grants; and a Victorian Endowment for Science, Knowledge, and Innovation (VESKI) Fellowship (to R.A.D.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.240416.114.
References
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. 2012. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill MA, Goetz CA, Prlic M, O’Neil JJ, Harmon IR, Bensinger SJ, Turka LA, Brennan P, Jameson SC, Farrar MA 2003. Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells. J Immunol 171: 5853–5864 [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M 2007. Pax5: the guardian of B cell identity and function. Nat Immunol 8: 463–470 [DOI] [PubMed] [Google Scholar]
- Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M 2006. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity 24: 269–281 [DOI] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW 2005. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 37: 1289–1295 [DOI] [PubMed] [Google Scholar]
- Dorfler P, Busslinger M 1996. C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5), Pax-2 and Pax-8. EMBO J 15: 1971–1982 [PMC free article] [PubMed] [Google Scholar]
- Duy C, Yu JJ, Nahar R, Swaminathan S, Kweon SM, Polo JM, Valls E, Klemm L, Shojaee S, Cerchietti L, et al. 2010. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med 207: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawunder U, Leu TM, Schatz DG, Werner A, Rolink AG, Melchers F, Winkler TH 1995. Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity 3: 601–608 [DOI] [PubMed] [Google Scholar]
- Habib T, Park H, Tsang M, de Alboran IM, Nicks A, Wilson L, Knoepfler PS, Andrews S, Rawlings DJ, Eisenman RN, et al. 2007. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J Cell Biol 179: 717–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med 173: 1213–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltemes-Harris LM, Willette MJ, Ramsey LB, Qiu YH, Neeley ES, Zhang N, Thomas DA, Koeuth T, Baechler EC, Kornblau SM, et al. 2011. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. J Exp Med 208: 1135–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog S, Reth M, Jumaa H 2009. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol 9: 195–205 [DOI] [PubMed] [Google Scholar]
- Hoffmann R, Seidl T, Neeb M, Rolink A, Melchers F 2002. Changes in gene expression profiles in developing B cells of murine bone marrow. Genome Res 12: 98–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba H, Greaves M, Mullighan CG 2013. Acute lymphoblastic leukaemia. Lancet 381: 1943–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata N, Pennella MA, Woo JL, Berk AJ, Koeffler HP 2012. Dominant-negative mechanism of leukemogenic PAX5 fusions. Oncogene 31: 966–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WI, Wiesner SM, Largaespada DA 2007. Vav promoter–tTA conditional transgene expression system for hematopoietic cells drives high level expression in developing B and T cells. Exp Hematol 35: 1231–1239 [DOI] [PubMed] [Google Scholar]
- Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN, Hoogerbrugge PM 2007. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia 21: 1258–1266 [DOI] [PubMed] [Google Scholar]
- Kumagai M, Coustan-Smith E, Murray DJ, Silvennoinen O, Murti KG, Evans WE, Malavasi F, Campana D 1995. Ligation of CD38 suppresses human B lymphopoiesis. J Exp Med 181: 1101–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK 2014. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W 2013. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W 2014. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Mackay F, Figgett WA, Saulep D, Lepage M, Hibbs ML 2010. B-cell stage and context-dependent requirements for survival signals from BAFF and the B-cell receptor. Immunol Rev 237: 205–225 [DOI] [PubMed] [Google Scholar]
- Malin S, McManus S, Busslinger M 2010. STAT5 in B cell development and leukemia. Curr Opin Immunol 22: 168–176 [DOI] [PubMed] [Google Scholar]
- Mangum DS, Downie J, Mason CC, Jahromi MS, Joshi D, Rodic V, Muschen M, Meeker N, Trede N, Frazer JK, et al. 2014. VPREB1 deletions occur independent of λ light chain rearrangement in childhood acute lymphoblastic leukemia. Leukemia 28: 216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne CD, Fleming HE, Paige CJ 2004. IL-7 does not prevent pro-B/pre-B cell maturation to the immature/sIgM+ stage. Eur J Immunol 34: 2647–2655 [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446: 758–764 [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui C-H, et al. 2008. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 453: 110–114 [DOI] [PubMed] [Google Scholar]
- Nakayama J, Yamamoto M, Hayashi K, Satoh H, Bundo K, Kubo M, Goitsuka R, Farrar M, Kitamura D 2008. BLNK suppresses pre-B-cell leukemogenesis through inhibition of JAK3. Blood 113: 1483–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebral K, Denk D, Attarbaschi A, Konig M, Mann G, Haas OA, Strehl S 2009. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia 23: 134–143 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Kee BL 2007. The transcriptional regulation of B cell lineage commitment. Immunity 26: 715–725 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Urbánek P, Rolink A, Busslinger M 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev 11: 476–491 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink AG, Busslinger M 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401: 556–562 [DOI] [PubMed] [Google Scholar]
- Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, Rodig SJ, Kung AL, Bradner JE, Weinstock DM 2012. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood 120: 2843–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan G 2002. c-MYC: more than just a matter of life and death. Nat Rev Cancer 2: 764–776 [DOI] [PubMed] [Google Scholar]
- Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, Scuoppo C, Zuber J, Dickins RA, Kogan SC, et al. 2011. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell 145: 145–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridans C, Holmes ML, Polli M, Wettenhall JM, Dakic A, Corcoran LM, Smyth GK, Nutt SL 2008. Identification of Pax5 target genes in early B cell differentiation. J Immunol 180: 1719–1728 [DOI] [PubMed] [Google Scholar]
- Revilla-I-Domingo R, Bilic I, Vilagos B, Tagoh H, Ebert A, Tamir IM, Smeenk L, Trupke J, Sommer A, Jaritz M et al. 2012. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J 31: 3130–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, Chen SC, Payne-Turner D, Churchman ML, Harvey RC, et al. 2012. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell 22: 153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink A, Grawunder U, Winkler TH, Karasuyama H, Melchers F 1994. IL-2 receptor α chain (CD25, TAC) expression defines a crucial stage in pre-B cell development. Int Immunol 6: 1257–1264 [DOI] [PubMed] [Google Scholar]
- Rolink AG, Nutt SL, Melchers F, Busslinger M 1999. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature 401: 603–606 [DOI] [PubMed] [Google Scholar]
- Rolink AG, Winkler T, Melchers F, Andersson J 2000. Precursor B cell receptor-dependent B cell proliferation and differentiation does not require the bone marrow or fetal liver environment. J Exp Med 191: 23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaniel C, Gottar M, Roosnek E, Melchers F, Rolink AG 2002. Extensive in vivo self-renewal, long-term reconstitution capacity, and hematopoietic multipotency of Pax5-deficient precursor B-cell clones. Blood 99: 2760–2766 [DOI] [PubMed] [Google Scholar]
- Schebesta M, Pfeffer PL, Busslinger M 2002. Control of pre-BCR signaling by Pax5-dependent activation of the BLNK gene. Immunity 17: 473–485 [DOI] [PubMed] [Google Scholar]
- Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M 2007. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity 27: 49–63 [DOI] [PubMed] [Google Scholar]
- Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, Wechsler J, Yang J, Hayes J, Klein RJ, et al. 2013. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet 45: 1226–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Huang C, Geng H, Chen Z, Harvey R, Kang H, Ng C, Titz B, Hurtz C, Sadiyah MF, et al. 2013. BACH2 mediates negative selection and p53-dependent tumor suppression at the pre-B cell receptor checkpoint. Nat Med 19: 1014–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiguchi M, Dow LE, Prier JE, Carmichael CL, Kile BT, Turner SJ, Lowe SW, Huang DC, Dickins RA 2013. Variability of inducible expression across the hematopoietic system of tetracycline transactivator transgenic mice. PLoS ONE 8: e54009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, Taylor B, Dillon N, Georgopoulos K, Jumaa H, et al. 2007. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity 26: 335–344 [DOI] [PubMed] [Google Scholar]
- Trageser D, Iacobucci I, Nahar R, Duy C, von Levetzow G, Klemm L, Park E, Schuh W, Gruber T, Herzog S, et al. 2009. Pre-B cell receptor-mediated cell cycle arrest in Philadelphia chromosome-positive acute lymphoblastic leukemia requires IKAROS function. J Exp Med 206: 1739–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbánek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79: 901–912 [DOI] [PubMed] [Google Scholar]
- van Loo PF, Dingjan GM, Maas A, Hendriks RW 2007. Surrogate-light-chain silencing is not critical for the limitation of pre-B cell expansion but is for the termination of constitutive signaling. Immunity 27: 468–480 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW 2013. Cancer genome landscapes. Science 339: 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Lim E, Vaillant F, Asselin-Labat M-L, Visvader JE, Smyth GK 2010. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26: 2176–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]