Abstract
Considerable advances have been recently made in understanding the molecular aspects of pathogenesis and in developing therapeutic approaches for polyglutamine (polyQ) diseases. Studies on pathogenic mechanisms have extended our knowledge of mutant protein toxicity, confirmed the toxicity of mutant transcript and identified other toxic RNA and protein entities. One very promising therapeutic strategy is targeting the causative gene expression with oligonucleotide (ON) based tools. This straightforward approach aimed at halting the early steps in the cascade of pathogenic events has been widely tested for Huntington's disease and spinocerebellar ataxia type 3. In this review, we gather information on the use of antisense oligonucleotides and RNA interference triggers for the experimental treatment of polyQ diseases in cellular and animal models. We present studies testing non-allele-selective and allele-selective gene silencing strategies. The latter include targeting SNP variants associated with mutations or targeting the pathologically expanded CAG repeat directly. We compare gene silencing effectors of various types in a number of aspects, including their design, efficiency in cell culture experiments and pre-clinical testing. We discuss advantages, current limitations and perspectives of various ON-based strategies used to treat polyQ diseases.
INTRODUCTION
Expansions of short tandem repeat sequences in different genes are responsible for numerous human hereditary neurological diseases. Most of these disorders are caused by the expansion of repeated trinucleotides and are called triplet repeat expansion diseases (1). Their largest subgroup is polyQ diseases, which are caused by the expansion of CAG repeats present in open reading frames (ORFs) of specific functionally unrelated genes. These disorders include Huntington's disease (HD), dentatorubral-pallidoluysian atrophy (DRPLA), spinal bulbar muscular atrophy (SBMA) and spinocerebellar ataxia (SCA) types 1, 2, 3, 6, 7 and 17 (Table 1). Additionally, SCA8 shares some features with polyQ diseases due to the antisense transcription of non-protein-coding gene containing CTG expansion and the translation of antisense transcripts to polyQ proteins (2). The common feature of polyQ diseases is their late onset, as initial symptoms usually appear in affected subjects in their 30s or 40s. The age at onset and the severity of polyQ disorders correlate with the size of the CAG repeat expansion. Typically, normal alleles of polyQ disease genes contain 10–30 CAG repeats, and mutant alleles contain 40–60 repeated units. However, repeats as short as 21 CAG tracts in the CACNA1A gene can cause SCA6, and expansions reaching more than 100 repeated units may occur in HD and SCA7 (Table 1). PolyQ diseases also share some pathogenic pathways leading to neurodegeneration. The mutant genes are ubiquitously expressed in the central nervous system (CNS) and peripheral tissues (3), but the pathology develops primarily in distinct brain areas characteristic of each disorder (Table 1). Interestingly, the expression of the mutant gene is usually not much higher in the brain areas mainly affected by the disease than in other brain areas and peripheral tissues. This result suggests that additional factors are required to stimulate pathogenesis.
Table 1. Brief characteristics of polyQ diseases.
| Disease | Ref. | Gene | Repeat tract length (normal/mutant) | Expression | Protein/size/function | Main site of pathogenesis | mutCAG-binding factors |
|---|---|---|---|---|---|---|---|
| DRPLA: dentatorubral-pallidoluysian atrophy | (288,289) | ATN1 | 6–36/49–88 | Ubiquitous | Atrophin-1/∼125 kDa/transcription regulation | Globus pallidus, subthalamic nucleus, dentate nucleus, white matter | ? |
| HD: Huntington's disease | (290) | HTT | 6–35/36–121 | Ubiquitous, high in neurons | Huntingtin/∼350 kDa/embryonic development, neurogenesis | Striatum, globus pallidus, substantia nigra | MBNL-1 SRSF6, U2AF65 |
| SBMA: spinal and bulbar muscular atrophy | (291) | AR | 6–37/38–62 | Ubiquitous | Androgen receptor/∼110 kDa/steroid-hormone activated transcription | Spinal anterior horn, facial nucleus, skeletal muscle | ? |
| SCA1: spinocerebellar ataxia type 1 | (292) | ATXN1 | 8–44/45–82 | Ubiquitous | Ataxin-1/∼85 kDa/transcription regulation, alternative splicing | Purkinje cells, dentate nucleus, brainstem, spinal cord | ? |
| SCA2: spinocerebellar ataxia type 2 | (293–295) | ATXN2 | 13–31/36–63 | Ubiquitous, high in Purkinje cells | Ataxin-2/∼140 kDa/mRNA maturation and translation, stress-granule formation, endocytosis, Ca-mediated signaling | Cerebellum (Purkinje cells), pons, inferior olives, thalamus, substantia nigra | ? |
| SCA3: spinocerebellar ataxia type 3 | (296) | ATXN3 | 12–44/60–84 | Ubiquitous | Ataxin-3/∼30 kDa/ubiquitin-mediated proteolysis, chromatin remodeling | Substantia nigra, cranial nerve motor nuclei, striatum | MBNL-1, U2AF65, NCL |
| SCA6: spinocerebellar ataxia type 6 | (297) | CACNA1A | 4–18/21–33 | Ubiquitous, but predominant in brain | Calcium channel alpha 1A subunit/∼30 kDa/calcium-dependent processes | Purkinje cells, dentate nucleus, inferior olive | ? |
| SCA7: spinocerebellar ataxia type 7 | (298) | ATXN7 | 4–34/37–306 | Ubiquitous, high in brain and testis | Ataxin-7/∼90 kDa/transcription regulation | Purkinje cells, pons, dentate nucleus, inferior olive, retina | ? |
| SCA17: spinocerebellar ataxia type 17 | (299,300) | TBP | 29–42/45–63 | Ubiquitous | TATA-binding protein/∼40 kDa/transcription regulation | Purkinje cells, striatum, cerebral cortex | ? |
Although the polyQ disorders remain incurable and only symptomatic treatment is offered to patients, many drugs are currently being tested to reverse the disease or to slow its progress. The most direct therapeutic strategy is silencing the casual gene expression. For some of these approaches, pre-clinical testing in rodent models is well advanced, and small-scale clinical testing is just beginning. In this review, we briefly refer to the pathogenesis of polyQ diseases and present the current status of therapeutic strategies, especially the oligonucleotide (ON) based techniques. We also discuss issues important for the design and further testing of various treatment approaches and conclude with our thoughts on their clinical perspectives.
PATHOGENESIS OF POLYQ DISEASES
In the traditional view of gene expression, the expanded CAG*CTG repeats present in mutant polyQ disease genes are transcribed into CAG repeats in RNA and translated into polyQ tracts in the encoded proteins (Figure 1). The abnormal protein–protein interactions triggered by mutant proteins were long thought to be the only factor responsible for the pathogenesis of polyQ diseases. In recent years, however, several examples of mutant transcript toxicity were demonstrated, and additional toxic RNA and protein entities were identified.
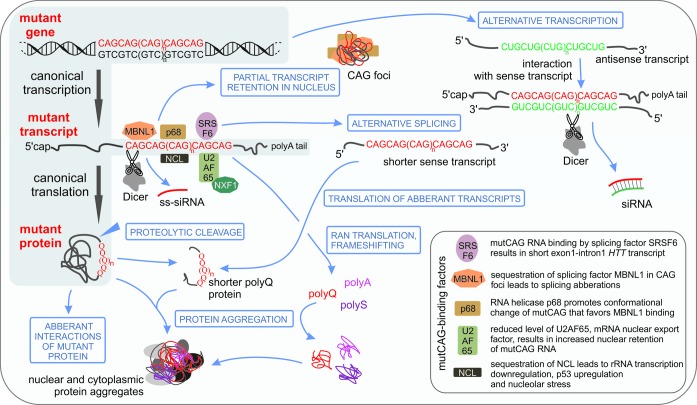
Figure 1.
Toxic entities in the pathogenesis of polyQ diseases. The main products of the mutant gene are the mutant transcript containing the expanded CAG repeats and the mutant protein containing the expanded polyQ tract. The hallmarks of primary toxic events are nuclear aggregates containing mutant RNAs (CAG foci) or mutant proteins (full length or fragments). The interactions and events leading to the production of additional toxic entities from the mutant transcript and mutant protein are indicated. The antisense transcription of the mutant polyQ gene may result in transcripts containing expanded CUG repeats. siRNAs generated by the RNase Dicer from the expanded tracts may interact with complementary sequences in the transcriptome and cause the downregulation of the expression of numerous genes. Aberrant translation may lead to peptides containing polyA, polyS or polyQ tracts. See the text for more details.
Protein toxicity
Proteins implicated in polyQ diseases differ in size and cellular function and are mainly involved in the regulation of transcription (Table 1) (4). The toxicity of mutant proteins in polyQ diseases is thought to result primarily from a gain-of-function mechanism triggered by protein misfolding and aggregation (5) (Figure 1). The nuclear ubiquitin-positive aggregates of mutant proteins are the hallmark of polyQ disorders (6–9). There is no clear answer yet as to whether neuronal intranuclear inclusions (NIIs) play an essential role in the pathogenesis or partially protect cells from cytotoxicity. Several studies on cellular and animal models showed no clear correlation between inclusion formation and cell death (10–12). Importantly, mutant protein fragments containing polyQ tracts are more toxic than full-length proteins and might be crucial for pathogenesis (13). Such fragments, which are mainly the products of cleavages by caspases, were identified for huntingtin, atrophin-1, ataxin-2, ataxin-3 and ataxin-7 (14–16). In agreement with these observations, more severe phenotypes are observed in animal models of polyQ diseases that express fragments of mutant genes than in models expressing full-length proteins (17).
Post-translational modifications are also significant contributors to polyQ protein toxicity (18). The phosphorylation of the polyQ protein may affect its proteolytic cleavage, as well as influence SUMOylation and ubiquitinylation. Interestingly, specific post-translational modifications can affect the toxicity of polyQ proteins in opposite ways, e.g. huntingtin phosphorylation at Ser421 results in a decrease in NII formation and reduces toxicity (19), whereas phosphorylation of ataxin-1 at Ser776 is required for SCA1 pathology (20). In the case of ataxin-1, the importance of Ser776 phosphorylation comes from the fact that it triggers a structural change in the region in which several functional signals are localized (21,22).
Not only new protein–protein interactions are triggered by mutant proteins, interactions characteristic of wild-type proteins can also be either enhanced or attenuated in case of mutant polyQ proteins (23). The beta-sheet conformation and coiled-coil structure are thought to mediate aggregation, as well as interactions with other proteins (24–28). Such altered protein–protein interactions were characterized very extensively for huntingtin (29–32), and they are thought to be crucial for the vulnerability of specific types of neurons to degeneration. The other examples include the following: ataxin-1 association with transcription factor RORalpha (33) and translational repressor Capicua (34) and ataxin-7 interactions with transcription coactivator complex STAGA (35,36). The formation of additional toxic protein products may be triggered by the expanded CAG repeat tracts in transcripts. These extra proteins include frame-shifted polypeptides (37–39) and products of repeat-associated non-AUG (RAN) translation (40,41) (Figure 1).
RNA toxicity
Growing evidence supports the importance of mutant transcript toxicity in the pathogenesis of polyQ diseases. Expanded CAG repeats in transcripts form hairpin structures containing periodic A-A mismatches (42–47). A similar hairpin structure, formed by CUG repeats (48), triggers the major pathogenic mechanism in myotonic dystrophy type 1 (DM1) (49), the disease caused by a CTG expansion in the 3' UTR of the DMPK gene (50). The hallmark of CUG repeat toxicity is the formation of nuclear foci by mutant transcripts and sequestered MBNL1 protein (51). Ribonucleoprotein foci formation and MBNL1-dependent deregulation of alternative splicing were also observed in HD and SCA3 cells (52). The toxicity caused by expanded CAG repeat RNA was demonstrated using genetic constructs containing mutant CAG repeat tracts expressed in different model organisms (53). Experiments performed in Drosophila compared the effects of transcripts that were translated with those that were not and contained pure or CAA-interrupted CAG repeats encoding polyQ tracts (54–56). Significant toxicity was reported for translated and untranslated CAG repeat tracts, but it was not observed for untranslated CAA-interrupted tracts, which do not form stable hairpin structures (57). Pathogenic features were also observed in a transgenic mouse model in which the expression of an expanded untranslated CAG repeat tract was directed to muscle (58). A comparison of two HD mouse models, which contained different patterns of CAA-interrupted CAG repeat tracts, supported the contribution of CAG RNA toxicity to the pathogenesis of polyQ disorders (59). Using a Drosophila SCA3 model and a HD mouse model, the involvement of the NXF1/U2AF65 RNA export pathway in RNA-mediated toxicity was demonstrated (60). The interaction of mutant CAG repeats with nucleolin was shown to induce nucleolar stress, leading to apoptosis in Drosophila and human cellular models of SCA3 as well as in HD mouse model (61). Helicase p68 was also shown to colocalize with expanded CAG repeats and increase MBNL1 binding to mutant transcripts (62). Moreover, splicing factor SRSF6 was reported to interact with expanded tracts in HTT transcripts, which results in short HTT sense transcripts being translated into toxic peptides (63). Furthermore, other toxic RNA entities were identified: antisense transcripts (64) and short CAG repeat RNAs (65,66) (Figure 1).
THERAPEUTIC TARGETING OF MUTANT GENES AND THEIR EXPRESSION PRODUCTS—AVENUES FOR POLYQ DISEASES
Taking advantage of the fact that each polyQ disease is monogenic, a rational therapeutic strategy could be designed to lower the causative gene expression. As proof of the concept, the inducible expression of a mutant transgene was turned off in rodent models of HD and SCA1, and recovery from the disease could be seen (67–70). This result included the reversal of aggregate formation and improvement in the motor phenotype.
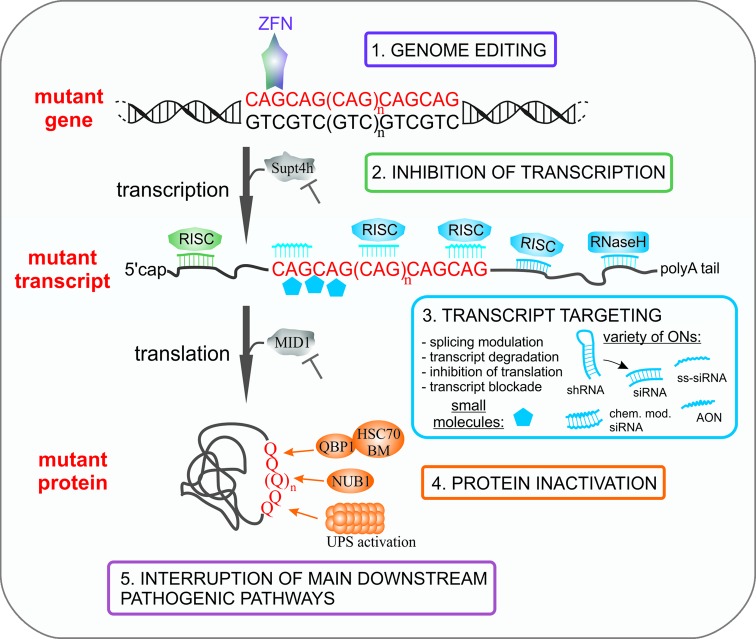
The possible ways to block the pathogenic pathway in polyQ diseases include the following: genome editing, transcription inhibition, transcript degradation or modification, translation arrest, and inhibition or degradation of toxic protein (Figure 2). To prevent the activation of pathogenic pathways, the mutation should be eliminated or blocked at the earliest step of mutant gene expression, i.e. in the DNA sequence. The technologies designed for DNA editing are developing rapidly (71–73) and were already tested for therapeutic use (74–79). In the case of polyQ diseases, gene editing tools can be directly targeted to expanded CAG sequences in genomic DNA. Zinc finger nucleases that recognize and cleave CAG repeat sequences were tested in mammalian cells (80). More recently, the CAG repeat-targeting zinc finger proteins (ZFPs) were used in a R6/2 HD mouse model to achieve efficient and specific repression of mutant HTT (81). The ZFP-mediated mutant transgene repression likely involved steric blockade formation for RNA polymerase during transcription and resulted in a substantial correction of the molecular and behavioral pathogenic hallmarks. In attempts to therapeutically regulate gene transcription, the promoter sequence may be targeted with short RNA duplexes (82–85). The feasibility of this approach is supported by growing evidence that core RNA interference (RNAi) proteins are present and operate in the nucleus (86–89). It is not clear whether the observed changes in the histone code are caused by short RNAs targeting DNA or RNA (sense or antisense) covering the promoter sequence, but clearly Argonaute proteins are involved in these processes (90,91). In the case of polyQ diseases, this strategy would likely result in silencing both alleles of the gene.
Figure 2.
Strategies directed at the elimination of toxic entities in polyQ diseases. The main steps of mutant gene expression at which therapeutic intervention may be applied are indicated. The possible interventions include: 1. editing the CAG expansion in the mutant gene, 2. inhibiting mutant gene transcription, 3. interacting of potential drugs with mutant transcript leading to its degradation or inhibition of translation, 4. inactivating the mutant protein by its degradation or blockage and 5. targeting the main downstream pathways. See the text for more details.
The prevailing strategy for the therapeutic regulation of gene expression is transcript targeting with ON-based tools (Figure 2). This regulation takes advantage of various mechanisms, which are determined by the ON type, its features such as structure, chemical modification and target localization and its dependence on specific cellular proteins (92–94).
Antisense oligonucleotides (AONs) may be designed to target pre-mRNA for its correction. Specific AONs can modulate splicing, e.g. exclusion of exon containing mutation or skipping transcript region encoding protein portion implicated in pathogenesis (95–97). This strategy was tested for SCA3 and HD in cell culture and initially assessed in vivo with AONs modified with 2'-O-methyl (2'OMe) and containing a phosphorothioate (PS) backbone. The AONs directed the exclusion of two ATXN3 exons that contained the CAG repeat tract while the reading frame and functional domains of ataxin-3 were preserved (98). In the HD model, the AONs induced the skipping of the exon 12 fragment of the HTT transcript that encodes a crucial cleavage site (99). This resulted in a protein resistant to caspase-6 cleavage, which is responsible for generating more toxic truncated huntingtin.
ON-based approaches that have been developed to promote specific mRNA cleavage include mainly RNase H-activating AONs and RNAi-based tools. AONs directing transcript degradation by RNase H now constitute the majority of ON-based drugs in clinical trials (100). RNase H is expressed at relatively low levels, localizes to the nucleus and cytoplasm and mediates the cleavage of the RNA in an RNA–DNA heteroduplex (101). In mammalian cells, the 15–20 nt long chemically modified AONs are used for RNase H1 activation, and the cleavage site is located between the 8th and 12th nucleotides from the 3' DNA terminus of the heteroduplex substrate (102–104).
RNAi has been widely explored for post-transcriptional gene silencing in functional assays as well as for therapeutic purposes. The RNAi effectors are short interfering RNAs (siRNAs), which are ∼21 nt duplexes (105,106). siRNAs share many features with microRNAs (miRNAs), which regulate the expression of the majority of genes in human cells (107). Both siRNAs and miRNAs are activated through the RNA-induced silencing complex (RISC), which contains the core Argonaute (AGO) protein (108,109). siRNAs are usually fully complementary duplexes containing 2-nt 3' overhangs. Also siRNAs typically show perfect complementarity to the target sequence and activate AGO2-directed transcript cleavage. This cleavage site is located opposite of the internucleotide bond between the 10th and 11th nucleotides from the 5' terminus of the siRNA strand (110). miRNAs are usually not fully complementary duplexes; they form mismatches in the interaction with target, and trigger mainly mRNA deadenylation and degradation or translation inhibition (111–113).
Another type of regulatory ONs are those that do not induce mRNA degradation but are transcript blockers and may affect translation, the local structure or interactions with proteins. One possibility is that translation arrest by AONs targets translation initiation, which results in the formation of a steric blockade to mRNA scanning (114,115). Translational inhibition may also be induced by miRNA-like siRNAs, in a RISC-dependent process, which was tested for polyQ diseases and will be described in detail in the next section (‘CAG repeat-targeting’). Moreover, the ON-based blockers may be designed to inhibit toxic interactions of specific proteins with mutant transcripts. Various tools targeting the expanded CUG repeat tract in DMPK transcripts were tested. Morpholino oligomers and AONs containing 2'OMe and a PS backbone or locked nucleic acid (LNA) residues caused the reversion of splicing defects and myotonia in DM1 mouse models (116–118). This reversion was achieved by blocking the expanded CUG repeat interactions with the splicing factor MBNL1.
In another strategy, structure-specific small-molecule drugs are designed or selected for targeting expanded repeats in transcripts (119). An extensive high-throughput search was performed for CUG repeat-binding drugs as potential therapeutics for DM1, taking advantage of the stable hairpin structure formed by the expanded CUG repeats (120–126). The crystal structures of the CUG and CAG repeat duplexes (127–130) were very useful in the design of this type of drugs. The bis-benzimidazole-derivatives were shown to efficiently bind to expanded CAG repeats in the transcript (131). More recently, the benzylguanidine-containing small molecule was described as an efficient inhibitor of the expanded CAG repeat RNA interaction with MBNL1, which resulted in the reversal of the alternative splicing defects in cell culture (132). Nevertheless, it seems that transcript targeting by small-molecule drugs could only inhibit RNA toxicity in polyQ disorders.
Another way of interfering with the expression of a mutant gene is targeting specific protein factors involved in the transcription and translation of the mutant entities. In the case of polyQ diseases, the transcription elongation factor Supt4h was shown to be required for the transcription of long CAG repeat tracts (133). The downregulation of Supt4h by siRNAs in cultured neuronal cells resulted in a decrease in the mutant huntingtin level. In another recent study, the translation regulatory protein complex containing the MID1 protein was demonstrated to stimulate the translation of CAG repeat expansions and was suggested to be a potential therapeutic target (134).
Some other therapeutic strategies, which will be only briefly mentioned here, are focused on the elimination of protein toxicity by targeting mutant proteins. These approaches include (i) inducing protein degradation and (ii) inhibiting processes of aggregation, proteolytic cleavage and post-translational modifications of mutant protein (135,136). In spite of the inconsistent data regarding the toxicity of polyQ aggregates, the therapeutic potential of decreasing polyQ aggregation was shown upon increasing chaperone activity (137) or using specific inhibitors (138–140). Small molecule drugs were screened for altering polyQ aggregation and enhancing the clearance pathway (141). Direct polyQ protein degradation may be enhanced by stimulating the ubiquitin–proteasome system (142) or autophagy (143). For the degradation of mutant huntingtin, the polyglutamine binding peptide 1 (QBP1) was fused with the binding motif of heat shock cognate protein 70 (HSC70) (144). The fusion protein was intrastriatally delivered in an rAAV vector and ameliorated the disease phenotype in the HD mouse model. Recently, the NUB1 factor was shown to lower the mutant huntingtin level by enhancing its proteasomal degradation (145). The stimulation of NUB1 was achieved by the treatment of cell culture with interferon. Structural motifs such as beta-sheet conformation of the polyQ tract or coiled-coil structure, which are common in polyQ proteins, could also be targeted to diminish polyQ pathology. In the case of particular polyQ proteins, specific domains or structural motifs could also be blocked to prevent abnormal interactions, e.g. blocking the AXH domain of ataxin-1 (146,147). Apart from targeting mutant transcript and protein in polyQ diseases, it can be advantageous to overexpress the normal allele of a mutant gene as was shown for ataxin-1-like (Atxn-1l) overexpression in SCA1 mouse models (148,149). Elevated Atxn1l levels suppressed SCA1 neuropathology likely by displacing mutant ataxin-1 from its native complex with Capicua (34).
Therapeutic approaches that focus on the inhibition of the main downstream pathogenic pathways in polyQ diseases are described elsewhere (150–154). These neuroprotective strategies include the correction of mitochondrial dysfunction, calcium dysregulation or neuroinflammation.
ON-BASED STRATEGIES FOR SILENCING MUTANT GENES CAUSING POLYQ DISEASES
Allele-selective versus non-allele-selective strategies
Therapeutic strategies for suppressing causative gene expression in polyQ diseases may be designed to target both alleles of the gene or only the mutant allele. In considering the requirement of allele selectivity in therapeutic gene downregulation, the cellular function of the wild-type protein is an important factor (Table 1). The key question is whether partial loss of the wild-type protein could be tolerated in adult life? Some light on this issue was shed by studies employing conditional knock-out mouse model of HTT. Using this model, an essential role of huntingtin, mainly in the CNS, was shown (155,156). Thus, decreasing the level of wild-type protein may pose a risk to patients. Moreover, the wild-type protein function may decrease the toxicity of the mutant protein, as demonstrated for SCA3, in a mechanism involving ubiquitin-mediated proteolysis (157), and for HD (158). In the study of HD mouse model, loss of wild-type Htt was shown to negatively affect neuropathology, motor dysfunction and lifespan of mice (158). These results could explain the disease phenotype in rare homozygous SCA3 and HD patients with two mutant alleles, whose phenotype is more severe than that of patients with a single mutant allele (159,160). However, the overexpression of wild-type ataxin-3 in SCA3 rat model was found not to mitigate the pathology (161).
In the next subsections, we first present the results of targeting a sequence that is present in both alleles of polyQ-related transcripts, which may be referred to as non-allele-selective targeting. Then, we continue with the description of single nucleotide polymorphism- (SNP-) and CAG repeat-targeting approaches, which are designed to preferentially silence the mutant allele.
Non-allele-selective targeting
The pre-clinical testing of various ON-based therapeutic tools, performed in rodent models of polyQ diseases, was the most extensive testing so far for the non-allele-selective strategy (Table 2, ‘Non-allele-selective targeting’). In the majority of cases, the mutant human transgene was targeted and silenced, taking advantage of sequence differences between mouse and human homologues. In these studies, much attention was paid to demonstrate the reversal of the molecular and behavioral pathogenic hallmarks.
Table 2. Testing ONs in therapeutic strategies against polyQ diseases.
| Disease/Gene | Silencing agent | Model | Main results | Ref. Year |
|---|---|---|---|---|
| Non-allele-selective targeting | ||||
| SCA1/ATXN1 | shRNA in AAV1 vector | Transgenic mouse model (human ataxin-1 with 82Q) | ICB injection resulted in a reduction in the accumulation of mutant ataxin-1 and an improvement of motor coordination | (162) 2004 |
| HD/HTT | shRNA in AAV1 vector | Transgenic mouse model (HD-N171–82Q) | Reduced inclusions in stratum, correction of disease phenotype after IST injection | (163) 2005 |
| HD/HTT | siRNA | Transgenic mouse model R6/2 | IVT infusion of siRNAs in the liposome complex reduced huntingtin inclusions, improved motor coordination and prolonged lifespan | (167) 2005 |
| HD/HTT | shRNA in AAV5 vector | Transgenic mouse model R6/1 | Reduced NIIs and disease phenotype ameliorated after IST injection | (164) 2005 |
| HD/HTT | shRNA in AV vector | Transgenic mouse model R6/2 and model induced by the AAV vector containing the human HTT fragment | IST injection resulted in a reduction in huntingtin inclusions | (165) 2007 |
| HD/HTT | Cholesterol-conjugated siRNA | Model induced by the AAV vector containing the human HTT fragment | Inhibition of neurodegeneration and improvement in motor coordination by IST injection | (168) 2007 |
| HD/HTT | miRNA-based shRNA in AAV2/1 vector | Knock-in mouse model CAG140 and a transgenic mouse model (N171–82Q) | miRNA-based constructs were found to be safer than typical shRNAs after IST injection and caused decreased striatal toxicity, improvement in motor coordination and a prolonged life span | (166,247) 2008 2009 |
| HD/HTT | shRNA in LV vector | Mouse and rat models induced by the injection of the LV vector containing a human HTT fragment (with 82Q) | The silencing of mHTT only or together with normal Htt resulted in a decrease of huntingtin inclusions after IST injections | (172) 2009 |
| SCA3/ATXN3 | shRNA in LV vector | Rat model induced by the injection of the LV vector containing human ATXN3 (with 72Q) | Silencing of both, normal endogenous Atxn3 and mutant transgene, by IST injections of vectors resulted in neuropathology reduction and did not cause any significant side effects | (161) 2010 |
| HD/HTT | siRNA | Non-human primates | Convection-enhanced delivery of siHtt by IST (putamen) injection resulted in the widespread distribution throughout the striatum and efficient huntingtin silencing | (175) 2011 |
| HD/HTT | miRNA-based shRNA in AAV2/1 vector | Non-human primates | HTT suppression after IST (putamen) injection did not cause any neuronal degeneration or behavioral abnormalities, and no immune response to AAV vector was detected | (174) 2011 |
| HD/HTT | shRNA in AAV vector | Non-human primates | The efficient downregulation of normal htt in the striatum after IST (putamen) injection, without adverse behavioral effects and histopathological changes in brain tissue | (176) 2012 |
| HD/HTT | AON | Transgenic mouse models: YAC128, BACHD, R6/2 and non-human primates | RNase H-activating AONs, delivered by IVT infusion, mediated HTT transgene silencing throughout the CNS and improvement in motor coordination | (169) 2012 |
| SCA1/ATXN1 | Artificial miRNA in AAV vector | B05 mouse model | The efficient knockdown of a transgene in the cerebellum, after ICB injections of vectors, improved neuropathology and rescued the behavioral phenotype | (149) 2013 |
| SCA3/ATXN3 | Artificial miRNA mimics in AAV2/1 vector | Transgenic mouse model (SCA3/MJD84.2) | Effective downregulation of ataxin-3 in the cerebellum after ICB injections of vectors, but no corrected phenotype was reported | (170,171) 2013 |
| HD/HTT | shRNA in AAV9 vector | Transgenic mouse models: BACHD and N171-82Q | Systemic delivery of viral vectors by intra-jugular vein injection reduced mHTT expression in brain and peripheral tissues, prevented inclusion formation in key brain regions and prevented severe weight loss | (274) 2014 |
| HD/HTT | miRNA-based shRNA in AAV2/1 vector | Transgenic mouse model YAC128 | Reduction of striatal huntingtin aggregates and improvements in behavioral deficits after IST injections | (173) 2014 |
| Polymorphism site-targeting | ||||
| SCA3/ATXN3 | siRNA, shRNA in AV vector | Cos-7 and HeLa cells transfected with plasmids containing a SNP site | Selective suppression of mutant ataxin-3 by targeting a SNP site located just after a CAG repeat tract | (183) 2003 |
| SCA3/ATXN3 | siRNA | HEK293T cells transfected with plasmids containing a SNP site | Inhibition of mutant ataxin-3 by targeting a SNP site located just after a CAG repeat tract | (184) 2004 |
| HD/HTT | siRNA | HeLa cells transfected with plasmids containing a SNP site | Five SNPs targeted, mismatch position 16 described as providing high discrimination | (190) 2006 |
| HD/HTT | siRNA | Patient-derived fibroblast cell lines | One SNP targeted and allele-selective siRNA selected | (191) 2008 |
| SCA3/ATXN3 | shRNA in LV vector | Rat model induced by the injection of the LV vector with human ATXN3 | Selective silencing of mutant transgene, after IST injections of vectors, resulted in a decrease of neuropathological abnormalities | (186) 2008 |
| HD/HTT | siRNA | Patient-derived fibroblast cell lines | Selective suppression of mutant huntingtin by targeting a polymorphic site of a 3-nt deletion | (301) 2009 |
| HD/HTT | siRNA | NIH3T cells transfected with plasmids containing a SNP site, patient-derived fibroblast cell lines | Three SNPs targeted and allele-selective siRNAs selected | (192) 2009 |
| HD/HTT | siRNA | Patient-derived fibroblast cell lines | Three SNPs targeted, five allele-selective siRNAs were estimated to be useful for three-quarters of HD patients | (193) 2009 |
| SCA7/ATXN7 | shRNA, pri-miRNA-based hairpins | HEK293T cells transfected with plasmids containing a SNP site | pri-miRNA mimics targeted mutant ATXN7 transcript efficiently and decreased mutant protein aggregation | (188) 2009 |
| HD/HTT | siRNA | HeLa cells transfected with plasmids containing a SNP site and patient-derived lymphoblast cell lines | Four SNPs targeted and allele-selective siRNAs selected | (194) 2010 |
| HD/HTT | AON | Patient-derived fibroblast cell lines, cultured primary neurons and BACHD and YAC128 mouse models | Effective and selective (∼5-fold) silencing of HTT transgene in a model containing a targeted SNP variant using AON modified with PS, MOE, S-cEt and delivered by IST injections | (195) 2011 |
| SCA3/ATXN3, HD/HTT, SCA1/ATXN1 | siRNA | Patient-derived fibroblast cell lines | For each SNP targeted allele-selective siRNAs were identified that preferentially inhibited the expression of the mutant allele | (179) 2012 |
| SCA3/ATXN3 | shRNA in LV vector | Transgenic mouse model (expressing truncated ataxin-3 with 69Q in Purkinje cells) | Reduction in Purkinje cell pathology and improvement in motor coordination after allele-selective transgene silencing using vectors delivered by ICB injections | (187) 2013 |
| HD/HTT | AON | Transgenic mouse model Hu97/18 | AON modified with PS, S-cEt and 2'MOE, showed >100-fold selectivity in the silencing of mutant HTT and efficient and selective huntingtin suppression in CNS after delivery by IVT infusion | (196) 2013 |
| CAG repeat-targeting | ||||
| SBMA/AR | dsRNA (81 bp) | Drosophila S2 cells and HEK293T cells | Non-allele-selective silencing of normal and mutant AR transgenes | (302) 2002 |
| SCA1/ATXN1 | shRNA in AAV vector | HEK293T or PC6–3 neuronal cells transfected with plasmids encoding normal and mutant ataxin-1 | shCAG, targeting CAG repeat region was used as a positive control of silencing, both alleles were silenced efficiently | (162) 2004 |
| HD/HTT, SCA3/ATXN3 | AON | Patient-derived fibroblast cell lines | Selective inhibition of mutant huntingtin and ataxin-3 with PNA- and LNA-modified AONs (selectivity fold > 6) | (201,303) 2009 |
| HD/HTT | AON | Patient-derived fibroblast cell lines | Selectivity of mutant HTT silencing even >6-fold for AONs with modifications LNA, cEt, carba-LNA, cEt-PS, MOE-2'F-PS | (202) 2010 |
| HD/HTT | siRNA | Patient-derived fibroblast cell lines | Selectivity of mutant HTT silencing > 40-fold for siRNA containing mismatches with the target sequence | (199) 2010 |
| HD/HTT | siRNA | Patient-derived fibroblast cell lines | Selective inhibition of mutant huntingtin using CAG/CUG duplexes with specific mutations | (200) 2011 |
| SCA3/ATXN3 | AON, siRNA | Patient-derived fibroblast cell lines | Selectivity of mutant ataxin-3 silencing 5-fold for PNA modified AON and 16-fold for siRNAs | (204) 2011 |
| HD/HTT | AON | Patient-derived fibroblast cell lines | AONs modified with LNA and containing oligospermine conjugate for efficient delivery | (304) 2011 |
| HD/HTT | AON | Patient-derived fibroblast cell lines | High allele selectivity was demonstrated for morpholino-modified AON | (179) 2012 |
| HD/HTT | ss-siRNA chemically modified | Patient-derived fibroblast cell lines and knock-in mouse model HdhQ150/Q7 | Selectivity of mutant HTT silencing >30-fold in cell culture, specific HTT silencing in mouse brain by ss-siRNAs delivered by IVT infusion | (206) 2012 |
| HD/HTT, SCA3/ATXN3 | siRNA with abasic substitutions | Patient-derived fibroblast cell lines | RNA duplexes containing centrally located abasic residues (and additional 2'OMe and PS modifications) showed allele selectivity in HTT and ATXN3 silencing | (305) 2013 |
| HD/HTT, SCA3/ATXN3 | siRNA with UNA substitutions | Patient-derived fibroblast cell lines | siRNAs containing single UNA-modified nucleotides in antisense strand showed >40 and >10-fold selectivity for mutant HTT and mutant ATXN3, respectively | (306) 2013 |
| SCA3/ATXN3 | ss-siRNA chemically modified | Patient-derived fibroblast cell lines | Selectivity of mutant ATXN3 silencing >35-fold for selected ON | (207) 2013 |
| HD/HTT | sd-siRNA | Patient-derived fibroblast cell lines | Guide strand-only siRNAs showed high selectivity, >60-fold, in HTT silencing | (208) 2013 |
The table is divided into the three parts, depending on the strategy type, which include the targeting of the following: the sequence harboring the polymorphism site, CAG expansion or another region of the transcript sequence. In each part of the table, the studies are placed in the order from the oldest studies to the most recent. The specific ON-based tools and models used in the studies are listed, together with brief descriptions of the main results. The selectivity of the silencing is based on the ratio of IC50 for inhibiting the normal allele versus IC50 for the mutant allele. For description of ON administration in vivo, the abbreviations are used: IST, intrastriatal; IVT, intraventricular; ITH, intrathecal; ICB, intracerebellar.
A study by the Davidson group in the SCA1 mouse model provided the first in vivo evidence for efficient gene silencing in a polyQ disorder (162). A few studies were performed using transgenic HD models and HTT transgene-specific RNAi triggers: shRNAs delivered in AAV vectors (163–166) or siRNAs (167,168). Locally delivered RNAi triggers caused a substantial reduction in transgene expression, which resulted in reduced neuropathy and positively affected the behavioral phenotypes of the mice. In a very extensive study, the mutant HTT transgene was silenced with AONs in three mouse models of HD (169). Treatment with a 2'-O-methoxyethyl- (2'MOE-) modified gapmer (Figure 3A) caused a reduction in the huntingtin level throughout the CNS up to ∼25% and resulted in the improvement of motor coordination. Recently, artificial miRNA mimics targeting the 3' UTR of the mutant ATXN3 transgene and delivered in AAV were tested in a transgenic SCA3 model (170,171). Constructs based on miRNA precursors, which are considered safer, were effective in transgene silencing in the cerebellum, but no correction of the phenotype was reported, likely due to the insufficient distribution of the inhibitor across the brain (170).
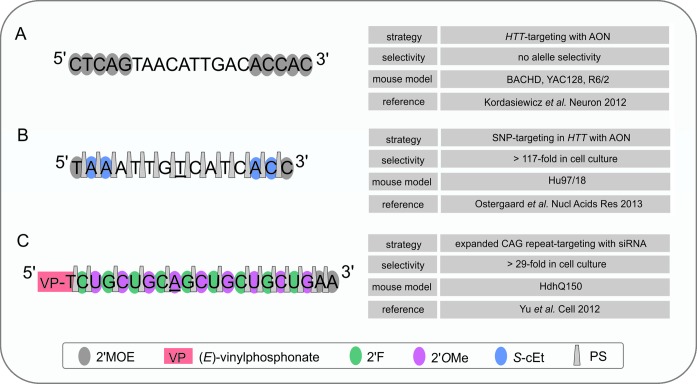
Figure 3.
The most advanced ON-based tools for silencing the HTT gene tested in HD mouse models. The sequences with a schematic representation of chemical modification patterns are given for three selected ON-based silencing tools (A, B, C) tested as potential therapeutics for HD. (A) represents the non-allele-selective approach for HTT silencing, while (B) and (C) are designed to preferentially target the mutant allele by the SNP-targeting strategy (B) or by CAG-targeting (C) (nucleotides essential for the selective activity of these ONs are underlined). (A) and (B) are AONs that activate RNase H for transcript degradation, while (C) is ss-siRNA that activates an RNAi-based mechanism. In the experiments in HD mouse models, all ONs were delivered by intraventricular infusion. See the text for more details.
In several experiments, the silencing of the normal endogenous HTT and ATXN3 was performed in order to test the non-allele-selective strategy straightforwardly. In rodent models, normal Htt or Atxn3 was inhibited usually together with the mutant human transgene, while only endogenous HTT was silenced in non-human primates. Studies performed in HD mice treated with RNAi triggers demonstrated that lowering the wild-type protein to 30–40% of control level was well tolerated (166,172,173). Similarly, in a study performed in SCA3 rat model, silencing of endogenous Atxn3 to ∼30%, together with mutant ATXN3, was shown to reduce neurodegeneration and suggested feasibility of inhibition of both alleles (161). Moreover, it was shown that silencing of endogenous Atxn3 in wild-type and SCA3 rats did not cause additional neuropathology (161). The RNAi-mediated silencing of endogenous HTT in non-human primates was described in three independent studies (174–176). A partial reduction in huntingtin after the injection of shRNA in the AAV vector to putamen was well tolerated, as shown by the analyses performed up to 6 weeks after treatment (174). siRNA delivery for a period of 4 weeks to the Rhesus brain by infusion pump, resulting in a 50% decrease in the huntingtin level, was shown not to cause any significant histopathological changes (175). shRNA delivered in the AAV vector caused a 45% reduction in HTT expression, while no histopathological changes occurred in the brain and no motor abnormalities were detected even 6 months after the treatment (176). HTT silencing was also performed in non-human primates by AON administration. Huntingtin downregulation up to 30% of the control level was achieved, but the long-term safety of this suppression was not analyzed (169). In spite of the lack of adverse effects on the phenotype observed after silencing the endogenous HTT in animal models, a serious concern remains regarding human cases in which longer-lasting gene inhibition will be required.
In another strategy, the silencing of both alleles of the causative gene is combined with the exogenous expression of a version of the normal allele that has been engineered to be resistant to the silencing agent (177). This gene-knockdown and replacement strategy was tested for SCA6 and SCA3 in cell culture models (178,179). In these studies, the expression of both alleles of the CACNA1A and ATXN3 endogenes was silenced in human cultured cells using siRNAs. For other disorders, such as retinitis pigmentosa, there are reports of testing the replacement strategy in vivo (180). In principle, with the use of this approach, a similar effect could be achieved as with the allele-selective approaches, i.e. the elimination of the mutant allele only. However, the replacement strategy is more demanding as both the silencing agent and the expression vector for the normal allele must be efficiently delivered, and the exogenous protein has to be expressed at an appropriate level.
SNP-targeting
Allele-selective silencing may be achieved by targeting ONs to transcript regions harboring SNP variants that distinguish between the normal and mutant alleles of the causative gene. It is advantageous if the SNP is highly heterozygous in the population and/or is associated with the expansion mutation. This configuration allows the design of a single ON inhibitor for a group of patients carrying such a variant. The therapeutic ON is designed to form a canonical base pair with the SNP variant present in the mutant allele, which triggers transcript cleavage and degradation, whereas the mismatch present in the interaction with the normal allele should render it resistant to cleavage.
Design rules for the SNP-targeting ONs that were described for siRNAs refer mainly to mismatch type and localization (181,182). The central positioning of a purine–purine mismatch is recommended in the interaction of the antisense strand of the siRNA with the normal transcript. However, the sequence flanking the SNP variant may strongly influence the silencing efficiency and selectivity. Therefore, the design rules are rather rough guidelines, and often a set of siRNAs needs to be tested to find efficient and allele-selective inhibitors.
In the majority of studies that used SNP-targeting to silence the polyQ disease genes, the ONs were designed to function using the RNAi pathway (Table 2, ‘Polymorphism site-targeting’). The first studies of this type were performed using cellular models of SCA3 (183,184). The targeted SNP is located at the 3' end of the CAG repeat tract, and its G and C variants are strongly associated with the normal and mutant ATXN3 alleles, respectively (43,185). The designed siRNAs included strong G-C pairing in the interaction with the mutant allele and a strong G-G mismatch in the interaction with the normal allele, which allowed for highly preferential mutant allele silencing (179,183,184). The SNP-targeting strategy was also tested using shRNA in rat and mouse models of SCA3 (186,187). shRNAs were delivered in LV vectors by injection to the striatum. Two months after the injections, a significant decrease in the formation of disease-associated inclusions and the correction of neuronal dysfunction was observed in the rat model (186). The substantial reversal of the neuropathological phenotype was also observed in a severely affected mouse model (187). The SNP site was also targeted in a cellular model of SCA7 (188). The exogenous ATXN7 transcripts containing different SNP variants were selectively targeted by siRNAs expressed from pri-miRNA-based hairpin constructs.
SNP-targeting was most extensively investigated for HD, for which approximately 10 different SNPs were targeted to silence mutant HTT expression. The relatively long sequence of the HTT gene gives the opportunity to identify many SNP variants in patients’ cells (189). High allele-discriminatory properties of siRNA were observed for two localizations of mismatch with the target: the central and 16th nt positions (counting from the 5'-end of the antisense strand) (179,190–194). In a comprehensive study, five allele-selective siRNAs were designed for common variants of three SNPs to cover approximately three-quarters of the HD population (193). Furthermore, RNase H-activating SNP-targeting AONs were described by the Hayden group (195) and the Seth group (196) as selective inhibitors of mutant HTT expression. In the first study, the most efficient AONs were PS substituted 19-mers with nine DNA residues in the gap and five S-constrained-ethyl (S-cEt) motifs on each wing (195). The AONs were injected into the striatum of animals from two HD mouse models, in which different SNP variants were identified in the transgenes. The expression of mutant human HTT was efficiently silenced only in the BACHD model, in which the targeted SNP variant was present and the selectivity of silencing was assessed to be ∼5-fold (195). More recently, the researchers from Isis Pharmaceuticals in collaboration with the Hayden group demonstrated the power of chemistry in designing an AON that targeted the same SNP variant with >100-fold selectivity in suppressing mutant huntingtin (196). Such a dramatic improvement in allele selectivity was achieved by iterative redesigning of the specific chemical modification pattern in AON, which included the base, backbone and sugar modifications. The best selectivity was achieved with a 15-mer containing 2'MOE, S-cEt and PS modifications (Figure 3B). This ON was tested in a new humanized mouse model of HD (197), and efficient HTT silencing across brain tissue was reported, together with high tolerability for AON administration.
CAG repeat-targeting
Allele-selective CAG repeat-targeting takes advantage of differences in the length of the repeat tracts in normal and mutant alleles. This approach is challenging because typical ON-based silencing tools are relatively short, and their binding sites are present not only in the mutant transcript but also in the normal allele and numerous other transcripts containing CAG repeat tracts. Furthermore, transcripts containing CUG repeats may be targeted by the sense strand of siRNA duplexes composed of CAG and CUG repeat strands. There are ∼200 and ∼100 mRNAs in human cells that contain repeat tracts composed of at least six units of CAG and CUG repeats, respectively (198). Due to this fact, it was thought that CAG repeat-targeting strategy cannot be selective enough to be used for therapeutic purposes. This pessimistic scenario was realized when RNAi triggers composed of fully complementary CAG/CUG repeats, shRNA (162) and siRNA (199,200) were used, which resulted in the non-selective downregulation of normal and mutant transcripts. These ONs activated the typical RNAi pathway, i.e. the mechanism of specific transcript cleavage, which turned out to be indiscriminatory for longer and shorter repeat tracts. However, recent studies have shown that high selectivity for mutant allele silencing can be achieved, and this approach is now regarded as feasible (Table 2, ‘CAG repeat-targeting’).
The allele-selective silencing of polyQ genes by repeat-targeting was achieved by different types of ONs, which reduced the mutant protein level without transcript degradation. The first inhibitors of this type were chemically modified translation blockers described by the Corey group (201). The most efficient inhibitors were CTG PNA oligomers conjugated with lysine residues, which showed >6-fold selectivity for mutant huntingtin and ataxin-3 inhibition. In further studies, similar effects were obtained with ONs modified with LNA, cEt, carba-LNA, MOE-2'F-PS, 2'OMe-PS (202) and morpholino oligomers (179). These extensively chemically modified oligomers are thought to act as efficient blockers for the translation of the mutant polyQ protein (Figure 4).
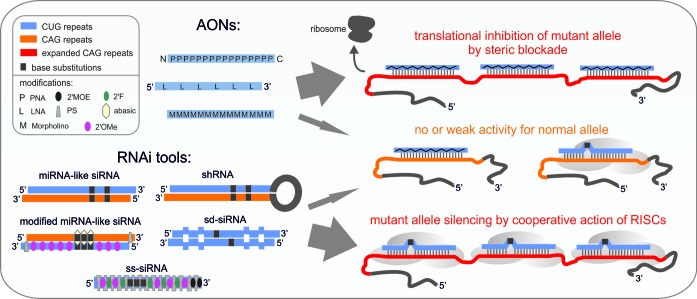
Figure 4.
The variety of CAG repeat-targeting ON-based tools showing preferential activity for the mutant allele. On the left side, various CAG repeat-targeting ONs and oligomers are presented. These molecules include a group of AONs, PNA and morpholino oligomers, as well as LNAs and RNAi-based tools including siRNAs, which are delivered as synthetic RNAs or expressed as shRNAs in cells. Specific chemical modifications, as well as the positions of base substitutions, are marked (see figure legend in the left upper corner). In some ONs, the CAG strand must have been included, while other ONs are composed of the CUG repeats only. On the right side, the interactions of ONs with CAG tracts in normal (marked in orange) and mutant alleles (marked in red) are presented. Only the binding of several ONs to the expanded CAG repeat tract in mRNA results in efficient gene silencing. The major mechanism is translational inhibition, which may occur by a steric blockade formed by AONs or by the RISC machinery recruited by siRNAs. See the text for more details.
Furthermore, various siRNAs targeting CAG repeat expansion were described, and some of them showed significant preference in the silencing of mutant alleles (Table 2, ‘CAG repeat-targeting’). The idea of Corey's group was to weaken the siRNA-target interaction by introducing base substitutions to the siRNA, resulting in one or more base mismatches with the target sequence (199). The substitutions were of different types, and in the most selective siRNAs, they were located in the central region. The same effect was obtained by our group by placing base substitutions in the 3' portions of the CUG repeat strands (200). In our design, the wobble base pairs were formed in the interaction of the siRNA antisense strand with its CAG repeat target. In both siRNA designs, the formation of mismatches with the target resulted in miRNA-like activity of these siRNAs (199,200,203). The CAG/CUG miRNA-like siRNAs were first described as an allele-selective tool for HD (199,200) and soon after for SCA3 (204).
Researchers from Isis Pharmaceuticals developed a pattern of chemical modifications that was introduced to single-stranded siRNAs (ss-siRNAs) to ensure their high stability and activity in vivo (205). The same pattern was used for the efficient inhibition of huntingtin expression by Corey et al. in collaboration with Isis (206). The ss-siRNAs composed of CUG repeats contained both base substitutions in the central region of ON, generating mismatches with targeted CAG repeats, as well as a specific pattern of 2'F, 2'OMe, PS and 2'MOE modifications (Figure 3C). Some ss-siRNAs showed high allele selectivity in silencing mutant huntingtin expression in HD fibroblasts and were successfully tested in a HD mouse model (206). More recently, similar ONs were tested for ATXN3 silencing in SCA3 fibroblasts, and allele selectivity was also achieved (207). Interestingly, the CAG repeat-targeting ss-siRNAs tested as RNAi-activating inhibitors of ataxin-3 translation were also shown to be RNAi-independent activators of the alternative splicing of the ATXN3 transcript, resulting in CAG repeat skipping.
Most recently, we have developed self-duplexing siRNAs (sd-siRNAs) as another type of allele-selective CAG repeat-targeting ONs for HD (208). The CUG repeat single strands to which one, two or three U to A or U to G substitutions were introduced in either the central or 3' region formed guide-strand-only duplexes functioning as miRNAs. In that study, the first repeat-targeting shRNA showing allele selectivity was also described (208).
CAG repeat-targeting siRNAs are not typical RNAi triggers, and their silencing mechanism, as well as basis for allele selectivity, needs further investigation. The involvement of the AGO2 and GW182 proteins in the silencing mechanism has already been established (203). The hairpin structure likely formed by the expanded CAG repeats in cells is not stable enough to resist the attack by RISC (47,199,200,209). The molecular basis of allele selectivity is thought to result from the cooperative action of multiple RISCs residing on expanded CAG repeats (203) (Figure 4). The single RISC bound to shorter repeats is easily stripped by translating ribosomes. This result also explains the minimal sequence-specific off-target effects in repeat-targeting, which was previously considered a serious issue.
CELLULAR AND RODENT MODELS USED TO EVALUATE THE ON-BASED THERAPEUTIC TOOLS
Cellular models of polyQ diseases are valuable in the initial screening of ONs. Fibroblast cells derived from polyQ disease patients are a very convenient model that is extensively used in evaluating the allele-selective approaches. These cells contain endogenous mutations, express both normal and mutant alleles and are relatively easy to culture and transfect. Even in testing the therapeutic reversal of the polyQ pathogenic hallmarks, fibroblasts may be a useful model, as some pathogenic events occur in these cells (52,210,211). Nevertheless, in recent trend, cultured neuronal cells are being used. These cells are more adequate, as they better represent brain cells, which should be mainly targeted in therapeutic treatment. The relevant neuronal cells may be derived from brain tissue of polyQ rodent models. Human neuronal cells may be obtained from patient fibroblasts by their reprogramming to induced pluripotent stem cells (iPSCs), followed by differentiation to neurons. The first example of the use of iPSC-derived neurons for modeling polyQ disorders was described in the study of SCA3 pathogenesis (212). In the most extensive study published to date, a set of iPSCs obtained from HD patient-derived fibroblast cells was generated and analyzed in the context of pathogenesis (213). Several studies on polyQ disorders were performed to characterize the iPSC models and the neuronal cells derived from them (214–219). With regard to testing therapeutic approaches, the cultured mouse neurons, as well as human neuronal cell models derived from embryonic stem cells and iPSCs, were used for the validation of specific polyQ protein-targeting strategy for HD (145). These cells were also used in cell-replacement strategies after their genetic correction and differentiation into neuronal cells (220–222). The use of neuronal cell culture models is anticipated to increase rapidly in the coming years, both for the studies of pathogenesis, as well as for testing therapeutic drugs.
The in vivo testing of ONs targeting polyQ diseases was performed mainly in transgenic rodent models. The experience gathered in testing ONs in rodent models of HD, SCA3 and SCA1 can be useful in the design of strategies for other polyQ diseases. There is a variety of models available that differ in their genetic and phenotypic characteristics and are used based on their validity for different approaches (17,223,224). PolyQ rodent models differ in the severity of their phenotypes, which depend on the specific features of the mutant exogene, mainly the length of the whole exogene and its repeated sequence, as well as its expression level. There are important issues to consider in testing the allele-selective silencing of causative genes in vivo. Due to sequence differences between rodent and human orthologs, the designed ONs are often specific only for human transgenes (186,187). A model best suited for testing the allele selectivity of targeting should contain two alleles of full-length human sequences, the normal version and the mutant with the pathogenic repeat tracts, as well as the relevant SNP variants. However, such a model might present a mild phenotype; therefore, the simultaneous use of a model with a faster and stronger phenotype may be required. Most of the full-length HD models contain only mutant transgenes and include knock-in types with chimeric sequences under mouse promoters or transgenic types with full-length human HTT (17). Recently, a transgenic mouse model of HD with two human HTT alleles (containing 18/97 CAG repeats and a human promoter) was created (197) and used in the successful testing of AON in an allele-selective strategy (196) (see the ‘SNP-targeting’ section). The development of new cellular and mouse models is underway for better evaluation of ON-based therapeutic strategies, and models of larger mammals are also under investigation (225–227).
COMPARISON OF DIFFERENT ON-BASED STRATEGIES
In the harnessing of ON-based tools for therapy, a number of important issues still need to be addressed. The high efficiency of ONs is required, which depends on their delivery, uptake and stability. Low doses and appropriate delivery are needed to prevent toxicity. Two strategies are being tested to ensure long-lasting activity of ONs: (i) incorporation of chemical modifications to increase ON stability and (ii) delivery of silencing reagents in genetic vectors (compared in Table 3). These strategies differ in the design of the relevant therapeutic tools and in the obstacles that need to be overcome in pre-clinical testing.
Table 3. Comparison of chemical and genetic approaches for ON delivery.
| Chemically modified ONs (AONs and siRNA) | Genetic vectors (viral) | |
|---|---|---|
| Design | • Variety of chemical modifications available | • Expression level can be determined by the choice of the promoter |
| • The chemical modification pattern must be carefully optimized for high stability, efficiency and lack of toxicity | • Heterogeneous cleavage of pri- or pre-miRNA-based constructs generates isomiRs, which is considered a drawback | |
| Neuronal tissue specificity | • Can be obtained by specific ligands or adaptors | • Can be obtained by vector tropism |
| Distribution in brain | • Free uptake by neuronal cells when delivered intra-CNS | • Brain transduction observed for some vectors in rodents and non-human primates but still less dispersed compared to synthetic ONs |
| Lifespan after delivery | • Much higher than for unmodified ONs but still transient | • Possibly long-term |
| • Require repetitive administration | • Appropriate for permanent treatment | |
| Safety | • Transient activity may be regarded as an advantage concerning the safety | • Dosage control is more problematic |
| • Direct control of dosage | • Possible mutagenesis and immunogenicity | |
| Other | • Requires large-scale chemical synthesis | • Difficulties in large-scale manufacturing |
Design
Various features determined by the nucleotide sequence of the ON are important for its activity. The extensive relevant information was gathered for siRNAs and used to establish rules for effective siRNA design (228). The guidelines for siRNAs involve their nucleotide composition, their duplex thermodynamics and the structure of the duplex ends (229–231). The stable structure formed by the siRNA guide strand or AON (232), as well as the stable structure of the target sequence (233–235), reduces the efficiency of transcript inhibition. However, some AONs were shown to be more sensitive to target structure than siRNAs (236), and finding active AONs required more effort, i.e. a larger set had to be tested. The siRNA hybridization with the target is facilitated by RISC, which contains helicase activity, but it is likely that the ability of AON to find the target sequence is also stimulated by yet unknown proteins (237).
Another aspect of ON design is the selection of the chemical modification pattern. The AONs require suitable chemical modifications for activity. Chemical modification not only increases the biological stability of the AON and the efficiency of its hybridization with the target sequence, but may also be critical for its effective delivery and low toxicity. These issues will be discussed in the following subsections. The variety of chemical modifications introduced into ON include internucleotide bonds (e.g. PS, boranophosphate), sugar units (e.g. 2'OMe, 2'OMOE, 2'F and LNA) and nucleobases (e.g. 5-bromouracil and diaminopurine). PS linkages were applied to the first generation of RNase H-activating AONs, and they improved their cellular stability and increased their binding to serum proteins in vivo. However, the efficiency of PS-modified AONs was not satisfactory due to decreased affinity for the target sequence and the need to use high doses, which often resulted in toxic effects. In the second generation of AONs, the modifications of the ribose moiety increased AON stability and its binding to the target sequence, which resulted in higher efficiency of those inhibitors (238,239). LNA modification, the representative of bicyclic sugar modifications, improves nuclease resistance, decreases immune stimulation and strongly increases binding affinity. The development of 2'MOE-modified AONs by Isis was crucial for the improvement in potency in the second-generation AONs. Furthermore, the efficiency of siRNAs was increased by their appropriate chemical modifications (240–242). The central regions of RNaseH1-activating AONs and AGO2-activating siRNAs are generally less extensively modified, as cleavages triggered by these enzymes are very sensitive to the modification status of the central nucleotides. PS, 2'F and 2'OMe modifications are well tolerated by RNase H and AGO (Figure 3). The unmodified siRNAs outperform first-generation AONs in their efficiency in cell culture experiments, but the second-generation AONs are as active as siRNAs (237).
Some AON modifications are still more drastic and have the sugar-phosphate backbone replaced by a peptide (PNA) or morpholino moiety. These oligomers are very resistant to cellular nucleases, and their activity results from strong binding to a complementary sequence in the transcript. They do not trigger transcript cleavage but form a blockade for translation or redirect splicing. Thus far, testing this alternative splicing modulation by morpholino oligomers is well advanced for disorders such as Duchenne muscular dystrophy (243).
The gene silencing reagents are very often expressed in cells from a genetic vector. However, this approach is applicable to RNAi tools only, as they are efficient as non-chemically modified RNAs. These gene inhibitors include shRNAs and shmiRs (pri-miRNA-based constructs) (244,245), which are processed in cells to short RNAi triggers by the miRNA biogenesis machinery. Typical shRNA were found more efficient than shmiRs (246), but they also exerted more toxic effects than shmiRs, what was directly compared in vivo in HD mouse model (247). Several types of viral vectors were used to deliver shRNA and shmiR expression cassettes to target polyQ-related gene expression: AAV, AV and LV (Table 2).
The increased understanding of the potential roles of mutant transcripts in the pathogenesis of polyQ diseases prompts researchers to develop therapeutic strategies leading to the elimination of toxic RNAs. AGO- and RNase-H-activating ONs (compared in Table 4) are typically designed to induce transcript degradation. In these cases, the level of toxic RNA and protein components are reduced in the cell. However, some of the CAG repeat-targeting reagents were described to decrease the mutant protein only, i.e. PNA and morpholino oligomers, as well as RNA duplexes acting as miRNAs. These reagents, however, bind to repeat tracts in the mRNA to induce translational inhibition, and this mechanism may diminish RNA toxicity, e.g. by competing with specific cellular proteins for the interaction with the CAG repeat expanded tract.
Table 4. Comparison of the mechanisms of gene expression downregulation in terms of their therapeutic utility.
| AONs (RNAse H-activating) | RNAi | |
|---|---|---|
| Activity | • Nuclear and cytoplasmic | • Mainly cytoplasmic |
| Design | • Chemical modifications required | • Active in vivo as chemically modified or in genetic vectors |
| • Can be more extensively chemically modified than siRNA | • More limited chemical modifications can be introduced | |
| • Single-stranded (smaller) | • Mainly double-stranded (larger molecules) | |
| • Relatively large set of ONs must be tested to find active ones | • Design rules are well established | |
| Efficiency | • Lower or equal to that of siRNAs in cell culture, depending on the chemical modification pattern used | • Very high efficiency in cell culture, due to typical catalytic mode of action |
| • Better efficiency in vivo than siRNAs | • Moderate sensitivity for target structure | |
| • Might be more sensitive for target structure than siRNAs | • RISC complex facilitates finding targets and hybridizing | |
| • It is unknown whether it finds and hybridizes to targets unassisted | ||
| Off-target (sequence specific) | • Moderate risk | • High risk as activity is based on ‘seed’ pairing |
| • Improvement in siRNA design overcame some off-targeting | ||
| • Heterogeneous cleavages in shRNAs and shmiRs generate unwanted siRNAs | ||
| Off-target (non-sequence specific) | • High risk of immunostimulatory responses through interactions with toll-like receptors (TLRs) | • Possible disturbance of the endogenous miRNA pathway |
| • Activation of pathways by dsRNAs | ||
| Strategy development | • Generally more advanced in clinical trials, as developed since the late 1970s | • Developed since 2000s |
| • For polyQ disorders tested in last few years | • Extensively tested in cell culture and rodent models of polyQ diseases |
Off-target effects
The required efficiency of gene inhibition is usually achieved by an excess of ON over the target. This approach raises specificity problems because in human transcriptomes, there are multiple sequences that are partially complementary to the ON sequence. Some of these unintended interactions are productive, causing sequence-specific off-target effects. The AONs are considered more selective than RNAi tools. The RNase H cleavage mechanism is very sensitive to base mismatches, and only one, two or at most three mismatches are tolerated. The AGO-mediated silencing mechanism is more tolerant for siRNA-target mismatches, as these are typically present in miRNA–mRNA interactions. For most miRNAs, base-pairing in the seed region (nucleotides 2–8 from the 5' end of short RNA) is crucial for activity (248,249), and therefore, complementarity in this region is mainly used for predictions of potential off-targeting by siRNA (250,251). The siRNA designs aimed at reducing off-target effects (252,253) included shortening the passenger strand (254,255) and introducing chemical modifications into the sense strand of the siRNA to render it inactive.
Another issue is off-targeting, which is independent of the ON hybridization to transcripts. The competition of exogenous RNAi triggers with endogenous miRNAs for RISC charging may cause alterations in natural miRNA regulatory networks (256,257). Moreover, ONs of various types may interact with other cellular and extracellular proteins and induce toxic effects. Many proteins are involved in the activation of the immunological response to exogenous nucleic acids (258,259). Furthermore, specific immunostimulatory sequence motifs were identified, which should be avoided in the ON design (260–263). This issue is the subject of several reviews (264–266), and relevant information is gathered in the RNAimmuno database (263).
Delivery
The efficient delivery of ONs can be facilitated by their specific chemical modification, conjugation or formulation and by using an appropriate administration route. Most of the described pre-clinical testing of the ON-based approaches for polyQ diseases focused on the local delivery of inhibitors to the brain. The tests were performed in rodent models of polyQ diseases and in non-human primates. The viral vectors containing shRNA-encoding cassettes, and likewise, synthetic siRNAs or AONs, were either locally injected or infused into CNS (Table 2). The specific injection site was typically the brain region primarily affected by the disease, i.e. the cerebellum in the case of SCA1 and the striatum in HD models. The brain distribution of the injected viral vectors showed relatively high transduction efficiency, although the vector was not dispersed widely and equally throughout the tissue (163,164). In several recent studies, chemically modified AON and ss-siRNA inhibitors without any formulation were infused into cerebrospinal fluid (CSF), which resulted in their more widespread and uniform distribution across the brain (169,196,206). In spite of this success, further research was conducted to enhance the delivery of various types of inhibitors directly to the brain (267,268,269).
Regarding the less invasive systemic or intrathecal delivery, the improved pharmacokinetic properties of ONs may prevent their rapid excretion and increase their uptake. The general barriers after systemic administration are: (i) clearance from blood via the kidney, (ii) degradation by nucleases in plasma, (iii) crossing the capillary endothelium (extravasation to other organs than liver, kidney and spleen), (iv) cellular uptake from the extracellular matrix, (v) escape from endosomes, (vi) reaching a specific cell compartment (270,271). Crossing the blood–brain barrier is the major hurdle for the ONs to be delivered systemically in the case of CNS diseases. In several studies, the methods were tested for the efficient delivery of the ONs to the brain by systemic administration, with the promising example of delivery in exosomes (272,273). Recently, the intra-jugular vein delivery of AAV9 vectors for HTT silencing was described by the McBride group (274). In this report, the effective transduction of multiple brain regions and peripheral tissues was demonstrated. This treatment resulted in the reduction of neuropathology in HD mouse models, although it did not prevent motor deficits.
CLINICAL ASPECTS
Still, some limitations must be overcome for therapeutic use of ONs. They are mainly due to shortcomings in the efficiency and safety of ONs in clinical trials. This comprises problems regarding efficient ON delivery to targeted tissues without causing any adverse effects.
There are additional issues to be considered while taking ONs from pre-clinical testing to human trails for the development of treatments available to patients. The basic evaluation of ON potency as a therapeutic agent is the analysis of targeted gene downregulation. For a more complete assessment of ON properties in vivo, specific analyses need to be performed to observe the potential reversal of selected markers of the molecular pathogenesis of polyQ diseases. From a clinical point of view, it is challenging to develop methods for the validation of therapeutic benefits that would not be harmful and could be used in human trials (275,276).
The question remains whether it is sufficient to suppress mutant gene expression locally in the regions where the disease primarily develops or whether it is necessary to target either the whole CNS or also the peripheral tissues. As significant dysfunctions in the periphery were reported in patients and in mouse models of HD and other polyQ disorders (277–282), systemic delivery seems to be the best option, provided it also successfully delivers ONs to the CNS.
Another important question is at what stage of disease is it necessary to intervene and silence the gene expression so the pathology will completely, or at least partially, reverse? Treatment at the late stage of disease poses the risk that some toxic events will become irreversible. Results from an inducible mouse model of SCA1 indicate that the pathogenic effects of the mutation in ataxin-1 accumulate starting from postnatal development (33). The recovery from disease was observed in several inducible rodent models when the mutant transgene was turned off at an early or middle stage of the polyQ disease (67,68,70,283,284). Nevertheless, full recovery was not achieved when the transgene expression was halted at a late stage of the disease (68,284). Similar results were obtained with ON-based silencing of the mutant transgenes in rodents (Table 2), but partial recovery even from a severe phenotype was reported (187). Considering the human lifespan, it is preferable that a single administration of a drug provides long-term mutant gene silencing. In this context, the result obtained by Cleveland et al. is very promising. Using HD mouse models, the authors demonstrated that AONs intraventricularly infused into the CSF caused phenotypic recovery lasting much longer than mutant gene downregulation (169). This finding, named ‘huntingtin holiday’ (285), is one of the major breakthroughs in HD research and a very good message for HD patients.
CONCLUSIONS AND FUTURE PERSPECTIVES
In this review, we gathered the existing information and discussed the progress of research aimed at developing therapeutic approaches for polyQ diseases. We presented and compared the two most advanced gene inhibition strategies, antisense and RNAi, which show many similarities, including their therapeutic potential.
The AONs directing transcript degradation by RNase H have been explored as potential drugs since the late 1970s, whereas RNAi has been used for post-transcriptional gene silencing since the early 2000s. Therefore, AON tools are more advanced in pre-clinical and clinical testing, but the experience gathered in testing AONs is in many respects useful for the development of RNAi-based strategies (286). This advantage refers to the general administration, distribution, metabolism and excretion characteristics of ONs, which can be relevant in many ways for both the AONs and siRNAs (259,287). As a result, rapid progress in advancing siRNAs to clinical trials is expected in coming years.
At this point, it is difficult to say which ON-based therapeutic strategy is best suited for polyQ diseases. The existing strategies offer great opportunities for therapy but also have limitations. The allele-selective SNP- and CAG repeat-targeting strategies (compared in Table 5) are regarded as more safe. CAG repeat-targeting strategy, which was considered some years ago a ‘mission impossible’, turned out to be not only feasible but also very promising according to the first in vivo studies in mouse models of HD (206). However, a full assessment of the CAG repeat-targeting strategy needs to be accomplished. The potential advantage of this strategy is its universality, as one drug could possibly be used to treat most of the polyQ disorders.
Table 5. Comparison of allele-selective approaches for polyQ diseases.
| SNP-targeting | CAG-targeting | |
|---|---|---|
| Available tools | • siRNAs/shRNAs | • siRNAs/shRNAs (miRNA-like, sd-siRNA, ss-siRNA) |
| • AONs (RNase H – activating) | • AONs (blockers of translation, splicing modifiers) | |
| Design | • Requires the identification of SNP variants | • The RNAi-based approach requires the positioning of base mismatches with target |
| Efficiency of silencing | • May be influenced by the sequence in the region of the SNP variant | • Comparable with specific sequence-targeting |
| • Based on multiple binding sites in the expanded repeat tract | ||
| Selectivity of silencing | • May depend on the types of SNP variants present in the mutant and normal alleles | • Depends on the difference between the repeat tract length in the normal and mutant alleles |
| Off-targeting | • Standard risk | • Risk of targeting numerous transcripts containing normal CAG repeat tracts |
The ON-based strategies are more advanced than other strategies capable of interfering with mutant gene expression. ONs may be designed for any transcript and may function by many mechanisms. Other strategies, such as genome editing, that are aimed at repairing a mutant gene or preventing its transcription are very attractive but still far from clinical applications. The clinical application seems to be still more distant for the cell therapy of polyQ diseases.
FUNDING
Ministry of Science and Higher Education [N N301 569340, N N302 633240 and IP2012032472]; European Regional Development Fund within Innovative Economy Programme [POIG.01.03.01-30-098/08]; National Science Centre [2012/06/A/NZ1/00094]. Source of open access funding: Polish Ministry of Science and Higher Education.
Conflict of interest statement. None declared.
REFERENCES
- 1.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Moseley M.L., Zu T., Ikeda Y., Gao W., Mosemiller A.K., Daughters R.S., Chen G., Weatherspoon M.R., Clark H.B., Ebner T.J., et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 3.Sharp A.H., Loev S.J., Schilling G., Li S.-H., Li X.-J., Bao J., Wagster M. V, Kotzuk J.A., Steiner J.P., Lo A., et al. Widespread expression of Huntington's disease gene (IT15) protein product. Neuron. 1995;14:1065–1074. doi: 10.1016/0896-6273(95)90345-3. [DOI] [PubMed] [Google Scholar]
- 4.Riley B.E., Orr H.T. Polyglutamine neurodegenerative diseases and regulation of transcription: assembling the puzzle. Genes Dev. 2006;20:2183–2192. doi: 10.1101/gad.1436506. [DOI] [PubMed] [Google Scholar]
- 5.Williams A.J., Paulson H.L. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 2008;31:521–528. doi: 10.1016/j.tins.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies S.W., Turmaine M., Cozens B.A., DiFiglia M., Sharp A.H., Ross C.A., Scherzinger E., Wanker E.E., Mangiarini L., Bates G.P. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 7.DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 8.Donaldson K.M., Li W., Ching K.A., Batalov S., Tsai C.C., Joazeiro C.A. Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8892–8897. doi: 10.1073/pnas.1530212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada M., Sato T., Tsuji S., Takahashi H. CAG repeat disorder models and human neuropathology: similarities and differences. Acta Neuropathol. 2008;115:71–86. doi: 10.1007/s00401-007-0287-5. [DOI] [PubMed] [Google Scholar]
- 10.Saudou F., Finkbeiner S., Devys D., Greenberg M.E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 11.Huynh D.P., Figueroa K., Hoang N., Pulst S.M. Nuclear localization or inclusion body formation of ataxin-2 are not necessary for SCA2 pathogenesis in mouse or human. Nat. Genet. 2000;26:44–50. doi: 10.1038/79162. [DOI] [PubMed] [Google Scholar]
- 12.Tarlac V., Turnbull V., Stefani D., Kelly L., Walsh R., Storey E. Inclusion formation by ataxins -1, -2, -3, and -7. Int. J. Neurosci. 2007;117:1289–1314. doi: 10.1080/00207450600936668. [DOI] [PubMed] [Google Scholar]
- 13.Graham R.K., Deng Y., Slow E.J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 14.Wellington C.L., Ellerby L.M., Hackam A.S., Margolis R.L., Trifiro M.A., Singaraja R., McCutcheon K., Salvesen G.S., Propp S.S., Bromm M., et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J. Biol. Chem. 1998;273:9158–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- 15.Kordasiewicz H.B., Thompson R.M., Clark H.B., Gomez C.M. C-termini of P/Q-type Ca2+ channel alpha1A subunits translocate to nuclei and promote polyglutamine-mediated toxicity. Hum. Mol. Genet. 2006;15:1587–1599. doi: 10.1093/hmg/ddl080. [DOI] [PubMed] [Google Scholar]
- 16.Young J.E., Gouw L., Propp S., Sopher B.L., Taylor J., Lin A., Hermel E., Logvinova A., Chen S.F., Chen S., et al. Proteolytic cleavage of ataxin-7 by caspase-7 modulates cellular toxicity and transcriptional dysregulation. J. Biol. Chem. 2007;282:30150–30160. doi: 10.1074/jbc.M705265200. [DOI] [PubMed] [Google Scholar]
- 17.Pouladi M.A., Morton A.J., Hayden M.R. Choosing an animal model for the study of Huntington's disease. Nat. Rev. Neurosci. 2013;14:708–721. doi: 10.1038/nrn3570. [DOI] [PubMed] [Google Scholar]
- 18.Pennuto M., Palazzolo I., Poletti A. Post-translational modifications of expanded polyglutamine proteins: impact on neurotoxicity. Hum. Mol. Genet. 2009;18:R40–R47. doi: 10.1093/hmg/ddn412. [DOI] [PubMed] [Google Scholar]
- 19.Humbert S., Bryson E.A., Cordelieres F.P., Connors N.C., Datta S.R., Finkbeiner S., Greenberg M.E., Saudou F. The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves Huntingtin phosphorylation by Akt. Dev. Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- 20.Chen H.K., Fernandez-Funez P., Acevedo S.F., Lam Y.C., Kaytor M.D., Fernandez M.H., Aitken A., Skoulakis E.M., Orr H.T., Botas J., et al. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- 21.De Chiara C., Menon R.P., Strom M., Gibson T.J., Pastore A. Phosphorylation of S776 and 14–3–3 binding modulate ataxin-1 interaction with splicing factors. PLoS One. 2009;4:e8372. doi: 10.1371/journal.pone.0008372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orr H.T. SCA1-phosphorylation, a regulator of Ataxin-1 function and pathogenesis. Prog. Neurobiol. 2012;99:179–85. doi: 10.1016/j.pneurobio.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr H.T. Polyglutamine neurodegeneration: expanded glutamines enhance native functions. Curr. Opin. Genet. Dev. 2012;22:251–255. doi: 10.1016/j.gde.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G.P., Davies S.W., Lehrach H., Wanker E.E. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 25.Poirier M.A., Jiang H., Ross C.A. A structure-based analysis of huntingtin mutant polyglutamine aggregation and toxicity: evidence for a compact beta-sheet structure. Hum. Mol. Genet. 2005;14:765–774. doi: 10.1093/hmg/ddi071. [DOI] [PubMed] [Google Scholar]
- 26.Nagai Y., Inui T., Popiel H.A., Fujikake N., Hasegawa K., Urade Y., Goto Y., Naiki H., Toda T. A toxic monomeric conformer of the polyglutamine protein. Nat. Struct. Mol. Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 27.Fiumara F., Fioriti L., Kandel E.R., Hendrickson W.A. Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell. 2010;143:1121–1135. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrakis S., Schaefer M.H., Wanker E.E., Andrade-Navarro M.A. Aggregation of polyQ-extended proteins is promoted by interaction with their natural coiled-coil partners. Bioessays. 2013;35:503–507. doi: 10.1002/bies.201300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bae B.-I., Hara M.R., Cascio M.B., Wellington C.L., Hayden M.R., Ross C.A., Ha H.C., Li X.-J., Snyder S.H., Sawa A. Mutant huntingtin: nuclear translocation and cytotoxicity mediated by GAPDH. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3405–3409. doi: 10.1073/pnas.0511316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaltenbach L.S., Romero E., Becklin R.R., Chettier R., Bell R., Phansalkar A., Strand A., Torcassi C., Savage J., Hurlburt A., et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savas J.N., Makusky A., Ottosen S., Baillat D., Then F., Krainc D., Shiekhattar R., Markey S.P., Tanese N. Huntington's disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10820–10825. doi: 10.1073/pnas.0800658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratovitski T., Chighladze E., Arbez N., Boronina T., Herbrich S., Cole R.N., Ross C.A. Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle. 2012;11:2006–2021. doi: 10.4161/cc.20423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serra H.G., Duvick L., Zu T., Carlson K., Stevens S., Jorgensen N., Lysholm A., Burright E., Zoghbi H.Y., Clark H.B., et al. RORα-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell. 2006;127:697–708. doi: 10.1016/j.cell.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 34.Lam Y.C., Bowman A.B., Jafar-Nejad P., Lim J., Richman R., Fryer J.D., Hyun E.D., Duvick L.A., Orr H.T., Botas J., et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 35.Palhan V.B., Chen S., Peng G.-H., Tjernberg A., Gamper A.M., Fan Y., Chait B.T., La Spada A.R., Roeder R.G. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helmlinger D., Hardy S., Abou-Sleymane G., Eberlin A., Bowman A.B., Gansmüller A., Picaud S., Zoghbi H.Y., Trottier Y., Tora L., et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toulouse A., Au-Yeung F., Gaspar C., Roussel J., Dion P., Rouleau G.A. Ribosomal frameshifting on MJD-1 transcripts with long CAG tracts. Hum. Mol. Genet. 2005;14:2649–2660. doi: 10.1093/hmg/ddi299. [DOI] [PubMed] [Google Scholar]
- 38.Davies J.E., Rubinsztein D.C. Polyalanine and polyserine frameshift products in Huntington's disease. J. Med. Genet. 2006;43:893–896. doi: 10.1136/jmg.2006.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stochmanski S.J., Therrien M., Laganiere J., Rochefort D., Laurent S., Karemera L., Gaudet R., Vyboh K., Van Meyel D.J., Di Cristo G., et al. Expanded ATXN3 frameshifting events are toxic in Drosophila and mammalian neuron models. Hum. Mol. Genet. 2012;21:2211–2218. doi: 10.1093/hmg/dds036. [DOI] [PubMed] [Google Scholar]
- 40.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A., et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U.S.A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cleary J.D., Ranum L.P. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum. Mol. Genet. 2013;22:R45–R51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobczak K., de Mezer M., Michlewski G., Krol J., Krzyzosiak W.J. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michlewski G., Krzyzosiak W.J. Molecular architecture of CAG repeats in human disease related transcripts. J. Mol. Biol. 2004;340:665–679. doi: 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 44.Sobczak K., Krzyzosiak W.J. Patterns of CAG repeat interruptions in SCA1 and SCA2 genes in relation to repeat instability. Hum. Mutat. 2004;24:236–247. doi: 10.1002/humu.20075. [DOI] [PubMed] [Google Scholar]
- 45.Sobczak K., Krzyzosiak W.J. Imperfect CAG repeats form diverse structures in SCA1 transcripts. J. Biol. Chem. 2004;279:41563–41572. doi: 10.1074/jbc.M405130200. [DOI] [PubMed] [Google Scholar]
- 46.Sobczak K., Michlewski G., de Mezer M., Kierzek E., Krol J., Olejniczak M., Kierzek R., Krzyzosiak W.J. Structural diversity of triplet repeat RNAs. J. Biol. Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Mezer M., Wojciechowska M., Napierala M., Sobczak K., Krzyzosiak W.J. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011;39:3852–3863. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Napierala M., Krzyzosiak W.J. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J. Biol. Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 49.Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T., et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell. 1992;69:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 51.Wojciechowska M., Krzyzosiak W.J. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mykowska A., Sobczak K., Wojciechowska M., Kozlowski P., Krzyzosiak W.J. CAG repeats mimic CUG repeats in the misregulation of alternative splicing. Nucleic Acids Res. 2011;39:8938–8951. doi: 10.1093/nar/gkr608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fiszer A., Krzyzosiak W.J. RNA toxicity in polyglutamine disorders: concepts, models, and progress of research. J. Mol. Med. (Berlin). 2013;91:683–691. doi: 10.1007/s00109-013-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLeod C.J., O'Keefe L.V., Richards R.I. The pathogenic agent in Drosophila models of “polyglutamine” diseases. Hum. Mol. Genet. 2005;14:1041–1048. doi: 10.1093/hmg/ddi096. [DOI] [PubMed] [Google Scholar]
- 55.Li L.-B., Yu Z., Teng X., Bonini N.M. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shieh S.Y., Bonini N.M. Genes and pathways affected by CAG-repeat RNA-based toxicity in Drosophila. Hum. Mol. Genet. 2011;20:4810–4821. doi: 10.1093/hmg/ddr420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sobczak K., Krzyzosiak W.J. CAG repeats containing CAA interruptions form branched hairpin structures in spinocerebellar ataxia type 2 transcripts. J. Biol. Chem. 2005;280:3898–3910. doi: 10.1074/jbc.M409984200. [DOI] [PubMed] [Google Scholar]
- 58.Hsu R.J., Hsiao K.M., Lin M.J., Li C.Y., Wang L.C., Chen L.K., Pan H. Long tract of untranslated CAG repeats is deleterious in transgenic mice. PLoS One. 2011;6:e16417. doi: 10.1371/journal.pone.0016417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pouladi M.A., Stanek L.M., Xie Y., Franciosi S., Southwell A.L., Deng Y., Butland S., Zhang W., Cheng S.H., Shihabuddin L.S., et al. Marked differences in neurochemistry and aggregates despite similar behavioural and neuropathological features of Huntington disease in the full-length BACHD and YAC128 mice. Hum. Mol. Genet. 2012;21:2219–2232. doi: 10.1093/hmg/dds037. [DOI] [PubMed] [Google Scholar]
- 60.Tsoi H., Lau C.K., Lau K.F., Chan H.Y.E. Perturbation of U2AF65/NXF1-mediated RNA nuclear export enhances RNA toxicity in polyQ diseases. Hum. Mol. Genet. 2011;20:3787–3797. doi: 10.1093/hmg/ddr297. [DOI] [PubMed] [Google Scholar]
- 61.Tsoi H., Lau T.C., Tsang S.Y., Lau K.F., Chan H.Y. CAG expansion induces nucleolar stress in polyglutamine diseases. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13428–13433. doi: 10.1073/pnas.1204089109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laurent F.X., Sureau A., Klein A.F., Trouslard F., Gasnier E., Furling D., Marie J. New function for the RNA helicase p68/DDX5 as a modifier of MBNL1 activity on expanded CUG repeats. Nucleic Acids Res. 2012;40:3159–3171. doi: 10.1093/nar/gkr1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sathasivam K., Neueder A., Gipson T.A., Landles C., Benjamin A.C., Bondulich M.K., Smith D.L., Faull R.L., Roos R.A., Howland D., et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2366–2370. doi: 10.1073/pnas.1221891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chung D.W., Rudnicki D.D., Yu L., Margolis R.L. A natural antisense transcript at the Huntington's disease repeat locus regulates HTT expression. Hum. Mol. Genet. 2011;20:3467–3477. doi: 10.1093/hmg/ddr263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krol J., Fiszer A., Mykowska A., Sobczak K., de Mezer M., Krzyzosiak W.J. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol. Cell. 2007;25:575–586. doi: 10.1016/j.molcel.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 66.Mateu-Huertas E., Porta S., Martí E., Pantano L., Guzmán M., Estivill X., Bañez-Coronel M., Kagerbauer B., Ferrer I. A pathogenic mechanism in Huntington's disease involves small CAG-repeated RNAs with neurotoxic activity. PLoS Genet. 2012;8:e1002481. doi: 10.1371/journal.pgen.1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto A., Lucas J.J., Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 68.Zu T., Duvick L.A., Kaytor M.D., Berlinger M.S., Zoghbi H.Y., Clark H.B., Orr H.T. Recovery from polyglutamine-induced neurodegeneration in conditional SCA1 transgenic mice. J. Neurosci. 2004;24:8853–8861. doi: 10.1523/JNEUROSCI.2978-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Régulier E., Trottier Y., Perrin V., Aebischer P., Déglon N. Early and reversible neuropathology induced by tetracycline-regulated lentiviral overexpression of mutant huntingtin in rat striatum. Hum. Mol. Genet. 2003;12:2827–2836. doi: 10.1093/hmg/ddg305. [DOI] [PubMed] [Google Scholar]
- 70.Díaz-Hernández M., Torres-Peraza J., Salvatori-Abarca A., Morán M.A., Gómez-Ramos P., Alberch J., Lucas J.J. Full motor recovery despite striatal neuron loss and formation of irreversible amyloid-like inclusions in a conditional mouse model of Huntington's disease. J. Neurosci. 2005;25:9773–9781. doi: 10.1523/JNEUROSCI.3183-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 72.Urnov F.D., Rebar E.J., Holmes M.C., Zhang H.S., Gregory P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 73.Perez-Pinera P., Gersbach C.a., Ousterout D.G. Advances in targeted genome editing. Curr. Opin. Chem. Biol. 2012;16:268–277. doi: 10.1016/j.cbpa.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rebar E.J., Huang Y., Hickey R., Nath A.K., Meoli D., Nath S., Chen B., Xu L., Liang Y., Jamieson A.C., et al. Induction of angiogenesis in a mouse model using engineered transcription factors. Nat. Med. 2002;8:1427–1432. doi: 10.1038/nm1202-795. [DOI] [PubMed] [Google Scholar]
- 75.Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 76.Zou J., Sweeney C.L., Chou B.K., Choi U., Pan J., Wang H., Dowey S.N., Cheng L., Malech H.L., et al. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells: functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood. 2011;117:5561–5572. doi: 10.1182/blood-2010-12-328161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Provasi E., Genovese P., Lombardo A., Magnani Z., Liu P.-Q., Reik A., Chu V., Paschon D.E., Zhang L., Kuball J., et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med. 2012;18:807–815. doi: 10.1038/nm.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H., Haurigot V., Doyon Y., Li T., Wong S.Y., Bhagwat A.S., Malani N., Anguela X.M., Sharma R., Ivanciu L., et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perez E.E., Wang J., Miller J.C., Jouvenot Y., Kim K.A., Liu O., Wang N., Lee G., Bartsevich V. V, Lee Y.-L., et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mittelman D., Moye C., Morton J., Sykoudis K., Lin Y., Carroll D., Wilson J.H. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9607–9612. doi: 10.1073/pnas.0902420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garriga-Canut M., Agustin-Pavon C., Herrmann F., Sanchez A., Dierssen M., Fillat C., Isalan M. Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E3136–E3145. doi: 10.1073/pnas.1206506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morris K. V, Chan S.W., Jacobsen S.E., Looney D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 83.Janowski B.A., Kaihatsu K., Huffman K.E., Schwartz J.C., Ram R., Hardy D., Mendelson C.R., Corey D.R. Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids. Nat. Chem. Biol. 2005;1:210–215. doi: 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- 84.Hawkins P.G., Santoso S., Adams C., Anest V., Morris K.V. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Janowski B.A., Huffman K.E., Schwartz J.C., Ram R., Hardy D., Shames D.S., Minna J.D., Corey D.R. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat. Chem. Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 86.Robb G.B., Brown K.M., Khurana J., Rana T.M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 87.Huang V., Li L.-C. miRNA goes nuclear. RNA Biol. 2012;9:269–273. doi: 10.4161/rna.19354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gagnon K.T., Corey D.R. Argonaute and the nuclear RNAs: new pathways for RNA-mediated control of gene expression. Nucleic Acid Ther. 2012;22:3–16. doi: 10.1089/nat.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gagnon K.T., Li L., Chu Y., Janowski B.A., Corey D.R. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim D.H., Villeneuve L.M., Morris K.V., Rossi J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 91.Janowski B.A., Huffman K.E., Schwartz J.C., Ram R., Nordsell R., Shames D.S., Minna J.D., Corey D.R. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 92.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 93.Sibley C.R., Seow Y., Wood M.J. Novel RNA-based strategies for therapeutic gene silencing. Mol. Ther. 2010;18:466–476. doi: 10.1038/mt.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kole R., Krainer A.R., Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu J., Hu J., Corey D.R. Expanding the action of duplex RNAs into the nucleus: redirecting alternative splicing. Nucleic Acids Res. 2012;40:1240–1250. doi: 10.1093/nar/gkr780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Prakash T.P., Bennett C.F., Rigo F., Chun S.J., Krainer A.R., Hua Y. Synthetic oligonucleotides recruit ILF2/3 to RNA transcripts to modulate splicing. Nat. Chem. Biol. 2012;8:555–561. doi: 10.1038/nchembio.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Van Ommen G.-J.B., Aartsma-Rus A. Advances in therapeutic RNA-targeting. N. Biotechnol. 2013;30:299–301. doi: 10.1016/j.nbt.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 98.Evers M.M., Tran H.D., Zalachoras I., Pepers B.A., Meijer O.C., den Dunnen J.T., van Ommen G.J., Aartsma-Rus A., van Roon-Mom W.M. Ataxin-3 protein modification as a treatment strategy for spinocerebellar ataxia type 3: removal of the CAG containing exon. Neurobiol. Dis. 2013;58C:49–56. doi: 10.1016/j.nbd.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 99.Evers M.M., Tran H.-D., Zalachoras I., Meijer O.C., den Dunnen J.T., van Ommen G.-J.B., Aartsma-Rus A., van Roon-Mom W.M.C. Preventing formation of toxic N-terminal Huntingtin fragments through antisense oligonucleotide-mediated protein modification. Nucleic Acid Ther. 2013;24:4–12. doi: 10.1089/nat.2013.0452. [DOI] [PubMed] [Google Scholar]
- 100.Martínez T., Wright N., López-Fraga M., Jiménez A.I., Pañeda C. Silencing human genetic diseases with oligonucleotide-based therapies. Hum. Genet. 2013;132:481–493. doi: 10.1007/s00439-013-1288-1. [DOI] [PubMed] [Google Scholar]
- 101.Cerritelli S.M., Crouch R.J. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lima W.F., Wu H., Nichols J.G., Prakash T.P., Ravikumar V., Crooke S.T. Human RNase H1 uses one tryptophan and two lysines to position the enzyme at the 3’-DNA/5’-RNA terminus of the heteroduplex substrate. J. Biol. Chem. 2003;278:49860–49867. doi: 10.1074/jbc.M306543200. [DOI] [PubMed] [Google Scholar]
- 103.Lima W.F., Nichols J.G., Wu H., Prakash T.P., Migawa M.T., Wyrzykiewicz T.K., Bhat B., Crooke S.T. Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J. Biol. Chem. 2004;279:36317–36326. doi: 10.1074/jbc.M405035200. [DOI] [PubMed] [Google Scholar]
- 104.Wu H., Lima W.F., Zhang H., Fan A., Sun H., Crooke S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 105.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 106.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 107.Friedman R.C., Farh K.K.-H., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gregory R.I., Chendrimada T.P., Cooch N., Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 110.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 113.Stroynowska-Czerwinska A., Fiszer A., Krzyzosiak W.J. The panorama of miRNA-mediated mechanisms in mammalian cells. Cell. Mol. Life Sci. 2014 doi: 10.1007/s00018-013-1551-6. 10.1007/s00018-013-1551-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baker B.F., Lot S.S., Condon T.P., Cheng-Flournoy S., Lesnik E.A., Sasmor H.M., Bennett C.F. 2’-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 115.Paessler S., Rijnbrand R., Stein D.A., Ni H., Yun N.E., Dziuba N., Borisevich V., Seregin A., Ma Y., Blouch R., et al. Inhibition of alphavirus infection in cell culture and in mice with antisense morpholino oligomers. Virology. 2008;376:357–370. doi: 10.1016/j.virol.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wheeler T.M., Sobczak K., Lueck J.D., Osborne R.J., Lin X., Dirksen R.T., Thornton C.a. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mulders S.A., van den Broek W.J., Wheeler T.M., Croes H.J., van Kuik-Romeijn P., de Kimpe S.J., Furling D., Platenburg G.J., Gourdon G., Thornton C.A., et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nakamori M., Gourdon G., Thornton C.A. Stabilization of expanded (CTG)•(CAG) repeats by antisense oligonucleotides. Mol. Ther. 2011;19:2222–2227. doi: 10.1038/mt.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Disney M.D. Rational design of chemical genetic probes of RNA function and lead therapeutics targeting repeating transcripts. Drug Discov. Today. 2013;18 doi: 10.1016/j.drudis.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gareiss P.C., Sobczak K., McNaughton B.R., Palde P.B., Thornton C.A., Miller B.L. Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA-MBNL1 interaction in vitro: discovery of lead compounds targeting myotonic dystrophy (DM1) J. Am. Chem. Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Arambula J.F., Ramisetty S.R., Baranger A.M., Zimmerman S.C. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc. Natl. Acad. Sci. U.S.A. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee M.M., Childs-Disney J.L., Pushechnikov A., French J.M., Sobczak K., Thornton C.A., Disney M.D. Controlling the specificity of modularly assembled small molecules for RNA via ligand module spacing: targeting the RNAs that cause myotonic muscular dystrophy. J. Am. Chem. Soc. 2009;131:17464–17472. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Disney M.D., Lee M.M., Pushechnikov A., Childs-Disney J.L. The role of flexibility in the rational design of modularly assembled ligands targeting the RNAs that cause the myotonic dystrophies. Chembiochem. 2010;11:375–382. doi: 10.1002/cbic.200900716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Warf M.B., Nakamori M., Matthys C.M., Thornton C.A., Berglund J.A. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Garcia-Lopez A., Llamusi B., Orzaez M., Perez-Paya E., Artero R.D. In vivo discovery of a peptide that prevents CUG-RNA hairpin formation and reverses RNA toxicity in myotonic dystrophy models. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11866–11871. doi: 10.1073/pnas.1018213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pushechnikov A., Lee M.M., Childs-Disney J.L., Sobczak K., French J.M., Thornton C.A., Disney M.D. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J. Am. Chem. Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kiliszek A., Kierzek R., Krzyzosiak W.J., Rypniewski W. Structural insights into CUG repeats containing the “stretched U-U wobble”: implications for myotonic dystrophy. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kiliszek A., Kierzek R., Krzyzosiak W.J., Rypniewski W. Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases. Nucleic Acids Res. 2010;38:8370–8376. doi: 10.1093/nar/gkq700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kumar A., Park H., Fang P., Parkesh R., Guo M., Nettles K.W., Disney M.D. Myotonic dystrophy type 1 RNA crystal structures reveal heterogeneous 1 × 1 nucleotide UU internal loop conformations. Biochemistry. 2011;50:9928–9935. doi: 10.1021/bi2013068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yildirim I., Park H., Disney M.D., Schatz G.C. A dynamic structural model of expanded RNA CAG repeats: a refined X-ray structure and computational investigations using molecular dynamics and umbrella sampling simulations. J. Am. Chem. Soc. 2013;135:3528–3538. doi: 10.1021/ja3108627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee M.M., Pushechnikov A., Disney M.D. Rational and modular design of potent ligands targeting the RNA that causes myotonic dystrophy 2. ACS Chem. Biol. 2009;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kumar A., Parkesh R., Sznajder L.J., Childs-Disney J.L., Sobczak K., Disney M.D. Chemical correction of pre-mRNA splicing defects associated with sequestration of muscleblind-like 1 protein by expanded r(CAG)-containing transcripts. ACS Chem. Biol. 2012;7:496–505. doi: 10.1021/cb200413a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu C.R., Chang C.R., Chern Y., Wang T.H., Hsieh W.C., Shen W.C., Chang C.Y., Chu I.C., Deng N., Cohen S.N., et al. Spt4 is selectively required for transcription of extended trinucleotide repeats. Cell. 2012;148:690–701. doi: 10.1016/j.cell.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 134.Krauss S., Griesche N., Jastrzebska E., Chen C., Rutschow D., Achmuller C., Dorn S., Boesch S.M., Lalowski M., Wanker E. Translation of HTT mRNA with expanded CAG repeats is regulated by the MID1-PP2A protein complex. Nat. Commun. 2013;4 doi: 10.1038/ncomms2514. doi:10.1038/ncomms2514. [DOI] [PubMed] [Google Scholar]
- 135.Appl T., Kaltenbach L., Lo D.C., Terstappen G.C. Targeting mutant huntingtin for the development of disease-modifying therapy. Drug Discov. Today. 2012;17:1217–1223. doi: 10.1016/j.drudis.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 136.Gabibov A., Skulachev V., Wieland F., Just W., Margulis B.A., Vigont V., Lazarev V.F., Kaznacheyeva E. V., Guzhova I. V. Pharmacological protein targets in polyglutamine diseases: mutant polypeptides and their interactors. FEBS Lett. 2013;587:1997–2007. doi: 10.1016/j.febslet.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 137.Chai Y., Koppenhafer S.L., Bonini N.M., Paulson H.L. Analysis of the role of heat shock protein (Hsp) molecular chaperones in polyglutamine disease. J. Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pollitt S.K., Pallos J., Shao J., Desai U.A., Ma A.A., Thompson L.M., Marsh J.L., Diamond M.I. A rapid cellular FRET assay of polyglutamine aggregation identifies a novel inhibitor. Neuron. 2003;40:685–694. doi: 10.1016/s0896-6273(03)00697-4. [DOI] [PubMed] [Google Scholar]
- 139.Desai U.A., Pallos J., Ma A.A., Stockwell B.R., Thompson L.M., Marsh J.L., Diamond M.I. Biologically active molecules that reduce polyglutamine aggregation and toxicity. Hum. Mol. Genet. 2006;15:2114–2124. doi: 10.1093/hmg/ddl135. [DOI] [PubMed] [Google Scholar]
- 140.Bauer P.O., Hudec R., Goswami A., Kurosawa M., Matsumoto G., Mikoshiba K., Nukina N. ROCK-phosphorylated vimentin modifies mutant huntingtin aggregation via sequestration of IRBIT. Mol. Neurodegener. 2012;7 doi: 10.1186/1750-1326-7-43. doi:10.1186/1750-1326-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fecke W., Gianfriddo M., Gaviraghi G., Terstappen G.C., Heitz F. Small molecule drug discovery for Huntington's disease. Drug Discov. Today. 2009;14:453–64. doi: 10.1016/j.drudis.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 142.Wong H.K., Bauer P.O., Kurosawa M., Goswami A., Washizu C., Machida Y., Tosaki A., Yamada M., Knopfel T., Nakamura T., et al. Blocking acid-sensing ion channel 1 alleviates Huntington's disease pathology via an ubiquitin-proteasome system-dependent mechanism. Hum. Mol. Genet. 2008;17:3223–3235. doi: 10.1093/hmg/ddn218. [DOI] [PubMed] [Google Scholar]
- 143.Sarkar S., Perlstein E.O., Imarisio S., Pineau S., Cordenier A., Maglathlin R.L., Webster J.A., Lewis T.A., O'Kane C.J., Schreiber S.L., et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat. Chem. Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bauer P.O., Goswami A., Wong H.K., Okuno M., Kurosawa M., Yamada M., Miyazaki H., Matsumoto G., Kino Y., Nagai Y., et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat. Biotechnol. 2010;28:256–263. doi: 10.1038/nbt.1608. [DOI] [PubMed] [Google Scholar]
- 145.Lu B., Al-Ramahi I., Valencia A., Wang Q., Berenshteyn F., Yang H., Gallego-Flores T., Ichcho S., Lacoste A., Hild M., et al. Identification of NUB1 as a suppressor of mutant Huntington toxicity via enhanced protein clearance. Nat. Neurosci. 2013;16:562–70. doi: 10.1038/nn.3367. [DOI] [PubMed] [Google Scholar]
- 146.De Chiara C., Menon R.P., Kelly G., Pastore A. Protein-protein interactions as a strategy towards protein-specific drug design: the example of ataxin-1. PLoS One. 2013;8:e76456. doi: 10.1371/journal.pone.0076456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.De Chiara C., Pastore A. Kaleidoscopic protein-protein interactions in the life and death of ataxin-1: new strategies against protein aggregation. Trends Neurosci. 2014;37:211–218. doi: 10.1016/j.tins.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bowman A.B., Lam Y.C., Jafar-Nejad P., Chen H.K., Richman R., Samaco R.C., Fryer J.D., Kahle J.J., Orr H.T., Zoghbi H.Y. Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nat. Genet. 2007;39:373–379. doi: 10.1038/ng1977. [DOI] [PubMed] [Google Scholar]
- 149.Keiser M.S., Geoghegan J.C., Boudreau R.L., Lennox K.A., Davidson B.L. RNAi or overexpression: alternative therapies for Spinocerebellar Ataxia Type 1. Neurobiol. Dis. 2013;56:6–13. doi: 10.1016/j.nbd.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bauer P.O., Nukina N. The pathogenic mechanisms of polyglutamine diseases and current therapeutic strategies. J. Neurochem. 2009;110:1737–1765. doi: 10.1111/j.1471-4159.2009.06302.x. [DOI] [PubMed] [Google Scholar]
- 151.Zuccato C., Valenza M., Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol. Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 152.Ross C.a., Tabrizi S.J. Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 153.Switonski P.M., Szlachcic W.J., Gabka A., Krzyzosiak W.J., Figiel M. Mouse models of polyglutamine diseases in therapeutic approaches: review and data table. Part II. Mol. Neurobiol. 2012;46:430–466. doi: 10.1007/s12035-012-8316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kaplan A., Stockwell B.R. Therapeutic approaches to preventing cell death in Huntington disease. Prog. Neurobiol. 2012;99:262–280. doi: 10.1016/j.pneurobio.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dragatsis I., Levine M.S., Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat. Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 156.Reiner A., Del Mar N., Meade C.A., Yang H., Dragatsis I., Zeitlin S., Goldowitz D. Neurons lacking huntingtin differentially colonize brain and survive in chimeric mice. J. Neurosci. 2001;21:7608–7619. doi: 10.1523/JNEUROSCI.21-19-07608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Warrick J.M., Morabito L.M., Bilen J., Gordesky-Gold B., Faust L.Z., Paulson H.L., Bonini N.M. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol. Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 158.Van Raamsdonk J.M., Pearson J., Rogers D.A., Bissada N., Vogl A.W., Hayden M.R., Leavitt B.R. Loss of wild-type huntingtin influences motor dysfunction and survival in the YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 2005;14:1379–1392. doi: 10.1093/hmg/ddi147. [DOI] [PubMed] [Google Scholar]
- 159.Lang A.E., Rogaeva E.A., Tsuda T., Hutterer J., George-Hyslop P., St Homozygous inheritance of the Machado-Joseph disease gene. Ann. Neurol. 1994;36:443–447. doi: 10.1002/ana.410360318. [DOI] [PubMed] [Google Scholar]
- 160.Squitieri F., Gellera C., Cannella M., Mariotti C., Cislaghi G., Rubinsztein D.C., Almqvist E.W., Turner D., Bachoud-Lévi A.-C., Simpson S.A., et al. Homozygosity for CAG mutation in Huntington disease is associated with a more severe clinical course. Brain. 2003;126:946–955. doi: 10.1093/brain/awg077. [DOI] [PubMed] [Google Scholar]
- 161.Alves S., Nascimento-Ferreira I., Dufour N., Hassig R., Auregan G., Nobrega C., Brouillet E., Hantraye P., Pedroso de Lima M.C., Deglon N., et al. Silencing ataxin-3 mitigates degeneration in a rat model of Machado-Joseph disease: no role for wild-type ataxin-3. Hum. Mol. Genet. 2010;19:2380–2394. doi: 10.1093/hmg/ddq111. [DOI] [PubMed] [Google Scholar]
- 162.Xia H., Mao Q., Eliason S.L., Harper S.Q., Martins I.H., Orr H.T., Paulson H.L., Yang L., Kotin R.M., Davidson B.L. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 163.Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I.H., Mao Q., Yang L., Kotin R.M., Paulson H.L., Davidson B.L. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rodriguez-Lebron E., Denovan-Wright E.M., Nash K., Lewin A.S., Mandel R.J. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol. Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Huang B., Schiefer J., Sass C., Landwehrmeyer G.B., Kosinski C.M., Kochanek S. High-capacity adenoviral vector-mediated reduction of huntingtin aggregate load in vitro and in vivo. Hum. Gene Ther. 2007;18:303–311. doi: 10.1089/hum.2006.160. [DOI] [PubMed] [Google Scholar]
- 166.Boudreau R.L., McBride J.L., Martins I., Shen S., Xing Y., Carter B.J., Davidson B.L. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang Y.L., Liu W., Wada E., Murata M., Wada K., Kanazawa I. Clinico-pathological rescue of a model mouse of Huntington's disease by siRNA. Neurosci. Res. 2005;53:241–249. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 168.DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R.K., Rajeev K.G., et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. U.S.A. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kordasiewicz H.B., Stanek L.M., Wancewicz E. V, Mazur C., McAlonis M.M., Pytel K.A., Artates J.W., Weiss A., Cheng S.H., Shihabuddin L.S., et al. Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Do Carmo Costa M., Luna-Cancalon K., Fischer S., Ashraf N.S., Ouyang M., Dharia R.M., Martin-Fishman L., Yang Y., Shakkottai V.G., Davidson B.L., et al. Toward RNAi therapy for the polyglutamine disease Machado-Joseph disease. Mol. Ther. 2013;21:1898–1908. doi: 10.1038/mt.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rodriguez-Lebron E., do Carmo Costa M., Molina-Luna K., Peron T.M., Fischer S., Boudreau R.L., Davidson B.L., Paulson H.L. Silencing mutant ATXN3 expression resolves molecular phenotypes in SCA3 transgenic mice. Mol. Ther. 2013;21:1909–1918. doi: 10.1038/mt.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Drouet V., Perrin V., Hassig R., Dufour N., Auregan G., Alves S., Bonvento G., Brouillet E., Luthi-Carter R., Hantraye P., et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 173.Stanek L.M., Sardi S.P., Mastis B.M., Richards A.R., Treleaven C.M., Taksir T. V, Misra K., Cheng S.H., Shihabuddin L.S. Silencing mutant huntingtin by AAV-mediated RNAi ameliorates disease manifestations in the YAC128 mouse model of Huntington's disease. Hum. Gene Ther. 2014 doi: 10.1089/hum.2013.200. 10.1089/hum.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.McBride J.L., Pitzer M.R., Boudreau R.L., Dufour B., Hobbs T., Ojeda S.R., Davidson B.L. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington's disease. Mol. Ther. 2011;19:2152–2162. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stiles D., Zhang Z., Ge P., Nelson B., Grondin R., Ai Y., Hardy P., Nelson P.T., Guzaev A.P., Butt M.T., et al. Widespread suppression of huntingtin with convection-enhanced delivery of siRNA. Exp. Neurol. 2011;233:463–471. doi: 10.1016/j.expneurol.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 176.Grondin R., Kaytor M.D., Ai Y., Nelson P.T., Thakker D.R., Heisel J., Weatherspoon M.R., Blum J.L., Burright E.N., Zhang Z., et al. Six-month partial suppression of Huntingtin is well tolerated in the adult rhesus striatum. Brain. 2012;135:1197–1209. doi: 10.1093/brain/awr333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kim D.H., Rossi J.J. Coupling of RNAi-mediated target downregulation with gene replacement. Antisense Nucleic Acid Drug Dev. 2003;13:151–155. doi: 10.1089/108729003768247619. [DOI] [PubMed] [Google Scholar]
- 178.Kubodera T., Yokota T., Ishikawa K., Mizusawa H. New RNAi strategy for selective suppression of a mutant allele in polyglutamine disease. Oligonucleotides. 2005;15:298–302. doi: 10.1089/oli.2005.15.298. [DOI] [PubMed] [Google Scholar]
- 179.Fiszer A., Olejniczak M., Switonski P.M., Wroblewska J.P., Wisniewska-Kruk J., Mykowska A., Krzyzosiak W.J. An evaluation of oligonucleotide-based therapeutic strategies for polyQ diseases. BMC Mol. Biol. 2012;13 doi: 10.1186/1471-2199-13-6. doi:10.1186/1471-2199-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.O'Reilly M., Palfi A., Chadderton N., Millington-Ward S., Ader M., Cronin T., Tuohy T., Auricchio A., Hildinger M., Tivnan A., et al. RNA interference-mediated suppression and replacement of human rhodopsin in vivo. Am. J. Hum. Genet. 2007;81:127–135. doi: 10.1086/519025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Du Q., Thonberg H., Wang J., Wahlestedt C., Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huang H., Qiao R., Zhao D., Zhang T., Li Y., Yi F., Lai F., Hong J., Ding X., Yang Z., et al. Profiling of mismatch discrimination in RNAi enabled rational design of allele-specific siRNAs. Nucleic Acids Res. 2009;37:7560–7569. doi: 10.1093/nar/gkp835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Miller V.M., Xia H., Marrs G.L., Gouvion C.M., Lee G., Davidson B.L., Paulson H.L. Allele-specific silencing of dominant disease genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li Y., Yokota T., Matsumura R., Taira K., Mizusawa H. Sequence-dependent and independent inhibition specific for mutant ataxin-3 by small interfering RNA. Ann. Neurol. 2004;56:124–129. doi: 10.1002/ana.20141. [DOI] [PubMed] [Google Scholar]
- 185.Gaspar C., Lopes-Cendes I., Hayes S., Goto J., Arvidsson K., Dias A., Silveira I., Maciel P., Coutinho P., Lima M., et al. Ancestral origins of the Machado-Joseph disease mutation: a worldwide haplotype study. Am. J. Hum. Genet. 2001;68:523–528. doi: 10.1086/318184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Alves S., Nascimento-Ferreira I., Auregan G., Hassig R., Dufour N., Brouillet E., Pedroso de Lima M.C., Hantraye P., Pereira de Almeida L., Deglon N. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS One. 2008;3:e3341. doi: 10.1371/journal.pone.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nóbrega C., Nascimento-Ferreira I., Onofre I., Albuquerque D., Hirai H., Déglon N., de Almeida L.P. Silencing mutant ataxin-3 rescues motor deficits and neuropathology in Machado-Joseph disease transgenic mice. PLoS One. 2013;8:e52396. doi: 10.1371/journal.pone.0052396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Scholefield J., Greenberg L.J., Weinberg M.S., Arbuthnot P.B., Abdelgany A., Wood M.J. Design of RNAi hairpins for mutation-specific silencing of ataxin-7 and correction of a SCA7 phenotype. PLoS One. 2009;4:e7232. doi: 10.1371/journal.pone.0007232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Warby S.C., Montpetit A., Hayden A.R., Carroll J.B., Butland S.L., Visscher H., Collins J.A., Semaka A., Hudson T.J., Hayden M.R. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am. J. Hum. Genet. 2009;84:351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schwarz D.S., Ding H., Kennington L., Moore J.T., Schelter J., Burchard J., Linsley P.S., Aronin N., Xu Z., Zamore P.D. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Van Bilsen P.H., Jaspers L., Lombardi M.S., Odekerken J.C., Burright E.N., Kaemmerer W.F. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington's disease patient-derived fibroblasts. Hum. Gene Ther. 2008;19:710–719. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- 192.Lombardi M.S., Jaspers L., Spronkmans C., Gellera C., Taroni F., Di Maria E., Donato S.D., Kaemmerer W.F. A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 193.Pfister E.L., Kennington L., Straubhaar J., Wagh S., Liu W., DiFiglia M., Landwehrmeyer B., Vonsattel J.P., Zamore P.D., Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr. Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Takahashi M., Watanabe S., Murata M., Furuya H., Kanazawa I., Wada K., Hohjoh H. Tailor-made RNAi knockdown against triplet repeat disease-causing alleles. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21731–21736. doi: 10.1073/pnas.1012153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Carroll J.B., Warby S.C., Southwell A.L., Doty C.N., Greenlee S., Skotte N., Hung G., Bennett C.F., Freier S.M., Hayden M.R. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the huntington disease gene/allele-specific silencing of mutant huntingtin. Mol. Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ostergaard M.E., Southwell A.L., Kordasiewicz H., Watt A.T., Skotte N.H., Doty C.N., Vaid K., Villanueva E.B., Swayze E.E., Frank Bennett C., et al. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013;41:9634–9650. doi: 10.1093/nar/gkt725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Southwell A.L., Warby S.C., Carroll J.B., Doty C.N., Skotte N.H., Zhang W., Villanueva E.B., Kovalik V., Xie Y., Pouladi M.A., et al. A fully humanized transgenic mouse model of Huntington disease. Hum. Mol. Genet. 2013;22:18–34. doi: 10.1093/hmg/dds397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kozlowski P., de Mezer M., Krzyzosiak W.J. Trinucleotide repeats in human genome and exome. Nucleic Acids Res. 2010;38:4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hu J., Liu J., Corey D.R. Allele-selective inhibition of Huntingtin expression by switching to an miRNA-like RNAi mechanism. Chem. Biol. 2010;17:1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fiszer A., Mykowska A., Krzyzosiak W.J. Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats. Nucleic Acids Res. 2011;39:5578–5585. doi: 10.1093/nar/gkr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hu J., Matsui M., Gagnon K.T., Schwartz J.C., Gabillet S., Arar K., Wu J., Bezprozvanny I., Corey D.R. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gagnon K.T., Pendergraff H.M., Deleavey G.F., Swayze E.E., Potier P., Randolph J., Roesch E.B., Chattopadhyaya J., Damha M.J., Bennett C.F., et al. Allele-selective inhibition of mutant Huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry. 2010;49:10166–10178. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hu J., Liu J., Yu D., Chu Y., Corey D.R. Mechanism of allele-selective inhibition of huntingtin expression by duplex RNAs that target CAG repeats: function through the RNAi pathway. Nucleic Acids Res. 2012;40:11270–11280. doi: 10.1093/nar/gks907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hu J., Gagnon K.T., Liu J., Watts J.K., Syeda-Nawaz J., Bennett C.F., Swayze E.E., Randolph J., Chattopadhyaya J., Corey D.R. Allele-selective inhibition of ataxin-3 (ATX3) expression by antisense oligomers and duplex RNAs. Biol. Chem. 2011;392:315–325. doi: 10.1515/BC.2011.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lima W.F., Prakash T.P., Murray H.M., Kinberger G.A., Li W., Chappell A.E., Li C.S., Murray S.F., Gaus H., Seth P.P., et al. Single-stranded siRNAs activate RNAi in animals. Cell. 2012;150:883–894. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 206.Yu D., Pendergraff H., Liu J., Kordasiewicz H.B., Cleveland D.W., Swayze E.E., Lima W.F., Crooke S.T., Prakash T.P., Corey D.R. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150:895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Liu J., Yu D., Aiba Y., Pendergraff H., Swayze E.E., Lima W.F., Hu J., Prakash T.P., Corey D.R. ss-siRNAs allele selectively inhibit ataxin-3 expression: multiple mechanisms for an alternative gene silencing strategy. Nucleic Acids Res. 2013;41:9570–9583. doi: 10.1093/nar/gkt693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fiszer A., Olejniczak M., Galka-Marciniak P., Mykowska A., Krzyzosiak W.J. Self-duplexing CUG repeats selectively inhibit mutant huntingtin expression. Nucleic Acids Res. 2013;41:10426–10437. doi: 10.1093/nar/gkt825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Krzyzosiak W.J., Sobczak K., Wojciechowska M., Fiszer A., Mykowska A., Kozlowski P. Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic Acids Res. 2012;40:11–26. doi: 10.1093/nar/gkr729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sathasivam K., Woodman B., Mahal a, Bertaux F., Wanker E.E., Shima D.T., Bates G.P. Centrosome disorganization in fibroblast cultures derived from R6/2 Huntington's disease (HD) transgenic mice and HD patients. Hum. Mol. Genet. 2001;10:2425–2435. doi: 10.1093/hmg/10.21.2425. [DOI] [PubMed] [Google Scholar]
- 211.Marchina E., Misasi S., Bozzato A., Ferraboli S., Agosti C., Rozzini L., Borsani G., Barlati S., Padovani A. Gene expression profile in fibroblasts of Huntington's disease patients and controls. J. Neurol. Sci. 2014;337:42–46. doi: 10.1016/j.jns.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 212.Koch P., Breuer P., Peitz M., Jungverdorben J., Kesavan J., Poppe D., Doerr J., Ladewig J., Mertens J., Tuting T., et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480:543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 213.Consortium. Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhang N., An M.C., Montoro D., Ellerby L.M. Characterization of human Huntington's disease cell model from induced pluripotent stem cells. PLoS Curr. 2010;2:RRN1193. doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Camnasio S., Delli Carri A., Lombardo A., Grad I., Mariotti C., Castucci A., Rozell B., Lo Riso P., Castiglioni V., Zuccato C., et al. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington's disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol. Dis. 2012;46:41–51. doi: 10.1016/j.nbd.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 216.Jeon I., Lee N., Li J.-Y., Park I.-H., Park K.S., Moon J., Shim S.H., Choi C., Chang D.-J., Kwon J., et al. Neuronal properties, in vivo effects, and pathology of a Huntington's disease patient-derived induced pluripotent stem cells. Stem Cells. 2012;30:2054–2062. doi: 10.1002/stem.1135. [DOI] [PubMed] [Google Scholar]
- 217.Chae J.-I., Kim D.-W., Lee N., Jeon Y.-J., Jeon I., Kwon J., Kim J., Soh Y., Lee D.-S., Seo K.S., et al. Quantitative proteomic analysis of induced pluripotent stem cells derived from a human Huntington's disease patient. Biochem. J. 2012;446:359–371. doi: 10.1042/BJ20111495. [DOI] [PubMed] [Google Scholar]
- 218.Nihei Y., Ito D., Okada Y., Akamatsu W., Yagi T., Yoshizaki T., Okano H., Suzuki N. Enhanced aggregation of androgen receptor in induced pluripotent stem cell-derived neurons from spinal and bulbar muscular atrophy. J. Biol. Chem. 2013;288:8043–8052. doi: 10.1074/jbc.M112.408211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Xia G., Santostefano K., Hamazaki T., Liu J., Subramony S.H., Terada N., Ashizawa T. Generation of human-induced pluripotent stem cells to model spinocerebellar ataxia type 2 in vitro. J. Mol. Neurosci. 2013;51:237–248. doi: 10.1007/s12031-012-9930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.An M.C., Zhang N., Scott G., Montoro D., Wittkop T., Mooney S., Melov S., Ellerby L.M. Genetic correction of Huntington's disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kaye J.A., Finkbeiner S. Modeling Huntington's disease with induced pluripotent stem cells. Mol. Cell. Neurosci. 2013;56:50–64. doi: 10.1016/j.mcn.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gao A., Peng Y., Deng Y., Qing H. Potential therapeutic applications of differentiated induced pluripotent stem cells (iPSCs) in the treatment of neurodegenerative diseases. Neuroscience. 2013;228:47–59. doi: 10.1016/j.neuroscience.2012.09.076. [DOI] [PubMed] [Google Scholar]
- 223.Crook Z.R., Housman D. Huntington's disease: can mice lead the way to treatment. Neuron. 2011;69:423–435. doi: 10.1016/j.neuron.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Figiel M., Szlachcic W.J., Switonski P.M., Gabka A., Krzyzosiak W.J. Mouse models of polyglutamine diseases: review and data table. Part I. Mol. Neurobiol. 2012;46:393–429. doi: 10.1007/s12035-012-8315-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Yang S.-H., Cheng P.-H., Banta H., Piotrowska-Nitsche K., Yang J.-J., Cheng E.C.H., Snyder B., Larkin K., Liu J., Orkin J., et al. Towards a transgenic model of Huntington's disease in a non-human primate. Nature. 2008;453:921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Jacobsen J.C., Bawden C.S., Rudiger S.R., McLaughlan C.J., Reid S.J., Waldvogel H.J., MacDonald M.E., Gusella J.F., Walker S.K., Kelly J.M., et al. An ovine transgenic Huntington's disease model. Hum. Mol. Genet. 2010;19:1873–1882. doi: 10.1093/hmg/ddq063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Yang D., Wang C.-E., Zhao B., Li W., Ouyang Z., Liu Z., Yang H., Fan P., O'Neill A., Gu W., et al. Expression of Huntington's disease protein results in apoptotic neurons in the brains of cloned transgenic pigs. Hum. Mol. Genet. 2010;19:3983–3994. doi: 10.1093/hmg/ddq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sciabola S., Cao Q., Orozco M., Faustino I., Stanton R. V. Improved nucleic acid descriptors for siRNA efficacy prediction. Nucleic Acids Res. 2013;41:1383–1394. doi: 10.1093/nar/gks1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 230.Matveeva O., Nechipurenko Y., Rossi L., Moore B., Saetrom P., Ogurtsov A.Y., Atkins J.F., Shabalina S.A. Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Res. 2007;35:e63. doi: 10.1093/nar/gkm088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Patrick Walton S., Wu M., Gredell J.A., Chan C. Designing highly active siRNAs for therapeutic applications. FEBS J. 2010;277:4806–4813. doi: 10.1111/j.1742-4658.2010.07903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Patzel V., Rutz S., Dietrich I., Koberle C., Scheffold A., Kaufmann S.H. Design of siRNAs producing unstructured guide-RNAs results in improved RNA interference efficiency. Nat. Biotechnol. 2005;23:1440–1444. doi: 10.1038/nbt1151. [DOI] [PubMed] [Google Scholar]
- 233.Westerhout E.M., Berkhout B. A systematic analysis of the effect of target RNA structure on RNA interference. Nucleic Acids Res. 2007;35:4322–4330. doi: 10.1093/nar/gkm437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Tafer H., Ameres S.L., Obernosterer G., Gebeshuber C.A., Schroeder R., Martinez J., Hofacker I.L. The impact of target site accessibility on the design of effective siRNAs. Nat. Biotechnol. 2008;26:578–583. doi: 10.1038/nbt1404. [DOI] [PubMed] [Google Scholar]
- 235.Gredell J.A., Berger A.K., Walton S.P. Impact of target mRNA structure on siRNA silencing efficiency: a large-scale study. Biotechnol. Bioeng. 2008;100:744–755. doi: 10.1002/bit.21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kretschmer-Kazemi Far R., Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 2003;31:4417–4424. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Crooke S.T., Vickers T.A., Lima W.F., Wu H. Mechanisms of antisense drug action, an introduction. In: Crooke ST, editor. Antisense Drug Technology: Principles, Strategies, and Applications. Boca Raton: CRC Press; 2008. pp. 3–46. [Google Scholar]
- 238.Geary R.S., Yu R.Z., Levin A.A. Pharmacokinetics of phosphorothioate antisense oligodeoxynucleotides. Curr. Opin. Investig. Drugs. 2001;2:562–573. [PubMed] [Google Scholar]
- 239.Henry S.P., Geary R.S., Yu R., Levin A.A. Drug properties of second-generation antisense oligonucleotides: how do they measure up to their predecessors? Curr. Opin. Investig. Drugs. 2001;2:1444–1449. [PubMed] [Google Scholar]
- 240.Sipa K., Sochacka E., Kazmierczak-Baranska J., Maszewska M., Janicka M., Nowak G., Nawrot B. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA. 2007;13:1301–1316. doi: 10.1261/rna.538907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Bramsen J.B., Laursen M.B., Nielsen A.F., Hansen T.B., Bus C., Langkjaer N., Babu B.R., Hojland T., Abramov M., Van Aerschot A., et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Engels J.W. Gene silencing by chemically modified siRNAs. New Biotechnol. 2013;30:302–307. doi: 10.1016/j.nbt.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 243.Cirak S., Arechavala-Gomeza V., Guglieri M., Feng L., Torelli S., Anthony K., Abbs S., Garralda M.E., Bourke J., Wells D.J., et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.McManus M.T., Petersen C.P., Haines B.B., Chen J., Sharp P.A. Gene silencing using micro-RNA designed hairpins. RNA. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Boudreau R.L., Monteys A.M., Davidson B.L. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA. 2008;14:1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B., et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S. V, Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 251.Boudreau R.L., Spengler R.M., Davidson B.L. Rational design of therapeutic siRNAs: minimizing off-targeting potential to improve the safety of RNAi therapy for Huntington's disease. Mol. Ther. 2011;19:2169–2177. doi: 10.1038/mt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K., et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sioud M. Deciphering the code of innate immunity recognition of siRNAs. Methods Mol. Biol. 2009;487:41–59. doi: 10.1007/978-1-60327-547-7_2. [DOI] [PubMed] [Google Scholar]
- 254.Sun X., Rogoff H.A., Li C.J. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat. Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- 255.Chang C.I., Yoo J.W., Hong S.W., Lee S.E., Kang H.S., Sun X., Rogoff H.A., Ban C., Kim S., Li C.J., et al. Asymmetric shorter-duplex siRNA structures trigger efficient gene silencing with reduced nonspecific effects. Mol. Ther. 2009;17:725–732. doi: 10.1038/mt.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 257.Khan A.A., Betel D., Miller M.L., Sander C., Leslie C.S., Marks D.S. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Olejniczak M., Galka P., Krzyzosiak W.J. Sequence-non-specific effects of RNA interference triggers and microRNA regulators. Nucleic Acids Res. 2010;38:1–16. doi: 10.1093/nar/gkp829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Dirin M., Winkler J. Influence of diverse chemical modifications on the ADME characteristics and toxicology of antisense oligonucleotides. Expert Opin. Biol. Ther. 2013;13:875–88. doi: 10.1517/14712598.2013.774366. [DOI] [PubMed] [Google Scholar]
- 260.Hornung V., Guenthner-Biller M., Bourquin C., Ablasser A., Schlee M., Uematsu S., Noronha A., Manoharan M., Akira S., de Fougerolles A., et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 261.Judge A.D., Sood V., Shaw J.R., Fang D., McClintock K., MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 262.Fedorov Y., Anderson E.M., Birmingham A., Reynolds A., Karpilow J., Robinson K., Leake D., Marshall W.S., Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Olejniczak M., Galka-Marciniak P., Polak K., Fligier A., Krzyzosiak W.J. RNAimmuno: a database of the nonspecific immunological effects of RNA interference and microRNA reagents. RNA. 2012;18:930–935. doi: 10.1261/rna.025627.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sioud M. RNA interference and innate immunity. Adv. Drug Deliv. Rev. 2007;59:153–163. doi: 10.1016/j.addr.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 265.Judge A., MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum. Gene Ther. 2008;19:111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- 266.Olejniczak M., Polak K., Galka-Marciniak P., Krzyzosiak W.J. Recent advances in understanding of the immunological off-target effects of siRNA. Curr. Gene Ther. 2011;11:532–543. doi: 10.2174/156652311798192770. [DOI] [PubMed] [Google Scholar]
- 267.Samaranch L., Salegio E.A., San Sebastian W., Kells A.P., Foust K.D., Bringas J.R., Lamarre C., Forsayeth J., Kaspar B.K., Bankiewicz K.S. Adeno-associated virus serotype 9 transduction in the central nervous system of nonhuman primates. Hum. Gen. Ther. 2012;23:382–389. doi: 10.1089/hum.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kenny G.D., Bienemann A.S., Tagalakis A.D., Pugh J.A., Welser K., Campbell F., Tabor A.B., Hailes H.C., Gill S.S., Lythgoe M.F., et al. Multifunctional receptor-targeted nanocomplexes for the delivery of therapeutic nucleic acids to the brain. Biomaterials. 2013;34:9190–9200. doi: 10.1016/j.biomaterials.2013.07.081. [DOI] [PubMed] [Google Scholar]
- 269.San Sebastian W., Samaranch L., Kells A.P., Forsayeth J., Bankiewicz K.S. Gene therapy for misfolding protein diseases of the central nervous system. Neurotherapeutics. 2013;10:498–510. doi: 10.1007/s13311-013-0191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Juliano R., Alam M.R., Dixit V., Kang H. Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 2008;36:4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.White P.J., Anastasopoulos F., Pouton C.W., Boyd B.J. Overcoming biological barriers to in vivo efficacy of antisense oligonucleotides. Expert Rev. Mol. Med. 2009;11:e10. doi: 10.1017/S1462399409001021. [DOI] [PubMed] [Google Scholar]
- 272.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 273.El Andaloussi S., Lakhal S., Mäger I., Wood M.J.A. Exosomes for targeted siRNA delivery across biological barriers. Adv. Drug Deliv. Rev. 2013;65:391–397. doi: 10.1016/j.addr.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 274.Dufour B.D., Smith C.A., Clark R.L., Walker T., McBride J.L. Intra-jugular vein delivery of AAV9-RNAi prevents neuropathological changes and weight loss in Huntington's disease mice. Mol. Ther. 2014 doi: 10.1038/mt.2013.289. 10.1038/mt.2013.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Weir D.W., Sturrock A., Leavitt B.R. Development of biomarkers for Huntington's disease. Lancet Neurol. 2011;10:573–590. doi: 10.1016/S1474-4422(11)70070-9. [DOI] [PubMed] [Google Scholar]
- 276.Georgiou-Karistianis N., Scahill R., Tabrizi S.J., Squitieri F., Aylward E. Structural MRI in Huntington's disease and recommendations for its potential use in clinical trials. Neurosci. Biobehav. Rev. 2013;37:480–490. doi: 10.1016/j.neubiorev.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 277.Strand A.D., Aragaki A.K., Shaw D., Bird T., Holton J., Turner C., Tapscott S.J., Tabrizi S.J., Schapira A.H., Kooperberg C., et al. Gene expression in Huntington's disease skeletal muscle: a potential biomarker. Hum. Mol. Genet. 2005;14:1863–1876. doi: 10.1093/hmg/ddi192. [DOI] [PubMed] [Google Scholar]
- 278.Anderson A.N., Roncaroli F., Hodges A., Deprez M., Turkheimer F.E. Chromosomal profiles of gene expression in Huntington's disease. Brain. 2008;131:381–388. doi: 10.1093/brain/awm312. [DOI] [PubMed] [Google Scholar]
- 279.Sassone J., Colciago C., Cislaghi G., Silani V., Ciammola A. Huntington's disease: the current state of research with peripheral tissues. Exp. Neurol. 2009;219:385–397. doi: 10.1016/j.expneurol.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 280.Bradford J.W., Li S., Li X.-J. Polyglutamine toxicity in non-neuronal cells. Cell Res. 2010;20:400–407. doi: 10.1038/cr.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Diamanti D., Mori E., Incarnato D., Malusa F., Fondelli C., Magnoni L., Pollio G. Whole gene expression profile in blood reveals multiple pathways deregulation in R6/2 mouse model. Biomark. Res. 2013;1 doi: 10.1186/2050-7771-1-28. doi:10.1186/2050-7771-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Träger U., Andre R., Lahiri N., Magnusson-Lind A., Weiss A., Grueninger S., McKinnon C., Sirinathsinghji E., Kahlon S., Pfister E.L., et al. HTT-lowering reverses Huntington's disease immune dysfunction caused by NFκB pathway dysregulation. Brain. 2014;137:819–833. doi: 10.1093/brain/awt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Boy J., Schmidt T., Wolburg H., Mack A., Nuber S., Böttcher M., Schmitt I., Holzmann C., Zimmermann F., Servadio A., et al. Reversibility of symptoms in a conditional mouse model of spinocerebellar ataxia type 3. Hum. Mol. Genet. 2009;18:4282–4295. doi: 10.1093/hmg/ddp381. [DOI] [PubMed] [Google Scholar]
- 284.Furrer S.A., Waldherr S.M., Mohanachandran M.S., Baughn T.D., Nguyen K.-T., Sopher B.L., Damian V.A., Garden G.A., La Spada A.R. Reduction of mutant ataxin-7 expression restores motor function and prevents cerebellar synaptic reorganization in a conditional mouse model of SCA7. Hum. Mol. Genet. 2013;22:890–903. doi: 10.1093/hmg/dds495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Lu X.-H., Yang X.W. “Huntingtin holiday”: progress toward an antisense therapy for Huntington's disease. Neuron. 2012;74:964–966. doi: 10.1016/j.neuron.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Corey D.R. RNA learns from antisense. Nat. Chem. Biol. 2007;3:8–11. doi: 10.1038/nchembio0107-8. [DOI] [PubMed] [Google Scholar]
- 287.Levin A.A., Yu R.Z., Geary R.S. Basic principles of the pharmacokinetics of antisense oligonucleotide drugs. In: Crooke ST, editor. Antisense Drug Technology: Principles, Strategies, and Applications. Boca Raton: CRC Press; 2008. pp. 183–215. [Google Scholar]
- 288.Koide R., Ikeuchi T., Onodera O., Tanaka H., Igarashi S., Endo K., Takahashi H., Kondo R., Ishikawa A., Hayashi T., et al. Unstable expansion of CAG repeat in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) Nat. Genet. 1994;6:9–13. doi: 10.1038/ng0194-9. [DOI] [PubMed] [Google Scholar]
- 289.Nagafuchi S., Yanagisawa H., Sato K., Shirayama T., Ohsaki E., Bundo M., Takeda T., Tadokoro K., Kondo I., Murayama N., et al. Dentatorubral and pallidoluysian atrophy expansion of an unstable CAG trinucleotide on chromosome 12p. Nat. Genet. 1994;6:14–18. doi: 10.1038/ng0194-14. [DOI] [PubMed] [Google Scholar]
- 290.MacDonald M.E., Ambrose C.M., Duyao M.P. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 291.La Spada A.R., Wilson E.M., Lubahn D.B., Harding A.E., Fischbeck K.H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 292.Orr H.T., Chung M.Y., Banfi S., Kwiatkowski T.J., Jr, Servadio A., Beaudet A.L., McCall A.E., Duvick L.A., Ranum L.P., Zoghbi H.Y. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 293.Imbert G., Saudou F., Yvert G., Devys D., Trottier Y., Garnier J.M., Weber C., Mandel J.L., Cancel G., Abbas N., et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat. Genet. 1996;14:285–291. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- 294.Pulst S.M., Nechiporuk A., Nechiporuk T., Gispert S., Chen X.N., Lopes-Cendes I., Pearlman S., Starkman S., Orozco-Diaz G., Lunkes A., et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- 295.Sanpei K., Takano H., Igarashi S., Sato T., Oyake M., Sasaki H., Wakisaka A., Tashiro K., Ishida Y., Ikeuchi T., et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat. Genet. 1996;14:277–284. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- 296.Kawaguchi Y., Okamoto T., Taniwaki M., Aizawa M., Inoue M., Katayama S., Kawakami H., Nakamura S., Nishimura M., Akiguchi I., et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- 297.Zhuchenko O., Bailey J., Bonnen P., Ashizawa T., Stockton D.W., Amos C., Dobyns W.B., Subramony S.H., Zoghbi H.Y., Lee C.C. Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat. Genet. 1997;15:62–69. doi: 10.1038/ng0197-62. [DOI] [PubMed] [Google Scholar]
- 298.David G., Abbas N., Stevanin G., Durr A., Yvert G., Cancel G., Weber C., Imbert G., Saudou F., Antoniou E., et al. Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat. Genet. 1997;17:65–70. doi: 10.1038/ng0997-65. [DOI] [PubMed] [Google Scholar]
- 299.Koide R., Kobayashi S., Shimohata T., Ikeuchi T., Maruyama M., Saito M., Yamada M., Takahashi H., Tsuji S. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease. Hum. Mol. Genet. 1999;8:2047–2053. doi: 10.1093/hmg/8.11.2047. [DOI] [PubMed] [Google Scholar]
- 300.Nakamura K., Jeong S.Y., Uchihara T., Anno M., Nagashima K., Nagashima T., Ikeda S., Tsuji S., Kanazawa I. SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum. Mol. Genet. 2001;10:1441–1448. doi: 10.1093/hmg/10.14.1441. [DOI] [PubMed] [Google Scholar]
- 301.Zhang Y., Engelman J., Friedlander R.M. Allele-specific silencing of mutant Huntington's disease gene. J. Neurochem. 2009;108:82–90. doi: 10.1111/j.1471-4159.2008.05734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Caplen N.J., Taylor J.P., Statham V.S., Tanaka F., Fire A., Morgan R.A. Rescue of polyglutamine-mediated cytotoxicity by double-stranded RNA-mediated RNA interference. Hum. Mol. Genet. 2002;11:175–184. doi: 10.1093/hmg/11.2.175. [DOI] [PubMed] [Google Scholar]
- 303.Hu J., Matsui M., Corey D.R. Allele-selective inhibition of mutant huntingtin by peptide nucleic acid-peptide conjugates, locked nucleic acid, and small interfering RNA. Ann. N.Y. Acad. Sci. 2009;1175:24–31. doi: 10.1111/j.1749-6632.2009.04975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Gagnon K.T., Watts J.K., Pendergraff H.M., Montaillier C., Thai D., Potier P., Corey D.R. Antisense and antigene inhibition of gene expression by cell-permeable oligonucleotide-oligospermine conjugates. J. Am. Chem. Soc. 2011;133:8404–8407. doi: 10.1021/ja200312y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Liu J., Pendergraff H., Narayanannair K.J., Lackey J.G., Kuchimanchi S., Rajeev K.G., Manoharan M., Hu J., Corey D.R. RNA duplexes with abasic substitutions are potent and allele-selective inhibitors of huntingtin and ataxin-3 expression. Nucleic Acids Res. 2013;41:8788–8801. doi: 10.1093/nar/gkt594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Aiba Y., Hu J., Liu J., Xiang Q., Martinez C., Corey D.R. Allele-selective inhibition of Huntingtin and Ataxin-3 expression by RNA duplexes containing unlocked nucleic acid (UNA) substitutions. Biochemistry. 2013;52:9329–9338. doi: 10.1021/bi4014209. [DOI] [PMC free article] [PubMed] [Google Scholar]