Background: Polyreactive antibodies can efficiently neutralize HIV.
Results: Heme induces polyreactivity of a human antibody, which recognizes distinct gp120 clades by an identical binding mechanism.
Conclusion: Antigen-binding promiscuity of a polyreactive antibody allows recognition of antigen with high molecular heterogeneity.
Significance: Characterization of the interaction of polyreactive antibodies with envelope proteins of HIV reveals novel strategies for control of the virus.
Keywords: Antibody, Heme, HIV-1 Protease, Protein-Protein Interaction, Thermodynamics, Antibody Polyreactivity, gp120
Abstract
Polyreactive antibodies play an important role for neutralization of human immunodeficiency virus (HIV). In addition to intrinsic polyreactive antibodies, the immune system of healthy individuals contains antibodies with cryptic polyreactivity. These antibodies acquire promiscuous antigen binding potential post-translationally, after exposure to various redox-active substances such as reactive oxygen species, iron ions, and heme. Here, we characterized the interaction of a prototypic human antibody that acquires binding potential to glycoprotein (gp) 120 after exposure to heme. The kinetic and thermodynamic analyses of interaction of the polyreactive antibody with distinct clades of gp120 demonstrated that the antigen-binding promiscuity of the antibody compensates for the molecular heterogeneity of the target antigen. Thus, the polyreactive antibody recognized divergent gp120 clades with similar values of the binding kinetics and quantitatively identical changes in the activation thermodynamic parameters. Moreover, this antibody utilized the same type of noncovalent forces for formation of complexes with gp120. In contrast, HIV-1-neutralizing antibodies isolated from HIV-1-infected individuals, F425 B4a1 and b12, demonstrated different binding behavior upon interaction with distinct variants of gp120. This study contributes to a better understanding of the physiological role and binding mechanism of antibodies with cryptic polyreactivity. Moreover, this study might be of relevance for understanding the basic aspects of HIV-1 interaction with human antibodies.
Introduction
The normal immune system contains antibodies (Abs)2 that bind to multiple structurally unrelated self-antigens and foreign antigens. These Abs are defined as polyreactive (1). The polyreactivity of Abs and B cell receptors is believed to contribute to the diversification of the immune repertoire and endows the immune system with the ability to bind, and respond, to virtually any potential antigen at a given time (2–4). Circulating polyreactive Abs found under physiological conditions, especially those also present in preimmune repertoires, have been proposed to constitute a first line of defense against pathogens and to participate in the maintenance of immune homeostasis (1, 5–8). Various pathological conditions, including different autoimmune diseases and viral infections, have been associated with an expansion of the repertoires of polyreactive Abs (9–12).
Of particular interest is the role of polyreactive Abs in infection by HIV-1. gp120 is a subunit of the HIV envelope protein complex that is responsible for virus attachment to host cells and thus represents an important target for neutralizing antibody responses (13–15). The enormous sequence heterogeneity of gp120 allows the virus to subvert the neutralizing antibody response (13, 15–17). After prolonged infection, however, some patients mount antibody responses that efficiently neutralize many different variants of HIV-1 (18–21). The repertoires of HIV-1-binding Abs from these patients contain abnormally high levels of autoreactive and polyreactive Abs (22–25). Moreover, some of the well characterized human monoclonal Abs that neutralize many variants of HIV-1 (referred to as “broadly neutralizing antibodies”, bNAbs) express a polyreactive antigen-binding behavior (20, 26–31). These findings imply that polyreactive Abs might play an important role for neutralization of HIV-1. In addition to intrinsically polyreactive Abs, normal immune repertoires contain Abs that can acquire antigen-binding polyreactivity post-translationally. Thus, exposure of these Abs to biologically relevant redox agents, such as reactive oxygen species, heme, iron ions, etc., results in a structural reorganization of their antigen-binding sites and acquisition of antigen-binding specificities to multiple unrelated antigens (32–34). Importantly, the substances that reveal the cryptic antibody polyreactivity are usually released in vivo at sites of inflammation or tissue damage and thus might modulate the antigen-binding properties of the susceptible Abs that are present in the immediate microenvironment (35). Biological functions and mechanism of antibodies with cryptic polyreactivity are not well understood. We observed that exposure of human polyclonal IgG, obtained from healthy donors, to heme results in acquisition of binding potential to HIV-1 gp120. Moreover, we identified a panel of human monoclonal antibodies isolated from seronegative individuals that acquire gp120-binding potential upon exposure to heme.3
The elucidation of interaction of cryptic polyreactive antibody with divergent variants of highly heterogeneous antigens, such as gp120, would contribute valuable information for understanding the binding mechanism of these antibodies. Furthermore, these studies might have relevance for understanding the basic aspects of the interaction of HIV-1 with antibodies.
Here, we provide biophysical characterization of the binding of a prototypic human IgG1 with cryptic polyreactivity. This antibody was cloned from a seronegative individual and acquires binding potential to HIV-1 gp120 only after interaction with heme. We compared the binding kinetics and thermodynamics of heme-induced Ab with the binding mechanism of two HIV-1-neutralizing antibodies as follows: F425 B4a1 and b12. Our results reveal that the polyreactive Ab has the potential to accommodate the molecular heterogeneity of gp120 and recognizes distinct variants of gp120 with an identical binding mechanism.
EXPERIMENTAL PROCEDURES
Viral Proteins and Antibodies
Recombinant envelope glycoprotein 120 (gp120) from HIV-1 strains BaL (clade B), CN54 (clade C), 96ZM651 (clade C), and 93TH975 (clade A/E) were obtained through the National Institutes of Health AIDS Reagent Program, Division of AIDS, NIAID. Recombinant gp120 from strains 92RW020 (clade A) and JRCSF (clade B) were purchased from Immune Technology Corp. (New York). Two monoclonal human gp120 V3 loop-specific antibodies (F425 B4a1 and 447-52D), two gp120 CD4-binding site-specific antibodies (b12 and VRC01), one V1/V2 region-specific antibody (PG9), and one gp120 glycan-specific antibody (2G12) were obtained through National Institutes of Health AIDS Reagent Program, Division of AIDS, NIAID. Antibody F425 B4a1 was contributed by Dr. Marshall Posner and Dr. Lisa Cavacini; antibody 447-52D was contributed by Dr. Susan Zolla-Pazner; antibody b12 was contributed by Dr. Denis Burton and Dr. Carlos Barbas; antibody VRC01 was contributed by Dr. John Mascola; antibody 2G12 was contributed by Dr. Hermann Katinger, and antibody PG9 was contributed by Dr. Denis Burton. The human monoclonal antibody (Ab21) was selected from a repertoire of monoclonal IgG1 antibodies. Ab21 was cloned from a memory B cell obtained from the synovium of a patient with rheumatoid arthritis (36, 37). Briefly, the variable region genes encoding the heavy and light chains were amplified by single cell PCR and cloned in PUC19 vector containing the genes encoding the constant Fc-γ1 or κ regions. The antibody was expressed by transient expression using HEK293 cells. Ab21 uses VH3–23 and Vκ1–27 variable gene families. This antibody is extensively mutated, with 22 and 20 mutations in the genes encoding the heavy and light chain variable regions, respectively. It also possesses long complementarity determining H3 region (22 amino acid residues).
Reagents
All reagents used in the study were of analytical grade quality. Stock solutions of oxidized heme (ferriprotoporphyrin IX) were prepared by dissolving hemin (Fluka, St. Louis, MO) in 0.05 n solution of NaOH or alternatively in pure DMSO. All metalloporphyrins were obtained from Frontier Scientific Inc. (Logan, UT). Stock solutions were prepared in DMSO (or water in the case of Mg(II)PP IX). The treatment of immunoglobulins was always performed with freshly prepared heme or other metalloporphyrins under dim light conditions.
ELISA, Binding to gp120
Ninety six well polystyrene plates (Nunc Maxisorp, Roskilde, Denmark) were coated with recombinant HIV-1 gp120 variants 92RW020, BaL, JRCSF, or CN54, diluted to 2.5 μg/ml in PBS. After incubation for 3 h at 22 °C, the residual binding sites on plates were blocked by PBS containing 0.25% Tween 20 (Sigma). In the first experimental setting, Ab21 (1 μm, 0.15 mg/ml) diluted in PBS was exposed to a fixed concentration (10 μm) of heme. After 30 min of incubation on ice, native and heme-exposed IgG were diluted serially in PBS containing 0.05% Tween 20 (PBS-T) to final concentrations of 50, 25, 12.5, 6.25, 3.125, 1.562, 0.781, 0.390, 0.195, and 0.0975 μg/ml for Ab21 and monoclonal gp120-specific antibodies 447-52D and F425 B4a1, and incubated with gp120 BaL-coated plates for 2 h at 22 °C. In the second experimental setting, a fixed concentration of Ab21 (6.7 μm, 1 mg/ml) was exposed for 30 min on ice to increasing concentrations of heme, 0, 0.25, 0.5, 1, 2, 4, 8, 16, 32, and 64 μm. Heme-exposed Ab21 was diluted in PBS-T to 50 μg/ml and incubated for 2 h at 22 °C with plates coated with the gp120 variants 92RW020, BaL, JRCSF, or CN54. After incubation with antibodies, in both experimental variants microtitration plates were washed extensively with PBS-T and incubated with a peroxidase-conjugated mouse anti-human IgG (clone JDC-10, Southern Biotech, Birmingham, AL) for 1 h at 22 °C. Immunoreactivity of IgG was revealed by measuring the absorbance at 492 nm after addition of peroxidase substrate, o-phenylenediamine dihydrochloride (Sigma) and stopping the reaction by addition of 2 n HCl.
ELISA, Effect of Metalloporphyrins on the Reactivity of Ab21
Ab21 was diluted to 1 μm in PBS and exposed to 10 μm different metalloporphyrins as follows: Zn(II)PP IX; Pd(II)PP IX; protoporphyrin IX (PP IX); Fe(III)PP IX (heme); Ni(II)PP IX; Co(III)PP IX; Cr(III)PP IX; Mg(II)PP IX; Mn(III)PP IX; Ga(III)PP IX, or Sn(IV)MP IX. After incubation for 5 min at 22 °C, samples were diluted in PBS-T to a final concentration of 50 μg/ml and then incubated with gp120 immobilized on ELISA plates for 2 h at 22 °C. The ability of metalloporphyrins to interfere with the binding of heme-exposed Ab21 to gp120 was studied by ELISA. gp120 (CN54) was immobilized on an ELISA plate at 2.5 μg/ml. After blocking, the protein was incubated for 5 min at 22 °C with PBS alone or PBS that contained 100 μm Pd(II)PP IX. Ab21 treated at 6.7 μm (1 mg/ml) with 20 μm heme was diluted to 50, 25, 12.5, 6.25, 3.125, 1.562, 0.781, 0.390, 0.195, and 0.0975 μg/ml in PBS-T and incubated with PBS or Pd(II)PP IX preincubated gp120 for 2 h at 22 °C. Next steps of the assay are identical to the ELISA protocol described above.
ELISA, Binding to a Panel of Antigens
The protocol utilized was essentially identical as described above, with small modifications. Each ELISA plate was coated with the following proteins or other macromolecules: human factor IX (LFB, France); human factor VIII (Kogenate FS, Bayer Healthcare); human factor H (CompTech, Tyler, TX); human C5 (CompTech); human C-reactive protein (Calbiochem); human hemoglobin (Sigma, apo-form prepared in-house), bovine histone type III (Sigma); HIV-1 gp120 96ZM651; HIV-1 gp41 MN (National Institutes of Health AIDS Research and Reagents Program); calf thymus DNA (Sigma), and Escherichia coli LPS, serotype O55:B5 (Sigma). All molecules were diluted in PBS at 5 μg/ml and incubated for 2 h at 22 °C. Ab21 was diluted to 1 μm (150 μg/ml) in PBS and exposed to 10 μm heme or to buffer only. Native and heme-exposed Ab21 were further diluted in T-PBS to 2 μg/ml and incubated with immobilized antigens for 2 h at 22 °C. The next steps of the assay are identical as those described above.
For evaluation of the effect of heme on Ab21 reactivity against the panel of unrelated antigens, the ratio of the binding intensity (A492 nm) of heme-treated versus the native Ab21 was obtained. This ratio was presented as the binding index. The value of the index equal to 1 indicates that heme does not influence the binding of Ab21 to the particular antigen. Index >1 indicates that the heme exposure causes an acquisition of a binding potential to the target antigen.
Absorbance Spectroscopy
Absorbance spectra were measured by using UNICAM Helios b, UV-visible spectrophotometer. Ab21 was diluted to 1 μm in PBS and titrated with increasing concentrations of heme (0, 0.25, 0.5, 1, 2, 4, 8, and 16 μm). Aliquots of hemin stock solution (1 mm) were added both to a cuvette containing Ab21 and to a reference cuvette containing only buffer. After addition of each heme aliquot and incubation for 2 min in dark, the absorbance spectra in the wavelength range 350–700 were recorded. The spectra were scanned at rate of 300 nm/min. All measurements were performed at 22 °C, in a quartz cuvette with optical path of 1 cm.
Fluorescence Spectroscopy
Quenching of intrinsic tryptophan fluorescence of Ab21 by heme was measured by using Hitachi F-2500 fluorescence spectrophotometer (Hitachi Instruments Inc., Wokingham, UK). Ab21 was diluted to 0.1 μm in PBS and titrated with increasing concentrations (0, 0.01, 0.025, 0.05, 0.1, 0.2, 0.4, 0.8, 1.6, and 3.2 μm) of heme, added as aliquots of hemin stock solutions. The emission spectra of Ab21 was recorded 1 min following addition of each aliquot of heme. A wavelength of 295 nm was used to selectively excite tryptophan residues. Excitation and emission slits were adjusted to 10 nm. The emission spectra of Ab21 were measured in the wavelength range 310–450 nm, at a scan speed of 300 nm/min. Quartz cuvette with a 1-cm optical path was used in the experiment. All measurements were done at 22 °C.
Size-exclusion Chromatography
Molecular composition of native and heme-exposed Ab21 was compared by using FPLC Akta Purifier (GE Healthcare), equipped with Superose 12 10/300 column. Ab21 was diluted to 100 μg/ml (670 nm) in PBS and exposed to 6.7 μm (10 m excess) of heme. One ml of each native or heme-exposed Ab21 was loaded onto a column equilibrated with PBS. The flow rate of 0.5 ml/min was used. Chromatograms were recorded by using UV detection of protein at wavelength of 280 nm.
Determination of Functional Affinity of Antibodies by ELISA
Ninety six well polystyrene plates were coated with recombinant HIV-1 gp120 BaL diluted to 2.5 μg/ml in PBS. After incubation for 3 h at 22 °C, the residual binding sites on plates were blocked by PBS containing 0.25% Tween 20. Ab21 was diluted to 1 μm and exposed to 10 μm heme (1 mm stock solution in 0.05 n NaOH). Heme-treated Ab21 was diluted to 2 μg/ml in T-PBS. Ab21 was incubated with gp120 BaL-coated plates for 2 h at 22 °C. After incubation, microtitration plates were washed extensively with T-PBS and incubated for 10 min at 37 °C with increasing concentrations of urea (0, 0.5, and 1–8 m) in PBS. After extensive washing of the plates with T-PBS, the immunoreactivity was developed as described above.
pH Dependence Analyses
Ninety six well polystyrene plates were coated with 2.5 μg/ml HIV-1 gp120 (BaL) in PBS for 3 h at 22 °C. The plates were blocked with 0.25% Tween 20 in PBS. The induction of cryptic polyreactivity of Ab21 was performed by exposure of 2 μm Ab21 to 20 μm hematin (1 mm stock in 0.05 n NaOH). To study the pH dependence of the interaction between gp120 and antibodies Ab21, F425 B4a1, and b12, buffers with different pH values were used as follows: acetate buffer (25 mm sodium acetate/acetic acid, 150 mm NaCl, 0.05% Tween 20) was used to maintain pH values of 4 and 5; MES buffer (25 mm MES, 150 mm NaCl, 0.05% NaCl), pH 6; HBS buffer (10 mm HEPES, 150 mm NaCl, 0.05% Tween 20), pH 7 and 8; TBS buffer (50 mm Tris, 150 mm NaCl, 0.05% Tween 20), pH values of 7–11. For study of binding to gp120, heme-treated Ab21 and antibodies F425 B4a1 and b12 were diluted to 15 and 1 μg/ml, respectively, in buffers with different pH values and incubated with immobilized gp120 for 2 h at an ambient temperature of 22 °C. After extensive washing, the plates were incubated for 1 h at 22 °C with a peroxidase-conjugated mouse anti-human IgG (clone JDC-10, Southern Biotech, Birmingham, AL). Immunoreactivities were revealed using the o-phenylenediamine substrate (Sigma). Absorbance at 492 nm was read after stopping of the reaction by addition of 2 n HCl.
Surface Plasmon Resonance Interaction Analyses
The binding kinetics and thermodynamics of interaction of human monoclonal IgG1 antibodies Ab21, F425 B4a1, and b12 with gp120 were determined by surface plasmon resonance-based technique (BIAcore 2000, BIAcore, Uppsala, Sweden).
Recombinant gp120 molecules from different virus strains 92RW020, BaL, JRCSF, CN54, 96ZM651, and 93TH975 were immobilized on a CM5 sensor chip (BIAcore) using amino-coupling kit (BIAcore), as described by the manufacturer. In brief, gp120 was diluted in 5 mm maleic acid, pH 5, to a final concentration of 10 μg/ml and injected over the preactivated sensor surface. The achieved immobilization levels for gp120 variants 92RW020, BaL, JRCSF, CN54, 96ZM651, and 93TH975 were 6483, 1558, 5685, 6757, 4316, and 2857 resonance units, respectively. Experiments were performed using HBS-EP (0.01 m HEPES, pH 7.4, containing 0.15 m NaCl, 3 mm EDTA, and 0.005% Tween 20). To study the effect of ionic strength, the binding was performed in modified HBS-EP, containing 0.05 m or 0.45 NaCl (low or high ionic strength). The 2-fold dilutions of antibody F425 B4a1 (50 to 0.049 nm), antibody b12 (20 to 0.039 (0.0049) nm), and of heme-exposed Ab21 (250 to 0.244 nm) were injected at flow rate of 30 μl/min. The association and dissociation phases of the interaction were monitored for 4 and 5 min, respectively. The binding to the surface of the control uncoated flow cell was subtracted from the binding to the gp120-coated flow cells. The regeneration of the binding surface after binding of antibodies F425 B4a1 and b12 was performed by injection of 4 m MgCl2. The regeneration of heme-exposed Ab21 was achieved by exposure of the sensor surface to 300 mm imidazole. The BIAevaluation version 4.1 software (BIAcore) was used for the estimation of the kinetic rate constants. Calculations were performed by global analysis of the experimental data using the Langmuir binding with a drifting baseline model included in the software. The highest injected antibody concentrations i.e. 250 nm (Ab21) and 50 nm (F425 B4a1) were not included in the kinetic analyses. In the case of antibody b12, the highest injected concentration (20 nm) was only included for evaluation of the binding kinetics to gp120 96ZM651.
Evaluation of Binding Thermodynamics
We applied Eyring's approach for evaluation of the activation thermodynamic parameters of the interactions of different variants of gp120 with antibodies Ab21, F425 B4a1, and b12. Kinetic measurements were performed independently at 10, 15, 20, 25, 30, and 35 °C. For evaluation of the activation thermodynamics, the kinetic rate constants were used to build Arrhenius plots. The values of slopes of the Arrhenius plots were calculated by using a linear regression analysis of the experimental kinetic data by using GraphPad Prism version 4 (GraphPad Prism Inc.) and substituted in Equations 1–4,
where the “slope” = ∂ln(ka/d/∂(1/T), and where Ea is the activation energy. The enthalpy, entropy, and Gibbs free energy changes characterizing the association phase were calculated using Equations 2–4,
where T is the temperature in Kelvin, k′ is the Boltzmann constant, and h is Planck's constant. All activation thermodynamic parameters were determined at the reference temperature of 25 °C (298.15 K).
Determination of Epitope of Ab21 on gp120
Surface plasmon resonance was used to determine the epitope(s) of Ab21 on gp120. gp120 (BaL) was immobilized on a CM5 sensor chip by using standard amino-coupling protocol (see above). Interactions of HIV-1-neutralizing antibodies that recognize different epitopes on gp120 (b12 (CD4bs); VRC01 (CD4bs); 447-52D (V3 loop); 2G12 (glycan-dependent epitope); and PG9 (glycan and protein epitope in V1/V2 region) were evaluated before and after the binding of heme-exposed Ab21.Ab21 at a concentration of 1 μm and then exposed to 10 μm hematin in PBS. Ab21 (250 nm) was allowed to interact with gp120 for 10 min. Each HIV-1-neutralizing antibody was injected for 5 min at the following concentrations: 5 nm (b12), 20 nm (VRC01), 100 nm (447-52D and 2G12), and 500 nm (PG9). All dilutions of antibodies were performed in HBS-EP. The ability of polyreactive Ab21 to reduce the binding of different HIV-1-neutralizing antibodies to gp120 was evaluated by estimating the ratio of the maximal binding response (in resonance units) of each of HIV-1-specific antibody in the presence and absence of bound polyreactive Ab21.
RESULTS
Ab21 Acquires Polyreactivity and a Binding Potential to gp120 after Exposure to Heme
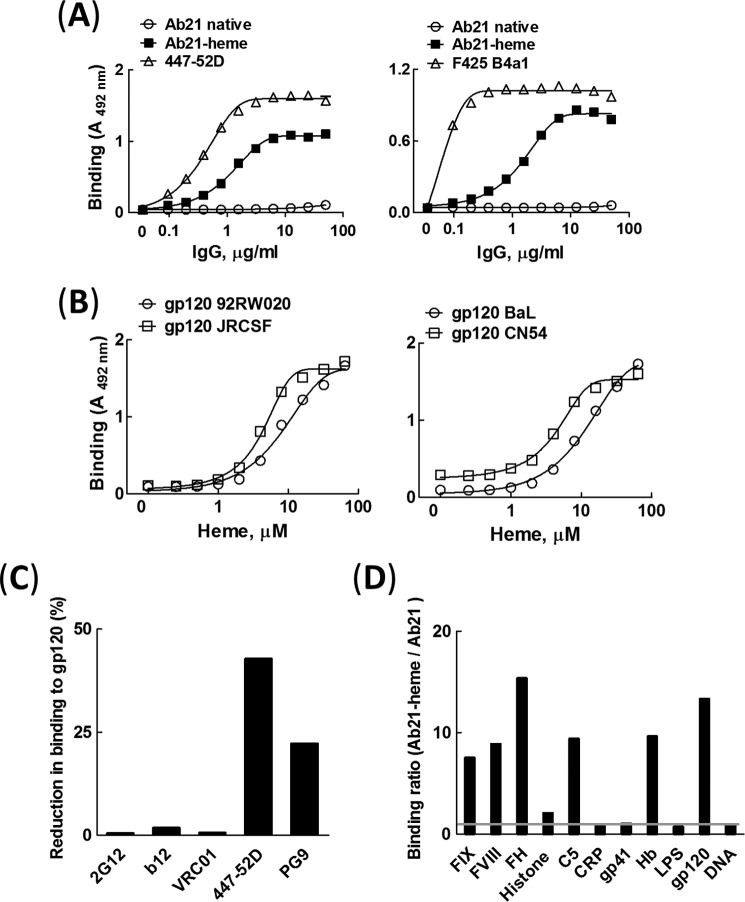
To elucidate the mechanism of interaction of gp120 with Abs with cryptic polyreactivity, a prototypic heme-sensitive antibody was selected from a repertoire of human monoclonal IgG1 Abs.3 The selected antibody, referred to in this study as Ab21, acquires a strong binding potential for gp120 only after exposure to heme (Fig. 1A). First, we studied the binding of Ab21 to gp120 BaL and compared it with the binding of the HIV-1 gp120 V3 loop-specific Abs, F425 B4a1 and 447-52D. These Abs were isolated from HIV-1 seropositive patients and have a potent neutralization activity against HIV-1 BaL virus (38–40). The binding of heme-exposed Ab21 to gp120 BaL was substantial but of a lower intensity as compared with the interactions of HIV-1 BaL-neutralizing Abs (Fig. 1A). The induction of binding to gp120 of Ab21 was dependent on heme concentration (Fig. 1B). Remarkably, heme-exposed Ab21 recognized divergent variants of gp120 with almost similar binding intensities (Fig. 1B).
FIGURE 1.
Characterization of antigen-binding properties of human heme-sensitive monoclonal IgG. A, comparison of binding of native Ab21 (open circles), heme-exposed Ab21 (closed squares), and patient-derived HIV-1-neutralizing antibodies 447-52D and F425 B4a1 (open triangles) to gp120. Monoclonal antibodies were diluted 2-fold (50 to 0.0975 μg/ml) and incubated with gp120 variant BaL immobilized on ELISA plate. Each data point represents the mean of optical density of duplicate wells ±S.E. Representative results of one of two independent experiments is shown. B, heme concentration dependence of reactivity of Ab21 to distinct variants of gp120. Ab21 (6.7 μm or 1 mg/ml) was exposed to increasing concentrations of heme (0–64 μm) and incubated at 0.335 μm (50 μg/ml) with different types of gp120 (92RW020, JRCSF, BaL, and CN54) immobilized on ELISA plates. Each data point represents mean optical density and ±S.E. from duplicate wells. Representative results of one of two independent experiments are shown. C, inhibition of binding of broadly neutralizing antibodies to gp120 (BaL) by heme-exposed Ab21. The interactions of HIV-1-neutralizing antibodies (20 nm VRC01; 5 nm b12; 10 nm 2G12; 100 nm 447-52D, and 500 nm PG9) were studied by surface plasmon resonance-based assay before and after binding of 250 nm heme-exposed Ab21 to immobilized gp120. The graph shows percentage reduction of binding of bNAbs to gp120 in the presence of bound polyreactive Ab21. D, binding of Ab21 to a panel of unrelated antigens. The effect of heme on the reactivity of Ab21 was evaluated by estimation of the ratio of binding intensity (absorbance at 492 nm) of the heme-treated Ab21 (1 μm Ab21/10 μm heme) versus the binding intensity of the native Ab21. The native and heme-exposed Ab21 were incubated at 2 μg/ml with a panel of antigens (indicated on the graph). Binding ratio of 1 (depicted as a gray line on the graph) indicates that binding intensity of native and of heme-exposed Ab21 does not differ.
To map the epitope(s) of polyreactive Ab21 on gp120, we performed competition assay with HIV-1 broadly neutralizing antibodies that target different regions of gp120 (Fig. 1C). The binding of heme-exposed Ab21 to gp120 did not influence the binding of CD4bs-specific antibodies (b12 and VRC01) (41, 42) or gp120 glycan-specific antibody (2G12) (43). However, the binding of polyreactive Ab21 resulted in reduction of the binding of antibodies that target the V1/V2 region (PG9) (44) and especially the V3 region (447-52D) (Fig. 1C).
The exposure of Ab21 to heme resulted in acquisition of polyreactivity as is evident from the binding to a panel of unrelated antigens (Fig. 1D). Importantly, Ab21 acquired marked reactivity only to a subset of antigens in the studied panel (factor IX, factor VIII, factor H, C5, hemoglobin, and gp120). This result implied that heme-exposed Ab21 is not nonspecifically binding to all possible antigens but still has a potential to discriminate between unrelated molecules/epitopes.
Heme Binds to Ab21
Our previous studies indicate that induction of polyreactivity of antibodies by heme occurs through direct binding of the macrocyclic cofactor to the immunoglobulin molecule (34). To figure out whether this is also valid for acquisition of binding polyreactivity by Ab21, we performed spectroscopy analyses (Fig. 2, A and B). Absorbance spectroscopy revealed that the exposure of Ab21 to heme resulted in an increase in the absorbance intensity of heme in high and low energy regions of the spectrum (Fig. 2A). Differential spectral analyses also indicated the presence of a substantial red shift (>10 nm) in the high energy region. These spectral changes of heme are consistent with binding of the cofactor molecule and coordination of its central iron ion to the IgG molecule. Furthermore, the ability of heme to interact with Ab21 was studied by quenching the intrinsic tryptophan fluorescence of the protein (Fig. 2B). Exposure to heme resulted in a dose-dependent decrease in the fluorescence signal of Ab21. Taken together, these results indicate that Ab21 binds heme.
FIGURE 2.
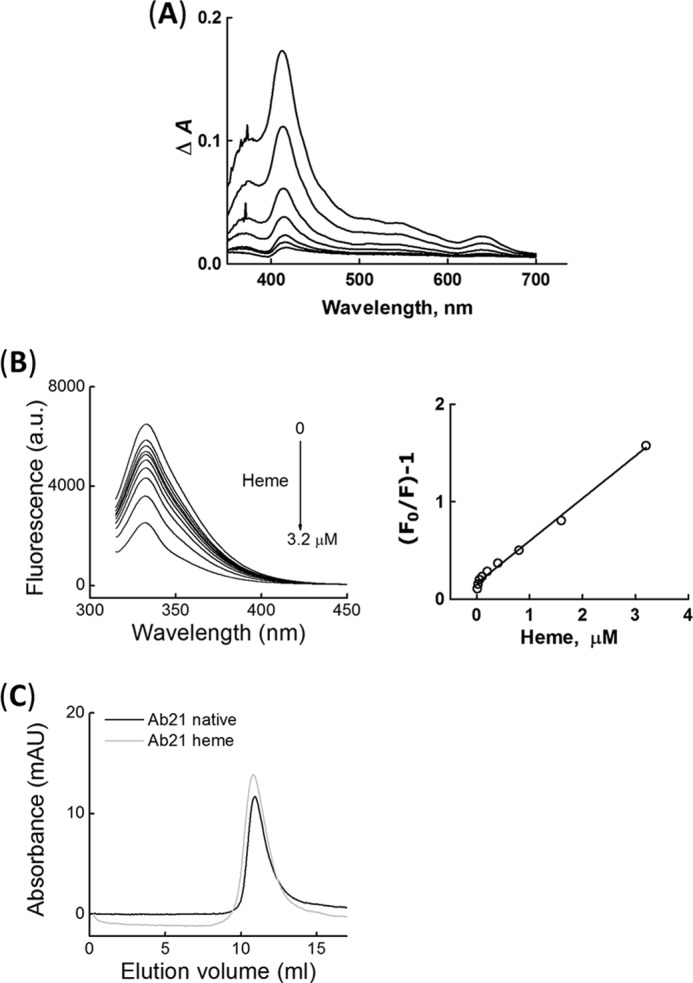
Characterization of heme binding to Ab21. A, differential spectra of heme obtained by absorbance spectroscopy after titration of 1 μm Ab21 with increasing concentrations of heme (0.25–16 μm). Differential spectra were obtained after subtraction of spectra of heme in PBS from the spectra of identical concentrations of heme in the presence of Ab21. B, quenching of tryptophan fluorescence of Ab21 by heme. Left panel, emission spectra of Ab21 (0.1 μm) obtained after titration with increasing concentrations of heme (0–3.2 μm). Right panel, Stern-Volmer plot that depicts the quenching of tryptophan fluorescence by increasing concentrations of heme. C, size-exclusion chromatography. Elution profiles of 1 μm native Ab21 (black line) and 1 μm Ab21, exposed to 10 μm heme (gray line).
Next, we tested the possibility that heme exposure induces formation of aggregates of Ab21, which might result in multivalent, nonspecific binding to gp120 or to other proteins. Size-exclusion chromatography profiles indicated that native Ab21 and Ab21 exposed to a 10-fold molar excess of heme elute as a single pick, which corresponds to an approximate molecular mass of 150 kDa (Fig. 2C). This result ruled out formation of aggregates of Ab21 upon exposure to an excess of heme.
Nature of Metal Ion in the Porphyrin Structure Plays an Essential Role for Induction of Specificity of Ab21 to gp120
We tested the ability of different metalloporphyrins IX (analogues of heme that differ by the nature of the central metal ion) to induce binding specificity of Ab21 to gp120. The nature of the metal ion in the porphyrin structure was found to determine the potential of uncovering the cryptic binding specificity of Ab21 to gp120. The most prominent effect on the reactivity of Ab21 was observed after exposure to heme (Fe(III) protoporphyrin IX). The antibody also demonstrated sensitivity to Co(III) protoporphyrin IX and Cr(III) protoporphyrin IX (Fig. 3A). However, most of the studied metalloporphyrins were unable to induce binding of Ab21 to gp120 (Fig. 3A).
FIGURE 3.
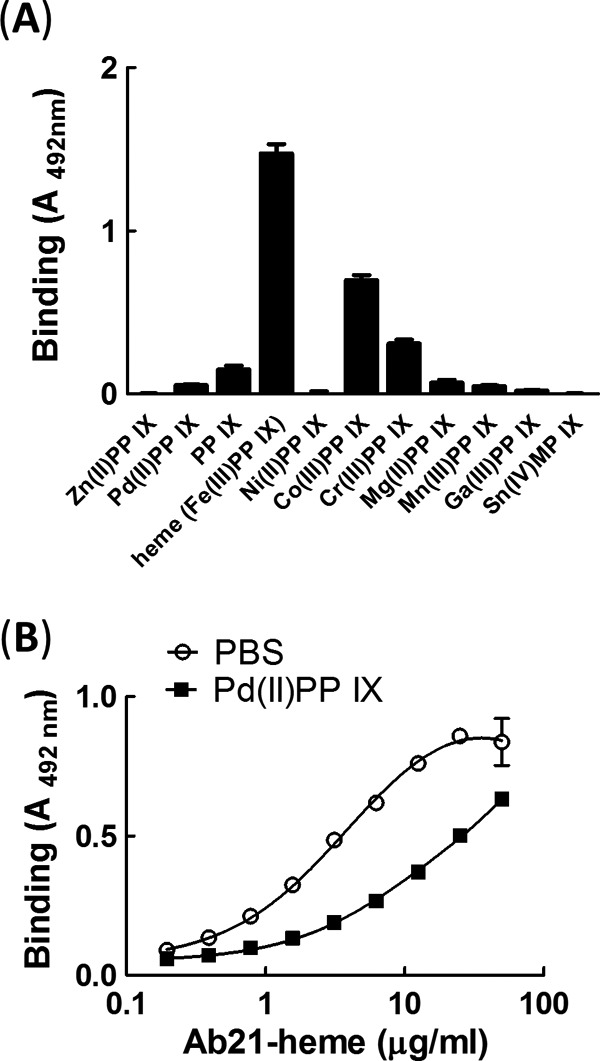
Effect of metalloporphyrins on reactivity of Ab21 to gp120. A, Ab21 at 1 μm was exposed to 10 μm Pd(II)PP IX, protoporphyrin IX, Fe(III)PP IX (heme), Ni(II)PP IX, Co(III)PP IX, Cr(III)PP IX, Mg(II)PP IX, Mn(III)PP IX, Ga(III)PP IX, or Sn(IV)MP IX. After dilution to IgG concentration of 50 μg/ml (0.33 μm), samples were incubated with immobilized gp120. B, inhibition of binding of Ab21-heme to gp120 exposed to Pd(II)PP IX. Immobilized gp120 was first treated with 100 μm Pd(II)PP IX in PBS or buffer only and after incubated with increasing concentrations of Ab21 exposed to excess of heme.
Previous studies have implied that heme might serve as an interfacial cofactor of cryptic polyreactive antibodies for binding to diverse antigens (34). To test this hypothesis in the case of Ab21, we preincubated immobilized gp120 with an excess of metalloporphyrin that was unable to induce gp120-binding specificity of Ab21, i.e. Pd(II) protoporphyrin IX (Fig. 3A). This treatment may saturate the binding sites for porphyrins on the surface of gp120 and prevent interaction of heme. Saturation of gp120 with inert porphyrin resulted in a reduction of binding of heme-exposed Ab21 (Fig. 3B).
Heme-exposed Ab21 Recognizes Distinct Variants of gp120 with Similar Binding Kinetics
The binding kinetics of the heme-sensitive antibody Ab21 with a panel of distinct gp120 variants was compared with that of F425 B4a1 and of a bNAb specific for the CD4-binding site on gp120, b12. An example of binding to gp120 BaL and gp120 93TH975 is depicted on Fig. 4. The kinetic analyses confirmed that Ab21 is able to recognize distinct variants of gp120 with a very narrow range of apparent binding affinities (KD values between 30.6 and 67.4 nm, see Table 1). Values of the kinetic rate constants (ka and kd) also clustered in a narrow range (Table 1). The measured association rate constants (ka in the range 6.8 × 104–1.4 × 105 m−1 s−1) reflect a rapid complex formation with all gp120 tested. However, these complexes are not very stable, as evident from the relatively high kd values (Table 1). In contrast to the uniform binding behavior of heme-Ab21, F425 B4a1 and b12, which are derived from seropositive patients, were much more discriminative in their interactions with gp120. Thus, surface plasmon resonance analyses revealed that F425 B4a1 and b12 formed very stable complexes with the gp120 variants, BaL and JRCSF, as evident by the absence of detectable dissociation (Fig. 4 and Table 1), whereas complexes with gp120 92RW020 were less stable, and lower affinity interactions were seen with gp120 96ZM651. Whereas Ab F425 B4a1 formed very stable complexes with gp120 variant CN54, b12 demonstrated lower stability of the complexes and bound this variant of gp120 with lower affinity (Table 1). F425 B4a1 and b12 were not able to interact at all with gp120 93TH975 (Fig. 4). F425 B4a1 recognized gp120 JRCSF, gp120 BaL, and gp120 CN54 with an apparent KD value (assuming a kd value of 10−5 s−1) in the picomolar range (Table 1). F425 B4a1 bound gp120 92RW020 and gp120 96ZM651 with KD values of 4.96 and 358 nm, respectively (Table 1). The bNAb b12 recognized gp120 variants BaL and JRCSF with low picomolar affinity (apparent KD values of 6 pm), although it bound other gp120 variants with nanomolar affinity (Table 1).
FIGURE 4.
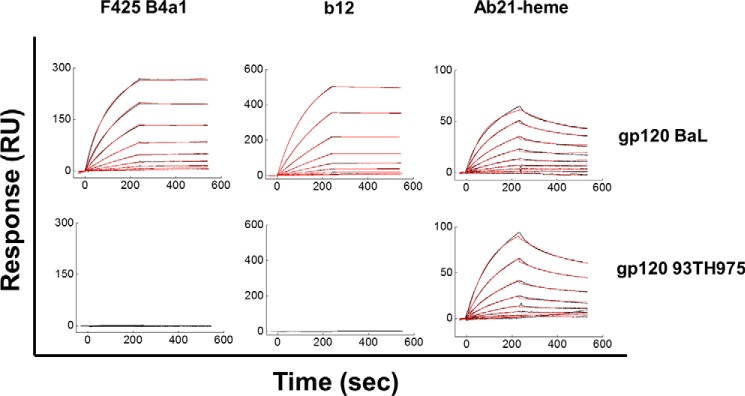
Representative real time interaction profiles obtained by surface plasmon resonance kinetic analyses of interaction of human monoclonal antibodies Ab21 and F425 B4a1 with gp120. Real time interaction profiles were obtained by binding increasing concentrations of Ab21-heme (0.244 (0.488) to 125 nm), F425 B4a1 (0.049 to 25 nm), and b12 (0.019 to 5 nm) to distinct variants of gp120 (BaL and 93TH975) immobilized on sensor chips. The kinetic analyses were performed at 25 °C. Each graph represents experimental curves (bleak lines) and curves obtained by fitting the data to the model of Langmuir binding with a drifting baseline by BIAevaluation (red line). The induction of reactivity of Ab21 to gp120 was performed by exposing 1 μm antibody to 10 μm hematin.
TABLE 1.
Kinetics of interaction of monoclonal antibodies with gp120
Values of the kinetic rate constants (ka and kd) and equilibrium dissociation constant (KD) were determined by global analyses of real time interaction profiles obtained after interaction of F425 B4a1 (0.049–25 nm), b12 (0.019 (0.039)–5 nm for binding to gp120 BaL, JRCSF, and CN54; 0.039–10 nm, for binding to gp120 92RW020; and 0.039–20 nm for bindings to gp120 96ZM651) and heme-exposed Ab21 (0.49–125 nm) with different variants of gp120. The absence of dissociation of F425 B4a1 in the case of binding to gp120 BaL, gp120 JRCSF, and gp120 CN54 and of b12 in the case of bindings to gp120 BaL and gp120 JRCSF precluded evaluation of reliable values of kd by applying global kinetic analyses. However, by using kinetics simulation software (BIAsimulation), it was estimated that the dissociation rate of F425 B4a1 and b12 in complex with these variants of gp120 is equal or lower than 10−5 s−1. The provisional values of kd (equal to 10−5 s−1) and resulting from the ratio kd/ka, arbitrary KD values are given in table in italics.
| F425 B4a1 |
B12 |
Ab21-heme |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ka ×105 m−1 s−1 (mean ± S.D.) | kd ×10−3 s−1 (mean ± S.D.) | KD | ka ×105 m−1 s−1 (mean ± S.D.) | kd ×10−3 s−1 (mean ± S.D.) | KD | ka ×105 m−1 s−1 (mean ± S.D.) | kd ×10−3 s−1 (mean ± S.D.) | KD | |
| nm | nm | nm | |||||||
| 92RW020 | 2.05 ± 0.01 | 1.02 ± 0.07 | 4.96 | 12.2 ± 0.001 | 4.8 ± 0.11 | 3.6 | 0.982 ± 0.013 | 6.01 ± 0.47 | 61.2 |
| BaL | 4.10 ± 0.07 | <0.01 | 0.024 | 15.2 ± 0.001 | <0.01 | 0.006 | 0.968 ± 0.012 | 2.96 ±0.22 | 30.6 |
| JRCSF | 3.19 ±0.026 | <0.01 | 0.031 | 15.5 ±0.01 | <0.01 | 0.006 | 0.907 ± 0.011 | 6.11 ± 0.15 | 67.4 |
| CN54 | 3.04 ± 0.055 | <0.01 | 0.033 | 6.44 ± 0.6 | 1.44 ± 0.06 | 2.23 | 0.775 ± 0.017 | 2.65 ± 0.078 | 34.2 |
| 96ZM651 | 0.56 ± 0.026 | 20 ± 0.28 | 358 | 5.16 ± 0.38 | 15 ± 0.45 | 29.1 | 1.43 ± 0.051 | 7.03 ± 0.36 | 49 |
| 93TH975 | #a | # | # | # | # | # | 0.681 ± 0.008 | 3.18 ± 0.12 | 46.6 |
a # means absence of detectable binding.
The marked differences in the affinity of F425 B4a1 or b12 for binding to different gp120 variants were mainly due to differences in the dissociation rate constants. In most of the cases, the antibody association rate was very high (Table 1). All interactions of b12 were characterized by an association rate constant that was greater than that of F425 B4a1 and Ab21 (Table 1 and Fig. 6).
FIGURE 6.
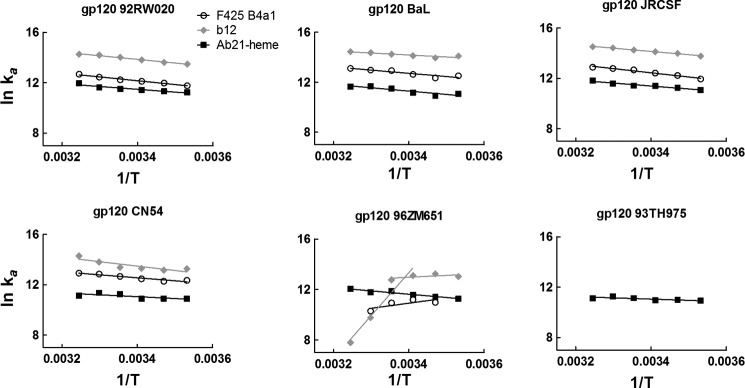
Temperature dependence of association of Ab21, F425 B4a1, and b12 with gp120. Arrhenius plots depict the temperature dependence of association rate constant of Ab21-heme (black squares), F425 B4a1 (white circles), and b12 (gray diamonds) obtained after interaction with different variants of gp120. The association rate constants were determined by global analysis of sensorograms generated after evaluation of binding kinetics of Ab21-heme, F425 B4a1, and b12 (as described under “Experimental Procedures”) at varying temperatures (10, 15, 20, 25, 30, and 35 °C). The slopes of the Arrhenius plots were determined by linear regression analyses. The temperature dependence of binding of b12 to 96ZM651 displayed complex behavior. The data were fitted by linear regression analyses in two alternative ranges, 10–25 and 25–35 °C.
By analyzing data with a model that assumes the presence of mass transfer limitations, we observed that the values of the association rate constants of the Abs were not biased by this possible binding artifact (data not shown).
Finally, we studied the stability at an equilibrium of the complexes formed between polyreactive Ab21 and gp120 by elution with increasing concentrations of urea, a method used for estimation of antibody avidity (45). As shown in Fig. 5, exposure to increasing concentrations (0–8 m) of urea dissociated only negligibly the complexes of Ab21 (at 8 m urea the binding was reduced by only 20%). These data imply that heme-induced polyreactive Ab21 interacts with the HIV envelope protein with a high functional affinity (avidity).
FIGURE 5.
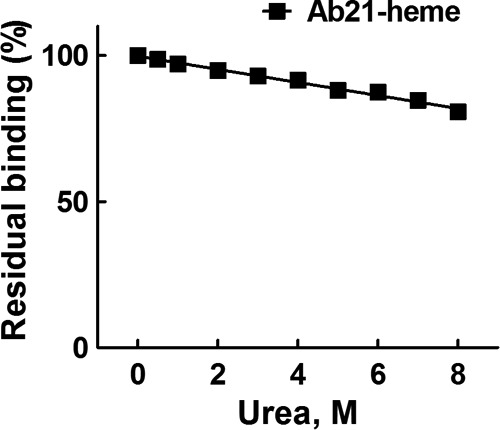
Estimation of antibody avidity to gp120 BaL. Ab21 was diluted to 1 μm and exposed to 10 μm hematin. Heme-treated Ab21 was diluted 2 μg/ml in T-PBS. After incubation with Ab21, the microtitration plate was washed extensively with T-PBS and incubated for 10 min at 37 °C with increasing concentrations of urea as follows: 0, 0.5, and 1–8 m in PBS. After extensive washing of the plate with T-PBS, the immunoreactivity was developed as described above.
Heme-induced Polyreactive Antibody Recognizes Distinct gp120 Variants with Similar Energetic Changes
To provide further understanding about the mechanism of promiscuous recognition of gp120 by cryptic polyreactive antibodies, we compared the binding thermodynamics of heme-exposed Ab21, F425 B4a1, and b12. Kinetic parameters of the interaction of the three Abs with different variants of gp120 were determined as a function of temperature (in the range 10–35 °C) and are presented as Arrhenius plots on Fig. 6. The association of heme-exposed Ab21 to all studied gp120 variants demonstrated identical temperature dependence; an increase in the temperature resulted in a concomitant increase in the association rate constant (Fig. 6). In contrast, the association of F425 B4a1 and b12 showed two modes of temperature sensitivity, depending on the gp120 variant. In the case of gp120 92RW020, BaL, JRCSF, and CN54, an increase in the temperature resulted in an increase in ka. The temperature sensitivity of the ka was nearly identical to that of the ka determined for heme-induced Ab21 (Fig. 6). In contrast, the association of F425 B4a1 with gp120 96ZM651 showed an increase of the temperature leading to a decrease in ka; at the highest temperature used (35 °C), the association was completely abrogated. Association of b12 with gp120 96ZM651 displayed complex behavior (Fig. 6). Thus, in range 10–25 °C, the temperature dependence of ka of b12 had a similar tendency as that observed for F425 B4a1. However, the association rate of b12 showed a very high temperature sensitivity in the 25–35 °C range (Fig. 6).
The dissociation rate of Ab21 from all variants of gp120 was not sensitive to changes in temperature (data not shown). In cases where a reliable estimation of the kd was possible for F425 B4a1 and b12, we observed a temperature sensitivity of the dissociation (data not show). Thus, an increase in the temperature resulted in an elevation of the dissociation rate. Temperature dependences of the KD value indicated that the overall binding affinity of cryptic polyreactive antibody was only negligibly influenced by changes in the interaction temperature. In contrast, the temperature sensitivities of KD of F425 B4a1 and b12 were more pronounced. In the case of F425 B4a1, an elevation of the interaction temperature resulted in a decrease in the binding affinity to gp120 92RW020 and especially to gp120 96ZM651 (data not shown). In the case of b12, the reduction of the binding affinity with an increase of temperature was pronounced for interactions with gp120 CN54 and gp120 96ZM651 (data not shown).
The Arrhenius plots were used for calculation of changes in the activation thermodynamics of Ab21, F425 B4a1, and b12 during binding to gp120. Energetic changes during association of cryptic Ab21 to different gp120 molecules were qualitatively identical (Fig. 7). Thus, changes in the activation enthalpy (ΔH‡) of binding of Ab21 to all variants of gp120 were positive (ΔH‡ varied from 5.8 to 19.7 kJ mol−1), indicating that the association of the antibody is an endothermic process characterized by an unfavorable contribution to the overall binding affinity. The changes in activation entropy (TΔS‡), observed during the association process of Ab21, were also qualitatively identical for all gp120 variants (Fig. 7). Thus, TΔS‡ had an unfavorable contribution to the overall affinity with values ranging from −24 to −39.6 kJ mol−1. These values of TΔS‡ are consistent with the presence of minimal structural changes in the interacting proteins during association. The activation thermodynamics analyses revealed that the HIV-1-neutralizing antibodies F425 B4a1 and b12 utilize different mechanisms for binding to different variants of gp120 (Fig. 7). Whereas the association of F425 B4a1 and b12 with gp120 from strains 92RW020, BaL, JRCSF, and CN54 was characterized with unfavorable ΔH‡ values (ranging from 17.1 to 25.2 kJ mol−1 in the case of F425 B4a1 and from 10.5 to 26.7 kJ mol−1 in the case of b12), the association of the same antibodies to gp120 96ZM651 presented with a negative sign and favorable contribution to the overall binding affinity. Importantly, the recognition of gp120 96ZM651 by b12 revealed marked changes in the association enthalpy when temperature ranged between 25 and 35 °C (Fig. 7). Qualitative differences in ΔH‡ reflect differences in the type of noncovalent interactions between the antibodies and different gp120. Although the changes in the activation entropy for all gp120 recognized by F425 B4a1 and b12 were unfavorable, the negative value of TΔS‡ observed for the interaction with gp120 96ZM651 was higher than for the other gp120 variants (especially in the case of b12) (Fig. 7). The increased unfavorable value of TΔS‡ diminished the favorable contribution of ΔH‡ for binding to gp120 96ZM651, as evident from similar values of activation ΔG‡ that represent the energetic barrier for successful association (Fig. 7).
FIGURE 7.
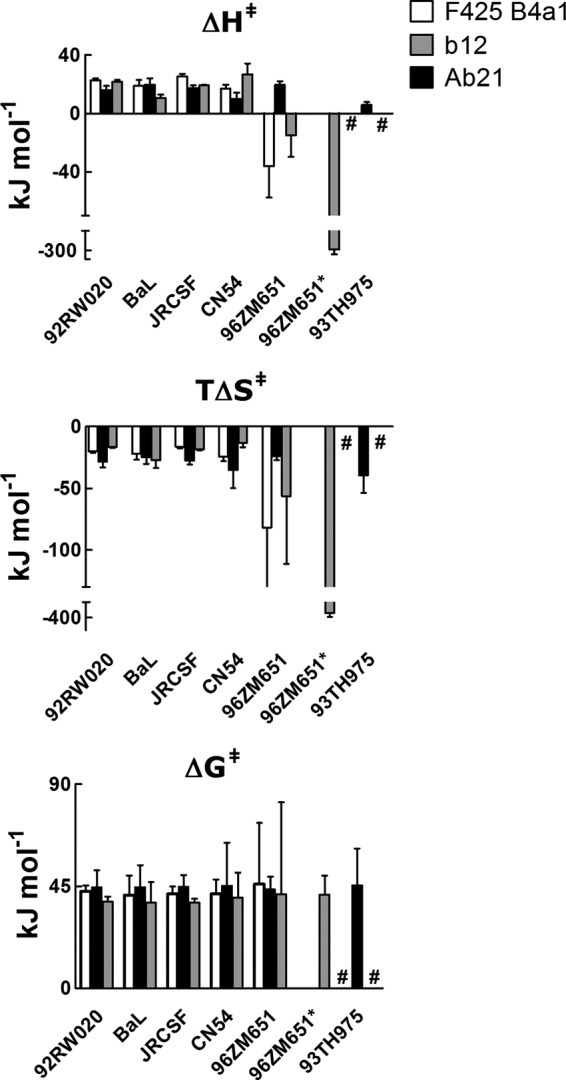
Activation thermodynamics of binding of Ab21, F425 B4a1, and b12 to gp120. Changes in the activation enthalpy (upper panel), activation entropy (middle panel), and activation free energy of Gibbs (lower panel) during the association of Ab21-heme (black bars), F425 B4a1 (white bars), and b12 (gray bars) to indicated variants of gp120. The changes in the thermodynamic parameters were evaluated by using Eyring's analyses and data from Arrhenius plots depicted on Fig. 5. Activation thermodynamic parameters were determined by using reference temperature of 298.15 K. Error bars depicts S.E. Because of complex temperature dependence, the binding thermodynamics of b12 to gp120 96ZM651 were evaluated by fitting the kinetic data in two temperature ranges, 10–25 and 25–35 °C. Thermodynamic data from high temperature range are marked by asterisk. #, absence of detectable interaction.
The results from thermodynamic analyses suggest that the polyreactive Ab21 recognizes different gp120 molecules by using an identical molecular mechanism. In contrast, F425 B4a1 and b12 utilize at least two distinct mechanisms for binding to different gp120 variants.
Role of Noncovalent Interactions in the Recognition of gp120 by Cryptic Abs
To understand the type of noncovalent forces that drive the association and stabilize the complexes of cryptic polyreactive antibody with gp120, nonequilibrium and equilibrium binding assays were applied (Fig. 8, Table 2, and data not shown). First, the ionic strength dependence of association of Ab21, F425 B4a1, and b12 was determined using surface plasmon resonance. The results indicate that the ionic strength has a very limited impact on the association of heme-exposed Ab21 to different gp120 variants (Fig. 8 and Table 2). Although, in the case of F425 B4a1, the association rate constants were more sensitive to ionic strength, the reduction of ka was only marginal, and even at the highest ionic strength, the rate of association remained very high with a ka in the order of 105 (except in the case of binding to gp120 96ZM651, see Table 2). Interestingly, b12 displayed marked ionic strength dependence upon recognition of all gp120 variants. Thus, an increase of NaCl from 50 to 450 mm resulted in a 12–143-fold reduction of association rate (Table 2). The binding to gp120 96ZM651 was completely abrogated at high ionic strength (Table 2). Polar and ionic interactions between proteins depend on ionization of charged groups and thus are also affected by pH. The pH dependence of Ab21 binding to gp120 indicated that this interaction is not sensitive to changes between pH 4 and 11 (data not shown). Taken together, the results from ionic strength and pH dependence assays imply that the binding of heme-exposed Ab21 to gp120 relies mainly on nonpolar interactions, such as hydrophobic and van der Waals contacts. Antibody F425 B4a1 also predominantly uses nonpolar interactions, whereas polar interactions play a more pronounced role for the formation of complexes of b12 with different gp120 variants.
FIGURE 8.
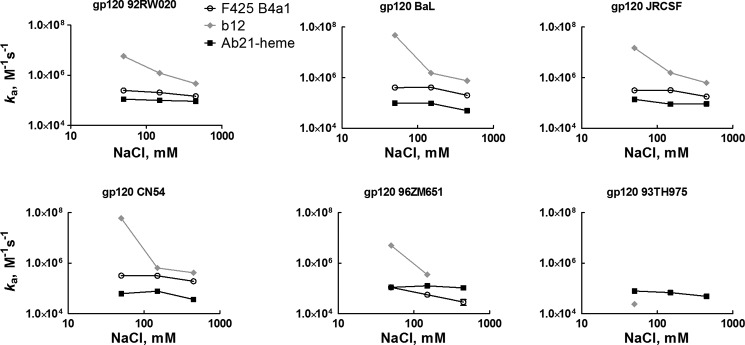
Ionic strength dependences of the association rate constant obtained by global kinetic analyses of binding of Ab21 exposed to heme (black squares), F425 B4a1 (white circles), and b12 (gray diamonds) to indicated variants of gp120. Kinetic analyses were performed at 25 °C in buffers with different NaCl concentrations, 0.05, 0.15, and 0.45 m. For further details see Table 2.
TABLE 2.
Ionic strength dependence of association rate constant of F425 B4a1, b12, and Ab21 for binding to distinct variants of gp120
Values of association rate constant ka were obtained by global analyses of sensorograms generated after injection of serial 2-fold dilutions of F425 B4a1 (25 to 0.049 nm), b12 (0.0049 (0.019)–5 nm, for binding to gp120 BaL, JRCSF, and CN54; 0.0049 (0.039)–10 nm, for binding to gp120 92RW020; and 0.039–20 nm, for binding to gp120 96ZM651), and of heme-exposed Ab21 (125 to 0.488 (0.244) nm) onto sensor chips with immobilized variants of gp120. The interactions were studied in HBS-EP buffer containing different concentration of NaCl, 0.05, 0.15, and 0.45 m. The global analyses were performed by fitting experimental data by using Langmuir binding with a drifting baseline kinetic model. In case of b12 binding to gp120 BaL, JRCSF, and CN54, at 0.05 m NaCl, due to possible involvement of mass transport, the experimental data were fitted by using Langmuir binding kinetic model with correction for mass transport artifacts. The values presented are mean ± S.D. of three independent fits of the experimental data.
| F425 B4a1 |
b12 |
Ab21-heme |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.05 m | NaCl 0.15 m | 0. 45 m | 0.05 m | NaCl 0.15 m | 0. 45 m | 0.05 m | NaCl 0.15 m | 0.45 m | |
| ka ×105m−1 s−1 (mean ± S.D.) | |||||||||
| 92RW020 | 2.5 ± 0.006 | 2.05 ± 0.01 | 1.47 ±0.006 | 57.3 ± 2.2 | 12.2 ± 0.001 | 4.6 ± 0.47 | 1.12 ± 0.021 | 0.99 ± 0.014 | 0.92 ± 0.012 |
| BaL | 4.05 ± 0.1 | 4.1 ± 0.069 | 2.01 ± 0.023 | 477 ± 4.6 | 15.2 ± 0.001 | 7.58 ± 0 | 0.98 ± 0.02 | 0.97 ± 0.012 | 0.49 ± 0.009 |
| JRCSF | 3.14 ± 0.023 | 3.19 ± 0.026 | 1.79 ± 0.011 | 150 ± 4.2 | 15.5 ± 0.01 | 6.22 ± 0.9 | 1.38 ± 0.023 | 0.9 ± 0.008 | 0.92 ± 0.018 |
| CN54 | 3.21 ± 0.021 | 3.04 ± 0.055 | 1.91 ± 0.01 | 600 ± 5.7 | 6.44 ± 0.6 | 4.17 ± 0.1 | 0.62 ± 0.014 | 0.77 ± 0.017 | 0.36 ± 0.003 |
| 96ZM651 | 1.1 ± 0.006 | 0.56 ± 0.026 | 0.28 ± 0.13 | 50 ± 1.2 | 5.16 ± 0.38 | # | 1.1 ± 0.006 | 1.26 ± 0 | 1.04 ± 0.02 |
| 93TH975 | # | # | # | 0.24 ± 0.04 | # | # | 0.78 ± 0.03 | 0.68 ± 0.008 | 0.48 ± 0.02 |
DISCUSSION
Here, we demonstrated that a human IgG1 cloned from a seronegative individual acquires polyreactivity and a strong binding potential to HIV-1 gp120 after exposure to heme. The polyreactive form of Ab21 was able to accommodate gp120 variants, belonging to divergent HIV-1 clades as follows: A, B, C, and A/E. Our data suggest that the epitope of heme-exposed Ab21 is most probably localized in the V3 region of the gp120 molecule. Kinetic and thermodynamic analyses of the interactions indicate that heme-exposed Ab21 utilizes an identical binding mechanism to recognize all distinct gp120 molecules. In contrast, HIV-1-neutralizing antibodies F425 B4a1 and b12 demonstrate high discriminatory activity to different variants of gp120. These antibodies also utilize alternative binding mechanisms for recognition of distinct gp120 variants.
Immune repertoires of all healthy individuals contain a fraction of antibodies that acquire polyreactivity following interaction with heme (34, 46, 47). The mechanism of heme-induced transition to promiscuous antigen binding of these Abs is not well understood. Our previous analyses revealed that antibodies sensitive to heme adopt antigen-binding polyreactivity upon direct binding of heme to the IgG molecule. Thus, we hypothesized that some Igs utilize heme as a cofactor to extend their antigen-binding potential. In this study, we also demonstrate that heme binds specifically to the human monoclonal Ab21. The changes in the absorbance spectra of the oxidized form of heme, namely a pronounced red shift in the high energy spectral region, suggest alterations of its electronic structure. These spectral changes are consistent with the alteration of the microenvironment of heme and coordination of its central iron ion by certain amino acid residue(s) (most probably histidine). The binding of heme to Ab21 was also confirmed by quenching of the intrinsic tryptophan fluorescence of Ab21. Importantly, the interaction of heme with Ab21 was not accompanied by aggregation of the IgG molecule, as is evident from the data obtained by size-exclusion chromatography.
We also tested the potential of other metalloporphyrins to induce gp120-binding reactivity of Ab21. Among the analyzed metalloporphyrins, only Fe(III)-, Co(III)-, or Cr(III)-containing protoporphyrins were able to potentiate the reactivity of Ab21. Similarities in the chemical and coordination properties of these metal ions may explain this observation.
Our results imply that heme bound to Ab21 might serve as an interfacial cofactor for the binding of gp120. Indeed, preincubation of gp120 with metalloporphyrin that does not influence the reactivity of Ab21 resulted in the reduction in the binding of heme-exposed polyreactive Ab21. Interestingly, gp120 has been demonstrated to bind different porphyrins, and this binding occurs to the V3 loop (48–51), a region also recognized by Ab21.
The induction of antigen-binding polyreactivity of Ab21 by heme was accompanied by acquisition of specificity to HIV-1 gp120. Enormous sequence heterogeneity of gp120 makes this protein an elusive target for neutralizing antibodies (13, 14). Antibodies that are able to bind and neutralize many divergent strains of HIV-1, i.e. bNAbs, usually recognize invariant functionally important regions of gp120 (14, 15). In this study, we tested the potential of a human antibody with cryptic polyreactivity to accommodate the considerable sequence heterogeneity of gp120. The kinetic and thermodynamic analyses reveal a capability of heme-induced polyreactive Abs to accommodate the relatively high affinity (KD <70 nm) of different variants of gp120 by using an identical physicochemical mechanism of molecular recognition. Thus, the association of the polyreactive antibody in all interactions was characterized by an unfavorable contribution of activation enthalpy and entropy. The low extent in the changes of entropy (ranging from −24 to −39.6 kJ mol−1) suggests that interaction of Ab21 with distinct gp120 variants is not accompanied by significant alterations in the conformation of the antigen-binding site or a considerable reorganization of solvent structure (52, 53). The observed thermodynamic behavior of Ab21 is typical for affinity-matured antibodies that possess preoptimized antigen-binding sites (2, 54). Although with differences in the absolute values, the changes in the activation enthalpy of binding of polyreactive Ab21 to distinct variants of gp120 had identical qualitative signature. The changes in the association enthalpy depend on formation of noncovalent contacts upon interaction (2, 52, 53). Indeed, our data from pH binding analyses and the study of the effect of ionic strength revealed that heme-exposed Ab21 forms complexes with all different variants of gp120 by predominantly using hydrophobic interactions. This result is in agreement with previous studies demonstrating that cryptic polyreactive antibodies use hydrophobic effect for binding to their targets (33, 34). In contrast to polyreactive Ab21, both F425 B4a1 and b12 utilized distinct molecular mechanisms for bindings to different gp120 variants. Moreover, both Abs were not able to recognize gp120 belonging to HIV-1 clade A/E. Interestingly, the binding of F425 B4a1 and b12 to the variant of gp120 with high sensitivity to neutralization by these antibodies (HIV-1 BaL) was characterized by a thermodynamic mechanism identical to that of polyreactive Ab21.
Different studies have suggested that antibody polyreactivity contributes to neutralization of HIV-1 (24, 27, 31, 55–58). Several mechanisms have been proposed to explain the role of antibody polyreactivity in HIV-1 neutralization. Thus, the autoreactivity and polyreactivity of gp41-specific Abs 2F5 and 4E10 (27) allow binding to membrane phospholipids through a long and hydrophobic complementarity determining H3 region, which ensures appropriate orientation of the antigen-binding site for further recognition of epitopes on gp41 (59–61). Similarly the polyreactivity of gp120-specific antibody 21c allows simultaneous recognition by a single antigen-binding site of self-protein, CD4 and the viral gp120 (62); simultaneous binding to both proteins markedly increases the affinity of the antibody. Other studies have proposed that polyreactive antibodies neutralize the virus by virtue of enhanced binding avidity (24, 25). Because of the sparse distribution of HIV spikes, simultaneous engagement of two identical epitopes on the envelope proteins by a single antibody is not possible (63). A model was proposed, referred to as heteroligation, which suggests that polyreactive antibodies neutralize HIV by gain in avidity through the simultaneous engagement of one antigen-binding site with envelope proteins and the second antigen-binding site with yet unidentified molecule(s) on the viral surface (24, 25). Yet our data suggest another possible role of Ab polyreactivity for recognition of highly heterogeneous viruses. By virtue of antigen-binding promiscuity, cryptic polyreactive Abs are more tolerant to variations in the antigen as compared with highly specific Abs. The antigen-recognition heterogeneity of cryptic polyreactive Abs thus compensates for the sequence heterogeneity of HIV-1 gp120. However, the penalty of promiscuous antigen recognition resides in a decreased binding affinity. The lower affinity of polyreactive Abs might nevertheless be compensated by an elevated functional affinity (avidity) (24, 25). Indeed, in this study, we observed that the polyreactive form of Ab21 binds gp120 with high avidity.
Our data might also have repercussions for understanding the physiopathology of HIV-1 infection. It is noteworthy that different studies have highlighted the inter-relation between iron metabolism of the host and HIV-1 pathogenesis (64). Many proteins use heme as a cofactor in gas transport and in catalysis of various redox reactions. Under physiological conditions, heme is tightly bound to proteins and is constrained intracellularly. However, as a result of various pathologies, accompanied by hemolysis and tissue damage, heme can be liberated and reach high concentrations (>20 μm) in plasma (65–68). Interestingly, it has been shown that free extracellular heme has a potent inhibitory effect on HIV-1 replication (69–71). The inhibitory effect of heme was proposed to be either due to direct interaction with the reverse transcriptase (72, 73) or through indirect effects of heme on host cells (such as up-regulation of heme oxygenase-1) (69–71). Importantly, a recent analysis that includes more than 400,000 patients with sickle cell anemia, a condition accompanied by high levels of intravascular hemolysis and heme release, demonstrated a 70% lower incidence of HIV-1 infection as compared with the normal population (74). In contrast, the incidence of other viral infections, such as hepatitis C virus and hepatitis B virus, was elevated, suggesting a specific protective effect of hemolytic disorders on HIV-1 infection. The latter study provides strong evidence for the high sensitivity of HIV-1 to free extracellular heme. Based on our data, we speculate that the induction by heme of the cryptic HIV-1-binding potential of circulating sensitive Igs could synergize with other mechanisms for heme-mediated inhibition of HIV-1 infection.
In conclusion, we demonstrated that a cryptic polyreactive antibody, induced by heme, has high potential to accommodate the structural diversity of a highly heterogeneous antigen such as gp120. Further studies are required for understanding the functional role of cryptic polyreactive antibodies in HIV-1 infection. Such investigations could contribute to the general understanding of antibody polyreactivity and the role of the inflammatory environment for HIV-1 infection and could provide novel strategies for combating highly heterogeneous pathogens.
Acknowledgments
We thank the National Institutes of Health AIDS Reagent Program, Division of AIDS, NIAID, for providing us vital materials (as listed under “Experimental Procedures”).
This work was supported by INSERM, CNRS, and UPMC-Paris 6 (France) and by grants from Centre de Recherche des Cordeliers (Prix Jeunes Chercheurs 2008), Agence Nationale de la Recherche Grant ANR-13-JCV1-006-01, and Laboratoire Français du Fractionnement et des Biotechnologies (Les Ulis, France).
M. Lecerf, T. Scheel, A. D. Pashov, A. Jarossay, D. Ohayon, C. Planchais, S. Mesnage, C. Berek, S. V. Kaveri, S. Lacroix-Desmazes, and J. D. Dimitrov, manuscript in preparation.
- Ab
- antibody
- bNAb
- broadly neutralizing antibody
- PP
- protoporphyrin
- gp
- glycoprotein.
REFERENCES
- 1. Notkins A. L. (2004) Polyreactivity of antibody molecules. Trends Immunol. 25, 174–179 [DOI] [PubMed] [Google Scholar]
- 2. Manivel V., Bayiroglu F., Siddiqui Z., Salunke D. M., Rao K. V. (2002) The primary antibody repertoire represents a linked network of degenerate antigen specificities. J. Immunol. 169, 888–897 [DOI] [PubMed] [Google Scholar]
- 3. Eisen H. N., Chakraborty A. K. (2010) Evolving concepts of specificity in immune reactions. Proc. Natl. Acad. Sci. U.S.A. 107, 22373–22380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dimitrov J. D., Planchais C., Roumenina L. T., Vassilev T. L., Kaveri S. V., Lacroix-Desmazes S. (2013) Antibody polyreactivity in health and disease: statu variabilis. J. Immunol. 191, 993–999 [DOI] [PubMed] [Google Scholar]
- 5. Ochsenbein A. F., Fehr T., Lutz C., Suter M., Brombacher F., Hengartner H., Zinkernagel R. M. (1999) Control of early viral and bacterial distribution and disease by natural antibodies. Science 286, 2156–2159 [DOI] [PubMed] [Google Scholar]
- 6. Zhou Z. H., Tzioufas A. G., Notkins A. L. (2007) Properties and function of polyreactive antibodies and polyreactive antigen-binding B cells. J. Autoimmun. 29, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ehrenstein M. R., Notley C. A. (2010) The importance of natural IgM: scavenger, protector and regulator. Nat. Rev. Immunol. 10, 778–786 [DOI] [PubMed] [Google Scholar]
- 8. Zhou Z. H., Zhang Y., Hu Y. F., Wahl L. M., Cisar J. O., Notkins A. L. (2007) The broad antibacterial activity of the natural antibody repertoire is due to polyreactive antibodies. Cell Host Microbe 1, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meffre E., Wardemann H. (2008) B-cell tolerance checkpoints in health and autoimmunity. Curr. Opin. Immunol. 20, 632–638 [DOI] [PubMed] [Google Scholar]
- 10. Yurasov S., Wardemann H., Hammersen J., Tsuiji M., Meffre E., Pascual V., Nussenzweig M. C. (2005) Defective B cell tolerance checkpoints in systemic lupus erythematosus. J. Exp. Med. 201, 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Warter L., Appanna R., Fink K. (2012) Human poly- and cross-reactive anti-viral antibodies and their impact on protection and pathology. Immunol. Res. 53, 148–161 [DOI] [PubMed] [Google Scholar]
- 12. Menard L., Saadoun D., Isnardi I., Ng Y. S., Meyers G., Massad C., Price C., Abraham C., Motaghedi R., Buckner J. H., Gregersen P. K., Meffre E. (2011) The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J. Clin. Invest. 121, 3635–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pantophlet R., Burton D. R. (2006) GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24, 739–769 [DOI] [PubMed] [Google Scholar]
- 14. Klein F., Mouquet H., Dosenovic P., Scheid J. F., Scharf L., Nussenzweig M. C. (2013) Antibodies in HIV-1 vaccine development and therapy. Science 341, 1199–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kwong P. D., Mascola J. R., Nabel G. J. (2013) Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat. Rev. Immunol. 13, 693–701 [DOI] [PubMed] [Google Scholar]
- 16. Burton D. R., Stanfield R. L., Wilson I. A. (2005) Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. U.S.A. 102, 14943–14948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bunnik E. M., Euler Z., Welkers M. R., Boeser-Nunnink B. D., Grijsen M. L., Prins J. M., Schuitemaker H. (2010) Adaptation of HIV-1 envelope gp120 to humoral immunity at a population level. Nat. Med. 16, 995–997 [DOI] [PubMed] [Google Scholar]
- 18. Stamatatos L., Morris L., Burton D. R., Mascola J. R. (2009) Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nat. Med. 15, 866–870 [DOI] [PubMed] [Google Scholar]
- 19. Burton D. R., Poignard P., Stanfield R. L., Wilson I. A. (2012) Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337, 183–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Corti D., Lanzavecchia A. (2013) Broadly neutralizing antiviral antibodies. Annu. Rev. Immunol. 31, 705–742 [DOI] [PubMed] [Google Scholar]
- 21. McCoy L. E., Weiss R. A. (2013) Neutralizing antibodies to HIV-1 induced by immunization. J. Exp. Med. 210, 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ditzel H. J., Barbas S. M., Barbas C. F., 3rd, Burton D. R. (1994) The nature of the autoimmune antibody repertoire in human immunodeficiency virus type 1 infection. Proc. Natl. Acad. Sci. U.S.A. 91, 3710–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ditzel H. J., Itoh K., Burton D. R. (1996) Determinants of polyreactivity in a large panel of recombinant human antibodies from HIV-1 infection. J. Immunol. 157, 739–749 [PubMed] [Google Scholar]
- 24. Mouquet H., Scheid J. F., Zoller M. J., Krogsgaard M., Ott R. G., Shukair S., Artyomov M. N., Pietzsch J., Connors M., Pereyra F., Walker B. D., Ho D. D., Wilson P. C., Seaman M. S., Eisen H. N., Chakraborty A. K., Hope T. J., Ravetch J. V., Wardemann H., Nussenzweig M. C. (2010) Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467, 591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mouquet H., Nussenzweig M. C. (2012) Polyreactive antibodies in adaptive immune responses to viruses. Cell. Mol. Life Sci. 69, 1435–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dimitrov J. D., Kazatchkine M. D., Kaveri S. V., Lacroix-Desmazes S. (2011) “Rational vaccine design” for HIV should take into account the adaptive potential of polyreactive antibodies. PLoS Pathog. 7, e1002095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haynes B. F., Fleming J., St Clair E. W., Katinger H., Stiegler G., Kunert R., Robinson J., Scearce R. M., Plonk K., Staats H. F., Ortel T. L., Liao H. X., Alam S. M. (2005) Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308, 1906–1908 [DOI] [PubMed] [Google Scholar]
- 28. Kwong P. D., Mascola J. R. (2012) Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37, 412–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klein F., Gaebler C., Mouquet H., Sather D. N., Lehmann C., Scheid J. F., Kraft Z., Liu Y., Pietzsch J., Hurley A., Poignard P., Feizi T., Morris L., Walker B. D., Fätkenheuer G., Seaman M. S., Stamatatos L., Nussenzweig M. C. (2012) Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J. Exp. Med. 209, 1469–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang G., Holl T. M., Liu Y., Li Y., Lu X., Nicely N. I., Kepler T. B., Alam S. M., Liao H. X., Cain D. W., Spicer L., VandeBerg J. L., Haynes B. F., Kelsoe G. (2013) Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J. Exp. Med. 210, 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liao H. X., Lynch R., Zhou T., Gao F., Alam S. M., Boyd S. D., Fire A. Z., Roskin K. M., Schramm C. A., Zhang Z., Zhu J., Shapiro L., NISC Comparative Sequencing Program, Mullikin J. C., Gnanakaran S., Hraber P., Wiehe K., Kelsoe G., Yang G., Xia S. M., Montefiori D. C., Parks R., Lloyd K. E., Scearce R. M., Soderberg K. A., Cohen M., Kamanga G., Louder M. K., Tran L. M., Chen Y., Cai F., Chen S., Moquin S., Du X., Joyce M. G., Srivatsan S., Zhang B., Zheng A., Shaw G. M., Hahn B. H., Kepler T. B., Korber B. T., Kwong P. D., Mascola J. R., Haynes B. F. (2013) Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McIntyre J. A. (2004) The appearance and disappearance of antiphospholipid autoantibodies subsequent to oxidation–reduction reactions. Thromb. Res. 114, 579–587 [DOI] [PubMed] [Google Scholar]
- 33. Dimitrov J. D., Ivanovska N. D., Lacroix-Desmazes S., Doltchinkova V. R., Kaveri S. V., Vassilev T. L. (2006) Ferrous ions and reactive oxygen species increase antigen-binding and anti-inflammatory activities of immunoglobulin G. J. Biol. Chem. 281, 439–446 [DOI] [PubMed] [Google Scholar]
- 34. Dimitrov J. D., Roumenina L. T., Doltchinkova V. R., Mihaylova N. M., Lacroix-Desmazes S., Kaveri S. V., Vassilev T. L. (2007) Antibodies use heme as a cofactor to extend their pathogen elimination activity and to acquire new effector functions. J. Biol. Chem. 282, 26696–26706 [DOI] [PubMed] [Google Scholar]
- 35. Mihaylova N. M., Dimitrov J. D., Djoumerska-Alexieva I. K., Vassilev T. L. (2008) Inflammation-induced enhancement of IgG immunoreactivity. Inflamm. Res. 57, 1–3 [DOI] [PubMed] [Google Scholar]
- 36. Scheel T. (2009) Mathematisch-Naturwissenschaftliche Fakultät I, Ph.D. thesis, Humboldt-Universität zu Berlin, Berlin [Google Scholar]
- 37. Scheel T., Gursche A., Zacher J., Häupl T., Berek C. (2011) V-region gene analysis of locally defined synovial B and plasma cells reveals selected B cell expansion and accumulation of plasma cell clones in rheumatoid arthritis. Arthritis Rheum. 63, 63–72 [DOI] [PubMed] [Google Scholar]
- 38. Gorny M. K., Conley A. J., Karwowska S., Buchbinder A., Xu J. Y., Emini E. A., Koenig S., Zolla-Pazner S. (1992) Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66, 7538–7542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cavacini L., Duval M., Song L., Sangster R., Xiang S. H., Sodroski J., Posner M. (2003) Conformational changes in env oligomer induced by an antibody dependent on the V3 loop base. AIDS 17, 685–689 [DOI] [PubMed] [Google Scholar]
- 40. Chomont N., Hocini H., Gody J. C., Bouhlal H., Becquart P., Krief-Bouillet C., Kazatchkine M., Bélec L. (2008) Neutralizing monoclonal antibodies to human immunodeficiency virus type 1 do not inhibit viral transcytosis through mucosal epithelial cells. Virology 370, 246–254 [DOI] [PubMed] [Google Scholar]
- 41. Zhou T., Georgiev I., Wu X., Yang Z. Y., Dai K., Finzi A., Kwon Y. D., Scheid J. F., Shi W., Xu L., Yang Y., Zhu J., Nussenzweig M. C., Sodroski J., Shapiro L., Nabel G. J., Mascola J. R., Kwong P. D. (2010) Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burton D. R., Pyati J., Koduri R., Sharp S. J., Thornton G. B., Parren P. W., Sawyer L. S., Hendry R. M., Dunlop N., Nara P. L. (1994) Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027 [DOI] [PubMed] [Google Scholar]
- 43. Trkola A., Purtscher M., Muster T., Ballaun C., Buchacher A., Sullivan N., Srinivasan K., Sodroski J., Moore J. P., Katinger H. (1996) Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70, 1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McLellan J. S., Pancera M., Carrico C., Gorman J., Julien J. P., Khayat R., Louder R., Pejchal R., Sastry M., Dai K., O'Dell S., Patel N., Shahzad-ul-Hussan S., Yang Y., Zhang B., Zhou T., Zhu J., Boyington J. C., Chuang G. Y., Diwanji D., Georgiev I., Kwon Y. D., Lee D., Louder M. K., Moquin S., Schmidt S. D., Yang Z. Y., Bonsignori M., Crump J. A., Kapiga S. H., Sam N. E., Haynes B. F., Burton D. R., Koff W. C., Walker L. M., Phogat S., Wyatt R., Orwenyo J., Wang L. X., Arthos J., Bewley C. A., Mascola J. R., Nabel G. J., Schief W. R., Ward A. B., Wilson I. A., Kwong P. D. (2011) Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gassmann C., Bauer G. (1997) Avidity determination of IgG directed against tick-borne encephalitis virus improves detection of current infections. J. Med. Virol. 51, 242–251 [PubMed] [Google Scholar]
- 46. McIntyre J. A., Faulk W. P. (2009) Redox-reactive autoantibodies: biochemistry, characterization, and specificities. Clin. Rev. Allergy Immunol. 37, 49–54 [DOI] [PubMed] [Google Scholar]
- 47. Dimitrov J. D., Planchais C., Kang J., Pashov A., Vassilev T. L., Kaveri S. V., Lacroix-Desmazes S. (2010) Heterogeneous antigen recognition behavior of induced polyspecific antibodies. Biochem. Biophys. Res. Commun. 398, 266–271 [DOI] [PubMed] [Google Scholar]
- 48. Dairou J., Vever-Bizet C., Brault D. (2004) Interaction of sulfonated anionic porphyrins with HIV glycoprotein gp120: photodamages revealed by inhibition of antibody binding to V3 and C5 domains. Antiviral Res. 61, 37–47 [DOI] [PubMed] [Google Scholar]
- 49. Vzorov A. N., Dixon D. W., Trommel J. S., Marzilli L. G., Compans R. W. (2002) Inactivation of human immunodeficiency virus type 1 by porphyrins. Antimicrob. Agents Chemother. 46, 3917–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Neurath A. R., Strick N., Debnath A. K. (1995) Structural requirements for and consequences of an antiviral porphyrin binding to the V3 loop of the human immunodeficiency virus (HIV-1) envelope glycoprotein gp120. J. Mol. Recognit. 8, 345–357 [DOI] [PubMed] [Google Scholar]
- 51. Debnath A. K., Jiang S., Strick N., Lin K., Haberfield P., Neurath A. R. (1994) Three-dimensional structure-activity analysis of a series of porphyrin derivatives with anti-HIV-1 activity targeted to the V3 loop of the gp120 envelope glycoprotein of the human immunodeficiency virus type 1. J. Med. Chem. 37, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 52. Janin J. (1995) Principles of protein-protein recognition from structure to thermodynamics. Biochimie 77, 497–505 [DOI] [PubMed] [Google Scholar]
- 53. Amzel L. M. (2000) Calculation of entropy changes in biological processes: folding, binding, and oligomerization. Methods Enzymol. 323, 167–177 [DOI] [PubMed] [Google Scholar]
- 54. Manivel V., Sahoo N. C., Salunke D. M., Rao K. V. (2000) Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity 13, 611–620 [DOI] [PubMed] [Google Scholar]
- 55. Haynes B. F., Nicely N. I., Alam S. M. (2010) HIV-1 autoreactive antibodies: are they good or bad for HIV-1 prevention? Nat. Struct. Mol. Biol. 17, 543–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mouquet H., Klein F., Scheid J. F., Warncke M., Pietzsch J., Oliveira T. Y., Velinzon K., Seaman M. S., Nussenzweig M. C. (2011) Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS One 6, e24078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhu Z., Qin H. R., Chen W., Zhao Q., Shen X., Schutte R., Wang Y., Ofek G., Streaker E., Prabakaran P., Fouda G. G., Liao H. X., Owens J., Louder M., Yang Y., Klaric K. A., Moody M. A., Mascola J. R., Scott J. K., Kwong P. D., Montefiori D., Haynes B. F., Tomaras G. D., Dimitrov D. S. (2011) Cross-reactive HIV-1-neutralizing human monoclonal antibodies identified from a patient with 2F5-like antibodies. J. Virol. 85, 11401–11408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Haynes B. F., Kelsoe G., Harrison S. C., Kepler T. B. (2012) B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 30, 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Alam S. M., McAdams M., Boren D., Rak M., Scearce R. M., Gao F., Camacho Z. T., Gewirth D., Kelsoe G., Chen P., Haynes B. F. (2007) The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178, 4424–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Alam S. M., Morelli M., Dennison S. M., Liao H. X., Zhang R., Xia S. M., Rits-Volloch S., Sun L., Harrison S. C., Haynes B. F., Chen B. (2009) Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U.S.A. 106, 20234–20239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Scherer E. M., Leaman D. P., Zwick M. B., McMichael A. J., Burton D. R. (2010) Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc. Natl. Acad. Sci. U.S.A. 107, 1529–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Diskin R., Marcovecchio P. M., Bjorkman P. J. (2010) Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat. Struct. Mol. Biol. 17, 608–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Klein J. S., Bjorkman P. J. (2010) Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 6, e1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Drakesmith H., Prentice A. (2008) Viral infection and iron metabolism. Nat. Rev. Microbiol. 6, 541–552 [DOI] [PubMed] [Google Scholar]
- 65. Muller-Eberhard U., Javid J., Liem H. H., Hanstein A., Hanna M. (1968) Plasma concentrations of hemopexin, haptoglobin and heme in patients with various hemolytic diseases. Blood 32, 811–815 [PubMed] [Google Scholar]
- 66. Balla J., Jacob H. S., Balla G., Nath K., Eaton J. W., Vercellotti G. M. (1993) Endothelial-cell heme uptake from heme proteins: induction of sensitization and desensitization to oxidant damage. Proc. Natl. Acad. Sci. U.S.A. 90, 9285–9289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wagener F. A., Volk H. D., Willis D., Abraham N. G., Soares M. P., Adema G. J., Figdor C. G. (2003) Different faces of the heme-heme oxygenase system in inflammation. Pharmacol. Rev. 55, 551–571 [DOI] [PubMed] [Google Scholar]
- 68. Kumar S., Bandyopadhyay U. (2005) Free heme toxicity and its detoxification systems in human. Toxicol. Lett. 157, 175–188 [DOI] [PubMed] [Google Scholar]
- 69. Devadas K., Dhawan S. (2006) Hemin activation ameliorates HIV-1 infection via heme oxygenase-1 induction. J. Immunol. 176, 4252–4257 [DOI] [PubMed] [Google Scholar]
- 70. Levere R. D., Gong Y. F., Kappas A., Bucher D. J., Wormser G. P., Abraham N. G. (1991) Heme inhibits human immunodeficiency virus 1 replication in cell cultures and enhances the antiviral effect of zidovudine. Proc. Natl. Acad. Sci. U.S.A. 88, 1756–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schmidt W. N., Mathahs M. M., Zhu Z. (2012) Heme and HO-1 inhibition of HCV, HBV, and HIV. Front. Pharmacol. 3, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Staudinger R., Abraham N. G., Levere R. D., Kappas A. (1996) Inhibition of human immunodeficiency virus-1 reverse transcriptase by heme and synthetic heme analogs. Proc. Assoc. Am. Physicians 108, 47–54 [PubMed] [Google Scholar]
- 73. Argyris E. G., Vanderkooi J. M., Venkateswaran P. S., Kay B. K., Paterson Y. (1999) The connection domain is implicated in metalloporphyrin binding and inhibition of HIV reverse transcriptase. J. Biol. Chem. 274, 1549–1556 [DOI] [PubMed] [Google Scholar]
- 74. Nouraie M., Nekhai S., Gordeuk V. R. (2012) Sickle cell disease is associated with decreased HIV but higher HBV and HCV comorbidities in U.S. hospital discharge records: a cross-sectional study. Sex. Transm. Infect. 88, 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]