Abstract
Background & Aims
Alterations in 5-hydroxytryptamine (5-HT) signaling have been implicated as a factor contributing to the altered bowel habit of irritable bowel syndrome (IBS) patients. Tryptophan hydroxylase 1 (TPH1) is the rate–limiting enzyme in enterochromaffin cell 5-HT biosynthesis. We hypothesized that genetic variants affecting TPH1 gene expression might alter intestinal 5-HT bioavailability and subsequently the propensity for distinct bowel habit subtypes in IBS. In this study, we assessed the only common TPH1 proximal promoter variant (-347C/A; rs7130929) and its association with bowel habit predominance in IBS.
Methods
Electrophoretic mobility shift assays were performed to assess whether the -347C/A allele variant affects the DNA-binding of nuclear factors. Genotype distribution was determined for 422 IBS patients subtyped using Rome III criteria and for 495 healthy controls recruited from two university medical centers. Association with bowel habit was tested using a multinomial logistic regression model controlling for race, anxiety, depression, and study site.
Results
Early growth response factor 1 (EGR-1) bound with higher affinity to a site comprising the minor A-allele of SNP -347C/A. TPH1 genotype frequencies did not differ between IBS patients and controls overall. The CC genotype was more prevalent in the IBS-D subtype (47%) than in the IBS-C (25%) and IBS-M (37%) subtypes (P=0.039) after adjusting for race and other covariates. Colonic biopsies from a small cohort of IBS patients from one center were tested for higher TPH1 mRNA expression in samples with CC compared to CA genotype, but the results did not reach statistical significance.
Conclusions
The TPH1 promoter SNP -347C/A differentially binds EGR1, correlates with IBS bowel habit subtypes and possibly colonic TPH1 expression consistent with its role in modulating intestinal 5-HT signaling.
Keywords: Egr1, Sp1, ZBP-89, 5HT, serotonin, SNP
INTRODUCTION
Abnormalities in serotonin (5-HT) signaling have been implicated in IBS pathogenesis. When induced by luminal chemical and mechanical signals, enterochromaffin (EC) cells release 5-HT, which stimulates 5-HT3 and 5-HT4 receptors on primary afferent neurons, to induce peristaltic and secretomotor reflexes that regulate intestinal motility and secretion (1). In patients with diarrhea-predominant IBS (IBS-D), platelet-depleted postprandial plasma 5-HT concentrations are higher compared to patients with constipation (IBS-C) and healthy controls (2) (3–5), while IBS-C patients have a reduced postprandial rise in plasma 5-HT compared to controls (3,6). Plasma 5-HT is almost exclusively derived from gut EC cells secreting 5-HT, which is not taken up by platelets and overflows into the circulation (6). Plasma 5-HT levels also positively correlate with gut motility under both fasting and fed conditions (7) suggesting that they parallel the mucosal bioavailability of 5-HT in the intestine. Together, these findings suggest that differences in mucosal 5-HT levels correlate with the IBS clinical bowel habit phenotype. This concept is indirectly supported by the beneficial effects of 5-HT4 receptor agonists in IBS-C and 5-HT3 antagonists in IBS-D (8).
One factor that could modulate mucosal 5-HT availability is the activity or expression of the 5-HT selective reuptake transporter (SERT), which terminates bioamine action and prevents receptor desensitization by removing 5-HT from the interstitial space. While SERT mRNA is widely expressed, the quantities found in the gut epithelium are very low, particularly in the colon. Thus it may not be surprising that conflicting results have been reported concerning colonic SERT mRNA in IBS (9–12).
Conceivably, 5-HT biosynthesis is another process potentially influencing mucosal signaling. Tryptophan hydroxylase-1 (TPH1) is the rate-limiting enzyme in the biosynthesis of 5-HT in EC and mast cells. While TPH1 and TPH2 both are expressed in the gut, TPH2 is expressed by enteric and central neurons, and TPH1 is the predominant enzyme in EC cells (13). Although the activity of TPH1 is controlled at multiple levels including posttranslational regulation (14), recent evidence from animal studies indicates that changes in TPH1 transcription can affect proportional changes in intestinal and plasma 5-HT levels (15). TPH1 is therefore an intriguing candidate gene for conditions with altered 5-HT bioavailability as proposed for the distinct bowel habit subtypes in IBS.
This idea is the basis for the development of an oral TPH1 inhibitor acting locally on the GI mucosa (15). A recent phase II clinical trial demonstrated the efficacy of this novel compound in relieving symptoms of non-constipating IBS (16–18). The clinical response to therapy correlated with a decrease in 24-h urine excretion of the metabolite 5-hydroxyindoleacetic acid (5-HIAA) reflecting reduced 5-HT biosynthesis, which is promising particularly since there is currently a lack of established biomarkers to predict treatment response in IBS.
We recently identified the murine Tph1 homolog as a target of the zinc finger transcription factor ZBP-89 (ZNF148) (19). Evaluating putative ZBP-89 binding sites in the human TPH1 promoter, we observed that a common single nucleotide polymorphism (SNP) adjacent to a GC-rich sequence motif altered DNA-binding of nuclear proteins to this site. We hypothesized that this promoter variant could affect the amount of 5-HT generated by the EC cells via a gene dosage effect and thus modulate the susceptibility to distinct bowel habit phenotypes in IBS. Here we report that this -347C/A promoter SNP (rs7130929) differentially binds early growth response factor 1 (EGR-1) in vitro and associates with bowel habit subtypes in IBS patients.
METHODS
Human Study Subjects
Male and female IBS patients and healthy control subjects, who were at least 18 years of age and participating in clinical research studies or were being evaluated in the gastroenterology clinic at two metropolitan university medical centers, were consecutively recruited to provide saliva or blood samples for DNA analysis. For the colonic biopsy study, IBS patients and healthy controls between the ages of 18 and 55 were recruited at one of the centers although subjects were not consecutively enrolled. The diagnosis of IBS and bowel habit subtyping was determined by Rome III criteria (20), the absence of other chronic gastrointestinal (GI) conditions that could explain IBS symptoms, and by a clinician with expertise in IBS. Healthy control subjects were recruited by advertisement and did not have a history of IBS, abdominal surgeries other than an appendectomy, cholecystectomy or hysterectomy without residual GI symptoms, or other chronic GI or pain conditions, and were not taking psychotropic medication or participating in psychotherapy. The controls did not meet IBS bowel habit subtyping criteria for diarrhea, constipation or mixed pattern (i.e., they did not have >25% hard/lumpy or loose/watery stools). Validated questionnaires were used to assess IBS symptoms. Depression and anxiety symptoms were measured using the Hospital Anxiety and Depression (HAD) scale (21) at one center and the Beck Depression and Anxiety scales at the other (22,23). Subjects were compensated for their participation in the study. The Institutional Review Boards at both study sites approved this study and informed consent was obtained from all subjects.
Genotyping
Blood or saliva samples (Oragene DNA Self-Collection Kit (DNA Genotek, Inc., Ottawa, Canada) collected for DNA extraction at Washington University were frozen and banked in the BioBank Core of the Digestive Diseases Research Center. DNA was extracted using Qiagen Gentra Puregene Reagents (Qiagen Inc., Valencia, CA), and genotyping was performed using Sequenom MassARRAY technology (Sequenom Inc, San Diego, CA.) in a single batch containing all of the study samples (cases and controls). The laboratory staff was blinded to both the study subject group assignment and the study hypotheses. DNA samples were also collected from a total of 575 subjects (IBS patients and controls) at the UCLA site. Saliva samples were collected in Oragene DNA Collection Kits (DNA Genotek, Inc., Ottawa, Canada) at the UCLA Center for Neurobiology of Stress. UCLA Biological Samples Processing Core extracted DNA from the saliva samples using the Autopure LS Nucleic Acid Purification instrument (Gentra Systems, Inc., Minneapolis, MN). Eight percent were excluded due to low amounts of DNA or subjects declining to share samples. DNA was sent to the University of Michigan for genotyping, which was successfully completed in 99.6% of samples. For genotyping of the -347C/A (rs7130929) variant by restriction fragment length polymorphism (RFLP), a 484 bp fragment was amplified by hotstart PCR (Qiagen) with the following primers: forward-5′-CTGCGTGTATCTGACTGGTGT-3′ and reverse-5′-GGGATAAGGAGCTAATCG-ACTGA-3′, digested with MslI restriction enzyme (New England Biolabs), then separated on 2% agarose gels. Only amplicons from the A-allele were cut by MslI into 207 bp and 277 bp fragments. The extended human TPH1 promoter region (24) was amplified using Platinum Taq DNA polymerase (Invitrogen) and oligonucleotide primers 5′-CTGCGTGTATCTGACTGGTGT-3′ and 5′-GAAAGGTCTCTCCCTGACCA-3′ in subsets of the IBS and control populations. Amplicons were sequenced bidirectionally at the University of Michigan DNA Sequencing Core.
Measurement of TPH1 mRNA in Colonic Biopsies
Flexible sigmoidoscopy to at least 40 cm from the anal verge was performed in the UCLA Medical Procedures Unit. Subjects were instructed to use two tap-water enemas as the bowel preparation. During the sigmoidoscopy, sigmoid colon biopsies were taken at 30 cm from the anal verge and immediately snap frozen in liquid nitrogen to avoid RNA degradation prior to tissue extraction.
Total RNA was isolated from non-pooled colonic biopsies and cDNA generated by reverse transcription (Applied Biosystems, Foster City, CA). Real-time PCR amplification was performed on a 7500 FAST Real-Time PCR System (Applied Biosystems, Foster City, CA) using Fast Universal PCR Master Mix and validated fluorogenic TaqMan probe sets (Applied Biosystems) for TPH1 (Hs00188220_m1) and 18S-rRNA (Hs99999901_s1). Relative expression of TPH1 was normalized to 18S-rRNA. Duplicate sample analysis for each patient showed excellent correlation throughout the study group.
Electrophoretic Mobility Shift Assay (EMSA)
Since the rs7130929 SNP resided within the proximal promoter of the TPH1 gene where transcription factors can bind, EMSA protein-DNA binding assays were performed. Protein extracts from nuclei of the human 5HT-producing BON cell line were prepared essentially as previously described (25). The concentration of total nuclear protein determined colorimetrically using the bicinchoninic acid method (BCA assay; Pierce Biotechnology). The EMSA probe was prepared by end labeling with radioactive 32P using T4 Polynucleotide Kinase the two allele specific pairs of polyacrylamide gel-purified complementary oligonucleotides corresponding to the −361 to −326 region of the TPH1 promoter (5′-CAGAAGCACAGAGA(g/t)GTGTGGGAGGTGGGGGGATTC-3′). Duplex DNA assembled by annealing of the 32P-labeled complementary oligonucleotides was purified on G50 spin columns. The specific competitor DNAs were prepared by annealing the unlabeled oligonucleotide pairs. Binding reactions contained 0.5 pmol probe, 2 μg nuclear protein extract, 0.5 μg sonicated salmon sperm DNA, and 0.5 μg poly(dI-dC) (Amersham Biosciences) in a total volume of 20 μl. The binding buffer consisted of 20 mM Hepes (pH 7.9), 5 mM MgCl2, 50 mM KCl, 100 μM ZnSO4, 0.5 mM dithiothreitol, and 10% glycerol. After incubation (10 min at room temperature followed by 20 min on ice), the samples were loaded onto a 5% polyacrylamide gel (acrylamide:bis-acrylamide 37.5:1) and separated at 10 V/cm in 0.5× TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA). Dried gels were analyzed by autoradiography. For antibody interference, the reaction mixtures lacking the probe were incubated for 20 min on ice with antibodies against Sp1 (clone H225), Sp3 (D20), EGR-3 (C-24), EGR-1 (rabbit polyclonal 588), GATA-1 (N6; all from Santa Cruz Biotechnology, Inc.), or ZBP-89 (26), before addition of the probe and further processing as above. For competition experiments, unlabeled competitor DNAs in 100-fold molar excess over the labeled probe were included in the binding reactions.
Luciferase Reporter Assays
To test the function of the two SNP alleles in the 5HT-producing BON cell line, the −568/+19 promoter region of the humanTPH1 gene was amplified by PCR using forward primer 5′-TATGGTACCTTTGGGATAAGGAGCTAATCGA-3′ and reverse primer 5′-TATCTCGAGTAGGTGCAGGCTGGGTCG-3′ from genomic DNA of subjects homozygous for either the A- or C-allele of rs7130929. The two products were cloned directionally into the promoter-less luciferase reporter vector pGL3-basic (Promega) to generate -347A-Luc and -347C-Luc, respectively. Both constructs were confirmed by sequencing.
BON cells were grown in 24-well plates to 90–95% confluence and transiently transfected in triplicate with 0.25 μg/well of the TPH1 reporter constructs using Lipofectamine 2000 transfection reagent (Invitrogen) diluted in serum-free OptiMEM I. Twenty ng/well of the pRL-Tk plasmid (Promega), expressing Renilla luciferase from a thymidine kinase promoter, was co-transfected as an internal control. The medium was replaced by fresh DME/F-12 8 h following transfection. To evaluate the effect of EGR-1 overexpression, cells were co-transfected with 0.2 μg/well of a human EGR-1 expression vector (Origene) or the empty pCMV6 vector as the control, and luciferase activity was measured 24 hours post transfection. In EGR1 knockdown experiments, BON cells were co-transfected with either 20 pmol/well of EGR1 siRNA (Silencer Select s4538; Invitrogen) or scrambled control siRNA (control siRNA No. 2; Invitrogen). Twenty-four hours following transfection with siRNA, the cells were stimulated for 4 h with 200 nM TPA before assaying for luciferase activity.
Western Blotting
BON cells were homogenized in lysis buffer (50 mM Tris/HCl (pH 8.0), 150 mM NaCl, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 1% NP40) containing a protease inhibitor cocktail (Roche, Indianapolis, IN). Fifty micrograms protein/well were separated on 4–20% SDS-polyacrylamide gradient gels under reducing conditions and blotted to PVDF membranes. Blots were probed with anti-EGR-1 (S-25, Santa Cruz Biotechnology, Inc.) and anti-GAPDH (MAB374, Millipore).
Statistical Analysis
The two subject groups (UCLA, Washington University) were combined in the analysis. Since current anxiety and depression symptoms were measured using two different validated questionnaires at the two study sites, we calculated Z-scores for anxiety and depression after combining the subject groups.
Hardy–Weinberg equilibrium was tested for the TPH1 SNP (at -347; rs7130929). All three genetic models were evaluated for the TPH1 SNP (additive, dominant, and recessive; where recessive was coded as homozygous for the major C-allele versus the other genotypes). An additive effect genetic model was tested among IBS cases and controls using a Cochran-Armitage test for trend. The dominant and recessive models were tested among IBS and controls using a Chi-square test. We utilized multinomial logistic regression models to determine the relationship between the TPH1 rs7130929 SNP with the odds of having a certain bowel habit within the IBS group and while controlling for potential confounders including race/ethnicity, study site and psychological symptoms (e.g. anxiety and depression symptoms) that differ between IBS bowel habit subgroups. Goodness of fit was evaluated using a test of deviance. We chose to control for psychological symptoms because a recent study showed that a TPH1 SNP (rs4537731) was associated with psychological distress (27). In addition, we only modeled using IBS-C, IBS-D, and IBS-M patients since there were only 12 IBS-U patients. Therefore, the outcome consists of 3 nominal categories (IBS-C, IBS-D, and IBS-M with IBS-C as the reference group).
Adjusted odds ratios (OR) and corresponding 95% confidence intervals were reported from the models. TPH1 mRNA data was log-transformed to achieve approximate normality and geometric means. The 95% confidence intervals (CI) were reported. A two-sample t-test was used to compare TPH1 mRNA expression between IBS cases and controls, ANOVA was used to compare TPH1 mRNA levels across bowel habit, and Spearman correlation was used to compare gene expression across additively coded TPH1 genotypes. All statistical analyses were performed using SAS version 9.3 (SAS Institute, Cary, NC, USA) or R version 2.15.1 (http://cran.r-project.org/). Significance was assessed at the 0.05 level.
RESULTS
Allele-Specific Nuclear Protein Binding to Proximal TPH1 Promoter Variant
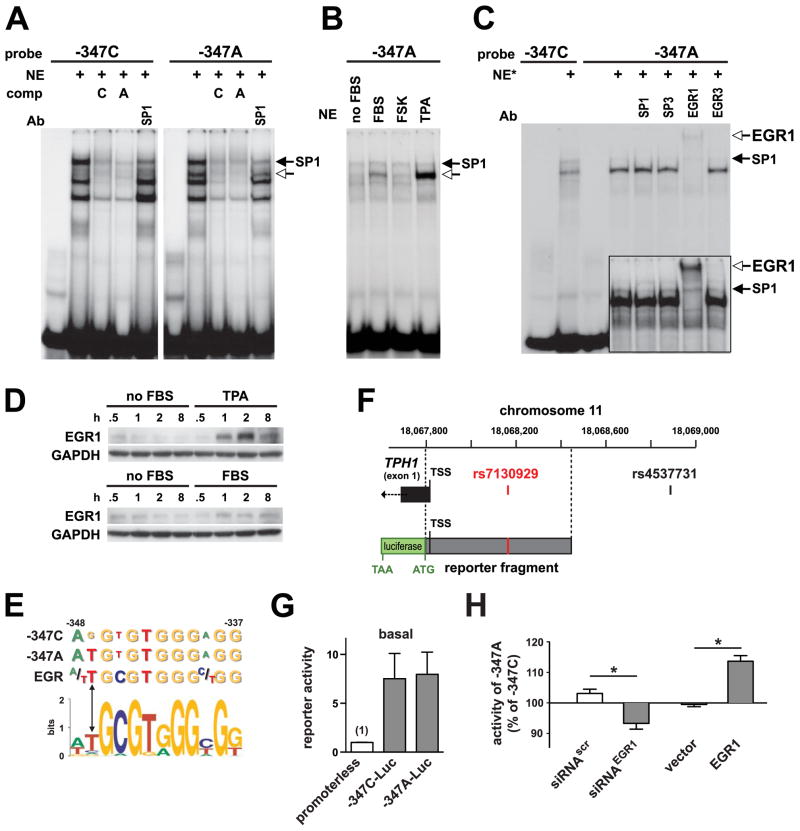
Since we recently discovered that transcription factor ZBP-89 regulates the mouse Tph1 promoter (19), we analyzed the human TPH1 promoter for ZBP-89 consensus regulatory elements. We identified several putative sites in the proximal promoter with one near a biallelic SNP (rs7130929) at position -347 relative to the transcriptional start site. Since the location of the SNP was within the proximal promoter of the TPH1 gene, we considered that the SNP might contain the binding site for transcription factors that bind DNA and modulate gene expression. Therefore we used electrophoretic mobility shift assays (EMSAs), a DNA binding assay, to evaluate whether nuclear proteins actually bind to the 36 base pair DNA sequence containing the -347C/A (rs7130929) SNP of the TPH1 promoter and whether the two different alleles modulated protein binding. The ancestral allele at this site was “C” and the minor variant (A-allele) occurred with a frequency of 35% in the general population (www.ncbi.nlm.nih.gov). Nuclear proteins were extracted from BON cells, a 5-HT producing human pancreatic carcinoid cell line. We found that radioactive EMSA probes containing either the C- or A-allele at -347 specifically bound three protein complexes (Figure 1A). In addition, the EMSA revealed a distinct protein complex that preferentially bound to the A-allele (-347A probe). Since a GC-rich site flanked the SNP-containing DNA element, we tested whether zinc finger transcription factors accounted for differential binding of the protein complexes to the A-allele. This was accomplished by adding different antibodies to determine if they disrupted transcription factor binding. Indeed Sp1 and to a lesser extent, Sp3 and ZBP-89 respectively decreased the intensity of the upper and lower complexes, while there appeared to be little effect of the GATA1 antibodies (Figure 1A, Suppl. Fig. S1). However, none of these factors accounted for the novel complex binding preferentially to the A-allele. To further characterize this complex, we examined whether DNA binding was induced by common signal transduction pathways, e.g., protein kinase A (PKA), activated by forskolin (FSK) or protein kinase C (PKC), activated by phorbol esters. We found that the allele-specific binding activity was robustly induced in extracts from serum-starved BON cells treated for one hour with fetal bovine serum (FBS) or phorbol esters (TPA), but not by FSK (Figure 1B).
Figure 1.
Allele-specific binding of EGR-1 to the -347C/A polymorphism of the TPH1 core promoter. (A) Electrophoretic mobility shift assay using allele-specific probes (-347C and -347A) and nuclear protein extract (NE) prepared from BON cells. The open arrow marks the position of a protein complex with preferential binding to the -347A-allele. Specific competition (comp) was performed by co-incubation with 100-fold molar excess of cold probes. (B) Expression of the -347A-allele specific binding activity in serum starved BON cells (no FBS) treated for one hour with 20% FBS, PKC-activating phorbol ester (TPA), or the adenylyl cyclase activator forskolin (FSK). (C) Supershift experiments to determine the TPA-induced (200 nM; 1 h) nuclear factor with preferential binding to the -347A-allele. (D) Western blot analysis of EGR-1 expression in BON cells treated with FBS or TPA. (E) Location of SNP -347C/A at position −1 relative to a consensus nonameric EGR recognition motif. The -347A-allele corresponds to the preferred nucleotide according to the extended EGR binding matrix determined by in vitro DNA binding site selection (depicted as sequence logo) (28). (F) Schematic of the TPH1 proximal promoter region on chromosome 11 depicting the location of the first (noncoding) exon with the transcription start site (TSS) mapped by Boularand et al. (24) and the position of the rs7130929 variant. Also indicated is the region contained in the allele-specific luciferase reporter constructs and the location of the upstream variant genotyped in the study by Jun et al. (27) (rs4537731). (G) Promoter activity of the two allele specific reporter constructs (-347C-Luc and -347A-Luc) in untreated BON cells. Activity of the promoterless luciferase vector is set to 1. (H) Relative activity of the A-allele under conditions of low or high EGR-1 expression level. BON cells were transfected with -347C-Luc or -347A-Luc, and cotransfected with either siRNA for EGR1 (control: scrambled siRNA) or EGR-1 expression vector (control: empty vector). For each experiment and within each treatment group, the relative activity of the minor A allele was normalized to the activity of the C-allele (set to 100%). Each bar represents mean ± standard error from three independent experiments performed in triplicate transfections. * P<0.05.
Early Growth Response Factor 1 Binds Differentially to TPH1 -347 SNP Site and Modulates TPH1 Promoter Activity
Since rapid induction to proliferative signals, e.g., FBS and TPA is a characteristic feature of immediate early genes, we considered the possibility that the novel protein complex binding to the -347A site would be formed by an immediate-early transcription factor that recognizes GC-rich sequences. Few zinc finger transcription factors are known to rapidly increase their binding in response to proliferative signals. However, members of the early growth response (EGR) family of transcription factors are known to meet these criteria. Indeed, EGR-1 but not EGR-3 antibodies, completely shifted the A-allele-specific complex confirming EGR-1 as the nuclear protein (Figure 1C). We demonstrated by Western blots that both TPA and FBS stimulate EGR-1 protein expression in BON cells (Figure 1D). EGR-1 was robustly induced within one hour of stimulation, especially with TPA, consistent with increased binding on the EMSAs. Therefore, the two alleles of the -347 TPH1 promoter SNP differentially bind EGR-1. When we reviewed in silico the extended weight matrix for EGR-1 binding sites, we found that the SNP lies adjacent (position −1) to a nonameric EGR family recognition site overlapping an Sp1 binding motif (Figure 1E). In vitro DNA binding site selection studies indicate a strong preference for adenine at −1 of the extended EGR-1 binding matrix (28), which is consistent with our EMSA results showing preferential binding of EGR-1 to the minor A-allele.
Having established that the common -347C/A SNP in the TPH1 promoter alters an EGR-1 binding site, we tested whether differential binding of EGR-1 can modulate TPH1 promoter activity using a cell line-based reporter assay. The human TPH1 (−568/+19) proximal promoter region (Figure 1F) containing either the C or A-alleles at position -347 was subcloned into a luciferase reporter plasmid and transfected into BON cells. Both the -347A-Luc and -347C-Luc reporter constructs drove significant luciferase expression compared to a promoterless control vector (Figure 1G). However, there was no significant difference at baseline between the promoter activities of the two alleles in BON cells using this type of cell line-based assay. Therefore, to compare the relative activity of the two alleles at low or high EGR-1 expression levels, an EGR-1 expression vector or siRNA for EGR1 was cotransfected with the TPH1 reporter construct containing either the A- or C-allele. The -347C/A SNP did not alter promoter activity when cotransfected with empty expression vector or scrambled sequence siRNA control (Figure 1H). However, when EGR1 expression was suppressed by RNA interference (siRNAEGR1), expression of the A allele was lower than the C-allele (P=0.036) (Figure 1G). Conversely, when EGR-1 was overexpressed, expression of the A-allele was slightly but significantly higher than the C-allele (P=0.009). Despite only a modest effect of the SNP on the activity of the isolated proximal TPH1 promoter fragment in vitro, our results indicate that modulating EGR-1 binding to the -347 site could potentially lead to differential TPH1 expression in vivo as a function of the two SNP alleles.
Genetic Association of -347C/A (rs7130929) TPH1 Variant with IBS Bowel Habit Subtype
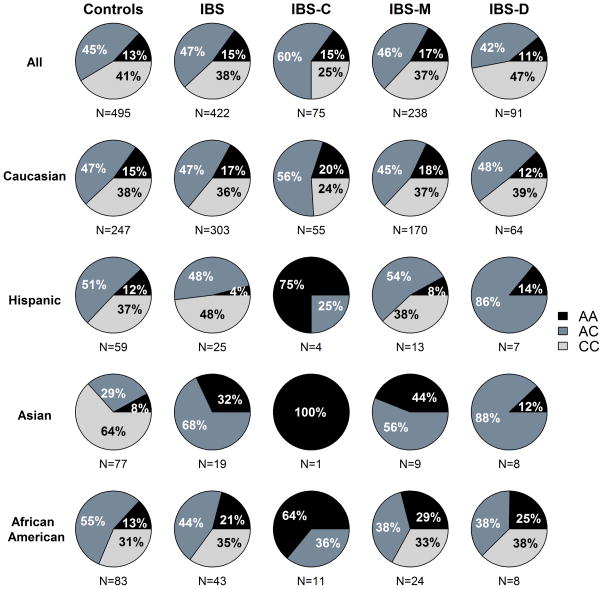
To determine whether the -347C/A TPH1 promoter variant was associated with distinct bowel habit subtypes in IBS, we genotyped 422 IBS patients classified by their predominant bowel habit phenotype according to Rome III criteria and also 495 healthy controls. The UCLA samples included 209 IBS patients and 302 controls (all salivary DNA) and the Washington University samples were comprised of 213 IBS patients (66% blood and 34% salivary DNA), and 193 controls (94% blood and 6% salivary DNA). Pertinent demographic and clinical characteristics of IBS and control groups are presented in Table 1. No significant departure from Hardy-Weinberg equilibrium was found for the -347C/A (rs7130929) SNP (P=0.75). Genotype frequencies did not differ between IBS patients (38% CC, 47% AC, 15% AA) and controls (41% CC, 45% AC, 13% AA) or within any of the race/ethnic groups (Figure 2) indicating that the -347C/A SNP was not a risk factor for IBS.
Table 1.
Clinical characteristic of IBS patients and healthy controls
| Controls (N=495) | IBS (N=422) | p-value | |
|---|---|---|---|
| Female: n (%) | 331 (66.9%) | 331 (78.4%) | <0.001 |
| Age: median (IQR) | 36.5 (31) | 44 (23) | <0.001 |
| Race/Ethnicity: n (%) | <0.001 | ||
| Caucasian | 247 (49.9%) | 303 (71.8%) | |
| Hispanic | 59 (11.9%) | 25 (5.9%) | |
| Asian | 77 (15.6%) | 19 (4.5%) | |
| African American | 83 (16.8%) | 43 (10.2%) | |
| Other/Multiracial | 17 (3.4%) | 13 (3.1%) | |
| Anxiety Z Score: mean (SEM) | −0.99 (0.94) | 0 (0.05) | <0.001 |
| Depression Z Score: mean (SEM) | −0.85 (0.57) | 0.99 (0.05) | p<0.001 |
| Bowel Habit Subtype: n (%) | p<0.001 | ||
| IBS-C | 75 (17.8%) | ||
| IBS-D | 91 (21.6%) | ||
| IBS-M | 238 (56.4%) | ||
| IBS-U | 12 (2.8%) |
Figure 2.
Genotype frequency distribution of the -347C/A TPH1 polymorphism in healthy controls and IBS patients, and within IBS bowel habit subtypes defined by Rome III criteria. Distribution of genotype frequencies with respect to race and ethnicity are also shown. Note that the Caucasian group is comprised of non-Hispanic whites only.
When comparing the TPH1 genotype frequencies between the IBS bowel habit subgroups, the IBS-U group was excluded because there were too few samples (n=12, 3%). The dominant, recessive and additive coding of the rs7130929 TPH1 SNP were evaluated in a multinomial regression model predicting bowel habit while controlling for race/ethnicity and the recessive model was found to yield the lowest P value. Thus, we focused on the recessively coded TPH1 SNP in the following results. The prevalence of the CC genotype was significantly different across all three IBS bowel habit subgroups (P=0.016). There was a significantly greater prevalence of the CC genotype in the IBS-D subtype (47%) compared to the IBS-C (25%) (P=0.004) and IBS-M (37%) (P=0.057) subtypes (Figure 2). These differences in bowel habit remained significant in a multinomial regression model predicting bowel habit from TPH1 genotypes controlling for race/ethnicity, anxiety, depression, and study site (P=0.039). The odds ratio for comparing the prevalence of the C-allele in IBS-D to IBS-C was 2.47 (95% CI 1.23–4.95, P=0.011). The odds ratio for comparing the prevalence of the C-allele in IBS-M to IBS-C was 1.77 (95% CI 0.97 to 3.23, P=0.063). As shown in Figure 2, the Caucasian group only included non-Hispanic whites since the genotype frequencies for Hispanics was shown separately. However, to determine if genotype frequencies differed within the Caucasian group, the largest racial group within the study population, we included both Hispanic and non-Hispanic Caucasians to improve power. In the IBS-D subgroup, 42.9%, 45.7% and 11.4% had the CC, AC and AA TPH1 genotypes. Comparable proportions in the IBS-C group were 23.7%, 57.6%, and 18.6% and in the IBS-M were 37.2%, 45.0% and 17.7%. The C-allele was more prevalent in the IBS-D group compared to the IBS-C group (P=0.015), which was greater than the IBS-M group (P=0.046) after controlling for anxiety, depression, and study site. The prevalence of the C-allele was similar in IBS-D and IBS-M (p=0.359). Both regression models (all race/ethnicity and within Caucasian) were evaluated for goodness of fit and had P-values ranging from 0.69–0.71, which indicated no evidence of dispersion. Overall, these results suggested that the -347C/A (rs7130292) polymorphism in the proximal TPH1 promoter is associated with bowel habit predominance within our IBS population.
Association of the -347C/A (rs7130929) TPH1 Variant with Colonic Mucosal TPH1 mRNA
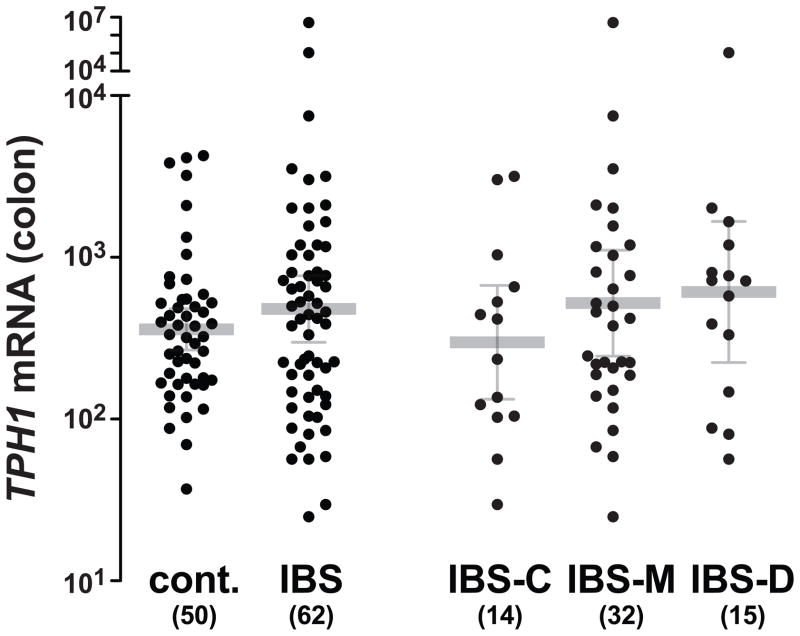
We measured TPH1 mRNA in colonic biopsies obtained from 62 patients with IBS (15 IBS-D, 32 IBS-M, 14 IBS-C, 1 IBS-U) and 50 healthy controls. The results were suggestive of higher TPH1 transcript levels in IBS patients compared to healthy control subjects (Figure 3) and specifically in IBS-D patients (geometric mean [95% CI]; 610 [224–1664]) compared to healthy control subjects (358 [267–481]; P=0.099) as expected. However within the IBS group, TPH1 mRNA levels were not statistically different in the patients with the -347C/C TPH1 genotype (geometric mean [95% CI]; 652 [214–1990]) compared to those with the -347C/A (388 [193–781]) and -347A/A (398 [32–5009]) genotypes (P=0.615).
Figure 3.
Colonic expression of TPH1 mRNA in IBS patients with distinct bowel habit subtypes. Relative TPH1 mRNA levels were normalized for expression of 18S ribosomal RNA and followed a log-normal distribution (Kologorov-Smirnov test). Average and error bars correspond to the geometrical means ± 95% CI.
DISCUSSION
In this study, we identified a functional variant in the human TPH1 promoter that associates with bowel habit subtype in IBS patients. These results support the role of TPH1 as a candidate gene for conditions with abnormal mucosal 5-HT bioavailability as previously inferred from gene dosage effects in animal models (15).
In our evaluation of the two alleles comprising the -347C/A (rs7130929) SNP site for binding to nuclear proteins, we found that EGR-1 preferentially bound to the A-allele sequence along with other zinc finger transcription factors, e.g., Sp1, Sp3 and ZBP-89. This finding was consistent with in vitro binding site selection studies showing that the −1 position adjacent to the canonical EGR consensus sequence impacts the affinity of EGR-1 binding (28). The differential binding of EGR-1 to the -347 (rs7130929) SNP suggested that the SNP could modulate DNA regulatory protein binding, TPH1 expression and ultimately 5-HT production. Recent studies support the notion that differences in 5-HT bioavailability in the colon might correlate with distinct bowel habit subtypes in IBS patients (29). Therefore, to understand the significance of this SNP in vivo, we examined the allele frequencies in IBS patients in which altered intestinal 5-HT bioavailability appears to be linked to distinct bowel habit phenotypes. We hypothesized that in IBS patients, the IBS-D subtype that has been linked to higher 5-HT bioavailability, would be associated with a transcriptionally more active TPH1 promoter allele. We found that in a small subset of our cohort, colonic TPH1 expression showed a trend towards higher expression in IBS-D compared to healthy controls although this difference was not statistically significant. In IBS patients, the presence of the -347A allele significantly reduced the odds for manifestation of the IBS-D over IBS-C subtype. Overall, these findings suggest that the A-allele reduces TPH1 transcriptional activity compared to the C-allele, at least in the context of IBS. Mean colonic TPH1 mRNA was lower in IBS patients with the CA genotype compared to those with the CC genotype but this difference did not reach statistical significance. Unfortunately, the low number of available colonic biopsies from IBS patients with the AA genotype precluded meaningful comparison with CC homozygotes. In addition, the wide variation in TPH1 mRNA levels suggests that there are other factors (e.g., environmental, dietary) that exert a significant effect in addition to the TPH1 genotype. Clearly, future studies in a larger number of colonic tissue samples in IBS are needed to further substantiate the relationship between the -347 SNP (rs7130929) and TPH1 expression in vivo.
The TPH1 -347 (rs7130929) SNP was not directly associated with the diagnosis of IBS overall. This finding is consistent with the results of a recent study by Jun et al. in Caucasian female IBS patients showing that the TPH1 -1066 (rs4537731) SNP, which is in linkage disequilibrium (i.e., highly correlated) to the -347 SNP (rs7130929) in this racial group (Supplemental Table 1), but does not predict IBS status (27). Rather, the -347 SNP (rs7130920) predicts bowel habit predominance. However, Jun et al. found that patients who were homozygous for the minor allele of the -1066 SNP had a higher severity in daily diarrhea symptoms compared to the other two genotypes but there were no differences in bowel habit subtype. These results differ from those in our study, although there are methodological differences. Our subject groups had diverse racial/ethnic backgrounds and were not comprised of only Caucasians as in the Jun et al. study. While we cannot make conclusions about the association of the TPH1 SNP and IBS-D vs. IBS-C within each racial group due to the relatively small sample sizes, this association was significant within the Caucasian group. Another methodological difference is that we did not measure daily IBS symptoms. Furthermore, in the Jun et al. study, the bowel habit subtypes were classified using Rome II criteria (personal communication S Jun and MM Heitkemper), while we used Rome III criteria. The Rome II bowel habit subclassification criteria (30) categorizes only IBS-D and IBS-C based on a combination of abdominal and bowel symptoms (including stool form). In contrast, the Rome III bowel habit subclassification criteria (20) identify IBS-D, IBS-C, IBS-M, and IBS-U (unsubtyped) based solely on the prevalence of stool form types. The Rome II and III bowel habit subtype classifications are based on recall of the prevalence of the GI symptoms over the past three months and do not require prospective daily stool diaries (9). Thus, there can be poor agreement between Rome II and Rome III subtyping of IBS bowel habits (31). In our study, we showed that patients who were homozygous for the major allele of the -347 (rs7130929) SNP were more likely to have IBS-D versus IBS-C. Similarly, IBS-D patients were more likely to have the major allele of the −1066 (rs4537731) SNP versus IBS-C in our IBS patient population (P=0.07). Thus, the differences in our studies are likely due to methodological differences in symptom assessment and bowel habit classification. Perhaps, objective measures of stool frequency and consistency and GI transit would have correlated better with TPH1 genotype and mRNA levels.
In summary, we have identified a proximal TPH1 promoter SNP (rs7130929) that segregates with stool consistency in a patient population with IBS (i.e., greater CC genotype frequency in the IBS-D subgroup). This variant differentially affects binding of the transcription factor EGR-1, an early response gene product that potentially modulates TPH1 gene expression in the context of the -347 (rs7130929) A-allele. As a result, the SNP genotype appears to correlate with TPH1 expression in vivo, and could play a valuable role in predicting an IBS patient’s response to medications targeting the 5-HT signaling system including recently developed TPH-1 antagonists (16).
Supplementary Material
STUDY HIGHLIGHTS.
WHAT IS THE CURRENT KNOWLEDGE
Plasma serotonin (5-hydroxytryptamine; 5-HT) levels correlate with the diarrhea predominant bowel habit in patients with irritable bowel syndrome (IBS).
TPH1 is the rate-limiting enzyme for 5-HT biosynthesis in enteroendocrine cells, the major source of 5-HT in the gut and systemic circulation.
TPH1 is subject to a gene dosage effect in a murine model suggesting that genetic variants affecting TPH1 expression could modulate colonic 5-HT bioavailability and possibly IBS bowel habit phenotype.
WHAT IS NEW HERE
The TPH1 -347C/A promoter polymorphism (rs7130929) differentially affects binding of the DNA regulatory protein called early growth response protein 1 (EGR-1)
The frequency of the TPH1 CC genotype is associated with the IBS-D subtype as defined by the Rome III criteria.
Acknowledgments
Grant Support: NIH grants R01 DK55732 (JLM), P50 DK64539 (LC, EAM) and R01 DK48351 (EAM), RC1 DK 086150-01 (CP), P30 DK52574 (RDN) and K23 DK84113 (GSS). IRB Approval for UCLA cohort: Original approval on January 22, 2007; current approval on July 19, 2012. IRB Approval for Washington University cohort: November 29, 2012
Financial support: NIH grants R01 DK55732 (JLM), P50 DK64539 (LC, EAM) and R01 DK48351 (EAM) and RC1 DK 086150-01 (CP) and K23 D0K84113 (GSS). The BioBank Core of the Washington University Digestive Diseases Research Center is supported by NIH grant P30 DK52574.
Footnotes
Potential competing interests: None.
Specific author contributions: study design analysis, SNP, RFLP analysis, molecular biology, data analysis, wrote manuscript: Helmut Grasberger; concept design, patient recruitment, medical history and physical examinations, collection of patient samples, data analysis, critical review of manuscript, IRB, wrote manuscript: Lin Chang; statistical analysis, critical manuscript editing: Wendy Shih; statistical analysis, critical analysis of manuscript and editing: Angela P. Presson; patient recruitment, medical history and physical examinations, collection of patient samples, critical analysis of the manuscript: Gregory Sayuk; IRB, sample processing, critical analysis of the manuscript, funding: Rodney Newberry prepared mRNA and TPH1 RT-qPCR from biopsies: Iordanis Karagiannides; qPCR materials, obtained funding, critical analysis of manuscript: Charalabos Pothoulakis; IRB, patient recruitment and analysis, obtained funding for patient studies: Emeran Mayer; experimental design, data analysis, manuscript draft and editing, funding: Juanita L. Merchant.
References
- 1.Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology. 2007;132:397–414. doi: 10.1053/j.gastro.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Bearcroft CP, Perrett D, Farthing MJ. Postprandial plasma 5-hydroxytryptamine in diarrhoea predominant irritable bowel syndrome: a pilot study. Gut. 1998;42:42–46. doi: 10.1136/gut.42.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson W, Lockhart S, Whorwell PJ, et al. Altered 5-hydroxytryptamine signaling in patients with constipation- and diarrhea-predominant irritable bowel syndrome. Gastroenterology. 2006;130:34–43. doi: 10.1053/j.gastro.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 4.Houghton LA, Atkinson W, Whitaker RP, et al. Increased platelet depleted plasma 5-hydroxytryptamine concentration following meal ingestion in symptomatic female subjects with diarrhoea predominant irritable bowel syndrome. Gut. 2003;52:663–670. doi: 10.1136/gut.52.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuo XL, Li YQ, Yang XZ, et al. Plasma and gastric mucosal 5-hydroxytryptamine concentrations following cold water intake in patients with diarrhea-predominant irritable bowel syndrome. J Gastroenterol Hepatol. 2007;22:2330–2337. doi: 10.1111/j.1440-1746.2006.04772.x. [DOI] [PubMed] [Google Scholar]
- 6.Dunlop SP, Coleman NS, Blackshaw E, et al. Abnormalities of 5-hydroxytryptamine metabolism in irritable bowel syndrome. Clin Gastroenterol Hepatol. 2005;3:349–357. doi: 10.1016/s1542-3565(04)00726-8. [DOI] [PubMed] [Google Scholar]
- 7.Houghton LA, Atkinson W, Lockhart C, et al. Sigmoid-colonic motility in health and irritable bowel syndrome: a role for 5-hydroxytryptamine. Neurogastroenterol Motil. 2007;19:724–731. doi: 10.1111/j.1365-2982.2007.00943.x. [DOI] [PubMed] [Google Scholar]
- 8.Ford AC, Talley NJ, Schoenfeld PS, et al. Efficacy of antidepressants and psychological therapies in irritable bowel syndrome: systematic review and meta-analysis. Gut. 2009;58:367–378. doi: 10.1136/gut.2008.163162. [DOI] [PubMed] [Google Scholar]
- 9.Coates MD, Mahoney CR, Linden DR, et al. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology. 2004;126:1657–1664. doi: 10.1053/j.gastro.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Camilleri M, Andrews CN, Bharucha AE, et al. Alterations in expression of p11 and SERT in mucosal biopsy specimens of patients with irritable bowel syndrome. Gastroenterology. 2007;132:17–25. doi: 10.1053/j.gastro.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerckhoffs AP, Ter Linde JJ, Akkermans LM, et al. Trypsinogen IV, serotonin transporter transcript levels and serotonin content are increased in small intestine of irritable bowel syndrome patients. Neurogastroenterol Motil. 2008;20:900–907. doi: 10.1111/j.1365-2982.2008.01100.x. [DOI] [PubMed] [Google Scholar]
- 12.Spiller R, Bennett A. Searching for the answer to irritable bowel syndrome in the colonic mucosa: SERTainty and unSERTainty. Gastroenterology. 2007;132:437–441. doi: 10.1053/j.gastro.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Chalazonitis A, Huang YY, et al. Essential roles of enteric neuronal serotonin in gastrointestinal motility and the development/survival of enteric dopaminergic neurons. J Neurosci. 2011;31:8998–9009. doi: 10.1523/JNEUROSCI.6684-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Z, Liu T, Chattoraj A, et al. Posttranslational regulation of TPH1 is responsible for the nightly surge of 5-HT output in the rat pineal gland. J Pineal Res. 2008;45:506–514. doi: 10.1111/j.1600-079X.2008.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Yang Q, Sun W, et al. Discovery and characterization of novel tryptophan hydroxylase inhibitors that selectively inhibit serotonin synthesis in the gastrointestinal tract. J Pharmacol Exp Ther. 2008;325:47–55. doi: 10.1124/jpet.107.132670. [DOI] [PubMed] [Google Scholar]
- 16.Brown PM, Drossman DA, Wood AJ, et al. The tryptophan hydroxylase inhibitor LX1031 shows clinical benefit in patients with nonconstipating irritable bowel syndrome. Gastroenterology. 2011;141:507–516. doi: 10.1053/j.gastro.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tack J, Janssen P, Wouters M, et al. Targeting serotonin synthesis to treat irritable bowel syndrome. Gastroenterology. 2011;141:420–422. doi: 10.1053/j.gastro.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 18.Camilleri M. LX-1031, a tryptophan 5-hydroxylase inhibitor, and its potential in chronic diarrhea associated with increased serotonin. Neurogastroenterol Motil. 2011;23:193–200. doi: 10.1111/j.1365-2982.2010.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Essien BE, Grasberger H, Romain RD, et al. ZBP-89 Regulates Expression of Tryptophan Hydroxylase I and Mucosal Defense Against Salmonella Typhimurium in Mice. Gastroenterology. 2013;144:1466–1477. e1469. doi: 10.1053/j.gastro.2013.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longstreth GF, Thompson WG, Chey WD, et al. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 21.Zigmond AS, Snaith RP. The hospital anxiety and depression scale. Acta Psychiatr Scand. 1983;67:361–370. doi: 10.1111/j.1600-0447.1983.tb09716.x. [DOI] [PubMed] [Google Scholar]
- 22.Beck AT, Ward CH, Mendelson M, et al. An inventory for measuring depression. Archives of general psychiatry. 1961;4:561–571. doi: 10.1001/archpsyc.1961.01710120031004. [DOI] [PubMed] [Google Scholar]
- 23.Beck AT, Epstein N, Brown G, et al. An inventory for measuring clinical anxiety: psychometric properties. Journal of consulting and clinical psychology. 1988;56:893–897. doi: 10.1037//0022-006x.56.6.893. [DOI] [PubMed] [Google Scholar]
- 24.Boularand S, Darmon MC, Mallet J. The human tryptophan hydroxylase gene. An unusual splicing complexity in the 5′-untranslated region. J Biol Chem. 1995;270:3748–3756. doi: 10.1074/jbc.270.8.3748. [DOI] [PubMed] [Google Scholar]
- 25.Schreiber E, Matthias P, Muller MM, et al. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merchant JL, Iyer GR, Taylor BR, et al. ZBP-89, a Kruppel-like zinc finger protein, inhibits epidermal growth factor induction of the gastrin promoter. Mol Cell Biol. 1996;16:6644–6653. doi: 10.1128/mcb.16.12.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jun S, Kohen R, Cain KC, et al. Associations of tryptophan hydroxylase gene polymorphisms with irritable bowel syndrome. Neurogastroenterol Motil. 2011;23:233–239. e116. doi: 10.1111/j.1365-2982.2010.01623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swirnoff AH, Milbrandt J. DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gershon MD. Review article: roles played by 5-hydroxytryptamine in the physiology of the bowel. Aliment Pharmacol Ther. 1999;13 (Suppl 2):15–30. [PubMed] [Google Scholar]
- 30.Thompson WG, Longstreth GF, Drossman DA, et al. Functional bowel disorders and functional abdominal pain. Gut. 1999;45(Suppl 2):II43–47. doi: 10.1136/gut.45.2008.ii43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ersryd A, Posserud I, Abrahamsson H, et al. Subtyping the irritable bowel syndrome by predominant bowel habit: Rome II versus Rome III. Aliment Pharmacol Ther. 2007;26:953–961. doi: 10.1111/j.1365-2036.2007.03422.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.