Abstract
Because modern humans originated in Africa and have adapted to diverse environments, African populations have high levels of genetic and phenotypic diversity. Thus, genomic studies of diverse African ethnic groups are essential for understanding human evolutionary history and how this leads to differential disease risk in all humans. Comparative studies of genetic diversity within and between African ethnic groups creates an opportunity to reconstruct some of the earliest events in human population history and are useful for identifying patterns of genetic variation that have been influenced by recent natural selection. Here we describe what is currently known about genetic variation and evolutionary history of diverse African ethnic groups. We also describe examples of recent natural selection in African genomes and how these data are informative for understanding the frequency of many genetic traits, including those that cause disease susceptibility in African populations and populations of recent African descent.
African populations have the highest levels of genetic variation among all humans. Studies of these populations are used to understand demographic changes in Africa and to identify disease-related genes (e.g., APOL1).
Africa is where modern humans evolved and is the starting place for the global expansion of our species (Stringer and Andrews 1988; Stringer 1994; Templeton 2002). African populations also have the highest levels of genetic and phenotypic variation among all humans. This variation is informative for characterizing demographic history in Africa, including times when populations increased in size, contracted, migrated, or when admixture between them occurred. Genetic variation also provides data that are useful in identifying local adaptation to diverse environments (Campbell and Tishkoff 2008). Characterizing human genetic variation and examining phenotypic variation in extant African populations is fundamental to the identification of genes that play a role in function, adaptation, and complex disease susceptibility in Africans and populations of recent African descent.
LINGUISTIC, CULTURAL, AND SUBSISTENCE DIVERSITY IN AFRICA
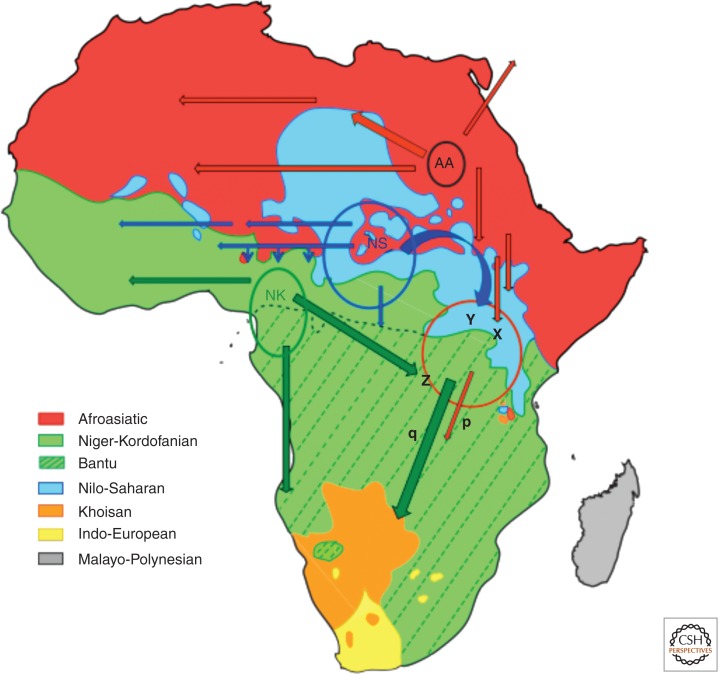
There are over 2000 indigenous languages spoken in Africa. They are classified into four major linguistic families—Niger-Kordofanian (spoken primarily by agriculturalist populations located in large contiguous regions of sub-Saharan Africa from West Africa to eastern and southern Africa), Nilo-Saharan (spoken predominantly by pastoralist populations in central and eastern Africa), Afroasiatic (spoken predominantly by agro-pastoralists and pastoralist populations in northern and eastern Africa), and Khoesan (a language family that contains click consonants, spoken by hunter–gatherer San populations in southern Africa as well as the Hadza and Sandawe hunter–gatherers in Tanzania) (Fig. 1).
Figure 1.
The geographic distribution of the major linguistic groups in Africa. The map was drawn using information (geographic locations of ethnic speakers in Africa that are based on published sources) from Haspelmath et al. (2008) (wals.info/), www.ethnologue.com (International HapMap 2005), Greenberg (1963, 1972), Vansina (1995), and Ehret (1971, 1993, 1995, 2001b). Geographic range occupied by Bantu speakers, the major linguistic subfamily within the Niger-Kordofanian phylum mentioned in the text, is also shown. Putative centers of origin and estimated time of initial expansion based on linguistic studies for three of the four language families are also listed: AA, Afroasiatic (14 kya) (Ehret 1995); NS, Nilo-Saharan (Blench 1993; Ehret 1993; Blench 2006); NK, Niger-Kordofanian (5 kya) (Nurse 1997; Ehret 2001a). Afroasiatic-speaking pastoralists were the first food-producing populations to migrate into East Africa circa 5 kya (X) (Leakey 1931; Butzer 1969; Robbins 1972; Barthelme 1977); followed by Nilo-Saharan-speaking pastoralists circa 3 kya (Y) (Leakey 1931; Bower 1973; Ambrose 1982; Distefano 1990), and later Bantu-speaking agriculturalists after circa 2.5 kya (Z) (Posnansky 1961a,b). P and q represent initial expansion of pastoralists (2.5 kya) and later Bantu-speaking agriculturalists (after 2 kya) to southern Africa from East Africa, respectively.
African populations also live in a diverse range of environments (including deserts, savannahs, and tropical environments), have different exposures to infectious disease, and have diverse diets and subsistence patterns. Because African populations exist in such a broad spectrum of environments, they may have regional or population-specific genetic variants that play a role in local adaptation to different selection pressures. The study of African genomic variation is uniquely interesting because of the large proportion of human genetic variation observed in African populations and the length of human history in the continent.
PATTERNS OF GENETIC DIVERSITY AND DEMOGRAPHIC HISTORY IN AFRICA
The characterization of human genetic variation began with studies that described human blood groups and protein polymorphisms (Cavalli-Sforza 1994). Since then, it has grown to include numerous studies of mtDNA (mitochondrial DNA) that is inherited maternally, Y chromosome variation that is inherited paternally, and loci from the full nuclear genome that include sequences of repetitive DNA, single nucleotide polymorphisms (SNPs), and structural variation (SVs) (Campbell and Tishkoff 2008).
The pattern of genetic variation at these loci in modern African populations reflects their demographic and evolutionary histories (Campbell and Tishkoff 2008). Modern humans evolved in Africa ∼200 kya (thousand years ago) and migrated from Africa within the past 50–100 kya, successfully colonizing most of the terrestrial parts of the globe (Campbell and Tishkoff 2008). This is called the “Out-of-Africa” migration. Currently, the precise location of the origin of modern humans remains a contentious issue. Some argue that South Africa is the location where our species originated (Tishkoff et al. 2009; Compton 2011; Henn et al. 2011), whereas others argue that East Africa is where modern humans originated (Prugnolle et al. 2005; Ray et al. 2005). However, these inferences are biased by the lack of archeological and fossil data in tropical areas of Africa and the fact that the geographic location of populations in the present may have differed in the past.
Genetic evidence also indicates that human populations underwent several major population expansions at different time periods over the last 100 ky (Rogers and Harpending 1992; Harpending et al. 1993; Excoffier and Schneider 1999; Atkinson et al. 2009; Cox et al. 2009): an initial major population expansion in Africa ∼110–70 kya (Rogers and Harpending 1992; Harpending et al. 1993; Excoffier and Schneider 1999; Atkinson et al. 2008, 2009) and subsequent migration and expansion events around the globe at ∼60–55 kya (Atkinson et al. 2009), ∼40–25 kya, and ∼12 kya (Harpending et al. 1993; Atkinson et al. 2009; Cox et al. 2009).
Prior to the domestication of plants and animals ∼10,000 years ago, all human populations practiced hunting-gathering/foraging for subsistence. Although we do not know their original homeland, ancestors of the extant African hunter–gatherer populations, including the current Khoesan speakers, migrated into central, eastern, and southern Africa probably >20 kya (Nurse 1997; Ehret 2002). It is speculated that speakers of Khoesan languages were widespread throughout a contiguous area that encompassed southern and eastern African regions (Sutton 1973; Smith 1992). Based on archaeological and linguistic data, it is hypothesized that recent migration events that occurred, due to changes in subsistence patterns, modified the genetic landscape in Africa through admixture of populations practicing pastoralism and/or agriculture with indigenous hunter–gatherer populations. For example, the expansion of agriculturalists from Central-West Africa and the migration of pastoralist populations from northeastern Africa have contributed to the current genetic landscape in Africa. One of the largest migration events in Africa that was mediated by a shift in subsistence patterns was the migration of Bantu-speaking populations, a subfamily of Niger-Kordofanian languages, from their proposed homeland in Nigeria/Cameroon across sub-Saharan Africa within the past 5000 years (Fig. 1) (Nurse 1997; Ehret 2001a). The result of this expansion is the current predominance of Bantu languages in many African countries. Other major migration events have included the migration of Nilo-Saharan pastoralists from their homeland in Chad/Sudan, both westward across the Sahel ∼7000 years ago and eastward into Kenya and Tanzania within the past ∼3000 years (Blench 1993; Ehret 1993). Afro-Asiatic-speaking agro-pastoralists migrated from their homeland, which encompassed the Nile Valley to the Ethiopian highlands, into western, northern, and eastern Africa within the past 8000–5000 years (Ehret 1995, 1998) (Fig. 1). Demographic history in Africa has also been influenced by the more recent migration of non-Africans into Africa. Most noteworthy is the migration of southwestern Asians into Northern and Eastern Africa within the past ∼3000 years (Van Beek 1967; Butzer 1981; Phillipson 2009). Europeans, southern and eastern Asians have also migrated into Southern Africa within the past several hundred years.
Consistent with the “Out of Africa” model of modern human origins, analyses of genetic data indicate that Africans have higher levels of genetic diversity than non-Africans (Cann et al. 1987, 2002; Tishkoff et al. 1998; Marth et al. 2004). Overall, the extant patterns of genetic variation in global populations suggests serial founder events, with increasing genetic distance correlated with geographic distance from East Africa, indicating the cumulative effect of genetic drift as humans expanded into the rest of the world (Prugnolle et al. 2005; Ramachandran et al. 2005; Hofer et al. 2009). Studies of autosomal genetic variation show that genetic distance between populations also increases with geographic distance within Africa (Prugnolle et al. 2005; Ramachandran et al. 2005; Hofer et al. 2009; Tishkoff et al. 2009). The greater genetic diversity in sub-Saharan Africans, as compared to other continental populations, may also be partially a result of ancient admixture with yet-to-be identified archaic population(s) (Zietkiewicz et al. 1998; Labuda et al. 2000; Plagnol and Wall 2006; Hammer et al. 2011; Lachance et al. 2012), analogous to the proposed admixture between Neanderthals and modern humans in Eurasia (Reich et al. 2010, 2011; Abi-Rached et al. 2011; Green et al. 2011; Yotova et al. 2011). Genomic introgression from Neanderthals has been detected in some East African populations but was inferred to be a signature of recent back-migrations from Eurasia (Wang et al. 2013).
Many of the recent population genetic studies of African populations are based on analysis of genetic markers that were genotyped in a small number of populations that are part of the CEPH-HGDP collection (Centre d’Étude du Polymorphisme Humain–Human Genome Diversity Panel) or the International Haplotype Map (HapMap) Project (Batzer and Deininger 2002; Rosenberg et al. 2002; Li et al. 2008). Only six African populations are included in the CEPH-HGDP panel, whereas the International HapMap Project includes just three African populations (Niger-Kordofanian-speaking Yoruba from Nigeria, Bantu-speaking Luhya from Kenya, and Nilo-Saharan-speaking Maasai from Kenya). A more recent international collaborative effort that aims to catalogue human genetic variation through whole genome resequencing, the “The 1000 Genomes Project” (2010), originally had only two African populations represented in the project but has since expanded to include the Esan from Nigeria, individuals from The Gambia, the Luhya from Kenya, and the Mende from Sierra Leone. However, it should be noted that all of these populations speak Niger-Kordofanian languages and share recent genetic ancestry. The 1000 Genomes project has also expanded to include people of recent African descent including African Americans from the southwestern United States and African Caribbean individuals from Barbados. Although these projects are important in their description of overall human genetic diversity, they are limited in their coverage of African populations, despite the recent additions (Consortium 2010). Because these projects have sampled a small number of African populations it is likely that a substantial portion of human genetic variation will be missed. Thus, it is important to continue to add African populations that are underrepresented in human genomic studies, particularly those speaking languages belonging to the other three language families in Africa. Analysis of genomic variation across a broad range of African populations will be useful for elucidating fine-scale population structure and demographic patterns in African populations.
The current studies of global human genetic variation suggest that human populations may have been divided into distinct subpopulations in Africa prior to the migration of modern humans Out-of-Africa ∼100 kya (Labuda et al. 2000; Satta and Takahata 2004; Adeyemo et al. 2005; Plagnol and Wall 2006; Bryc et al. 2009; Tishkoff et al. 2009; Wall et al. 2009; de Wit 2010; Patterson 2010; Sikora 2010; Henn et al. 2012; Pagani et al. 2012; Schlebusch et al. 2012). For example, we (Tishkoff et al. 2009) identified 14 ancestral population clusters in Africa with four predominant clusters that broadly represent populations from major African geographical regions and those that speak the four African language families mentioned above. The remaining 10 are mainly restricted to specific geographic regions, languages, or in some cases, individual populations (e.g., Hadza hunter–gatherers). Other studies of African genetic diversity (Adeyemo et al. 2005; Bryc et al. 2009; de Wit 2010; Patterson 2010; Sikora 2010; Henn et al. 2012; Pagani et al. 2012; Schlebusch et al. 2012) have largely recapitulated the regional clusters (Tishkoff et al. 2009) while also adding some fine-scale genetic structure between populations from different ethnicities or populations that speak different languages within the regions studied.
Studies of genetic variation in Africa suggest that even though high levels of mixed ancestry are observed in most African populations, the genetic variation observed in Africa is broadly correlated with geography, language classification (Adeyemo et al. 2005; Bryc et al. 2009; Tishkoff et al. 2009) and subsistence classifications (Tishkoff et al. 2009). For example, genetic variation among Nilo-Saharan and Afroasiatic-speaking populations from both Central and East Africa (Tishkoff et al. 2009; Bryc et al. 2009) reflect the geographic region from which they originated, and generally shows a complex pattern of admixture between these populations and the Niger-Kordofanian speakers who migrated into the region more recently. Consistent with linguistic evidence regarding the origin of Nilo-Saharan languages in the Chad/Sudan border, the highest proportion of Nilo-Saharan ancestry is observed among southern Sudanese populations (Tishkoff et al. 2009). Additionally, North/Northwest African populations are genetically differentiated from sub-Saharan African populations, but have considerable shared ancestry with East African Afroasiatic speakers (Tishkoff et al. 2009; Henn et al. 2012) and southwest Asian populations (Rosenberg et al. 2002; Behar et al. 2008, 2010; Li et al. 2008; Kopelman et al. 2009; Tishkoff et al. 2009; Atzmon et al. 2010; Bray 2010; Hunter-Zinck 2010; Henn et al. 2012), likely reflecting historic migrations from those source populations into northern Africa. Finally, several studies of genetic variation in the “colored” population from South Africa (Tishkoff et al. 2009; de Wit et al. 2010; Patterson et al. 2010) are consistent with the documented history of this population, which indicates that they have South African Khoesan, Niger-Kordofanian, European, South Asian, and Indonesian ancestors (Tishkoff et al. 2009; de Wit et al. 2010; Patterson et al. 2010).
mtDNA AND Y CHROMOSOME VARIATION
A refined view of uniparental genetic diversity in Africa has been hampered by a lack of extensive sampling in Africa and a lack of consensus on the proper method to estimate mutation rates that are essential to make inferences about human demography. The time to the most recent common ancestor (TMRCA) for mtDNA lineages, based on large datasets, ranges from 121.5 to 221.5 kya, (Ingman et al. 2000; Behar et al. 2008), which is older than most estimates of the Y chromosome TMRCA, which are based on patchy sampling within Africa (60–140 kya) (Thomason 1988; Cruciani et al. 2011; Wei et al. 2013). The fact that there has been limited sampling in Africa is further exemplified by recent results that include more African samples and infer that the TMRCA of the human Y chromosome is similar (120–200 kya) (Mendez et al. 2013) or even older (237–581 kya) than the TMRCA of mtDNA (Ingman et al. 2000; Behar et al. 2008; Francalacci et al. 2013; Mendez et al. 2013; Poznik et al. 2013). Generally, there is a lack of precision in TMRCA estimates reported by different studies using the same marker systems (Francalacci et al. 2013; Mendez et al. 2013; Poznik et al. 2013). Nonetheless, the consensus based on numerous studies, is that human populations exhibit structure (e.g., genetic differences among populations from different regions) based on both mtDNA and Y chromosome variation (Wallace et al. 1999; Ingman et al. 2000; Maca-Meyer et al. 2001; Herrnstadt et al. 2002; Mishmar et al. 2003).
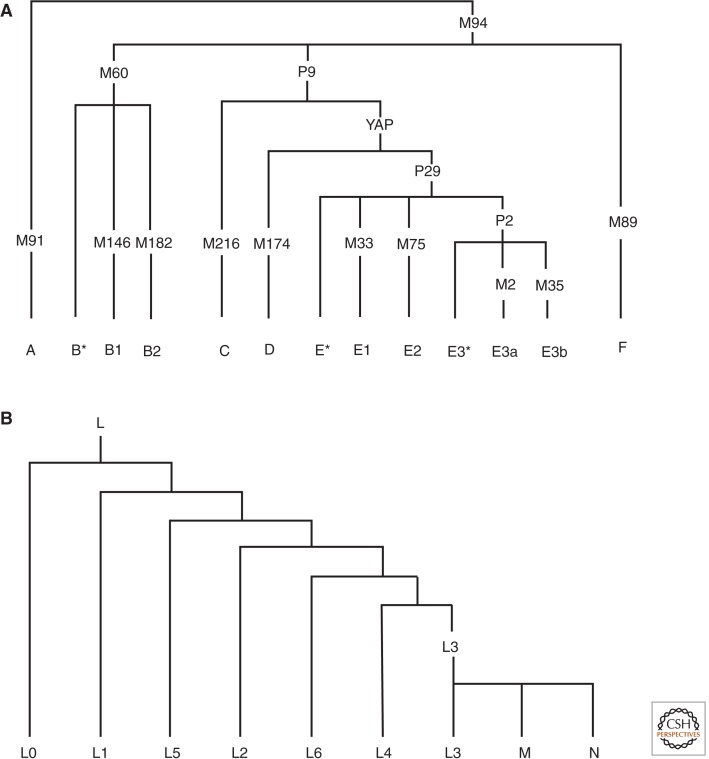
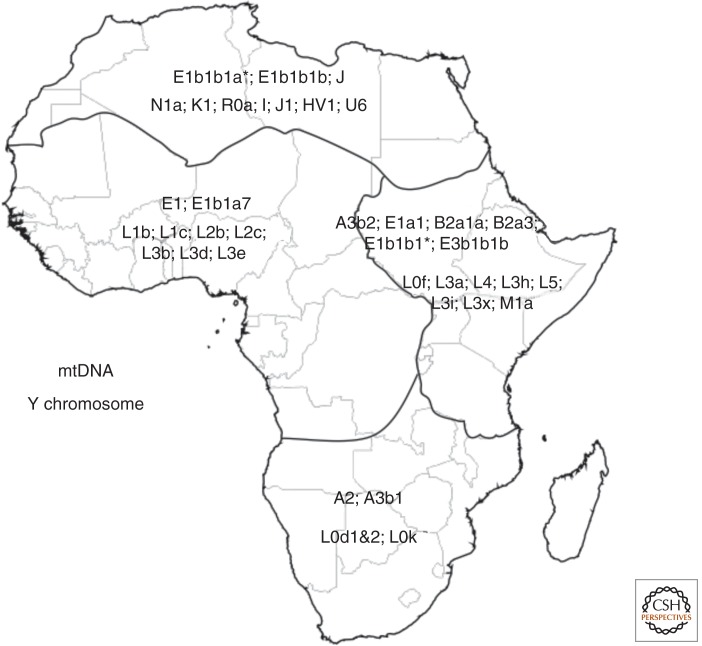
The distribution pattern of uniparental lineages in Africa suggests ancient geographical and cultural subdivision in African. African populations exhibit greater diversity in the Y chromosome than those of other continents (Hammer 2002). Based on the latest classification of Y chromosome haplogroups (Karafet et al. 2008), A, B, E, and J haplogroups are found in Africans, whereas C, D, and the lineages descending from haplogroup F are almost exclusively observed outside of sub-Saharan Africa (Fig. 2). The oldest lineages, A and B, with a TMRCA over ∼75 kya, are present only in Africans. The younger lineages, E and J haplogroups, with a TMRCA less than 75 kya, are present in both African and non-African populations (Fig. 2) (Seielstad et al. 1998; Scozzari et al. 1999). Haplogroup E represents the great majority of Y chromosome haplotypes across Africa (Cruciani et al. 2002). Furthermore, the sublineages within these haplogroups exhibit unique regional distribution patterns within Africa (Cruciani 2002, 2010, 2011; Hammer 2002; Luis et al. 2004; Wood et al. 2005; Batini 2007, 2011; Berniell-Lee et al. 2009), with some restricted to specific geographical regions and/or language families (Fig. 3), consistent with substructuring of diverse African populations.
Figure 2.
Evolutionary tree of Y chromosome and mtDNA haplogroups. (A) Nomenclature for major lineages of Y chromosome haplotypes. The M, P, and YAP labels leading to haplogroup/s are SNPs and indels that are used to define these haplogroups. Haplogroup F encompasses haplogroups F to T. Haplogroups A, B, and E are mainly found in Africa, whereas the rest are found mainly outside Africa. (B) Overview of mtDNA haplogroup phylogeny. In the mtDNA haplogroup nomenclature, the letter names of the haplogroups run from A to Z, with further subdivisions using numbers (from 0) and lowercase letters (from a). The naming was done in the order of their discovery and does not reflect the actual genetic relationships. Haplogroup M and N encompasses all of the haplogroups lettered A to Z excluding haplogroup L. Haplogroups L (L0–L6) are mainly found in Africa, whereas the rest are found mainly outside Africa (Richards et al. 1998; Macaulay et al. 1999; Quintana-Murci et al. 1999; Salas et al. 2002; Kivisild et al. 2004; Behar et al. 2008).
Figure 3.
Distribution of mtDNA and Y chromosome haplogroups across Africa. The black lines represent the approximate geographical boundaries of the distribution of each haplogroup cluster, with each of the clusters predominantly observed in the demarcated regions.
Analogous to variation observed on the Y chromosome, mtDNA haplotypes also show geographic structuring. African populations are characterized by having the oldest haplogroup lineages (L0–L5), with TMRCA over ∼80 kya (Bandelt et al. 1995, 2001; Chen et al. 1995, 2000; Graven et al. 1995; Soodyall et al. 1996; Watson et al. 1996, 1997; Alves-Silva et al. 2000; Torroni et al. 2001; Salas et al. 2002), as well as the M1 haplogroup, an L3 sublineage. Although lineages M, N, and all of their descending lineages with inferred TMRCA less than 80 kya are found in most global populations, they are observed almost exclusively outside of sub-Saharan Africa. There is also a unique distribution pattern of mtDNA haplotypes across Africa with some lineages restricted to specific geographical regions and/or language families (Fig. 3) (Newman 1980; Vigilant 1990, 1991; Chen et al. 1995, 2000; Ehret 1995, 2006; Watson et al. 1996, 1997; Rando et al. 1998; Krings et al. 1999; Richards et al. 2000; Pereira et al. 2001; Salas et al. 2002; Thomas et al. 2002; Destro-Bisol et al. 2004; Kivisild et al. 2004; Beleza et al. 2005; Coia et al. 2005; Jackson et al. 2005; Cerny et al. 2006, 2008, 2009; Gonzalez et al. 2006; Batini et al. 2007; Gonder et al. 2007; Behar et al. 2008; Coudray et al. 2008; Quintana-Murci et al. 2008, 2010; Castri et al. 2009; Coelho et al. 2009; Poloni et al. 2009; Saunier et al. 2009; Stefflova et al. 2009; Veeramah et al. 2010). Additionally, there is also a predominance of some Y chromosome and mtDNA lineages in specific regions of Africa. There is also sharing of many of these lineages between regions (Watson et al. 1996, 1997; Pereira et al. 2001; Salas et al. 2002; Beleza et al. 2005; Behar et al. 2008; Quintana-Murci et al. 2008), suggesting that there have been both recent and ancient migration events between these regions over the last 20 ky (Ehret 1967).
Despite the large amount of ethnic and genetic diversity observed in African relative to non-African populations, there has been limited sampling in Africa. Additional sampling and characterization of genetic diversity, particularly from underrepresented central, southern and eastern African countries will be crucial to deciphering fine-scale genetic history of genetically, culturally, and linguistically diverse African populations and for reconstructing both ancient and recent human evolutionary history. Comprehensive analysis of African genetic variation will help us address some of the outstanding questions about the origin of our species such as: (1) precisely where and when modern humans originated in Africa; (2) the number and age of ancestral structured populations in Africa; (3) the age and direction of ancient migration events both within and out of Africa (e.g., we do not yet have a consensus from genetic studies on the timing and route/s modern humans took out of Africa to populate the rest of the globe); and, (4) determination of the archaic source population(s) from which genetic introgression occurred into African populations and when these events took place.
DARWINIAN SELECTION AND GENETIC VARIATION
Demographic events such as those described above, that is changes in population size, short- and long-range migrations, and admixture or gene flow from one population to another, influence the levels and patterns of genetic variation across the genome (Campbell and Tishkoff 2008). In addition to demography, natural selection, recombination and mutation, which take place in specific parts of the genome, can also influence genetic variability.
Natural selection occurs when the fitness effects of genotypes are unequal (Nielsen 2005). For example, a beneficial genetic variant is likely to increase in frequency in a population over time because of positive selection. Likewise, if a genetic variant is deleterious, it may be selected against and its frequency in a population will decrease over time. The occurrence of natural selection can thus create substantial differences in the patterns of genetic variation between human populations (Aquadro et al. 2001). Identifying loci that exhibit patterns of genetic variation that are indicative of natural selection is, therefore, important because these loci are likely to be regions of the genome that are, or have been, functionally important and play a role in adaptation to local environments. Furthermore, understanding which regions of the genome have been subject to natural selection will aid in the identification of genetic variation that contributes to phenotypic variation among human populations, including disease susceptibility (Ronald and Akey 2005). Here, we discuss results of several studies of local adaptation to distinct environments and diets within Africa.
Lactase Persistence
Adult mammals lose the ability to effectively digest lactose, the main carbohydrate in milk, after weaning. The inability to digest lactose is a result of decreased production of the enzyme lactase-phlorizin hydrolase, or lactase (Swallow 2003; Ingram et al. 2009). Individuals who are unable to digest lactose have what is called the “lactase nonpersistent” (LNP) trait. Although the LNP trait was once considered to be pathologic, it is now well understood that this trait is “ancestral” (i.e., all ancestors of modern humans had this condition) and widespread among human populations (Auricchio et al. 1963; Newcomer et al. 1983; Segal et al. 1983). Individuals with the “lactase persistent” (LP) trait continue to have high levels of lactase production into adulthood. The LP trait is common only in populations whose ancestors practiced cattle domestication or pastoralism (Ingram et al. 2009). For example, LP occurs at high frequencies in northern European dairying populations (e.g., Finns, Swedes, and Danes), and decreases in frequency in southern Europe, the Middle East, and in most of Asia (Durham 1991). LP is also found to be common in some pastoral populations in Africa (Tishkoff et al. 2007). Thus, it has been hypothesized that LP is an adaptive trait in human populations that practice cattle domestication and dairying (Swallow 2003).
In Europe, a common regulatory variant outside of the gene that encodes lactase (LCT) has been identified as being strongly associated with the LP trait (Enattah et al. 2002; Ingram et al. 2007; Tishkoff et al. 2007; Enattah et al. 2008). Several studies also suggest that this variant is causal and is a target of recent natural selection (Enattah et al. 2002; Poulter et al. 2003; Bersaglieri et al. 2004; Coelho et al. 2005). This suggestion is based on several lines of evidence. First, experiments in small intestine cell lines suggest that the European mutation increases the expression of the lactase gene and causes an increase in the production of the lactase enzyme (Enattah et al. 2002; Olds and Sibley 2003; Troelsen et al. 2003; Lewinsky et al. 2005). Additionally, several different tests indicate that the LCT locus has experienced recent strong natural selection (Poulter et al. 2003; Bersaglieri et al. 2004; Coelho et al. 2005). These studies have identified unexpectedly long genomic regions of genetic similarity flanking the LCT locus, which is a signature of recent positive selection. When positive selection causes a variant or variants to increase in frequency, the selected variant will cause neighboring genetic variation to “hitchhike” as the selected variant increases in frequency, thereby causing large tracks of identical sequences surrounding the variant under selection. This genetic feature has been identified in many European individuals with the regulatory variant who have the LP trait (Poulter et al. 2003). Finally, the estimated age of the European LP-associated variant is between ∼20,000 and ∼2000 years ago (Bersaglieri et al. 2004; Coelho et al. 2005; Tishkoff et al. 2007). These dates are consistent with the origins of dairy farming in southwest Asia that have been estimated from the archaeological record at ∼8–9 kya (Evershed et al. 2008).
Although this is a convincing example of gene/culture co-evolution in Europe, the genetic basis of the LP phenotype was largely unknown in Africa for many years. Previous studies (Mulcare et al. 2004) indicated that the European regulatory variant was noticeably absent in many African populations, including populations that have a history of pastoralism and drinking milk. This observation called into question whether the European variant is truly the causal variant for lactase persistence (Ingram et al. 2007). However, genotype/phenotype association studies of LP in Africa (Swallow 2003; Tishkoff et al. 2007) have identified three novel variants upstream of LCT, near the European variant, that are significantly associated with LP in African populations. All three of the African sites enhance the expression of LCT (Tishkoff et al. 2007; Enattah et al. 2008), and the most common of the three African mutations also shows strong evidence of recent strong positive selection. The estimated age of this African variant is ∼3000–7000 years, consistent with the introduction of cattle domestication south of the Saharan Desert within the past ∼5500 years (Tishkoff et al. 2007). Together, these examples from Europe and Africa show us that strong selective pressure can increase the frequency of numerous rare mutations that arise independently and regulate lactase gene expression where people practice dairying. The occurrence of multiple mutations that evolved under similar selection pressures is a good example of convergent evolution in several different human populations.
Malaria
Examination of population-level genetic variation and recent natural selection caused by infectious diseases can help us understand differences in disease susceptibility and incidence, and why some genetic variants, especially those that are potentially deleterious, are common in specific populations. Malaria is one such example that has had an important impact on patterns of human genetic variation and the geographic distribution of a number of genetic disorders (Fortin et al. 2002).
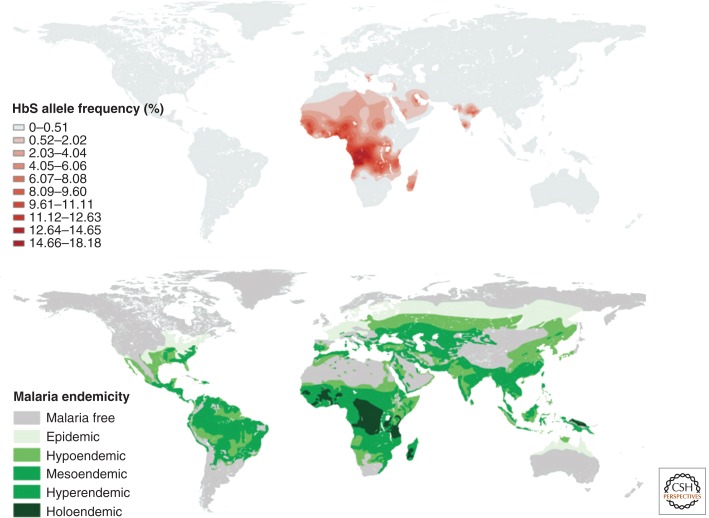
Malaria is a parasitic disease caused by species in the genus Plasmodium. There are several Plasmodium species that infect humans, including P. falciparum, P. ovale, P. malariae, and P. vivax. A large number of genetic variants that are associated with malaria-protective phenotypes have been identified and many of them cause serious disease (Ko et al. 2012; Gomez et al. 2013a). For example, variants associated with hemoglobinopathies, α and β thalassemias, and the structural hemoglobin variants S, C, and E, are classic examples of deleterious genetic variants that nevertheless confer protection from malaria (Weatherall 2001; Gomez et al. 2013b). Early studies of thalassemias (Haldane 1949; Flint et al. 1986) noted that these diseases have a geographic distribution that coincides with P. falciparum endemicity. In what is now called the “malaria hypothesis,” JBS Haldane suggested that the genetic variants responsible for the thalassemias may be maintained in some human populations because of an advantage associated with malaria resistance (Haldane 1949). In a recent analysis, Piel et al. (2010) revisited the global distribution of sickle cell anemia, a disease caused by a mutation in the β-globin gene (HbS). They showed strong geographical correlation between HbS allele frequency and malaria endemicity in Africa (Fig. 4), consistent with the malaria hypothesis.
Figure 4.
Global distribution of the HbS sickle cell anemia allele compared to the historic geographic distribution of malaria. (Top) Global distribution of HbS allele frequency predicted from Bayesian geostatistical modeling. (Bottom) Historical map of malaria endemicity. The classes are defined by PfPr2−10 (PfPr2−10 = proportion of 2- to 10-year olds with confirmed blood stage asexual parasites): malaria free, PfPr2−10 = 0; epidemic, PfPr2−10 ≈ 0; hypoendemic, PfPr2−10 < 0.10; mesoendemic, PfPr2−10 ≥ 0.10 and < 0.50; hyperendemic, PfPr2−10 ≥ 0.50 and < 0.75; holoendemic, PfPr0−1 − 1 ≥ 0.75 (this class was measured in children ages 0–1) (From Piel et al. (2010); reproduced, with express permission, from Nature Publishing Group © 2010.)
Additional studies of other genes have shed further light on our understanding of the co-evolution of humans and P. falciparum, and provide evidence of recent natural selection in the human genome. For example, studies of genetic variation at G6PD (glucose-6-phosphate dehyrogenase), a gene that is known to harbor mutations associated with G6PD enzyme deficiency and protection from P. falciparum infection, have revealed signatures of genetic variation consistent with recent natural selection (Tishkoff et al. 2001; Saunders et al. 2002; Verrelli et al. 2002; Sabeti et al. 2006). Among the G6PD variants that are associated with protection from malaria, the A-variant is most common in sub-Saharan Africa. Several studies, in African and non-African populations (Tishkoff et al. 2001; Sabeti et al. 2002; Saunders et al. 2002; Verrelli et al. 2002), have identified signatures of recent natural selection acting on the A-variant and inferred the TMRCA for this variant to range from ∼1200 to 12,000 ya, which is consistent with the idea that selective pressures caused by malaria increased in African populations as population densities increased after the introduction of domesticated plants and animals (Tishkoff et al. 2001).
Kidney Disease
In the United States, diseases like type 2 diabetes mellitus, hypertension, and prostate cancer are known to be more common in African Americans than European Americans (Landis et al. 1999; Brancati et al. 2000; Ong et al. 2007). This disparity in disease occurrence suggests that genetic factors mediating disease risk, together with environmental factors, may differ among European and African descent populations (Hunter 2005; Yang et al. 2005; Cooper 2013). It has been proposed that genetic risk factors could be more common in African populations because they may have been adaptive in past African environments but increase risk for disease in today’s Western environment (Campbell and Tishkoff 2008).
One example of such a disease at high prevalence in African Americans is chronic kidney disease (CKD). In 2009, the mortality rate of African Americans with CKD was significantly higher than the mortality rate in European Americans and patients of other ethnicities (United States Renal Data System 2011). Additionally, in 2009, end-stage renal disease (ESRD), a serious complication of CKD in which a patient has complete or near complete loss of renal function and requires either frequent dialysis or a kidney transplant, occurred at a rate that is 3.5 times greater in African Americans than European Americans (United States Renal Data System 2011). There are many different causes of CKD; however, the primary proximal causes include diabetes and hypertension. There are also inherited forms of CKD; some of these include polycystic kidney disease (PKD), focal segmental glomerulosclerosis (FSGS), and pediatric nephrotic syndrome (Friedman and Pollak 2011).
Most African Americans have mixed ancestry originating from Africa and Europe; studies have inferred the average amount of European ancestry in African Americans to be ∼20% and the remainder to be predominantly of western and central African origin (Smith et al. 2004; Bryc et al. 2009; Tishkoff et al. 2009). It should be noted, however, that there is substantial variation in the level of African ancestry in individual African Americans. The proportion of African ancestry in a given individual can range from 99% to 1%. Additionally, the level of African or European ancestry in African Americans not only varies by individual but also by genomic region (Bryc et al. 2009). To identify genetic factors mediating disease predisposition in populations of recent African descent, like African Americans, a number of different approaches have been employed (Dong et al. 1999; Smith et al. 2004; Malhotra et al. 2005; Leak et al. 2007; Adeyemo et al. 2009; Sale et al. 2009; McDonough et al. 2011). One of these, called admixture mapping, is based on the premise that if a trait or disease of interest has a different prevalence in the parental populations prior to admixture, then it is likely that the genetic variation causing the disease or trait of interest in the admixed population is associated with chromosomal segment(s) derived from that parental population where the disease or trait is more prevalent (Winkler et al. 2010).
The identification of genetic risk factors for CKD in African Americans is an important example of the success of admixture mapping (Kao et al. 2008; Kopp et al. 2008; Genovese et al. 2010; Tzur et al. 2010). In 2008, two studies independently identified MYH9 as a candidate locus associated with CKD or ESRD in African Americans. Kopp et al. (2008) analyzed FSGS African-American cases and controls and showed a significant association on chromosome 22 near the MYH9 locus. They also found a marked increase of African ancestry in FSGS cases. Because MYH9 is expressed in the kidney, the authors considered this to be a good causal candidate gene. In the second study, Kao et al. (2008) also used admixture mapping to search for genetic variants associated with CKD and identified a single location significantly associated with disease on chromosome 22 in a population of nondiabetic ESRD patients. They also showed that the amount of European ancestry in the ESRD cases at that region was significantly lower than the average across the genome. Similar to Kopp et al. (2008), Kao et al. (2008) identified MYH9 as the likely candidate gene responsible for the strong association signal, given the expression patterns of MYH9 in the kidney.
Following publication of these two papers, numerous studies were conducted to find the functional variants responsible for these admixture signals (Kao et al. 2008; Kopp et al. 2008; Freedman et al. 2009a,b; Reeves-Daniel et al. 2010; Rosset et al. 2011; Freedman and Murea 2012). In 2010, two groups (Genovese et al. 2010; Tzur et al. 2010) used the data from the 1000 Genomes Project to identify potential functional variants that differ in frequency between Africans and Europeans near the MYH9 locus. Genovese et al. (2010) performed association tests of genome-wide variants with FSGS using African-American cases and controls (a genome wide association study or GWAS). They showed that the strongest signal with FSGS was not at MYH9 but at APOL1, which is a gene that is a short distance from MYH9. The strongest association signal was observed at a genetic variant termed G1. When the authors controlled for the association of G1 with FSGS they found a second associated allele that they termed G2. The combined signal at G1 and G2 was 35 orders of magnitude greater than the signal found at MYH9. Tzur et al. (2010) also used the 1000 Genomes data to search for new variants near MYH9 that might be functional. They found four genetic variants that were potential candidates; two variants in APOL1 (also identified by Genovese et al. 2010), one in APOL3, located further upstream, and one in FOXRED2, located downstream from MYH9. They found that the APOL1 variants are more strongly associated with CKD than the MYH9 risk alleles in African-American and Hispanic-American ESRD cases.
In addition to the association study, Genovese et al. (2010) tested whether either the G1 or G2 gene variants exhibited a signature of recent positive selection. The results of their analyses suggest that at G1 there is a strong signal of recent positive selection and a weak, but noteworthy, signal of recent positive selection at G2. Because it is unlikely that G1 and G2 were positively selected for their deleterious role in the pathogenesis of kidney disease, Genovese et al. (2010), Tzur et al. (2010), and Oleksyk et al. (2010) speculated that these alleles must serve a separate beneficial role in an African environment. In Africa, APOL1 is an important factor in resistance to infection from Trypanosoma brucei brucei, one of the trypanosomes that cause African sleeping sickness. Genovese et al. hypothesized that this function may be the underlying reason for the apparent recent positive selection at APOL1 (also see Oleksyk et al. 2010; Tzur et al. 2010). To test this hypothesis, Genovese et al. (2010), examined whether the G1 or G2 alleles influence the resistance phenotypes conferred by APOL1 during infection caused by three subspecies of Trypanosoma brucei (Trypanosoma brucei brucei, Trypanosoma brucei rhodesiense, and Trypanosoma brucei gambiens). They showed that the G1 and G2 alleles effectively killed >60% of the T.b.rhodesiense samples and suggested that the signature of selection observed at these variants may be the result of the selective advantage conferred against Trypanosoma infection. However, it should be noted that T.b.rhodesiense is presently found in East Africa and the signature of selection identified by Genovese et al. was found in West African populations. Additionally, Tzur et al. (2010) did not find the APOL1 variants in people from Ethiopia, which implies that the G1 and G2 alleles may not be common in East African populations. The absence of the G1 and G2 variants in Ethiopia is difficult to reconcile with the observation that that G1 and G2 are most effective at killing Tryanosoma species that are common in East Africa. However, Genovese et al. (2010) suggest that changes in Trypanosoma biology and distribution and/or human migration could explain the lack of correlation between parasite range and the distribution of the G1 and G2 alleles today. They also propose that the G1/G2 variants at APOL1 could play a role in immunity to other pathogens common in western Africa.
More recently, Ko et al. (2013) further examined genetic variation at APOL1 and showed that the G2 variant has a similar frequency across diverse African populations (3%–8%) and that the G1 variant is only common in the Yoruba population (39%). These authors identified another variant, termed G3, which has a more widespread distribution than the G1 variant, and showed a signature of recent natural selection in a West African Fulani population. Further studies of G1/G2 allele frequencies in diverse African populations and additional tests of recent natural selection are necessary to better understand the evolutionary history of this locus and how selection has influenced allele frequencies at APOL1. It is also important to note that the functional mechanism(s) underlying the association of G1/G2 with CKD is currently unknown. In addition to further studies of kidney disease in African populations, additional studies are also needed to demonstrate the functional roles that the G1, G2, and G3 variants play in infectious disease and kidney disease.
The identification of regions in African genomes that are recent targets of natural selection has resulted in important insights into the genetic basis of many pathological and nonpathologic phenotypes common in African populations. However, despite the growth of evolutionary studies to understand the prevalence of disease phenotypes, there is a general lack of epidemiological studies that help us understand the prevalence of noncommunicable diseases in African countries and how genetic variation plays a role in presence and prevalence of these diseases. Dalal et al. (2011) reported that in 2004 about one-quarter of all deaths in sub-Saharan Africa were caused by noncommunicable diseases, and they estimate that by 2030 noncommunicable diseases will increase to 46% of all deaths in sub-Saharan Africa. Unfortunately, their review of the literature suggests that community-based epidemiologic studies of noninfectious diseases, such as diabetes, cardiovascular disease, and obesity, are lacking in many African nations compared to the focus on maternal–child health and infectious diseases. Thus, despite the current shift in the prevalence of noninfectious diseases, we are currently ill-equipped to describe who is being affected and what the genetic and environmental risk factors that contribute to noncommunicable disease susceptibility in Africa are.
In the coming decades, it will be crucial to focus on the prevalence of noninfectious diseases in African countries and to understand what genetic variants predispose African people to conditions like obesity and cardiovascular disease. The effort to understand the genetic basis of complex noninfectious diseases in Africa should be twofold—our efforts should be placed on large-scale studies of both families and unrelated people. These two approaches will allow us to better characterize how disease-causing variants are distributed among diverse populations, and family-based studies will also help us to identify rare genetic variants that are difficult to identify in studies of unrelated people. Comparison of African populations and those within the African diaspora across diverse environments may also help to disentangle genetic and environmental effects on variable phenotypes and disease risk. These efforts will help to elucidate factors contributing to complex diseases in Africa and may also elucidate the genetic basis of these conditions in populations that have recent African admixture, like African Americans, Hispanics, and peoples of the Caribbean.
CONCLUSIONS
The pattern of genetic variation observed at mtDNA, Y chromosome, and autosomal loci in African populations reflects the demographic history of these populations. These data indicate high levels of genetic diversity within and between African populations, and also show that the current pattern of genetic variation in Africa is a result of both ancient and recent migration and admixture events.
The genomes of Africans have also been impacted by natural selection, sometimes resulting in prevalence of genetic variants that cause disease. Studies of natural selection in African populations show that functionally important genetic variants may be common but geographically restricted within Africa because of local adaptation to a particular lifestyle or environment. Also, many common variants that are adaptive because of protection from an infectious disease may also result in susceptibility to a different, possibly noninfectious, disease in populations of recent African origin. This observation points to the importance of including ethnically diverse Africans in human genetic studies because these populations represent an important component of human genomic variation, and data from African populations help us to understand the context in which some genetic variants were selected.
Going forward, as the cost of whole genome sequencing decreases, it will become feasible to conduct large-scale genomic sequencing studies across ethnically diverse Africans. These data are necessary to understand the genetic basis of complex disease in Africa and also the genetic basis of complex disease in populations like African Americans. Currently, most large-scale studies that seek to identify genetic variants that are associated with complex disease risk are heavily biased toward the identification of genetic variants initially discovered in European populations. Large-scale sequencing studies in African populations will identify new non-European variants that can be added to the repertoire of variation that is included in large association studies. The inclusion of a more diverse panel of variants will increase what we know about the genetic basis of complex diseases because new variants will allow us to search for associations among sites that are common in people from many different ethnic backgrounds. These studies will shed light on modern human origins, African and African-American population history, and the genetic basis of nonpathologic traits as well as traits that affect disease susceptibility.
Footnotes
Editor: Aravinda Chakravarti
Additional Perspectives on Human Variation available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- The 1000 Genomes Project Consortium. 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, Gragert L, Babrzadeh F, Gharizadeh B, Luo M, Plummer FA, et al. 2011. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science 334: 89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemo AA, Chen G, Chen Y, Rotimi C 2005. Genetic structure in four West African population groups. BMC Genet 6: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, et al. 2009. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet 5: e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Silva J, da Silva Santos M, Guimaraes PE, Ferreira AC, Bandelt HJ, Pena SD, Prado VF 2000. The ancestry of Brazilian mtDNA lineages. Am J Hum Genet 67: 444–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose SH 1982. HHistorical and linguistic reconstructions in East Africa; The archeological evidence. In The archeological and linguistic reconstruction of African History (ed. Ehret C, Posnansky ME). Boston University African Studies Center, Berkeley, CA [Google Scholar]
- Aquadro CF, Bauer DuMont V, Reed FA 2001. Genome-wide variation in the human and fruitfly: A comparison. Curr Opin Genet Dev 11: 627–634 [DOI] [PubMed] [Google Scholar]
- Atkinson QD, Gray RD, Drummond AJ 2008. mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Mol Biol Evol 25: 468–474 [DOI] [PubMed] [Google Scholar]
- Atkinson QD, Gray RD, Drummond AJ 2009. Bayesian coalescent inference of major human mitochondrial DNA haplogroup expansions in Africa. Proc Biol Sci 276: 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzmon G, Hao L, Pe’er I, Velez C, Pearlman A, Palamara PF, Morrow B, Friedman E, Oddoux C, Burns E, et al. 2010. Abraham’s children in the genome era: Major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern ancestry. Am J Hum Genet 86: 850–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auricchio S, Rubino A, Landolt M, Semenza G, Prader A 1963. Isolated intestinal lactase deficiency in the adult. Lancet 2: 324–326 [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB 1995. Mitochondrial portraits of human populations using median networks. Genetics 141: 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Alves-Silva J, Guimaraes PE, Santos MS, Brehm A, Pereira L, Coppa A, Larruga JM, Rengo C, Scozzari R, et al. 2001. Phylogeography of the human mitochondrial haplogroup L3e: A snapshot of African prehistory and Atlantic slave trade. Ann Hum Genet 65: 549–563 [DOI] [PubMed] [Google Scholar]
- Barthelme JW 1977. Holocene sites north-east of Lake Turkana: Preliminary report. Azania 12: 33–41 [Google Scholar]
- Batini C, Coia V, Battaggia C, Rocha J, Pilkington MM, Spedini G, Comas D, Destro-Bisol G, Calafell F 2007. Phylogeography of the human mitochondrial L1c haplogroup: Genetic signatures of the prehistory of Central Africa. Mol Phylogenet Evol 43: 635–644 [DOI] [PubMed] [Google Scholar]
- Batini C, Lopes J, Behar DM, Calafell F, Jorde LB, van der Veen L, Quintana-Murci L, Spedini G, Destro-Bisol G, Comas D 2011. Insights into the demographic history of African Pygmies from complete mitochondrial genomes. Mol Biol Evol 28: 1099–1110 [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL 2002. Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379 [DOI] [PubMed] [Google Scholar]
- Behar DM, Villems R, Soodyall H, Blue-Smith J, Pereira L, Metspalu E, Scozzari R, Makkan H, Tzur S, Comas D, et al. 2008. The dawn of human matrilineal diversity. Am J Hum Genet 82: 1130–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, Rootsi S, Chaubey G, Kutuev I, Yudkovsky G, et al. 2010. The genome-wide structure of the Jewish people. Nature 466: 238–242 [DOI] [PubMed] [Google Scholar]
- Beleza S, Gusmao L, Amorim A, Carracedo A, Salas A 2005. The genetic legacy of western Bantu migrations. Hum Genet 117: 366–375 [DOI] [PubMed] [Google Scholar]
- Berniell-Lee G, Calafell F, Bosch E, Heyer E, Sica L, Mouguiama-Daouda P, van der Veen L, Hombert JM, Quintana-Murci L, Comas D 2009. Genetic and demographic implications of the Bantu expansion: Insights from human paternal lineages. Mol Biol Evol 26: 1581–1589 [DOI] [PubMed] [Google Scholar]
- Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN 2004. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet 74: 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blench R 1993. Recent developments in African language classification and their implications for prehistory. In The archaeology of Africa: Food, metals and towns (ed. Shaw PST, Andah B, Okpoko A), pp. 71–103 Routledge, London [Google Scholar]
- Blench R 2006. Archaeology, language, and the African past. AltaMira, Lanham, MD [Google Scholar]
- Bower JRF 1973. Seronera: Excavations at a stone bowl site in the Serengeti National Park, Tanzania. Azania 8: 71–104 [Google Scholar]
- Brancati FL, Kao WH, Folsom AR, Watson RL, Szklo M 2000. Incident type 2 diabetes mellitus in African American and white adults: The Atherosclerosis Risk in Communities Study. JAMA 283: 2253–2259 [DOI] [PubMed] [Google Scholar]
- Bray SM, Mulle JG, Dodd AF, Pulver AE, Wooding S, Warren ST 2010. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc Natl Acad Sci 107: 16222–16227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo JM, Wambebe C, Tishkoff SA, et al. 2009. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci 107: 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo JM, Wambebe C, Tishkoff SA, et al. 2010. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci 107: 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butzer KW 1981. Rise and fall of Axum, Ethiopia: A geo-archaeological interpretation. Am. Antiq. 46: 471–495 [Google Scholar]
- Butzer KW, Brown FH, Thurber DL 1969. Horizontal sediments of the lower Omo Basin: The Kibish formation. Quarternaria 11: 15–29 [Google Scholar]
- Campbell MC, Tishkoff SA 2008. African genetic diversity: Implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet 9: 403–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann RL, Stoneking M, Wilson AC 1987. Mitochondrial DNA and human evolution. Nature 325: 31–36 [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, et al. 2002. A human genome diversity cell line panel. Science 296: 261–262 [DOI] [PubMed] [Google Scholar]
- Castri L, Tofanelli S, Garagnani P, Bini C, Fosella X, Pelotti S, Paoli G, Pettener D, Luiselli D 2009. mtDNA variability in two Bantu-speaking populations (Shona and Hutu) from Eastern Africa: Implications for peopling and migration patterns in sub-Saharan Africa. Am J Phys Anthropol 140: 302–311 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL 1994, The history and geography of human genes. Princeton University Press, Princeton, NJ. [Google Scholar]
- Cerny V, Hajek M, Bromova M, Cmejla R, Diallo I, Brdicka R 2006. MtDNA of Fulani nomads and their genetic relationships to neighboring sedentary populations. Hum Biol 78: 9–27 [DOI] [PubMed] [Google Scholar]
- Cerny V, Mulligan CJ, Ridl J, Zaloudkova M, Edens CM, Hajek M, Pereira L 2008. Regional differences in the distribution of the sub-Saharan, West Eurasian, and South Asian mtDNA lineages in Yemen. Am J Phys Anthropol 136: 128–137 [DOI] [PubMed] [Google Scholar]
- Cerny V, Fernandes V, Costa MD, Hajek M, Mulligan CJ, Pereira L 2009. Migration of Chadic-speaking pastoralists within Africa based on population structure of Chad Basin and phylogeography of mitochondrial L3f haplogroup. BMC Evol Biol 9: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Torroni A, Excoffier L, Santachiara-Benerecetti AS, Wallace DC 1995. Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am J Hum Genet 57: 133–149 [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Olckers A, Schurr TG, Kogelnik AM, Huoponen K, Wallace DC 2000. mtDNA variation in the South African Kung and Khwe-and their genetic relationships to other African populations. Am J Hum Genet 66: 1362–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho M, Luiselli D, Bertorelle G, Lopes AI, Seixas S, Destro-Bisol G, Rocha J 2005. Microsatellite variation and evolution of human lactase persistence. Hum Genet 117: 329–339 [DOI] [PubMed] [Google Scholar]
- Coelho M, Sequeira F, Luiselli D, Beleza S, Rocha J 2009. On the edge of Bantu expansions: mtDNA, Y chromosome and lactase persistence genetic variation in southwestern Angola. BMC Evol Biol 9: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coia V, Destro-Bisol G, Verginelli F, Battaggia C, Boschi I, Cruciani F, Spedini G, Comas D, Calafell F 2005. Brief communication: mtDNA variation in North Cameroon: Lack of Asian lineages and implications for back migration from Asia to sub-Saharan Africa. Am J Phys Anthropol 128: 678–681 [DOI] [PubMed] [Google Scholar]
- Compton JS 2011. Pleistocene sea-level fluctuations and human evolution on the southern coastal plain of South Africa. Quat Sci Rev 30: 506–527 [Google Scholar]
- *.Cooper RS 2013. Race in biological and biomedical research. Cold Spring Harb Perspect Med 3: a008573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudray C, Olivieri A, Achilli A, Pala M, Melhaoui M, Cherkaoui M, El-Chennawi F, Kossmann M, Torroni A, Dugoujon JM 2008. The complex and diversified mitochondrial gene pool of Berber populations. Ann Hum Genet 73: 196–214 [DOI] [PubMed] [Google Scholar]
- Cox MP, Morales DA, Woerner AE, Sozanski J, Wall JD, Hammer MF 2009. Autosomal resequence data reveal Late Stone Age signals of population expansion in sub-Saharan African foraging and farming populations. PLoS ONE 4: e6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, et al. 2002. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 70: 1197–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Trombetta B, Sellitto D, Massaia A, Destro-Bisol G, Watson E, Beraud Colomb E, Dugoujon JM, Moral P, Scozzari R 2010. Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages. Eur J Hum Genet 18: 800–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Trombetta B, Massaia A, Destro-Bisol G, Sellitto D, Scozzari R 2011. A revised root for the human Y chromosomal phylogenetic tree: The origin of patrilineal diversity in Africa. Am J Hum Genet 88: 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal S, Beunza JJ, Volmink J, Adebamowo C, Bajunirwe F, Njelekela M, Mozaffarian D, Fawzi W, Willett W, Adami HO, et al. 2011. Non-communicable diseases in sub-Saharan Africa: What we know now. Int J Epidemiol 40: 885–901 [DOI] [PubMed] [Google Scholar]
- Destro-Bisol G, Coia V, Boschi I, Verginelli F, Caglia A, Pascali V, Spedini G, Calafell F 2004. The analysis of variation of mtDNA hypervariable region 1 suggests that Eastern and Western Pygmies diverged before the Bantu expansion. Am Nat 163: 212–226 [DOI] [PubMed] [Google Scholar]
- de Wit E, Delport W, Rugamika CE, Meintjes A, Moller M, van Helden PD, Seoighe C, Hoal EG 2010. Genome-wide analysis of the structure of the South African coloured population in the Western Cape. Hum Genet 128: 145–153 [DOI] [PubMed] [Google Scholar]
- Distefano JA 1990. Hunters or hunted? Towards a history of the Okiek of Kenya. Hist Africa 17: 41–57 [Google Scholar]
- Dong Y, Zhu H, Sagnella GA, Carter ND, Cook DG, Cappuccio FP 1999. Association between the C825T polymorphism of the G protein beta3-subunit gene and hypertension in blacks. Hypertension 34: 1193–1196 [DOI] [PubMed] [Google Scholar]
- Durham WH 1991. Coevolution. Genes, culture and human diversity. Stanford University Press, Stanford, CT. [Google Scholar]
- Ehret C 1967. Cattle-keeping and milking in Eastern and Southern African History: The linguistic evidence. J African Hist 8: 1–17 [Google Scholar]
- Ehret C 1971. Southern Nilotic history: Linguistic approaches to the study of the past. Northwestern University Press, Evanston, IL [Google Scholar]
- Ehret C 1993. Nilo-Saharans and the Saharo-Sudanese Neolithic. In The archaeology of Africa: Food, metals and towns (ed. Shaw PST, Andah B, Okpoko A), pp. 104–125 Routledge, London [Google Scholar]
- Ehret C 1995. Reconstructing proto-Afroasiatic (Proto-Afrasian): Vowels, tone, consonants, and vocabulary. University of California Press, Berkeley, CA [Google Scholar]
- Ehret C 1998. An African classical age: Eastern and Southern Africa in world history, 1000 B.C. to A.D. 400. University Press of Virginia, Charlottesville, VA [Google Scholar]
- Ehret C 2001a. Bantu expansions: Re-envisioning a central problem of early African History. Int J African Historic Stud 34: 5–41 [Google Scholar]
- Ehret C 2001b. Historical-comparative reconstruction of Nilo-Saharan. Rüdiger Köppe Verlag, Cologne, Germany [Google Scholar]
- Ehret C 2002. The civilizations of Africa: A history to 1800. University Press of Virginia, Charlottesville, VA [Google Scholar]
- Ehret C 2006. Linguistic stratigraphies and Holocene history in Northeastern Africa. In Studies in African archaeology, pp. 1019–1055 Poznan Archaeological Museum, Poznan, Poland [Google Scholar]
- Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I 2002. Identification of a variant associated with adult-type hypolactasia. Nat Genet 30: 233–237 [DOI] [PubMed] [Google Scholar]
- Enattah NS, Jensen TG, Nielsen M, Lewinski R, Kuokkanen M, Rasinpera H, El-Shanti H, Seo JK, Alifrangis M, Khalil IF, et al. 2008. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am J Hum Genet 82: 57–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evershed RP, Payne S, Sherratt AG, Copley MS, Coolidge J, Urem-Kotsu D, Kotsakis K, Ozdogan M, Ozdogan AE, Nieuwenhuyse O, et al. 2008. Earliest date for milk use in the Near East and southeastern Europe linked to cattle herding. Nature 455: 528–531 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Schneider S 1999. Why hunter–gatherer populations do not show signs of pleistocene demographic expansions. Proc Natl Acad Sci 96: 10597–10602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Hill AV, Bowden DK, Oppenheimer SJ, Sill PR, Serjeantson SW, Bana-Koiri J, Bhatia K, Alpers MP, Boyce AJ, et al. 1986. High frequencies of α-thalassaemia are the result of natural selection by malaria. Nature 321: 744–750 [DOI] [PubMed] [Google Scholar]
- Fortin A, Stevenson MM, Gros P 2002. Susceptibility to malaria as a complex trait: Big pressure from a tiny creature. Hum Mol Genet 11: 2469–2478 [DOI] [PubMed] [Google Scholar]
- Francalacci P, Morelli L, Angius A, Berutti R, Reinier F, Atzeni R, Pilu R, Busonero F, Maschio A, Zara I, et al. 2013. Low-pass DNA sequencing of 1200 Sardinians reconstructs European Y-chromosome phylogeny. Science 341: 565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman BI, Murea M 2012. Target organ damage in African American hypertension: Role of APOL1. Curr Hypertens Rep 14: 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DJ, Pollak MR 2011. Genetics of kidney failure and the evolving story of APOL1. J Clin Invest 121: 3367–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman BI, Hicks PJ, Bostrom MA, Cunningham ME, Liu Y, Divers J, Kopp JB, Winkler CA, Nelson GW, Langefeld CD, et al. 2009a. Polymorphisms in the non-muscle myosin heavy chain 9 gene (MYH9) are strongly associated with end-stage renal disease historically attributed to hypertension in African Americans. Kidney Int 75: 736–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman BI, Kopp JB, Winkler CA, Nelson GW, Rao DC, Eckfeldt JH, Leppert MF, Hicks PJ, Divers J, Langefeld CD, et al. 2009b. Polymorphisms in the nonmuscle myosin heavy chain 9 gene (MYH9) are associated with albuminuria in hypertensive African Americans: The HyperGEN study. Am J Nephrol 29: 626–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, et al. 2010. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez F, Ko WY, Davis A, Tishkoff SA 2013a. Malarial disease and human genetic variation: Evidence for natural selection at malaria susceptibility candidate loci. In Primates, pathogens and evolution (ed. Brinkworth J, Pechenkina E). Springer, New York [Google Scholar]
- Gomez F, Tomas G, Ko WY, Ranciaro A, Froment A, Ibrahim M, Lema G, Nyambo TB, Omar SA, Wambebe C, et al. 2013b. Patterns of nucleotide and haplotype diversity at ICAM-1 across global human populations with varying levels of malaria exposure. Hum Genet 132: 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder MK, Mortensen HM, Reed FA, de Sousa A, Tishkoff SA 2007. Whole-mtDNA genome sequence analysis of ancient African lineages. Mol Biol Evol 24: 757–768 [DOI] [PubMed] [Google Scholar]
- Gonzalez AM, Cabrera VM, Larruga JM, Tounkara A, Noumsi G, Thomas BN, Moulds JM 2006. Mitochondrial DNA variation in Mauritania and Mali and their genetic relationship to other Western Africa populations. Ann Hum Genet 70: 631–657 [DOI] [PubMed] [Google Scholar]
- Graven L, Passarino G, Semino O, Boursot P, Santachiara-Benerecetti S, Langaney A, Excoffier L 1995. Evolutionary correlation between control region sequence and restriction polymorphisms in the mitochondrial genome of a large Senegalese Mandenka sample. Mol Biol Evol 12: 334–345 [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. 2011. A draft sequence of the Neandertal genome. Science 328: 710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JH 1963. The languages of Africa. Indiana University, Bloomington [Google Scholar]
- Greenberg JH 1972. Linguistic evidence regarding Bantu origins. J Afr Hist 13: 189–216 [Google Scholar]
- Haldane JBS 1949. The rate of mutation in human genes. Hereditas 35: 267–273 [Google Scholar]
- Hammer MF, Zegura SL 2002. The human Y Chromosome haplogroup tree: Nomenclature and phylogeography of its major divisions. Annu Rev Anthropol 31: 303–321 [Google Scholar]
- Hammer MF, Woerner AE, Mendez FL, Watkins JC, Wall JD 2011. Genetic evidence for archaic admixture in Africa. Proc Natl Acad Sci 108: 15123–15128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpending HC, Sherry ST, Rogers AR, Stoneking M 1993. The genetic structure of ancient human populations. Curr Anthropol 34: 483–496 [Google Scholar]
- Haspelmath M, Dryer MS, Gil D, Comrie B, eds. 2008. The world atlas of language structures online. Max Planck Digital Library, Munich [Google Scholar]
- Henn BM, Gignoux CR, Jobin M, Granka JM, Macpherson JM, Kidd JM, Rodriguez-Botigue L, Ramachandran S, Hon L, Brisbin A, et al. 2011. From the cover: Feature article: Hunter–gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci 108: 5154–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Gravel S, Wang W, Brisbin A, Byrnes JK, Fadhlaoui-Zid K, Zalloua PA, Moreno-Estrada A, Bertranpetit J, et al. 2012. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet 8: e1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, et al. 2002. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70: 1152–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer T, Ray N, Wegmann D, Excoffier L 2009. Large allele frequency differences between human continental groups are more likely to have occurred by drift during range expansions than by selection. Ann Hum Genet 73: 95–108 [DOI] [PubMed] [Google Scholar]
- Hunter DJ 2005. Gene-environment interactions in human diseases. Nat Rev Genet 6: 287–298 [DOI] [PubMed] [Google Scholar]
- Hunter-Zinck H, Musharoff S, Salit J, Al-Ali KA, Chouchane L, Gohar A, Matthews R, Butler MW, Fuller J, Hackett NR, et al. 2010. Population genetic structure of the people of Qatar. Am J Hum Genet 87: 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Paabo S, Gyllensten U 2000. Mitochondrial genome variation and the origin of modern humans. Nature 408: 708–713 [DOI] [PubMed] [Google Scholar]
- Ingram CJ, Elamin MF, Mulcare CA, Weale ME, Tarekegn A, Raga TO, Bekele E, Elamin FM, Thomas MG, Bradman N, et al. 2007. A novel polymorphism associated with lactose tolerance in Africa: Multiple causes for lactase persistence? Hum Genet 120: 779–788 [DOI] [PubMed] [Google Scholar]
- Ingram CJ, Mulcare CA, Itan Y, Thomas MG, Swallow DM 2009. Lactose digestion and the evolutionary genetics of lactase persistence. Hum Genet 124: 579–591 [DOI] [PubMed] [Google Scholar]
- International HapMap C. 2005. A haplotype map of the human genome. Nature 437: 1299–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BA, Wilson JL, Kirbah S, Sidney SS, Rosenberger J, Bassie L, Alie JA, McLean DC, Garvey WT, Ely B 2005. Mitochondrial DNA genetic diversity among four ethnic groups in Sierra Leone. Am J Phys Anthropol 128: 156–163 [DOI] [PubMed] [Google Scholar]
- Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, et al. 2008. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet 40: 1185–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF 2008. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 18: 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Reidla M, Metspalu E, Rosa A, Brehm A, Pennarun E, Parik J, Geberhiwot T, Usanga E, Villems R 2004. Ethiopian mitochondrial DNA heritage: Tracking gene flow across and around the gate of tears. Am J Hum Genet 75: 752–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko WY, Gomez F, Tishkoff S 2012. Evolution of human erythrocyte-specific genes involved in malaria susceptibility. In Rapidly evolving genes and genetic systems (ed. Singh RS, Xu J, Kulathinal RJ). Oxford University Press, Oxford [Google Scholar]
- Kopelman NM, Stone L, Wang C, Gefel D, Feldman MW, Hillel J, Rosenberg NA 2009. Genomic microsatellites identify shared Jewish ancestry intermediate between Middle Eastern and European populations. BMC Genet 10: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, Oleksyk T, McKenzie LM, Kajiyama H, Ahuja TS, et al. 2008. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet 40: 1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings M, Salem AE, Bauer K, Geisert H, Malek AK, Chaix L, Simon C, Welsby D, Di Rienzo A, Utermann G, et al. 1999. mtDNA analysis of Nile River Valley populations: A genetic corridor or a barrier to migration? Am J Hum Genet 64: 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuda D, Zietkiewicz E, Yotova V 2000. Archaic lineages in the history of modern humans. Genetics 156: 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance J, Vernot B, Elbers CC, Ferwerda B, Froment A, Bodo JM, Lema G, Fu W, Nyambo TB, Rebbeck TR, et al. 2012. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter–gatherers. Cell 150: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis SH, Murray T, Bolden S, Wingo PA 1999. Cancer statistics, 1999. CA Cancer J Clin 49: 8–31 [DOI] [PubMed] [Google Scholar]
- Leak TS, Keene KL, Langefeld CD, Gallagher CJ, Mychaleckyj JC, Freedman BI, Bowden DW, Rich SS, Sale MM 2007. Association of the proprotein convertase subtilisin/kexin-type 2 (PCSK2) gene with type 2 diabetes in an African American population. Mol Genet Metab 92: 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leakey LSB 1931. The Stone Age cultures of Kenya Colony. The University Press, Cambridge [Google Scholar]
- Lewinsky RH, Jensen TG, Moller J, Stensballe A, Olsen J, Troelsen JT 2005. T-13910 DNA variant associated with lactase persistence interacts with Oct-1 and stimulates lactase promoter activity in vitro. Hum Mol Genet 14: 3945–3953 [DOI] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104 [DOI] [PubMed] [Google Scholar]
- Luis JR, Rowold DJ, Regueiro M, Caeiro B, Cinnioglu C, Roseman C, Underhill PA, Cavalli-Sforza LL, Herrera RJ 2004. The Levant versus the Horn of Africa: Evidence for bidirectional corridors of human migrations. Am J Hum Genet 74: 532–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maca-Meyer N, Gonzalez AM, Larruga JM, Flores C, Cabrera VM 2001. Major genomic mitochondrial lineages delineate early human expansions. BMC Genet 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Batsheva B-T, Sykes B, Torroni A 1999. The emerging tree of West Eurasian mtDNAs: A synthesis of control region sequences and RFLPs. Am J Hum Genet 64: 232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Coon H, Feitosa MF, Li WD, North KE, Price RA, Bouchard C, Hunt SC, Wolford JK 2005. Meta-analysis of genome-wide linkage studies for quantitative lipid traits in African Americans. Hum Mol Genet 14: 3955–3962 [DOI] [PubMed] [Google Scholar]
- Marth GT, Czabarka E, Murvai J, Sherry ST 2004. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics 166: 351–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough CW, Palmer ND, Hicks PJ, Roh BH, An SS, Cooke JN, Hester JM, Wing MR, Bostrom MA, Rudock ME, et al. 2011. A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int 79: 563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez Fernando L, Krahn T, Schrack B, Krahn A-M, Veeramah Krishna R, Woerner August E, Fomine Forka Leypey M, Bradman N, Thomas Mark G, Karafet Tatiana M, et al. 2013. An African American paternal lineage adds an extremely ancient root to the human Y chromosome phylogenetic tree. Am J Hum Genet 92: 454–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, et al. 2003. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci 100: 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcare CA, Weale ME, Jones AL, Connell B, Zeitlyn D, Tarekegn A, Swallow DM, Bradman N, Thomas MG 2004. The T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C-13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am J Hum Genet 74: 1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomer AD, Park HS, O’Brien PC, McGill DB 1983. Response of patients with irritable bowel syndrome and lactase deficiency using unfermented acidophilus milk. Am J Clin Nutr 38: 257–263 [DOI] [PubMed] [Google Scholar]
- Newman P 1980. The classification of Chadic within Afroasiatic. Leiden Universitaire Press, Leiden [Google Scholar]
- Nielsen R 2005. Molecular signatures of natural selection. Annu Rev Genet 39: 197–218 [DOI] [PubMed] [Google Scholar]
- Nurse D 1997. The contributions of linguistics to the study of history in Africa. Journal Afr Hist 38: 359–391 [Google Scholar]
- Olds LC, Sibley E 2003. Lactase persistence DNA variant enhances lactase promoter activity in vitro: Functional role as a cis regulatory element. Hum Mol Genet 12: 2333–2340 [DOI] [PubMed] [Google Scholar]
- Oleksyk TK, Nelson GW, An P, Kopp JB, Winkler CA 2010. Worldwide distribution of the MYH9 kidney disease susceptibility alleles and haplotypes: Evidence of historical selection in Africa. PLoS ONE 5: e11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KL, Cheung BM, Man YB, Lau CP, Lam KS 2007. Prevalence, awareness, treatment, and control of hypertension among United States adults 1999–2004. Hypertension 49: 69–75 [DOI] [PubMed] [Google Scholar]
- Pagani L, Kivisild T, Tarekegn A, Ekong R, Plaster C, Gallego Romero I, Ayub Q, Mehdi SQ, Thomas MG, Luiselli D, et al. 2012. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am J Hum Genet 91: 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Petersen DC, van der Ross RE, Sudoyo H, Glashoff RH, Marzuki S, Reich D, Hayes VM 2010. Genetic structure of a unique admixed population: Implications for medical research. Hum Mol Genet 19: 411–419 [DOI] [PubMed] [Google Scholar]
- Pereira L, Macaulay V, Torroni A, Scozzari R, Prata MJ, Amorim A 2001. Prehistoric and historic traces in the mtDNA of Mozambique: Insights into the Bantu expansions and the slave trade. Ann Hum Genet 65: 439–458 [DOI] [PubMed] [Google Scholar]
- Phillipson L 2009. Lithic artefacts as a source of cultural, social and economic information: The evidence from Aksum, Ethiopia. Afr Archaeol Rev 26: 45–58 [Google Scholar]
- Piel FB, Patil AP, Howes RE, Nyangiri OA, Gething PW, Williams TN, Weatherall DJ, Hay SI 2010. Global distribution of the sickle cell gene and geographical confirmation of the malaria hypothesis. Nat Commun 1: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V, Wall JD 2006. Possible ancestral structure in human populations. PLoS Genet 2: e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poloni ES, Naciri Y, Bucho R, Niba R, Kervaire B, Excoffier L, Langaney A, Sanchez-Mazas A 2009. Genetic evidence for complexity in ethnic differentiation and history in East Africa. Ann Hum Genet 73: 582–600 [DOI] [PubMed] [Google Scholar]
- Posnansky M 1961a. 168. Dimple-based pottery from Uganda. Man 61: 141–142 [Google Scholar]
- Posnansky M 1961b. Pottery types from archaeological sites in East Africa. J Afr Hist 2: 177–198 [Google Scholar]
- Poulter M, Hollox E, Harvey CB, Mulcare C, Peuhkuri K, Kajander K, Sarner M, Korpela R, Swallow DM 2003. The causal element for the lactase persistence/non-persistence polymorphism is located in a 1 Mb region of linkage disequilibrium in Europeans. Ann Hum Genet 67: 298–311 [DOI] [PubMed] [Google Scholar]
- Poznik GD, Henn BM, Yee M-C, Sliwerska E, Euskirchen GM, Lin AA, Snyder M, Quintana-Murci L, Kidd JM, Underhill PA, et al. 2013. Sequencing Y chromosomes resolves discrepancy in time to common ancestor of males versus females. Science 341: 562–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, Manica A, Balloux F 2005. Geography predicts neutral genetic diversity of human populations. Curr Biol 15: R159–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS 1999. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23: 437–441 [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L, Quach H, Harmant C, Luca F, Massonnet B, Patin E, Sica L, Mouguiama-Daouda P, Comas D, Tzur S, et al. 2008. Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter–gatherers and Bantu-speaking farmers. Proc Natl Acad Sci 105: 1596–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Harmant C, Quach H, Balanovsky O, Zaporozhchenko V, Bormans C, van Helden PD, Hoal EG, Behar DM 2010. Strong maternal Khoisan contribution to the South African coloured population: A case of gender-biased admixture. Am J Hum Genet 86: 611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, Feldman MW, Cavalli-Sforza LL 2005. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci 102: 15942–15947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando JC, Pinto F, Gonzalez AM, Hernandez M, Larruga JM, Cabrera VM, Bandelt HJ 1998. Mitochondrial DNA analysis of northwest African populations reveals genetic exchanges with European, near-eastern, and sub-Saharan populations. Ann Hum Genet 62: 531–550 [DOI] [PubMed] [Google Scholar]
- Ray N, Currat M, Berthier P, Excoffier L 2005. Recovering the geographic origin of early modern humans by realistic and spatially explicit simulations. Genome Res 15: 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves-Daniel AM, Iskandar SS, Bowden DW, Bostrom MA, Hicks PJ, Comeau ME, Langefeld CD, Freedman BI 2010. Is collapsing C1q nephropathy another MYH9-associated kidney disease? A case report. Am J Kidney Dis 55: e21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PLF, et al. 2011. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468: 1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards MB, Macaulay VA, Bandelt HJ, Sykes BC 1998. Phylogeography of mitochondrial DNA in western Europe. Ann Hum Genet 62: 241–260 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, Sellitto D, Cruciani F, Kivisild T, et al. 2000. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67: 1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Robbins LH 1972. Archeology in the Turkana District, Kenya. Science 176: 359–366 [DOI] [PubMed] [Google Scholar]
- Rogers AR, Harpending H 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9: 552–569 [DOI] [PubMed] [Google Scholar]
- Ronald J, Akey JM 2005. Genome-wide scans for loci under selection in humans. Hum Genomics 2: 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW 2002. Genetic structure of human populations. Science 298: 2381–2385 [DOI] [PubMed] [Google Scholar]
- Rosset S, Tzur S, Behar DM, Wasser WG, Skorecki K 2011. The population genetics of chronic kidney disease: Insights from the MYH9-APOL1 locus. Nat Rev Nephrol 7: 313–326 [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837 [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES 2006. Positive natural selection in the human lineage. Science 312: 1614–1620 [DOI] [PubMed] [Google Scholar]
- Salas A, Richards M, De la Fe T, Lareu MV, Sobrino B, Sanchez-Diz P, Macaulay V, Carracedo A 2002. The making of the African mtDNA landscape. Am J Hum Genet 71: 1082–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale MM, Lu L, Spruill IJ, Fernandes JK, Lok KH, Divers J, Langefeld CD, Garvey WT 2009. Genome-wide linkage scan in Gullah-speaking African American families with type 2 diabetes: The Sea Islands Genetic African American Registry (Project SuGAR). Diabetes 58: 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satta Y, Takahata N 2004. The distribution of the ancestral haplotype in finite stepping-stone models with population expansion. Mol Ecol 13: 877–886 [DOI] [PubMed] [Google Scholar]
- Saunders MA, Hammer MF, Nachman MW 2002. Nucleotide variability at G6pd and the signature of malarial selection in humans. Genetics 162: 1849–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunier JL, Irwin JA, Strouss KM, Ragab H, Sturk KA, Parsons TJ 2009. Mitochondrial control region sequences from an Egyptian population sample. Forensic Sci Int Genet 3: e97–103 [DOI] [PubMed] [Google Scholar]
- Schlebusch CM, Skoglund P, Sjodin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MG, et al. 2012. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science 338: 374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scozzari R, Cruciani F, Santolamazza P, Malaspina P, Torroni A, Sellitto D, Arredi B, Destro-Bisol G, De Stefano G, Rickards O, et al. 1999. Combined use of biallelic and microsatellite Y-chromosome polymorphisms to infer affinities among African populations. Am J Hum Genet 65: 829–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal I, Gagjee PP, Essop AR, Noormohamed AM 1983. Lactase deficiency in the South African black population. Am J Clin Nutr 38: 901–905 [DOI] [PubMed] [Google Scholar]
- Seielstad MT, Minch E, Cavalli-Sforza LL 1998. Genetic evidence for a higher female migration rate in humans. Nat Genet 20: 278–280 [DOI] [PubMed] [Google Scholar]
- Sikora M, Laayouni H, Calafell F, Comas D, Bertranpetit J 2010. A genomic analysis identifies a novel component in the genetic structure of sub-Saharan African populations. Eur J Hum Genet 19: 84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AB 1992. Origins and spread of pastoralism in Africa. Annu Rev Anthropol 21: 125–141 [Google Scholar]
- Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, et al. 2004. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet 74: 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soodyall H, Vigilant L, Hill AV, Stoneking M, Jenkins T 1996. mtDNA control-region sequence variation suggests multiple independent origins of an “Asian-specific” 9-bp deletion in sub-Saharan Africans. Am J Hum Genet 58: 595–608 [PMC free article] [PubMed] [Google Scholar]
- Stefflova K, Dulik MC, Pai AA, Walker AH, Zeigler-Johnson CM, Gueye SM, Schurr TG, Rebbeck TR 2009. Evaluation of group genetic ancestry of populations from Philadelphia and Dakar in the context of sex-biased admixture in the Americas. PLoS ONE 4: e7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C 1994. Out of Africa—A personal history. In Origins of anatomically modern humans (ed. Nitecki M, Nitecki D). Plenum, New York [Google Scholar]
- Stringer CB, Andrews P 1988. Genetic and fossil evidence for the origin of modern humans. Science 239: 1263–1268 [DOI] [PubMed] [Google Scholar]
- Sutton J 1973. The settlement of East Africa. In Zamani (ed. Ogot BA), pp. 70–97 Longmans, Nairobi [Google Scholar]
- Swallow DM 2003. Genetics of lactase persistence and lactose intolerance. Annu Rev Genet 37: 197–219 [DOI] [PubMed] [Google Scholar]
- Templeton A 2002. Out of Africa again and again. Nature 416: 45–51 [DOI] [PubMed] [Google Scholar]
- Thomas MG, Weale ME, Jones AL, Richards M, Smith A, Redhead N, Torroni A, Scozzari R, Gratrix F, Tarekegn A, et al. 2002. Founding mothers of Jewish communities: Geographically separated Jewish groups were independently founded by very few female ancestors. Am J Hum Genet 70: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason SGaK, T. 1988. Language contact, creolization, and genetic linguistics. University of California Press, Berkeley and Los Angeles [Google Scholar]
- Tishkoff SA, Goldman A, Calafell F, Speed WC, Deinard AS, Bonne-Tamir B, Kidd JR, Pakstis AJ, Jenkins T, Kidd KK 1998. A global haplotype analysis of the myotonic dystrophy locus: Implications for the evolution of modern humans and for the origin of myotonic dystrophy mutations. Am J Hum Genet 62: 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, Destro-Bisol G, Drousiotou A, Dangerfield B, Lefranc G, Loiselet J, et al. 2001. Haplotype diversity and linkage disequilibrium at human G6PD: Recent origin of alleles that confer malarial resistance. Science 293: 455–462 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M, et al. 2007. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet 39: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo J-M, Doumbo O, et al. 2009. The genetic structure and history of Africans and African Americans. Science 324: 1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Calderon FL, Simionati B, Valle G, Richards M, et al. 2001. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet 69: 1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelsen JT, Olsen J, Moller J, Sjostrom H 2003. An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology 125: 1686–1694 [DOI] [PubMed] [Google Scholar]
- Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, et al. 2010. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet 128: 345–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- United States Renal Data System 2011. USRDS 2011 annual data report: Atlas of chronic kidney disease and end-stage renal disease in the United States. National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD [Google Scholar]
- Van Beek GW 1967. Monuments of Axum in the light of South Arabian archeology. J Am Orient Soc 87: 113–122 [Google Scholar]
- Vansina J 1995. New linguistic evidence and “the Bantu Expansion.” J Afr His 36: 173–195 [Google Scholar]
- Veeramah KR, Connell BA, Pour NA, Powell A, Plaster CA, Zeitlyn D, Mendell NR, Weale ME, Bradman N, Thomas MG 2010. Little genetic differentiation as assessed by uniparental markers in the presence of substantial language variation in peoples of the Cross River region of Nigeria. BMC Evol Biol 10: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli BC, McDonald JH, Argyropoulos G, Destro-Bisol G, Froment A, Drousiotou A, Lefranc G, Helal AN, Loiselet J, Tishkoff SA 2002. Evidence for balancing selection from nucleotide sequence analyses of human G6PD. Am J Hum Genet 71: 1112–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigilant L 1990. Control region sequences from African populations and the evolution of human mitochondrial DNA. University of California, Berkeley, CA: [DOI] [PubMed] [Google Scholar]
- Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC 1991. African populations and the evolution of human mitochondrial DNA. Science 253: 1503–1507 [DOI] [PubMed] [Google Scholar]
- Wall JD, Lohmueller KE, Plagnol V 2009. Detecting ancient admixture and estimating demographic parameters in multiple human populations. Mol Biol Evol 26: 1823–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Brown MD, Lott MT 1999. Mitochondrial DNA variation in human evolution and disease. Gene 238: 211–230 [DOI] [PubMed] [Google Scholar]
- Wang S, Lachance J, Tishkoff SA, Hey J, Xing J 2013. Apparent variation in Neanderthal admixture among African populations is consistent with gene flow from non-African populations. Genome Biol Evol 5: 2075–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, Bauer K, Aman R, Weiss G, von Haeseler A, Paabo S 1996. mtDNA sequence diversity in Africa. Am J Hum Genet 59: 437–444 [PMC free article] [PubMed] [Google Scholar]
- Watson E, Forster P, Richards M, Bandelt HJ 1997. Mitochondrial footprints of human expansion in Africa. Am J Hum Genet 61: 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherall DJ 2001. Phenotype-genotype relationships in monogenic disease: Lessons from the thalassaemias. Nat Rev Genet 2: 245–255 [DOI] [PubMed] [Google Scholar]
- Wei W, Ayub Q, Chen Y, McCarthy S, Hou Y, Carbone I, Xue Y, Tyler-Smith C 2013. A calibrated human Y-chromosomal phylogeny based on resequencing. Genome Res 23: 388–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler CA, Nelson GW, Smith MW 2010. Admixture mapping comes of age. Annu Rev Genomics Hum Genet 11: 65–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ET, Stover DA, Ehret C, Destro-Bisol G, Spedini G, McLeod H, Louie L, Bamshad M, Strassmann BI, Soodyall H, et al. 2005. Contrasting patterns of Y chromosome and mtDNA variation in Africa: Evidence for sex-biased demographic processes. Eur J Hum Genet 13: 867–876 [DOI] [PubMed] [Google Scholar]
- Yang Q, Khoury MJ, Friedman J, Little J, Flanders WD 2005. How many genes underlie the occurrence of common complex diseases in the population? Int J Epidemiol 34: 1129–1137 [DOI] [PubMed] [Google Scholar]
- Yotova V, Lefebvre J-F, Moreau C, Gbeha E, Hovhannesyan K, Bourgeois S, Bédarida S, Azevedo L, Amorim A, Sarkisian T, et al. 2011. An X-linked haplotype of Neandertal origin is present among all non-African populations. Mol Biol Evol 28: 1957–1962 [DOI] [PubMed] [Google Scholar]
- Zietkiewicz E, Yotova V, Jarnik M, Korab-Laskowska M, Kidd KK, Modiano D, Scozzari R, Stoneking M, Tishkoff S, Batzer M, et al. 1998. Genetic structure of the ancestral population of modern humans. J Mol Evol 47: 146–155 [DOI] [PubMed] [Google Scholar]