Abstract
Regions of close apposition between two organelles, often referred to as membrane contact sites (MCSs), mostly form between the endoplasmic reticulum and a second organelle, although contacts between mitochondria and other organelles have also begun to be characterized. Although these contact sites have been noted since cells first began to be visualized with electron microscopy, the functions of most of these domains long remained unclear. The last few years have witnessed a dramatic increase in our understanding of MCSs, revealing the critical roles they play in intracellular signaling, metabolism, the trafficking of metabolites, and organelle inheritance, division, and transport.
Introduction
The compartmentalization of cells allows the segregation and regulation of the myriad reactions that occur within them. The tremendous benefits of intracellular compartmentalization also come at a price; to function optimally, cells must transmit signals and exchange material between compartments. Numerous mechanisms have evolved to facilitate these exchanges. One that has not been well appreciated until the last few years is the transmission of signals and molecules between organelles that occurs at regions where the organelles are closely apposed, often called membrane contact sites (MCSs). These sites were first characterized because of their critical roles in the intracellular exchange of lipids and calcium, which can be directly channeled between organelles via MCSs. More recently, it has also become apparent that MCSs are important sites for intracellular signaling, organelle trafficking, and inheritance, and that MCSs are specialized regions where regulatory complexes are assembled (English and Voeltz, 2013; Helle et al., 2013).
A hallmark of MCSs is that membranes from two organelles (or compartments of the same organelle) are tethered to one another, but not all instances in which membranes interact with or are tethered to one another are considered MCSs. True MCSs have four properties: (1) membranes from two intracellular compartments are tethered in close apposition, typically within 30 nm, (2) the membranes do not fuse (though they may transiently hemi-fuse), (3) specific proteins and/or lipids are enriched at the MCS, and (4) MCS formation affects the function or composition of at least one of the two organelles in the MCS.
This review will discuss what we know about proteins that tether organelles, the exchange of small molecules at MCSs, and other emerging functions of MCSs.
MCS tethers
An MCS tether is a protein or complex of proteins (Fig. 1) that simultaneously binds the two apposing membranes at an organelle contact site and plays a role in maintaining the site (English and Voeltz, 2013; Helle et al., 2013). In many cases it is not yet clear if these proteins and complexes are genuine tethers, which are necessary to maintain MCSs, or function at MCSs but are not necessary to sustain contacts. Distinguishing between these possibilities is an important challenge for the field, especially when more than one protein or complex of proteins independently hold together the membranes at an MCS.
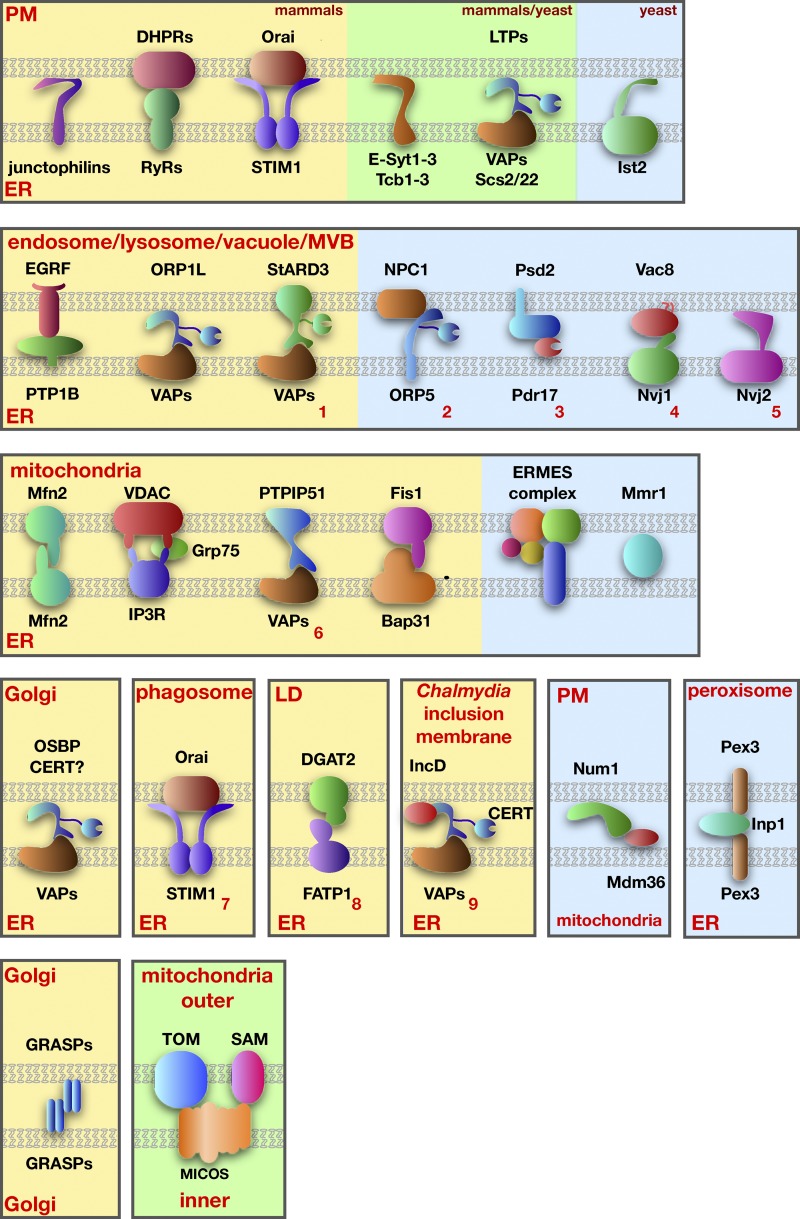
Figure 1.
Proteins proposed to mediate tethering at MCSs. Mammalian proteins are shown on a yellow background, yeast proteins on a blue background, and proteins found in both mammals and yeast are on a green background. Tethering complexes not described in the text are indicated with red numbers: (1) StARD3-VAPs (Alpy et al., 2013), (2) NPC1-ORP5 (Du et al., 2011), (3) Psd2-Pdr17 (Riekhof et al., 2014), (4) Vac8-Nvj1 (Pan et al., 2000), (5) Nvj2 (Toulmay and Prinz, 2012), (6) PTPIP51-VAPs (De Vos et al., 2012), (7) Orai1-STIM1 (Nunes et al., 2012), (8) DGAT2-FATP1 (Xu et al., 2012), and (9) IncD-CERT-VAPs (Derré et al., 2011; Elwell et al., 2011).
As a growing number of potential tethers are identified, three trends are emerging. First, most MCSs are maintained by several tethers. One of the best-characterized examples of this is the junction of the ER and plasma membrane (PM) in Saccharomyces cerevisiae. Recent work showed that it was necessary to eliminate six ER resident proteins to dramatically reduce the normally extensive interactions between the ER and PM (Manford et al., 2012; Stefan et al., 2013). This suggests that these six proteins mediate tethering independently of each other. Four of the six proteins (three calcium and lipid-binding domain proteins 1–3, also called Tcb1–3, and Ist2) are integral ER membrane proteins that have cytosolic domains that bind the plasma membranes (Fischer et al., 2009; Toulmay and Prinz, 2012). The other two proteins, Scs2 and Scs22 (Scs, suppressor of Ca2+ sensitivity), are homologues of mammalian VAPs (vesicle-associated membrane protein–associated proteins). VAPs are integral membrane tail-anchored proteins in the ER that bind proteins containing FFAT (phenylalanines in an acid tract) motifs (Loewen et al., 2003). A number of proteins that contain these motifs also have domains that bind lipids and proteins in the PM, allowing them to simultaneously bind and tether the ER and PM. For example, some oxysterol-binding protein (OSBP)–related proteins (ORPs) have FFAT motifs and pleckstrin homology (PH) domains that bind phosphoinositides (PIPs) in the plasma membrane (Levine and Munro, 1998; Weber-Boyvat et al., 2013). Thus, ORPs and other FFAT motif-containing proteins can mediate ER–PM tethering via VAPs. It should be noted that VAPs and proteins bound by VAPs also mediate tethering between the ER and organelles in addition to the PM. These are shown in Fig. 1.
A second emerging trend is that tethering seems to be a dynamic, regulated process, and we are beginning to understand the mechanisms of dynamic apposition of membranes at MCSs by tethers. One example is ER–PM tethering mediated by proteins called extended synaptotagmins (E-Syts), which are homologues of the yeast Tcb tethers. The tethering of the ER and PM by E-Syts is regulated by Ca2+ and the PM-enriched lipid PI(4,5)P2 (Chang et al., 2013; Giordano et al., 2013). Binding of these molecules by E-Syts may control both the extent of ER–PM contact and the distance between these organelles at MCSs. A second example of regulated MCS formation is provided by a recent study on OSBP. This protein and other FFAT motif-containing proteins have been thought to mediate ER–Golgi tethering by simultaneously binding VAPs in the ER and PIPs in the Golgi complex (Kawano et al., 2006; Peretti et al., 2008). In an elegant set of experiments, Mesmin et al. (2013) showed that OSBP regulates its own ability to mediate ER–Golgi tethering by modulating PI4P levels in the Golgi complex. When PI4P levels in the Golgi complex are high, OSBP tethers the ER and Golgi complex and also transports PI4P from the Golgi to the ER. When the PI4P reaches the ER, it is hydrolyzed by the phosphatase Sac1, preventing it from being transferred back to the Golgi. The reduction in Golgi complex PI4P levels by OSBP causes OSBP to dissociate from the Golgi, decreasing ER–Golgi tethering. Thus, OSBP negatively regulates its own tethering of the ER and Golgi membranes. Lipid transport by OSBP and similar proteins will be discussed in more detail in the section on lipid transport at MCSs.
The third important feature of many MCS tethering complexes is that most have functions in addition to tethering. This is well illustrated by complexes proposed to mediate ER–mitochondria tethering in mammalian cells, where four such complexes have been described (Fig. 1). For example, Mfn2 (mitofusin-2) acts as a tether (de Brito and Scorrano, 2008), but the primary function of this dynamin-like protein is to mediate mitochondrial fusion. Although Mfn2 is largely in the outer mitochondrial membrane (OMM), a small fraction also resides the ER, and it has been proposed that the interaction of Mfn2 in the ER with Mfn2 in the OMM tethers the ER and mitochondria (de Brito and Scorrano, 2008). The other ER–mitochondria tethering complexes proposed in mammals (Fig. 1) also have additional functions—either Ca2+ signaling or apoptotic signaling between these organelles.
Tethers within organelles
MCSs may form not only between organelles but also between compartments of the same organelle. In two cases, proteins necessary for these intra-organelle contacts are known. The Golgi complex is divided into a number of cisternae that remain closely apposed in some cell types, forming stacked compartments. Two tethering proteins maintain connections between Golgi cisternae. Golgi reassembly stacking protein 65 (GRASP65) forms contacts between cis- and medial-Golgi cisternae and GRASP55 mediates medial- to trans-cisternal interactions (Fig. 1; Barr et al., 1997; Shorter et al., 1999). The Golgi stack disassembles when both GRASPs are depleted, indicating that they are the primary or sole tethers (Xiang and Wang, 2010). Tethering by these proteins is regulated by kinases to allow Golgi cisternal disassembly during the cell cycle. Whether the inter-Golgi contacts formed by GRASPs mediate signaling or lipid exchange between cisternae is not yet known (Tang and Wang, 2013).
MCSs also form inside organelles with internal membranes: mitochondria, chloroplasts, and multivesicular bodies. These MCSs may form between membranes within these organelles or between internal membranes and the outer membrane of the organelle. Recently, three groups discovered a tethering complex involved in forming contacts between mitochondrial cisternae and between cisternae and the mitochondrial outer membrane (Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011). This complex, called the mitochondrial contact site and cristae organizing system (MICOS), is conserved from yeast to humans and contains at least six proteins (Fig. 1). It is necessary to maintain inner membrane organization and also interacts with protein complexes in the outer membrane, including the translocase of the outer membrane (TOM) complex and the sorting and assembly machinery (SAM) complex (van der Laan et al., 2012; Zerbes et al., 2012).
Lipid exchange at MCSs
Lipid exchange between organelles at MCSs may serve a number of important functions. One is that it allows cells to rapidly modulate the lipid composition of an organelle independently of vesicular trafficking. In addition, some organelles, such as mitochondria and chloroplasts, must obtain most of the lipids they require for membrane biogenesis by nonvesicular lipid trafficking that almost certainly occurs at MCSs (Osman et al., 2011; Wang and Benning, 2012; Horvath and Daum, 2013). Finally, and perhaps most importantly, lipid transfer at MCSs may play an important role in lipid metabolism by channeling lipids to or away from enzymes in different compartments.
Some lipid exchange at MCSs is facilitated by soluble lipid transport proteins (LTPs), which can shuttle lipid monomers between membranes (Fig. 2 A). In other cases, known LTPs do not seem to be required and lipids may be exchanged at MCSs by other mechanisms (Fig. 2, B and C), which will be discussed next.
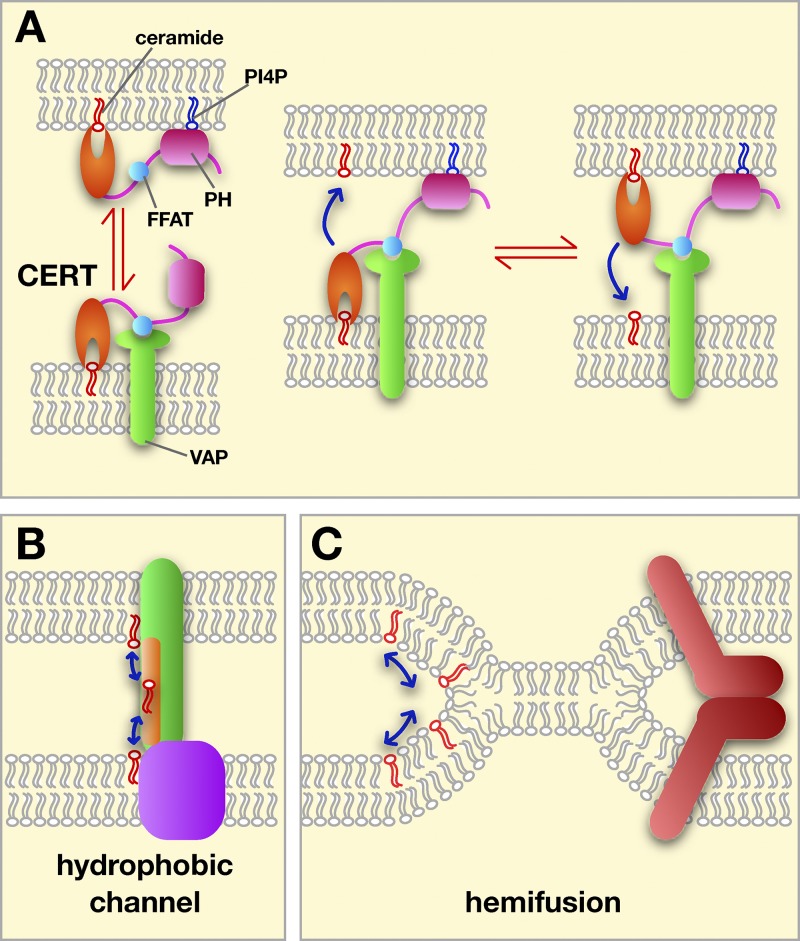
Figure 2.
Possible mechanisms of lipid exchange at MCSs. (A) Transfer by LTPs using CERT as an example. The targeting PH domain (pink) and FFAT motif (blue) are shown. CERT could shuttle between membranes (left) or transfer while binding both membranes (right). (B) Some transfer could occur through hydrophobic channels or tunnels (in green) bridging the two membranes at a MCS. (C) Lipid exchange between hemifused membranes. Hemifusion could be promoted and regulated by proteins (red).
Most LTPs fall into at least five superfamilies that differ structurally but that all have a hydrophobic pocket or groove that can bind a lipid monomer, and often have a lid domain that shields the bound lipid from the aqueous phase (D’Angelo et al., 2008; Lev, 2010). This allows LTPs to shuttle lipid monomers between membranes. LTPs probably transfer lipids between organelles in cells most efficiently at MCSs, where they have only a short distance to diffuse between membranes. LTPs that may transfer lipids at contact sites are: OSBP, ceramide transport protein (CERT), the yeast OSBP homologues Osh6 and Osh7, protein tyrosine kinase 2 N-terminal domain–interacting receptor 2 (Nir2), and Ups1 (Hanada, 2010; Connerth et al., 2012; Chang et al., 2013; Maeda et al., 2013; Mesmin et al., 2013).
LTPs could function by shuttling between membranes at MCSs or while simultaneously bound to both membranes (Fig. 2 A). Many LTPs have domains that target them to the two membranes at an MCS. For example, OSBP and CERT have FFAT motifs, which bind ER resident VAPs, and PH domains that bind PIPs in the Golgi complex or PM.
Another important emerging aspect of lipid exchange by some LTPs is that it may be driven by their ability to exchange one lipid for another. For example, OSBP can transfer both cholesterol and PI4P. At ER–Golgi MCSs, OSBP may facilitate the net movement of cholesterol from the ER to the Golgi and PI4P in the opposite direction (Mesmin et al., 2013). The difference in the PI4P concentrations in the ER and Golgi (lower in the ER than in the Golgi) may drive the net transfer of cholesterol to the Golgi. The ability to exchange one lipid for another has been found for other LTPs (Schaaf et al., 2008; de Saint-Jean et al., 2011; Kono et al., 2013) and may be critical for driving directional lipid exchange at MCSs.
Some lipid exchange at MCSs does not seem to be facilitated by LTPs. The best evidence for this comes from studies on lipid transfer between the ER and mitochondria. It has long been known that lipids are exchanged between these two organelles; mitochondria must acquire most of the lipid it requires for membrane biogenesis from the rest of the cell. Lipid exchange at ER–mitochondria MCSs occurs by a mechanism that does not require energy, at least in vitro, and does not require any cytosolic factors (Osman et al., 2011; Vance, 2014).
How this lipid transfer occurs is not known, and two possible types of mechanism are shown in Fig. 2, B and C. One is that some MCS proteins form a hydrophobic channel that allows lipids to move between membranes. Such a channel would be similar to an LTP, but whereas lipids enter and exit LTPs by the same opening, they enter and exit channels by different openings. This difference could allow lipid exchange by a channel to be regulated and, if the channel could bind two different lipids simultaneously, it might couple the transfer of the lipids. A domain that may form channels at MCSs has been identified. Called the synaptotagmin-like mitochondrial lipid-binding protein (SMP) domain, it has been predicted to be part of a superfamily of proteins that includes cholesterol ester transfer protein (CETP; Kopec et al., 2010). CETP has a tubular lipid-binding domain that transfers lipids between high-density and low-density lipoproteins, probably while simultaneously bound to both (Qiu et al., 2007; Zhang et al., 2012). SMP domains could transfer lipids between membranes by a similar mechanism. Consistent with this possibility, all SMP-containing proteins in budding yeast localize to MCSs and many mammalian SMP-containing proteins do as well (Toulmay and Prinz, 2012). Interestingly, SMP domains are present in three of the five proteins in a yeast ER–mitochondria tethering complex called ERMES (Kornmann et al., 2009). Whether ERMES facilitates lipid exchange between the ER and mitochondria is not yet clear. Mitochondria derived from cells missing ERMES have altered lipid composition (Osman et al., 2009; Tamura et al., 2012; Tan et al., 2013), indicating that lipid exchange between the ER and mitochondria could be altered in these strains. On the other hand, little or no defect in the rates of phospholipid exchange between ER and mitochondria were found in ERMES mutants (Kornmann et al., 2009; Nguyen et al., 2012; Voss et al., 2012). Thus, whether proteins that contain SMP domains actually facilitate lipid exchange remains to be determined.
As second possible mechanism of lipid transfer at MCSs that does not require LTPs is membrane hemifusion (Fig. 2 C), which could allow rapid exchange of large amounts of lipids between compartments. Recent indirect evidence suggests that hemifusion may occur between the ER and chloroplasts (Mehrshahi et al., 2013). This is consistent with an earlier study using optical tweezers that found the ER and chloroplasts remained attached to one another even when a stretching force of 400 pN was applied (Andersson et al., 2007). Whether hemifusion occurs at MCSs in animal cells remains to be determined.
Calcium signaling at MCSs
MCSs between the ER and PM and the ER and mitochondria play central roles in intracellular Ca2+ storage, homeostasis, and signaling in mammalian cells. MCSs between the ER and lysosomes may also be important, though they are less well understood (Helle et al., 2013; Lam and Galione, 2013).
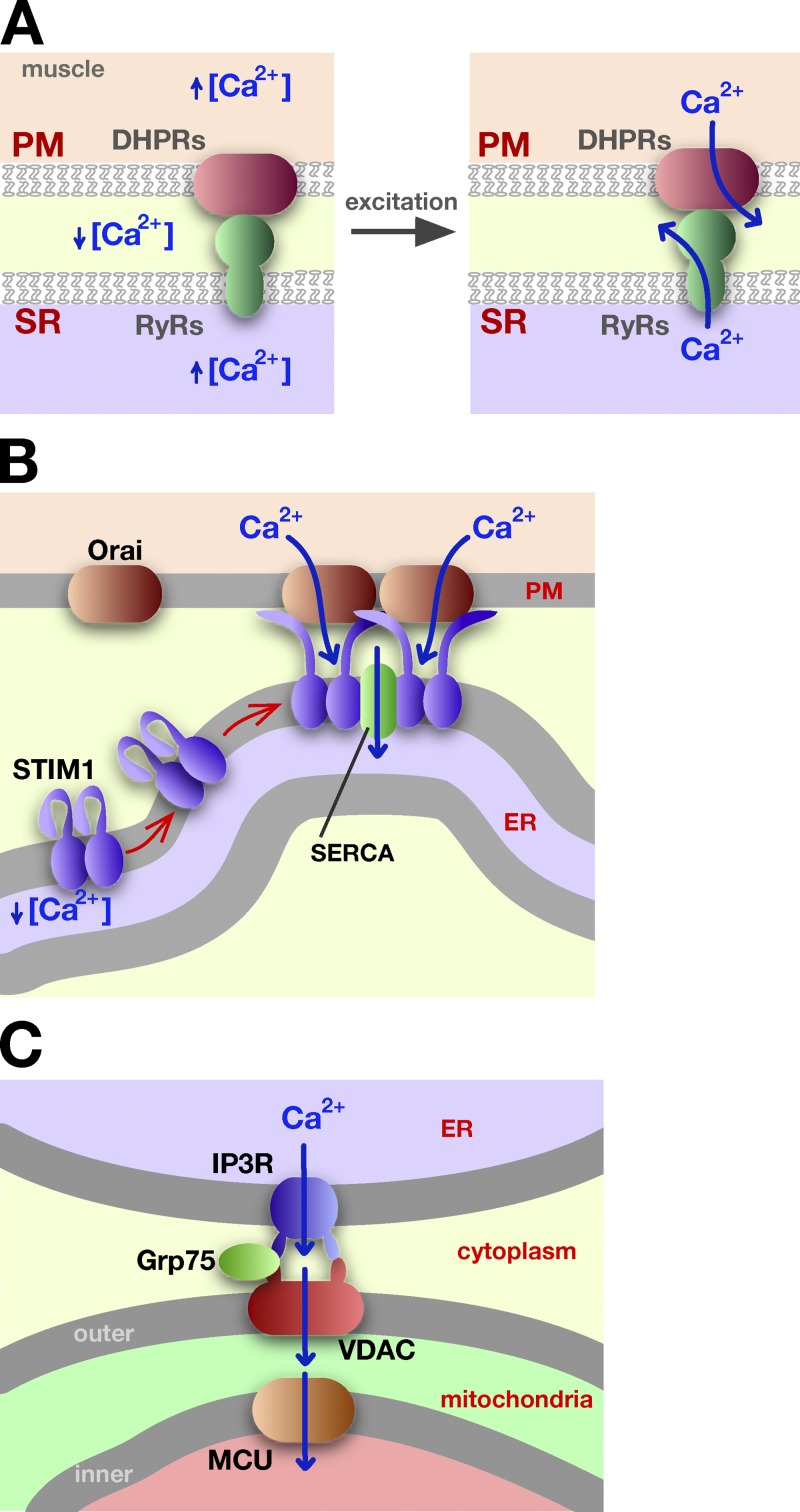
One of the best-characterized MCSs is the one formed between the PM and ER in muscle cells. In both cardiac and skeletal muscle cells, deep invaginations of the PM, called T (transverse)-tubules, allow it to form extensive contacts with the ER, called the sarcoplasmic reticulum (SR) in muscle cells. These contacts are essential for coupling excitation and contraction. Before excitation, Ca2+ levels in the cytoplasm of muscle cells are low, whereas the Ca2+ concentrations in the SR and outside muscle cells are high. During muscle excitation, Ca2+ rapidly flows into the cytosol through channels in the PM and the SR (Fig. 3 A). The channels in the PM, called dihydropyridine receptors (DHPRs), and those in the SR, known ryanodine receptors RyRs, directly interact with each other where the SR and PM are closely apposed, allowing the opening of both types of channels to be coordinated (Fabiato, 1983; Bannister, 2007; Beam and Bannister, 2010; Rebbeck et al., 2011).
Figure 3.
Ca2+ trafficking at ER–PM MCSs. (A) In muscle cells, the interaction of the RyR in the SR and with DHPR in the PM allows the coordinated release of Ca2+ during muscle excitation and contraction. See text for details. (B) When STIM1 senses low Ca2+ concentration in the ER, it undergoes a conformational change that allows it to oligomerize and bind to the PM, to the protein Orai1, and to accumulate at ER–PM MCSs. Ca2+ influx at these sites facilitates Ca2+ import into the ER by sarco/endoplasmic reticulum Ca2+-ATPase (SERCA). (C) Calcium channeling from the ER lumen to the mitochondrial matrix. Calcium exits the ER through the inositol trisphosphate receptor (IP3R) channel, enters mitochondria via VDAC, and then uses the mitochondrial Ca2+ uniporter (MCU) to move into the mitochondrial matrix.
The extensive contacts between the SR and PM in muscle cells are largely maintained by tethering proteins called junctophilins, which have a single transmembrane domain in the SR and a large cytosolic domain that interacts with the PM. Expression of junctophilins in cells lacking them induces ER–PM contacts (Takeshima et al., 2000) and cells lacking junctophilins have abnormal SR–PM MCSs and defects in Ca2+ signaling (Ito et al., 2001; Komazaki et al., 2002; Hirata et al., 2006). Thus, junctophilins are both necessary and sufficient for generating functional SR–PM contacts. However, cells lacking junctophilins still maintain some SR–PM contacts, indicating that other proteins also tether the SR and the PM. Some of this residual tethering probably comes from the interaction of DHPRs and RyRs.
ER–PM contacts also play a role in regulating intracellular Ca2+ levels in non-excitable cells. When the Ca2+ concentration in the ER lumen is low it triggers Ca2+ entry into the cytosol and ER from outside cells (Fig. 3 B), a process known as store-operated Ca2+ entry (SOCE). The PM channel responsible for Ca2+ entry is Orai1, and the sensor of Ca2+ concentration in the ER lumen is the integral membrane protein stromal interaction molecule-1 (STIM1). When STIM1 senses that the Ca2+ concentration in the ER is low, it oligomerizes and undergoes a conformational change that exposes a basic cluster of amino acids in its C terminus that binds PIPs in the PM (Stathopulos et al., 2006, 2008; Liou et al., 2007; Muik et al., 2011). STIM1 also binds to Orai1 in the PM and activates it (Kawasaki et al., 2009; Muik et al., 2009; Park et al., 2009; Wang et al., 2009). Activation of STIM1 causes it to shift from being relatively evenly distributed on the ER to forming a number of puncta, which are regions were the ER and PM are closely apposed. It seems likely that STIM1 accumulates at and expands preexisting ER–PM MCSs and may also drive the formation of new MCSs (Wu et al., 2006; Lur et al., 2009; Orci et al., 2009).
The interaction of STIM1 and Orai1 at ER–PM contacts during SOCE is an elegant mechanism for channeling both signals and small molecules at an MCS. The signal that ER luminal Ca2+ concentration is low is transmitted directly from STIM1 in the ER to Orai1 in the PM. The close contact of PM and ER also allows Ca2+ to move from outside the cell into the lumen of the ER without significantly increasing cytosolic Ca2+ levels (Jousset et al., 2007). During SOCE, ER Ca2+ levels are restored by the sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) pump (Sampieri et al., 2009; Manjarrés et al., 2011). This pump is enriched in ER–PM contacts with STIM1 and may interact directly with it, suggesting how Ca2+ can be effectively channeled from outside cells directly into the ER lumen at ER–PM MCSs (Fig. 3 B).
Interestingly, it has become clear that proteins that are not part of the SOCE pathway also facilitate ER–PM connections during Ca2+ signaling. The E-Syts have multiple domains that probably bind Ca2+. They have been shown to regulate both the number of the ER–PM contacts and the distance between the ER and PM at MCSs during Ca2+ signaling (Chang et al., 2013; Giordano et al., 2013).
MCSs between the ER and mitochondria similarly facilitate Ca2+ movement from the ER lumen to mitochondria (Rizzuto et al., 1998; Csordás et al., 2006). Ca2+ channels in the ER and OMM interact with each other at MCSs (Fig. 3 C). The channel in the ER is called the inositol trisphosphate receptor (IP3R), while the voltage-dependent anion channel (VDAC) in the outer mitochondrial membrane is a nonspecific pore that allows Ca2+ entry into mitochondria. These proteins, together with the cytosolic chaperone Grp75, form a complex that links the ER and mitochondria and facilitates Ca2+ exchange (Szabadkai et al., 2006).
More evidence that Ca2+ transfer from the ER to mitochondria occurs at MCSs came from studies on the channel that allows Ca2+ to move across the inner mitochondrial membrane, called the mitochondrial Ca2+ uniporter (MCU). Surprisingly, this channel has an affinity for Ca2+ that is lower than the typical Ca2+ concentration in the cytosol (Kirichok et al., 2004). However, Ca2+ release by the ER at ER–mitochondrial MCSs suggests a solution to this puzzle; the local Ca2+ concentration at these MCSs is probably high enough for MCU to function (Csordás et al., 2010). Close contacts between the ER and mitochondria are therefore essential for channeling Ca2+ from the ER lumen to the mitochondrial matrix.
It is thought that MCSs between the ER (or SR) and lysosomes regulate Ca2+ release by lysosomes, but the mechanism is not yet understood (Kinnear et al., 2004, 2008; Galione et al., 2011; Morgan et al., 2011).
Enzymes working in trans and signaling at MCSs
MCSs allow rapid and efficient signaling between intracellular compartments. We are still just beginning to understand the mechanisms and functions of this signaling. One way that signals are transmitted between the two compartments at an MCS is for an enzyme in one compartment to modify substrates in the second; that is, for the enzyme to work in trans. Although there are currently only a few examples of this, which are discussed here, it seems likely that many more will be uncovered.
The protein tyrosine phosphatase PTP1B regulates a number of receptor tyrosine kinases. PTP1B resides on the surface of the ER with its active site in the cytosol, and yet the receptor tyrosine kinases it modifies are in the PM. Although this was initially puzzling, it was found that PTP1B probably encounters its substrates at MCSs, either at ER–PM junctions or at contacts between the ER and endocytic recycling compartments (Haj et al., 2002; Boute et al., 2003; Anderie et al., 2007; Eden et al., 2010; Nievergall et al., 2010). Interestingly, in some cases the interaction of PTP1B with substrates in the PM occurs on portions of the PM that are part of cell–cell contacts (Haj et al., 2012), suggesting that ER–PM contacts could play a role in signaling, not only between the ER and PM but between cells as well. Dephosphorylation of receptor tyrosine kinases by PTP1B at contact sites probably allows their kinase activity to be regulated in response to changes in the ER or changes in cellular architecture that alter MCSs. For example, the dephosphorylation of epidermal growth factor receptor (EGFR) by PTP1B occurs at regions of close contact between the ER and multivesicular bodies, causing EGFR to become sequestered with multivesicular bodies (Eden et al., 2010). This may provide a mechanism for cells to regulate EGFR levels on the PM in response to signals in the ER.
Lipid metabolism enzymes can also work in trans at MCSs. In two cases, both in yeast, enzymes that reside in the ER have been found to modify lipids in the PM at MCSs. In one instance, the phosphatase Sac1, which is on the surface of the ER, can dephosphorylate PIPs in the PM (Stefan et al., 2011). In the second, the ER enzyme Opi3 methylates phosphatidylethanolamine in the PM, a reaction that is required for the conversion of phosphatidylethanolamine to phosphatidylcholine (Tavassoli et al., 2013). Remarkably, the PIP-binding protein Osh3 (Tong et al., 2013) regulates both reactions, suggesting that lipid metabolism at ER–PM junctions is regulated by PIPs. It seems likely that ER–PM junctions play important roles in integrating lipid metabolism in both organelles.
MCSs and organelle trafficking and inheritance
In addition to being sites at which signals and small molecules are exchanged between cellular compartments, there is growing evidence that MCS formation also regulates organelle trafficking and inheritance.
In budding yeast, organelle transport is polarized from the mother cell to the growing bud and is required for proper organelle inheritance. The transport of peroxisomes and mitochondria to the bud is regulated by their association with the ER or PM.
Knoblach et al. (2013) found that tethering of the ER to peroxisomes requires Pex3, an integral membrane protein that resides in both compartments, and Inp1, a cytosolic protein that binds to Pex3. This tether keeps peroxisomes in mother cells. When peroxisomes divide they are transferred to the bud by the myosin V motor Myo2 and become attached to the ER in the bud. In cells lacking the ER–peroxisome tether, peroxisomes accumulate in daughter cells. Thus, tethering plays a critical role in ensuring that some peroxisomes are retained in mother cells and that both cells inherit peroxisomes.
Mitochondrial inheritance in yeast is regulated by close contacts with both the ER and PM. Mitochondria–PM contacts mediated by a complex containing Num1 and Mdm36 ensure that mitochondria are properly distributed between mother and daughter cells and seem to be particularly important for retaining mitochondria in the mother cells (Klecker et al., 2013; Lackner et al., 2013). Interestingly, Num1–Mdm36-mediated contacts also associate with the ER (Lackner et al., 2013), suggesting that three membranes may somehow associate at these MCSs. An ER–mitochondria tether containing the protein Mmr1, which anchors mitochondria to bud tips, also plays a role in mitochondrial inheritance (Swayne et al., 2011). Thus, the Num1-tethering complex and Mmr1-tethering complex seem to play antagonistic roles in mitochondrial distribution; the Num1 complex promotes mitochondrial retention in the mother, whereas the Mmr1 complex favors retention in the bud.
MCSs also play a role in endosomal trafficking in mammalian cells. One of the complexes that tethers the ER to endosomes contains VAPs and ORP1L, which is an OSBP homologue that can bind cholesterol (Fig. 1). ORPlL can also binds the p150Glued subunit of the dynein–dynactin motor that participates in endosome transport along microtubules (Johansson et al., 2007). When cellular cholesterol levels are high, ORP1L associates with p150Glued but not VAPs and endosomes are transported on microtubules. However, when cholesterol levels decrease, ORP1L undergoes a conformation change that dissociates it from p150Glued and allows it to bind to VAPs on the surface of the ER, thus forming a tether between endosomes and the ER (Rocha et al., 2009). Under these conditions, endosome transport on microtubules is blocked. ORPlL is therefore a cholesterol sensor that regulates a switch between the association of endosomes with either motors or the ER.
MCSs and organelle division
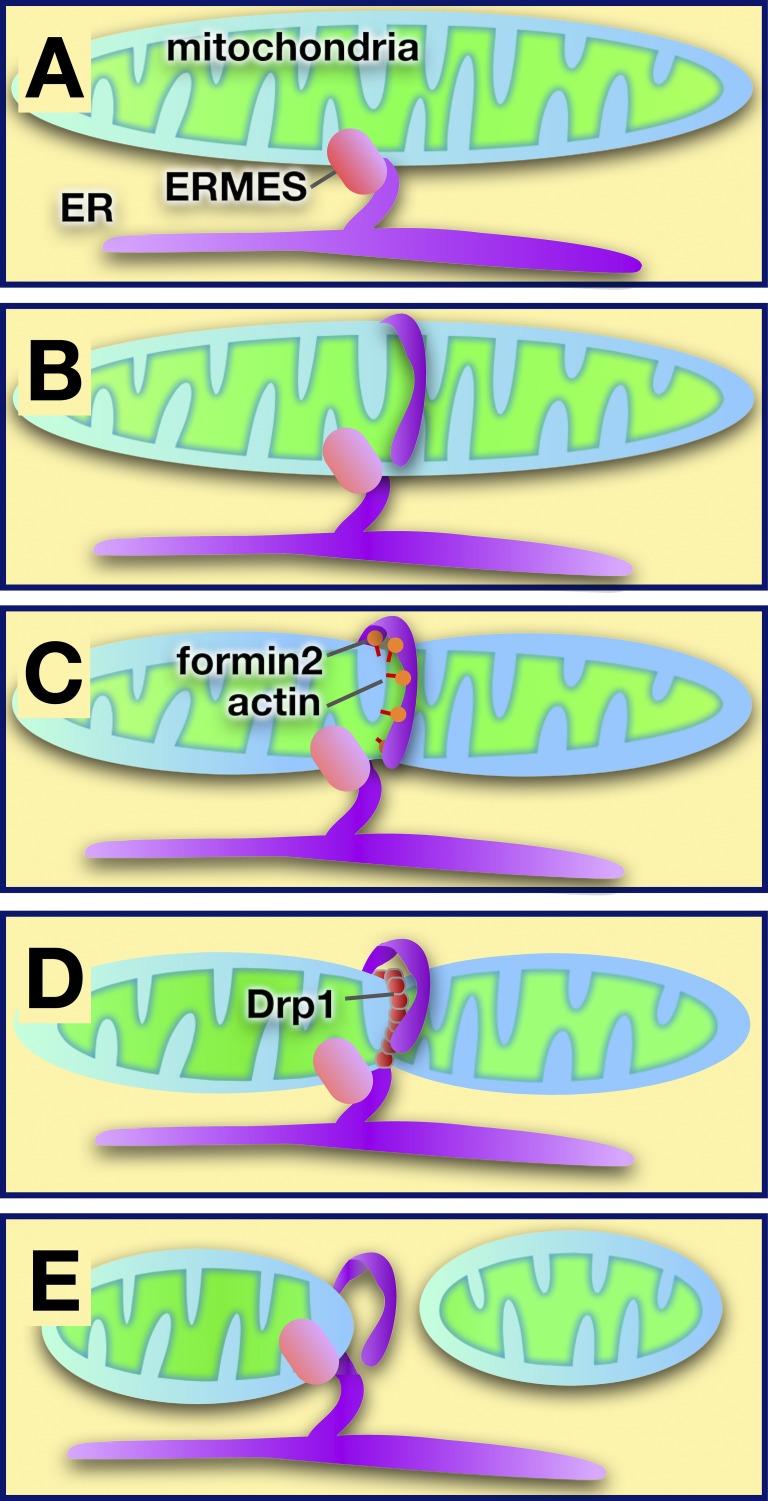
A groundbreaking study revealed a new and unexpected role for MCSs between the ER and mitochondria: the ER regulates mitochondrial fission (Friedman et al., 2011). Although a mechanistic understanding of how ER participates in mitochondrial fission is not yet available, the sequence of events is beginning to come into focus (Fig. 4). The ER encircles mitochondria at sites where scission will occur. The ERMES complex is present at these sites (Murley et al., 2013). Because mammalian cells lack ERMES, another tethering complex must perform the same function in higher eukaryotes. Mitochondrial division requires membrane scission by the dynamin-like protein Dnm1/Drp1, which multimerizes on the outer mitochondria membrane. Close contacts between the ER and mitochondria occur before Dnm1/Drp1 assembly, suggesting that these contacts promote or regulate the association of Dnm1/Drp1 with mitochondria and hence mitochondrial division. It is possible that when the ER encircles mitochondria it causes mitochondria to constrict to a diameter that allows Dnm1/Drp1 to assemble. The force necessary to drive constriction may come from actin polymerization. A recent study found that the ER protein, inverted formin-2, probably drives actin polymerization at these sites and is necessary for mitochondria fusion (Korobova et al., 2013).
Figure 4.
Model of ER-mediated regulation of mitochondrial fission at sites of contact. (A) The ER and mitochondria are tethered by ERMES in yeast (other tethers are used in higher eukaryotes). (B) The ER encircles mitochondria at sites where division will occur. (C) Actin polymerization facilitated by formin 2 may cause mitochondrial constriction. (D) The dynamin-like protein Drp1 is recruited to the mitochondrial surface, where it multimerizes and causes mitochondrial scission. (E) After fission, the ER remains associated with the mitochondrion that retains the ERMES complex.
Understanding the assembly and regulation of the mitochondrial division machinery at ER–mitochondria MCSs and how this is linked to mitochondrial and perhaps ER function remain fascinating questions for the future. Another interesting question is whether other MCSs play roles in the fission of other organelles.
Proposed functions of ER–mitochondrial MCSs
A growing number of studies have suggested that ER–mitochondria MCSs play critical roles in autophagy, apoptosis, inflammation, reactive oxygen species signaling, and metabolic signaling. ER–mitochondria MCSs have also been implicated in Alzheimer’s disease, Parkinson’s disease, and some viral infections. These topics have been recently reviewed (Eisner et al., 2013; Raturi and Simmen, 2013; Marchi et al., 2014; Vance, 2014) and will not be discussed in detail here.
One issue with most of the studies on the functions of ER–mitochondria junctions is that they rely, at least in part, on density gradient purification of the ER that associates with mitochondria. These operationally defined membranes, often called mitochondrial-associated membranes (MAMs), remain poorly defined. In fact, a significant number of proteins that are enriched in MAMs do not seem to be enriched at ER–mitochondria junctions when their localization is determined by other methods (Helle et al., 2013; Vance, 2014). Therefore, it remains unclear why some proteins and lipids are enriched in MAMs.
Here, two interesting findings will be discussed that suggest the importance of ER–mitochondrial junctions in signaling in addition to their well-known role in Ca2+ signalling.
The induction of apoptosis requires signal transmission between the ER and mitochondria. Part of this signaling process occurs through an interaction between the ER protein Bap31 and the mitochondrial fission protein Fission 1 homologue (Fis1; Iwasawa et al., 2011). This interaction occurs at ER–mitochondria MCSs and results in the cleavage of Bap31 by caspase-8 to form p20Bap31, which is pro-apoptotic. Both Bap31 and Fis1 are parts of larger complexes that are still being characterized. Interestingly, it has recently been found that a protein called cell death–involved p53 target-1 (CDIP1) binds to Bap31 during ER stress and promotes apoptotic signaling from the ER to mitochondria (Namba et al., 2013), suggesting how ER stress signals are transmitted from the ER to mitochondria through MCSs.
Another important connection between ER–mitochondrial MCSs and signaling has to do with the target of rapamycin (TOR) kinase complexes, which are critical regulators of growth and metabolism. The mammalian TOR complex 2 (mTORC2) was found to interact with the IP3R–Grp75–VDAC complex that tethers the ER and mitochondria (Betz et al., 2013). Remarkably, this study presents evidence that mTORC regulates both the formation of ER–mitochondrial MCSs and mitochondrial function, suggesting an interesting new mechanism for how metabolic signaling can impact mitochondrial function via MCSs.
Conclusions and perspectives
The potential of MCSs to facilitate Ca2+ signaling and channel lipids between organelles was recognized some time ago (Levine and Loewen, 2006), but it has only been in the last few years that we have finally begun to have some mechanistic insight into how these processes occur and how MCSs are formed. Many fundamental questions remain to be addressed. How lipid exchange at MCSs that does not require soluble LTPs occurs or whether transient hemifusion of membranes at MCS ever occurs remain open questions. Another is the mechanisms by which Ca2+ regulates MCS formation between the ER and other organelles. One major challenge for the field will be devising better methods to visualize MCSs and identify proteins and lipids enriched at these sites. It is particularly important to better understand what the MAM fraction is and what it means for proteins and lipids to be enriched in this fraction.
One of the most exciting developments in the study of MCSs in the last few years has been the discovery of the role of MCSs in organelle trafficking, inheritance, and dynamics. These studies have revealed that MCSs not only play critical roles in signaling and metabolism, but also modulate the intracellular distribution of organelles and organelle architecture. Understanding how MCSs perform these functions will probably shed light on the connection between the still murky relationship between organelle structure and function as well as the role of the ER as a regulator of other organelles. Given the current pace of discovery, it seems likely that in the next few years our knowledge of the functions of MCSs will grow dramatically.
Acknowledgments
Thank you to Orna Cohen-Fix and Alex Toulmay for helpful discussions and critically reading the manuscript.
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
The author declares no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CERT
- ceramide transport protein
- DHPR
- dihydropyridine receptor
- E-Syt
- extended synaptotagmin
- FFAT
- phenylalanines in an acid tract
- LTP
- lipid transport protein
- MAM
- mitochondrial-associated membrane
- MCS
- membrane contact site
- OMM
- outer mitochondrial membrane
- ORP
- oxysterol-binding protein–related protein
- OSBP
- oxysterol-binding protein
- PH
- pleckstrin homology
- PIP
- phosphoinositide
- PM
- plasma membrane
- SOCE
- store-operated Ca2+ entry
- SR
- sarcoplasmic reticulum
- STIM1
- stromal interaction molecule-1
- VAP
- vesicle-associated membrane protein–associated protein
- VDAC
- voltage-dependent anion channel
References
- Alpy F., Rousseau A., Schwab Y., Legueux F., Stoll I., Wendling C., Spiegelhalter C., Kessler P., Mathelin C., Rio M.C., et al. 2013. STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J. Cell Sci. 126:5500–5512 10.1242/jcs.139295 [DOI] [PubMed] [Google Scholar]
- Anderie I., Schulz I., Schmid A. 2007. Direct interaction between ER membrane-bound PTP1B and its plasma membrane-anchored targets. Cell. Signal. 19:582–592 10.1016/j.cellsig.2006.08.007 [DOI] [PubMed] [Google Scholar]
- Andersson M.X., Goksör M., Sandelius A.S. 2007. Membrane contact sites: physical attachment between chloroplasts and endoplasmic reticulum revealed by optical manipulation. Plant Signal. Behav. 2:185–187 10.4161/psb.2.3.3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister R.A. 2007. Bridging the myoplasmic gap: recent developments in skeletal muscle excitation-contraction coupling. J. Muscle Res. Cell Motil. 28:275–283 10.1007/s10974-007-9118-5 [DOI] [PubMed] [Google Scholar]
- Barr F.A., Puype M., Vandekerckhove J., Warren G. 1997. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 91:253–262 10.1016/S0092-8674(00)80407-9 [DOI] [PubMed] [Google Scholar]
- Beam K.G., Bannister R.A. 2010. Looking for answers to EC coupling’s persistent questions. J. Gen. Physiol. 136:7–12 10.1085/jgp.201010461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C., Stracka D., Prescianotto-Baschong C., Frieden M., Demaurex N., Hall M.N. 2013. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc. Natl. Acad. Sci. USA. 110:12526–12534 10.1073/pnas.1302455110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boute N., Boubekeur S., Lacasa D., Issad T. 2003. Dynamics of the interaction between the insulin receptor and protein tyrosine-phosphatase 1B in living cells. EMBO Rep. 4:313–319 10.1038/sj.embor.embor767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.L., Hsieh T.S., Yang T.T., Rothberg K.G., Azizoglu D.B., Volk E., Liao J.C., Liou J. 2013. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 5:813–825 10.1016/j.celrep.2013.09.038 [DOI] [PubMed] [Google Scholar]
- Connerth M., Tatsuta T., Haag M., Klecker T., Westermann B., Langer T. 2012. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 338:815–818 10.1126/science.1225625 [DOI] [PubMed] [Google Scholar]
- Csordás G., Renken C., Várnai P., Walter L., Weaver D., Buttle K.F., Balla T., Mannella C.A., Hajnóczky G. 2006. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174:915–921 10.1083/jcb.200604016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordás G., Várnai P., Golenár T., Roy S., Purkins G., Schneider T.G., Balla T., Hajnóczky G. 2010. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell. 39:121–132 10.1016/j.molcel.2010.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo G., Vicinanza M., De Matteis M.A. 2008. Lipid-transfer proteins in biosynthetic pathways. Curr. Opin. Cell Biol. 20:360–370 10.1016/j.ceb.2008.03.013 [DOI] [PubMed] [Google Scholar]
- de Brito O.M., Scorrano L. 2008. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 456:605–610 10.1038/nature07534 [DOI] [PubMed] [Google Scholar]
- de Saint-Jean M., Delfosse V., Douguet D., Chicanne G., Payrastre B., Bourguet W., Antonny B., Drin G. 2011. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195:965–978 10.1083/jcb.201104062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos K.J., Mórotz G.M., Stoica R., Tudor E.L., Lau K.F., Ackerley S., Warley A., Shaw C.E., Miller C.C. 2012. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 21:1299–1311 10.1093/hmg/ddr559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derré I., Swiss R., Agaisse H. 2011. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog. 7:e1002092 10.1371/journal.ppat.1002092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Kumar J., Ferguson C., Schulz T.A., Ong Y.S., Hong W., Prinz W.A., Parton R.G., Brown A.J., Yang H. 2011. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J. Cell Biol. 192:121–135 10.1083/jcb.201004142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E.R., White I.J., Tsapara A., Futter C.E. 2010. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat. Cell Biol. 12:267–272 [DOI] [PubMed] [Google Scholar]
- Eisner V., Csordás G., Hajnóczky G. 2013. Interactions between sarco-endoplasmic reticulum and mitochondria in cardiac and skeletal muscle - pivotal roles in Ca2+ and reactive oxygen species signaling. J. Cell Sci. 126:2965–2978 10.1242/jcs.093609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell C.A., Jiang S., Kim J.H., Lee A., Wittmann T., Hanada K., Melancon P., Engel J.N. 2011. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog. 7:e1002198 10.1371/journal.ppat.1002198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English A.R., Voeltz G.K. 2013. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb. Perspect. Biol. 5:a013227 10.1101/cshperspect.a013227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiato A. 1983. Calcium-induced release of calcium from the cardiac sarcoplasmic reticulum. Am. J. Physiol. 245:C1–C14 [DOI] [PubMed] [Google Scholar]
- Fischer M.A., Temmerman K., Ercan E., Nickel W., Seedorf M. 2009. Binding of plasma membrane lipids recruits the yeast integral membrane protein Ist2 to the cortical ER. Traffic. 10:1084–1097 10.1111/j.1600-0854.2009.00926.x [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galione A., Parrington J., Funnell T. 2011. Physiological roles of NAADP-mediated Ca2+ signaling. Sci China Life Sci. 54:725–732 10.1007/s11427-011-4207-5 [DOI] [PubMed] [Google Scholar]
- Giordano F., Saheki Y., Idevall-Hagren O., Colombo S.F., Pirruccello M., Milosevic I., Gracheva E.O., Bagriantsev S.N., Borgese N., De Camilli P. 2013. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 153:1494–1509 10.1016/j.cell.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haj F.G., Verveer P.J., Squire A., Neel B.G., Bastiaens P.I. 2002. Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science. 295:1708–1711 10.1126/science.1067566 [DOI] [PubMed] [Google Scholar]
- Haj F.G., Sabet O., Kinkhabwala A., Wimmer-Kleikamp S., Roukos V., Han H.M., Grabenbauer M., Bierbaum M., Antony C., Neel B.G., Bastiaens P.I. 2012. Regulation of signaling at regions of cell-cell contact by endoplasmic reticulum-bound protein-tyrosine phosphatase 1B. PLoS ONE. 7:e36633 10.1371/journal.pone.0036633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K. 2010. Intracellular trafficking of ceramide by ceramide transfer protein. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 86:426–437 10.2183/pjab.86.426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M., Körner C., Walther D., Mokranjac D., Kaesmacher J., Welsch U., Griffith J., Mann M., Reggiori F., Neupert W. 2011. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30:4356–4370 10.1038/emboj.2011.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle S.C., Kanfer G., Kolar K., Lang A., Michel A.H., Kornmann B. 2013. Organization and function of membrane contact sites. Biochim. Biophys. Acta. 1833:2526–2541 10.1016/j.bbamcr.2013.01.028 [DOI] [PubMed] [Google Scholar]
- Hirata Y., Brotto M., Weisleder N., Chu Y., Lin P., Zhao X., Thornton A., Komazaki S., Takeshima H., Ma J., Pan Z. 2006. Uncoupling store-operated Ca2+ entry and altered Ca2+ release from sarcoplasmic reticulum through silencing of junctophilin genes. Biophys. J. 90:4418–4427 10.1529/biophysj.105.076570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Collins S.R., Cassidy-Stone A., Hummel E., Devay R.M., Lackner L.L., Westermann B., Schuldiner M., Weissman J.S., Nunnari J. 2011. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195:323–340 10.1083/jcb.201107053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S.E., Daum G. 2013. Lipids of mitochondria. Prog. Lipid Res. 52:590–614 10.1016/j.plipres.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Ito K., Komazaki S., Sasamoto K., Yoshida M., Nishi M., Kitamura K., Takeshima H. 2001. Deficiency of triad junction and contraction in mutant skeletal muscle lacking junctophilin type 1. J. Cell Biol. 154:1059–1067 10.1083/jcb.200105040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasawa R., Mahul-Mellier A.L., Datler C., Pazarentzos E., Grimm S. 2011. Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 30:556–568 10.1038/emboj.2010.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V.M., Neefjes J. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J. Cell Biol. 176:459–471 10.1083/jcb.200606077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousset H., Frieden M., Demaurex N. 2007. STIM1 knockdown reveals that store-operated Ca2+ channels located close to sarco/endoplasmic Ca2+ ATPases (SERCA) pumps silently refill the endoplasmic reticulum. J. Biol. Chem. 282:11456–11464 10.1074/jbc.M609551200 [DOI] [PubMed] [Google Scholar]
- Kawano M., Kumagai K., Nishijima M., Hanada K. 2006. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J. Biol. Chem. 281:30279–30288 10.1074/jbc.M605032200 [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Lange I., Feske S. 2009. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem. Biophys. Res. Commun. 385:49–54 10.1016/j.bbrc.2009.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnear N.P., Boittin F.X., Thomas J.M., Galione A., Evans A.M. 2004. Lysosome-sarcoplasmic reticulum junctions. A trigger zone for calcium signaling by nicotinic acid adenine dinucleotide phosphate and endothelin-1. J. Biol. Chem. 279:54319–54326 10.1074/jbc.M406132200 [DOI] [PubMed] [Google Scholar]
- Kinnear N.P., Wyatt C.N., Clark J.H., Calcraft P.J., Fleischer S., Jeyakumar L.H., Nixon G.F., Evans A.M. 2008. Lysosomes co-localize with ryanodine receptor subtype 3 to form a trigger zone for calcium signalling by NAADP in rat pulmonary arterial smooth muscle. Cell Calcium. 44:190–201 10.1016/j.ceca.2007.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirichok Y., Krapivinsky G., Clapham D.E. 2004. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 427:360–364 10.1038/nature02246 [DOI] [PubMed] [Google Scholar]
- Klecker T., Scholz D., Förtsch J., Westermann B. 2013. The yeast cell cortical protein Num1 integrates mitochondrial dynamics into cellular architecture. J. Cell Sci. 126:2924–2930 10.1242/jcs.126045 [DOI] [PubMed] [Google Scholar]
- Knoblach B., Sun X., Coquelle N., Fagarasanu A., Poirier R.L., Rachubinski R.A. 2013. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 32:2439–2453 10.1038/emboj.2013.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komazaki S., Ito K., Takeshima H., Nakamura H. 2002. Deficiency of triad formation in developing skeletal muscle cells lacking junctophilin type 1. FEBS Lett. 524:225–229 10.1016/S0014-5793(02)03042-9 [DOI] [PubMed] [Google Scholar]
- Kono N., Ohto U., Hiramatsu T., Urabe M., Uchida Y., Satow Y., Arai H. 2013. Impaired α-TTP-PIPs interaction underlies familial vitamin E deficiency. Science. 340:1106–1110 10.1126/science.1233508 [DOI] [PubMed] [Google Scholar]
- Kopec K.O., Alva V., Lupas A.N. 2010. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 26:1927–1931 10.1093/bioinformatics/btq326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., Walter P. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Ramabhadran V., Higgs H.N. 2013. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 339:464–467 10.1126/science.1228360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner L.L., Ping H., Graef M., Murley A., Nunnari J. 2013. Endoplasmic reticulum-associated mitochondria-cortex tether functions in the distribution and inheritance of mitochondria. Proc. Natl. Acad. Sci. USA. 110:E458–E467 10.1073/pnas.1215232110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam A.K., Galione A. 2013. The endoplasmic reticulum and junctional membrane communication during calcium signaling. Biochim. Biophys. Acta. 1833:2542–2559 10.1016/j.bbamcr.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Lev S. 2010. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat. Rev. Mol. Cell Biol. 11:739–750 10.1038/nrm2971 [DOI] [PubMed] [Google Scholar]
- Levine T., Loewen C. 2006. Inter-organelle membrane contact sites: through a glass, darkly. Curr. Opin. Cell Biol. 18:371–378 10.1016/j.ceb.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Levine T.P., Munro S. 1998. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol. 8:729–739 10.1016/S0960-9822(98)70296-9 [DOI] [PubMed] [Google Scholar]
- Liou J., Fivaz M., Inoue T., Meyer T. 2007. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc. Natl. Acad. Sci. USA. 104:9301–9306 10.1073/pnas.0702866104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C.J., Roy A., Levine T.P. 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22:2025–2035 10.1093/emboj/cdg201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lur G., Haynes L.P., Prior I.A., Gerasimenko O.V., Feske S., Petersen O.H., Burgoyne R.D., Tepikin A.V. 2009. Ribosome-free terminals of rough ER allow formation of STIM1 puncta and segregation of STIM1 from IP(3) receptors. Curr. Biol. 19:1648–1653 10.1016/j.cub.2009.07.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K., Anand K., Chiapparino A., Kumar A., Poletto M., Kaksonen M., Gavin A.C. 2013. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature. 501:257–261 10.1038/nature12430 [DOI] [PubMed] [Google Scholar]
- Manford A.G., Stefan C.J., Yuan H.L., Macgurn J.A., Emr S.D. 2012. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 23:1129–1140 10.1016/j.devcel.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Manjarrés I.M., Alonso M.T., García-Sancho J. 2011. Calcium entry-calcium refilling (CECR) coupling between store-operated Ca(2+) entry and sarco/endoplasmic reticulum Ca(2+)-ATPase. Cell Calcium. 49:153–161 10.1016/j.ceca.2011.01.007 [DOI] [PubMed] [Google Scholar]
- Marchi S., Patergnani S., Pinton P. 2014. The endoplasmic reticulum-mitochondria connection: one touch, multiple functions. Biochim. Biophys. Acta. 1837:461–469 10.1016/j.bbabio.2013.10.015 [DOI] [PubMed] [Google Scholar]
- Mehrshahi P., Stefano G., Andaloro J.M., Brandizzi F., Froehlich J.E., DellaPenna D. 2013. Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proc. Natl. Acad. Sci. USA. 110:12126–12131 10.1073/pnas.1306331110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., Antonny B. 2013. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 155:830–843 10.1016/j.cell.2013.09.056 [DOI] [PubMed] [Google Scholar]
- Morgan A.J., Platt F.M., Lloyd-Evans E., Galione A. 2011. Molecular mechanisms of endolysosomal Ca2+ signalling in health and disease. Biochem. J. 439:349–374 10.1042/BJ20110949 [DOI] [PubMed] [Google Scholar]
- Muik M., Fahrner M., Derler I., Schindl R., Bergsmann J., Frischauf I., Groschner K., Romanin C. 2009. A cytosolic homomerization and a modulatory domain within STIM1 C terminus determine coupling to ORAI1 channels. J. Biol. Chem. 284:8421–8426 10.1074/jbc.C800229200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik M., Fahrner M., Schindl R., Stathopulos P., Frischauf I., Derler I., Plenk P., Lackner B., Groschner K., Ikura M., Romanin C. 2011. STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation. EMBO J. 30:1678–1689 10.1038/emboj.2011.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P., Nunnari J. 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife. 2:e00422 10.7554/eLife.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T., Tian F., Chu K., Hwang S.Y., Yoon K.W., Byun S., Hiraki M., Mandinova A., Lee S.W. 2013. CDIP1-BAP31 complex transduces apoptotic signals from endoplasmic reticulum to mitochondria under endoplasmic reticulum stress. Cell Rep. 5:331–339 10.1016/j.celrep.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.T., Lewandowska A., Choi J.Y., Markgraf D.F., Junker M., Bilgin M., Ejsing C.S., Voelker D.R., Rapoport T.A., Shaw J.M. 2012. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 13:880–890 10.1111/j.1600-0854.2012.01352.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievergall E., Janes P.W., Stegmayer C., Vail M.E., Haj F.G., Teng S.W., Neel B.G., Bastiaens P.I., Lackmann M. 2010. PTP1B regulates Eph receptor function and trafficking. J. Cell Biol. 191:1189–1203 10.1083/jcb.201005035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes P., Cornut D., Bochet V., Hasler U., Oh-Hora M., Waldburger J.M., Demaurex N. 2012. STIM1 juxtaposes ER to phagosomes, generating Ca²⁺ hotspots that boost phagocytosis. Curr. Biol. 22:1990–1997 10.1016/j.cub.2012.08.049 [DOI] [PubMed] [Google Scholar]
- Orci L., Ravazzola M., Le Coadic M., Shen W.W., Demaurex N., Cosson P. 2009. From the Cover: STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 106:19358–19362 10.1073/pnas.0911280106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C., Merkwirth C., Langer T. 2009. Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 122:3823–3830 10.1242/jcs.037655 [DOI] [PubMed] [Google Scholar]
- Osman C., Voelker D.R., Langer T. 2011. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192:7–16 10.1083/jcb.201006159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S., Goldfarb D.S. 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell. 11:2445–2457 10.1091/mbc.11.7.2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.Y., Hoover P.J., Mullins F.M., Bachhawat P., Covington E.D., Raunser S., Walz T., Garcia K.C., Dolmetsch R.E., Lewis R.S. 2009. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 136:876–890 10.1016/j.cell.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretti D., Dahan N., Shimoni E., Hirschberg K., Lev S. 2008. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell. 19:3871–3884 10.1091/mbc.E08-05-0498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mistry A., Ammirati M.J., Chrunyk B.A., Clark R.W., Cong Y., Culp J.S., Danley D.E., Freeman T.B., Geoghegan K.F., et al. 2007. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat. Struct. Mol. Biol. 14:106–113 10.1038/nsmb1197 [DOI] [PubMed] [Google Scholar]
- Raturi A., Simmen T. 2013. Where the endoplasmic reticulum and the mitochondrion tie the knot: the mitochondria-associated membrane (MAM). Biochim. Biophys. Acta. 1833:213–224 10.1016/j.bbamcr.2012.04.013 [DOI] [PubMed] [Google Scholar]
- Rebbeck R.T., Karunasekara Y., Gallant E.M., Board P.G., Beard N.A., Casarotto M.G., Dulhunty A.F. 2011. The β(1a) subunit of the skeletal DHPR binds to skeletal RyR1 and activates the channel via its 35-residue C-terminal tail. Biophys. J. 100:922–930 10.1016/j.bpj.2011.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof W.R., Wu W.I., Jones J.L., Nikrad M., Chan M.M., Loewen C.J., Voelker D.R. 2014. An assembly of proteins and lipid domains regulates transport of phosphatidylserine to phosphatidylserine decarboxylase 2 in Saccharomyces cerevisiae. J. Biol. Chem. 289:5809–5819 10.1074/jbc.M113.518217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R., Pinton P., Carrington W., Fay F.S., Fogarty K.E., Lifshitz L.M., Tuft R.A., Pozzan T. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 280:1763–1766 10.1126/science.280.5370.1763 [DOI] [PubMed] [Google Scholar]
- Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., Neefjes J. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 185:1209–1225 10.1083/jcb.200811005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampieri A., Zepeda A., Asanov A., Vaca L. 2009. Visualizing the store-operated channel complex assembly in real time: identification of SERCA2 as a new member. Cell Calcium. 45:439–446 10.1016/j.ceca.2009.02.010 [DOI] [PubMed] [Google Scholar]
- Schaaf G., Ortlund E.A., Tyeryar K.R., Mousley C.J., Ile K.E., Garrett T.A., Ren J., Woolls M.J., Raetz C.R., Redinbo M.R., Bankaitis V.A. 2008. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol. Cell. 29:191–206 10.1016/j.molcel.2007.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Watson R., Giannakou M.E., Clarke M., Warren G., Barr F.A. 1999. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18:4949–4960 10.1093/emboj/18.18.4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopulos P.B., Li G.Y., Plevin M.J., Ames J.B., Ikura M. 2006. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: An initiation mechanism for capacitive Ca2+ entry. J. Biol. Chem. 281:35855–35862 10.1074/jbc.M608247200 [DOI] [PubMed] [Google Scholar]
- Stathopulos P.B., Zheng L., Li G.Y., Plevin M.J., Ikura M. 2008. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell. 135:110–122 10.1016/j.cell.2008.08.006 [DOI] [PubMed] [Google Scholar]
- Stefan C.J., Manford A.G., Baird D., Yamada-Hanff J., Mao Y., Emr S.D. 2011. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 144:389–401 10.1016/j.cell.2010.12.034 [DOI] [PubMed] [Google Scholar]
- Stefan C.J., Manford A.G., Emr S.D. 2013. ER-PM connections: sites of information transfer and inter-organelle communication. Curr. Opin. Cell Biol. 25:434–442 10.1016/j.ceb.2013.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne T.C., Zhou C., Boldogh I.R., Charalel J.K., McFaline-Figueroa J.R., Thoms S., Yang C., Leung G., McInnes J., Erdmann R., Pon L.A. 2011. Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast. Curr. Biol. 21:1994–1999 10.1016/j.cub.2011.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G., Bianchi K., Várnai P., De Stefani D., Wieckowski M.R., Cavagna D., Nagy A.I., Balla T., Rizzuto R. 2006. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 175:901–911 10.1083/jcb.200608073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H., Komazaki S., Nishi M., Iino M., Kangawa K. 2000. Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell. 6:11–22 [DOI] [PubMed] [Google Scholar]
- Tamura Y., Onguka O., Hobbs A.E., Jensen R.E., Iijima M., Claypool S.M., Sesaki H. 2012. Role for two conserved intermembrane space proteins, Ups1p and Ups2p, [corrected] in intra-mitochondrial phospholipid trafficking. J. Biol. Chem. 287:15205–15218 10.1074/jbc.M111.338665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T., Ozbalci C., Brügger B., Rapaport D., Dimmer K.S. 2013. Mcp1 and Mcp2, two novel proteins involved in mitochondrial lipid homeostasis. J. Cell Sci. 126:3563–3574 10.1242/jcs.121244 [DOI] [PubMed] [Google Scholar]
- Tang D., Wang Y. 2013. Cell cycle regulation of Golgi membrane dynamics. Trends Cell Biol. 23:296–304 10.1016/j.tcb.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoli S., Chao J.T., Young B.P., Cox R.C., Prinz W.A., de Kroon A.I., Loewen C.J. 2013. Plasma membrane—endoplasmic reticulum contact sites regulate phosphatidylcholine synthesis. EMBO Rep. 14:434–440 10.1038/embor.2013.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J., Yang H., Yang H., Eom S.H., Im Y.J. 2013. Structure of Osh3 reveals a conserved mode of phosphoinositide binding in oxysterol-binding proteins. Structure. 21:1203–1213 10.1016/j.str.2013.05.007 [DOI] [PubMed] [Google Scholar]
- Toulmay A., Prinz W.A. 2012. A conserved membrane-binding domain targets proteins to organelle contact sites. J. Cell Sci. 125:49–58 10.1242/jcs.085118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M., Bohnert M., Wiedemann N., Pfanner N. 2012. Role of MINOS in mitochondrial membrane architecture and biogenesis. Trends Cell Biol. 22:185–192 10.1016/j.tcb.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Vance J.E. 2014. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim. Biophys. Acta. 1841:595–609 10.1016/j.bbalip.2013.11.014 [DOI] [PubMed] [Google Scholar]
- von der Malsburg K., Müller J.M., Bohnert M., Oeljeklaus S., Kwiatkowska P., Becker T., Loniewska-Lwowska A., Wiese S., Rao S., Milenkovic D., et al. 2011. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell. 21:694–707 10.1016/j.devcel.2011.08.026 [DOI] [PubMed] [Google Scholar]
- Voss C., Lahiri S., Young B.P., Loewen C.J., Prinz W.A. 2012. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J. Cell Sci. 125:4791–4799 10.1242/jcs.105635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Deng X., Zhou Y., Hendron E., Mancarella S., Ritchie M.F., Tang X.D., Baba Y., Kurosaki T., Mori Y., et al. 2009. STIM protein coupling in the activation of Orai channels. Proc. Natl. Acad. Sci. USA. 106:7391–7396 10.1073/pnas.0900293106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Benning C. 2012. Chloroplast lipid synthesis and lipid trafficking through ER-plastid membrane contact sites. Biochem. Soc. Trans. 40:457–463 10.1042/BST20110752 [DOI] [PubMed] [Google Scholar]
- Weber-Boyvat M., Zhong W., Yan D., Olkkonen V.M. 2013. Oxysterol-binding proteins: functions in cell regulation beyond lipid metabolism. Biochem. Pharmacol. 86:89–95 10.1016/j.bcp.2013.02.016 [DOI] [PubMed] [Google Scholar]
- Wu M.M., Buchanan J., Luik R.M., Lewis R.S. 2006. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 174:803–813 10.1083/jcb.200604014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Wang Y. 2010. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J. Cell Biol. 188:237–251 10.1083/jcb.200907132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Zhang S.O., Cole R.A., McKinney S.A., Guo F., Haas J.T., Bobba S., Farese R.V.J., Jr, Mak H.Y. 2012. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 198:895–911 10.1083/jcb.201201139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbes R.M., van der Klei I.J., Veenhuis M., Pfanner N., van der Laan M., Bohnert M. 2012. Mitofilin complexes: conserved organizers of mitochondrial membrane architecture. Biol. Chem. 393:1247–1261 10.1515/hsz-2012-0239 [DOI] [PubMed] [Google Scholar]
- Zhang L., Yan F., Zhang S., Lei D., Charles M.A., Cavigiolio G., Oda M., Krauss R.M., Weisgraber K.H., Rye K.A., et al. 2012. Structural basis of transfer between lipoproteins by cholesteryl ester transfer protein. Nat. Chem. Biol. 8:342–349 10.1038/nchembio.796 [DOI] [PMC free article] [PubMed] [Google Scholar]