Abstract
Gentamicin doses of 2 and 10 μg/ml were bactericidal against 64% and 100%, respectively, of gentamicin-susceptible KPC-2-producing Klebsiella pneumoniae strains. Treatment with the combination of doripenem (8 μg/ml) plus colistin (2 μg/ml) was inferior to treatment with gentamicin (2 μg/ml), doripenem-gentamicin, gentamicin-colistin, and doripenem-gentamicin-colistin against strains with glycine and aspartic acid insertions in OpmK36 porin at amino acid (aa) positions 134 and 135 (n = 9). Doripenem-colistin was comparable to other 2- or 3-drug regimens and superior to single drugs against wild-type/minor ompK36 mutants (n = 5). An algorithm incorporating ompK36 genotypes and susceptibility to gentamicin and doripenem may predict antimicrobial activity against KPC-producing K. pneumoniae.
TEXT
Carbapenem-resistant Klebsiella pneumoniae strains have emerged as major nosocomial pathogens capable of causing infections that are generally unresponsive to conventional antimicrobial therapy and are associated with high mortality rates (1, 2). Observational studies from our center and others suggest that outcomes are improved with carbapenem-containing combination regimens (3). However, these findings have not been validated in clinical trials, and the optimal combinations have not been defined.
Carbapenem resistance is mediated through several mechanisms, including the production of metallo-β-lactamases and non-metallo-carbapenemases (such as Klebsiella pneumoniae carbapenemase [KPC] and OXA-type carbapenemase), with or without disturbances of outer membrane proteins (OMPs), such as porins. KPC subtype 2 (KPC-2)-producing sequence type 258 (ST258) K. pneumoniae strains predominate in U.S. hospitals and have spread worldwide. At our center and many others, ST258 K. pneumoniae strains carry a mutant ompK35 porin gene, which results in a premature stop codon at amino acid (aa) position 89 (STOP-aa89) (4–6). We previously demonstrated that strains with the mutant ompK35 gene and a wild-type or minor mutant ompK36 gene were highly susceptible to the combination of doripenem and colistin (DOR+COL) in time-kill assays in vitro. In contrast, DOR+COL was inactive against strains carrying the mutant ompK35 and major ompK36 mutations, such as an IS5 insertion in the promoter and a 6-bp insertion that encodes glycine and aspartic acid at amino acid positions 134 and 135 (ins aa134-135GD) (7). The ins aa134-135GD mutation is particularly important since the mutated site falls within a transmembrane β-strand loop 3 that constitutes the porin channel eyelet. The ins aa134-135GD mutant strains exhibit diminished carbapenem uptake due to porin channel constriction (8).
In an earlier time-kill study, we demonstrated that treatments with doripenem and gentamicin (DOR+GENT) were ineffective against GENT-resistant KPC-producing K. pneumoniae (KPC-K. pneumoniae) strains (9). At our center, however, ∼60% of ST258 KPC-K. pneumoniae strains are GENT susceptible (6). The objectives of this study were to compare the in vitro activities of GENT, DOR+GENT, DOR+COL, GENT+COL, and DOR+GENT+COL against 14 GENT-susceptible ST258 KPC-2 K. pneumoniae strains with various ompK36 genotypes. In a previous study, the strains were shown to be indistinguishable by pulsed-field gel electrophoresis typing (6).
The resistance mechanisms for the strains were assessed by PCR and DNA sequencing, and the MICs of each agent were determined by broth microdilution (10–15) (Table 1). Each strain harbored the STOP-aa89 ompK35 mutation, blaKPC-2, blaSHV-12, and blaTEM-1 but was negative for blaCTX-M, blaIMP, blaNDM, blaVIM, blaOXA-48, and blaAmpC (blaACT-1, blaACC, blaBIL-1, blaCMY, blaDHA, blaFOX, blaLAT, blaMIR-1, and blaMOX). Sixty-four percent (9/14), 21% (3/14), and 14% (2/14) of the strains had ins aa134-135GD ompK36 mutations, wild-type ompK36, or minor ompK36 mutations (a guanine insertion at nucleotide [nt] position 382 [ins nt382G], and an asparagine-asparagine-threonine-glutamic acid [NNTE] deletion at amino acid positions 84 to 87 [del aa84-87NNTE]), respectively. Since we previously showed that DOR+COL responses were similar among ins nt382G del aa84-87NNTE ompK36 mutants and wild-type ompK36 strains, we considered them together in this study (7). Interpretive breakpoints for colistin against Enterobacteriaceae have not been established by the CLSI, but we considered 21% (3/14) of the strains to be resistant (MICs, >2 μg/ml).
TABLE 1.
Killing activity of DOR, GENT, and COL alone or in combinations against 14 GENT-susceptible KPC-K. pneumoniae clinical strains
| Strain | MIC (μg/ml) of: |
ompK36 genotype | Killing activity (log10 CFU/ml)a of: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DOR | GENT | COL | DOR | GENT (10 μg/ml) | GENT (2 μg/ml) | COL | DOR+COL | DOR+GENTb | GENT+COL | DOR+GENT+COL | ||
| 216 | 4 | 0.5 | 8 | WTc | 3.71 | −6.08 | 1.19 | 3.91 | 0.72 | −6.09 | −6.11 | −6.11 |
| 347 | 8 | 2 | 0.25 | WT | 3.90 | −6.03 | −3.95 | 2.07 | −4.57 | −6.06 | −3.74 | −6.09 |
| 709 | 8 | 0.5 | 0.5 | WT | 4.06 | −5.90 | −5.88 | −5.86 | −5.87 | −5.92 | −5.92 | −5.89 |
| 539 | 32 | 2 | 1 | ins 382G | 3.71 | −6.02 | 1.02 | −1.00 | −6.08 | 2.10 | −6.13 | −6.10 |
| 41 | 2 | 1 | 0.25 | del 84-87NNTE | 3.68 | −6.05 | −4.79 | 3.60 | −6.03 | −6.02 | −6.05 | −6.03 |
| 115 | 32 | 0.5 | 0.125 | ins aa134-135GD | 3.93 | −6.04 | −6.14 | 3.75 | 3.65 | −6.14 | −6.15 | −6.16 |
| 743 | 64 | 0.5 | 0.25 | ins aa134-135GD | 4.25 | −5.71 | 1.73 | 1.73 | −2.81 | −3.86 | −5.77 | −5.75 |
| 155 | 32 | 1 | 0.25 | ins aa134-135GD | 3.70 | −6.08 | −6.08 | 1.68 | 2.77 | −6.10 | −6.05 | −6.09 |
| 484 | 128 | 1 | 0.25 | ins aa134-135GD | 3.95 | −6.02 | −4.03 | 1.48 | 2.15 | −4.08 | −6.02 | −6.02 |
| 807 | 64 | 0.5 | 0.5 | ins aa134-135GD | 3.93 | −6.01 | −6.05 | 3.89 | 3.83 | −6.10 | −6.05 | −6.06 |
| 615 | 128 | 2 | 0.5 | ins aa134-135GD | 3.86 | −6.06 | 3.92 | 3.93 | 3.87 | 3.69 | −2.62 | −0.68 |
| 436 | 32 | 2 | 0.5 | ins aa134-135GD | 3.82 | −6.08 | −6.11 | 3.79 | 3.78 | −6.11 | −6.09 | −6.10 |
| 669 | 64 | 1 | 4 | ins aa134-135GD | 3.75 | −6.08 | −2.01 | 1.09 | −0.08 | −1.68 | −6.08 | −6.05 |
| 184 | 32 | 0.5 | 8 | ins aa134-135GD | 3.83 | −6.08 | −6.12 | 3.96 | 3.80 | −6.11 | −6.09 | −6.13 |
Killing activity was defined as the difference between the log10 concentration of KPC-K. pneumoniae after 24 h of incubation with drug(s) and the log10 of starting inoculum. Positive numbers denote growth of strains compared with starting inoculum.
Gentamicin concentration was 2 μg/ml during combination testing.
WT, wild type.
We performed standard time-kill assays for DOR, COL, and GENT as single agents and in two- and three-drug combinations using previously described methods (6, 7). In general, GENT is administered once a day in order to exploit its concentration-dependent bactericidal activity against Gram-negative bacteria. For life-threatening infections, the targeted peak and trough serum concentrations are 8 to 10 and 1 to 2 μg/ml, respectively (16, 17). In single-drug time-kill studies, we first used a GENT concentration of 10 μg/ml. GENT exerted bactericidal activity (≥3-log10 reduction in CFU/ml) within minutes against each of the 14 strains, which were maintained without regrowth through 24 h (Table 1). Since these high-level activities limited our ability to show additional effects with the addition of DOR or COL, we performed subsequent time-kill assays with GENT at 2 μg/ml. DOR (8 μg/ml) and COL (2 μg/ml) concentrations that are achieved in human sera were used in the single-drug and combination studies (7, 9).
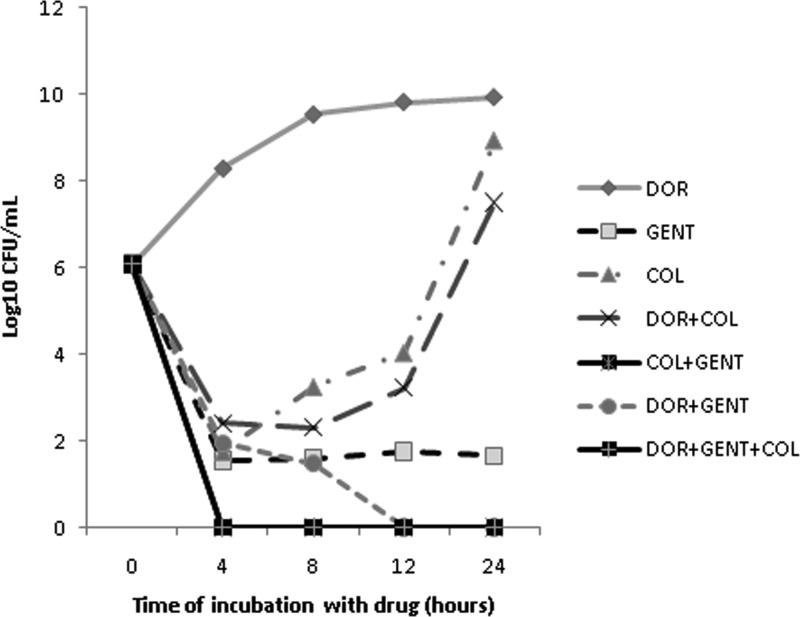
Kill curves showing the median kills by the single drugs and the 2- and 3-drug combinations are presented in Fig. 1. GENT (2 μg/ml) achieved bactericidal activity and complete kills at 24 h against 64% (9/14) and 36% (5/14) of the strains, respectively (Table 1). COL exerted bactericidal activity against only one strain (7%). DOR was not active against any of the strains. To compare the activities of the drugs over the 24-h period, we measured areas under the bactericidal curves (AUBCs); greater killing resulted in lower AUBCs. GENT (median AUBC, 53-log10 CFU/ml · h) was more bactericidal than COL (115-log10 CFU/ml · h; P = 0.0008) and DOR (221.3-log10 CFU/ml · h; P < 0.0001). COL was more bactericidal than DOR (P < 0.0001).
FIG 1.
Kill curves of KPC-K. pneumoniae strains by single drugs and combinations. Data represent the median log10 CFU/ml for specific drug regimens over time. There were no differences in growth at 12 and 24 h between strains incubated with DOR and those with no drug control. Note that the kill curves for COL+GENT and DOR+GENT+COL overlap.
The addition of DOR to GENT or COL improved the rates of bactericidal activity from 64% (9/14) to 79% (11/14) and 7% (1/14) to 29% (4/14), respectively (P = 0.02 for DOR+GENT versus DOR+COL). GENT+COL and DOR+GENT+COL each had bactericidal activity against 93% (13/14) of the strains (P = 0.0013 versus DOR+COL). DOR+GENT, DOR+COL, and GENT+COL were synergistic against 21% (3/14), 36% (5/14), and 36% (5/14) of the strains, respectively (≥2-log10-greater kills at 24 h with the combinations than with the best single agent). DOR+GENT+COL did not provide synergy compared to the most active two-drug combination.
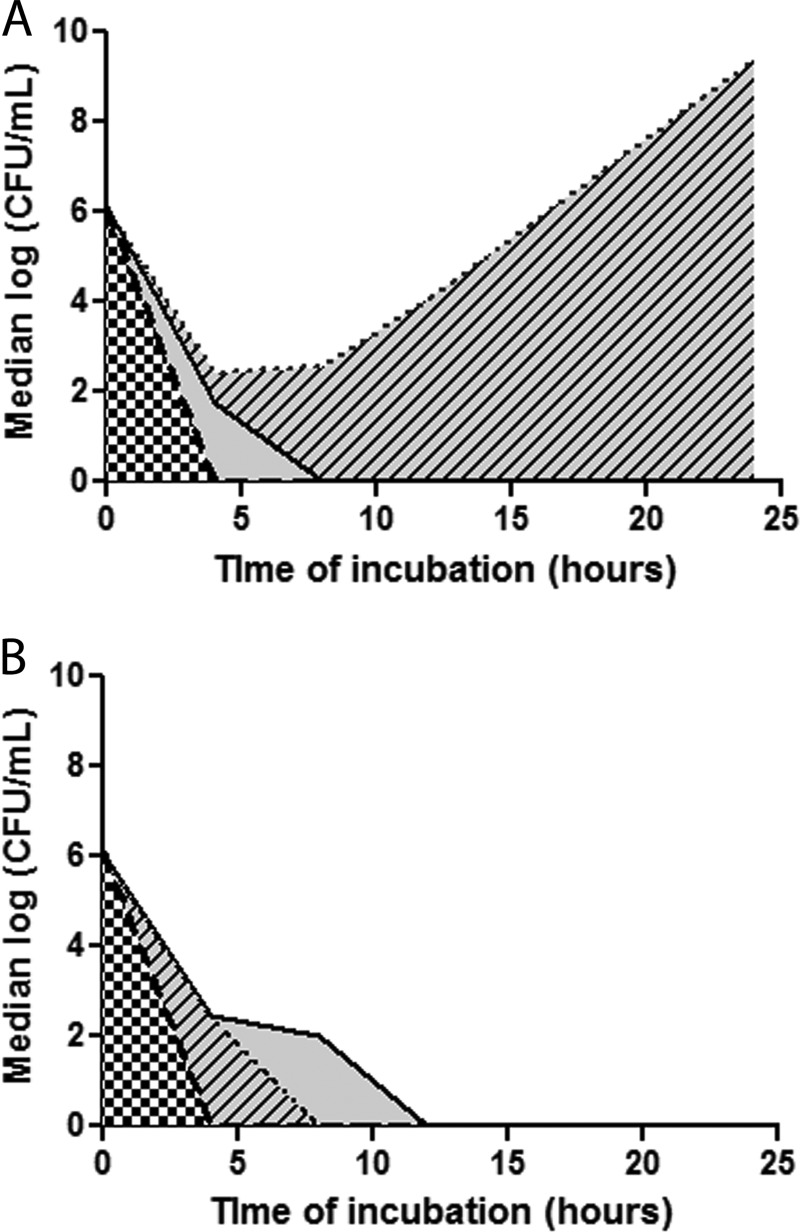
DOR+COL exerted greater bactericidal activity against the wild-type/minor ompK36 mutants than the ins aa134-135GD mutants (P = 0.0002) (Fig. 2 and 3). DOR+COL did not differ significantly from GENT (2 μg/ml) alone, DOR+GENT, GENT+COL, or DOR+GENT+COL against the wild-type/minor ompK36 mutants. However, DOR+GENT, GENT+COL, DOR+GENT+COL, and GENT alone were significantly more bactericidal than DOR+COL against the ins aa134-135GD mutants; for these strains, the activity of DOR+COL did not differ from that of COL alone. Note that the GENT+COL combination was synergistic against 40% (2/5) and 33% (3/9) of the wild-type/minor ompK36 and ins aa134-135GD mutants, respectively (P = 1.0).
FIG 2.
Median AUBCs for 2-drug combinations against KPC-K. pneumoniae isolates, stratified by ompK36 genotypes. Shown are time-kill results against ins aa134-135GD mutants (A) and wild-type/minor mutants (B). Data represent the median log10 CFU/ml for specific combinations over time. The black dotted line represents median time-kills by DOR+COL. The solid black line and the thick broken black lines represent median time-kills by DOR+GENT and GENT+COL, respectively. Therefore, the AUBCs highlighted by black-and-white checkers depict the activity of GENT+COL, and AUBCs highlighted by solid gray and gray with black diagonal lines depict the differences observed with DOR+GENT and DOR+COL, respectively.
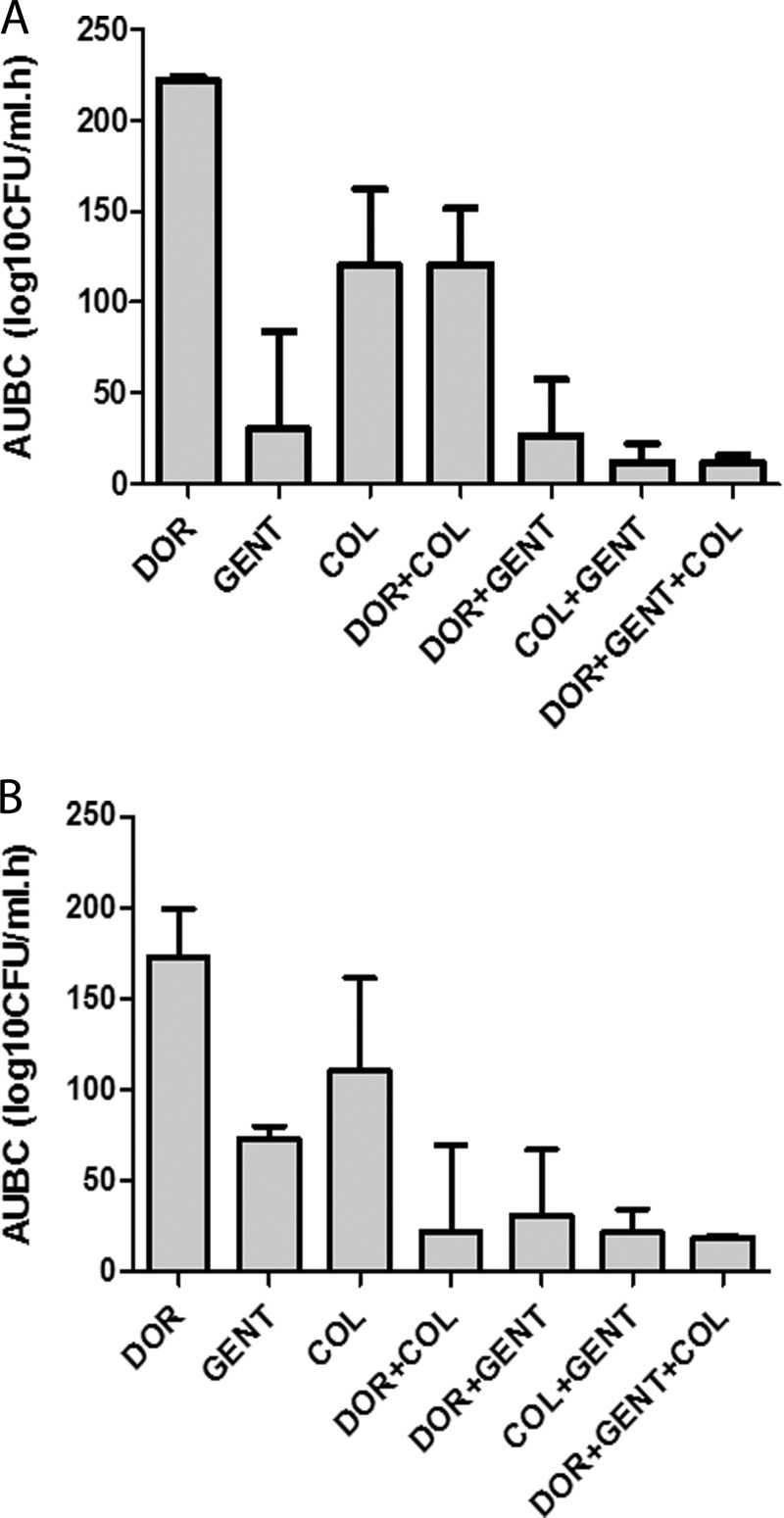
FIG 3.
AUBCs for single drugs and combinations against KPC-K. pneumoniae strains, stratified by ompK36 genotypes. Shown are time-kill results against ins aa134-135GD (A) and wild-type/minor (B) mutants. Data represent median AUBCs with interquartile ranges for specific regimens over time. For ins aa134-135GD mutants, the median AUBCs were higher for DOR+COL than for GENT (P = 0.007), DOR+GENT (P = 0.004), GENT+COL (P = 0.002), and DOR+GENT+COL (P < 0.001). For wild-type/minor mutants, the median AUBCs were not significantly different between DOR+COL, DOR+GENT, GENT+COL, and DOR+GENT+COL.
This study is the latest in a series of studies from our group to identify effective antimicrobial regimens against KPC-K. pneumoniae strains (7, 9, 12). We demonstrated that GENT and GENT-containing combination regimens were highly effective in time-kill assays against GENT-susceptible KPC-2-producing ST258 strains. GENT concentrations of 10 and 2 μg/ml were rapidly bactericidal against 100% (14/14) and 63% (9/14) of the strains, respectively. The activity of GENT as a single agent limited our ability to demonstrate positive interactions with other drugs. Nevertheless, DOR+GENT (2 μg/ml) was synergistic against 3 strains, achieved bactericidal rather than bacteriostatic activity against 2 strains, and suppressed the regrowth of 2 strains (data not shown). GENT+COL was even more effective than DOR+GENT in vitro, but the clinical utility of this combination may be limited by the nephrotoxicity of both agents. The use of DOR+GENT+COL did not offer advantages over two-drug combinations, which may reflect the strong activity of GENT+COL and the limited activity of DOR by itself. These data suggest that GENT is a useful treatment option against infections caused by GENT-susceptible KPC-K. pneumoniae.
DOR+COL was inferior to GENT alone against GENT-susceptible ins aa134-135GD ompK36 mutant strains in vitro. Since porin channel constriction resulting from the ins aa134-135GD mutation results in diminished carbapenem uptake, DOR+GENT is unlikely to offer much advantage over GENT alone in treating infections by these strains. For strains with minor ompK36 mutations or wild-type ompK36, DOR+COL, GENT, and DOR+GENT were equally bactericidal in the time-kill studies. Therefore, the DOR+COL combination remains an option for treating infections caused by these strains. In such cases, we have insufficient data to conclude that GENT, DOR+GENT, or DOR+COL is superior. We feel that DOR+GENT is the preferred regimen for two reasons. First, the combination may achieve synergy against at least some GENT-susceptible wild-type/minor mutant ompK36 strains. Second, GENT by itself exerts significantly greater activity against GENT-susceptible strains than does COL. However, clinicians may consider DOR+COL for the treatment of infections in which GENT is potentially limited by pharmacokinetic considerations. In diseases like pneumonia or in cases of abscesses, for example, the efficacy of GENT may be compromised by suboptimal penetration into infected lung tissue or reduced activity at an acidic pH, respectively (18, 19). In certain cases of severe infection, clinicians may choose to initiate treatment with DOR+GENT+COL as they await MIC data. This strategy will increase the likelihood that both GENT-susceptible and GENT-resistant strains are covered. GENT or COL can be discontinued when data become available, which may ameliorate the increased risk of nephrotoxicity with the agents combined.
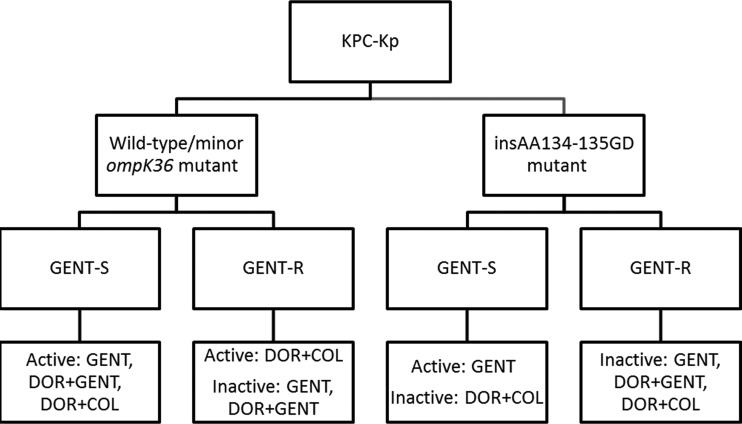
Considering the current data and our previous findings that GENT and DOR+GENT were ineffective against GENT-resistant KPC-2-producing ST258 K. pneumoniae strains (9), we propose a paradigm that uses GENT susceptibility data and ompK36 genotypes to identify antimicrobial combinations that are likely to be active (Fig. 4). DOR MICs may be incorporated into the paradigm as proxies for ompK36 genotypes, since ompK36 genotypes determine the level of carbapenem resistance. Our data must be corroborated and extended using KPC-K. pneumoniae strains with various genetic backgrounds from different centers and geographical locations. All in vitro findings should be validated in animal models of KPC-K. pneumoniae infections as a prelude to testing treatment paradigms in human clinical trials. The present study and others from our group further support the development of molecular assays that can rapidly identify the antimicrobial regimens that are best-suited to treat infections due to KPC-K. pneumoniae or other extensively resistant bacteria. Improved outcomes among patients with these infections will require multifaceted strategies that integrate advances in molecular biology and informatics, optimized pharmacokinetics and pharmacodynamics of antimicrobials, new drugs, and judicious antimicrobial usage.
FIG 4.
Proposed algorithm for predicting active antimicrobial regimens against KPC-K. pneumoniae strains. A doripenem MIC of ≤8 μg/ml can be used as a proxy for the presence of wild-type/minor ompK36 mutation, and a doripenem MIC of >8 μg/ml can be used as a proxy for the presence of an ins aa134-135GD mutation. DOR+GENT may offer synergy against some GENT-susceptible wild-type/minor ompK36 mutant strains, but it is not likely to offer synergy over GENT alone against GENT-susceptible ins aa134-135GD mutant strains, since constriction of the porin channel will restrict DOR uptake.
ACKNOWLEDGMENTS
This study was fully supported by the University of Pittsburgh Medical Center XDR Pathogen Laboratory.
R.K.S. is supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award KL2TR000146.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print 24 February 2014
REFERENCES
- 1.Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, Schlaeffer F, Sherf M. 2009. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect. Control Hosp. Epidemiol. 30:972–976. 10.1086/605922 [DOI] [PubMed] [Google Scholar]
- 2.Bratu S, Landman D, Haag R, Recco R, Eramo A, Alam M, Quale J. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch. Intern. Med. 165:1430–1435. 10.1001/archinte.165.12.1430 [DOI] [PubMed] [Google Scholar]
- 3.Qureshi ZA, Paterson DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E, Polsky B, Adams-Haduch JM, Doi Y. 2012. Treatment outcome of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of combination antimicrobial regimens. Antimicrob. Agents Chemother. 56:2108-2113. 10.1128/AAC.06268-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domenech-Sanchez A, Martinez-Martinez L, Hernandez-Alles S, del Carmen Conejo M, Pascual A, Tomas JM, Alberti S, Benedi VJ. 2003. Role of Klebsiella pneumoniae OmpK35 porin in antimicrobial resistance. Antimicrob. Agents Chemother. 47:3332–3335. 10.1128/AAC.47.10.3332-3335.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landman D, Bratu S, Quale J. 2009. Contribution of OmpK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J. Med. Microbiol. 58:1303–1308. 10.1099/jmm.0.012575-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clancy CJ, Chen L, Shields RK, Zhao Y, Cheng S, Chavda KD, Hao B, Hong JH, Doi Y, Kwak EJ, Silveira FP, Abdel-Massih R, Bogdanovich T, Humar A, Perlin DS, Kreiswirth BN, Nguyen MH. 2013. Epidemiology and molecular characterization of bacteremia due to carbapenem-resistant Klebsiella pneumoniae in transplant recipients. Am. J. Transplant. 13:2619-2633. 10.1111/ajt.12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clancy CJ, Hong JH, Cheng S, Hao B, Shields RK, Farrell AN, Doi Y, Zhao Y, Perlin DS, Kreiswirth BN, Nguyen MH. 2013. Mutations of the ompK36 porin gene and promoter impact responses of ST258, KPC-2-producing Klebsiella pneumoniae strains to doripenem and doripenem-colistin. Antimicrob. Agents Chemother. 57:5258-5265. 10.1128/AAC.01069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberti S, Rodriquez-Quinones F, Schirmer T, Rummel G, Tomas JM, Rosenbusch JP, Benedi VJ. 1995. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional model, and complement binding. Infect. Immun. 63:903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jernigan MG, Press EG, Nguyen MH, Clancy CJ, Shields RK. 2012. The combination of doripenem and colistin is bactericidal and synergistic against colistin-resistant, carbapenemase-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 56:3395–3398. 10.1128/AAC.06364-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI M100–S22. CLSI, Wayne, PA [Google Scholar]
- 11.Chen L, Chavda KD, Mediavilla JR, Zhao Y, Fraimow HS, Jenkins SG, Levi MH, Hong T, Rojtman AD, Ginocchio CC, Bonomo RA, Kreiswirth BN. 2012. Multiplex real-time PCR for detection of an epidemic KPC-producing Klebsiella pneumoniae ST258 clone. Antimicrob. Agents Chemother. 56:3444–3447. 10.1128/AAC.00316-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong JH, Clancy CJ, Cheng S, Shields RK, Chen L, Doi Y, Zhao Y, Perlin DS, Kreiswirth BN, Nguyen MH. 2013. Characterization of porin expression in Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae identifies isolates most susceptible to the combination of colistin and carbapenems. Antimicrob. Agents Chemother. 57:2147-2153. 10.1128/AAC.02411-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- 14.Essack SY, Hall LM, Pillay DG, McFadyen ML, Livermore DM. 2001. Complexity and diversity of Klebsiella pneumoniae strains with extended-spectrum beta-lactamases isolated in 1994 and 1996 at a teaching hospital in Durban, South Africa. Antimicrob. Agents Chemother. 45:88–95. 10.1128/AAC.45.1.88-95.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 16.Thomson AH, Campbell KC, Kelman AW. 1990. Evaluation of six gentamicin nomograms using a Bayesian parameter estimation program. Ther. Drug Monit. 12:258–263. 10.1097/00007691-199005000-00008 [DOI] [PubMed] [Google Scholar]
- 17.Thomson AH, Duncan N, Silverstein B, Alcock S, Jodrell D. 1996. Antimicrobial practice: development of guidelines for gentamicin dosing. J. Antimicrob. Chemother. 38:885–893. 10.1093/jac/38.5.885 [DOI] [PubMed] [Google Scholar]
- 18.Vaudaux P. 1981. Peripheral inactivation of gentamicin. J. Antimicrob. Chemother. 8(Suppl A):17–25 [DOI] [PubMed] [Google Scholar]
- 19.Panidis D, Markantonis SL, Boutzouka E, Karatzas S, Baltopoulos G. 2005. Penetration of gentamicin into the alveolar lining fluid of critically ill patients with ventilator-associated pneumonia. Chest 128:545–552. 10.1378/chest.128.2.545 [DOI] [PubMed] [Google Scholar]