Abstract
We developed a multiplex PCR assay capable of identifying two capsular polysaccharide synthesis sequence types (sequence type 258 [ST258] cps-1 and cps-2) in epidemic Klebsiella pneumoniae ST258 strains. The assay performed with excellent sensitivity (100%) and specificity (100%) for identifying cps types in 60 ST258 K. pneumoniae sequenced isolates. The screening of 419 ST258 clonal isolates revealed a significant association between cps type and K. pneumoniae carbapenemase (KPC) variant: cps-1 is largely associated with KPC-2, while cps-2 is primarily associated with KPC-3.
TEXT
Carbapenem-resistant Enterobacteriaceae (CRE), especially Klebsiella pneumoniae carbapenemase (KPC)-harboring K. pneumoniae, are rapidly emerging as significant international public health concerns (1–3). Although KPCs have been found in numerous K. pneumoniae clones as well as in other Gram-negative species, a significant proportion of the global KPC burden is associated with a single K. pneumoniae multilocus sequence type (ST), ST258 (1–3). Currently, little is known about the genomic markers of this epidemiologically successful clone, and the genetic determinants of its spread, colonization, and disease progression remain unclear. Recent comparative genomic studies revealed two ST258 sublineages with two distinct capsular polysaccharide gene (cps) clusters (4). Here, we describe a multiplex PCR assay used to distinguish the two cps regions in KPC-harboring K. pneumoniae ST258 strains, providing a novel molecular tool for tracking their dissemination and studying their epidemiology.
We recently sequenced the genomes of 85 ST258 K. pneumoniae strains and single-locus variants (SLV) (ST379, ST418, and ST512) using Illumina HiSeq and Roche 454 GS-FLX platforms; two ST258 genomes were completely closed (those of K. pneumoniae strains NJST258_1 and NJST258_2) (4). One epidemiologically important finding was that ST258 strains carry two distinct cps gene clusters. One cps gene cluster is highly similar to that from a KPC-producing K. pneumoniae strain, Kp13 (cpsKp13), an ST442 clinical isolate obtained from a patient in southern Brazil in 2009 (5, 6). The second cps gene cluster is novel, harboring several unique genes compared to others in GenBank (ncbi.nlm.nih.gov/GenBank/). We propose to name this novel cps gene cluster ST258 cps-1 and the cpsKp13-like cluster ST258 cps-2. The capsular polysaccharide is one of the major determinants of antigenic variation observed in K. pneumoniae and contributes to its success as a pathogen. Our preliminary human neutrophil studies revealed little or no killing effects between cps-1- and cps-2-harboring ST258 isolates (4), supporting the notion that the capsular variation observed in ST258 strains is likely involved in the survival of the pathogen and its ability to escape the host immune system. In this study, the sequences of ST258 cps-1 and cps-2 from two representative strains (K. pneumoniae BK30660 and BK32192) were generated by de novo assembly of the Illumina HiSeq reads using the CLC Genomics Workbench software (version 5.5.1; CLC bio, Aarhus, Denmark).
The multiplex PCR scheme to identify and distinguish the two ST258 cps types comprises three primer pairs. Primer pairs I and II were designed to target the K. pneumoniae capsular polymerase gene (wyz-I and wyz-II) in ST258 cps-1 and cps-2, respectively (Fig. 1A and Table 1). The third primer pair was developed to detect the K. pneumoniae species-specific Klebsiella hemolysin gene (khe) (7) and provides an internal positive control to confirm DNA template quality and PCR efficiency (Table 1).
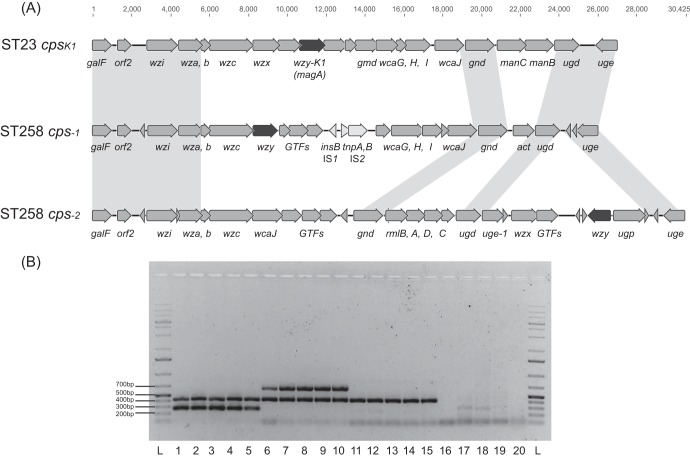
FIG 1.
(A) cps region genetic structures of ST258 cps-1 (GenBank accession no. KF793262), ST258 cps-2 (GenBank accession no. KF793263), and cpsK1 (from ST23 strain K. pneumoniae NTUH-K2044, GenBank accession no. AB198423). Gray shading denotes shared regions of homology of >95% nucleotide similarity. Open reading frames are portrayed by arrows, and the K-antigen polymerase gene, wzy, is highlighted in black. (B) Examples of ST258 cps multiplex PCR. Lanes L, ladder; lanes 1 to 5, five cps-1-carrying ST258 K. pneumoniae isolates; lanes 6 to 10, five cps-2-carrying ST258 K. pneumoniae isolates; lanes 11 to 15, five non-ST258 K. pneumoniae isolates; lanes 16 to 20, five non-K. pneumoniae Gram-negative isolates (K. oxytoca, E. coli, E. cloacae, Citrobacter freundii, and P. mirabilis).
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′–3′) | Length (bp) | Target |
|---|---|---|---|
| wzy258-I-f | TACGGGGATTCCGGGAACAGCA | 269 | Gene wzy in ST258 cps-1 |
| wzy258-I-r | ACAAAACCTCAATTGCTCTTCGGCT | ||
| wzy258-II-f | GCACAAGAGAAATTGGATCTGACAACG | 638 | Gene wzy in ST258 cps-2 |
| wzy258-II-r | ACTTCCAAATCCCATTGCAACTGCT | ||
| khe-f | ACCATGTCCGATTTAATCACAACACGC | 423 | K. pneumoniae-specific gene khe (KPHS_02720 in HS11286) |
| khe-r | GCAGACGAACTTCCTGCTCGGT |
The DNA template was isolated from bacterial colonies using the boiling lysis method described elsewhere (8). Multiplex PCR was performed in 15-μl reaction volumes consisting of 0.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), 2 mM MgCl2, 250 μM each deoxynucleoside triphosphate (dNTP), 0.1 μM primer for khe-f and khe-r, 0.2 μM primer for ST258wzy-I-f and ST258wzy-I-r, and 0.4 μM primer for ST258wzy-II-f and ST258wzy-II-r in 1× PCR buffer (Applied Biosystems). The optimal cycling conditions were as follows: an initial denaturation step of 95°C for 10 min, followed by 35 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 1 min, and a final extension step of 72°C for 5 min. Amplicons were visualized after running at 120 V for 1.5 h on 1% agarose gels containing GelStar nucleic acid gel stain (Ionza Ltd., Allendale, NJ) (Fig. 1B).
This multiplex PCR assay was initially validated using 60 ST258 clonal group K. pneumoniae strains that had been characterized by whole-genome sequencing (WGS) (4). The cps gene clusters of these 60 isolates were evaluated by de novo and reference assembly against sequences of ST258 cps-1 and cps-2. Among them, 28 isolates carried cps-1, while the remaining 32 harbored cps-2. The K. pneumoniae-specific khe target was amplified for all 60 isolates, generating a 423-bp amplification product. Twenty-eight ST258 cps-1 isolates were each positive for a 269-bp amplicon, while 32 ST258 cps-2 isolates were all positive for a 638-bp amplicon, and there was 100% concordance with the WGS results. The assay showed excellent sensitivity (100%) and specificity (100%) for identifying cps-1 and cps-2 in this collection of 60 ST258 K. pneumoniae isolates.
This multiplex PCR was further evaluated using 120 Gram-negative and Gram-positive clinical isolates comprising a wide variety of different species and K. pneumoniae sequence types, selected from our strain collection at the Public Health Research Institute (PHRI) Tuberculosis Center. Among them, 55 non-ST258 K. pneumoniae isolates comprising 40 different STs were selected from our previous study (9). The remaining 65 isolates included the most common clinical Gram-negative and Gram-positive species: the Gram-negative bacterial isolates included Acinetobacter spp. (n = 5), Citrobacter spp. (n = 4), Enterobacter spp. (n = 8), Escherichia coli (n = 10), Klebsiella oxytoca (n = 2), Morganella morganii (n = 2), Proteus mirabilis (n = 2), Pseudomonas spp. (n = 5), Salmonella spp. (n = 2), Serratia spp. (n = 2), and Shigella sp. (n = 1); the Gram-positive isolates included Enterococcus spp. (n = 4), Staphylococcus spp. (n = 8), Streptococcus spp. (n = 8), and Micrococcus spp. (n = 2).
All non-ST258 K. pneumoniae isolates were khe positive by multiplex PCR, while all other Gram-negative and Gram-positive isolates were negative for the khe target, demonstrating both its sensitivity and specificity for identifying and distinguishing K. pneumoniae from other organisms. Among the 55 non-ST258 K. pneumoniae isolates, 54 were PCR positive only for khe, as the ST258 cps-1 and cps-2 primers failed to generate PCR amplicons; one ST42 K. pneumoniae isolate from a hospital in New York in 2001 produced a PCR amplicon for ST258 cps-1. The wyz and wzi genes of this isolate were further amplified (wyz1-f, TGGCCTTAGCAATACCAACT; wzy1-r, TTTTCATTTCTATATATTTTTCATCAATGC; wzi-f4, TAACAAGCAGCTTAGGGTAA; wzi-r, TTCTTCATAATGTCACATCAT), and the sequences of the two genes showed 100% identity compared with the ST258 cps-1 genes in strain BK32192, strongly suggesting that this ST42 isolate harbors the same ST258 cps-1 region. This finding indicates that ST258 cps regions are not limited to ST258 and may be found in other K. pneumoniae backgrounds.
Following validation of the multiplex PCR assay, an additional 327 K. pneumoniae ST258 clonal strains from 10 hospitals in New York and New Jersey were also screened by this multiplex PCR method. As part of an ongoing surveillance project, hospitals in New York and New Jersey routinely submit carbapenem-resistant and -susceptible Enterobacteriaceae to our laboratory for genotyping. The K. pneumoniae ST258 clonal type and blaKPC variants of all strains were characterized by two multiplex real-time PCR methods previously developed in our lab (8, 9). Among them, 146 carried blaKPC-2 (44.6%), while 167 harbored blaKPC-3 (51.1%), and the remaining 14 K. pneumoniae strains were negative for blaKPC (4.3%) (Table 2). Among these 327 isolates, 132 (40.4%) carried cps-1, while 186 harbored cps-2. Nine ST258 isolates could not be assigned to either cps-1 or cps-2, strongly suggesting that they harbor either a different cps region or that the cps region is deleted in these strains. There was a significant association between KPC variant and cps type. In this study, 91% of the ST258 cps-1 isolates carried blaKPC-2, which is significantly higher than the proportion of isolates with blaKPC-3 (3.0%) or lacking KPC (6.1%) (P < 0.01 for each versus blaKPC-2). Conversely, 87.1% of the ST258 cps-2 isolates carried blaKPC-3, which is significantly higher than the proportion of isolates with blaKPC-2 (9.7%) (P < 0.01) or lacking KPC (3.2%) (P < 0.01).
TABLE 2.
Distribution of different cps types in clinical ST258 clonal K. pneumoniae isolates
| ST258 cps type | No. (%) of isolates with indicated KPC variant from: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PHRI (United States) |
PHE (United Kingdom) |
CLS (Canada)a |
|||||||||
| KPC-2 | KPC-3 | KPC−b | Total | KPC-2 | KPC-3 | Total | KPC-2 | KPC-3 | KPC-11 | Total | |
| cps-1 | 120 (90.9) | 4 (3.0) | 8 (6.1) | 132 | 34 (100) | 0 (0) | 34 | 13 (68.4) | 0 (0) | 6 (31.6) | 19 |
| cps-2 | 18 (9.7) | 162 (87.1) | 6 (3.2) | 186 | 3 (9.4) | 29 (90.6) | 32 | 0 (0) | 7 (100) | 0 (0) | 7 |
| Non-cps-1 or -2 | 8 (88.9) | 1 (11.1) | 0 (0) | 9 | 0 (0) | 0 (0) | 0 | 0 (0) | 0 (0) | 0 (0) | 0 |
Isolates were from Greece and Israel (10).
KPC−, negative for blaKPC.
Further evaluation of the assay was undertaken at the Antimicrobial Resistance and Healthcare Associated Infections Reference Unit (AMRHAI) of Public Health England (PHE) and Calgary Laboratory Services (CLS) in Canada. The PHE collection includes 92 blaKPC-harboring Enterobacteriaceae isolates, with 56 ST258 and 10 ST512 K. pneumoniae isolates. The K. pneumoniae-specific target khe was amplified from 89 K. pneumoniae strains but not from single isolates of E. coli, K. oxytoca, or Enterobacter cloacae in the test panel. Among the 66 ST258/ST512 isolates, the cps-1 target was identified in 34 isolates, all of which expressed KPC-2, whereas 29 of 32 isolates expressing KPC-3 were positive for cps-2. Ten ST512 isolates, which are SLVs of ST258, expressed KPC-3 and were positive for cps-2. Twenty-three non-ST258 K. pneumoniae isolates and the three other Enterobacteriaceae isolates were all PCR negative for both cps-1 and cps-2. The CLS collection includes 26 ST258 K. pneumoniae isolates from Greece (n = 18) and Israel (n = 8) obtained from the Study for Monitoring Antimicrobial Resistance Trends (SMART) worldwide surveillance program (10). Thirteen isolates harboring KPC-2 or KPC-11 (blaKPC-11 is a single nucleotide variant to blaKPC-2) were all positive for cps-1, whereas the remaining seven KPC-3-bearing isolates were all positive for cps-2, which is consistent with the association described above (Table 2).
In summary, we have developed and validated in three centers a novel multiplex PCR which can rapidly identify the two dominant cps types in epidemiological K. pneumoniae ST258 isolates. A screening study from New Jersey and New York hospitals and analyses at the PHE AMRHAI reference unit and Canada CLS confirmed the robustness of the assay and revealed the wide spread of cps-1- and cps-2-harboring ST258 isolates in geographically distinct regions. The significance of the strong association between KPC-2 and KPC-3 variants with cps-1 and cps-2, respectively, among ST258 K. pneumoniae isolates and our finding that cps-1 resides in an ST42 strain are currently being investigated to better understand the molecular epidemiology and evolution of these spreading carbapenem-resistant strains.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of ST258 cps-1 (from strain BK32192) and cps-2 (from strain BK30660) were deposited as GenBank accession no. KF793262 and KF793263, respectively.
ACKNOWLEDGMENTS
This study was supported by a grant (to B.N.K.) from the National Institutes of Health (1R01AI090155) and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. This work was also supported by Public Health Service grants R01AI072219 and R01AI063517 (to R.A.B.) from the National Institutes of Health and funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program, and the Geriatric Research Education and Clinical Center VISN 10 (to R.A.B.).
The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
B.N.K. discloses that he holds two patents that focus on using DNA sequencing to identify bacterial pathogens.
Footnotes
Published ahead of print 14 April 2014
REFERENCES
- 1.Patel G, Bonomo RA. 2013. “Stormy waters ahead”: global emergence of carbapenemases. Front. Microbiol. 4:48. 10.3389/fmicb.2013.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13:785–796. 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25:682–707. 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLeo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, Chavda KD, Jacob MR, Mathema B, Olsen RJ, Bonomo RA, Musser JM, Kreiswirth BN. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 111:4988–4993. 10.1073/pnas.1321364111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramos PIP, Picão RC, Vespero EC, Pelisson M, Zuleta LFG, Almeida LGP, Gerber AL, Vasconcelos ATR, Gales AC, Nicolás MF. 2012. Pyrosequencing-based analysis reveals a novel capsular gene cluster in a KPC-producing Klebsiella pneumoniae clinical isolate identified in Brazil. BMC Microbiol. 12:173. 10.1186/1471-2180-12-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramos PIP, Picão RC, de Almeida LGP, Lima NCB, Girardello R, Vivan ACP, Xavier DE, Barcellos FG, Pelisson M, Vespero EC, Médigue C, de Vasconcelos ATR, Gales AC, Nicolás MF. 2014. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genomics 15:54. 10.1186/1471-2164-15-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartman LJ, Selby EB, Whitehouse CA, Coyne SR, Jaissle JG, Twenhafel NA, Burke RL, Kulesh DA. 2009. Rapid real-time PCR assays for detection of Klebsiella pneumoniae with the rmpA or magA genes associated with the hypermucoviscosity phenotype: screening of nonhuman primates. J. Mol. Diagn. 11:464–471. 10.2353/jmoldx.2009.080136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (blaKPC) variants. J. Clin. Microbiol. 49:579–585. 10.1128/JCM.01588-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Chavda KD, Mediavilla JR, Zhao Y, Fraimow HS, Jenkins SG, Levi MH, Hong T, Rojtman AD, Ginocchio CC, Bonomo RA, Kreiswirth BN. 2012. Multiplex real-time PCR for detection of an epidemic KPC-producing Klebsiella pneumoniae ST258 clone. Antimicrob. Agents Chemother. 56:3444–3447. 10.1128/AAC.00316-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JDD. 2013. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob. Agents Chemother. 57:130–136. 10.1128/AAC.01686-12 [DOI] [PMC free article] [PubMed] [Google Scholar]