Abstract
Pyrosequencing of the 16S rRNA gene, community-level physiological profiles determined by the use of Biolog EcoPlates, and proteolysis analyses were used to characterize Canestrato Pugliese Protected Designation of Origin (PDO) cheese. The number of presumptive mesophilic lactococci in raw ewes' milk was higher than that of presumptive mesophilic lactobacilli. The numbers of these microbial groups increased during ripening, showing temporal and numerical differences. Urea-PAGE showed limited primary proteolysis, whereas the analysis of the pH 4.6-soluble fraction of the cheese revealed that secondary proteolysis increased mainly from 45 to 75 days of ripening. This agreed with the concentration of free amino acids. Raw ewes' milk was contaminated by several bacterial phyla: Proteobacteria (68%; mainly Pseudomonas), Firmicutes (30%; mainly Carnobacterium and Lactococcus), Bacteroidetes (0.05%), and Actinobacteria (0.02%). Almost the same microbial composition persisted in the curd after molding. From day 1 of ripening onwards, the phylum Firmicutes dominated. Lactococcus dominated throughout ripening, and most of the Lactobacillus species appeared only at 7 or 15 days. At 90 days, Lactococcus (87.2%), Lactobacillus (4.8%; mainly Lactobacillus plantarum and Lactobacillus sakei), and Leuconostoc (3.9%) dominated. The relative utilization of carbon sources by the bacterial community reflected the succession. This study identified strategic phases that characterized the manufacture and ripening of Canestrato Pugliese cheese and established a causal relationship between mesophilic lactobacilli and proteolysis.
INTRODUCTION
Cheeses are the most diverse group of dairy products. Italy is one of the countries with the largest and most diverse production of cheeses, which are made with cows', goats', buffalos', and especially ewes' milk (1). Pecorino is the trivial name given to Italian cheeses made with raw or heated ewes' milk, which are manufactured mainly in Central and South Italy according to ancient and unique techniques. Some of these cheeses are known worldwide because they have the prestigious Protected Designation of Origin (PDO) recognition.
Canestrato Pugliese is an Italian ewes' milk PDO cheese (CEE Regulation 1107/96), which is manufactured in the Apulia region. The cheese derives its name and traditional cylindrical shape from the rush basket, canestro, where the curd is ripened. Raw, whole ewes' milk of 1 or 2 daily milkings is generally used, although pasteurized milk may also be processed. The addition of natural whey cultures is facultative, and liquid or powdered calf rennet is used. Dry salting lasts 1 to 2 days, and during ripening (3 to 12 months) in the canestro, the cheese is turned regularly and rubbed with a mixture of oil and vinegar. The cheese weighs 1 to 5 kg (2). New cheese varieties, ripened for only a few days, were recently introduced into the market, and in 2012, the production of Canestrato Pugliese was estimated to be ca. 25 tons (t) (http://www.clal.it/).
Overall, the PDO recognition is given to agriculture products whose features are linked essentially or exclusively to specific geographic areas. Ancient livestock and cheese practices are adopted within the PDO area, which are responsible for the peculiar characteristics of Canestrato Pugliese cheese. Previously, mainly culture-dependent approaches were used to describe the microbiological features of ripened Canestrato Pugliese cheese (3–5) and the possibility of supporting viable probiotic bacteria (6). However, the microbial dynamics that lead from raw ewes' milk to ripened cheese were never thoroughly described, and deep sequencing approaches were never used. Overall, environmental and technological (e.g., shaping, salting, temperature, and time of ripening) factors select for the specific cheese microbiota, which in turn influence the biochemical changes that occur during ripening and contribute to the unique cheese flavor (7). Adventitious microorganisms, which are represented mainly by nonstarter lactic acid bacteria (NSLAB), are derived from raw milk (8) or from the dairy environment and equipment surfaces (9) and play a pivotal role during manufacture and ripening of raw milk cheeses. In particular, facultatively heterofermentative lactobacilli represent the largest and the most diverse proportion of the NSLAB population of almost all ripened cheese varieties (10, 11). The most recent literature (12–17) shows how the structure and evolution of the cheese microbiota could be highlighted through a deep sequencing approach. Nevertheless, none of those studies established the causal relationship between microbiota composition and cheese characteristics, based mainly on proteolysis during ripening. The combined compositional, microbiological, and biochemical characterization of the cheese is fundamental to finding the above-described causal relationship under in situ conditions, which may highlight the peculiar traits of the cheese variety. In the case of Canestrato Pugliese cheese, the above-mentioned combined approach may provide new insights regarding (i) the influence of raw ewes' milk as source of microbially diverse populations, (ii) the microbial dynamics that occur before the cheese is ripened, (iii) the presence of bacterial subpopulations, and (iv) the correlations among technology, microbiota, and cheese features.
First, this study used the above-described complementary approach, which was based on pyrosequencing of the 16S rRNA gene, community-level physiological profiles determined by the use of Biolog EcoPlates, and proteolysis analyses, to characterize Canestrato Pugliese PDO cheese.
MATERIALS AND METHODS
Manufacture of cheese.
PDO Canestrato Pugliese cheese was manufactured with raw ewes' milk (Gentile di Puglia ewe breed), without using commercial or natural whey cultures. Manufacturing was carried out at an industrial plant (Molino a Vento) located in Biccari, Foggia, Apulia region, Italy. Cheese making was carried out on two consecutive days (total of 2 batches), using ewes' milk from 2 daily milkings. All the results were the averages for 2 batches, which were analyzed in triplicate (total of 6 samples analyzed). Raw ewes' milk was heated at 37°C, and liquid calf rennet (25 ml 100 liters−1) was added, and coagulation took place within 30 min. After cutting (size of ca. 0.5 to 1.0 cm), the curd-whey mixture was held at 37°C for ca. 10 min. After whey drainage and molding in the canestro, the curd was stored for approximately 24 h at room temperature and was then dry salted (cheese post-dry salting). Ripening was done at ca. 11°C with a relative humidity of 70% for 90 days. The weight of the cheese was approximately 1 kg. Raw ewes' milk, curd immediately after molding, and cheese samples after 1 day (post-dry salting) (C1), 3 days (C3), 7 days (C7), 15 days (C15), 30 days (C30), 45 days (C45), 60 days (C60), 75 days (C75), and 90 days (C90) of ripening were collected from each batch. All samples were transported to the laboratory under refrigerated conditions (ca. 4°C) and analyzed immediately or frozen (−80°C).
Compositional, microbiological, and biochemical analyses.
Samples of milk, curd, and cheese were analyzed for protein (18), fat (19), moisture (oven drying at 102°C) (20), and salt (21) contents. The pH was measured by using a Foodtrode electrode (Hamilton, Bonaduz, Switzerland). The raw ewes' milk used had the following composition: pH 6.61, 5.6% fat, 4.3% protein, and 0.09% salt. No significant (P > 0.05) differences were found between the 2 batches.
Microbiological analyses were carried out as described previously (22). Twenty grams of sample was homogenized with 180 ml of a sterile sodium citrate (2% [wt/vol]) solution. Presumptive mesophilic lactobacilli and cocci were enumerated in MRS broth supplemented with cycloheximide (0.1 g liter−1) and on M17 agar (Oxoid), respectively, under conditions of anaerobiosis at 30°C for 48 h. Presumptive thermophilic cocci were enumerated on M17 agar (Oxoid, Basingstoke, Hampshire, United Kingdom) under conditions of anaerobiosis at 42°C for 48 h. Enterococci were counted on Slanetz-Barteley agar (Oxoid) at 37°C for 48 h. The number of yeast cells was estimated at 30°C for 48 h by using Sabouraud dextrose agar (SDA) medium (Oxoid) supplemented with chloramphenicol (0.1 g liter−1). The number of molds on Wort agar (Oxoid) at 25°C for 5 days was estimated. Total coliforms were counted by using Violet Red bile lactose (Oxoid) at 37°C for 24 h. Except for enterococci, the media for plating of bacteria were supplemented with cycloheximide at 0.17 g liter−1.
The pH 4.6-insoluble and -soluble nitrogen fractions of the samples were analyzed by urea-polyacrylamide gel electrophoresis (PAGE) and reversed-phase high-pressure liquid chromatography (RP-HPLC), as described previously by Andrews (23) and Gobbetti et al. (24), respectively. Total and individual free amino acids (FAA) from the water-soluble extracts were determined by using a Biochrom series 30 amino acid analyzer (Biochrom Ltd., Cambridge Science Park, United Kingdom), as described previously by Di Cagno et al. (25).
Extraction of total bacterial genomic RNA.
Ninety milliliters of potassium phosphate (50 mM; pH 7.0) buffer was added to 10 g of sample and homogenized for 5 min, and total RNA extraction was carried out by using the RiboPure-Bacteria kit (Ambion RNA, Life Technologies Co., Carlsbad, CA, USA), according to the manufacturer's instructions. Quality of RNA was checked by agarose gel electrophoresis. The RNA concentration was measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE). The purified RNA (100 ng) (final volume, 20 μl) was incubated at 42°C for 2 min in 2 μl of 7× gDNA Wipeout buffer (QuantiTect reverse transcription kit; Qiagen SRL, Milan, Italy) and RNase-free water (final volume, 14 μl). The cDNA was obtained by using the QuantiTect reverse transcription kit (Qiagen), according to the manufacturer's instructions. All reactions were set up in a Rotor Gene 6000 instrument (Corbett Life Science, New South Wales, Australia) equipped with a 36-well reaction rotor.
Amplicon library preparation and pyrosequencing.
cDNA was used to study bacterial diversity through pyrosequencing of the amplified V1-V3 region (amplicon size, 520 bp) (13). PCRs were carried out by using cDNA as the template, as previously described (13). PCR products were purified twice by using an Agencourt AMPure kit (Beckman Coulter, Milan, Italy) and then quantified by using the QuantiFluor system (Promega, Milan, Italy) prior to further processing. Amplicons were used for pyrosequencing on the GS Junior platform (454 Life Sciences, Roche Diagnostics, Milan, Italy), according to the manufacturer's instructions, using titanium chemistry.
Bioinformatics.
A first filtering of the results was performed by using 454 amplicon signal processing, and sequences were then analyzed by using QIIME 1.5.0 software (26). After the split library script was performed with QIIME, the reads were excluded from the analysis if they had an average quality score of <25, if they were <300 bp, and if there were ambiguous base calls. Sequences that passed the quality filter were denoised, and singletons were excluded. Operational taxonomic units (OTUs) were defined by 97%; the taxonomy assignment and alpha and beta diversity analyses were performed by using QIIME, as previously described (27).
Community-level catabolic profiles.
To assess the functional diversity of the microbial communities in milk, curd, and cheese during ripening, Biolog Eco-Microplates (Biolog, Inc., Hayward, CA, USA) were used to acquire bacterial community-level catabolic profiles (CLCPs) (28). Microplates contained 31 carbon sources grouped for chemical class (carbohydrates, carboxylic acids, polymers, amino acids, and amines) and the control, without a carbon source, in triplicate. Ten grams of sample was homogenized with 90 ml of a sterile sodium chloride (0.9% [wt/vol]) solution and centrifuged at 10,000 × g for 15 min at 4°C. The pellet was washed with sterile 50 mM Tris-HCl (pH 7.0), washed further with sterile sodium chloride solution, and then centrifuged again. The cell suspension was diluted (1:1,000) into a sterile sodium chloride solution and dispensed (150 μl) into each of 96 wells of the Biolog Eco-Microplates. Incubation was done at 30°C in the dark, and color development was measured at 590 nm with a microplate reader (Biolog Microstation) every 24 h up to 120 h. Three indices were determined (29, 30). Shannon's diversity index (H′) indicates the substrate utilization pattern, H′ = −∑pi ln(pi), where pi is the ratio of the activity of a particular substrate to the sum of all substrate activities at 120 h; Substrate richness (S), measuring the number of different substrates used, was calculated as the number of wells with a corrected absorbance of >0.25. Substrate evenness (E) was defined as the equitability of activities across all utilized substrates, E = H′/log S.
Statistical analyses.
All cheese analyses were carried out three times for each of the two batches (total of 6 analyses for each type of cheese). Data were subjected to one-way analysis of variance (ANOVA), and pair comparisons of treatment means were achieved with Tukey's procedure at a P value of <0.05, using Statistica for Windows statistical software (version 7.0).
Nucleotide sequence accession number.
The sequence data were submitted to the sequence read archive database of the National Center for Biotechnology Information under accession no. SRP038100.
RESULTS
Cheese compositional and microbiological analyses.
The main chemical composition of Canestrato Pugliese cheese during manufacture and ripening is shown in Table 1. The pH value markedly decreased from the curd during manufacture (pH 6.81 ± 0.2) to 3 days of ripening (pH 5.17 ± 0.2) and then varied slightly. The highest increase in the concentration of NaCl was also found during this same interval of time, even though the concentration progressively increased throughout ripening. As expected, cheese moisture decreased progressively during ripening, and it was 35.4% ± 1.4% at 90 days. The concentrations of fat and protein inversely followed the trend for moisture, thus increasing during ripening. After 90 days, the cheese had fat and protein contents of 25.7% ± 0.6% and 29.5% ± 0.7%, respectively. Presumptive mesophilic lactobacilli were present in raw ewes' milk (4.8 ± 0.2 log CFU g−1) and attained the highest level (8.9 ± 0.3 log CFU g−1) after 3 days of ripening (Table 2). The number of lactobacilli remained elevated throughout ripening, even though it progressively decreased (ca. 1.8 log cycles after 90 days). Compared to presumptive mesophilic lactobacilli, raw ewes' milk contained a slightly but significantly (P < 0.05) high number of mesophilic cocci, which had already markedly increased (ca. 3 log cycles) at 1 day of ripening (post-dry salting). The increase continued to up to 30 days of ripening (9.1 ± 0.3 log CFU g−1), and the number then decreased to ca. 8.0 log CFU g−1. High numbers of thermophilic cocci were found throughout ripening. Nevertheless, the level was 1 or 2 log cycles lower than that found for mesophilic cocci. Enterococci were found in raw ewes' milk (4.8 ± 0.1 log CFU g−1), the numbers of which had already increased after 1 day of ripening (7.0 ± 0.2 log CFU g−1), remained almost constant up to 30 days, and then progressively decreased (ca. 2 log cycles). Initially, yeasts and molds were present at similar levels, with a common tendency to decrease (2.6 ± 0.1 and 3.4 ± 0.1 log CFU g−1). Total coliforms in raw ewes' milk were counted. The number significantly (P < 0.05) increased after 1 and 3 days of ripening, but total coliforms progressively disappeared at the end of ripening.
TABLE 1.
Main chemical composition during manufacture and ripening of Canestrato Pugliese cheesea
| Day(s) of ripening | Mean pH ± SD | Mean moisture content (%) ± SD | Mean fat content (%) ± SD | Mean protein content (%) ± SD | Mean NaCl content (%) ± SD |
|---|---|---|---|---|---|
| Curdb | 6.81 ± 0.2A | 71.2 ± 1.1A | 9.1 ± 0.4E | 12.6 ± 0.5F | 0.3 ± 0.0H |
| 1c | 5.88 ± 0.2B | 59.3 ± 1.6B | 13.8 ± 0.5D | 17.7 ± 0.5E | 1.6 ± 0.1G |
| 3 | 5.17 ± 0.2C | 54.7 ± 0.3C | 16.3 ± 0.6C | 22.7 ± 0.8D | 2.9 ± 0.1F |
| 7 | 5.16 ± 0.1C | 52.8 ± 1.1C | 16.7 ± 0.3C | 23.3 ± 0.7CD | 3.1 ± 0.1DE |
| 15 | 5.07 ± 0.1D | 50.6 ± 1.5CD | 17.2 ± 0.4C | 24.2 ± 0.4C | 3.2 ± 0.1D |
| 30 | 5.09 ± 0.2D | 45.4 ± 1.2D | 19.1 ± 0.6B | 27.2 ± 1.0B | 3.3 ± 0.1C |
| 45 | 5.11 ± 0.2C | 41.5 ± 1.7DE | 22.0 ± 0.2AB | 28.1 ± 0.6AB | 3.4 ± 0.2C |
| 60 | 5.13 ± 0.3C | 37.5 ± 1.6E | 23.3 ± 0.9A | 28.9 ± 1.3A | 3.5 ± 0.2C |
| 75 | 5.12 ± 0.2C | 36.3 ± 1.3F | 24.6 ± 0.5A | 29.2 ± 0.2A | 3.9 ± 0.1B |
| 90 | 5.10 ± 0.6D | 35.4 ± 1.4F | 25.7 ± 0.6A | 29.5 ± 0.7A | 4.2 ± 0.2A |
Shown are mean values ± standard deviations for two batches of each type of cheese, analyzed in triplicate. Data in the same column with different letters (A to H) are significantly different (P < 0.05).
Curd after molding.
Curd after dry salting.
TABLE 2.
Cell numbers of microbial groups during manufacture and ripening of Canestrato Pugliese cheesea
| Microbial group | Mean no. of cells (log CFU g−1) ± SD |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Milk | Curdb | Day(s) of ripening |
|||||||||
| 1c | 3 | 7 | 15 | 30 | 45 | 60 | 75 | 90 | |||
| Mesophilic lactobacilli | 4.8 ± 0.2F | 5.6 ± 0.1E | 7.6 ± 0.1C | 8.9 ± 0.3A | 8.1 ± 0.3B | 8.0 ± 0.3B | 8.0 ± 0.1B | 8.0 ± 0.5B | 7.5 ± 0.2C | 7.3 ± 0.2CD | 7.1 ± 0.3D |
| Mesophilic cocci | 5.0 ± 0.1E | 6.0 ± 0.3D | 8.3 ± 0.4BC | 8.4 ± 0.4B | 8.3 ± 0.2BC | 9.0 ± 0.1A | 9.1 ± 0.3A | 8.7 ± 0.4AB | 8.5 ± 0.3B | 8.1 ± 0.2C | 8.0 ± 0.1C |
| Thermophilic cocci | 4.3 ± 0.1G | 5.3 ± 0.2F | 7.6 ± 0.4A | 7.3 ± 0.2A | 7.4 ± 0.2A | 7.4 ± 0.3A | 7.1 ± 0.1B | 6.8 ± 0.2C | 6.4 ± 0.2D | 6.3 ± 0.3D | 6.0 ± 0.1E |
| Enterococci | 4.8 ± 0.1D | 5.0 ± 0.2D | 7.0 ± 0.2A | 7.2 ± 0.1A | 6.9 ± 0.2A | 7.2 ± 0.2A | 7.1 ± 0.1A | 6.6 ± 0.1A | 6.1 ± 0.2C | 5.9 ± 0.2C | 5.0 ± 0.2D |
| Yeasts | 3.5 ± 0.2D | 3.2 ± 0.1E | 3.3 ± 0.1DE | 4.6 ± 0.2A | 4.6 ± 0.1A | 4.8 ± 0.2A | 4.0 ± 0.1B | 3.7 ± 0.1C | 3.4 ± 0.1DE | 3.3 ± 0.1DE | 2.6 ± 0.1F |
| Molds | 3.1 ± 0.0F | 3.3 ± 0.1E | 3.6 ± 0.2D | 4.4 ± 0.1B | 4.2 ± 0.2BC | 5.2 ± 0.1A | 4.0 ± 0.1C | 3.2 ± 0.0F | 3.1 ± 0.1F | 3.9 ± 0.2CD | 3.4 ± 0.1DE |
| Total coliforms | 4.3 ± 0.2C | 4.3 ± 0.1C | 5.9 ± 0.2A | 5.7 ± 0.2A | 5.3 ± 0.1B | 4.1 ± 0.1C | 3.4 ± 0.2D | 2.9 ± 0.1E | 2.1 ± 0.2F | 1.70 ± 0.0G | <1H |
Shown are mean values ± standard deviations for two batches of each type of cheese, analyzed in triplicate. Means within rows with different letters (A to H) are significantly different (P < 0.05).
Curd after molding.
Curd after dry salting.
Proteolysis.
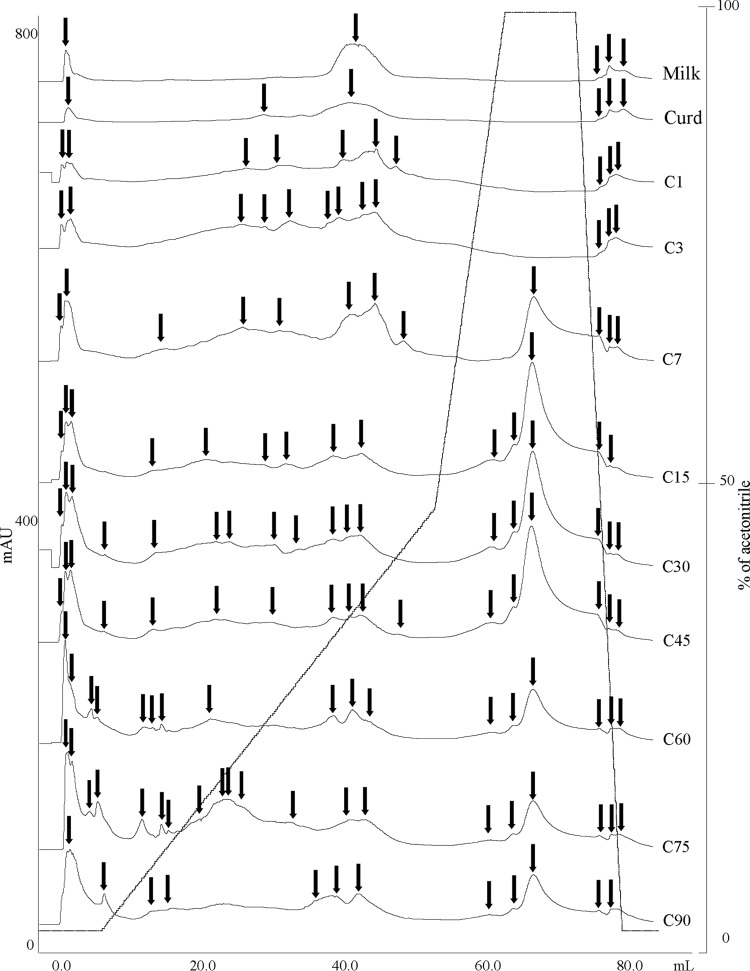
The pH 4.6-insoluble and -soluble nitrogen fractions of the cheese were analyzed by urea-PAGE (see Fig. S1A and S1B in the supplemental material). αs1-Casein (CN) persisted to the end of ripening, and its main degradation began from 60 days onwards. The urea-PAGE electrophoretograph of the pH 4.6-soluble fraction showed some differences during this time. Characteristic polypeptide bands appeared at 1 day of ripening. They persisted up to 60 days, and the profile then simplified. Complementary information emerged with RP-HPLC analysis (Fig. 1). The number of peaks, which were recognized and matched visually with the Unicorn program (Amersham Biosciences), varied during manufacture and ripening of Canestrato Pugliese cheese. Five peaks (raw ewes' milk) to 20 peaks (75 days) were detected, which decreased to 12 peaks at 90 days of ripening. An increase of the area of hydrophilic and hydrophobic peptide peaks was found up to 45 days. These results agreed with the concentration of total free amino acids (FAA) (Table 3). As expected, raw ewes' milk and curd after molding had the lowest concentrations of FAA (115.3 ± 4.1 and 133 ± 4.0 mg kg−1, respectively). Subsequently, the concentration of FAA markedly (P < 0.05) increased and showed the highest rate from 7 days (1,275.2 ± 38.6 mg kg−1) to 75 days (6,509.0 ± 325.5 mg kg−1) of ripening. Overall, the FAA found at the highest concentrations were Asp, Glu, Val, Leu, and Phe.
FIG 1.
Reversed-phase fast protein liquid chromatography of the pH 4.6-soluble nitrogen fractions during manufacture and ripening of Canestrato Pugliese cheese. Shown are raw ewes' milk (Milk), curd after molding (Curd), cheese post-dry salting (C1), and cheese during ripening (3, 7, 15, 30, 45, 60, 75, and 90 days [C3 to C90]). Arrows refer to hydrophilic and hydrophobic peptide peaks.
TABLE 3.
Mean FAA levels found in Canestrato Pugliese cheese during manufacture and ripeninga
| FAA | Mean concn of FAA (mg kg−1) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Milk | Curdb | Day(s) of ripening |
|||||||||
| 1c | 3 | 7 | 15 | 30 | 45 | 60 | 75 | 90 | |||
| Asp | 0.0 | 0.0 | 21.5 | 40.9 | 42.4 | 100.9 | 169.0 | 236.2 | 346.8 | 483.4 | 491.1 |
| Thr | 0.0 | 0.0 | 28.1 | 18.3 | 20.0 | 41.8 | 63.7 | 133.5 | 108.4 | 153.0 | 158.2 |
| Ser | 0.0 | 0.0 | 46.7 | 29.4 | 40.6 | 58.4 | 47.6 | 42.5 | 53.7 | 51.1 | 53.3 |
| Glu | 8.4 | 11.1 | 159.6 | 175.8 | 207.9 | 330.8 | 452.7 | 541.8 | 800.7 | 1,176.7 | 1,187.8 |
| Gly | 10.5 | 10.3 | 22.2 | 6.7 | 14.5 | 14.9 | 26.2 | 48.2 | 58.8 | 84.0 | 89.5 |
| Ala | 6.7 | 5.2 | 53.8 | 68.5 | 63.5 | 93.7 | 135.8 | 212.6 | 212.5 | 284.3 | 298.2 |
| Cys | 0.0 | 0.0 | 8.7 | 13.8 | 30.0 | 63.0 | 66.2 | 84.1 | 93.2 | 99.2 | 103 |
| Val | 26.0 | 25.4 | 80.1 | 151.2 | 171.3 | 260.8 | 350.2 | 478.3 | 597.3 | 773.1 | 791.1 |
| Met | 4.6 | 17.0 | 35.1 | 56.8 | 55.6 | 101.6 | 128.9 | 229.0 | 251.1 | 316.0 | 322.8 |
| Ile | 2.5 | 0.0 | 22.7 | 40.9 | 41.3 | 95.6 | 126.8 | 232.1 | 269.5 | 327.1 | 330.7 |
| Leu | 13.3 | 12.5 | 75.6 | 155.4 | 173.2 | 296.9 | 407.9 | 641.1 | 778.5 | 945.6 | 951.4 |
| Tyr | 5.5 | 21.8 | 36.8 | 48.7 | 32.3 | 49.7 | 36.9 | 97.0 | 150.6 | 92.6 | 94.8 |
| Phe | 3.1 | 3.9 | 31.3 | 100.1 | 108.3 | 177.8 | 260.2 | 335.3 | 504.3 | 569.6 | 579.1 |
| His | 1.0 | 2.5 | 18.9 | 20.0 | 34.1 | 36.5 | 49.2 | 56.1 | 56.8 | 24.1 | 25.2 |
| Trp | 0.0 | 2.2 | 19.3 | 17.3 | 25.4 | 40.2 | 45.8 | 59.0 | 71.3 | 78.0 | 80.9 |
| Orn | 15.8 | 16.3 | 26.2 | 31.6 | 42.9 | 93.3 | 139.8 | 210.7 | 360.0 | 448.5 | 458.3 |
| Lys | 9.4 | 7.9 | 18.9 | 8.7 | 8.9 | 18.4 | 67.3 | 135.3 | 175.7 | 247.5 | 249.1 |
| Arg | 0.0 | 0.0 | 19.0 | 40.5 | 53.9 | 72.6 | 49.3 | 28.4 | 81.2 | 60.1 | 62.5 |
| Pro | 8.7 | 0.0 | 65.2 | 135.6 | 109.7 | 129.3 | 148.2 | 198.3 | 233.1 | 295.5 | 298.2 |
| Total | 115.3 ± 4.1H | 135.9 ± 4.0H | 789.1 ± 39.45G | 1,159 7 ± 45.5F | 1,275 2 ± 38.6F | 2,075.7 ± 103.8E | 2,771.1 ± 110.8D | 3,999.1 ± 159.9C | 5,203.1 ± 260.1B | 6,509.0 ± 325.5A | 6,625.2 ± 331.6A |
Shown are mean values ± standards deviations for two batches of each type of cheese, analyzed in triplicate. Means with different letters (A to H) are significantly different (P < 0.05).
Curd after molding.
Curd after dry salting.
Microbial community structure and dynamics.
16S rRNA gene amplicons of the bacterial communities of raw ewes' milk and cheeses were sequenced. No significant (P > 0.05) differences were found between the two batches analyzed. After 454 amplicon signal processing, a total of 66,924 raw sequence reads of 16S rRNA gene amplicons were obtained (average length, 485 bp). The calculated alpha diversity parameters are reported in Table S1 in the supplemental material. The lowest Chao1 richness and Shannon diversity index values were found for raw ewes' milk (17.33 and 1.43, respectively), but these values then markedly increased in the curd after molding (46.57 and 3.05, respectively). After 1 and 3 days of ripening, both indices decreased. A further increase was found throughout ripening. Good's estimated sample coverage (ESC) was >99% for all the samples, indicating a satisfactory description of the microbial diversity. The beta diversity parameter did not show significant differences between samples (data not shown).
The bacterial sequences from RNA, which were assigned to bacterial phyla, and their relative abundance (percent) varied during cheese manufacture and ripening (data not shown). RNA from raw ewes' milk included mainly Proteobacteria (68%) and Firmicutes (30%), followed by Bacteroidetes (0.05%) and Actinobacteria (0.02%). Almost the same structure was found in the curd after molding (46, 52, 0.3, and 0.2%, respectively). From day 1 of ripening (after dry salting) onwards, the phylum Firmicutes dominated (ca. 97 to 99%). After 1 day of ripening, the relative abundance of Proteobacteria markedly decreased to ca. 3%. A further decrease was found over time (0.02 to 0.8%).
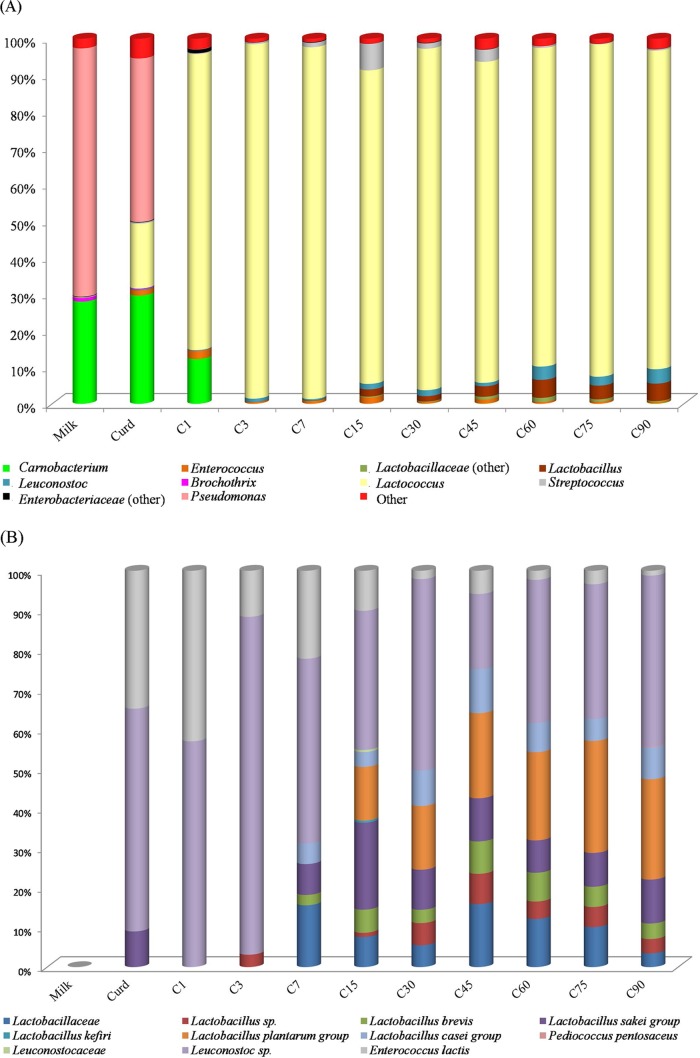
The distribution of the OTUs that were classified to the genus level is reported in Fig. 2A and Table 4. The microbial community diversified during manufacture and ripening of Canestrato Pugliese cheese. Pseudomonas (68%) and Carnobacterium (28%) were the main genera found in raw ewes' milk. Brochothrix (1.04%), Lactococcus (0.15%), Flavobacterium (0.05%), Enterobacteriaceae (0.16%), Luteococcus (0.02%), and Delftia (0.02%) were also found at low incidences (Table 4). As expected, the bacterial profile changed and was enriched in the curd after molding (Fig. 2A; see also Table S2 in the supplemental material). The genera Pseudomonas (44.7%) and Carnobacterium (29.7%) still persisted, but Lactococcus (17.8%) markedly emerged. At this time, other genera, mostly absent in raw milk, were also found: Enterococcus (1.4%), Staphylococcus (0.9%), Streptococcus (0.4%), and Leuconostoc (0.24%). Lactobacillus was shown to be present at a very low incidence (0.03%). After 1 day of ripening, which corresponded to the curd after dry salting, Lactococcus (81%) dominated, followed by Carnobacterium (12%) and Enterococcus (2.2%). Most of the bacteria that were found in low abundances in the curd after molding disappeared. Other bacteria belonging to the genera Citrobacter, Raoultella, and Escherichia replaced them. In agreement with the cell number of presumptive mesophilic cocci (Table 2), Lactococcus dominated throughout ripening. Other genera flanked Lactococcus but at lower abundances. In particular, Enterococcus, Leuconostoc, Streptococcus, Lactobacillus, and Staphylococcus were found. Abundances of Lactobacillus and the family Lactobacillaceae increased throughout ripening. The same was found for Leuconostoc. The Streptococcus abundance increased up to 15 days, decreasing afterwards. At 90 days, the bacterial profile of Canestrato Pugliese cheese was characterized by Lactococcus (87.2%), followed by Lactobacillus (4.8%) and Leuconostoc (3.9%). Within the Lactococcus genus, Lactococcus lactis was the dominant species, followed by Lactococcus garvieae and Lactococcus raffinolactis (data not shown). Taxonomic details up to the species level within the Streptococcus genus revealed that Streptococcus thermophilus was the most abundant OTU, followed by Streptococcus agalactiae (data not shown).
FIG 2.
Incidence of OTUs assigned to the genus level (A) and to genera belonging to the Enterococcaceae, Lactobacillaceae, and Leuconostocaceae families (B) based on 16S rRNA gene pyrosequencing analysis of all RNA samples directly from raw ewes' milk (Milk), curd after molding (Curd), cheese post-dry salting (C1), and cheese during ripening (3, 7, 15, 30, 45, 60, 75, and 90 days [C3 to C90]). Only OTUs with an incidence above 1% in at least one sample are shown.
TABLE 4.
Incidences of OTUs assigned to the genus level at a value below 1% in at least one samplea
| Taxon | Incidence of OTU (%) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Milk | Curdb | Days of ripening |
|||||||||
| 1c | 3 | 7 | 15 | 30 | 45 | 60 | 75 | 90 | |||
| Corynebacterium | 0 | 0.05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Microbacterium | 0 | 0.17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rothia | 0 | 0.047 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Luteococcus | 0.017 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Sanguibacter | 0 | 0.047 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Flavobacteriaceae (other) | 0 | 0.038 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chryseobacterium | 0 | 0.015 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Flavobacterium | 0.048 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Sejongia | 0 | 0.076 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Carnobacteriaceae (other) | 0 | 0 | 0.044 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Marinilactibacillus | 0 | 0 | 0 | 0 | 0 | 0.022 | 0 | 0.032 | 0 | 0 | 0 |
| Enterococcaceae (other) | 0 | 0 | 0.014 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lactobacillaceae (other) | 0 | 0 | 0 | 0 | 0 | 0.022 | 0.018 | 0 | 0.081 | 0.065 | 0.13 |
| Pediococcus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.018 |
| Leuconostocaceae (other) | 0 | 0 | 0 | 0 | 0 | 0.022 | 0 | 0 | 0 | 0 | 0 |
| Macrococcus | 0 | 0.038 | 0.15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Staphylococcus | 0 | 0.87 | 0.11 | 0 | 0.23 | 0.19 | 0.018 | 0.54 | 0.18 | 0.019 | 0.084 |
| Bosea | 0 | 0.038 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Caulobacteraceae (other) | 0 | 0.17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mycoplana | 0 | 0.07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Shinella | 0 | 0.038 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Comamonadaceae (other) | 0 | 0.07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acidovorax | 0 | 0.038 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Comamonas | 0 | 0.038 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Delftia | 0.017 | 0.076 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hylemonella | 0 | 0.038 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Xenophilus | 0 | 0.038 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Citrobacter | 0 | 0 | 0.62 | 0.14 | 0.084 | 0 | 0.029 | 0.032 | 0 | 0 | 0.055 |
| Enterobacter | 0 | 0 | 0.055 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Escherichia | 0 | 0 | 0.29 | 0 | 0 | 0 | 0.029 | 0.13 | 0 | 0.019 | 0.072 |
| Raoultella | 0 | 0 | 0.62 | 0.61 | 0.25 | 0 | 0.058 | 0.065 | 0 | 0.019 | 0 |
| Halomonas | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.065 | 0 | 0 | 0 |
| Moraxellaceae | 0 | 0.076 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acinetobacter | 0 | 0.076 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Psychrobacter | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.032 | 0 | 0 | 0 |
| Pseudomonadaceae (other) | 0.051 | 0.13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Based on 16S rRNA gene pyrosequencing analysis of all RNA samples directly from Canestrato Pugliese cheese during manufacture and ripening.
Curd after molding.
Curd after dry salting.
Lc. lactis dominated throughout manufacture and ripening. To highlight the succession of nonstarter lactic acid bacteria (NSLAB), the evolution of Firmicutes, with the exclusion of Lc. lactis, is displayed in Fig. 2B. When possible, taxonomic details up to the species level were assigned. Leuconostoc sp. already dominated in curd after molding (86%) and persisted up to 90 days (43%). The Lactobacillus sakei group was found in the curd after molding (13%). On the contrary, all the other species appeared only at 7 or 15 days of ripening. The Lactobacillus plantarum group and the Lactobacillus sakei group were the dominant species of mesophilic lactobacilli, which persisted up to 90 days of ripening. Although less abundant, the Lactobacillus casei group and Lactobacillus brevis also stably occurred in all cheeses. Pediococcus pentosaceus and Lactobacillus kefiri were found only occasionally.
Changes in community-level catabolic profiles.
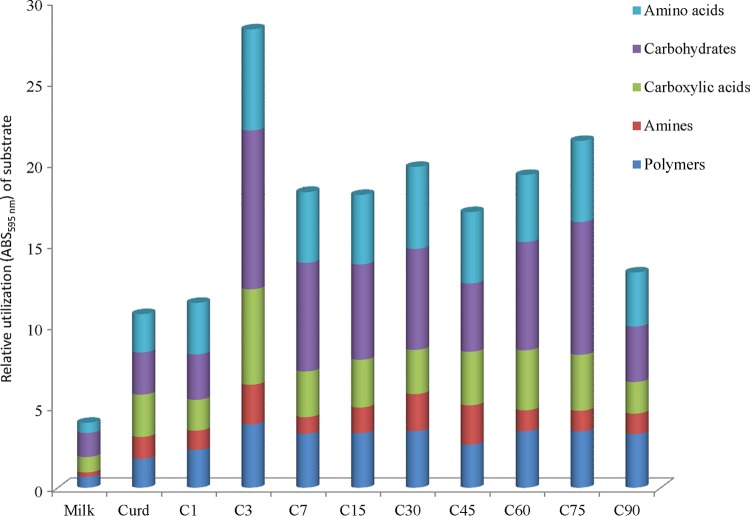
The relative utilization of carbon sources by the bacterial community showed that all 5 chemical classes (amino acids, carbohydrates, carboxylic acids, amines, and polymers) were variously degraded during manufacture and ripening of Canestrato Pugliese cheese (Fig. 3). In particular, carbohydrates and amino acids were the chemical classes used mainly throughout time. At 3 days of ripening, the most intense degradation of all the chemical classes occurred. The capacity for using the carbon sources stabilized over time and slightly decreased at the end of ripening. The catabolic profiles were also determined by calculating the indices H′, S, and E (see Table S2 in the supplemental material). Raw ewes' milk showed the lowest substrate utilization pattern (H′) (2.68 ± 0.2) and substrate richness (S) (12.33 ± 1.5) values. These indices significantly (P < 0.05) increased during cheese manufacture and ripening, and the highest values were found after 3 days (3.25 ± 0.1 and 20.33 ± 0.2, respectively). The E value, giving a measure of the statistical significance (equitability) of the H′ and S values, confirmed the significant (P < 0.05) differences over time.
FIG 3.
Relative utilization of different carbon sources, grouped for chemical classes (carbohydrates, carboxylic acids, polymers, amino acids, and amines), during manufacture and ripening of Canestrato Pugliese cheese. Shown are data for raw ewes' milk (Milk), curd after molding (Curd), cheese post-dry salting (C1), and cheese during ripening (3, 7, 15, 30, 45, 60, 75, and 90 days [C3 to C90]).
Correlations among microbiota, catabolic profiles, and proteolysis.
All the OTUs were considered in order to find correlations. Some correlations were found between the relative abundance of bacteria, proteolysis (FAA; Asp, Glu, Val, Leu, and Phe; and area of hydrophilic peptide peaks), and relative utilization of the carbon sources during manufacture and ripening of Canestrato Pugliese cheese (see Table S3 in the supplemental material). Only positive correlations (false discovery rate [FDR] < 0.05; r > 0.6) are listed. A correlation was found between FAA and the abundance of L. plantarum group, L. casei group, Lactobacillus sp., L. sakei group, Lactobacillaceae family, and L. brevis populations (FDR < 0.05). In particular, the highest correlation (r > 0.89) was found for L. plantarum. Regarding individual amino acids, the concentrations of Asp, Glu, Leu, Phe, and Val were also highly correlated with L. plantarum (r value of >0.85 to >0.89), followed by L. casei, L. sakei, and Lactobacillus sp. The levels of L. sakei, L. casei, Lactobacillus sp., Lactobacillaceae, L. brevis, and especially L. plantarum (r > 0.86) were also correlated with the area of hydrophilic peptide peaks. The abundance of Lc. lactis was positively correlated with the utilization of polymers, amines, carbohydrates, and amino acids (FDR < 0.05).
DISCUSSION
This study aimed to give new insights into how microbial ecology evolves during manufacture and ripening of Canestrato Pugliese PDO cheese made from raw ewes' milk without the addition of starter and how microbial dynamics affect proteolysis.
Mean values for the gross composition approached those for Canestrato Pugliese cheese (5, 31) and other Italian ewes' milk cheeses (32). The highest decrease of the pH value and the highest increase of the NaCl content, which might have had an effect on microbial succession, were found within 3 days of ripening. The microbial cell density reflected the succession of communities that frequently characterize cheeses during manufacturing and ripening. Raw ewes' milk harbored high levels of adventitious lactic acid bacteria, especially mesophilic cocci. As usual for ewes' milk cheeses, the use of mature milk, which was stored for 24 h at 4°C before manufacture, probably had an influence on the increased levels of mesophilic and psychrotrophic bacteria (33). The cell density of thermophilic cocci and especially those of mesophilic lactobacilli and cocci increased during manufacture and continued to increase up to 15 to 30 days of cheese ripening. At 90 days, mesophilic cocci had the highest cell density, followed by mesophilic lactobacilli. Similarly to other artisan raw milk cheeses (3, 34–36), enterococci represent a significant component of the adventitious microbiota of Canestrato Pugliese cheese, even though their numbers decreased at the end of ripening. The hostile ripening conditions did not permit the survival of total coliforms during ripening.
Proteolysis is the most complex and important biochemical event, which occurs during ripening of most cheese varieties (10). Primary proteolysis of Canestrato Pugliese cheese showed a slight and late hydrolysis of αs1-CN, probably due to the residual activity of chymosin. Unlike other Italian ewes' milk cheeses (e.g., Pecorino Romano), the manufacture of Canestrato Pugliese cheese did not include cooking of the curd, which allowed chymosin activity. A considerable amount of β-CN also persisted at the end of ripening. Nevertheless, γ-CN was found, which indicated the activity of plasmin. Chymosin activity toward β-CN is usually limited, mainly because of hydrophobic interactions between salt and proteins (37). Almost the same primary proteolysis was described previously (5). The urea-PAGE electrophoretograph of the pH 4.6-soluble fraction, the chromatogram data for the same fraction, and the concentration of FAA showed a certain extent of secondary proteolysis. The number and area of hydrophilic and hydrophobic peptide peaks increased up to 45 to 75 days of cheese ripening. At this time, the concentration of FAA became significantly high, further increasing and approaching the values found for other Italian ewes' milk cheeses (32, 38). Asp, Glu, Val, Leu, and Phe were found at the highest concentrations, which is typical for several Italian semihard and extrahard cheese varieties (5, 32, 38, 39).
Culture-independent analysis was carried out by using RNA as the template, and the OTUs found during processing are meant to be members of the active microbial populations in cheese (40). Alpha diversity analysis indicated that the microbial diversity increased during manufacture. Cheese manufacture, defined as those operations carried out during the first ca. 24 h, includes steps that have a direct effect on cheese microorganisms or influence the environment in which they are grown. Raw ewes' milk was contaminated by bacterial phyla that are surely the outcome of environmental contamination. Usually, Bacteroidetes, Actinobacteria, and especially Proteobacteria and Firmicutes dominate different areas throughout the farm, including teat surfaces, milking parlors, hay, air, and dust (41). Genera belonging to the Proteobacteria (e.g., Pseudomonas) and Firmicutes (e.g., Carnobacterium and Lactococcus) were mainly identified. The psychrotrophic Pseudomonas spp., which dominated raw ewes' milk, are the most common cause of milk spoilage (42) and may constitute 70 to 90% of the microbial population during milk storage at low temperatures (43). Lactococcus spp. also adapt well and grow at low temperatures, and Carnobacterium is frequently isolated from dairy environments (41). Carnobacteria are slow acidifiers and unsuitable as dairy starters, but they may positively contribute to cheese flavor (44). Pseudomonas, Carnobacterium, and Lactococcus still dominated in the curd after molding, but many other genera occurred at this time at lower relative abundances (Fig. 2A). Most of these genera required only 1 day of ripening (after dry salting) to be inhibited. Lactococcus lactis dominated the bacterial community throughout ripening, followed by Carnobacterium and Enterococcus lactis. The Gram-negative organisms Citrobacter sp. and Raoultella sp., belonging to the Enterobacteriaceae family, were part of the subdominant population. A high level of diversity of Gram-negative bacteria in raw milk and dairy products was suggested previously to have a potential role in dairy fermentations (41, 45). Lactobacillaceae family, Leuconostoc sp., Lactobacillus casei group, Lactobacillus brevis, and especially Lactobacillus sakei group and Lactobacillus plantarum group populations were found after 3 days of ripening, and their relative abundances increased at between 7 and 15 days (Fig. 2B). At the end of ripening, the bacterial profile of Canestrato Pugliese cheese was dominated by Lc. lactis, which was flanked by L. plantarum, L. sakei, L. casei, and Leuconostoc sp. Pediococcus pentosaceus and Lactobacillus kefiri occasionally appeared as subdominant species. Recent studies of Polish Oscypek and Croatian raw ewes' milk cheeses, also made without starters (12, 16), showed similar bacterial successions. The above-described adventitious NSLAB are a significant proportion of the microbial population of many ripened cheese varieties (10). NSLAB grow at low temperatures and adapt to the lack of fermentable carbohydrates, low pH and water activity (aw), and the presence of bacteriocins, which altogether make hostile environmental conditions during cheese ripening. The role of NSLAB during secondary proteolysis of cheeses was largely described. Usually, the use of NSLAB as adjunct cultures increases the levels of peptides and FAA, which enhance flavor intensity and accelerate cheese ripening (7, 10). Despite the lower relative abundance of mesophilic lactobacilli than lactococci, only this group of bacteria, especially L. plantarum, was positively correlated with the total concentration of FAA. The same correlation was found for the concentrations of Asp, Glu, Val, Leu, and Phe and the area of hydrophilic peptide peaks. Together with Lactobacillus paracasei, L. plantarum was the species most frequently isolated from cheeses during ripening and used as an adjunct starter (46). As shown by the CLCP (Fig. 3), microbial succession reflected the overall physiological diversity. The lowest substrate utilization pattern value was found for raw ewe's milk (lowest H′ and S values). The highest capacity to degrade carbon sources and the highest H′ value were found at 3 days of ripening, which coincided with environmental conditions that became particularly hostile (e.g., lowest pH value, increased concentration of NaCl, and, probably, lack of fermentable carbohydrates). The abundance of Lc. lactis throughout ripening was positively correlated with each substrate utilization pattern value.
This study allowed the identification of strategic phases that characterized the manufacture and ripening of Canestrato Pugliese cheese. Within 3 days of ripening, when the environment changed and became more hostile, the bacterial diversity simplified, being dominated by Lactococcus and showing the highest capacity to degrade various carbon sources. Once established under these conditions and in the time following (7 and 15 days), the population of mainly mesophilic lactobacilli increased, which led to a causal relationships with proteolysis (45 to 75 days). The approach of this study could be considered a model system to find relationships under in situ conditions, which may allow cheese characterization and the selection of adventitious NSLAB to be used as adjunct cultures to guarantee high-quality standards for typical/traditional cheeses.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Molino a Vento industrial plant, located in Biccari, Foggia, Apulia region, Italy, for the supply of ewes' milk, cheese manufacture, and technical support.
Footnotes
Published ahead of print 25 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00757-14.
REFERENCES
- 1.Gobbetti M. 2004. Extra-hard varieties, p 51–70 In Fox PF, McSweeney PLH, Cogan TM, Guinee TP. (ed), Cheese: chemistry, physics and microbiology. Elsevier, London, United Kingdom [Google Scholar]
- 2.Di Cagno R, Gobbetti M. 2011. Hard Italian cheeses, p 728–736 In Fuquay JW, Fox PF, McSweeney PLH. (ed), Encyclopedia of dairy sciences, 2nd ed, vol 1 Elsevier Academic Press, San Diego, CA [Google Scholar]
- 3.Aquilanti L, Dell'Aquila L, Zannini E, Zocchetti A, Clementi F. 2006. Resident lactic acid bacteria in raw milk Canestrato Pugliese cheese. Lett. Appl. Microbiol. 43:161–167. 10.1111/j.1472-765X.2006.01935.x [DOI] [PubMed] [Google Scholar]
- 4.Aquilanti L, Zannini E, Zocchetti A, Osimani A, Clementi F. 2007. Polyphasic characterization of indigenous lactobacilli and lactococci from PDO Canestrato Pugliese cheese. LWT 40:1146–11552. 10.1016/j.lwt.2006.09.001 [DOI] [Google Scholar]
- 5.Albenzio M, Corbo MR, Rehman SU, Fox PF, De Angelis M, Corsetti A, Sevi A, Gobbetti M. 2001. Microbiological and biochemical characterization of Canestrato Pugliese cheese made from raw milk, pasteurized milk or by heating the curd in hot whey. Int. J. Food Microbiol. 67:35–48. 10.1016/S0168-1605(00)00533-X [DOI] [PubMed] [Google Scholar]
- 6.Corbo MR, Albenzio M, De Angelis M, Sevi A, Gobbetti M. 2001. Microbiological and biochemical properties of Canestrato Pugliese hard cheese supplemented with bifidobacteria. J. Dairy Sci. 84:551–561. 10.3168/jds.S0022-0302(01)74507-9 [DOI] [PubMed] [Google Scholar]
- 7.Fox PF, Guinee TP, Cogan TM, McSweeney PLH. 2000. Fundamentals of cheeses sciences. Aspen Publishers, Gaithersburg, MD [Google Scholar]
- 8.Berthier F, Beuvier E, Dasen A, Grappin R. 2001. Origin and diversity of mesophilic lactobacilli in Comtè cheese, as revealed by PCR with repetitive and species-specific primers. Int. Dairy J. 11:293–305. 10.1016/S0958-6946(01)00059-0 [DOI] [Google Scholar]
- 9.Somers EB, Johnson ME, Wong ACL. 2001. Development of amino acids and organic acids in Norvegia, influence of milk treatment and adjunct Lactobacillus. J. Dairy Sci. 84:1926–1936. 10.3168/jds.S0022-0302(01)74634-6 [DOI] [PubMed] [Google Scholar]
- 10.Gobbetti M, De Angelis M, Di Cagno R, Rizzello CG. 2007. Relative contributions of starter cultures and non-starter bacteria to flavour of cheese, p 121–156 In Weimer BC. (ed), Improving the flavour of cheese. CRC Press, Boca Raton, FL [Google Scholar]
- 11.Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2011. Molecular approaches to analyzing the microbial composition of raw milk and raw milk cheese. Int. J. Food Microbiol. 150:81–94. 10.1016/j.ijfoodmicro.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 12.Alegria A, Szczenny P, Mayo B, Bardowski J, Kowalczyk M. 2012. Biodiversity in Oscypek, a traditional Polish cheese, determined by culture-dependent and independent approaches. Appl. Environ. Microbiol. 78:1890–1898. 10.1128/AEM.06081-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ercolini D, De Filippis F, La Storia A, Iacono M. 2012. “Remake” by high-throughput sequencing of the microbiota involved in the production of water buffalo mozzarella cheese. Appl. Environ. Microbiol. 78:8142–8145. 10.1128/AEM.02218-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2012. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Environ. Microbiol. 78:5717–5723. 10.1128/AEM.00918-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lusk TS, Ottesen AR, White JR, Allard MW, Brown EW, Kase JA. 2012. Characterization of microflora in Lati-style cheeses by next-generation sequencing technology. BMC Microbiol. 12:254–264. 10.1186/1471-2180-12-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuka MM, Wallish S, Engel M, Welzl G, Havranek J, Schloter M. 2013. Dynamics of bacterial communities during the ripening process of different Croatian cheese types derived from raw ewe's milk cheeses. PLoS One 8:e80734. 10.1371/journal.pone.0080734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Filippis F, La Storia A, Stellato G, Gatti M, Ercolini D. 2014. A selected core microbiome drives the early stages of three popular Italian cheese manufactures. PLoS One 9:e89680. 10.1371/journal.pone.0089680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.IDF. 1964. Determination of the protein content of processed cheeses products. Standard 25 International Dairy Federation, Brussels, Belgium [Google Scholar]
- 19.IIRS. 1955. Determination of the percentage of fat in cheese. Irish standard 69. Institute for Industrial Research and Standards, Dublin, Ireland [Google Scholar]
- 20.IDF. 1982. Cheese and processed cheese. Determination of the total solid content. IDF standard 4A International Dairy Federation, Brussels, Belgium [Google Scholar]
- 21.Fox PF. 1963. Potentiometric determination of salt in cheese. J. Dairy Sci. 46:744–745. 10.3168/jds.S0022-0302(63)89134-1 [DOI] [Google Scholar]
- 22.Di Cagno R, Quinto M, Corsetti A, Minervini F, Gobbetti M. 2006. Assessing the proteolytic and lypolytic activities of single strains of mesophilic lactobacilli as adjunct cultures using a Caciotta cheese model system. Int. Dairy J. 16:119–130. 10.1016/j.idairyj.2005.01.012 [DOI] [Google Scholar]
- 23.Andrews AT. 1983. Proteinases in normal bovine milk and their action on caseins. J. Dairy Res. 50:45–55. 10.1017/S0022029900032519 [DOI] [PubMed] [Google Scholar]
- 24.Gobbetti M, Morea A, Baruzzi F, Corbo MR, Matarante A, Considine T, Di Cagno R, Guinee T, Fox PF. 2002. Microbiological, compositional, biochemical and textural characterization of Caciocavallo Pugliese cheese during ripening. Int. Dairy J. 12:511–523. 10.1016/S0958-6946(02)00042-0 [DOI] [Google Scholar]
- 25.Di Cagno R, Buchin S, de Candia S, De Angelis M, Fox PF, Gobbetti M. 2007. Characterization of Italian cheeses ripened under nonconventional conditions. J. Dairy Sci. 90:2689–2704. 10.3168/jds.2006-654 [DOI] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Filippis F, La Storia A, Villani F, Ercolini D. 2013. Exploring the sources of bacterial spoilers in beefsteaks by culture-independent high-throughput sequencing. PLoS One 8:e70222. 10.1371/journal.pone.0070222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siragusa S, Di Cagno R, Ercolini D, Minervini F, Gobbetti M, De Angelis M. 2009. Taxonomic structure and monitoring of the dominant population of lactic acid bacteria during wheat flour sourdough type I propagation using Lactobacillus sanfranciscensis starters. Appl. Environ. Microbiol. 75:1099–1109. 10.1128/AEM.01524-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon CE. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27:379–423. 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- 30.Shannon CE. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27:623–656. 10.1002/j.1538-7305.1948.tb00917.x [DOI] [Google Scholar]
- 31.Di Cagno R, Upadhyay VK, McSweeney PLH, Corbo MR, Faccia M, Gobbetti M. 2004. Microbiological, compositional and biochemical characterization of PDO Canestrato Pugliese cheese. Ital. J. Food Sci. 16:45–58 http://cat.inist.fr/?aModele=afficheN&cpsidt=15596840 [Google Scholar]
- 32.Coda R, Brechany E, De Angelis M, De Candia S, Di Cagno R, Gobbetti M. 2006. Comparison of the compositional, microbiological, biochemical, and volatile profile characteristics of nine Italian ewes' milk cheeses. J. Dairy Sci. 89:4126–4143. 10.3168/jds.S0022-0302(06)72458-4 [DOI] [PubMed] [Google Scholar]
- 33.de Garnica ML, Santos JA, Gonzalo C. 2011. Short communication: influence of storage and preservation on microbiological quality of silo ovine milk. J. Dairy Sci. 94:1922–1927. 10.3168/jds.2010-3787 [DOI] [PubMed] [Google Scholar]
- 34.Cogan TM, Barbosa M, Beuvier E, Bianchi-Salvadori B, Cocconcelli PS, Fernandes I, Gomez J, Gomez R, Kalantzpoulos G, Ledda A, Medina M, Rea MC, Rodriguez E. 1997. Characterization of the lactic acid bacteria in artisanal dairy products. J. Dairy Res. 64:409–421. 10.1017/S0022029997002185 [DOI] [Google Scholar]
- 35.Prodromou K, Thasitou P, Haritonidou E, Tzanetakis N, Litopoulou-Tzanetaki E. 2001. Microbiology of “Orinotyri,” a ewe's cheese from the Greek mountains. J. Food Microbiol. 18:319–328. 10.1006/fmic.2001.0403 [DOI] [Google Scholar]
- 36.Dolci P, Alessandria V, Rantsiou K, Rolle L, Zeppa G, Cocolin L. 2008. Microbial dynamics of Castelmagno PDO, a traditional Italian cheese, with a focus on lactic acid bacteria ecology. Int. J. Food Microbiol. 122:302–311. 10.1016/j.ijfoodmicro.2007.12.018 [DOI] [PubMed] [Google Scholar]
- 37.Fox PF. 1989. Proteolysis during cheese manufacture and ripening. J. Dairy Sci. 72:1379–1383. 10.3168/jds.S0022-0302(89)79246-8 [DOI] [Google Scholar]
- 38.Di Cagno R, Banks J, Sheehan L, Fox PF, Brechany EY, Corsetti A, Gobbetti M. 2003. Comparison of the microbiological, compositional, biochemical, volatile profile and sensory characteristics of three Italian PDO ewes' milk cheeses. Int. Dairy J. 13:961–972. 10.1016/S0958-6946(03)00145-6 [DOI] [Google Scholar]
- 39.Gobbetti M, Folkertsma B, Fox PF, Corsetti A, Smacchi E, De Angelis M, Rossi J, Kilcawley K, Cortini M. 1999. Microbiology and biochemistry of Fossa (pit) cheese. Int. Dairy J. 9:763–773. 10.1016/S0958-6946(99)00147-8 [DOI] [Google Scholar]
- 40.Ercolini D. 2013. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 79:3148–3155. 10.1128/AEM.00256-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quigley L, McCarthy R, O'Sullivan O, Beresford TP, Fitzgerald GF, Ross RP, Stanton C, Cotter PD. 2013. The microbial content of raw and pasteurized cow milk as determined by molecular approaches. J. Dairy Sci. 96:4928–4937. 10.3168/jds.2013-6688 [DOI] [PubMed] [Google Scholar]
- 42.Ercolini D, Russo F, Ferrocino I, Villani F. 2009. Molecular identification of mesophilic and psychrotrophic bacteria from raw cow's milk. Food Microbiol. 26:228–231. 10.1016/j.fm.2008.09.005 [DOI] [PubMed] [Google Scholar]
- 43.Raats D, Offek M, Minz D, Halpern M. 2011. Molecular analysis of bacterial communities in raw cow milk and the impact of refrigeration on its structure and dynamics. Food Microbiol. 28:465–471. 10.1016/j.fm.2010.10.009 [DOI] [PubMed] [Google Scholar]
- 44.Afzal MI, Jacquet T, Delaunay S, Borges F, Millière JB, Revol-Junelles AM, Cailliez-Grimal C. 2010. Carnobacterium maltaromaticum: identification, isolation tools, ecology and technological aspects in dairy products. Food Microbiol. 27:573–579. 10.1016/j.fm.2010.03.019 [DOI] [PubMed] [Google Scholar]
- 45.Delbès-Paus C, Pochet S, Helinck S, Veisseire P, Bord C, Lebecque A, Coton M, Desmasures N, Coton E, Irlinger F, Montel MC. 2012. Impact of Gram-negative bacteria in interaction with a complex microbial consortium on biogenic amine content and sensory characteristics of an uncooked pressed cheese. Food Microbiol. 30:74–82. 10.1016/j.fm.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 46.Lynch CM, Muir DD, Banks JM, McSweeney PLH. 1999. Influence of adjunct cultures of Lactobacillus paracasei ssp. paracasei or Lactobacillus plantarum on cheddar cheese ripening. J. Dairy Sci. 82:1618–1628. 10.3168/jds.S0022-0302(99)75390-7 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.