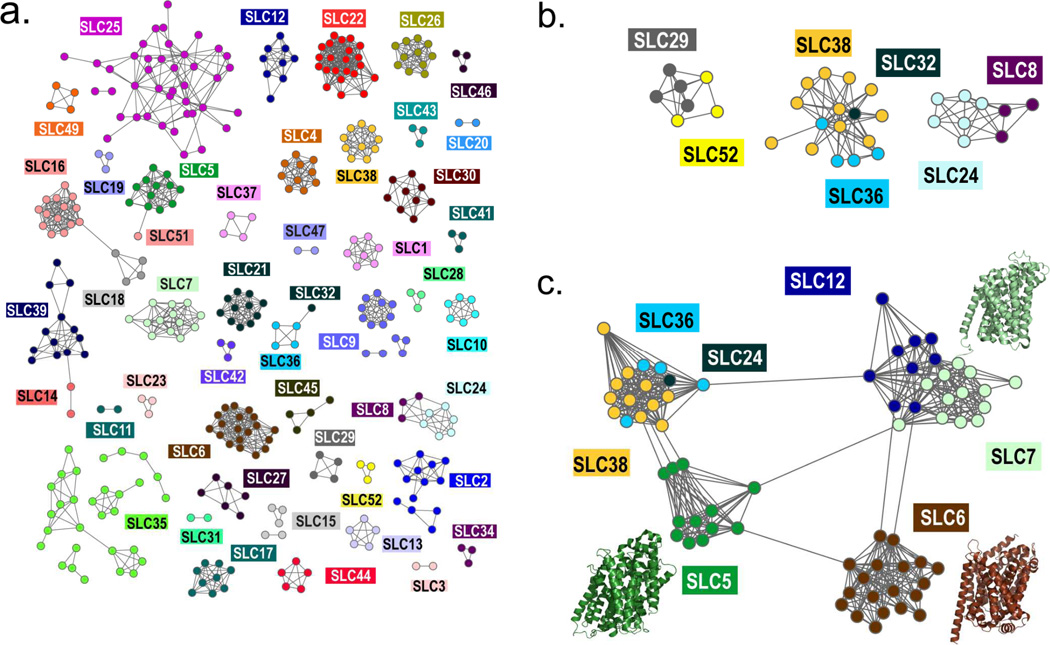
Sequence-based similarity networks
A sequence similarity network is made up of links corresponding to pairwise relationships that score better than a defined cutoff1,2 (Fig. 1). Pairwise sequence alignment scores, including percent sequence identity and Expectation Value (E-Value), were computed using SALIGN3. The E-value of a match is the number of sequences in the queried database that are expected to match by chance the query sequence at least as well as the assessed match; smaller values indicate more statistically significant alignments3. The E-value cutoffs for the final similarity networks were selected manually, similarly to our previous analysis1,2. Because of the small database that was used for the analysis (i.e., 386 sequences), E-value cutoffs that typically do not represent meaningful relationship among sequences when using large databases (e.g., E-value of 1) were also considered. Finally, the graphs representing the similarity networks were visualized using Cytoscape 2.8.14. We used the yFiles organic layout algorithm, which maintains all the connections between the nodes to illustrate relationships. Groups of nodes that are inter-connected usually cluster together in the network.
Figure 1.
Sequence similarity between human SLC sequences and PDB structures
For each transporter structure, we retrieved the amino acid sequence from the UniProt database5. We then ran the alignment server HHpred6 against the human proteome, using the default parameters. Finally, we selected the alignment between the query sequence of known structure and the human transporter protein with the highest sequence identity, and also retrieved the E-value for the alignment (Table 1 and Supplementary Table 1).
Table 1.
Drug ADME SLC families that can be modeled based on atomic resolution structures from other organisms.
| Familya | Functionb | Template Structurec |
Percent Sequence Identityd |
Representative Drug Substratese |
|---|---|---|---|---|
| SLC7 (14) | Cationic amino acid transporter/glycoprotein-associated family |  |
21 (1.4 × 10−47) | Melphalan, gabapentin, levodopa, baclofen |
| SLC10 (7) | Na+ bile salt co-transporters | 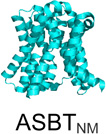 |
26 (1.8 × 10−42) | Rosuvastatin, atorvastatin, fluvastatin |
| SLC15 (4) | Proton oligopeptide co-transporters | 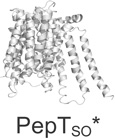 |
34 (2.2 × 10−28) | Valacyclovir, cephalexin, cefadroxil, enalapril, captopril |
| SLC22 (26) | Organic cation/anion/zwitterion transporters |  |
20 (4.9 × 10−34) | Metformin, acyclovir, methotrexate, olmesartan, ipratropium, oxaliplatin, cimetidine |
| SLC28 (3) | Na+-coupled nucleoside transporters | 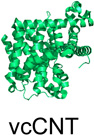 |
40 (6.4 × 10−130) | Fludarabine, gemcitabine, cytarabine |
| SLC47 (2) | Multidrug and toxin extrusion (MATE) transporters |  |
23 (4.8 × 10−51) | Metformin, trospium, fexofenadine |
Family marks the human SLC family, as annotated by the Bioparadigms database2. The number of human protein sequences in the family is provided in parenthesis.
Function gives the function of the human family, as described in the Bioparadigms database
Template Structure describes the most related atomic structure to the family. Structures with the MFS and NSS folds are marked with ‘*’ and ‘#‘, respectively. Detailed description of the structures, including the full name of the proteins and the corresponding references are described in the Supplementary Material.
Percent Sequence Identity provides the percent sequence identity of the best scoring hit from each family; E-value is given in parenthesis (Supplementary Material)
Representative Drug Substrates gives examples of key prescription drugs that are substrates of the transporter.
Atomic structures of homologs of drug ADME SLC transporters
The structures described in Table 1 are of the amino acid antiporter AdiC from Escherichia coli7, the, homolog of the apical sodium-dependent bile acid transporter from Neisseria meningitidis (ASBTNM)8, the peptide transporter from Shewanella oneidensis (PepTSO)9, the high-affinity phosphate importer PiPT from Piriformospora indica10, the concentrative nucleoside transporter from Vibrio cholera (vcCNT)11, and the multidrug and toxic compound extrusion transporter NorM from Vibrio cholera12.
Supplementary Material
REFERENCES
- 1.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlessinger A, et al. Comparison of human solute carriers. Protein science : a publication of the Protein Society. 2010;19:412–428. doi: 10.1002/pro.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madhusudhan MS, Webb BM, Marti-Renom MA, Eswar N, Sali A. Alignment of multiple protein structures based on sequence and structure features. Protein Eng Des Sel. 2009;22:569–574. doi: 10.1093/protein/gzp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Universal Protein Resource (UniProt) in 2010. Nucleic acids research. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic acids research. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao X, et al. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature. 2010;463:828–832. doi: 10.1038/nature08741. [DOI] [PubMed] [Google Scholar]
- 8.Hu NJ, Iwata S, Cameron AD, Drew D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature. 2011;478:408–411. doi: 10.1038/nature10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newstead S, et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. The EMBO journal. 2011;30:417–426. doi: 10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedersen BP, Kumar H, Waight AB, Risenmay AJ, Roe-Zurz Z, Chau BH, Schlessinger A, Bonomi M, Harries W, Sali A, Johri AK, Stroud RM. Crystal structure of a eukaryotic phosphate transporter. Nature. 2013 doi: 10.1038/nature12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson ZL, Cheong CG, Lee SY. Crystal structure of a concentrative nucleoside transporter from Vibrio cholerae at 2.4 A. Nature. 2012;483:489–493. doi: 10.1038/nature10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He X, et al. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467:991–994. doi: 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 14.Nugent T, Jones DT. Transmembrane protein topology prediction using support vector machines. BMC bioinformatics. 2009;10:159. doi: 10.1186/1471-2105-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: consensus prediction of membrane protein topology. Nucleic acids research. 2009;37:W465–W468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 17.Sun L, et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature. 2012;490:361–366. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 18.Faham S, et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321:810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 20.Mancusso R, Gregorio GG, Liu Q, Wang DN. Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter. Nature. 2012;491:622–626. doi: 10.1038/nature11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berardi MJ, Shih WM, Harrison SC, Chou JJ. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature. 2011;476:109–113. doi: 10.1038/nature10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 23.Hediger MA, et al. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteinsIntroduction. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.