Abstract
Objective
This meta-analysis aimed to comprehensively examine the relationship between the clinicopathological and demographical characteristics and ALK rearrangements in patients with non-small cell lung cancer (NSCLC).
Methods and Main Findings
In total, 62 qualified articles including 1178 ALK rearranged cases from 20541 NSCLC patients were analyzed, and the data were extracted independently by two investigators. NSCLC patients with ALK rearrangements tended to be younger than those without (mean difference: −7.16 years; 95% confidence interval (95% CI): −9.35 to −4.96; P<0.00001), even across subgroups by race. Compared with female NSCLC patients, the odds ratio (OR) of carrying ALK rearrangements was reduced by 28% (95% CI: 0.58–0.90; P = 0.004) in males, and this reduction was potentiated in Asians, yet in opposite direction in Caucasians. Likewise, smokers were less likely to have ALK rearrangements than never-smokers (OR = 0.33; 95% CI: 0.25–0.44; P<0.00001), even in race-stratified subgroups. Moreover, compared with NSCLC patients with tumor stage IV, ALK rearrangements were underrepresented in those with tumor stage I–III (OR = 0.58; 95% CI: 0.44–0.78; P = 0.0002). Patients with lung adenocarcinomas had a significantly higher rate of ALK rearrangements (7.2%) than patients with non-adenocarcinoma (2.0%) (OR = 2.25; 95% CI: 1.54–3.27; P<0.0001).
Conclusion
Our findings demonstrate that ALK rearrangements tended to be present in NSCLC patients with no smoking habit, younger age and tumor stage IV. Moreover, race, age, gender, smoking status, tumor stage and histology might be potential sources of heterogeneity.
Introduction
Lung cancer is the leading cause of cancer deaths worldwide. Most of lung cancer patients are diagnosed at an advanced stage with extremely poor prognoses. Non-small cell lung cancer (NSCLC) accounts for approximately 80% of all lung cancers. With the advancements of medical science, much hope has been laid on pharmacogenomics as a novel approach to circumvent problems in individualized medical therapy for cancer. For example, NSCLC patients with activating mutations in epidermal growth factor receptor (EGFR) gene had a good response to its tyrosine kinase inhibitors [1]. In 2007, Soda and colleagues first identified a tyrosine kinase as a promising therapeutic target and diagnostic molecular marker for NSCLC, and this kinase accelerates the formation of a fusion gene comprising the portions of echinoderm microtubule-associated protein-like 4 (EML4) and the anaplastic lymphoma kinase (ALK) in NSCLC cells [2]. This formation is biologically important as EML4 activates ALK kinase via mediating ligand-independent oligomerization of ALK [3], [4], and the activated ALK is responsible for the growth and survival of lung cancer cell lines, the process being highly sensitive to ALK kinase inhibitors [5]. Several clinical data have reported that administration of crizotinib, an inhibitor of ALK tyrosine kinases, was beneficial to lung cancer patients with ALK rearrangements [6], [7]. Epidemiologic studies showed that ALK rearrangements tended to occur in younger patients, never or light smokers and patients with adenocarcinoma rather than squamous cell or large cell carcinoma [8], [9]. In addition, there was evidence for a mutually exclusive condition between ALK rearrangements and EGFR or KRAS mutations [10]. It is estimated that the incidence of ALK rearrangements in unselected NSCLC populations is 2%–7% [9], [11], [12], indicating that only a small proportion of NSCLC patients will benefit from ALK kinase inhibitors, and the accurate and timely identification of these patients will have important therapeutic implications. Therefore, understanding the clinicopathological characteristics of ALK rearrangements will be a major requirement for optimal management of NSCLC patients. However, a comprehensive evaluation of these characteristics so far is lacking in medical literature. Given the accumulating data, there is an urgent need to synthesize available articles by means of a meta-analysis to comprehensively examine the relationship between the clinicopathological and demographical characteristics and ALK rearrangements in NSCLC patients.
Materials and Methods
We carried out this meta-analysis of cross-sectional studies in accordance with the guidelines set forth by the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) statement (see Checklist S1) [13].
Search strategy for identification of studies
We searched PubMed and EMBASE (Excerpta Medica database) for articles published before December 10, 2013. Subject terms included anaplastic lymphoma kinase or ALK and lung cancer. Search results were expressed in Boolean expression: ((anaplastic lymphoma kinase) OR ALK) AND (lung cancer)) AND English [Language].
Study selection
Two investigators (L.F. and Y.F.) independently obtained the full texts of potentially eligible articles based on their titles and abstracts. To avoid the double counting of the patients recruited in more than one publication, the authors were contacted for inquiry when necessary. Where more than one publication of a study population existed, we extracted information from the most recent or most complete publication.
Inclusion/exclusion criteria
Articles were included if they met the following criteria: (1) NSCLC was diagnosed based on either histological or cytological results; (2) ALK rearrangements were determined in ALK-status unknown NSCLC patients by using FISH or IHC or PCR method; (3) one or more clinicopathological or demographical characteristics including age, gender, smoking habit, tumor stage, histology and EGFR/KRAS mutation status were provided between ALK-rearranged and ALK negative NSCLC patients. Articles were excluded if they lacked valid data for comparisons between patients with and without ALK rearrangements across clinicopathological or demographical characteristics, or if they were conference abstracts/proceedings, case reports/series, editorials, narrative reviews and the non-English articles.
Data extraction
Two authors (L.F. and Y.F.) of this study independently extracted the following information: the first author's name, year of publication, sample size, the method to detect ALK rearrangements, tumor histology and stage, age, gender, race and smoking status if available. The discrepancies were resolved by the discussion and review of original articles, and a consensus was reached finally.
Statistical analysis
The meta-analysis was conducted using the open-source Review Manager (RevMan) Software (version 5.2.4, available at the website http://ims.cochrane.org/revman/download). Irrespective of the presence of heterogeneity between studies, the random-effects model was employed to combine individual effect-size estimates. The relationship between clinicopathological or demographical profiles including gender, smoking status, tumor stage, histology and ALK rearrangements were assessed by inverse variance (IV) method, and effect estimates were expressed as odds ratio (OR) [14] or weighted mean difference (WMD) and 95% confidence interval (95% CI). Age difference was estimated with inverse variance method and contrasts were expressed in the form of mean difference and 95% CI. For those articles with only median age and age range, we estimated mean age and standard deviation using the methods described by Hozo [15].
Heterogeneity was examined using the inconsistency index (I 2) statistic, which ranges from 0% to 100% and is defined as the percentage of the observed between-trial variability that is due to heterogeneity rather than chance. Generally, I 2>50% was used as a threshold indicating significant heterogeneity. Meta-regression analysis was carried out to evaluate the extent to which different study-level variables including clinicopathological or demographical characteristics as mentioned above explained the heterogeneity of pooled effect estimates.
Publication bias was assessed by the fail-safe number (Nfs). If the Nfs was smaller than the number of observed studies for the same comparison, this was interpreted as meaning that mete-analysis result might have a significant publication bias as previously described [16]. The Nfs was calculated as 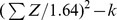 , where k is equal to the number of articles involved in calculation.
, where k is equal to the number of articles involved in calculation.
Results
Qualified articles
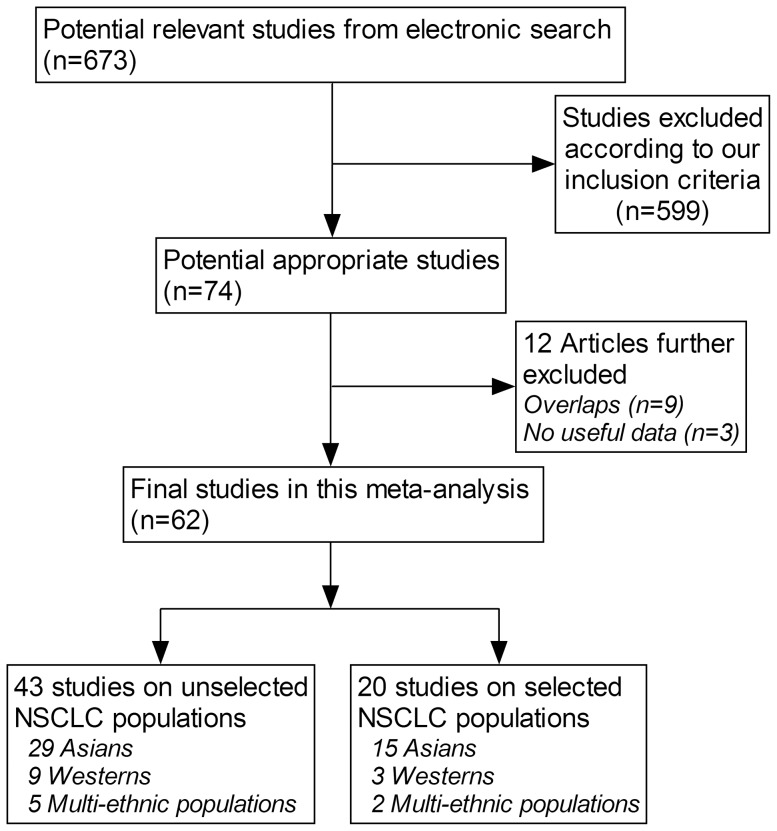
Based on the search strategy, a total of 20541 NSCLC patients were analyzed from 62 qualified articles [4],[8],[11],[12],[17]–[74], and of them 1178 patients (5.7%) had ALK rearrangements. 43 of 62 articles were conducted in Asians (19 in Chinese, 13 in Japanese, 10 in Koreans and 1 in Indians), 12 articles in Caucasians (7 in Americans and 5 in Europeans) and 7 articles in multi-ethnic populations. 43 articles involved unselected populations [4], [8], [11], [12], [17]–[55], and 20 articles enrolled specific groups of NSCLC patients according to either clinicopathological characteristics or genetic makeup [46], [56]–[74], with one article [46] provided data from both unselected and selected NSCLC groups. A flow diagram schematizing the process of excluding articles with specific reasons is presented in Figure 1.
Figure 1. Flow diagram of search strategy and study selection.
Characteristics
The characteristics of all qualified articles are summarized in Table 1 and Table S1. The incidence of ALK rearrangements ranged from 0.99% to 15.00% in unselected NCSLC groups based on 43 articles (4.77% in 29 East Asian groups; 3.91% in 9 Caucasian groups; 5.15% in 5 multi-ethnic groups). For selected NSCLC groups according to gender, smoking status, tumor stage, or EGFR mutation, the incidence of ALK rearrangements ranged from 4.30% to 34.78% based on 20 articles (15 in East Asians; 3 in Caucasians; 2 in multi-ethnic groups).
Table 1. Clinicopathological and demographical characteristics of NSCLC patients with ALK rearrangements.
| Comparisons | Subgroups | Articles | Characteristics | Effect estimate (95% CI) | p value | df(p) | I 2 | |
| Gender | NSCLC | Western | 9 | males (59/559, 10.6%) vs. females (54/536, 10.1%) | OR: 1.12 (0.55, 2.28) | 0.76 | 8(p = 0.05) | 49% |
| Asian | 36 | males (282/6492, 4.3%) vs. females (381/5070, 7.5%) | OR: 0.59 (0.49, 0.73) | <0.00001 | 36(p = 0.09) | 24% | ||
| Mixed | 5 | males (56/638, 8.8%) vs. females (72/1041, 6.9%) | OR: 1.51 (0.86, 2.64) | 0.15 | 4(p = 0.10) | 49% | ||
| Total | 50 | males (397/7689, 5.2%) vs. females (507/6647, 7.6%) | OR: 0.72 (0.58, 0.90) | 0.004 | 50(p<0.0001) | 51% | ||
| Ad only | Western | 4 | males (25/207, 12.1%) vs. females (20/281, 7.2%) | OR: 1.70 (0.64, 4.50) | 0.29 | 3(p = 0.18) | 39% | |
| Asian | 13 | males (136/2130, 6.4%) vs. females (160/2196, 7.3%) | OR: 0.83 (0.65, 1.06) | 0.13 | 12(0.97) | 0% | ||
| Mixed | 2 | males (30/305, 9.8%) vs. females (48/670, 7.2%) | OR: 1.44 (0.66, 3.13) | 0.36 | 1(p = 0.13) | 56% | ||
| Total | 19 | males (191/2642, 7.2%) vs. females (228/3147, 7.2%) | OR: 1.00 (0.79, 1.25) | 0.97 | 18(p = 0.29) | 13% | ||
| Smoking status | NSCLC | Western | 6 | smoker (27/668, 3.6%) vs. non-smoker (57/243, 23.5%) | OR: 0.16 (0.08, 0.32) | <0.00001 | 5(p = 0.25) | 24% |
| Asian | 26 | smoker (142/4363, 3.3%) vs. non-smoker (309/3724, 8.3%) | OR: 0.42 (0.31, 0.58) | <0.00001 | 25(p = 0.01) | 43% | ||
| Mixed | 4 | smoker (25/829, 3.0%) vs. non-smoker (67/492, 13.6%) | OR: 0.19 (0.12, 0.32) | <0.00001 | 3(0.99) | 0% | ||
| Total | 36 | smoker (194/5860, 3.3%) vs. non-smoker (433/4459, 9.7%) | OR: 0.33 (0.25, 0.44) | <0.00001 | 35(p = 0.0007) | 49% | ||
| Ad only | Western | 2 | smoker (6/296, 2.0%) vs. non-smoker (19/97, 19.6%) | OR: 0.06 (0.01, 0.52) | <0.00001 | 1(p = 0.15) | 52% | |
| Asian | 11 | smoker (63/1448, 4.4%) vs. non-smoker (160/1922, 8.3%) | OR: 0.54 (0.39, 0.75) | = 0.0002 | 10(p = 0.41) | 3% | ||
| Mixed | 1 | smoker (9/382, 2.4%) vs. non-smoker (35/293, 11.9%) | OR: 0.18 (0.08, 0.38) | <0.00001 | NA | NA | ||
| Total | 14 | smoker (78/2126, 3.7%) vs. non-smoker (214/2312, 9.3%) | OR: 0.36 (0.23, 0.57) | <0.0001 | 13(p = 0.008) | 54% | ||
| Tumor stage | NSCLC | Western | 3 | stage I–III (13/453, 2.9%) vs. stage IV (18/131, 13.7%) | OR: 0.21 (0.04, 0.21) | 0.05 | 2(p = 0.13) | 51% |
| Asian | 19 | stage I–III (248/5652, 4.4%) vs. stage IV (101/929, 10.9%) | OR: 0.70 (0.51, 0.98) | 0.04 | 18(p = 0.95) | 0% | ||
| Mixed | 4 | stage I–III (41/639, 6.4%) vs. stage IV (43/345, 12.5%) | OR: 0.57 (0.30, 1.11) | 0.10 | 3(p = 0.65) | 0% | ||
| Total | 26 | stage I–III (302/6744, 4.5%) vs. stage IV (162/1405, 11.5%) | OR: 0.58 (0.44, 0.78) | 0.0002 | 25(p = 0.43) | 2% | ||
| Ad only | Western | 1 | stage I–III (4/265, 1.5%) vs. stage IV (16/93, 17.2%) | OR: 0.07 (0.02, 0.23) | <0.00001 | NA | NA | |
| Asian | 7 | stage I–III (87/1727, 5.0%) vs. stage IV (22/222, 9.9%) | OR: 0.73 (0.40, 1.33) | 0.31 | 6(p = 0.83) | 0% | ||
| Mixed | 1 | stage I–III (28/253, 11.1%) vs. stage IV (6/47, 12.8%) | OR: 0.85 (0.33, 2.18) | 0.74 | NA | NA | ||
| Total | 9 | stage I–III (119/2245, 5.3%) vs. stage IV (44/362, 12.1%) | OR: 0.53 (0.25, 1.09) | 0.09 | 8(p = 0.03) | 52% | ||
| Histology | Ad vs. non-Ad | Western | 7 | Ad (100/1654, 6.0%) vs. non-Ad (23/677, 3.4%) | OR: 1.11 (0.59, 2.07) | 0.75 | 6(p = 0.30) | 18% |
| Asian | 23 | Ad (302/3997, 7.6%) vs. non-Ad (28/1898, 1.5%) | OR: 2.92 (1.88, 4.53) | <0.00001 | 22(p = 0.27) | 14% | ||
| Mixed | 3 | Ad (47/549, 8.6%) vs. non-Ad (3/149, 2.0%) | OR: 2.44 (0.79, 7.48) | 0.12 | 2(p = 0.63) | 0% | ||
| Total | 33 | Ad (449/6200, 7.2%) vs. non-Ad (54/2724, 2.0%) | OR: 2.25 (1.54, 3.27) | <0.0001 | 32(p = 0.09) | 26% | ||
| Ad vs. SCC | Western | 5 | Ad (24/443, 5.4%) vs. Scc (5/241, 2.1%) | OR: 1.33 (0.48, 3.68) | 0.58 | 4(p = 0.60) | 0% | |
| Asian | 19 | Ad (227/3594, 6.3%) vs. Scc (11/1415, 0.8%) | OR: 3.64 (2.17, 6.09) | <0.00001 | 18(p = 0.67) | 0% | ||
| Mixed | 3 | Ad (47/549, 8.6%) vs. Scc (1/98, 1.0%) | OR: 1.46 (0.33, 6.54) | 0.62 | 2(p = 0.37) | 0% | ||
| Total | 27 | Ad (298/4586, 6.5%) vs. Scc (17/1754, 1.0%) | OR: 2.79 (1.80, 4.33) | <0.00001 | 26(p = 0.57) | 0% | ||
| Age | NSCLC | Western | 8 | mean age: ALK(+) (54.5, 104) vs. ALK(−) (64.2, 872) | WMD: −6.18 (−15.13, 2.77) | 0.18 | 7(p<0.00001) | 96% |
| Asian | 28 | mean age: ALK(+) (54.6, 519) vs. ALK(−) (62.9, 8647) | WMD: −6.99 (−8.91, −5.08) | <0.00001 | 27(p<0.00001) | 74% | ||
| Mixed | 4 | mean age: ALK(+) (57.3, 120) vs. ALK(−) (66.0, 1248) | WMD: −9.57 (−13.28, −5.86) | <0.00001 | 3(p = 0.06) | 59% | ||
| Total | 40 | mean age: ALK(+) (55.1, 743) vs. ALK(−) (63.4, 10767) | WMD: −7.16 (−9.35, −4.96) | <0.00001 | 39(p<0.00001) | 87% | ||
| Ad only | Western | 4 | mean age: ALK(+) (57.0, 45) vs. ALK(−) (66.1, 444) | WMD: −8.62 (−19.27, 2.01) | 0.11 | 3(p<0.00001) | 92% | |
| Asian | 13 | mean age: ALK(+) (57.9, 242) vs. ALK(−) (62.9, 3826) | WMD: −5.40(−7.35, −3.46) | <0.00001 | 12(p = 0.01) | 52% | ||
| Mixed | 3 | mean age: ALK(+) (58.2, 101) vs. ALK(−) (66.2, 1126) | WMD: −8.98 (−13.52, −4.44) | 0.0001 | 2(p = 0.02) | 74% | ||
| Total | 20 | mean age: ALK(+) (57.9, 388) vs. ALK(−) (63.8, 5396) | WMD: −6.81 (−9.18, −4.45) | <0.00001 | 19(p<0.00001) | 80% | ||
| ALK vs. EGFR | Western | 2 | mean age: ALK(+) (51.9, 48) vs. EGFR mutated (61.5, 52) | WMD: −8.26 (−15.94, −0.57) | 0.04 | 1(p = 0.17) | 47% | |
| Asian | 9 | mean age: ALK(+) (54.1, 110) vs. EGFR mutated (59.7, 891) | WMD: −5.89 (−9.62, −2.16) | 0.002 | 8(p = 0.0005) | 72% | ||
| Mixed | 4 | mean age: ALK(+) (56.5, 161) vs. EGFR mutated (64.4, 582) | WMD: −8.17 (−11.17, −5.17) | <0.00001 | 3(p = 0.13) | 46% | ||
| Total | 15 | mean age: ALK(+) (55.0, 319) vs. EGFR mutated (61.5, 1525) | WMD: −6.95 (−9.32, −4.59) | <0.00001 | 14(p = 0.0003) | 64% | ||
| ALK vs. KRAS | Western | 1 | mean age: ALK(+) (51, 41) vs. KRAS mutated (59.5, 49) | WMD: −8.50 (−14.09, −2.91) | 0.003 | NA | NA | |
| Asian | 6 | mean age: ALK(+) (56.3, 67) vs. KRAS mutated (60.6, 66) | WMD: −4.26 (−7.70, −0.82) | 0.02 | 5(p = 0.46) | 0% | ||
| Mixed | 3 | mean age: ALK(+) (57.1, 142) vs. KRAS mutated (65.8, 707) | WMD: −8.54 (−11.53, −5.56) | <0.00001 | 2(p = 0.16) | 46% | ||
| Total | 10 | mean age: ALK(+) (55.9, 250) vs. KRAS mutated (65.0, 822) | WMD: −6.94 (−9.27, −4.60) | <0.00001 | 9(p = 0.13) | 35% | ||
Abbreviations: NSCLC, non-small-cell lung cancer; Ad, adenocarcinoma; SCC, squamous cell carcinoma; OR, odds ratio; WMD, weighted mean difference; CI, confidence interval.
ALK rearrangements and age
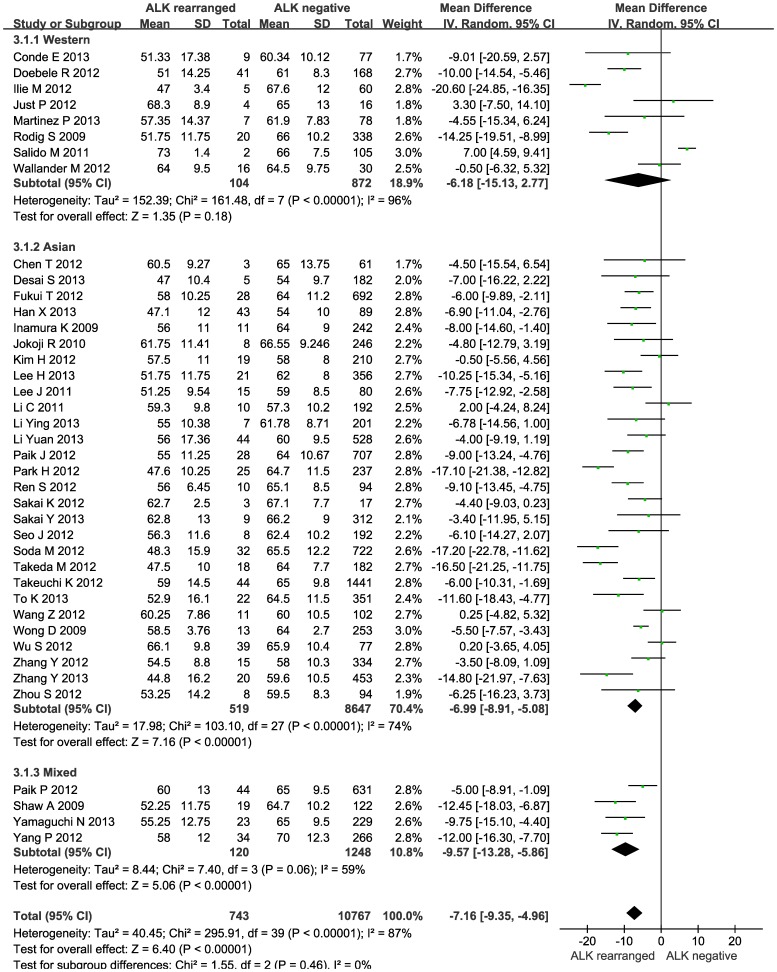
NSCLC patients with ALK rearrangements tended to be younger than those without (WMD: −7.16 years; 95% CI: −9.35 to −4.96; P<0.00001), and this tendency was persisted in Asians (WMD: −6.99; 95% CI: −8.91 to −5.08; P<0.00001) and Caucasians (WMD: −6.18, 95% CI: −15.13 to 2.77; P = 0.18), albeit strong evidence of heterogeneity (Figure 2). Analyzing patients with only lung adenocarcinomas observed similar results with significant heterogeneity for age in Asians (WMD: −5.40; 95% CI: −7.35 to −3.46; P<0.00001) and Caucasians (WMD: −8.62; 95% CI: −19.27 to 2.01, P = 0.11) (Figure S1). As indicated by the Nfs, there was no observable publication bias.
Figure 2. Forest plots of the mean difference of age between NSCLC patients with and without ALK rearrangements by race.
ALK rearrangements and gender
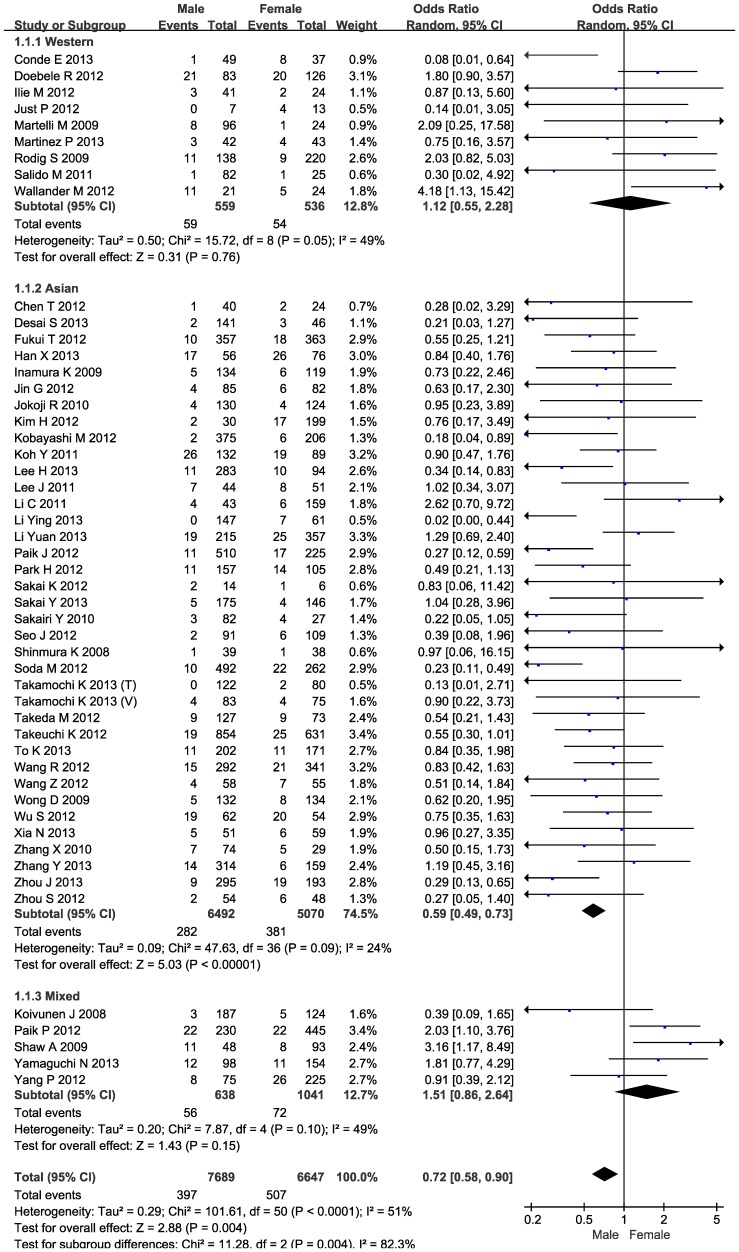
The frequencies of ALK rearrangements ranged respectively from 0% to 30.65% and from 2.63% to 37.04% in male (397/7689) and female (507/6627) patients with NSCLC based on 50 articles. Compared with female patients with NSCLC, the odds of carrying ALK rearrangements was reduced by 28% (95% CI: 0.58–0.90; P = 0.004) in males (I 2 = 51%) (Figure 3). In subgroup analysis by race, further significant reduction in odds was observed in male patients of Asian descent (OR = 0.59; 95% CI: 0.49–0.73; P<0.00001), but there was an elevated yet nonsignificant risk in male patients of Caucasian descent (OR = 1.12; 95% CI: 0.55–2.28; P = 0.76) with no heterogeneity or publication bias (Figure 3). After limiting articles to patients with lung adenocarcinomas (n = 19), risk estimates were similar in direction for male patients of Asian descent (OR = 0.83; 95% CI: 0.65–1.06; P = 0.13) and Caucasian descent (OR = 1.70; 95% CI: 0.64–4.50; P = 0.29), and there was no observable heterogeneity (Figure S2); however, the Nfs values were less than the number of articles in each comparison, indicating the presence of publication bias.
Figure 3. Forest plots of gender difference between NSCLC patients with and without ALK rearrangements by race.
ALK rearrangements and smoking status
The frequencies of ALK rearrangements in NSCLC patients ranged from 0% to 19.44% and from 0% to 41.67% among smokers and never-smokers, respectively. Smokers were less likely to have ALK rearrangements than never-smokers (OR = 0.33; 95% CI: 0.25–0.44; P<0.00001), even in subgroups by race, such as in Asians (OR = 0.42; 95% CI: 0.31–0.58; P<0.00001) and Caucasians (OR = 0.16; 95% CI: 0.08–0.32; P<0.00001) (Figure S3), and there was no heterogeneity and low probabilities of publication bias as reflected by the Nfs. Similarly in patients with only lung adenocarcinomas (14 qualified articles), smokers still had reduced risk compared with never-smokers (OR = 0.36; 95% CI: 0.23–0.57; P<0.00001; I 2 = 54%), even in East Asian patients (OR = 0.54; 95% CI: 0.38–0.75; P<0.00001; I 2 = 0%) (Figure S4), and still heterogeneity and publication bias were unlikely for these comparisons.
ALK rearrangements and tumor stage
ALK rearrangements were detected in 3.85%, 4.31%, 5.72% and 12.39% of NSCLC patients with tumor stage of I (30 articles: 199/5168), II (25 articles: 50/1161), III (27 articles: 119/2080) and IV (28 articles: 214/1727), respectively (Table S2). Compared with NSCLC patients with stage IV, ALK rearrangements were underrepresented in NSCLC patients with stage I–III (OR = 0.58; 95% CI: 0.44–0.78; P = 0.0002), without heterogeneity (I 2 = 2%). The ORs were 0.7 (95% CI: 0.51–0.98; P = 0.04) and 0.21 (95% CI: 0.04–0.21; P = 0.05) in East Asians (I 2 = 0%) and Caucasians (I 2 = 51%), respectively (Figure S5), and this finding was unlikely explained by publication bias. When only lung adenocarcinomas was concerned, NSCLC patients with stage I–III had lower yet nonsignificant incidence of ALK rearrangements than those with stage IV (OR = 0.53; 95% CI: 0.25–1.09; P = 0.09), with moderate heterogeneity (I 2 = 52%) and evident publication bias (Nfs = −1.14) (Figure S6).
ALK rearrangements and histology
ALK rearrangements were observed in 6.92% (55 articles: 918/13275), 0.92% (26 articles: 16/1746), 11.54% (13 articles: 9/78) and 5.97% (13 articles: 4/67) patients with lung adenocarcinomas, squamous carcinomas, adenosquamous carcinomas and large cell carcinomas, respectively (Table S3). Patients with lung adenocarcinomas had a significantly higher rate of ALK rearrangements (449/6200; 7.2%) than patients with non-adenocarcinoma (54/2724; 2.0%) (OR = 2.25; 95% CI: 1.54–3.27; P<0.0001; I 2 = 26%). In subgroup analysis by race, significance was only noted in Asians (OR = 2.92; 95% CI: 1.88–4.53; P<0.00001), without heterogeneity (I 2 = 14%) or publication bias (Nfs = 249.5) (Figure S7). Compared with squamous carcinomas, lung adenocarcinomas was associated with an increased rate of ALK rearrangements in Asians (OR = 3.64; 95% CI: 2.17–6.09; P<0.00001) with no heterogeneity (I 2 = 0%) and publication bias (Nfs = 168.2), whereas this increase was not obvious in Caucasians (OR = 1.33; 95% CI: 0.48–3.68; P = 0.58; I 2 = 0%) with evident publication bias (Nfs = −0.83) (Figure S8).
ALK rearrangements and EGFR/KRAS mutations
Generally, EGFR/KRAS mutations were more common than ALK rearrangements in NSCLC patients, and they rarely coexisted according to available reports in Table S4. Never-smokers tended to have EGFR mutations and ALK rearrangements, and smokers more likely had KRAS mutations. Moreover, ALK rearrangements tended to occur in younger NSCLC patients compared with EGFR (Figure S9) and KRAS (Figure S10) mutations, and there was no observable publication bias.
Meta-regression analysis
To further explore the extent to which study-level variables explain heterogeneity among individual effect estimates, we performed a set of meta-regression analyses, and none of clinicopathological or demographical characteristics examined can significantly explain the changes of ALK rearrangements in NSCLC patients (data not shown).
Discussion
Via a meta-analysis of the data from 62 articles and on 20541 NSCLC patients, we examined the relationship between a panel of clinicopathological and demographical characteristics and ALK rearrangements. The most notable finding of this study was that ALK rearrangements tended to be present in NSCLC patients with no smoking habit, younger age and tumor stage IV. Moreover, race, age, gender, smoking status, tumor stage and histology might be potential sources of between-study heterogeneity. Although other sources of heterogeneity cannot be easily ruled out, this study, to the best of our knowledge, is so far the largest meta-analysis examining the relationship between clinicopathological or demographical characteristics and ALK rearrangements in NSCLC patients.
A substantial body of evidence supports the prognostic and predictive value of ALK rearrangements in NSCLC patients [75]. Given the low prevalence of ALK rearrangements in NSCLC patients, the present meta-analysis was designed to resolve inconsistent results based on sparse data on clinicopathological or demographical characteristics from studies that may have been inconclusive due to the small sample size involved. A recent meta-analysis by Li and colleagues in 14 articles involving 125 ALK rearranged cases from 2580 NSCLC patients indicated high rate of ALK rearrangements in never-smokers with adenocarcinomas [50]. The present study is more comprehensive than the study by Li and colleagues in terms of the following aspects: first, we focused on a broader spectrum of clinicopathological or demographical characteristics in relation with ALK rearrangements in NSCLC patients; second, a more comprehensive subgroup analyses were conducted by age, gender, race, smoking status, tumor stage and histology, the existence of EGFR/KRAS mutations; third, our study involved more relevant articles (62 qualified articles: 1178 ALK rearranged cases from 20541 NSCLC patients), as the large number of articles provided sufficient power to assess a modest effect estimate. Consistent with the results of study by Li and colleagues and others [30], [50], never-smokers had a low occurrence rate of ALK rearrangements compared with smokers among NSCLC patients. In case of gender, conflicting findings were reported [9], [11], [38], [69], as well as in the meta-analysis by Li and colleagues [50], whereas our data showed remarkably lower risk of carrying ALK rearrangements in males than females.
Besides the smoking status- and gender-specific differences in clinicopathological or demographical features between NSCLC patients with and without ALK rearrangements, ethnic difference merits special consideration. Previous studies found similar overall distributions of ALK rearrangements between NSCLC patients of Asian and Caucasian descent [43], [76]. However in this meta-analysis, the prevalence of ALK rearrangements was heterogeneous between East Asians and Caucasians in terms of age, gender and histology. For example, ALK rearrangements tended to be prevalent in female and non-squamous patients in East Asians; however, there were no obvious clinical differences in Caucasians, indicating that tumorigenesis may be at least in part explained by race. As such, the findings presented in this meta-analysis must be evaluated with caution, as there is a danger of extrapolating the findings of Asians to other ethnic groups with high prevalence of ALK rearrangements.
Our findings also confirmed the view that ALK rearrangements are more common in patients at advanced NSCLC [69], [72] and in patients with lung adenocarcinoma than with non-adenocarcinoma especially squamous cell carcinoma [8], [9]. In agreement with the results of most previous reports, our meta-analysis indicated that ALK rearrangements were most frequently occurred in NSCLC patients of stage IV. Additionally extending this view, we in this meta-analysis found that the relationship between ALK rearrangements and lung cancer histology was race-dependent. In NSCLC patients of East Asian descent, lung adenocarcinomas had a markedly higher rate of ALK rearrangements than squamous cell carcinomas, while this was not the case in Caucasians, likely due to more cases of squamous cell carcinoma in Caucasians than East Asians (2.1% versus 0.8%).
Except for ALK rearrangements, other mutations such as EGFR/KRAS mutations were also commonly seen in NSCLC patients [43], [44]. However, there were rare coexistences of ALK rearrangements with EGFR/KRAS mutations. Considering that a considerable proportion of NSCLC patients had EGFR/KRAS mutations, it could be expected that ALK rearrangements will be more common among patients with EGFR/KRAS wild-type mutations, as reported by some observations [60], [62], [77]. It is of interest to note that Asian NSCLC patients who had ALK rearrangements shared similar features to those with EGFR mutations in terms of gender, smoking status and adenocarcinoma [1]. Therefore, improved understanding of EGFR/KRAS mutation status may facilitate the identification of NSCLC patients with ALK rearrangements, and further the development of more targeted therapy.
Some limitations should be considered when interpreting our results. First, only published studies in English language were retrieved in this meta-analysis, and publication bias might be possible. Second, ALK rearrangements were rare in NSCLC patients, which may limit the statistical power to detect publication bias. Third, the detection methods of ALK rearrangements were heterogeneous across retrieved articles, which may increase the risk of between-study heterogeneity. Last but not the least, there were many other clinicopathological or demographical characteristics that were poorly known, such as high frequencies of signet-ring cell and mucinous cribriform patterns in ALK rearranged adenocarcinomas [17], [78].
Taken together, our findings demonstrate that ALK rearrangements tended to be present in NSCLC patients with no smoking habit, younger age and tumor stage IV. Moreover, race, gender, age, smoking status, tumor stage and histology might be potential sources of between-study heterogeneity. Nevertheless, for practical reasons, we hope that this study will not remain just another endpoint of research but instead would encourage more validation studies of our findings in other independent large populations, which would acquiring a better understanding of ALK rearrangements in NSCLC patients.
Supporting Information
The PRISMA checklist.
(DOC)
Forest plots of the mean difference of age between lung adenocarcinomas patients with and without ALK rearrangements by race.
(PDF)
Forest plots of gender difference between lung adenocarcinomas patients with and without ALK rearrangements by race.
(PDF)
Forest plots of smoking difference between NSCLC patients with and without ALK rearrangements by race.
(PDF)
Forest plots of smoking difference between lung adenocarcinomas patients with and without ALK rearrangements by race.
(PDF)
Forest plots of tumor stage difference between NSCLC patients with and without ALK rearrangements by race.
(PDF)
Forest plots of tumor stage difference between lung adenocarcinomas patients with and without ALK rearrangements by race.
(PDF)
Forest plots of distribution difference in ALK rearrangements between patients with and without lung adenocarcinomas by race.
(PDF)
Forest plots of distribution difference in ALK rearrangements between patients with lung adenocarcinomas and squamous carcinomas by race.
(PDF)
Forest plots of the mean difference of age between NSCLC patients with ALK rearrangements and with EGFR mutations by race.
(PDF)
Forest plots of the mean difference of age between NSCLC patients with ALK rearrangements and with KRAS mutations by race.
(PDF)
The baseline characteristics of all qualified studies in this meta-analysis.
(DOC)
ALK rearrangements and tumor stage.
(DOC)
ALK rearrangements and NSCLC histology.
(DOC)
The baseline characteristics of all qualified articles assessing both ALK rearrangements and EGFR/KRAS mutations.
(DOC)
Funding Statement
This study was supported by the National Natural Science Foundation of China (81201837). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, et al. (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- 2. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, et al. (2007) Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566. [DOI] [PubMed] [Google Scholar]
- 3. Mano H (2008) Non-solid oncogenes in solid tumors: EML4-ALK fusion genes in lung cancer. Cancer Sci 99: 2349–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shinmura K, Kageyama S, Tao H, Bunai T, Suzuki M, et al. (2008) EML4-ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer 61: 163–169. [DOI] [PubMed] [Google Scholar]
- 5. Chen Z, Sasaki T, Tan X, Carretero J, Shimamura T, et al. (2010) Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res 70: 9827–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, et al. (2010) Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363: 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, et al. (2011) Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 12: 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paik JH, Choi CM, Kim H, Jang SJ, Choe G, et al. (2012) Clinicopathologic implication of ALK rearrangement in surgically resected lung cancer: a proposal of diagnostic algorithm for ALK-rearranged adenocarcinoma. Lung Cancer 76: 403–409. [DOI] [PubMed] [Google Scholar]
- 9. Soda M, Isobe K, Inoue A, Maemondo M, Oizumi S, et al. (2012) A Prospective PCR-Based Screening for the EML4-ALK Oncogene in Non-Small Cell Lung Cancer. Clin Cancer Res 11: 11. [DOI] [PubMed] [Google Scholar]
- 10. Takahashi T, Sonobe M, Kobayashi M, Yoshizawa A, Menju T, et al. (2010) Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol 17: 889–897. [DOI] [PubMed] [Google Scholar]
- 11. Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, et al. (2012) RET, ROS1 and ALK fusions in lung cancer. Nat Med 18: 378–381. [DOI] [PubMed] [Google Scholar]
- 12. Dai Z, Kelly JC, Meloni-Ehrig A, Slovak ML, Boles D, et al. (2012) Incidence and patterns of ALK FISH abnormalities seen in a large unselected series of lung carcinomas. Mol Cytogenet 5: 1755–8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moher D, Liberati A, Tetzlaff J, Altman DG (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 151: 264–269, W264. [DOI] [PubMed] [Google Scholar]
- 14. Sweeting MJ, Sutton AJ, Lambert PC (2004) What to add to nothing? Use and avoidance of continuity corrections in meta-analysis of sparse data. Stat Med 23: 1351–1375. [DOI] [PubMed] [Google Scholar]
- 15. Hozo SP, Djulbegovic B, Hozo I (2005) Estimating the mean and variance from the median, range, and the size of a sample. BMC medical research methodology 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Niu W, Qi Y, Qian Y, Gao P, Zhu D (2009) The relationship between apolipoprotein E epsilon2/epsilon3/epsilon4 polymorphisms and hypertension: a meta-analysis of six studies comprising 1812 cases and 1762 controls. Hypertens Res 32: 1060–1066. [DOI] [PubMed] [Google Scholar]
- 17. Zhou JX, Yang H, Deng Q, Gu X, He P, et al. (2013) Oncogenic driver mutations in patients with non-small-cell lung cancer at various clinical stages. Ann Oncol 24: 1319–1325. [DOI] [PubMed] [Google Scholar]
- 18. Conde E, Angulo B, Izquierdo E, Munoz L, Suarez-Gauthier A, et al. (2013) The ALK translocation in advanced non-small-cell lung carcinomas: preapproval testing experience at a single cancer centre. Histopathology 62: 609–616. [DOI] [PubMed] [Google Scholar]
- 19. Wang R, Hu H, Pan Y, Li Y, Ye T, et al. (2012) RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol 30: 4352–4359. [DOI] [PubMed] [Google Scholar]
- 20. Soda M, Isobe K, Inoue A, Maemondo M, Oizumi S, et al. (2012) A prospective PCR-based screening for the EML4-ALK oncogene in non-small cell lung cancer. Clin Cancer Res 18: 5682–5689. [DOI] [PubMed] [Google Scholar]
- 21. Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, et al. (2012) The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 22: 2109–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakai K, Okamoto I, Takezawa K, Hirashima T, Kaneda H, et al. (2012) A novel mass spectrometry-based assay for diagnosis of EML4-ALK-positive non-small cell lung cancer. J Thorac Oncol 7: 913–918. [DOI] [PubMed] [Google Scholar]
- 23. Rimkunas VM, Crosby KE, Li D, Hu Y, Kelly ME, et al. (2012) Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: identification of a FIG-ROS1 fusion. Clin Cancer Res 18: 4449–4457. [DOI] [PubMed] [Google Scholar]
- 24. Cardarella S, Ortiz TM, Joshi VA, Butaney M, Jackman DM, et al. (2012) The introduction of systematic genomic testing for patients with non-small-cell lung cancer. J Thorac Oncol 7: 1767–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paik PK, Johnson ML, D'Angelo SP, Sima CS, Ang D, et al. (2012) Driver mutations determine survival in smokers and never-smokers with stage IIIB/IV lung adenocarcinomas. Cancer 118: 5840–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee JK, Kim TM, Koh Y, Lee SH, Kim DW, et al. (2012) Differential sensitivities to tyrosine kinase inhibitors in NSCLC harboring EGFR mutation and ALK translocation. Lung Cancer 77: 460–463. [DOI] [PubMed] [Google Scholar]
- 27. Kanaji N, Bandoh S, Ishii T, Tadokoro A, Watanabe N, et al. (2012) Detection of EML4-ALK fusion genes in a few cancer cells from transbronchial cytological specimens utilizing immediate cytology during bronchoscopy. Lung Cancer 77: 293–298. [DOI] [PubMed] [Google Scholar]
- 28. Jin G, Jeon HS, Lee EB, Kang HG, Yoo SS, et al. (2012) EML4-ALK fusion gene in Korean non-small cell lung cancer. J Korean Med Sci 27: 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ilie M, Long E, Butori C, Hofman V, Coelle C, et al. (2012) ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 23: 2907–2913. [DOI] [PubMed] [Google Scholar]
- 30. Fukui T, Yatabe Y, Kobayashi Y, Tomizawa K, Ito S, et al. (2012) Clinicoradiologic characteristics of patients with lung adenocarcinoma harboring EML4-ALK fusion oncogene. Lung Cancer 77: 319–325. [DOI] [PubMed] [Google Scholar]
- 31. Chen TD, Chang IC, Liu HP, Wu YC, Wang CL, et al. (2012) Correlation of anaplastic lymphoma kinase overexpression and the EML4-ALK fusion gene in non-small cell lung cancer by immunohistochemical study. Chang Gung Med J 35: 309–317. [DOI] [PubMed] [Google Scholar]
- 32. An SJ, Chen ZH, Su J, Zhang XC, Zhong WZ, et al. (2012) Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status. PLoS One 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salido M, Pijuan L, Martinez-Aviles L, Galvan AB, Canadas I, et al. (2011) Increased ALK gene copy number and amplification are frequent in non-small cell lung cancer. J Thorac Oncol 6: 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X, Zhang S, Yang X, Yang J, Zhou Q, et al. (2010) Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer 9: 1476–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sakairi Y, Nakajima T, Yasufuku K, Ikebe D, Kageyama H, et al. (2010) EML4-ALK fusion gene assessment using metastatic lymph node samples obtained by endobronchial ultrasound-guided transbronchial needle aspiration. Clin Cancer Res 16: 4938–4945. [DOI] [PubMed] [Google Scholar]
- 36. Jokoji R, Yamasaki T, Minami S, Komuta K, Sakamaki Y, et al. (2010) Combination of morphological feature analysis and immunohistochemistry is useful for screening of EML4-ALK-positive lung adenocarcinoma. J Clin Pathol 63: 1066–1070. [DOI] [PubMed] [Google Scholar]
- 37. Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, et al. (2009) The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer 115: 1723–1733. [DOI] [PubMed] [Google Scholar]
- 38. Rodig SJ, Mino-Kenudson M, Dacic S, Yeap BY, Shaw A, et al. (2009) Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res 15: 5216–5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martelli MP, Sozzi G, Hernandez L, Pettirossi V, Navarro A, et al. (2009) EML4-ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am J Pathol 174: 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Inamura K, Takeuchi K, Togashi Y, Hatano S, Ninomiya H, et al. (2009) EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol 22: 508–515. [DOI] [PubMed] [Google Scholar]
- 41. Boland JM, Erdogan S, Vasmatzis G, Yang P, Tillmans LS, et al. (2009) Anaplastic lymphoma kinase immunoreactivity correlates with ALK gene rearrangement and transcriptional up-regulation in non-small cell lung carcinomas. Hum Pathol 40: 1152–1158. [DOI] [PubMed] [Google Scholar]
- 42. Zhang YG, Jin ML, Li L, Zhao HY, Zeng X, et al. (2013) Evaluation of ALK rearrangement in Chinese non-small cell lung cancer using FISH, immunohistochemistry, and real-time quantitative RT- PCR on paraffin-embedded tissues. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamaguchi N, Vanderlaan PA, Folch E, Boucher DH, Canepa HM, et al. (2013) Smoking status and self-reported race affect the frequency of clinically relevant oncogenic alterations in non-small-cell lung cancers at a United States-based academic medical practice. Lung Cancer 7: 013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xia N, An J, Jiang QQ, Li M, Tan J, et al. (2013) Analysis of EGFR, EML4-ALK, KRAS, and c-MET mutations in Chinese lung adenocarcinoma patients. Exp Lung Res 6: 6. [DOI] [PubMed] [Google Scholar]
- 45. To KF, Tong JH, Yeung KS, Lung RW, Law PP, et al. (2013) Detection of ALK rearrangement by immunohistochemistry in lung adenocarcinoma and the identification of a novel EML4-ALK variant. J Thorac Oncol 8: 883–891. [DOI] [PubMed] [Google Scholar]
- 46. Takamochi K, Takeuchi K, Hayashi T, Oh S, Suzuki K (2013) A Rational Diagnostic Algorithm for the Identification of ALK Rearrangement in Lung Cancer: A Comprehensive Study of Surgically Treated Japanese Patients. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sakai Y, Nakai T, Ohbayashi C, Imagawa N, Yanagita E, et al. (2013) Immunohistochemical Profiling of ALK Fusion Gene-Positive Adenocarcinomas of the Lung. Int J Surg Pathol 20: 20. [DOI] [PubMed] [Google Scholar]
- 48. Martinez P, Hernandez-Losa J, Montero MA, Cedres S, Castellvi J, et al. (2013) Fluorescence in situ hybridization and immunohistochemistry as diagnostic methods for ALK positive non-small cell lung cancer patients. PLoS One 8: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li Y, Pan Y, Wang R, Sun Y, Hu H, et al. (2013) ALK-Rearranged Lung Cancer in Chinese: A Comprehensive Assessment of Clinicopathology, IHC, FISH and RT-PCR. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Y, Li Y, Yang T, Wei S, Wang J, et al. (2013) Clinical significance of EML4-ALK fusion gene and association with EGFR and KRAS gene mutations in 208 Chinese patients with non-small cell lung cancer. PLoS One 8: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perner S, Wagner PL, Demichelis F, Mehra R, Lafargue CJ, et al. (2008) EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia 10: 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, et al. (2008) EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 14: 4275–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee HY, Ahn HK, Jeong JY, Kwon MJ, Han JH, et al. (2013) Favorable clinical outcomes of pemetrexed treatment in anaplastic lymphoma kinase positive non-small-cell lung cancer. Lung Cancer 79: 40–45. [DOI] [PubMed] [Google Scholar]
- 54. Gainor JF, Varghese AM, Ou SH, Kabraji S, Awad MM, et al. (2013) ALK Rearrangements Are Mutually Exclusive with Mutations in EGFR or KRAS: An Analysis of 1,683 Patients with Non-Small Cell Lung Cancer. Clin Cancer Res 19: 4273–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Desai SS, Shah AS, Prabhash K, Jambhekar NA (2013) A year of anaplastic large cell kinase testing for lung carcinoma: Pathological and technical perspectives. Indian J Cancer 50: 80–86. [DOI] [PubMed] [Google Scholar]
- 56. Laszlo A, Thotala D, Hallahan DE (2013) Membrane phospholipids, EML4-ALK, and Hsp90 as novel targets in lung cancer treatment. Cancer J 19: 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang Y, Sun Y, Pan Y, Li C, Shen L, et al. (2012) Frequency of driver mutations in lung adenocarcinoma from female never-smokers varies with histologic subtypes and age at diagnosis. Clin Cancer Res 18: 1947–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yoon HJ, Lee HY, Lee KS, Choi YL, Ahn MJ, et al. (2012) Repeat biopsy for mutational analysis of non-small cell lung cancers resistant to previous chemotherapy: adequacy and complications. Radiology 265: 939–948. [DOI] [PubMed] [Google Scholar]
- 59. Yang P, Kulig K, Boland JM, Erickson-Johnson MR, Oliveira AM, et al. (2012) Worse disease-free survival in never-smokers with ALK+ lung adenocarcinoma. J Thorac Oncol 7: 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu SG, Kuo YW, Chang YL, Shih JY, Chen YH, et al. (2012) EML4-ALK translocation predicts better outcome in lung adenocarcinoma patients with wild-type EGFR. J Thorac Oncol 7: 98–104. [DOI] [PubMed] [Google Scholar]
- 61. Wang Z, Zhang X, Bai H, Zhao J, Zhuo M, et al. (2012) EML4-ALK rearrangement and its clinical significance in Chinese patients with advanced non-small cell lung cancer. Oncology 83: 248–256. [DOI] [PubMed] [Google Scholar]
- 62. Wallander ML, Geiersbach KB, Tripp SR, Layfield LJ (2012) Comparison of reverse transcription-polymerase chain reaction, immunohistochemistry, and fluorescence in situ hybridization methodologies for detection of echinoderm microtubule-associated proteinlike 4-anaplastic lymphoma kinase fusion-positive non-small cell lung carcinoma: implications for optimal clinical testing. Arch Pathol Lab Med 136: 796–803. [DOI] [PubMed] [Google Scholar]
- 63. Takeda M, Okamoto I, Sakai K, Kawakami H, Nishio K, et al. (2012) Clinical outcome for EML4-ALK-positive patients with advanced non-small-cell lung cancer treated with first-line platinum-based chemotherapy. Ann Oncol 23: 2931–2936. [DOI] [PubMed] [Google Scholar]
- 64. Shaozhang Z, Xiaomei L, Aiping Z, Jianbo H, Xiangqun S, et al. (2012) Detection of EML4-ALK fusion genes in non-small cell lung cancer patients with clinical features associated with EGFR mutations. Genes Chromosomes Cancer 51: 925–932. [DOI] [PubMed] [Google Scholar]
- 65. Ren S, Kuang P, Zheng L, Su C, Li J, et al. (2012) Analysis of driver mutations in female non-smoker Asian patients with pulmonary adenocarcinoma. Cell Biochem Biophys 64: 155–160. [DOI] [PubMed] [Google Scholar]
- 66. Park HS, Lee JK, Kim DW, Kulig K, Kim TM, et al. (2012) Immunohistochemical screening for anaplastic lymphoma kinase (ALK) rearrangement in advanced non-small cell lung cancer patients. Lung Cancer 77: 288–292. [DOI] [PubMed] [Google Scholar]
- 67. Kim HR, Shim HS, Chung JH, Lee YJ, Hong YK, et al. (2012) Distinct clinical features and outcomes in never-smokers with nonsmall cell lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement. Cancer 118: 729–739. [DOI] [PubMed] [Google Scholar]
- 68. Just PA, Cazes A, Audebourg A, Cessot A, Pallier K, et al. (2012) Histologic subtypes, immunohistochemistry, FISH or molecular screening for the accurate diagnosis of ALK-rearrangement in lung cancer: a comprehensive study of Caucasian non-smokers. Lung Cancer 76: 309–315. [DOI] [PubMed] [Google Scholar]
- 69. Doebele RC, Lu X, Sumey C, Maxson DA, Weickhardt AJ, et al. (2012) Oncogene status predicts patterns of metastatic spread in treatment-naive nonsmall cell lung cancer. Cancer 118: 4502–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li C, Fang R, Sun Y, Han X, Li F, et al. (2011) Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee JO, Kim TM, Lee SH, Kim DW, Kim S, et al. (2011) Anaplastic lymphoma kinase translocation: a predictive biomarker of pemetrexed in patients with non-small cell lung cancer. J Thorac Oncol 6: 1474–1480. [DOI] [PubMed] [Google Scholar]
- 72. Koh Y, Kim DW, Kim TM, Lee SH, Jeon YK, et al. (2011) Clinicopathologic characteristics and outcomes of patients with anaplastic lymphoma kinase-positive advanced pulmonary adenocarcinoma: suggestion for an effective screening strategy for these tumors. J Thorac Oncol 6: 905–912. [DOI] [PubMed] [Google Scholar]
- 73. Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, et al. (2009) Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 27: 4247–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kobayashi M, Sonobe M, Takahashi T, Yoshizawa A, Kikuchi R, et al. (2012) Detection of ALK fusion in lung cancer using fluorescence in situ hybridization. Asian Cardiovasc Thorac Ann 20: 426–431. [DOI] [PubMed] [Google Scholar]
- 75. Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, et al. (2012) Non-small cell lung cancer. J Natl Compr Canc Netw 10: 1236–1271. [DOI] [PubMed] [Google Scholar]
- 76. Shaw AT, Engelman JA (2013) ALK in lung cancer: past, present, and future. J Clin Oncol 31: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ren S, Kuang P, Zheng L, Su C, Li J, et al. (2012) Analysis of Driver Mutations in Female Non-Smoker Asian Patients with Pulmonary Adenocarcinoma. Cell Biochem Biophys 17: 17. [DOI] [PubMed] [Google Scholar]
- 78. Yoshida A, Tsuta K, Nakamura H, Kohno T, Takahashi F, et al. (2011) Comprehensive histologic analysis of ALK-rearranged lung carcinomas. Am J Surg Pathol 35: 1226–1234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The PRISMA checklist.
(DOC)
Forest plots of the mean difference of age between lung adenocarcinomas patients with and without ALK rearrangements by race.
(PDF)
Forest plots of gender difference between lung adenocarcinomas patients with and without ALK rearrangements by race.
(PDF)
Forest plots of smoking difference between NSCLC patients with and without ALK rearrangements by race.
(PDF)
Forest plots of smoking difference between lung adenocarcinomas patients with and without ALK rearrangements by race.
(PDF)
Forest plots of tumor stage difference between NSCLC patients with and without ALK rearrangements by race.
(PDF)
Forest plots of tumor stage difference between lung adenocarcinomas patients with and without ALK rearrangements by race.
(PDF)
Forest plots of distribution difference in ALK rearrangements between patients with and without lung adenocarcinomas by race.
(PDF)
Forest plots of distribution difference in ALK rearrangements between patients with lung adenocarcinomas and squamous carcinomas by race.
(PDF)
Forest plots of the mean difference of age between NSCLC patients with ALK rearrangements and with EGFR mutations by race.
(PDF)
Forest plots of the mean difference of age between NSCLC patients with ALK rearrangements and with KRAS mutations by race.
(PDF)
The baseline characteristics of all qualified studies in this meta-analysis.
(DOC)
ALK rearrangements and tumor stage.
(DOC)
ALK rearrangements and NSCLC histology.
(DOC)
The baseline characteristics of all qualified articles assessing both ALK rearrangements and EGFR/KRAS mutations.
(DOC)