Abstract
Although many computer programs can perform population genetics calculations, they are typically limited in the analyses and data input formats they offer; few applications can process the large data sets produced by whole-genome resequencing projects. Furthermore, there is no coherent framework for the easy integration of new statistics into existing pipelines, hindering the development and application of new population genetics and genomics approaches. Here, we present PopGenome, a population genomics package for the R software environment (a de facto standard for statistical analyses). PopGenome can efficiently process genome-scale data as well as large sets of individual loci. It reads DNA alignments and single-nucleotide polymorphism (SNP) data sets in most common formats, including those used by the HapMap, 1000 human genomes, and 1001 Arabidopsis genomes projects. PopGenome also reads associated annotation files in GFF format, enabling users to easily define regions or classify SNPs based on their annotation; all analyses can also be applied to sliding windows. PopGenome offers a wide range of diverse population genetics analyses, including neutrality tests as well as statistics for population differentiation, linkage disequilibrium, and recombination. PopGenome is linked to Hudson’s MS and Ewing’s MSMS programs to assess statistical significance based on coalescent simulations. PopGenome’s integration in R facilitates effortless and reproducible downstream analyses as well as the production of publication-quality graphics. Developers can easily incorporate new analyses methods into the PopGenome framework. PopGenome and R are freely available from CRAN (http://cran.r-project.org/) for all major operating systems under the GNU General Public License.
Keywords: population genomics, software, single-nucleotide polymorphisms
Introduction
Recent sequencing technologies allow to map genetic variation across hundreds of individual genomes (Harrison 2012); notable examples are the 1000 genomes project in humans (1000genomes.org) and the 1001 genomes project in Arabidopsis thaliana (1001genomes.org). These technological developments have shifted the bottleneck in population genetics from data acquisition to data analysis.
Different software packages for population genetics (or population genomics) analyses typically have limited overlap in implemented statistics and accepted input formats. This diversity hampers both efficient data analysis and the quick dispersion of new statistical approaches. Many widely used software packages, such as DnaSP (Rozas et al. 2003), cannot handle the data formats developed for massive resequencing projects. Only few programs support the use of genomic annotation, and thus users interested in specific regions have to preprocess the data using other tools.
To address these issues, a new software tool for population genomic data analysis should:
read data in a variety of input formats, including both traditional formats and those used by the major resequencing projects;
implement a comprehensive range of population genetics/genomics analyses and statistics;
read associated annotation files and allow to systematically select regions of interest;
be able to analyze individual loci, multiple loci, and sliding windows;
be open source and be easily extendable by the scientific community to incorporate new types of analyses;
be integrated with powerful numerical and graphical capabilities; and
be platform independent.
We implemented these features in PopGenome, a package embedded in the freely available, platform-independent, statistical, and graphical computing environment R (http://cran.r-project.org/, last accessed April 30, 2014).
Description of PopGenome
Summary
To fully exploit the capabilities of the R statistical and graphical environment, and to allow the creation of stable workflows (scripts), all processes in PopGenome are executed as command line functions. However, we anticipate that a graphical user interface implementing the most important functionalities will be available in the near future.
PopGenome is designed to facilitate the easy integration of virtually all major types of population genetics and population genomics analyses, and, as outlined below, its current version includes a large array of different statistics (see overview in table 2). The emphasis on extensibility was inspired by the way Bioconductor (bioconductor.org) has come to dominate the analysis of microarray data, where newly developed methods for specific tasks are easily integrated into a pre-existing larger framework, thereby obviating the need to recreate tasks shared with existing analysis pipelines. We hope that PopGenome may become the kernel of a similar paradigm in the analysis of population genomics data, enabling researchers to effortlessly implement and share new or modified statistics.
Table 2.
Population Genetics Statistics Implemented in PopGenome’s Modules.
| Module | Statistics |
|---|---|
| Neutrality statistics | Tajima’s D (Tajima 1989), Fu and Li’s F* & D* (Fu and Li 1993), Fay and Wu’s H (Fay and Wu 2000), Zeng’s E (Zeng et al. 2006), Strobeck’s S (Strobeck 1987), Achaz’s Y (Achaz 2009), Fu’s FS (Fu 1997), Ramos-Onsins’ and Rozas’ R2 (Ramos-Onsins and Rozas 2002), as well as all corresponding theta values |
| Linkage disequilibrium | ZnS (Kelly 1997), B/Q (Wall 1999), ZA/ZZ (Rozas et al. 2001), and correlation coefficient r2 for each pair of SNPs within or between windows/regions |
| Recombination statistics | Four-gamete test (Hudson and Kaplan 1985) |
| Diversities | Nucleotide and haplotype diversity (Hudson, Boos et al. 1992); (Nei 1979); see “Neutrality statistics” for a list of calculated Theta values |
| Selective sweeps | CL, CLR (Nielsen et al. 2005) |
| FST estimates | GST (Nei 1973); FST (Hudson, Slatkin et al. 1992); GST, HST, KST (Hudson, Boos et al. 1992); Snn (Hudson 2000); PhiST (Excoffier and Smouse 1992) |
| MKT | McDonald–Kreitman test (McDonald and Kreitman 1991) |
| Mixed statistics | Site frequency spectrum; fixed and shared polymorphisms; biallelic structure |
| BayeScanR | Bayesian estimation of FST (Foll and Gaggiotti 2008) |
That PopGenome is embedded within the R framework not only facilitates the easy integration of extensions and stable workflows but also allows immediate and effortless postprocessing of analysis results with R’s powerful numerical and graphical capabilities.
Data Organization
PopGenome can read data both as full alignments and in single-nucleotide polymorphism (SNP) formats such as those generated by large resequencing projects. PopGenome’s ability to simultaneously manage large numbers of loci, which allows for variation in sequence and population coverage, provides a convenient framework for multilocus analyses.
After reading data, PopGenome first converts it into a biallelic matrix, that is, a matrix whose rows correspond to sequences and whose columns correspond to SNP positions in the alignment. Entries are either 0, indicating the major allele, or 1, indicating the minor allele (with entries corresponding to unknown variants labeled NA). In PopGenome, this matrix is stored as part of a GENOME object, which contains additional data needed for downstream analyses; this includes information on missing data, as well as annotations for individual SNPs (e.g., if these are transitions/transversions, coding/noncoding, or synonymous/nonsynonymous in the case of coding sequences).
Large input files are split automatically into smaller chunks, with the resulting partial biallelic matrices stored on the hard disk. For this temporary storage, we use ff objects (Adler et al. 2013). These data structures are stored on disk but behave (almost) as if they were in RAM, by transparently mapping only a section (pagesize) in main memory. An ff object needs about 3 kB of RAM to store SNP data from 1 million SNPs across 50 individuals. When analyzing larger data sets, PopGenome concatenates the partial biallelic matrices into a single temporary file.
This strategy allows to simultaneously process whole-chromosome or whole-genome SNP data from hundreds of individuals, as collected in the 1000 human genomes (1000genomes.org) and 1001 Arabidopsis genomes (1001genomes.org) projects. To further speed up the reading process, we employ the R-package parallel (Vera et al. 2008) that facilitates parallel computations on computers with multiple cores/CPUs.
Most functions in PopGenome are implemented in C or C++ to speed up computations and to limit memory requirements. This also applies to the reading of genome-scale alignments and SNP data. Supported alignment formats include FASTA, NEXUS, MEGA, MAF, and Phylip. An almost de facto standard for whole-genome variation data is the Variant Call Format (VCF), used, among others, by the 1000 genomes project (1000genomes.org) and the UK10k project (uk10k.org). PopGenome can read large SNP data sets stored in this format very efficiently, using indexes created with Tabix (Li 2011). SNPs in defined regions can be extracted directly from the corresponding file without time-consuming search operations over the entire file, with input speeds exceeding 6,000 variant positions per second even on older desktop computers.
The implementations of PopGenome’s data access functions conform to a set of optimization guidelines to guarantee a minimal reading time. To the best of our knowledge, there is currently no general-purpose population genetics software capable of directly accessing VCF files with comparable speed. Typical times required to read large data sets are listed in table 1.
Table 1.
Times Required to Read Large Data Sets.
| Data Set | Individuals | SNPs | Format | Time for Readinga |
|---|---|---|---|---|
| Arabidopsis (Chr 1) | 80 | 1,200,000 | SNP (1001 Genomes) | <1 minb |
| ∼3 min | ||||
| Human (Chr2: 100–150 Mb) | 1,094 | 660,000 | VCF (1000 Genomes) | ∼5 min |
| 3450 individual alignments | 25 | 200,000 | FASTA | ∼15 s |
aIntel® Core™ i3-2130 CPU @ 3.40 GHz × 4, 8 GB RAM, with data stored in temporary files.
bWithout temporary files, if sufficient RAM is available.
PopGenome sessions can be saved on hard disk; thus, the conversion to the biallelic matrix needs to be performed only once per data set. The function region.as.fasta() can be used to export data of a specific region, a group of subsites, or the entire data set (i.e., all SNPs or even complete genome alignments) as a FASTA file. A parameter include.unknown indicates if unknown positions should be included in the analyses.
Implemented Methods
To structure the rich landscape of population genetics and genomics analysis methods, PopGenome partitions the implemented methods into modules. Currently, PopGenome provides nine modules (table 2). All modules use the GENOME object created when the input data were read, and store their results in the same GENOME object. For analyses that require to distinguish between ancestral and derived alleles, outgroups can be specified.
As a default, all analyses packaged into one module are performed simultaneously when the module is executed. To accelerate calculations on large data sets, individual methods can be switched off using additional arguments. We plan to integrate more methods in the future, and welcome requests for the implementation of specific statistics. In the next release of PopGenome, we aim to incorporate methods for detecting recent selective sweeps, such as the algorithm implemented in the software SweeD (Pavlidis et al. 2013).
Apart from many “standard” statistics (e.g., neutrality and linkage disequilibrium statistics), PopGenome offers several tests for the detection of nonneutral evolution. So far, we have implemented the McDonald–Kreitmann test (McDonald and Kreitman 1991) and a wide range of FST measurements, including an implementation of a previously published method based on Bayesian statistics (Foll and Gaggiotti 2008). PopGenome also includes a calculation of r2 correlation coefficients and the corresponding P values (Fisher’s exact test) for interregion calculations. By concatenating the corresponding GENOME objects, statistics that rely on comparisons between regions can be calculated even when these are located on different chromosomes. Details of the implemented methods are given in table 2 and in the PopGenome documentation.
PopGenome is fully integrated with two widely used coalescent simulation tools: Hudson’s MS (Hudson 2002), as well as Ewing’s MSMS (Ewing and Hermisson 2010), which incorporates selection. The PopGenome function MS() compares the statistics calculated for the observed data with corresponding data simulated by the coalescent method. PopGenome supports the full coalescent simulation capabilities of MS and MSMS. Parameters can be specified as vectors in the dedicated class “cs.stats,” and thus different models (mutation rates, migration rates, etc.) can be applied to different windows or regions of the genome. PopGenome’s MS() function stores the calculated statistics of coalescent simulations in a dedicated R object. Direct comparison to coalescent simulations is currently implemented for the modules Neutrality statistics, Linkage, and FST. If statistics from other modules need to be compared with coalescent simulations, PopGenome can directly read MS output files and then process these data using the method of interest (readMS()).
R is an efficient environment for large-scale computations. However, although most native R functions are implemented in C or Fortran, R itself is an interpreted and vector-oriented language. As a consequence, some types of calculations tend to be slow when applied to large objects. To avoid major bottlenecks, we implemented several specialized calculations in C++. PopGenome finishes the calculation of most statistics in minutes even for very large data sets (table 3).
Table 3.
Calculation Speed for Haplotype and Nucleotide Diversity in Sliding Windows.
| Data | Sliding Window (nucleotides) | Running Timea |
|---|---|---|
| Arabidopsis (Chr 1) | Window size = 10,000 | ∼30 s |
| 80 individuals | Jump size = 10,000 | |
| 1,200,000 SNPs | Number of windows = 3,042 | |
| Human (Chr 2: 100–150 Mb) | Window size = 1,000 | ∼5 min |
| 1,094 individuals | Jump size = 1,000 | |
| 660,000 SNPs | Number of windows = 50,000 | |
| 3,450 alignments | 3,450 windows (alignments) | ∼7 s |
| 25 individuals | ||
| 5,086,953 sites | ||
| 200,097 SNPs |
aIntel® Core™ i3-2130 CPU @ 3.40 GHz × 4, 8 GB RAM.
Partitioning and Interpreting SNPs
Users can restrict analyses to subregions specified either by genomic coordinates or positions in SNP files. When an annotation file in GFF format is present (GFF v2 or v3), PopGenome will automatically label SNPs located in genes, exons, coding regions, and UTRs. Other annotations of interest in the GFF file can be read using the function getgffinfo(). The user can apply the full range of implemented methods to all SNPs observed in a specific class (e.g., all exonic SNPs), or to each region specified in the GFF file separately (e.g., all introns individually).
If a GFF file is present, PopGenome can also classify synonymous and nonsynonymous sites; for SNP data formats, this additionally requires a reference genome in FASTA format. PopGenome stores the codons internally as numerical values, coded from a polynomial function as in the PGE Toolbox (Cai 2008). Based on the GFF file, the function get.codons() will provide information about the nature of amino acid changes resulting from the observed SNPs (encoded amino acids, charges, hydrophobicities, size, and polarity changes). Per default, PopGenome assumes the standard genetic code, but alternative codes can be specified.
Most multipurpose population genetics software tools are geared toward the analysis of discrete loci (Rozas et al. 2003; Excoffier et al. 2005), restricting their utility for the analysis of whole-genome SNP data sets. One widely used approach to apply population genetics methods to whole-genome data is the analysis of sliding windows (Rozas et al. 2001). In PopGenome, users can freely choose window and jump sizes for sliding windows, measured either in nucleotides or in numbers of SNPs. The underlying algorithm copies the information (mostly pointer) stored in the GENOME object to another object of the same class, where the data are reorganized into the specified windows. The full spectrum of PopGenome methods is thus available both for arbitrarily large sets of individual loci and for systematic genomic scans.
Easy Integration of New Methods
To be used by the scientific community at large, a new algorithm ideally has to be implemented in a framework that allows its efficient application to data in the diverse file formats commonly used in different resequencing projects. PopGenome is geared toward making this task as easy as possible. All information required for the calculation of population genetics statistics, including the biallelic matrix as well as genomic annotation, is stored in a GENOME object, which new methods can directly access.
To further simplify the integration of new methods, we implemented the function create.PopGenome.method(), which generates a skeleton of a typical PopGenome function. New methods thus implemented are fully embedded in the PopGenome framework and can be applied to sliding windows or subsites in the same way as the existing modules. This approach frees developers of new population genetics or population genomics algorithms from the need to implement many auxiliary functions, such as efficient data input and output, data conversion, and region subsetting.
To enable PopGenome to work with additional data formats, users can write a simple parser that converts the data to a binary R object. The mechanism to integrate new methods or data formats is documented extensively in a tutorial, accessible by typing “vignette(“Integration_of_new_Methods”)” in R (see also supplementary file S1, Supplementary Material online).
Results
To illustrate the usage of PopGenome, we show two exemplary analyses for human and A. thaliana whole-genome SNP data. More details are found in the PopGenome documentation and in supplementary file S2, Supplementary Material online.
Diversity on A. thaliana Chromosome 1
The 1001 genomes project (1000genomes.org) stores all SNP calls from one individual A. thaliana plant in one (.SNP) file. We downloaded .SNP files for 80 individuals (Cao et al. 2011) into a subdirectory named “Arabidopsis.” After starting R and loading the PopGenome library, we read in the data for chromosome 1 to analyze diversity and population differentiation:
> library(PopGenome)
> genome <- readSNP(“Arabidopsis,” CHR=1)
We define the populations as a list of character vectors containing the individuals of each population:
> Central_Asia <- c(“ICE127,” “ICE130,” “ICE134,” “ICE138,” “ICE150,” “ICE152,” “ICE153” , “Sha”)
> … (analogous for the other populations) …
> populations <- list(Central_Asia, Caucasus, N_Europe, N_Africa, S_Italy, S_Russia, S_Tyrol, Swabia)
The population definitions are now added to the GENOME object:
> genome <- set.populations(genome, populations)
We then transform these data into consecutive 10-kb-sliding windows:
> genome.slide <- sliding.window.transform(genome, width=10000, jump=10000)
The nucleotide and haplotype diversities for each window are calculated in the module diversity.stats. We store the results also in the GENOME object:
> genome.slide <- diversity.stats(genome.slide)
The “slot” of the GENOME object that stores the nucleotide diversities of the individual populations is called nuc.diversity.within. Slots of such objects are accessed by appending the slot name to the object name, separated by an @ symbol: genome.slide@nuc.diversity.within. These data can be analyzed further using the built-in statistical and graphical capabilities of R. Here, we plot the sliding window nucleotide diversities (fig. 1A) with a specialized function:
> PopGplot(genome.slide@nuc.diversity.within, colours)
where colors is a vector listing the R names of the colors used for the eight populations in the figure. The haplotype diversity per 10-kb window (fig. 1B), as well as Hudson’s fixation index, FST (fig. 1C), is produced with similar function calls.
Fig. 1.
Diversity statistics for Arabidopsis thaliana chromosome 1. Data from the 1001 genomes project website (1001genomes.org) was analyzed in consecutive 10-kb windows. (A) Nucleotide diversity, (B) haplotype diversity, (C) fixation index (Hudson’s FST), contrasting one population against all other individuals. Each line corresponds to one population (see legend in panel [A]). Lines were smoothed using spline interpolation. The black bars around 15-Mb mask the centromere.
The data thus displayed in figure 1 indicate a lower level of diversity in the Central Asia population of A. thaliana (dark blue lines) along the whole chromosome. As reported earlier (Cao et al. 2011), we observe a dip in diversity in the region around 20 Mb in all populations, suggesting a recent selective sweep in this region.
We find two additional, even stronger candidate regions for selective sweeps around position 8 and 22 Mb. Closer inspection of these two regions shows that they are devoid of polymorphisms between positions 8765643 and 8831390, and between positions 21766562 and 21823063. This observation suggests additional recent, species-wide selective sweeps in these two regions. Furthermore, we find a strong decrease in diversity in plants from Asia and Southern Russia between genomic positions 11870001 and 11900000, suggesting a population-specific selective sweep in this region.
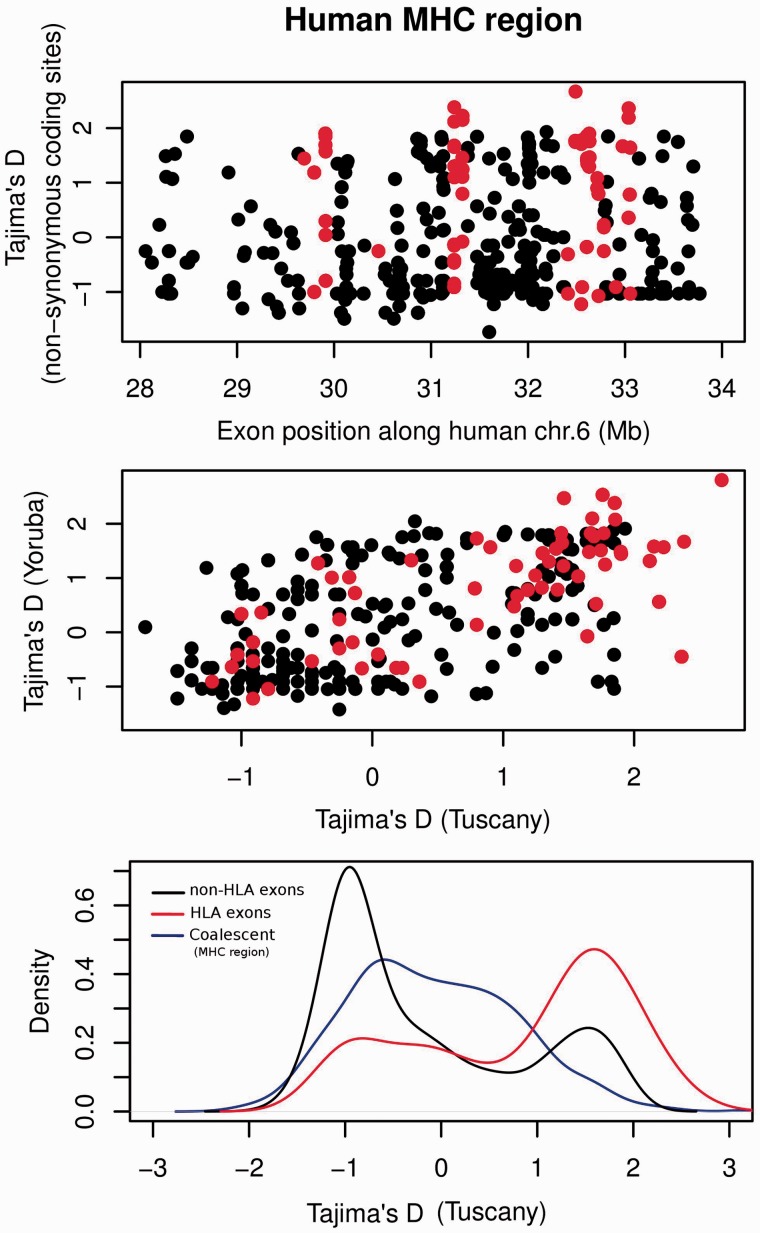
Tajima’s D across Human MHC Exons
As a second illustration of the PopGenome usage, we calculated Tajima’s D around the human MHC (Major Histocompatibility Complex) region. The 1000 genomes project stores SNP calls from all examined individuals in gzipped VCF format, together with a Tabix (Li 2011) index (.tbi) file. To read data for the MHC region on human chromosome 6 (including the corresponding annotation in the GFF file), we use the following line:
> genome <- readVCF(“chr6.vcf.gz,” numcols=10000, tid=“6,” from=28000000, to=34000000, gffpath=“chr6.gff”)
where “numcols” is the number of SNPs read in simultaneously, “tid” is a chromosome identifier, “from/to” delimit the nucleotide positions of the SNPs read in, and “gffpath” specifies the position of the GFF annotation file. Based on the annotations read from the GFF file and the chromosomal reference sequence in FASTA format, the next command labels positions in protein-coding regions as either synonymous or nonsynonymous:
> genome <- set.synnonsyn(genome, ref.chr=“chr6.fas”)
As in the Arabidopsis example, we define the populations via the vectors “Africa” and “Europe,” which contain the identifiers of the corresponding individuals:
> genome <- set.populations(genome, list(Africa, Europe))
Here, we want to calculate statistics for each exon individually, considering only nonsynonymous SNPs. To do this, we first split the region into individual loci, where each locus stores the SNP information from the coding sequence part contained in one exon, as annotated in the GFF file:
> genome.exons <- splitting.data(genome, subsites=“coding”)
Next, we calculate Tajima’s D across all nonsynonymous positions of each exon. This calculation is performed in the module “neutrality.stats”:
> genome.exons <- neutrality.stats(genome.exons, subsites=”nonsyn”)
We thus obtain the data displayed in figure 2. The high Tajima’s D values in some loci likely reflect balancing selection (Hedrick 1998), for example, due to frequency-dependent selection in pathogen recognition.
Fig. 2.
Tajima’s D calculated across nonsynonymous coding sites of exons in the human MHC region on chromosome 6. Each data point in (A) and (B) represents one exon; HLA type I and type II exons are shown in red. (A) Tajima’s D of a Tuscan population (117 individuals), plotted along chr. 6. (B) Comparison of Tajima’s D between a Tuscan (117 individuals) and a Yoruba (229 individuals) population. (C) Distribution (density curves) of the Tajima’s D values in (A) for MHC (red) and non-MHC exons (black). The blue curve displays the distribution of neutral values from coalescent simulations with Hudson’s MS based on all SNPs in the MHC region. Data from 1000genomes.org.
We can use coalescent simulations to derive the expected neutral distribution of Tajima’s D values across the MHC region. For this, we first calculate Theta across all SNPs in the MHC region:
> genome <- neutrality.stats(genome)
We then use the genome object as input for a call to Hudson’s MS program:
> ms <- MS(genome, thetaID=“Tajima,” neutrality=TRUE)
If no additional parameters are specified for the MS simulations, PopGenome will use the standard neutral model (SNM).
The simulated Tajima’s D values are then extracted:
> MS.get.stats(ms)
Figure 2C compares the distributions of expected Tajima’s D values under the SNM with values observed for Human Leukocyte Antigen (HLA, red) and non-HLA (black) exons. The distribution of Tajima’s D values is strongly shifted toward higher values in HLA exons compared with non-HLA exons, indicating strong balancing selection; a deviation from neutral expectations for HLA exons is supported by a comparison to the simulated data.
The highest Tajima’s D values—suggesting strong balancing selection—are seen in HLA type I and type II genes (marked in red in fig. 2). Tajima’s D values are correlated between the African (Yoruba) and European (Tuscany) populations (fig. 2B; Spearman’s R2 = 0.33). One notable outlier is the coding exon 2 of the HLA-DPA1 gene, which shows evidence of purifying selection in Yoruba, but strong evidence of balancing selection in Tuscans.
Discussion
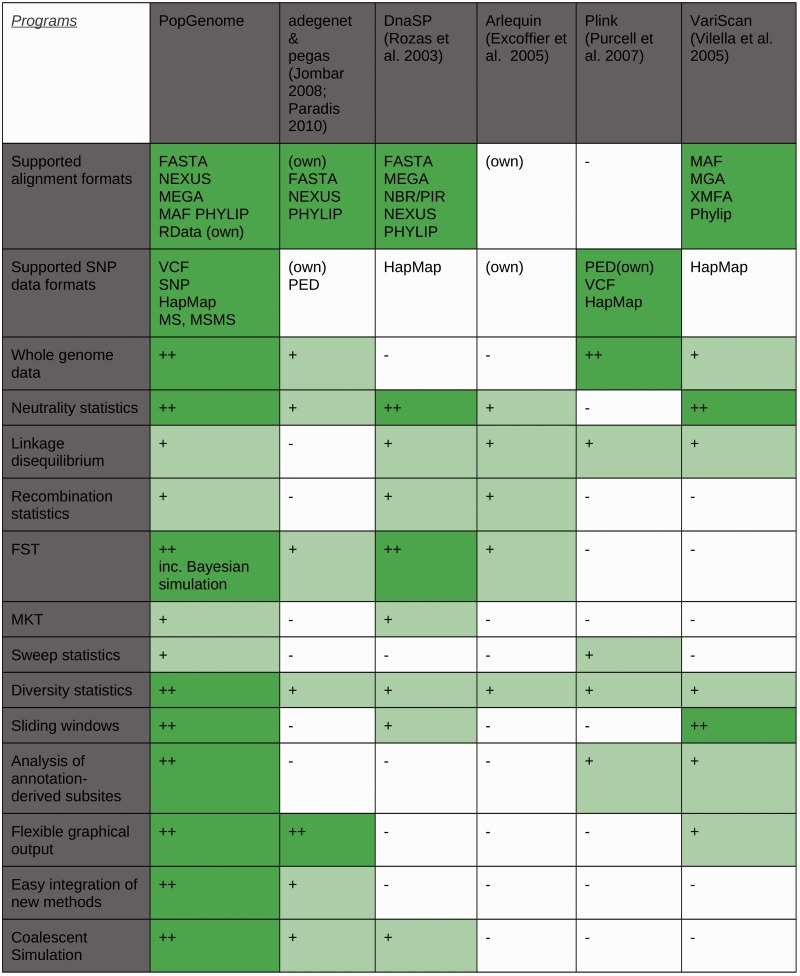
Several computer programs for population genetics or population genomics analyses are publicly available. However, these tend to specialize on specific subsets of analyses (e.g., Vilella 2005; Rozas et al. 2001; Purcell et al. 2007; Jombart 2008; Paradis 2010) or cannot process whole-genome data (e.g., Rozas et al. 2003; Excoffier et al. 2005). Figure 3 compares major features of PopGenome with five other widely used software packages.
Fig. 3.
Comparison of PopGenome with existing software for population genetics and population genomics analyses. Symbols reflect the breadth of the implemented functionalities: ++, broad; +, limited; −, nonexistent. Details on the criteria used for assignment to the breadth classes are given in supplementary table S1, Supplementary Material online.
PopGenome can not only read data in several common alignment formats but also understands the widest choice of SNP data formats; this includes data from the HapMap as well as the 1000 and 1001 genomes projects. PopGenome’s ability to work efficiently with temporary files allows calculations on very large SNP data sets. No other available program offers a similar combination of flexible data input options with a broad toolbox of population genetics and genomics statistics, including the ability to perform analyses in sliding windows.
PopGenome can read GFF annotation files, which permits high plasticity in the variability analysis of different regions of the genome. PopGenome can link this annotation automatically to the SNP data, which can thus be restricted to specific annotated features or feature groups (e.g., all introns vs. all exons). This feature also allows to discriminate synonymous and nonsynonymous codon positions in whole-genome SNP data sets, which is necessary, for example, for McDonald–Kreitman tests. Like adegenet/pegas (which are more limited in scope), PopGenome is fully integrated with the powerful graphical and data analysis capabilities of the R environment (http://cran.r-project.org/, last accessed April 30, 2014), thus simplifying downstream analyses and graphics as well as the development of stable work flows.
Supplementary Material
Supplementary files S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Juliette de Meaux, Laura Rose, Kristian Ulrich, Thorsten Klösges, and William Martin for helpful discussions. This work was supported by the Spanish Ministerio de Ciencia e Innovación Grant CGL2009-09346 to S.E.R.O. and by the German Research Foundation DFG grants EXC 1028 and CRC 680 to M.J.L.
References
- Achaz G. Frequency spectrum neutrality tests: one for all and all for one. Genetics. 2009;183(1):249–258. doi: 10.1534/genetics.109.104042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler D, Gläser C, Nenadic O, Oehlschlägel J, Zucchini W. 2013. ff: memory-efficient storage of large data on disk and fast access functions [R package version 2.2-11]. [cited 2013 Dec] Available from: http://CRAN.R-project.org/package=ff. [Google Scholar]
- Cai J. PGEToolbox: a Matlab toolbox for population genetics and evolution. J Hered. 2008;99(4):438–440. doi: 10.1093/jhered/esm127. [DOI] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43(10):956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- Ewing G, Hermisson J. MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics. 2010;26(16):2064–2065. doi: 10.1093/bioinformatics/btq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131(2):479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J, Wu C. Hitchhiking under positive Darwinian selection. Genetics. 2000;155(3):1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180(2):977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147(2):915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Li W. Statistical tests of neutrality of mutations. Genetics. 1993;133(3):693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R. Understanding genetic variation and function—the applications of next generation sequencing. Semin Cell Dev Biol. 2012;23(2):230–236. doi: 10.1016/j.semcdb.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Hedrick Balancing selection and MHC. Genetica. 1998;104(3):207–214. doi: 10.1023/a:1026494212540. [DOI] [PubMed] [Google Scholar]
- Hudson R. A new statistic for detecting genetic differentiation. Genetics. 2000;155(4):2011–2014. doi: 10.1093/genetics/155.4.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R, Boos D, Kaplan N. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9(1):138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- Hudson R, Kaplan N. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111(1):147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics Appl Note. 2002;18(2):337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hudson R, Slatkin M, Maddison W. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132(2):583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24(11):1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Kelly J. A test of neutrality based on interlocus associations. Genetics. 1997;146:1197–1206. doi: 10.1093/genetics/146.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011;27(5):718–719. doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351(6328):652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A. 1973;70(12):3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Williamson S, Kim Y, Hubisz M, Clark A, Bustamante C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26(3):419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- Pavlidis P, Zivkovic D, Stamatakis A, Alachiotis N. SweeD: likelihood-based detection of selective sweeps in thousands of genomes. Mol Biol Evol. 2013;30:2224–2234. doi: 10.1093/molbev/mst112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Onsins S, Rozas J. Statistical properties of new neutrality tests against population growth. Mol Biol Evol. 2002;19(12):2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034. [DOI] [PubMed] [Google Scholar]
- Rozas J, Gullaud M, Blandin G, Aguade M. DNA variation at the rp49 gene region of Drosophila simulans: evolutionary inferences from an unusual haplotype structure. Genetics. 2001;158(3):1147–1155. doi: 10.1093/genetics/158.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio J, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19(18):2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Strobeck C. Average number of nucleotide differences in a sample from a single subpopulation: a test for population subdivision. Genetics. 1987;117(1):149–153. doi: 10.1093/genetics/117.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera G, Jansen R, Suppi R. R/parallel—speeding up bioinformatics analysis with R. BMC Bioinformatics. 2008;9:390. doi: 10.1186/1471-2105-9-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella J, Blanco-Garcia A, Hutter S, Rozas J. VariScan: analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics. 2005;21(11):2791–2793. doi: 10.1093/bioinformatics/bti403. [DOI] [PubMed] [Google Scholar]
- Wall J. Recombination and the power of statistical tests of neutrality. Genet Res. 1999;74(1):65–79. [Google Scholar]
- Zeng K, Fu Y, Shi S, Wu C-I. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics. 2006;174(3):1431–1439. doi: 10.1534/genetics.106.061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.