Abstract
Study of antibiotic resistance was done among the metal tolerant E. coli isolates from hospital wastewater at Lucknow city. Metal tolerance was determined in terms of visible growth on metal amended plates at their varying concentrations. MICs were also determined among all metal tolerant E. coli isolates. All the isolates showed their MIC in between 100-2000 µg/ml while maximum isolates demonstrated their MICs at 400, 800 and 1600 µg/ml against all the metal tested. 23.07% of the isolates showed their MIC at 2000 µg/ml against Ni3+. Multiple antibiotic resistances were recorded among all the metal resistant E.coli isolates. A high level of resistance was observed against Methicillin (86.53%) followed by penicillin (73.07%), Cephradin (57.69%), Rifampicin (34.61%), Erythromycin (26.92%), Nalidixic acids (25%), Chloramphenicol (3.84%) and least to Gentamycine (1.92%). Streptomycin was recorded most effective against E.coli isolates among the entire antibiotic tested. Antimicrobial resistance observed among the bacteria from the aquatic system contaminated with hospital wastes may be threatful for the environment and public health both.
Keywords: Heavy metals, Antibiotics, Escherichia coli, Hospital wastewater
Background
Due to generous use of antibiotics and disinfectant in hospitals, their presence and level is increasing continuously in the discharged wastewater [1]. In most of aquatic environments the concentrations of antibiotics are very low but they have tendency to convert non pathogenic bacteria to pathogenic bacteria by increasing the resistance against antibiotics [2]. The wastewater has a high content of both organic and inorganic matter, as well as high densities of living organisms, including pathogenic, commensal and environmental bacteria. This characteristic makes a suitable condition for the spread of antibiotic resistance [3]. Numerous studies have indicated that hospital waste water receive inputs of heavy metal from the increased use of pharmaceuticals. Redionucleocides and other antimicrobial solvents [4]. There has been considerable speculation about possible genetic association between bacterial tolerance for these metals and multiple antibiotic resistances [5, 6, 7]. It has been suggested that genes encoding resistance to heavy metals can be located together with antibiotic resistance genes on either the same genetic structure (eg. plasmid), or different genetic structures within the same bacterial strain [8, 9] had suggested that metal and antibiotic resistance among bacteria are linked very closely together and that expression of antibiotic resistance may be dependent on exposure to metals. Heavy metals used in industry and in household products are along with antibiotics creating a selective pressure in the environment that leads to the mutations in microorganisms that will allowed them better survive and multiply [10]. Bacterial resistance to toxic heavy metals is a widespread phenomenon and reported to enhance the antibiotic resistance ability of microorganisms [11]. Antibiotic resistance is increasing among the bacteria due to excess use of the drugs and improper management of the hospital wastewater containing heavy metals, toxic chemicals, and radioactive elements. Heavy metals played a key role in increased antibiotic resistance among the bacteria from the environmental samples. Our research work has revealed the multiple antibiotic resistance and patterns among the metal tolerant E.coli from hospital wastewater. It may provide new ways in treating the infectious diseases especially by E.coli and important for risk assessment as well as risk management related to hospital effluents
Methodology
Sample collection:
Composite sample of hospital wastewater were collected from three different department i.e Medicine and Surgery (Site-1), Trauma centre (site-2) and Gynecology (site-3) during month of December to February from King George Medical University, Lucknow (KGMU). Sample was collected from 15cm depths of sewage in 250 ml sterilized glass bottles and transferred to laboratory (in ice box at 4°C) immediately , so the elapsed time between the sample collection and initial processing did not exceed more than 8 hours.
Isolation and Identification of metal tolerant E. coli:
Isolation of metal tolerant E. coli isolates from hospital wastewater samples were done on metal (Cr, Cd and Ni) amended EMB agar plates at 100 µg/ml concentration. Serial dilutions of the water samples were plated by spreading 0.1 ml on EMB medium for metal tolerant E. coli. Plates were incubated at 37°C for 24 h. Greenish with metallic sheen colonies were identified as E. coli These isolates were finally identified on the basis of morphological, cultural and IMVic tests (indole, methyl red, voges proskeur and citrate utilization tests) [12].
Determination of minimum inhibitory concentration (MIC) of heavy metal among E. coli isolates:
The heavy metal resistance was determined by the minimum inhibitory concentration (MIC) against the test bacterial strain by spot plate method [13]. Nutrient plates of each heavy metal (Chromium, Cadmium and Nickel) of different concentrations (100 µg/ml to 2000 µg/ml) were prepared. Inoculum of test strain (3×106 CFU/ml) was spotted on heavy metal amended plates and control plates in duplicate with the help of platinum loop of 5mm diameter. The plates were incubated at 37° C for 24 hr to observe the growth of bacterial strain on the spotted area. The MIC was defined as the minimum inhibitory concentration of the heavy metal that inhibits the visible growth of test strain. Metal concentration range bellow MIC was considered as sub MIC of the isolates.
Antibiotic Sensitivity Test:
All the isolates of E. coli were tested for their sensitivity to anti microbial agents by disc diffusion method [14]. Petri plates containing approximately 25-30 ml of Mueller-Hinton medium were spreader with 0.1 ml of 24 hour old culture of the isolates with sterile glass spreader and then impregnated with the antibiotic discs. The plates were incubated at 37c for 24 hours. The zone of inhibition was recorded in millimeter and interpreted as sensitive or resistant according to recommendation by the National committee for clinical laboratory standards NCCLS. (2008). The E. Coli ATCC 25922 was used as a control strain. The antimicrobial discs tested for all isolates were Nalidixic acids(NA) 30µg, Chloramphenicol (C) 30µg, Erythromycin (E)15µg, Cephradin (CH) 25µg, Streptomycin (S) 25µg, penicillin ( p) 10µg, Neomycin (N) 30µg , Methicillin (Met) 5µg, Gentamycin ( Gen) 30µg, Rifampicin ( Rif ) 2µg.
Multiple antibiotic resistances (MAR) indexing:
The MAR index profile based on isolate and sampling site was performed to evaluate the health risk of the environment. MAR index for test isolates was calculated according to the formula: No. of antibiotics to which all isolates were resistant/No. of antibiotics tested x No. of isolates as recommended by [15]. Sampling site based MAR index was calculated by the same formula modified by the total number of isolates from a sampling site as described [16].
Results
All the E. coli isolates were tested for their resistance to Cr6+, Cd2+ and Ni3+ at their varying concentrations (100-2000 µg/ml) in term of their visible growth. Growth patterns of the isolates were recorded at their sub MIC concentrations. The MICs of the heavy metals were also determined among all metal tolerant E. coli isolates. A varied trend of full, moderate and less growth patterns was observed among all the E. coli isolates at different MIC level of the metals tested. Maximum 48, 36 and 25 of the isolates showed less (+) growth pattern at different sub MICs of Cd,Cr and Ni while full growth (+++) was obtained by 8 and 5 of the isolates at sub MIC of Ni and Cr respectively. No full growth was observed against Cd. The highest number of isolates showed moderate growth (++) against Ni at its variable sub MIC levels as compared to Cr and Cd Table 1 (see supplementary material).
All the E. coli isolates showed their MIC range 100-1600 µg/ml against Cd2+ and Cr6+ while 200-2000 µg/ml against Ni3+. No MIC was observed at lower (100µg/ml) concentration against Ni. Most of the isolates demonstrated their MIC at 1200 µg/ml against Ni3+ while 800, 400 µg/ml against Cr6+ and Cd2+ respectively. 34.61% of the isolates showed their MIC at 800 µg/ml against Cr6+ while 25% isolates demonstrated MIC at 400 and 1200 µg/ml against Cd2+ and Ni3+ respectively (Figure 1). All metal tolerant 52 E. coli isolates were tested for their multidrug resistance and resistance patterns. A high level of resistance was observed against Methicillin (86.53%) and penicillin (73.07%) followed by Cephradin (57.69%), Rifampicin (34.61%), Erythromycin (26.92%) Nalidixic acids (25%), Chloramphenicol (3.84%) and least to Gentamycine (1.92%). No resistance was found against streptomycin among the total isolates Table 2 (see supplementary material). A varied trend of resistance patterns was observed among all the E. coli isolates against 10 antibiotics tested. The resistance patterns were recorded for 1 to 6 antibiotics tested. 7.69% of the isolates showed resistant patterns against 6 antibiotics at a time in 4 different combinations. 13.46% and 25% of the isolates showed their resistance patterns against 5 and 4 antibiotics at a time in 3 and 5 different combinations respectively Multi drug resistance index was also obtained in between 0.02-0.15. Among the all E. coli isolates tested are shown in Table 3 (see supplementary material). In our observation only five isolates were found to be sensitive for all antibiotics tested.
Figure 1.
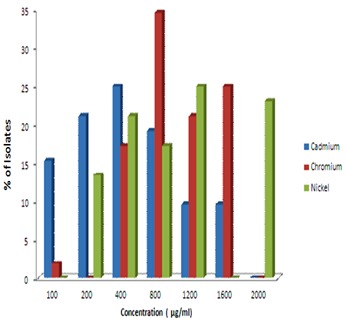
MIC of heavy metal among E. coli isolates from hospital wastewater.
Discussion
Studies have shown that the release of wastewater from hospitals was associated with an increase in the prevalence of antibiotic resistance [17]. Even, exposure to low concentrations level over long periods of time may results in selection and consequent spread of resistance to pharmaceuticals. Antimicrobial resistant gram negative bacterial species were identified in final hospital effluents [18, 19]. Antibiotic resistance in bacteria is a serious threat to society today, and one of the reasons responsible for this problem is over use of antibiotics in humans [20, 21]. Quantified antibiotic residues in waters associated with a hospital in India and assessed their association with quantities of antibiotics prescribed in the hospital and the susceptibility of E. coli found in the hospital effluent [22]. A positive correlation was found between the quantity of antibiotics prescribed in the hospital and antibiotic residue levels in the hospital wastewater. Increased introduction of antimicrobial agents into the environment via medical therapy, agriculture and animal husbandry has resulted in selective pressures on bacterial populations [23]. Microorganisms that are resistant to both antibiotics and metals have been isolated from nosocomial and burn wound infection [24, 25]. A high frequency of resistance among E. coli isolates isolated from hospital wastewater to antibiotics was observed in the present study. E. coli isolates showed a high resistance against methicillin (86.53%), penicillin (73.07%), cephradin (57.69%) and lower against rifampicin (34.61%), erythromycin (26.92%), nalidixic acid (25%),, chloramphenicol (3.84%) and least to gentamycin (1.92%) (Table 2). The resistance patterns were also recorded against 6 antibiotics in different combinations among the total E. coli isolates. Similar results have also been obtained in other studies [26, 27, 28, 29, 30, 31, 32]. They have also reported on transferable plasmids encoding resistance to various heavy metals and antibiotics in gram negative bacteria. All the E. coli isolates showed their MIC range 100-1600µg/ml against Cd2+ and Cr6+ while MIC 200-2000 µg/ml was recorded against Ni (Table 2). Similar findings were also reported by other authors [33, 34]. The situation concerning antibiotics and their resistances resembles in some aspects to heavy metal contamination. Like antibiotics, heavy metals are natural compounds present in different ecosystems. In this study the incidence of antibiotic and metal resistance among E. coli isolates was investigated. Hospital wastewater contained a higher number of single and multiple antibiotic resistances among coliform species [35]. The finding of this study presents a potential health problem as the predominant coliform species have increasingly been associated with outbreaks of hospital infections [36]. It is recommended that hospital waste must be treated before its release into the environment.
Conflict of interest
The authors declare that there is no conflict of interest.
Supplementary material
Acknowledgments
The authors express gratefulness to Syed Waseem Akhtar Vice - Chancellor of Integral University, Lucknow for providing the facilities to execute the research work and thankful to the Heads of the departments of Biosciences and Bioengineering respectfully.
Footnotes
Citation:Alam & Imran, Bioinformation 10(5): 267-272 (2014)
References
- 1.Zhang Q, et al. Science. 2011;333:1764. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 2.Kim S, Aga DS. J Toxicol Environ Health B Crit Rev. 2007;10:559. doi: 10.1080/15287390600975137. [DOI] [PubMed] [Google Scholar]
- 3.Guardabassi L, et al. Water Res. 2002;36:1955. doi: 10.1016/s0043-1354(01)00429-8. [DOI] [PubMed] [Google Scholar]
- 4.McArthur JV, Tuckfieldn RC. Appl Environ Microbiol. 2000;66:3722. doi: 10.1128/aem.66.9.3722-3726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhakephalkar PK, Chopade BA. Biometals. 1994;7:67. doi: 10.1007/BF00205197. [DOI] [PubMed] [Google Scholar]
- 6.Roane TM, Kellogg ST. Can J Microbiol. 1996;42:593. doi: 10.1139/m96-080. [DOI] [PubMed] [Google Scholar]
- 7.Wireman J, et al. Appl Environ Microbiol. 1997;63:4494. doi: 10.1128/aem.63.11.4494-4503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guardabassi L, et al. J Med Microbiol. 2000;49:929. doi: 10.1099/0022-1317-49-10-929. [DOI] [PubMed] [Google Scholar]
- 9.McArthur JV, Tuckfield RC. Appl Environ Microbiol. 2000;66:3722. doi: 10.1128/aem.66.9.3722-3726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calomiris JJ, et al. Appl Environ Microbiol. 1984;47:1238. doi: 10.1128/aem.47.6.1238-1242.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edlund C, et al. Clin Infect Dis. 1996;22:944. doi: 10.1093/clinids/22.6.944. [DOI] [PubMed] [Google Scholar]
- 12.Cappuccino JG, Sherman N. Inc California America. 1995 [Google Scholar]
- 13.Malik A, Jaiswal R. World J Microbiol Biotechnol. 2000;16:177. [Google Scholar]
- 14.Bauer AW, et al. Amer J Clinical. Pathol. 1966;36:492. [Google Scholar]
- 15.Downing T, et al. Genome Res. 2011;21:2143. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riaz S, et al. Afr J Biotechnol. 10:6325. [Google Scholar]
- 17.Elmanama A, et al. Journal of Al-Aqsa University (natural sciences series) 2006;10:108. [Google Scholar]
- 18.Atef M, et al. JTUSCI. 2008;1:23. [Google Scholar]
- 19.Watkinson Andrew, et al. Queensland Australia: University of Queensland; 2008. Antibiotics and Antibiotic Resistant Bacteria in the Aquatic Environment: A Global Issue, an Australian Perspective. [Google Scholar]
- 20.Okeke IN, et al. Emerg Infect Dis. 2007;13:1640. doi: 10.3201/eid1311.070674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diwan V, et al. BMC Public Health. 2010;10:414. doi: 10.1186/1471-2458-10-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. http://academy.asm.org/images/stories/
- 23.Col NF, O׳Connor RW. Rev Infect Dis. 1987;9:S232. doi: 10.1093/clinids/9.supplement_3.s232. [DOI] [PubMed] [Google Scholar]
- 24.Calomiris JJ, et al. Appl Environ Microbiol. 1984;47:1238. doi: 10.1128/aem.47.6.1238-1242.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poiata A, et al. Roum Arch Microbiol Immunol. 2000;59:71. [PubMed] [Google Scholar]
- 26.Murdoch DA, et al. J Trop Med Hyg. 1995;98:25. [PubMed] [Google Scholar]
- 27.Araque M, et al. Int J Antimicrob Agents. 2000;15:34. doi: 10.1016/s0924-8579(99)00168-5. [DOI] [PubMed] [Google Scholar]
- 28.Khan AU, Zaman MS. Biomedical Research. 2006;17:179. [Google Scholar]
- 29.Dash SK, et al. International journal of life science and pharma research. 2012;2250:0480. [Google Scholar]
- 30.Tankovic J, et al. Antimicrob Agents Chemother. 1996;40:2558. doi: 10.1128/aac.40.11.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.George DF, et al. I S R Network ISRN Microbiology. 2012;65:8470. [Google Scholar]
- 32.Ghosh A, et al. Biochem Biophys Res Commun. 2000;272:6. doi: 10.1006/bbrc.2000.2727. [DOI] [PubMed] [Google Scholar]
- 33.Karbasizaed, Naser B. Iran African Journal of Biotechnology. 2003;2:379. [Google Scholar]
- 34.Allen HK, et al. Nat Rev Microbiol. 2010;8:251. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 35.Walter MV, Vennes JW. Appl Environ Microbiol. 1985;50:930. doi: 10.1128/aem.50.4.930-933.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curie K, et al. J Hyg (Lond) 1978;80:115. doi: 10.1017/s0022172400053432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


