Abstract
A glycosidic flavanone miconioside C (1) has been isolated from the methanolic extract of the stems of Miconia prasina, together with 7-O-β-D-apiofuranosyl-(1→6)-β-D-glucopyranosylmatteucinol (2), miconioside B (3), matteucinol (4), farrerol (5) and desmethoxymatteucinol (6). Their structures were mainly established by extensive NMR studies (1H NMR, 13C NMR, DEPT, 1H-1H COSY, HSQC, HMBC) and mass spectrometry. The compounds 1- 3 were evaluated for in vitro binding assays using cannabinoid receptors (CB1 and CB2).
Keywords: Miconia prasina, Melastomataceae, flavanone, flavanone glycoside, cannabinoids receptors
1. Introduction
The Miconia genus (Melastomataceae) consists of approximately 700 species distributed throughout the tropical and subtropical regions of the Americas. Several Miconia species, as well as the compounds isolated from this genus, have shown biological activities, including genotoxic and mutagenic effects (Serpeloni et al., 2008). Moreover, Miconia myriantha has been reported as an enzyme inhibitor of secreted aspartic proteases (SAPs) from Candida albicans (Li et al., 2001).
As a part of our continuing search for novel biological agents (Wang et al., 2011; Gao et al., 2013; Tarawneh et al., 2013), Miconia prasina was evaluated for its biological properties. Previous phytochemical studies of the Miconia genus revealed the presence of several triterpenes (Peixoto et al., 2011), and flavanones (Zhang et al., 2003). This paper reports the fractionation, isolation and structural elucidation of a new flavanone glycoside, miconioside C 1 and five known flavanones 2-6, (Figure 1) from the methanolic extract of the stems of M. prasina. Compounds 1- 3 were evaluated for their in vitro binding affinity for cannabinoid receptors (CB1 and CB2). These compounds showed weak binding affinity for the cannabinoid receptors.
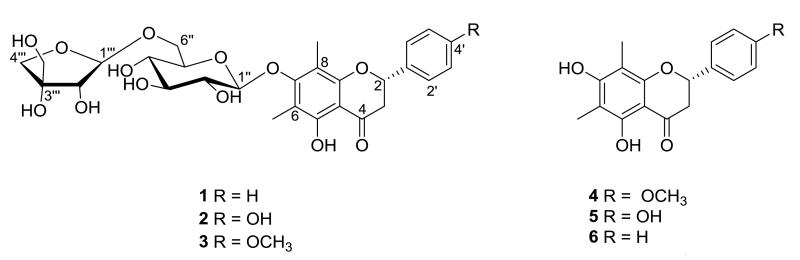
Figure 1.
Flavanones isolated from Miconia prasina
2. Results and discussion
Compound 1, was isolated as a yellowish amorphous solid, and its molecular formula was determined to be C28H34O13 by HRESIMS. The 1H NMR showed six protons singlet signals at δH 2.14, suggesting two aromatic methyls groups, a non-substituted phenyl moiety at δH 7.53 (2H, d, J = 7.2 Hz), 7.42 (2H, t, J = 7.2 Hz) and 7.35 (1H, t, J = 7.2 Hz) related with the B-ring. A flavanone skeleton was evident from the coupling pattern of the C-ring protons δH 2.85 (1H, dd, J = 2.9, 17.0 Hz), 3.15 (1H, dd, J = 12.9, 17.0 Hz) and 5.47 (1H, dd, J = 2.9, 12.9 Hz). Analysis of the 13C NMR spectrum and DEPT experiments, confirmed a flavanone moiety with the presence of a typical ketone carbonyl signal at δC 199.5, and the presence of sugar signals corresponding to one pentose and one hexose. The 1H NMR and 13C NMR spectroscopic data of 1 were found to be similar to those of miconioside B (Zhang et al., 2003) except for the absence of a hydroxyl group at C-4′. The nature and identity of this flavanone was deduced from the NMR experiments (COSY, NOESY, TOCSY, and HSQC). HMBC correlations allowed the complete assignments thus: the anomeric proton of the glucose at δH 4.69 (1H, d, J = 7.7 Hz, H-1″) showed correlation with the carbon at δC 162.7 (C-7), as well as, the correlation between the signal at δH 3.60 (1H, d,d J = 5.6 and 11.2 Hz, Ha-6″) and the signal at δC 110.9 (C-1‴), which together with published data (Zhang et al., 2003; Takahashi et al., 2001), identified 1 as 7-O-β-D- apiofuranosyl-(1→6)-β-D-glucopyranosyldemethoxymatteucinol. The absolute configuration at C-2 at the aglycone was established as S from the strong negative Cotton effect at 271 nm observed in the CD spectrum (Gaffield, 1970). Compound 1 is a newly reported flavanone glycoside 7-O-β-D-apiofuranosyl-(1→6)-β-D-glucopyranosyldemethoxymatteucinol, and is named miconioside C, the next consecutive designation in this nomenclature.
Compounds 2-6 were identified as the known compounds 7-O-β-D-apiofuranosyl-(1→6)-β-D-glucopyranosylmatteucinol 2 (Takahashi et al., 2001), miconioside B 3 (Zhang et al., 2003), matteucinol 4 (Wollenweber et al., 2000), farrerol 5 (Youssef et al., 1998) and demethoxymatteucinol 6 (Basnet et al., 1993). The structures of the known compounds were confirmed by comparison of their spectroscopic properties with published data. To date, this is the first chemical or biological studies dealing with Miconia prasina.
Compounds 1-3 were evaluated at a concentration of 10 μM for their affinity to bind with cannabinoid receptors (CB1 and CB2), following the methods described previously (Gao et al., 2011). The compounds 1-3 exhibited weak inhibition 26.5%, 13.1%, and 18.2% respectively for CB2, not activity for CB1. This is the first report of the evaluation of the binding affinity for the cannabinoid receptors of this kind of compounds.
3. Experimental
3.1. General experimental procedures
Optical rotations were recorded using a Rudolph Research Analytical Autopol V Polarimeter. UV was obtained using a Perkin-Elmer Lambda 3B UV/vis-spectrophotomer. CD spectra were recorder on an Aviv 202SF spectrometer.1H and 13C NMR spectra were obtained on Bruker model AMX 500 NMR spectrometer with standard pulse sequences, operating at 500 MHz in 1H and 125 MHz in 13C. The chemical shift values were reported in parts per million units (ppm) from trimethylsilane (TMS) using known solvent chemical shifts. Coupling constants were recorded in Hertz (Hz). Standard pulse sequences were used for COSY, HMQC, HMBC, TOCSY, NOESY and DEPT. High-resolution mass spectra (HRMS) were measured on a Micromass Q-Tof Micro mass spectrometer with a lock spray source. Column chromatography was carried out on silica gel (70-230 mesh, Merck) and Sephadex LH-20 (Mitsubishi Kagaku, Tokyo, Japan). TLC (silica gel 60 F254) was used to monitor fractions from column chromatography. Visualization of the TLC plates was achieved with a UV lamp (λ = 254 and 365 nm) and anisaldehyde/acid spray reagent (MeOH-acetic acid-anisaldehyde-sulfuric acid, 85:9:1:5). All HPLC analyses were performed on a Waters LC Module I equipped with a UV detector 486 utilizing the Millenium 32 Chromatography Manager software (Waters). An ODS column (Phenomenex Luna C18, 10 × 250 mm, 5 μm) was used. Solvents were HPLC grade, filtered through appropriate filters (water through 0.45 μm and organic solvents through 0.22 μm filters) and purged prior to and during analysis with nitrogen gas at a flow rate of 5 mL/min. All chemicals used were purchased from Sigma-Aldrich (St. Louis, Mo) with the following exceptions: for the binding experiments, [3H]-CP-55,940 (144 Ci/mmol), was purchased from Perkin-Elmer Life Sciences Inc. (Boston, MA, U.S.A.). CP-55,940 was purchased from Tocris Bioscience (Ellisville, Missouri, U.S.A.).
3.2. Plant material
The plant Miconia prasina was collected in Puerto Rico near Caguas in March of 2006 and identified by Gregory Gust. A voucher specimen (Gust 1009 MO) has been deposited in the Missouri Botanical Garden.
3.3. Extraction and isolation
The dried and powered stems of M. prasina (140 g) were extracted with methanol after maceration for three days. Removal of the solvent afforded a viscous residue (7.5 g), which was fractionated by Si gel Vacuum Liquid Chromatography (VLC) stepwise from hexane to methanol, yielding nine fractions (hexane; 3:1 hexane-EtOAc; 1:1 hexane-EtOAc; 1:3 hexane-EtOAc; EtOAc; 3:1 EtOAc-MeOH; 2:2 EtOAc-MeOH; 1:3 EtOAc-MeOH and MeOH). Fractions eluted with hexane-EtOAc (1:1) and hexane-EtOAc (1:3) were combined and subsequently chromatographed by solid-phase extraction (SPE) column initially with hexane and stepwise elution to MeOH. The subfraction eluted with 60% EtOAc in hexane was rechromatographed using Sephadex LH-20 eluted with methanol to furnish compounds 4 (8 mg) and 6 (6 mg); subfraction eluted with 100 % EtOAc was purified by semipreparative HPLC (phenomenex C-18 Luna 5μm, 21 mm × 250 mm, step gradient elution with 30-70% MeOH/H2O) to yield compound 5 (11 mg). Fractions eluted with EtOAc; and 3:1 EtOAc-MeOH were combined and rechromatographed by SPE column initially with 3:1 hexane/EtOAc and stepwise elution to MeOH yielding eight subfractions (50 mL each) (3:1 hexane-EtOAc; 1:1 hexane-EtOAc; 1:3 hexane-EtOAc; EtOAc; 3:1 EtOAc-MeOH; 2:2 EtOAc-MeOH; 1:3 EtOAc-MeOH and MeOH). Subfraction 4 (3:1 EtOAc-MeOH) was purified by preparative HPLC (step gradient elution with 15-85% MeOH/H2O) to furnish compounds 1 (3 mg) and 3 (5 mg). Compound 2 (8 mg) was purified with sephadex LH-20 eluted with CH2Cl2/MeOH (1:1) from subfraction 5 (2:2 EtOAc-MeOH).
3.4. Miconioside C (1)
Yellow gum. [α]25D: −39.3 (c 0.03, MeOH); UV/Vis λmax (MeOH) nm (log ε): 230 (3.83), 280 (4.82), 360 (3.29); IR (KBr): 3358, 2923, 1627, 1450, 1367, 1281, 1124, 1061, 668 cm−1; CD (MeOH, c 0.14): [θ]205 + 22086 (max), [θ]231 - 206 (max), [θ]271 - 19680 (max), [θ]340- 617 (max); 1H NMR (500 MHz, CD3OD): see Table 1. 13C NMR (125 MHz, CD3OD): see Table 1. HRESIMS m/z [M +Na]+ 601.1819 (calcd. for C28H34NaO13, 601.1897).
Table 1.
1H (500 MHz) and 13C NMR (125 MHz) spectral data for 1 in CD3OD.
| Position | Miconioside C (1) | ||||
|---|---|---|---|---|---|
|
| |||||
| δ c | δ h | δ c | δ h | ||
| Aglycone | Glc | ||||
| 2 | 80.2 | 5.47 dd (3.0, 12.9) | 1″ | 105.3 | 4.69 d (7.7) |
| 3 | 44.5 | 2.85 dd (3.0, 17.0) 3.15 dd (12.9, 17.0) |
2″ | 75.7 | 3.51 t br (8.7) |
| 4 | 199.5 | 3″ | 78.0 | 3.41 t (8.5) | |
| 5 | 159.9 | 4″ | 71.7 | 3.25-3.35* | |
| 6 | 113.2 | 5″ | 77.1 | 3.25-3.35* | |
| 7 | 162.7 | 6″ | 68.6 | 3.60 dd (5.6, 11.2) 3.84 dd (1.7, 11.2) |
|
| 8 | 112.0 | Api | |||
| 9 | 159.0 | 1‴ | 110.9 | 4.89 d (2.0) | |
| 10 | 106.4 | 2‴ | 77.9 | 3.80 d (2.0) | |
| 1′ | 140.6 | 3‴ | 80.5 | ||
| 2′, 6′ | 127.2 | 7.53 d (7.2) | 4‴ | 74.9 | 3.67 d (9.6) 3.74 d (9.6) |
| 3′, 5′ | 129.7 | 7.42 t (7.2) | 5‴ | 65.9 | 3.49 d (3.1) |
| 4' | 129.5 | 7.35 t (7.2) | |||
| 6-Me | 9.8 | 2.14 s br | |||
| 8-Me | 9.2 | 2.14 s br | |||
overlapped signals.
4.4. Cell Culture
HEK293 cells (ATCC) were stably transfected via electroporation with full length human recombinant cDNA for cannabinoid receptor subtypes 1 and 2 (obtained from Origene). These cells were maintained at 37 °C and 5% CO2 in a Dulbecco’s Modified Eagles’s medium (DMEM) nutrient mixture F-12 HAM supplemented with 29 mM sodium bicarbonate, 10% fetal bovine serum, penicillin-streptomycin, and G418 antibiotic solutions.
4.5. Radio-ligand Binding for Cannabinoid Receptor Subtypes
Compounds evaluated in the assay were run in competition binding against both the cannabinoid receptor subtypes, CB1 and CB2 (Ross et al., 1999). Cannabinoid receptor binding assays were performed following the methods described previously (Gao et al., 2011).
Supplementary Material
Highlights.
Phytochemical study of Miconia prasina.
One new flavanone glycoside and five known flavanones were isolated.
in vitro binding assays using cannabinoid receptors (CB1 and CB2) were evaluated.
Acknowledgements
The authors thank Dr. Melissa Jacob, for the description of the plant material and Dr. Susan Pedigo for running the CD spectra. The project described was supported by Grant Number 9P20GM104932 from the National Institute of General Medical Sciences a component of the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Furthermore, this investigation was conducted in a facility constructed with support from research facilities improvement program C06RR14503 from the NIH National Center for Research Resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version,
References
- Basnet P, Kadota S, Shimizu M, Xu HX, Namba T. 2′-Hydroxymatteucinol, a new C-methyl flavanone derivative from Mattecia orientalis; potent hypoglycemic activity in streptozotocin (STZ)-induces diabetic rat. Chem. Pharm. Bull. 1993;41:1790–1795. doi: 10.1248/cpb.41.1790. [DOI] [PubMed] [Google Scholar]
- Gaffield W. Circular dichroism, optical rotatory dispersion and absolute configuration of flavanones, 3-hydroxyflavanones and their glycosides. Tetrahedron. 1970;26:4093–4108. [Google Scholar]
- Gao J, Leon F, Radwan MM, Dale OR, Husni AS, Manly SP, Lupien S, Wang X, Hill RA, Dugan FM, Cutler HG, Cutler SJ. Benzyl derivatives with in vitro binding affinity for human opioids and cannabinoid receptors from the fungus Eurotium repens. J. Nat. Prod. 2011;74:1636–1639. doi: 10.1021/np200147c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Radwan MM, León F, Dale OR, Husni AS, Wu Y, Lupien S, Wang X, Manly SP, Hill RA, Dugan FM, Cutler HG, Cutler SJ. Neocosmospora sp.-derived resorcylic acid lactones with in vitro binding affinity for human opioid and cannabinoids receptors. J. Nat. Prod. 2013;76:824–828. doi: 10.1021/np300653d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Jacob MR, Pasco DS, ElSohly HN, Nimrod AC, Walker LA, Clark AM. Phenolic compounds from Miconia myriantha inhibiting Candida aspartic proteases. J. Nat. Prod. 2001;64:1282–1285. doi: 10.1021/np010172p. [DOI] [PubMed] [Google Scholar]
- Peixoto JA, Silva MLA, Crotti AEM, Veneziani RCS, Gimenez VMM, Januario AH, Groppo M, Magalhães LG, dos Santos FF, Alburquerque S, Filho AAS, Cunha WR. Antileishmanial activity of the hydroalcoholic extract of Miconia langsdorffii, isolated compounds and semi-synthetic derivatives. Molecules. 2011;16:1825–1833. doi: 10.3390/molecules16021825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross RA, Gibson TM, Stevenson LA, Saha B, Crocker P, Razdan RK, Pertwee RG. Structural determinants of the partial agonist-inverse agonist properties of 6′-azidohex-2′-yne-Δ8-tetrahydrocannabinol at cannabinoid receptors. Br. J. Pharmacol. 1999;128:735–743. doi: 10.1038/sj.bjp.0702836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpeloni JM, dosReis MB, Rodrigues J, dosSantos LC, Vilegas W, Varanda EA, Dokkedal AL, Cólus IMS. In vivo assessment of DNA damage and protective effects of extracts from Miconia species using the comet assay and micronucleus test. Mutagenesis. 2008;23:501–507. doi: 10.1093/mutage/gen043. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Hirata S, Minami H, Fukuyama Y. Triterpene and flavanone glycoside from Rhododendron simsii. Phytochemistry. 2001;56:875–879. doi: 10.1016/s0031-9422(00)00493-3. [DOI] [PubMed] [Google Scholar]
- Tarawneh A, León F, Radwan MM, Rosa LH, Cutler SJ. Secondary Metabolites from the Fungus Emericella nidulans. Nat. Prod. Commun. 2013;8:1285–1288. [PMC free article] [PubMed] [Google Scholar]
- Wang X, Habib E, León F, Radwan MM, Tabanca N, Gao J, Wedge DE, Cutler SJ. Antifungal metabolites from the roots of Diospyros virginiana by overpressure layer chromatography. Chem. Biodiversity. 2011;8:2331–2340. doi: 10.1002/cbdv.201000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenweber E, Wehde R, Dörr M, Lang G, Stevens JF. C-Methyl-flavonoids from the leaf waxes of some Myrtaceae. Phytochemistry. 2000;55:965–970. doi: 10.1016/s0031-9422(00)00348-4. [DOI] [PubMed] [Google Scholar]
- Youssef DTA, Ramadan MA, Khalifa AA. Acetophenones, a chalcone, a chromone and flavonoids from Pancratium maritimum. Phytochemistry. 1998;49:2579–2583. [Google Scholar]
- Zhang Z, ElSohly HN, Li XC, Khan SI, Broedel SE, Raulli RE, Cihlar RL, Walker LA. Flavanone glycosides from Miconia trailii. J. Nat. Prod. 2003;66:39–41. doi: 10.1021/np020429z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.