Abstract
Internal loops play an important role in structure and folding of RNA and in RNA recognition by other molecules such as proteins and ligands. An understanding of internal loops with propensities to form a particular structure will help predict RNA structure, recognition, and function. The structures of internal loops and from helix 40 of the large subunit rRNA in Deinococcus radiodurans and Escherichia coli, respectively, are phylogenetically conserved, suggesting functional relevance. The energetics and NMR solution structure of the loop were determined in the duplex, The internal loop forms a different structure in solution than in the crystal structures of the ribosomal subunits. In particular, the crystal structures have a bulged out adenine at the equivalent of position A15 and a reverse Hoogsteen UA pair (trans Watson-Crick/Hoogsteen UA) at the equivalent of U4 and A14, whereas the solution structure has a single hydrogen bond UA pair (cis Watson-Crick/sugar edge A15U4) between U4 and A15 and a sheared AA pair (trans Hoogsteen/sugar edge A14A5) between A5 and A14. There is cross-strand stacking between A6 and A14 (A6/A14/A15 stacking pattern) in the NMR structure. All three structures have a sheared GA pair (trans Hoogsteen/sugar edge A6G13) at the equivalent of A6 and G13. The internal loop has contacts with ribosomal protein L20 and other parts of the RNA in the crystal structures. These contacts presumably provide the free energy to rearrange the base pairing in the loop. Evidently, molecular recognition of this internal loop involves induced fit binding, which could confer several advantages. The predicted thermodynamic stability of the loop agrees with the experimental value, even though the thermodynamic model assumes a Watson–Crick UA pair.
INTRODUCTION
Internal loops are important words in the language of RNA. They have implications for folding and stability, and often form tertiary and quaternary interactions. An understanding of internal loop structure and energetics in the presence and absence of contacts is important for understanding fundamental aspects of RNA folding and molecular recognition. Predictions of RNA secondary structure often rely on free energy minimization algorithms that assume relatively simple structural models for estimating energetics (1–3). Crystal structures of ribosomal subunits from Deinococcus radiodurans (4), Escherichia coli (5), Haloarcula marismortui (6) and Thermus thermophilus (7, 8) provide insight into whether these structural models reflect the final structure of the RNA. Differences between models and crystal structures may reveal structural rearrangements important for function or suggest revisions in structural models.
Here we report the solution structure of a duplex, , that contains an internal loop derived from domain II helix 40, E. coli helix numbering (9), of the D. radiodurans large ribosomal subunit (LSU). At an equivalent position, the crystal structure of E. coli LSU contains an internal loop with the same sequence, (5). This internal loop is highly conserved in the secondary structures of large ribosomal subunits (Figure 1) and is a part of the binding site of L20 ribosomal protein (10). Other structurally similar internal loops are present in the D. radiodurans, E. coli and H. marismortui LSUs (Figure 1) (6). They adopt similar structure despite different locations and tertiary interactions. In D. radiodurans internal loop residues A1011 and A1012 engage in ribose zipper interactions and this is also seen for equivalent residues in H. marismortui (11, 12). In the D. radiodurans crystal, the loop has contacts with the hairpin between helix 39 and 40 of the ribosomal RNA and with the L20 protein. L20 is one of nine core proteins (13, 14) that bind to 23S rRNA during an early stage of ribosomal assembly in E. coli (15). The C-terminal domain of L20 represses the translation of its own mRNA and it has been suggested that its binding site on mRNA mimics the binding site on the rRNA (10, 16). The residues at the interface of the protein with helix 40 of 23S rRNA are conserved and involved in recognition of rRNA (17).
Figure 1.
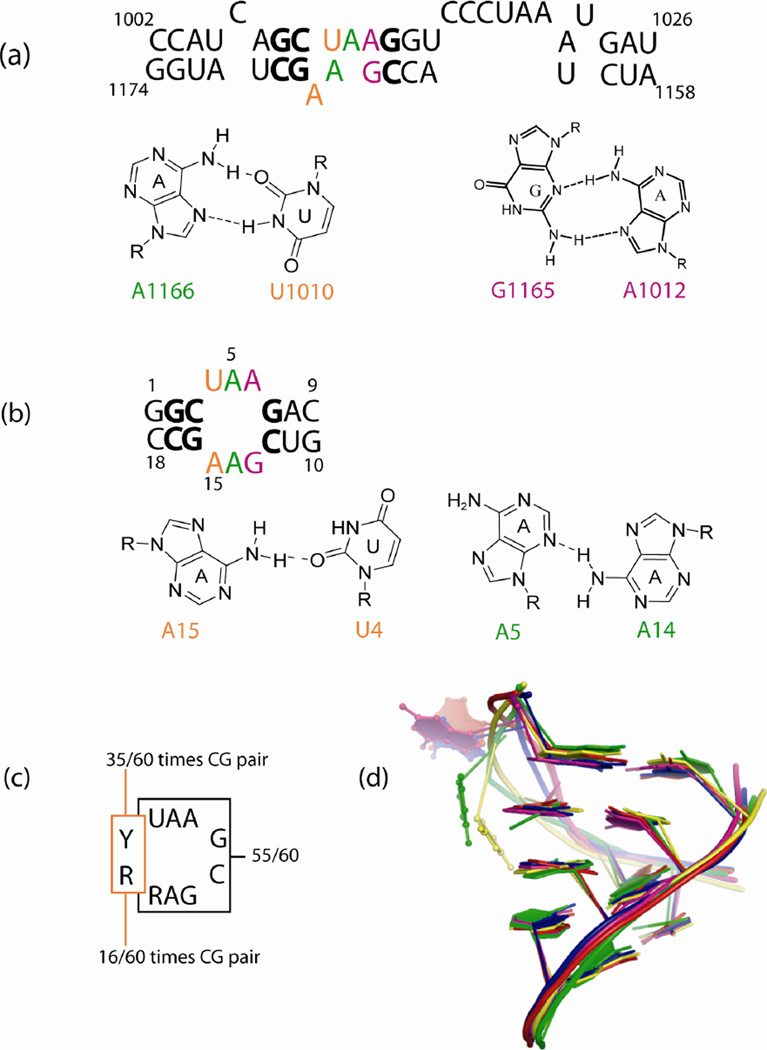
(a) Secondary structure of helix 40 in domain II of the large subunit ribosomal RNA of D. radiodurans. In the crystal structure, U1010 forms a reverse Hoogsteen pair with A1166, A1011 is stacked on A1012 and forms a cross-strand stack with A1166. A1012 forms a sheared pair with G1165. A1167 is flipped out of the helix (4).
(b) The duplex studied by NMR. Nucleotides in bold and color are identical to the natural sequence. Secondary structure prediction of the internal loop predicts it to be a 2 × 2 nucleotide internal loop with U4 forming a Watson Crick pair with A15. The NMR structure reveals a cis Watson-Crick/sugar edge UA pair with a single hydrogen bond.
(c) Conserved features of this internal loop. Out of the 60 secondary structures in Robin Gutell’s secondary structure database, http://www.rna.icmb.utexas.edu/ (37), 55 have the sequence in the box with R being a purine. 35/60 times YR is a CG pair and 16/60 times a UA pair.
(d) Overlap of all heavy atoms for the five similar internal loops observed in D. radiodurans ( in red, in green), E. coli ( in blue, in yellow) and H. marismortui ( in magenta). The bulged bases are shown in ball and stick. Green and yellow bulged bases are uracils.
Structure prediction programs such as RNAstructure 4.11 (1, 3) predict a 2 × 2 nucleotide internal loop with a UA closing pair, but in the crystal structures of the large ribosomal subunits of D. radiodurans (4) and E. coli (5) the adenine corresponding to A15 is flipped out from the helix and the adenine corresponding to A14 forms a reverse Hoogsteen pair with U4 (Figure 1a). In contrast, the NMR structure has a single hydrogen bonded UA pair between U4 and A15, and a sheared AA pair between A5 and A14 (Figure 1b). A sheared GA pair between G13 and A6 in the NMR structure is consistent with the crystal structures. Except for the non–Watson–Crick UA pair, the solution structure agrees with the model used to predict thermodynamic stability. The solution structure suggests that the internal loop undergoes a conformational change when it interacts with ribosomal proteins and rRNA. This internal loop thus exhibits an induced fit (18) rather than lock and key mechanism (19) of binding which may be important for function.
MATERIALS AND METHODS
Oligoribonucleotide synthesis and purification
Oligonucleotides were synthesized on an Applied Biosystems 392 DNA/RNA synthesizer using standard phosphoramidite chemistry (20, 21). The phosphoramidites and the CPG support were bought from Glen Research. For each 1 µmole synthesis, CPG support and the base protecting groups were removed by incubation in 2 mL ammonia/ethanol solution 3:1 (v/v) at 55 °C for 12 h (22). Solid support was removed by filtration and the filtrate was lyophilized followed by removal of silyl protecting groups on the 2' hydroxyls by incubation in 9:1 (v/v) TEA-3HF (triethylamine trihydrofluoride)/DMF (N, N-dimethyl formamide) at 55 °C for ∼ 2 h. RNA was precipitated with 2-butanol and the sample was centrifuged at 12000 rpm. The pellet was washed with 70% ethanol and left to dry at room temperature. The pellet was redissolved in 2 mL of 5 mM ammonium bicarbonate at pH 7 and the sample was loaded onto a Waters Sep-Pak C-18 reverse phase column to remove excess salts. The eluted sample was then applied onto a preparative Baker Si500F silica gel plate (20 × 20 cm, 500 µm thick). The plates were kept in a sealed TLC chamber with a running solution of 1-propanol/ammonia/water in the ratio of 50:40:10 (v/v/v). The slowest running band on the plate was identified with UV light and scraped off. RNase free water was used to dissolve the RNA and then a Waters Sep-Pak C18 reverse phase column was used to remove salts. The mass of the RNA was verified by ESI MS with a Hewlett-Packard 1100 LC/MS Chemstation and the purity was verified by analytical reverse phase HPLC. All the samples were >95% pure.
NMR sample preparation
Sample preparation was similar to Chen et al. (23) with minor modifications. An equal number of moles of each strand was mixed in 300 µL of RNase free water and then dialyzed overnight against 1 L filtered autoclaved water in a Gibco Life Technologies microdialysis system with a 1000 MWCO Spectro-por dialysis membrane and a Rainin Dynamax peristaltic pump. After dialysis, the sample was lyophilized and dissolved in 250 – 300 µL of NMR buffer (80 mM NaCl, 10 mM sodium phosphate and 0.5 mM Na2EDTA at pH 6.5). The sample was dried and reconstituted with 10% D2O in water to provide a lock signal for the exchangeable proton spectrum. For non-exchangeable proton spectra, D2O exchange was done by three repetitions of lyophilization with 99.96% D20 as the solvent and the sample was finally dissolved in 300 µL of 99.96% D2O from Cambridge Isotope Laboratories. The total duplex concentration was approximately 2 mM. The sample was placed in Shigemi tubes for collection of the spectra. Later, in order to observe the effect of Mg2 + ion on the structure, 10 mM MgCl2 was added to the NMR sample (80 mM NaCl, 10 mM sodium phosphate, 0.5 mM Na2EDTA and 10 mM MgCl2 at pH 6.5).
NMR spectroscopy
NMR spectra were collected on Varian Inova 500 and 600 MHz spectrometers. One-dimensional and two-dimensional imino proton spectra at different temperatures were acquired with an S-shaped excitation pulse (24) with spectral width of 12 kHz. SNOESY spectra were recorded in 90:10 (v/v) H2O/D2O at 15 and 25 °C with mixing times of 100 and 150 ms. Each spectrum was acquired with 256 t1 increments with 2k complex points and 70 scans per FID and a recycle delay of 1.2 s.
NOESY spectra of the sample in D2O were acquired at 15, 25 and 30 °C with mixing times of 100, 200, 400, and 600 ms. FIDs were acquired with spectral width of 4200 Hz and 2k complex points. A total of 360 FIDs were obtained with 32 scans per FID and a recycle delay of 2.8 s. TOCSY spectra were collected with mixing times of 8, 18, 30 and 100 ms at 15, 20, and 30 °C. These spectra were acquired with 2k complex points and a spectral width of 4000 Hz. A total of 256–512 FIDs were acquired with 24–64 scans per FID. Known temperature dependent chemical shifts of H2O or HDO relative to 3-(trimethylsilyl) tetradeutero sodium propionate (TSP) were used to reference the proton spectra. A natural abundance 1H-13C HSQC spectrum at 25 °C was acquired on a Varian Inova 600 MHz spectrometer with 6000 Hz spectral width for 1H dimension and 16600 Hz for 13C dimension. The spectrum has 96 increments, and was collected with 1k complex points with 672 scans per FID. Natural abundance 1H-13C HMQC and 1H-13C HSQC spectra were acquired at 25 and 30 °C, respectively. The HMQC was acquired with a spectral width of 5000 Hz for the 1H dimension and 14000 Hz for 13C dimension. The HSQC was acquired with a spectral width of 5000 Hz for the 1H dimension and 15000 Hz for 13C dimension. The HMQC spectrum consisted of 60 increments each having 1k complex points and 544 scans per FID. The HSQC spectrum consisted of 64 increments each having 1k complex points and 512 scans per FID. T1 relaxation rates for the base protons were measured by the inversion recovery method. The 1H-31P HETCOR spectrum was acquired with a spectral width of 1400 Hz for 1H and 2000 Hz for 31P. Two-dimensional spectra were processed using NMRpipe (25). Similar one-dimensional imino spectra at different temperatures, SNOESY at 15 °C with 150 ms mixing time and NOESY spectra (in D2O) with 100 ms and 200 ms mixing time at 25 and 30 °C were collected after addition of 10 mM MgCl2.
Restraint generation
NOESY cross-peak volumes were obtained with the Sparky software package (26) using box integration. Distance restraints were generated for the duplex from 100 and 200 ms mixing time NOESY spectra at 25 °C and a 150 ms SNOESY spectrum at 15 °C. The Sparky output was converted to Discover input by using a C ++ program, which converted the volume for each peak to a distance restraint using 1/r6 scaling and a two-spin approximation. Average H5-H6 peak volumes were used as a reference with a distance of 2.45 Å.
NOE derived distances were assigned error limits of ± 30% (100 ms mixing time) and ± 40% (200 ms mixing time and SNOESY spectra) to allow for relaxation, spin diffusion, baseline distortions, water exchange and noise. Restraints were estimated or discarded in case of extreme overlaps. No restraints were used from H5' and H5" protons. A total of 203 interproton distance restraints were used with 106 internucleotide and 97 intranucleotide restraints (Table S2, supporting information). NMR data are consistent with the formation of Watson-Crick pairs. Thus, 17 hydrogen bond distance restraints (1.8–2.5 Å between hydrogen and acceptor) were used for the six Watson-Crick pairs of the stem. Backbone dihedral angles for the Watson-Crick stem residues were loosely restrained: α (0 ± 120°), β (180 ± 30°), γ (60 ± 30°), δ (85 ± 30°), ε (−140 ± 40°), ζ (0 ± 120°), and χ (−170 ± 40°). No hydrogen bonding or dihedral angle restraints were used for the loop nucleotides, except for δ and χ. TOCSY and DQFCOSY spectra were used to identify residues in a C2' endo sugar pucker. A5, terminal residues (C9, C18) and G13 showed H1'-H2' cross-peaks in DQFCOSY spectra, and therefore were restrained to cover both the C3' and C2' endo conformation with δ (122.5 ± 67.5°). The remaining loop residues (U4, A6, A14, A15) were restrained to be in C3' endo conformation, δ (85 ± 30°). The glycosidic torsion angles were restrained in anti conformation, χ (−120 ± 90°), for all loop residues since the intensity of the intranucleotide cross-peak between H1'-H8/H6 was not comparable to the H5-H6 cross-peak. Supporting information has tables of distance, dihedral angle, and endocyclic sugar torsion angle restraints.
Simulated annealing
The Discover 98 software package running on a Silicon Graphics Octane computer was used to perform NMR-restrained molecular dynamics and energy minimization. Standard A-form starting structure was generated using Biosym Insight II software. Simulations were done with the AMBER 95 force field (27) using flat bottom restraint potentials, with force constants of 25 kcal/(mol A2) for distance restraints and 50 kcal/(mol rad2) for torsion-angle restraints and a maximum force of 1000 kcal/mol. Simulations were done in vacuo with the NMR derived restraints. For van der Waals interactions, group based summation was used with an 18 Å cutoff. For the electrostatic interactions, the cell-multipole method (28) was used with a distance-dependent dielectric constant, ε = 2r, where r is the distance in Angstroms. There were a total of 14 steps in the simulations (29, 30): (1) van der Waals and electrostatic energy scaled to 1% and 0%, respectively, and dihedral, NOE and covalent bond energy scaled to 100%; (2) steepest decent minimization started with 500 iterations; (3) restrained molecular dynamics (rMD) at 1000 K for 4 ps with 1 fs time steps; (4) 2 ps rMD at 900 K; (5) 2 ps rMD at 800 K; (6) van der Waals and electrostatic energy increased to 33%; (7) 2 ps rMD at 700 K; (8) van der Waals and electrostatic energy increased to 67%; (9) 2 ps rMD at 600 K; (10) van der Waals and electrostatic energy increased to 100%; (11) 2 ps rMD at 500 K; (12) 2 ps rMD at 400 K; (13) 2 ps rMD at 300 K; and (14) 40,000 iterations of conjugate energy minimization. Steps 3–5 effectively randomize the starting structure. A total of 50 structures were generated and used for analysis.
UV Melting Experiments and Thermodynamics
Oligonucleotides were lyophilized and dissolved in 1.0 M NaCl, 20 mM sodium cacodylate and 0.5 mM Na2EDTA at pH 6.5 (Melt buffer) or in 100 mM KCl, 10 mM MgCl2, 20 mM sodium cacodylate at pH 6.5. The concentration of single stranded RNA was calculated from the absorbance at 260 nm at 80 °C prior to dissolving in Melt buffer. Extinction coefficients were predicted from those of dinucleotide monophosphates and nucleosides (31, 32). Small mixing errors for non-complementary duplexes do not affect the thermodynamics appreciably (33). Absorbance vs. temperature melting curves for the duplex were acquired at 260 nm at heating rates of 1 °C/min with a Beckman Coulter DU640C spectrophotometer having a Peltier temperature controller cooled with flowing water.
Melting curves were fit to a two state model with Meltwin software, assuming linear sloping baselines and temperature independent ΔH° and ΔS° (2, 34, 35). Melting temperature of the duplex in kelvins, TM, at different concentrations, where CT is the total strand concentration, were used to calculate the thermodynamic parameters from (36):
| (1) |
The equation ΔG°37 = ΔH° - (310.15) ΔS° was used to calculate the free energy change at 37 °C (310.15 K). The ΔH° values were also obtained from averaging fits of individual melting curves and were within 6% of the values derived from the TM−1 vs ln CT/4 plots suggesting that a two state model is a good approximation for this duplex.
RESULTS
Sequence and structure conservation
A survey of secondary structures of 60 large ribosomal subunits (37) showed that 55/60 structures had the conserved sequence where Y represents a pyrimidine, either U or C and R represents a purine, either G or A (Figure 1c). YR is a CG pair in 35/60 cases and a UA in 16/60 cases. The crystal structures of the D. radiodurans (PDB: 1NKW), E. coli (PDB: 2AWB) and H. marismortui (PDB: 1JJ2) large subunit rRNAs show that this conserved internal loop adopts a very similar three dimensional structure (4–6) (Figure 1d).
In the D. radiodurans internal loop, , A1167 is flipped out of the helix. U1010 forms a reverse Hoogsteen pair with A1166 (Figure 1a). A1011 is stacked between A1166 and A1012 making a cross-strand as well as an intrastrand stacking pattern. All the internal loops in Figure 1d have similar three dimensional structural features, which suggests functional relevance.
The structural similarity between these internal loops from different organisms is striking especially since each is in a different context of tertiary and quaternary contacts (4–6). The structural similarity motivated the NMR studies presented below. The duplex was designed to provide a model system for the internal loop (Figure 1b). NMR studies were done in the absence and presence of Mg2 +. The spectra observed were very similar and did not indicate any change in structure upon addition of 10 mM MgCl2.
Assignments of exchangeable protons
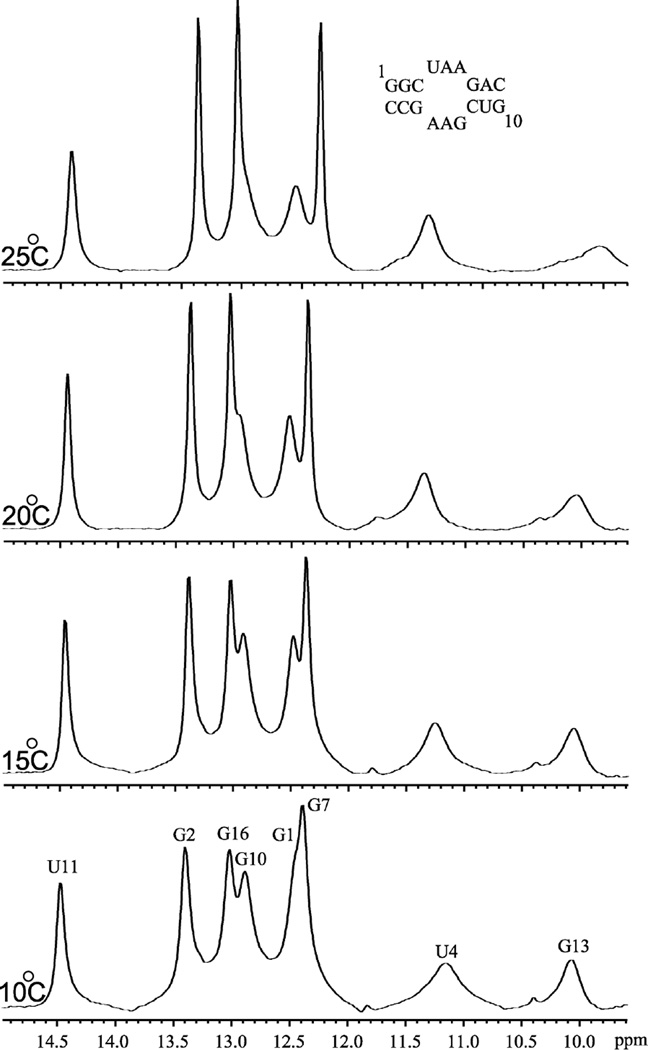
The RNAstructure 4.11 program predicts seven Watson–Crick base pairs (2 UA pairs and 5 GC pairs) in the stems. Imino proton (10–15 ppm) spectra of the duplex (Figure 2) provide information about base pairing. Five GC pairs were identified (11.5–13.5 ppm) but only the U11H3 peak was found in the usual UA Watson–Crick pair region (13.5–14.5 ppm). Evidently, U4 does not form a Watson–Crick pair. Two broad peaks at 10.1 and 11.2 ppm are from G13H1 and U4H3, respectively. Two weak peaks resonating at 10.4 and 11.8 ppm may be due to imino protons of G13 and U4 in minor conformations. G1H1 and G7H1 are overlapped at 10 °C but are resolved above 10 °C. In contrast, G10H1 and G16H1 are resolved at 10 °C but overlap at higher temperatures. SNOESY spectra were used to assign the imino resonances and to confirm the secondary structure (see Supporting Information). The cross-peak between G1H1 and G2H1 in the SNOESY spectrum helps assign G1H1 (12.57 ppm). G7H1 (12.35 ppm) was assigned on the basis of its cross-peak to U11H3 (14.46 ppm). The G2H1 (13.37 ppm) was assigned on the basis of a cross-peak to G16H1 (13.04 ppm). C3 and C17 amino protons gave cross-peaks to both G2H1 and G16H1. U4H1' gave a cross-peak to G16H1 and C18H1' gave a cross-peak to G2H1 which helped confirm these assignments. The remaining peak in the GC imino region (12.95 ppm) is assigned as G10H1 by exclusion.
Figure 2.
One-dimensional imino proton spectra of 5'GGCUAAGAC/3'CCGAAGCUG at temperatures ranging from 10 °C to 25 °C at pH 6.5. Assignments are given on the bottom spectrum.
Assignments of nonexchangeable protons
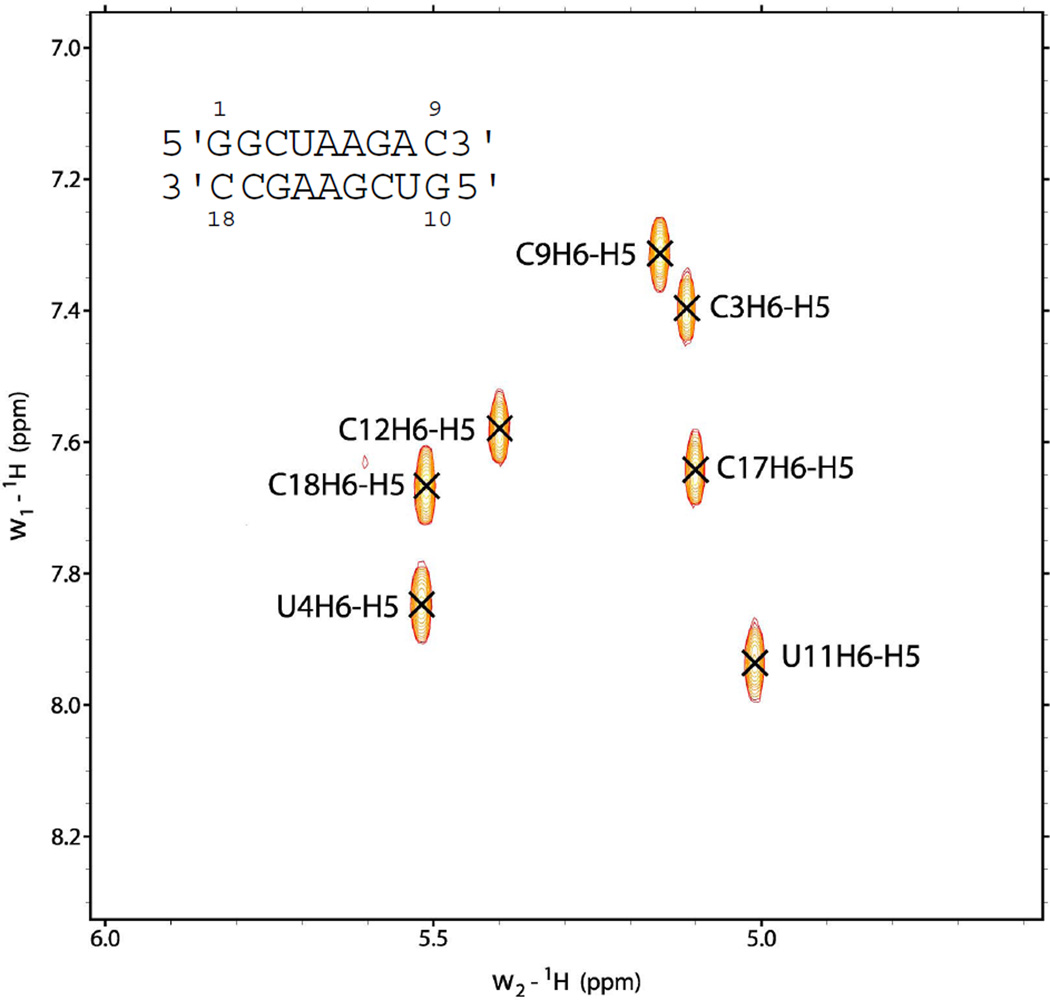
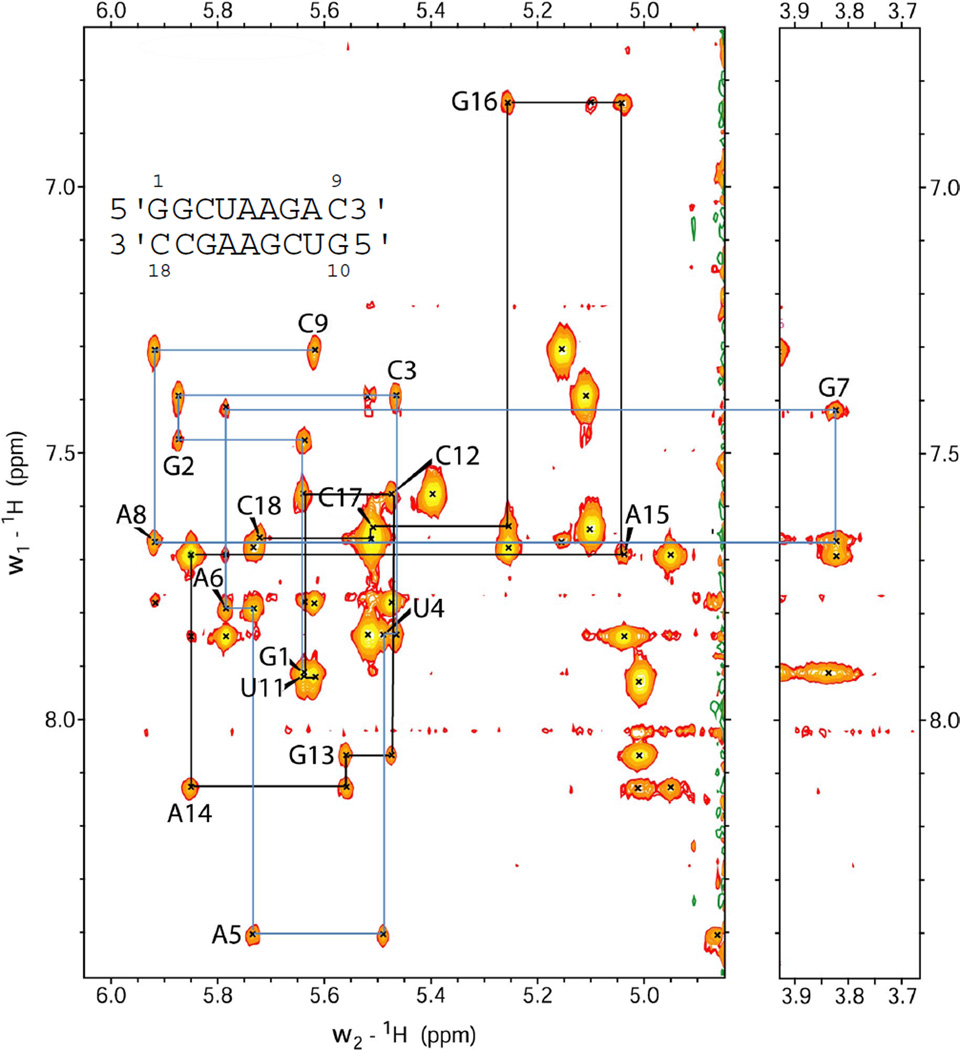
Resonances for non-exchangeable protons were assigned as described previously (38, 39). Supporting information has details of the chemical shift assignments at 25 °C. A TOCSY spectrum (Figure 3) at 25 °C with 30 ms mixing time was used to identify the seven H5-H6 cross-peaks. Lack of extra H5-H6 peaks in the TOCSY spectrum implies one predominant structure. The cross-peaks resulting from H5-H6 were used to identify the pyrimidine peaks in NOESY spectra and linked cross-peaks were used to identify consecutive pyrimidines C3, U4, C17 and C18. The (H8/H6/H2)-(H1'/H5) region of the 100 ms NOESY spectrum is shown in Figure 4. Sugar systems associated with the NOESY walk peaks were identified from strong H1'-H2' (∼ 2.8–3.0 Å) and H1'-H3' (∼ 3.5–4.0 Å) cross-peaks. Assignments follow standard connectivity from G1-C9. The G7H1' peak is shifted upfield to 3.82 ppm (25 °C) and was confirmed by natural abundance 1H-13C HSQC at 30 °C (Figure S6, Supporting Information). U11-C18 also followed a standard NOESY walk but G10H8 could not be identified because of overlap. Overlap between some H8 peaks caused difficulty in assignment of sugar protons, and such peaks were not used for modeling. H5' and H5″ are not assigned stereospecifically, but H5' protons usually resonate downfield from the H5″ protons due to the negative charge on the phosphate (40).
Figure 3.
TOCSY spectrum of at 25 °C showing H5-H6 cross-peaks for U's and C's.
Figure 4.
NOESY walk region, (H8/H6/H2)-(H1'/H5), of the 100 ms mixing time NOESY spectrum at 25 °C. Blue and black lines trace the walk for the top and bottom strands of the duplex, respectively. The H8/H6-H1' peaks are labeled.
A 1H-13C HSQC spectrum (Figure S4, Supporting Information) and measurement of base proton relaxation times (data not shown) helped identify and confirm the AH2 resonances. Cross-peaks to A6H1', G7H1' and A14H1' helped assign A6H2. An overlap at 25 °C between the A6H2-A14H1' and A15H8-A14H1' cross-peaks was resolved at 30 °C. A8H2 was assigned based on its cross-peaks to G7H1, A8H1', C9H1', and C12H1'. Assignment of A14H2 was based on its cross-peaks with A6H1', A6H2, A14H1', A14H2', A15H1' and A15H8. A15H2 was assigned on the basis of its cross-peaks to A5H1' and G16H1'. The assignment of A5H2 was based on A5H2-G13H2' and A5H2-A14H8 cross-peaks.
DQFCOSY, TOCSY and 1H-31P-HETCOR spectra were used to assign the sugar protons. The H2' and H3' peaks were identified from strong cross-peaks to H1'. They were confirmed by walking from strong to medium sequential cross-peaks (n) H8/H6–(n-1) H2'/H3'. Significant overlap in the sugar regions hindered generation of some standard internucleotide restraints for the modeling studies. 31P resonances for all residues except A5 (0.97 ppm) were observed within a 1.5 ppm (∼ −1.0 to 0.5 ppm) range in the HETCOR spectra (see Supporting Information), which is consistent with A-form geometry (41) as the predominant conformation of the duplex. The 31P resonance for A6 is about 0.5 ppm downfield from the next most downfield 31P resonance (G7, 0.05 ppm).
Spectra collected upon addition of 10 mM MgCl2
The addition of 10 mM Mg2 + resulted in largely unchanged imino and NOESY spectra (Figures S8, S9 in Supporting Information), including frequencies and intensities of cross-peaks. Changes in chemical shifts of imino, H8 and H1' resonances were mostly ≤ 0.02 ppm but always within 0.06 ppm (See Supporting Information, Table S4). Adenine H2s had cross-peaks similar to those in spectra without MgCl2. In the absence of any relevant changes, all the calculations were performed on spectra obtained without MgCl2.
Structural determination
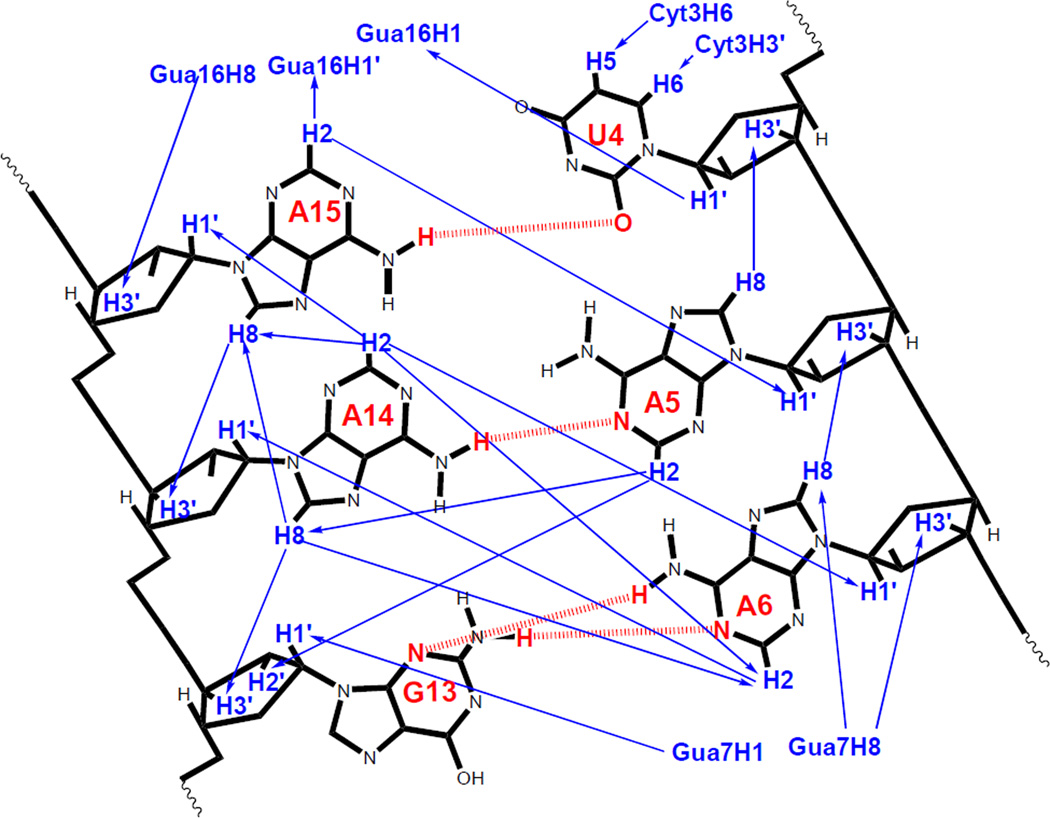
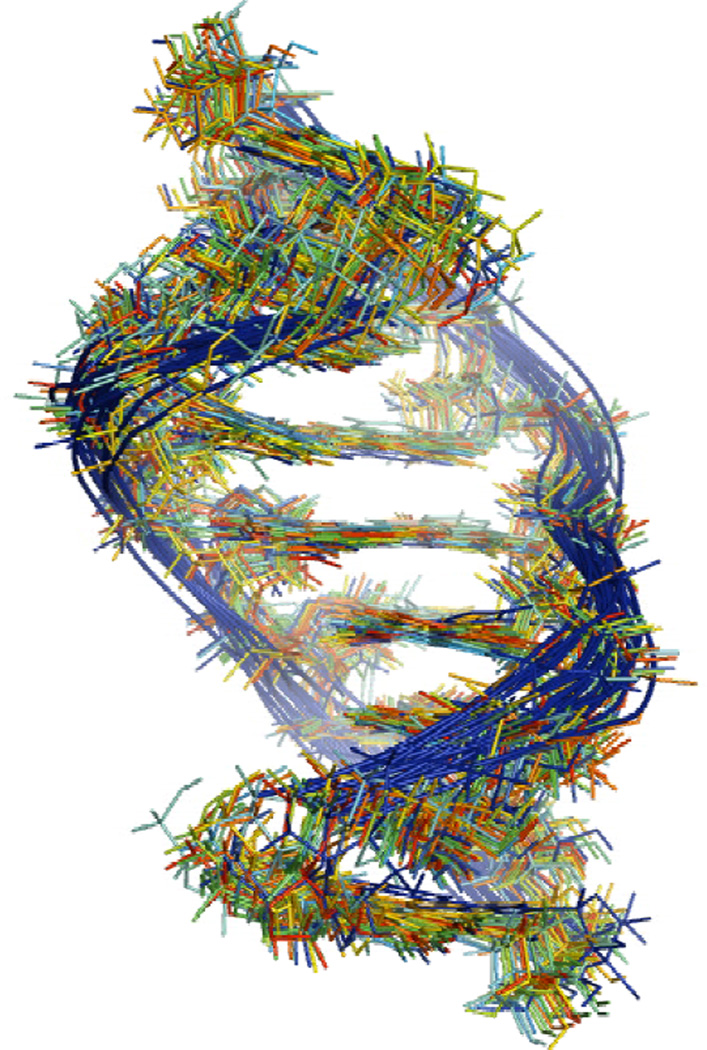
Distance and dihedral angle restraints derived from the NMR spectra (see Supporting Information) were used to model the structure of the duplex. Figure 5 summarizes some of the most important NOEs. A total of 50 structures were generated in the absence of solvent using the simulated annealing protocol described in Materials and Methods. The 50 structures converged to satisfy all NMR derived distance and dihedral angle restraints within 0.1 Å and 2°, respectively. Superposition of the 20 lowest energy structures reveals that the overall structure and local features are consistently reproduced (Figure 6). The average RMSD for all atom and heavy atom pairwise superposition of these 20 structures to the average structure is 0.49 and 0.45 Å, respectively. All 20 structures have an A15U4 pair with a single hydrogen bond (cis Watson-Crick/Sugar edge A15U4), a sheared A14A5 pair (trans Hoogsteen/sugar edge A14A5) (42) (see Figure 1b) and a sheared A6G13 pair (trans Hoogsteen/sugar edge A6G13) (see Figure 1a). Within the loop there is cross-strand stacking between A6 and A14 and intrastrand stacking between A14 and A15.
Figure 5.
Some interstrand and internucleotide NOEs which are important for the structure of the internal loop are shown with blue arrows. Hydrogen bonds between the bases (aromatic) are shown in red.
Figure 6.
Superposition of 20 lowest energy structures for derived from restrained molecular dynamics. The backbone is distorted between A5 and A6 and between G13 and A14.
In solution, A15 (A1167) is in a cis Watson–Crick /Sugar edge pair with U4 and not bulged out
A15, which was designed to mimic the bulged out base in the crystal structure, is not flipped out in the predominant conformation determined by NMR (Figures 1b, 6, 7). Cross-peaks in the NOESY spectrum between A15H2-A5H1' and A15H2-G16H1' suggest this adenine is inside the helix (Figure 5). The H8 resonance of A15 is no broader than the H8 resonances of other bases and thus does not show evidence for chemical exchange suggesting that A15 is held in a single conformation. A14H8-A15H8 and A15H8-G16H8 cross-peaks also indicate that the A15 base is located between the A14H8 and G16H8 protons, suggesting stacking of the three purines (Figure 5). The A14H2-A15H1' NOE is also consistent with A15 stacked on A14. In solution, we find no evidence for A15 being flipped out as in the crystal structure.
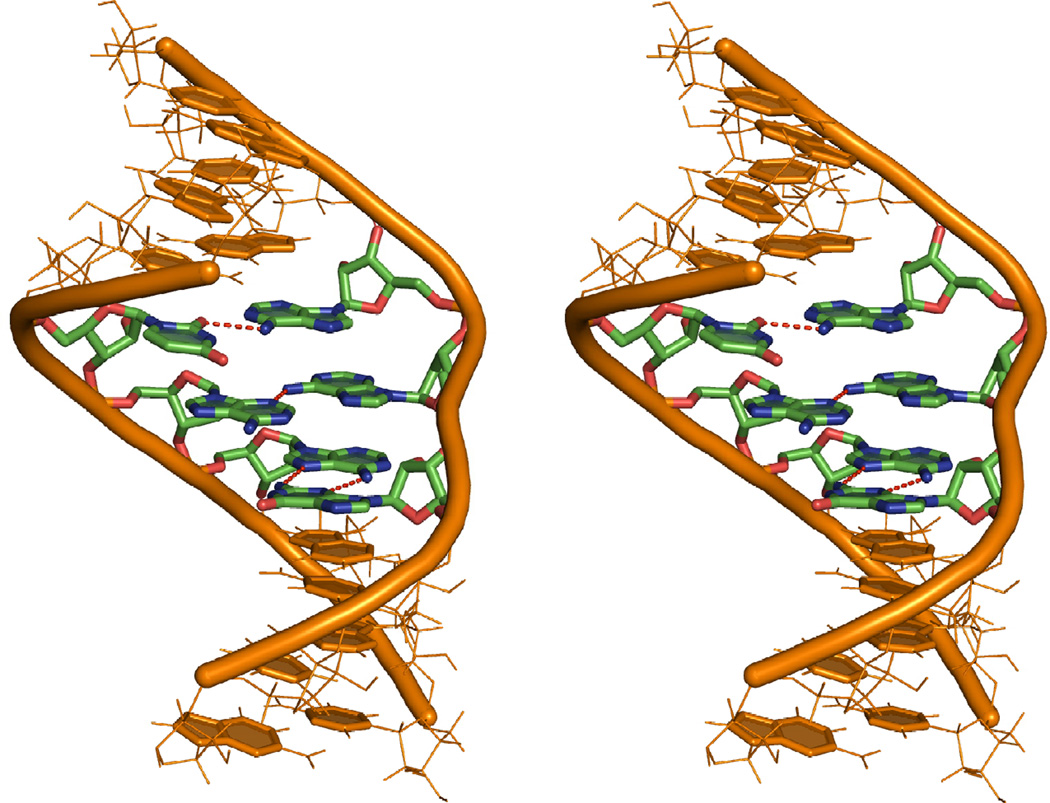
Figure 7.
Stereo view of the average modeled low energy structure for . The stem regions are shown in orange lines. The loop region is shown in sticks where red is for oxygen, blue for nitrogen, green for carbon and orange cartoon for the backbone. The 5'G1 is at the top and 3'C8 is at the bottom. Dotted red lines show hydrogen bonds between bases. Hydrogen atoms are hidden for clarity.
The NMR structure has a single hydrogen bond between the A15 amino and the U4 carbonyl O2 (cis Watson-Crick/sugar edge A15U4). The 1D imino region of the 1H spectrum has peaks at 10.2 and 11.6 ppm, either of which could be the U4 imino peak and both of which are outside the usual range for a Watson–Crick AU pair. Typically, 3' dangling U's are seen at around 10.9 ppm (23) and most G imino peaks in a GA sheared pair are seen at ∼ 10 ppm (38, 43–46) . The peak at 10.2 ppm becomes broader with increasing temperature while the non-terminal Watson–Crick peaks become sharper (Figure 2). This suggests that the peak at 10.2 ppm is the G13 imino proton, which is in chemical exchange with water. By exclusion, the peak at 11.6 ppm is from the U4 imino proton. In contrast, A8-U11 has an imino proton resonance at 14.46 ppm, which is in the expected region for a Watson–Crick pair. Heus et al. (47) observed that a UA pair with Watson–Crick geometry but longer than usual hydrogen bonds had the U imino proton resonance shifted upfield to 11.75 ppm. This supports opening of the A15U4 pair in the duplex such that it does not have both the hydrogen bonds of a regular Watson–Crick pair. Several distances are near or outside of the NOE-derived distance limits if the structure is modeled with A15U4 restrained to be a Watson–Crick pair. If A15U4 is in a Watson– Crick pair, then the A15H2-A5H1' distance (3.26 Å) should be shorter (2.88 Å), the A15H2-G16H1' distance (2.73 Å) should be longer (3.55 Å), the U4H1'-G16H1 distance (2.59 Å) must be longer (3.73 Å, a restraint violation) and the A15H8-A14H3' distance (3.03 Å) should be shorter (2.17 Å). These restraints however, are consistent with the proposed model with a single hydrogen bond between A15 and U4.
The unusual UA pair (cis Watson-Crick/sugar edge A15U4) may be stabilized due to stacking of U4 on C3. A 3' dangling U in a motif has a ΔG°37,Stack averaging –1.1 kcal/mol (48, 49) and is typically stacked with the C amino-H41 closely overlapping the carbonyl-O4 (∼ 3.39 Å). In the NMR structure, the U4 carbonyl-O4 is stacked on C3's amino in a similar way. Additionally, the U4 carbonyl-O4 is stacked on the A5 amino-H62 (∼ 2.99 Å) which would also favor this unusual UA pair (Figure S7, Supporting Information). Quantum mechanical studies of nucleic acid bases have suggested that amino groups are intrinsically non-planar (50, 51), but empirical potentials like AMBER consider amino groups to be planar (27). The A5 amino group may not be in the planar sp2 geometry but rather in a pyramidal geometry with the amino hydrogens out of plane. This would decrease the distance between the A5 amino hydrogens and U4 carbonyl-O4 and increase the stability of this interaction.
The crystal structure of the 58 nucleotide RNA from large ribosomal subunit and L11 protein has a similar UA pair between U1065 and A1073 (52). This base pair, however, might be stabilized by a tertiary contact in the major groove. The crystal structures of the specificity domain of ribonuclease P (53) and of the large ribosomal subunit of D. radiodurans (4) have similar UA pairs with a single hydrogen bond except that the Hoogsteen edge of A interacts with the sugar edge of U (cis Hoogsteen/sugar edge AU). The hydrogen bonds are between A124H61-U119O2 and A911H62-U890O2, respectively. In all cases, the carbonyl-O4 of U overlaps a C amino group. The positioning of U on C at the end of the helix may be energetically more important than forming a second hydrogen bond in a UA pair. The face of A that interacts with U may not be a big constraint as long as the hydrogen bond between AH61 and UO2 can form.
Sugar puckers of sheared purine-purine pairs
It has been suggested that the sugar pucker of G in a sheared GA pair may depend on the 5' base (47). In the case of a 5' U, the adjacent G in a sheared GA pair has a C2' endo conformation whereas with a 5' C, the adjacent G has a C3' endo conformation. In the internal loop described in this work, G13 has a C3' endo conformation as predicted. The loop also has a 5' U adjacent to an AA sheared pair. The A5 sugar has a dynamic conformation indicated by ∼ 5 Hz coupling between H1' and H2'. Although both C2' endo and C3' endo conformations were allowed in the modeling, all modeled structures have a 3' endo sugar conformation. Forcing the A5 sugar to a C2' endo conformation did not change the base pairing pattern. The results suggest that rules for the sugar pucker of an A in a sheared AA pair differ from those for a G in a sheared GA pair.
There is a sheared A5A14 pair
In the 20 lowest energy structures, A5 and A14 form a sheared AA pair (AA trans Hoogsteen/sugar edge A14A5, see Figure 1b) similar to that observed in the structure of the P4-P6 domain of the T. thermophila group I intron (42). Several observations are consistent with the formation of a sheared AA pair between A5 and A14. Cross-peaks are observed from A5H2 to A14H8 and G13H2' (Figure 5). The A5H2 to A14H8 and G13H2' cross-peaks give NMR derived distances of 4.4 and 4.1 Å, respectively. This indicates that the adenosines are not in a face-to-face geometry, which would result in interstrand A5H2 to A14H8 and G13H2' distances of at least 7 Å. Also, A6H2 to A14H1' has an NMR derived distance of 2.6 Å, which indicates that the minor groove is narrower than typically observed for an A-form helix. For equivalent protons in an A-form helix, the distance can range from 3.2 Å to 4.1 Å (54, 55). Therefore, the observed distances suggest that the minor groove does not widen to accommodate a face to face adenine pair.
The sheared G13A6 pair is consistent with the crystal structure
The G13A6 sheared pair is characterized by four pieces of evidence: (1) a shifted G imino peak in the 1D spectrum indicating the absence of an imino proton hydrogen bond, (2) a medium intensity cross-peak between G7H1 and G13H1' (2.89 Å ± 40% ), (3) An NOE cross-peak between A6H2 and G7H1' (2.96 Å ± 40%) indicating that the Watson–Crick edge of A6 is in the minor groove, and (4) an upfield shift of G7H1' to 3.82 ppm. An upfield shift of the H1' for the nucleotide 3' of A is expected for a sheared GA pair (43, 44, 47, 56–59). This upfield chemical shift can be explained by the large ring current effect of adenosine above the H1' proton of the 3′ residue (60, 61).
Thermodynamics
Thermodynamic parameters for the duplex were measured in the presence of 1 M NaCl and also in the presence of 0.15 M KCl and 10 mM MgCl2 (Table 1). Results for the two buffers were the same within experimental error (Table 1). The free energy increment at 37 °C for the loop was calculated as −1.78 kcal/mol (Table 2) from:
| (2) |
In this equation, and are the free energy changes for duplex formation with and without the internal loop at 37 °C. The is obtained from TM vs. ln (CT/4) plots. The was calculated from nearest neighbor parameters (1, 2).
Table 1.
Thermodynamic parameters for duplex formation by 5'GGCUAAGAC/3'CCGAAGCUG a
| ΔH° (kcal/mol) |
ΔS° (cal/K. mol) |
AG°37 (kcal/mol) |
Tmb (°C) |
|
|---|---|---|---|---|
| Curve fit Parameters |
−83.39 ± 10.63 (−85.70 ± 5.59) |
−240.04 ± 34.55 (−245.69 ± 17.74) |
−8.94 ± 0.24 (−9.50 ± 0.23) |
46.2 (48.1) |
| 1/TM vs ln CT Parameters c |
−80.83 ± 9.65 (−80.79 ± 7.92) |
−231.73 ± 30.54 (−230.27 ± 24.90) |
−8.96 ± 0.30 (−9.37 ± 0.25) |
46.6 (48.3) |
| Predicted d | −56.08 | −154.16 | −8.23 | 46.9 |
Buffer conditions are 1 M NaCl, 20 mM sodium cacodylate, 0.5 mM Na2EDTA, pH 6.5. Values in parenthesis are for buffer conditions with 0.15 M KCl, 20 mM sodium cacodylate, 10 mM MgCl2, pH 6.5.
Calculated for 10−4 M oligonucleotide concentration.
Plots of TM−1 versus ln CT /4 resulted in an r-value of 0.97.
Table 2.
Free energies for loop formation
| Secondary structure of the loop a |
Source of structural model | ΔG°37 LOOP (kcal/mol) |
|---|---|---|
| C UAA G G AAG C |
NMR b | −1.78 c |
| CUAAG GAAGC |
Predicted | −1.06 d |
| C UAAG G AA GC |
Crystal structure of D. radiodurans | 3.53d 1.09 e |
Non-Watson–Crick pairs are in bold.
UA forms a single hydrogen bonded base pair in the NMR structure.
Measured ΔG°37LOOP as calculated from eq. 2
Value for ΔG°LOOP as predicted from updated nearest neighbor parameters that incorporate previous data (1, 2, 96) and assuming a single Watson–Crick UA pair.
Value for ΔG°LOOP was predicted assuming a 3 × 3 nucleotide internal loop (73) with the non-Watson–Crick UA pair treated as part of the loop.
On the basis of thermodynamics, the internal loop is predicted to be a 2 × 2 nucleotide internal loop closed on one side with a Watson–Crick UA pair. The free energy increment for forming the loop at 37 °C can be predicted by averaging and (1, 62) and adding the nearest neighbor parameter for . The value of −1.06 kcal/mol is similar to the experimental value of −1.78 kcal/mol (Table 2). The predicted for the loop structure in the crystal structures of LSU rRNA is 3.53 kcal/mol if considered as a single nucleotide bulge and a 1 × 2 nucleotide internal loop separated by a UA pair, or 1.09 kcal/mol if treated as a 3 × 3 nucleotide internal loop. These estimated values are 5.31 and 2.87 kcal/mol, respectively less favorable than that measured for the 2 × 2 loop with AA and GA pairs (Table 2).
DISCUSSION
Secondary or three-dimensional structures are known for only a fraction of known RNA sequences. An understanding of the sequence dependence of the energetics and flexibility of various structural elements would facilitate modeling of structures and contribute to determination of structure-function relationships. Internal loops are a common structural element in RNAs. This study shows that the solution structure of a phylogenetically conserved internal loop, 5'CUAAG/3' GAAGC, is close to that predicted by a nearest neighbor energetic model, but differs from that found in the crystal structures of ribosomal subunits (4–6). Evidently, tertiary and quaternary interactions with rRNA and protein are strong enough to remodel the three-dimensional structure of the loop suggesting induced fit binding (63).
The crystal and solution structures differ
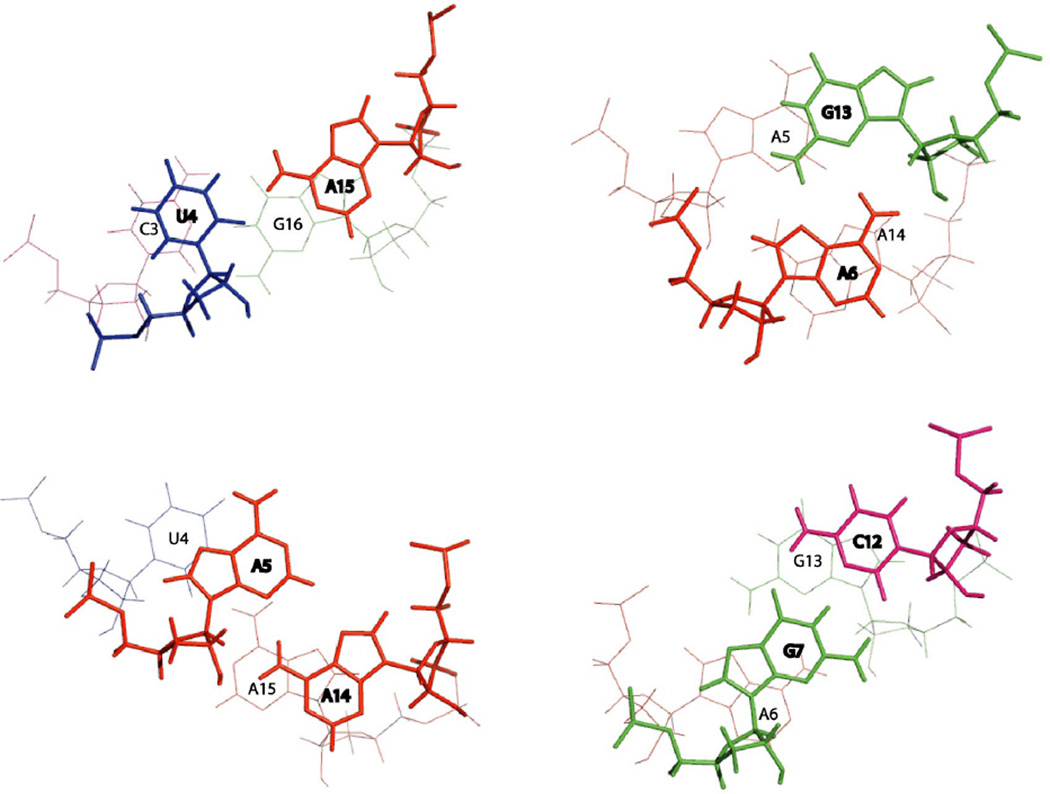
The solution structure of the internal loop differs from that in the crystal structures of large ribosomal subunits from D. radiodurans and E. coli but is close to the 2 × 2 nucleotide internal loop structure expected from a model used to approximate energetics (Figure 1). For example, in the D. radiodurans crystal structure, A1167 is flipped out of the helix. U1010 forms a reverse Hoogsteen pair with A1166 (Figure 1a). A1011 forms a cross-strand stack between A1012 and A1166. A1012 forms a sheared GA pair with G1165. The solution structure differs from the crystal structure in that A15, which corresponds to the bulged out A1167 in the crystal structure, forms a single hydrogen bonded pair with U4, while A5 and A14 form an AA sheared pair adjacent to the GA sheared pair. Cross-peaks such as A15H2-A5H1' and A15H2-G16H1' suggest that A15 is not flipped out (Figure 5). A6 forms a cross-strand stack with A14 and A14 makes an intrastrand stack on A15 (Figure 8). Cross-peaks such as A6H2 to A14H2, A6H2 to A14H1' and A14H2 to A6H1' suggest that A6 is stacking on A14. A14H2-A15H1', A14H8-A15H8, A14H2-A15H8 cross-peaks provide evidence for stacking of A14 on A15 (Figure 5).
Figure 8.
Stacking patterns of the residues in the loop generated from the average of the 20 lowest free energy structures using 3DNA (97). Adenines are colored red, cytosines are pink, guanines are green and uracils are blue. The base pair closer to the viewer is in bold.
Magnesium binding
No Mg2 + that might stabilize the internal loop structure is observed in the crystal structure of D. radiodurans (1NKW) but a Mg2 + is present between the phosphates of U999 and C1153 (equivalent to U4 and C12) in the E. coli crystal structure (2AWB). A Mg2+ ion is also observed at a similar position in the H. marismortui crystal structure (1JJ2). The distance between the U999 and C1153 phosphates is ∼ 6.8 Å in the crystal which is much less than the ∼ 18 Å in the NMR model. Addition of 10 mM MgCl2 to the NMR sample does not induce any significant change in the NMR spectra. Evidently, Mg2+ alone cannot induce a conformational switch to flip out A15.
Tertiary and quaternary interactions may stabilize the bulged A structure
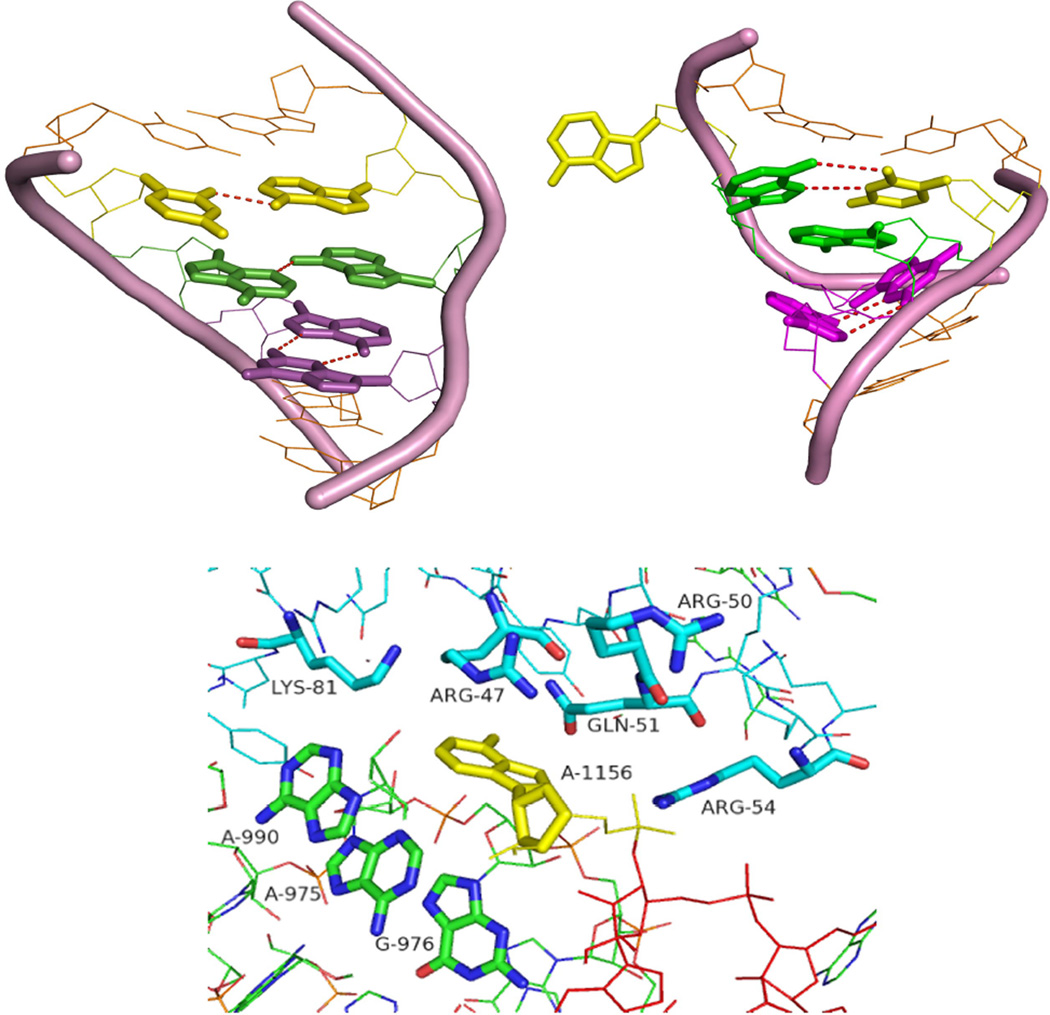
In D. radiodurans (), the bulged A1167 is close to the alpha carbons of Arg48, Arg51 and Asn52 of ribosomal protein L20 (residues within 6 Å radius) and to A986, G987 and A1001 of the rRNA (residues within 5 Å radius). The bulged G1260 in the H. marismortui internal loop is close to Lys158 and Gly156 from ribosomal protein L32E and to the A1073, G1074 and A1088 residues of the rRNA (residues within 5 Å). As shown in the lower panel of Figure 9, the E. coli internal loop () is close to Arg47, Arg50, Gln51and Arg54 of ribosomal protein L20 and Lys81 from ribosomal protein L22 (residues within 5 Å). It is also close to A975, A976 and A990 of the rRNA (residues within 5 Å). Thus different tertiary and quaternary interactions may stabilize the structures of these loops, rather than preorganized interactions within the loops themselves.
Figure 9.
A comparison between internal loop structures obtained by NMR (left upper panel) and crystallography of the D. radiodurans large ribosomal subunit (right upper panel) (4). Analogous base pairs are colored the same. The flanking pairs and the backbone of the loop are shown in orange. In the NMR structure, A15 and U4 (yellow) are paired in a cis Watson-Crick/sugar edge pair. In the crystal structure, A15 is bulged out and U4 forms a reverse Hoogsteen pair with A14. In the NMR structure, A5 and A14 (green) form a sheared pair, but A5 and A14 are stacked in the crystal structure. G13 and A6 (pink) form a sheared pair in both cases. Lower panel shows the tertiary contacts close to the bulged adenine (A1156) in the E. coli crystal structure (residues within 5 Å). The overall folding of the loop in E. coli is the same as for D. radiodurans (RMSD =0.67 Å)
Effects of non-Watson–Crick pairs on global structure
Tandem AA and GA pairs are commonly found in biologically functional RNA (64–66). One of the features of the isolated loop in solution is that the backbone is slightly narrower at two positions. The C1' to C1' distance in the typical Watson–Crick pairs in this structure is ∼ 10.7 Å. The distance between A5C1' and A14C1' is 9.4 Å and the distance between A6C1' and G13C1' is 9.5 Å because the backbone is drawn in due to formation of sheared pairs. In the crystals, the GA sheared pair has an average C1'- C1' distance of 9.7 Å. The NMR derived distance between A6H2 and A14H1' is 2.6 Å compared to 3.2–4.1 Å in a typical A-form helix so the minor groove is narrowed. Similar constriction or kinking of the backbone has been observed in previous structures involving purine purine sheared pairs (23, 55). The U4A15 pair in the NMR structure has a C1'- C1' distance of 10.9 Å whereas the reverse Hoogsteen pair in the crystals has an average C1'- C1' distance of 9.6 Å.
The switch in conformation may be facilitated by A5 dynamics
NMR spectra of this internal loop indicate structural dynamics at A5. The A5H2 peak is broad (∼ 21 Hz) and is weak in the HSQC spectrum suggesting dynamics (Figure S4). This is consistent with the ∼ 5 Hz coupling between A5H1' and A5H2' suggesting that the ribose ring is interchanging between C2' and C3' endo conformations. Dynamics at this residue are also consistent with the lack of A5 base stacking interactions with neighboring residues in the modeled structures (Figures 7, 8). A5 is only held by a single hydrogen bond in a sheared pair with A14 and A15 is only held by a single hydrogen bond to U4 (Figure 1b). Dynamics associated with A5 may provide a pathway for this internal loop to switch between two very different conformations. Rapid switching between syn and anti guanosines in GG pairs has been observed (67) and is thought to be facilitated by dynamics in sugar puckers (68). In the crystal structures, the equivalent of A5 forms a cross-strand stack with the equivalent of A14 (Figure 9). This stack allows the Hoogsteen edge of A14 to form two hydrogen bonds with U4 in a reverse Hoogsteen pair thus compensating for the hydrogen bonds lost between U4 with A15 and A5 with A14 and helping to stabilize the induced fit conformation.
The solution structure and energetics are similar to those predicted from simple models
While the solution structure of the 5'CUAAG/3'GAAGC loop differs from the crystal structures, it is very similar to that expected from models used to approximate the structures and energetics of 2 × 2 nucleotide internal loops (1, 69). In the energetic model, the free energy increment for a sequence non-symmetric loop, 5'PXYS/3'QWZT, is the average of that measured for the sequence symmetric loops, 5'PXWQ/3'QWXP and 5'TZYS/ 3'SYZT, when the non-Watson–Crick pairs, XW and ZY are sterically matched. Here PQ and TS are Watson–Crick pairs. NMR structures of the internal loops, CGAG (44), CAAG (29, 70), and UGAA (47) all have sheared purine purine pairs so that the same is expected for 5' UAAG/ 3' AAGC. The average of the measured free energy increments at 37 °C for 5'UAAA/ 3' AAAU (62, 71) and 5' CGAG/3' GAGC (62, 72) is 1.02 kcal/mol, which when added to the −2.08 kcal/mol free energy increment for 5'CU/3'GA predicts a free energy increment of −1.06 kcal/mol for the 5'CUAAG/3'GAAGC sequence. This is within experimental error of the measured value of −1.78 kcal/mol (Table 2). Evidently, there are no unexpected interactions between the sheared AA and GA pairs and the unusual UA pair has similar stability as a Watson–Crick pair in this context, or the two potential effects cancel.
Potential functional advantages of the difference between solution and crystal structures
The size symmetric loops, and , in D. radiodurans (4) have A1167 and U1392 flipped out, respectively, and similar structures are observed for the equivalent loops in E. coli (5). This base flip is not expected on the basis of the sequence dependence of internal loop stability (73) and is not observed in the solution structure of the internal loop. This internal loop is highly conserved, which suggests a need for it to facilitate a structural switch. The symmetric loop in D. radiodurans also has G631 flipped out. It is surprising that these loops that are expected to be structured are involved in base flipping. Presumably, an unstructured loop would undergo conformational changes more easily. Perhaps the structure of the isolated loop is important for initial folding of the RNA and/or initial recognition by protein while the induced structure is necessary for specific binding by protein or RNA. Thus induced fit binding is likely important for other internal loops predicted to be preorganized.
Induced fit binding (63) is common in RNA-protein complexes, but the reasons for this universality are not known (74–78). The differences between the NMR and crystal structures of the 5'CUAAG/3'GAAGC loop suggest some possible advantages for this induced fit binding. One potential reason for a bulged nucleotide to initially be enclosed in a structured loop is that it may be protected from chemical cleavage. It is known that backbone cleavage is enhanced at bulged nucleotides due to increased in-line orientation of the 2'OH with the phosphate (79, 80). A structured loop incorporating a nucleotide to be bulged would also provide increased stability for the initial folding of the RNA. The nucleotide can later be bulged in order to provide RNA/RNA tertiary interactions or a binding site for protein. The L20 protein is one of the first to bind to the rRNA and is among the six proteins that are required for formation of the first key intermediate in ribosomal assembly (13–15). The binding of protein could allow temporal ordering of a long range conformational change required for assembly of a large complex such as the ribosome. Internal loops with flipped bases can lead to considerable misalignment of helix axes (81).
The initial structure of the loop may also provide a kinetically important intermediate that allows the protein to rapidly find the location requiring base flipping, i.e. the initial structure may serve as a “book mark” identifying the site to be opened. Interestingly, Restrictocin–a sarcin like toxin recognizes a bulged-G type S-turn in the sarcin/ricin tetraloop of rat 23S rRNA and flips a G on the 5' side of the cleavage site facilitating in-line orientation of the 2'OH of G with the P-O5 of the 3' A (82, 83).
The nearest neighbor model (2, 3) predicts a free energy increment of 3.53 kcal/mol (Table 2) for the bulged internal loop structure found in crystals of the LSU of D. radiodurans (4) and E. coli (5). The predicted free energy is 5.3 kcal/mol less favorable than measured for the oligonucleotide mimic (Table 2). This predicted free energy difference between the crystal and solution structures suggests that tertiary or quaternary contacts with RNA or protein provide the necessary free energy difference to stabilize the crystal structure. If this was a case of lock and key binding (19), the structures in crystal and solution would be identical and fewer tertiary and/or quaternary contacts would be needed to achieve the required association constant. The increased number of favorable contacts with the protein and RNA required for induced fit binding may provide increased specificity.
Single base bulges are commonly occurring secondary structural elements (84) that play vital roles in RNA folding and recognition (85, 86). Base flipping is also commonly induced by nucleic acid modifying enzymes to gain access to the site of modification (87–89). For example, tRNA pseudouridine synthetase, TruB, flips target base U55 by disrupting a GU base pair (90). Modeling studies done on 23S rRNA methyltranseferase RrmJ suggest that the substrate nucleotide U2552 has to bulge out to be methylated at the 2'-O position (91). Initiation factor IF1 binds 16S rRNA and induces flipping of A1492 and A1493 from helix 44 (92). Evidently, many proteins are able to provide sufficient free energy to compensate for the cost of flipping a base.
Molecular dynamics and thermodynamics calculations on an interaction between the N terminal RNP domain of U1A mutant and stem loop 2 of U1 snRNA suggest that stacking interactions between a protein residue and RNA bases could contribute as much as ∼ 4.4 kcal/mol to binding (93). This suggests that a single protein contact like stacking could compensate for base flipping and subsequent base pairing rearrangement. Many enzymes insert a protein residue into the cavity created by a flipped out base in order to stabilize the structure (90, 94). It is also likely that the RNA contacts from other parts of the ribosome play a role in this induced fit recognition.
With increasing evidence for base flipping as a functional mechanism used by RNA, prediction of sites and loops where base flipping can occur will become important. Identification of sequences with base flipping propensities may help drug design by identifying sites where small molecules could compete with tertiary and quaternary binding by targeting the stable structure formed in the absence of these interactions (95).
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr. Brent Znosko for his help during setup of NMR experiments. We thank Mr. Rahul Tyagi for his help in writing the C++ program used to convert Sparky output to Discover Input.
Glossary
- CT
total concentration of all strands of oligonucleotides in solution
- LSU
large subunit
- N
any nucleotide, i.e. A, C, G or U
- Y
any pyrimidine i.e. U or C
- R
any purine, i.e. G or A
- TM
melting temperature in kelvin
- Tm
melting temperature in degrees Celsius
Footnotes
This work was supported by NIH grant GM22939 (D. H. T.).
Protein Data Bank entry: 2H49
SUPPORTING INFORMATION AVAILABLE
A table of NMR restraints, a table with the summary of NOE distance restraints for each residue, a table listing chemical shift assignments and figures of 31P HETCOR, SNOESY, HSQC, DQCOSY spectra are available. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Nat. Acad. Sci. U.S.A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xia T, SantaLucia J, Jr., Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 3.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 4.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic Eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 5.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the Bacterial Ribosome at 3.5 A Resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 6.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 angstrom resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 7.Clemons WM, Jr., May JL, Wimberly BT, McCutcheon JP, Capel MS, Ramakrishnan V. Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 8.Wimberly BT, Brodersen DE, Clemons WM, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 9.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal Structure of the Ribosome at 5.5 A Resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 10.Guillier M, Allemand F, Raibaud S, Dardel F, Springer M, Chiaruttini C. Translational feedback regulation of the gene for L35 in Escherichia coli requires binding of ribosomal protein L20 to two sites in its leader mRNA: a possible case of ribosomal RNA-messenger RNA molecular mimicry. Rna. 2002;8:878–889. doi: 10.1017/s1355838202029084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klosterman PS, Tamura M, Holbrook SR, Brenner SE. SCOR: a Structural Classification of RNA database. Nucleic Acids Res. 2002;30:392–394. doi: 10.1093/nar/30.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura M, Holbrook SR. Sequence and structural conservation in RNA ribose zippers. J. Mol. Biol. 2002;320:455–474. doi: 10.1016/s0022-2836(02)00515-6. [DOI] [PubMed] [Google Scholar]
- 13.Herold M, Nierhaus KH. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J. Biol. Chem. 1987;262:8826–8833. [PubMed] [Google Scholar]
- 14.Rohl R, Nierhaus KH. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc. Natl. Acad. Sci. U. S. A. 1982;79:729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowotny V, Nierhaus KH. Protein L20 from the large subunit of Escherichia coli ribosomes is an assembly protein. J. Mol. Biol. 1980;137:391–399. doi: 10.1016/0022-2836(80)90164-3. [DOI] [PubMed] [Google Scholar]
- 16.Raibaud S, Vachette P, Guillier M, Allemand F, Chiaruttini C, Dardel F. How bacterial ribosomal protein L20 assembles with 23 S ribosomal RNA and its own messenger RNA. J. Biol. Chem. 2003;278:36522–36530. doi: 10.1074/jbc.M304717200. [DOI] [PubMed] [Google Scholar]
- 17.Raibaud S, Lebars I, Guillier M, Chiaruttini C, Bontems F, Rak A, Garber M, Allemand F, Springer M, Dardel F. NMR structure of bacterial ribosomal protein L20: implications for ribosome assembly and translational control. J. Mol. Biol. 2002;323:143–151. doi: 10.1016/s0022-2836(02)00921-x. [DOI] [PubMed] [Google Scholar]
- 18.Koshland DE. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc. Nat. Acad. Sci. U.S.A. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer E. Einfluss der Configuration auf die Wirkung der Enzyme. Ber. Dtsch. Chem. Ges. 1894;27:2984–2993. [Google Scholar]
- 20.Wincott F, DiRenzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Usman N, Ogilvie KK, Jiang MY, Cedergren RJ. Automated chemical synthesis of long oligoribonucleotides using 2'-O-silylated ribonucleoside 3'-O-phosphoramidites on a controlled-pore glass support: Synthesis of a 43-nucleotide sequence similar to the 3'-half molecule of an Escherichia coli formylmethionine tRNA. J. Amer.Chem. Soc. 1987;109:7845–7854. [Google Scholar]
- 22.Stawinski J, Stromberg R, Thelin M, Westman E. Evaluation of the use of the tert-butyldimethylsilyl group for 2'-protection in RNA: Synthesis via the H-phosphonate approach. Nucleosides Nucleotides. 1988;7:779–782. [Google Scholar]
- 23.Chen G, Znosko BM, Kennedy SD, Krugh TR, Turner DH. Solution structure of an RNA internal loop with three consecutive sheared GA pairs. Biochemistry. 2005;44:2845–2856. doi: 10.1021/bi048079y. [DOI] [PubMed] [Google Scholar]
- 24.Smallcombe SH. Solvent suppression with symmetrically-shifted pulses. J. Amer.Chem. Soc. 1993;115:4776–4785. [Google Scholar]
- 25.Johnson BA, Blevins RA. Nmr View - A computer-program for the visualization and analysis of Nmr data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 26.Goddard TD, Kneller DG. Sparky - NMR assignment and integration software. 2004 [Google Scholar]
- 27.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A 2nd generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Amer. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 28.Ding HQ, Karasawa N, Goddard WA., III Atomic level simulations on a million particles: The cell multipole method for Coulomb and London nonbond interactions. J. Chem. Phys. 1992;97:4309–4315. [Google Scholar]
- 29.Znosko BM, Burkard ME, Schroeder SJ, Krugh TR, Turner DH. Sheared Aanti.Aanti base pairs in a destabilizing 2 × 2 internal loop: the NMR structure of 5'(rGGCAAGCCU)2. Biochemistry. 2002;41:14969–14977. doi: 10.1021/bi020326f. [DOI] [PubMed] [Google Scholar]
- 30.Burkard ME, Turner DH. NMR structures of r(GCAGGCGUGC)2 and determinants of stability for single guanosine-guanosine base pairs. Biochemistry. 2000;39:11748–11762. doi: 10.1021/bi000720i. [DOI] [PubMed] [Google Scholar]
- 31.Borer PN. Optical properties of nucleic acids, absorption and circular dichroism spectra. In: Fasman GD, editor. Handbook of Biochemistry and Molecular Biology: Nucleic Acids. 3rd Ed. Cleveland, OH: CRC Press; 1975. pp. 589–595. [Google Scholar]
- 32.Richards EG. Use of tables in calculation of absorption, optical rotatory dispersion and circular dichroism of polyribonucleotides. In: Fasman GD, editor. Handbook of Biochemistry and Molecular Biology: Nucleic Acids. 3rd Ed. Cleveland, OH: CRC Press; 1975. pp. 596–603. [Google Scholar]
- 33.Peritz AE, Kierzek R, Sugimoto N, Turner DH. Thermodynamic study of internal loops in oligoribonucleotides: symmetric loops are more stable than asymmetric loops. Biochemistry. 1991;30:6428–3436. doi: 10.1021/bi00240a013. [DOI] [PubMed] [Google Scholar]
- 34.Petersheim M, Turner DH. Base-stacking and base-pairing contributions to helix stability: Thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983;22:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 35.McDowell JA, Turner DH. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: Solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 36.Borer PN, Dengler B, Tinoco I, Jr., Uhlenbeck OC. Stability of ribonucleic acid double-stranded helices. J. Mol. Biol. 1974;86:843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- 37.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varani G, Aboulela F, Allain FHT. NMR investigation of RNA structure. Prog. Nucl. Magn. Reson. Spectrosc. 1996;29:51–127. [Google Scholar]
- 39.Varani G, Tinoco I. RNA structure and NMR spectroscopy. Q. Rev. Biophys. 1991;24:479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- 40.Varani G, Cheong C, Tinoco I., Jr. Structure of an unusually stable RNA hairpin. Biochemistry. 1991;30:3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- 41.Gorenstein D. 31P NMR, Principles and Applications. New York: Academic Press; 1984. [Google Scholar]
- 42.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 43.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 44.SantaLucia J, Jr., Turner DH. Structure of (rGGCGAGCC)2 in solution from NMR and restrained molecular dynamics. Biochemistry. 1993;32:12612–12623. doi: 10.1021/bi00210a009. [DOI] [PubMed] [Google Scholar]
- 45.Wu M, SantaLucia J, Jr., Turner DH. Solution structure of (rGGCAGGCC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry. 1997;36:4449–4460. doi: 10.1021/bi9625915. [DOI] [PubMed] [Google Scholar]
- 46.Wu M, Turner DH. Solution structure of (rGCGGACGC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry. 1996;35:9677–9689. doi: 10.1021/bi960133q. [DOI] [PubMed] [Google Scholar]
- 47.Heus HA, Wijmenga SS, Hoppe H, Hilbers CW. The detailed structure of tandem G·A mismatched base-pair motifs in RNA duplexes is context dependent. J. Mol. Biol. 1997;271:147–158. doi: 10.1006/jmbi.1997.1158. [DOI] [PubMed] [Google Scholar]
- 48.Burkard ME, Kierzek R, Turner DH. Thermodynamics of unpaired terminal nucleotides on short RNA helixes correlates with stacking at helix termini in larger RNAs. J. Mol. Biol. 1999;290:967–982. doi: 10.1006/jmbi.1999.2906. [DOI] [PubMed] [Google Scholar]
- 49.Freier SM, Sugimoto N, Sinclair A, Alkema D, Neilson T, Kierzek R, Caruthers MH, Turner DH. Stability of XGCGCp, GCGCYp, and XGCGCYp helixes: an empirical estimate of the energetics of hydrogen bonds in nucleic acids. Biochemistry. 1986;25:3214–3219. doi: 10.1021/bi00359a020. [DOI] [PubMed] [Google Scholar]
- 50.Sponer J, Burcl R, Hobza P. Interactions between amino groups in DNA. An ab initio study and a comparison with empirical potentials. J. Biomol. Struct. Dyn. 1994;11:1357–1376. doi: 10.1080/07391102.1994.10508073. [DOI] [PubMed] [Google Scholar]
- 51.Sponer J, Mokdad A, Sponer JE, Spackova N, Leszczynski J, Leontis NB. Unique tertiary and neighbor interactions determine conservation patterns of cis Watson-Crick A/G base-pairs. J. Mol. Biol. 2003;330:967–978. doi: 10.1016/s0022-2836(03)00667-3. [DOI] [PubMed] [Google Scholar]
- 52.Conn GL, Draper DE, Lattman EE, Gittis AG. Crystal structure of a conserved ribosomal protein-RNA complex. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- 53.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 54.Klosterman PS, Shah SA, Steitz TA. Crystal structures of two plasmid copy control related RNA duplexes: An 18 base pair duplex at 1.20 A resolution and a 19 base pair duplex at 1.55 A resolution. Biochemistry. 1999;38:14784–14792. doi: 10.1021/bi9912793. [DOI] [PubMed] [Google Scholar]
- 55.Znosko BM, Kennedy SD, Wille PC, Krugh TR, Turner DH. Structural features and thermodynamics of the J4/5 loop from the Candida albicans and Candida dubliniensis group I introns. Biochemistry. 2004;43:15822–15837. doi: 10.1021/bi049256y. [DOI] [PubMed] [Google Scholar]
- 56.Flinders J, Dieckmann T. A pH controlled conformational switch in the cleavage site of the VS ribozyme substrate RNA. J. Mol. Biol. 2001;308:665–679. doi: 10.1006/jmbi.2001.4627. [DOI] [PubMed] [Google Scholar]
- 57.Hoffmann B, Mitchell GT, Gendron P, Major F, Andersen AA, Collins RA, Legault P. NMR structure of the active conformation of the Varkud satellite ribozyme cleavage site. Proc. Nat. Acad. Sci. U.S.A. 2003;100:7003–7008. doi: 10.1073/pnas.0832440100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jucker FM, Heus HA, Yip PF, Moors EHM, Pardi A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 59.Michiels PJA, Schouten CHJ, Hilbers CW, Heus HA. Structure of the ribozyme substrate hairpin of Neurospora VS RNA: A close look at the cleavage site. Rna-a Publication of the Rna Society. 2000;6:1821–1832. doi: 10.1017/s1355838200001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Case DA. Calibration of ring-current effects in proteins and nucleic acids. J. Biomol. NMR. 1995;6:341–346. doi: 10.1007/BF00197633. [DOI] [PubMed] [Google Scholar]
- 61.Nowakowski J, Miller JL, Kollman PA, Tinoco I. Time evolution of NMR proton chemical shifts of an RNA hairpin during a molecular dynamics simulation. J. Amer. Chem. Soc. 1996;118:12812–12820. [Google Scholar]
- 62.Schroeder SJ, Turner DH. Thermodynamic stabilities of internal loops with GU closing pairs in RNA. Biochemistry. 2001;40:11509–11517. doi: 10.1021/bi010489o. [DOI] [PubMed] [Google Scholar]
- 63.Koshland DE, Jr., Neet KE. The catalytic and regulatory properties of enzymes. Annu. Rev. Biochem. 1968;37:359–410. doi: 10.1146/annurev.bi.37.070168.002043. [DOI] [PubMed] [Google Scholar]
- 64.Leontis NB, Westhof E. Conserved geometrical base-pairing patterns in RNA. Q. Rev. Biophys. 1998;31:399–455. doi: 10.1017/s0033583599003479. [DOI] [PubMed] [Google Scholar]
- 65.Nagaswamy U, Larios-Sanz M, Hury J, Collins S, Zhang ZD, Zhao Q, Fox GE. NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res. 2002;30:395–397. doi: 10.1093/nar/30.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gautheret DF, Konings D, Gutell RR. A major family of motifs involving G·A mismatches in ribosomal RNA. J. Mol. Biol. 1994;242:1–8. doi: 10.1006/jmbi.1994.1552. [DOI] [PubMed] [Google Scholar]
- 67.Burkard ME, Turner DH. NMR Structures of r(GCAGGCGUGC)2 and Determinants of Stability for Single Guanosine-Guanosine Base Pairs. Biochemistry. 2000;39:11748–11762. doi: 10.1021/bi000720i. [DOI] [PubMed] [Google Scholar]
- 68.Mathews DH, Case DA. Nudged Elastic Band Calculation of Minimal Energy Paths for the Conformational Change of a GG Non-canonical Pair. J. Mol. Biol. 2006;357:1683–1693. doi: 10.1016/j.jmb.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 69.Xia T, McDowell JA, Turner DH. Thermodynamics of nonsymmetric tandem mismatches adjacent to G·C base pairs in RNA. Biochemistry. 1997;36:12486–12497. doi: 10.1021/bi971069v. [DOI] [PubMed] [Google Scholar]
- 70.Znosko BM, Burkard ME, Krugh TR, Turner DH. Molecular recognition in purine-rich internal loops: thermodynamic, structural, and dynamic consequences of purine for adenine substitutions in 5'(rGGCAAGCCU)2. Biochemistry. 2002;41:14978–14987. doi: 10.1021/bi0203278. [DOI] [PubMed] [Google Scholar]
- 71.Wu M, McDowell JA, Turner DH. A periodic table of symmetric tandem mismatches in RNA. Biochemistry. 1995;34:3204–3211. doi: 10.1021/bi00010a009. [DOI] [PubMed] [Google Scholar]
- 72.Walter AE, Wu M, Turner DH. The stability and structure of tandem GA mismatches in RNA depend on closing base pairs. Biochemistry. 1994;33:11349–11354. doi: 10.1021/bi00203a033. [DOI] [PubMed] [Google Scholar]
- 73.Chen G, Znosko BM, Jiao X, Turner DH. Factors affecting thermodynamic stabilities of RNA 3 × 3 internal loops. Biochemistry. 2004;43:12865–12876. doi: 10.1021/bi049168d. [DOI] [PubMed] [Google Scholar]
- 74.Williamson JR. Induced fit in RNA-protein recognition. Nat. Struc. Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 75.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 76.Lee BM, Xu J, Clarkson BK, Martinez-Yamout MA, Dyson HJ, Case DA, Gottesfeld JM, Wright PE. Induced Fit and “Lock and Key” Recognition of 5 S RNA by Zinc Fingers of Transcription Factor IIIA. J. Mol. Biol. 2006;357:275–291. doi: 10.1016/j.jmb.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 77.Blanchard SC, Puglisi JD. Solution structure of the A loop of 23S ribosomal RNA. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3720–3725. doi: 10.1073/pnas.051608498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshizawa S, Fourmy D, Puglisi JD. Recognition of the codon-anticodon helix by ribosomal RNA. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- 79.Nakano S, Uotani Y, Uenishi K, Fujii M, Sugimoto N. Site-selective RNA cleavage by DNA bearing a base pair-mimic nucleoside. J. Am. Chem. Soc. 2005;127:518–519. doi: 10.1021/ja045445s. [DOI] [PubMed] [Google Scholar]
- 80.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. Rna. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luebke KJ, Tinoco I. Sequence Effects on RNA Bulge-Induced Helix Bending and a Conserved Five-Nucleotide Bulge from the Group I Introns. Biochemistry. 1996;35:11677–11684. doi: 10.1021/bi960914r. [DOI] [PubMed] [Google Scholar]
- 82.Correll CC, Yang X, Gerczei T, Beneken J, Plantinga MJ. RNA recognition and base flipping by the toxin sarcin. J. Synchrotron. Radiat. 2004;11:93–96. doi: 10.1107/s0909049503023896. [DOI] [PubMed] [Google Scholar]
- 83.Yang X, Gerczei T, Glover LT, Correll CC. Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping. Nat. Struc. Biol. 2001;8:968–973. doi: 10.1038/nsb1101-968. [DOI] [PubMed] [Google Scholar]
- 84.Wyatt JR, Tinoco I., Jr. In: RNA structural elements and RNA function; The RNA World. Gesteland RF, Atkins JF, editors. 1993. pp. 465–496. [Google Scholar]
- 85.Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO Journal. 1990;9:4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hermann T, Patel DJ. RNA bulges as architectural and recognition motifs. Structure Folding Des. 2000;8:R47–R54. doi: 10.1016/s0969-2126(00)00110-6. [DOI] [PubMed] [Google Scholar]
- 87.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O'Gara M, Horton JR, Roberts RJ, Cheng X. Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nat. Struc. Biol. 1998;5:872–877. doi: 10.1038/2312. [DOI] [PubMed] [Google Scholar]
- 89.Roberts RJ, Cheng X. Base flipping. Annu. Rev. Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 90.Hoang C, Ferre-D'Amare AR. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 91.Hager J, Staker BL, Jakob U. Substrate binding analysis of the 23S rRNA methyltransferase RrmJ. J. Bacteriol. 2004;186:6634–6642. doi: 10.1128/JB.186.19.6634-6642.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carter AP, Clemons WM, Jr., Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 93.Blakaj DM, McConnell KJ, Beveridge DL, Baranger AM. Molecular dynamics and thermodynamics of protein-RNA interactions: mutation of a conserved aromatic residue modifies stacking interactions and structural adaptation in the U1A–stem loop 2 RNA complex. J. Am. Chem. Soc. 2001;123:2548–2551. doi: 10.1021/ja005538j. [DOI] [PubMed] [Google Scholar]
- 94.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 95.Tok JBH, Bi L, Saenz M. Specific recognition of napthyridine-based ligands toward guanine-containing bulges in RNA duplexes and RNA-DNA heteroduplexes. Bioorg. Med. Chem. Lett. 2005;15:827. doi: 10.1016/j.bmcl.2004.10.059. [DOI] [PubMed] [Google Scholar]
- 96.Schroeder SJ, Burkard ME, Turner DH. The energetics of small internal loops in RNA. Biopolymers. 1999;52:157–167. doi: 10.1002/1097-0282(1999)52:4<157::AID-BIP1001>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 97.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.