Abstract
Despite extensive revisions over recent decades, the taxonomy of benthic octopuses (Family Octopodidae) remains in a considerable flux. Among groups of unresolved status is a species complex of morphologically similar shallow-water octopods from subtropical Australasia, including: Allopatric populations of Octopus tetricus on the eastern and western coasts of Australia, of which the Western Australian form is speculated to be a distinct or sub-species; and Octopus gibbsi from New Zealand, a proposed synonym of Australian forms. This study employed a combination of molecular and morphological techniques to resolve the taxonomic status of the ‘tetricus complex’. Phylogenetic analyses (based on five mitochondrial genes: 12S rRNA, 16S rRNA, COI, COIII and Cytb) and Generalised Mixed Yule Coalescent (GMYC) analysis (based on COI, COIII and Cytb) distinguished eastern and Western Australian O. tetricus as distinct species, while O. gibbsi was found to be synonymous with the east Australian form (BS = >97, PP = 1; GMYC p = 0.01). Discrete morphological differences in mature male octopuses (based on sixteen morphological traits) provided further evidence of cryptic speciation between east (including New Zealand) and west coast populations; although females proved less useful in morphological distinction among members of the tetricus complex. In addition, phylogenetic analyses suggested populations of octopuses currently treated under the name Octopus vulgaris are paraphyletic; providing evidence of cryptic speciation among global populations of O. vulgaris, the most commercially valuable octopus species worldwide.
Introduction
Taxonomy within the benthic octopuses (Family Octopodidae) continues to be a source of confusion and controversy and despite extensive revisions in recent decades, the true taxonomy of this family remains unresolved [1], [2], [3]. The most widely studied and economically significant group of cephalopods worldwide is the ‘Octopus vulgaris group’ of octopods. The type species of this group is the common octopus, Octopus vulgaris Cuvier, 1797. Octopus vulgaris alone accounts for >50% of the world's total octopod fisheries catch, exceeding 380,000 tonnes and has an international export value of >US$1 billion [4]. The Octopus vulgaris species group is comprised of tropical, sub-tropical and temperate species from the Americas, Europe, Africa, Asia and Australasia. Members of this group are large muscular octopuses that display similar morphological and behavioural traits as well as occupying similar ecological niches.
Within the subtropical waters of Australasia there is a group of morphologically, behaviourally and functionally similar Octopus species, closely related to Octopus vulgaris [3], [5]. These species, currently treated under the names Octopus tetricus on the east and west coasts of Australia and O. gibbsi in New Zealand, have been suggested to be a species complex; the taxonomy of which remains unresolved [3]. We treat these taxa collectively herein as the ‘tetricus complex’, after the first formally described species within this group, Octopus tetricus Gould, 1852; the common Sydney octopus.
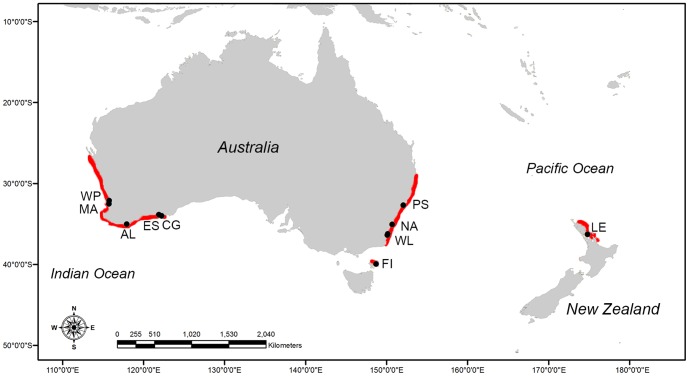
The tetricus complex comprises three geographically distinct member taxa (Figure 1). Octopus tetricus was originally described from New South Wales and occurs along the east Australian coastline, ranging from Eden in southern New South Wales to Moreton Bay in southern Queensland [6]. Octopus tetricus comprises a major portion of the small-scale commercial octopod fisheries landings in New South Wales [7], and is also often caught as by-catch in prawn and finfish trawls [8]. Recently O. tetricus has been reported in Tasmania, significantly south of its previous known range [9] although this has not been verified by molecular data.
Figure 1. Known distributions (shown in red) and sample locations (shown in black) for Octopus tetricus, (east Australia), O. cf. tetricus (Western Australia) and O. gibbsi (New Zealand).
Location acronyms: WP = Woodman's Point, MA = Mandurah, AL = Albany, ES = Esperance, CG = Cape Le Grand, FI = Flinders Island, Tasmania, WL = Wallaga Lake, NA = Narooma, PS = Port Stephens, LE = Leigh, New Zealand.
A second taxon, known as the common Perth octopus, occurs in Western Australia from Esperance to Shark Bay. This population has extensively been treated under the name Octopus tetricus [10], [11], [12], [13], [14] due to close similarities in morphological, behavioural and functional attributes between east and west coast forms. More recently however, the common Perth octopus has been treated under the name O. cf. tetricus; a reflection of the proposal that disjunct east and west populations may be sufficiently isolated and therefore represent sub- or distinct species [2], [15]. Joll [11] estimated that 250 tonnes of O. cf. tetricus were harvested annually from Western Australian waters, primarily as by-catch from lobster fisheries. Octopus cf. tetricus often preys upon lobsters caught in craypots, and is considered to negatively impact this economically important fisheries resource.
A third nominal species, Octopus gibbsi O'Shea, 1999, was coined to describe a benthic octopus of unknown relation found within the shallow coastal waters off northern New Zealand. Prior to description by O'Shea [16], O. gibbsi had been treated under the name O. tetricus [17], and more recently the validity of O. gibbsi as a distinct species has been questioned [2]. Examination of museum specimens showed strong morphological similarities between O. gibbsi and Australian forms, leading to the proposal that O. gibbsi is synonymous with O. tetricus [2].
A phylogenetic analysis of the sub-family Octopodinae using amino acid sequences from two mitochondrial (cytochrome oxidase subunit III and cytochrome b) and a single nuclear genetic marker (elongation factor-1α) assigned Octopus tetricus and O. cf. tetricus as sister taxa [3]. Analyses of genetic distance (Kimura 2 Parameter) between these two representatives showed 2.0% and 2.6% sequence divergence within each mitochondrial gene fragment respectively. However, only single representatives from both Western Australia and New South Wales were sequenced in this study. Consequently, analyses of Guzik et al., [3] were insufficient to detect the occurrence of speciation between disjunct east and west populations, and no traditional morphological based studies comparing the two populations have been conducted. Furthermore, no molecular work to date has investigated the phylogenetic status of O. gibbsi, thus its taxonomy remains unresolved.
This study aims to resolve the taxonomic status and phylogenetic relationships of the Octopus tetricus species complex, using a combination of molecular and morphological techniques. Due to the emerging fisheries value and the lack of species-level resolution within the tetricus complex, taxonomic resolution within this group will aid in the management of these marine resources.
Materials and Methods
All tissue samples and DNA extracts were loaned from existing museum/university collections. Thus, no animals were harmed or killed in conducting this study. All appropriate permissions were obtained from the relevant institutions prior to accessing their collections.
Molecular analyses
Sampling
Tissue samples of the ingroup (Octopus tetricus [n = 13], O. cf. tetricus [n = 17] and O. gibbsi [n = 4]) were sourced from collections at Museum Victoria, or provided by researchers associated with The University of Adelaide, the Western Australian Fisheries and Marine Research Laboratories Department and the University of Tasmania (Table S1 in File S1). Tissue samples (as arm or mantle tissue ∼1 cm in length) were taken from individuals collected from the Australian mainland, Flinders Island (Tasmania) and New Zealand (Figure 1). All tissue samples were stored at −20°C in 70–90% ethanol until processing.
Sequencing
DNA was extracted from mantle or tentacle tissue using the ‘High Salt Method’ [18]. Partial sequences of five mitochondrial genes were targeted; including12S ribosomal RNA (12S) [19], 16S ribosomal RNA (16S), and cytochrome oxidase subunits one (COI) [20], three (COIII) and cytochrome b (Cytb) [3]. 25 µL reactions comprised 0.1 µL Taq (Onetaq, New England Biolabs), 2.5 µL 10 x buffer (Paq5000™), 2 µL dNTP mix (10 µM, Bioline), 0.5 µL forward primer (10 µM), 0.5 µL reverse primer (10 µM), 17.4 µL ddH2O and 2 µL DNA (diluted to between 1–5 ng/µL). Reaction conditions are detailed elsewhere [21]. PCR products were sequenced by Macrogen Inc, Seoul, Korea. Genetic sequences generated in this study are accessible from GenBank under accession numbers KJ605215-KJ605347.
Octopus mimus and O. oculifer were selected as outgroup taxa on the basis that they are morphologically very similar to, and the closest known available relatives of the ingroup [2], [5], [22]. Sequences of the outgroup and additional sequences of ingroup taxa from previously published work were downloaded from GenBank (Table S2 in File S1). Multiple sequence alignments were performed using Geneious Muscle Alignment feature using the ClustalW default settings [23].
Phylogenetic analyses
jModelTest v0.1.1 [24] was used to carry out statistical selection of best-fit models of nucleotide substitution on the concatenated alignments and also for the COI alignment alone. The appropriate model was selected on the basis of ‘goodness of fit measure’ via the Akaike Information Criterion (AIC) [25].
Maximum likelihood (ML) topologies were constructed using PhyML v3.1 [26]. Full heuristic searches were undertaken and model parameter values were treated as unknown and were estimated. Strength of support for internal nodes of ML construction was measured using 1000 bootstrap replicates. Bayesian marginal posterior probabilities were calculated using MrBayes v3.2 [27]. Model parameter values were treated as unknown and were estimated. Random starting trees were used and the analysis was run for 15 million generations, sampling the Markov chain every 1000 generations. The program Tracer v1.3 [28] was used to ensure Markov chains had reached stationarity, and to determine the correct ‘burn-in’ for the analysis (the number of additional generations that must be discarded before stationarity is reached).
Genetic distance
Molecular Evolutionary Genetic Analysis (MEGA) v5.2 [29] was used to calculate genetic distances for populations of Octopus tetricus, O. gibbsi and O. cf. tetricus using the Tamura-Nei model [30]. Genetic distance was calculated using MEGA default settings (with the exceptions of the model and ‘pairwise deletion of missing data’ option). Mean values ± SE of interspecific and intraspecific variations in number of mutations per site were calculated for the barcoding mitochondrial gene COI to allow comparison with published literature.
Timing of divergence
Divergence time between clades were calculated based on an estimated rate of evolution of cephalopods; 3.81 substitutions per site per billion years (with 95% highest posterior density around this mean of 2.43–5.24; [31]) within a generalised molecular clock.
Coalescent delimitation
Potential species delimitation among Octopus tetricus, O. gibbsi and O. cf. tetricus was investigated using a Generalised Mixed Yule Coalescent (GMYC) model [32] applied to the molecular/phylogenetic data. Partitioned sequence data from the mitochondrial genes COI, COIII and Cytb were prepared into XML files using the software program BEAUti v1.7.5 [33]. 12S and 16S regions were excluded from the analysis due to low comparable sample representation (see Table S1 in File S1). A coalescent prior and relaxed molecular clock [34] were set as parameters before Bayesian analysis was performed using BEAST v1.7.5 [33]. Each analysis was performed independently twice and log/tree files were combined using LogCombiner v1.7.5 [33]. The data was then analysed via a single threshold model [35] in the software package Splits [36] available in R v3.0.1 [37], whereby clades with posterior probability values greater than 0.9 were acknowledged.
Morphological analyses
Morphological data was obtained from preserved whole specimens sourced from Museum Victoria, Australian Museum (Sydney) and the University of Tasmania. Samples were collected from south west (n = 15) and south east (n = 32) of the Australian mainland (between the years 1980–2007) as well as Flinders Island, Tasmania (n = 11; 2011) (Table S3 in File S1). All specimens had been initially fixed in 10% formalin and transferred to 70–90% ethanol for preservation. Morphological data for O. gibbsi (n = 6) was sourced from the published work of O' Shea [16].
Specimens were sexed based on three factors which allowed confident classification: 1) presence of terminal organ in males, 2) presence of hectocotylised arm in males and 3) number of genital glands present within the mantle (1 = male, 2 = female) [38]. Maturity in males was determined on the basis of the presence or absence of enlarged suckers (for mature and immature specimens, respectively) [39]. Maturity in females was determined by the state of egg development [40]. All specimens were weighed using digital scales to the nearest 0.1 gram after being removed from ethanol and patted dry with absorbent tissue.
Standard morphological characters were measured following Norman and Sweeney [41] (Table 1). Dorsal mantle length (MLd), mantle width (MW), head width (HW), arm width (AW), and the greatest non-enlarged sucker diameter (SDn) were recorded using digital callipers to the nearest 0.1 mm. In males, the greatest enlarged sucker diameter (SDe), the length of hectocotylised arm components (i.e. ligula [LL] and calamus [CL]) and terminal organ length (TOL); following dissection of the mantle, were also measured using digital callipers to the nearest 0.1 mm. For all specimens, third right (ALR3) and third left (ALL3) arm lengths were measured from arm tip to the beak opening using non-stretch string to the nearest 1 mm. The numbers of suckers occurring on the third right (SCR3) and third left (SCL3) arms were counted with the aid of a dissecting microscope. In cases where damage to an arm was perceived to inhibit growth, suckers appeared damaged, or arm regeneration was evident, arm length and sucker counts were not recorded. Where sucker and arm damage was minor, and sucker scars or remnants were visible, suckers and arm lengths were recorded. All missing values for individual traits were replaced with the global mean of that trait across the whole dataset.
Table 1. Description of morphological measurements recorded.
| Abbreviations | Description |
| MLd | Dorsal mantle length |
| MW | Greatest width of mantle |
| HW | Greatest width of head at the level of eyes |
| AW | Width of stoutest arm |
| SDn | Diameter of largest non-enlarged sucker on any arm |
| WD | Measurement of deepest web sector, from beak to midpoint of sector |
| ALL3/R3 | Length from beak to tip of third left/right arm |
| SDeL2/R2* | Largest enlarged sucker diameter on the second left/right arm |
| SDeL3/R3* | Largest enlarged sucker diameter on the third left/right arm |
| SCL3/R3 | Entire number of suckers along intact third left/right arm |
| LL* | Length from distal most sucker to tip of hectocotylised arm |
| CL* | Length from distal most sucker to tip of calamus |
| TOL* | Length of male terminal organ |
* Denotes morphological trait only recorded for male octopuses.
All morphological analyses were performed using Systat v13 [42]. Differences in morphological traits between tetricus complex taxa were investigated using a multivariate General Linear Model (GLM), in which location was treated as a fixed factor, morphological counts were all treated as dependent variables and MLd was entered as a co-variate [43]. Inclusion of MLd as a co-variate controlled for the effect of body size, and therefore allowed investigation of size free shape variation in morphological traits. MLd was considered an appropriate proxy for an individual's body size as it was found to be highly correlated with body mass (R2 = 0.8467, data not shown), is more often provided in the literature compared to total body length, and is a standardized measurement when compared to body weight (which can be obtained from fresh or preserved specimens) [44]. The presence or absence of an interaction between locations and MLd was investigated via GLM. A non-significant or weak significant result indicated individuals across all locations were of a similar size class and were therefore comparable.
Males and females were analysed separately to allow the inclusion of male reproductive organs in morphological analyses. Mean scaling was performed on all dependant variables prior to analyses as per Berner [43] using the software package R v3.0.1 [37]. The co-variate (MLd) was either log transformed (male) or mean scaled (female) to conform with homogeneity of variance and linearity. Only a single female of appropriate size class/maturity was available from New Zealand, which was excluded from female morphological analyses.
Following multivariate GLM analyses on each of the sexes, principle component (PC) loadings were calculated for each individual by multiplying the mean scaled raw data of each trait by the canonical loading of that trait (supplied by the GLM output) and summing the products for all traits [45]. Principle components were then plotted for visualisation and canonical correlations used to calculate the eigenvalues and proportion of variance explained by each PC (Tables S4-S12 in File S1).
The importance of each morphological character in delineation between tetricus complex taxa was further investigated by Roy-Bargman step-down analysis [46], which has the advantage of retaining information on correlations between multivariate variables compared with univariate F-tests. Following a significant result from GLM analysis morphological traits were ranked in theoretical order of importance by multiplying the first and second canonical loadings (CL1 and CL2) for each trait by the total variance explained by PC1 and PC2, respectively. The resulting values were added together, and traits displaying the highest joint CL were ranked as having the highest priority. Each trait was then investigated sequentially in order of descending ‘importance’ via regression analyses; in which location was a categorical predictor and MLd a co-variate (for size-correction) for all analyses. Higher priority traits were added as co-variates in each successive analysis. Tukey's post-hoc tests were performed for each significant step-down analysis to determine differences in morphological traits among locations. Step-down analysis was continued until tests yielded an insignificant effect. Probability values were adjusted via the Bonferroni correction method to account for multiple testing.
To further explore classification of tetricus populations into taxonomic groups, Discriminant Function Analysis (DFA) was performed. As DFA cannot incorporate co-variates, analyses were conducted on calculated principle component loadings for each sex. Principle components were used for DFAs as they were calculated from the original multivariate GLM, and were therefore size corrected. In addition, PCs are composite variables calculated for each individual, and consequently encompass any correlations between morphological traits [45]. For all DFAs, Jackknifed correlation matrices were used as they are considered a more reliable estimator of group membership assignment [47].
Results
Molecular analyses
Phylogenetic analyses
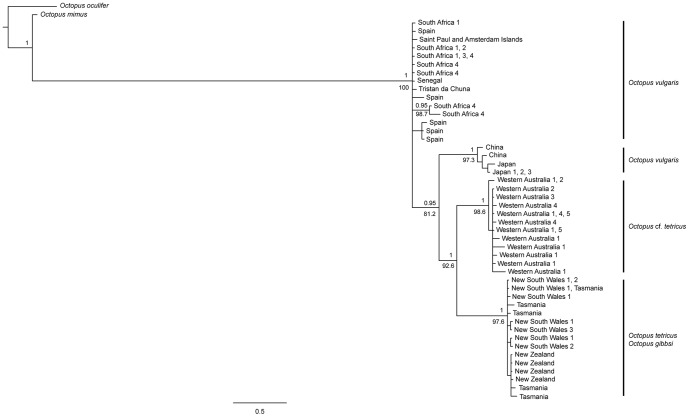
The AIC indicated that TrN+G was the preferred evolutionary model for the concatenated alignment and this was utilised within ML and Bayesian phylogenetic analyses. Topologies resulting from ML and Bayesian analyses were identical, recovering a highly supported clade containing Octopus tetricus from east Australia and Tasmania, as well as O. gibbsi from New Zealand (bootstrap value [BS] = 97.6, posterior probability [PP] = 1; Figure 2). All individuals collected from Western Australia fell within a highly supported monophyletic clade (BS = 98.6, PP = 1). A sister-taxon relationship was supported between the Western Australian and east coast (east Australia, Tasmania and New Zealand) clades (BS = 92.6, PP = 1).
Figure 2. Bayesian topology depicting the phylogenetic relationships among five currently accepted species of Octopoda.
Analyses are based on five combined partial mitochondrial genes (12s rRNA, 16s rRNA, COI, COIII and Cytb) showing bootstrap values ≥ 50 below the node and posterior probability values ≥ 0.7 above the node. Outgroup is comprised of Octopus oculifer and O. mimus. Node labels reflect locations represented by individuals contributing to node (Western Australia, 1 = Mandurah, 2 = Woodman's Point, 3 = Albany, 4 = Cape Le Grand, 5 = Esperance; East Australia, 1 = Wallaga Lake, 2 = Port Stephens, 3 = Narooma; South Africa, 1 = Port Elizabeth, 2 = Umhlanga, 3 = Hout Bay, 4 = Durban).
All Octopus vulgaris individuals collected from the waters off Japan and China formed a highly supported monophyletic clade (BS = 97.3, PP = 1). The Japanese and Chinese O. vulgaris and the tetricus complex were supported as a monophyletic clade (BS = 81.2, PP = 0.95). This clade fell within a larger clade containing O. vulgaris individuals from Spain (type location; Mediterranean Sea), South Africa, St Paul and Amsterdam Islands, thereby rendering the O. vulgaris clade to be paraphyletic.
Genetic distance
Octopus gibbsi was treated as O. tetricus in genetic distance calculations on COI sequence data based on high support values of phylogenetic analyses previously described. Comparisons of within species (i.e. within O. cf. tetricus or within O. tetricus/O. gibbsi) and between species TrN genetic distance for O. tetricus (including O. gibbsi) and O. cf. tetricus showed that mean between species divergence (3.34%) was approximately 17.5 times greater than mean within species divergence (0.19%).
Timing of divergence
Based on TrN distances, a date of divergence of ∼3.2–6.9 million years ago (ma) was estimated between Octopus tetricus from the east coast of Australia (inclusive of O. gibbsi) and O. cf. tetricus from Western Australia (Table S13 in File S1). Furthermore, the Australian tetricus complex clades and the Japanese/Chinese O. vulgaris clade were estimated as being separated by ∼5.4–11.6 million years (Table S14 in File S1).
Coalescent delimitation
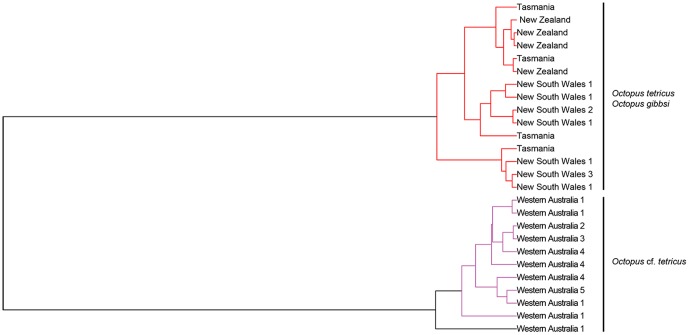
Two ML clusters and three entities (i.e. species) were supported via GMYC analysis (p = 0.01). All individuals from the east coast of Australia, Tasmania (Octopus tetricus) and New Zealand (previously O. gibbsi) comprised a single monophyletic clade, whilst the second monophyletic clade was comprised entirely of individuals from Western Australia (Figure 3). A third clade was supported by the GMYC analysis and comprised a single individual from Western Australia, although this clade was paraphyletic, forming a monophyletic clade with other Western Australian individuals.
Figure 3. Generalised Mixed Yule Coalescent (GMYC) Bayesian topology depicting the phylogenetic relationships of Octopus tetricus (east Australia and Tasmania), O. cf. tetricus (Western Australia) and O. gibbsi (New Zealand).
Analysis is based on three concatenated partial mitochondrial genes (COI, COIII and Cytb). Three species clades were supported via GMYC analysis; East Australia and New Zealand (red) and Western Australia (purple and black). Node labels reflect locations represented by individuals contributing to node (Western Australia, 1 = Mandurah, 2 = Woodman's Point, 3 = Albany, 4 = Cape Le Grand, 5 = Esperance; East Australia, 1 = Wallaga Lake, 2 = Port Stephens, 3 = Narooma).
Morphological analyses
Males
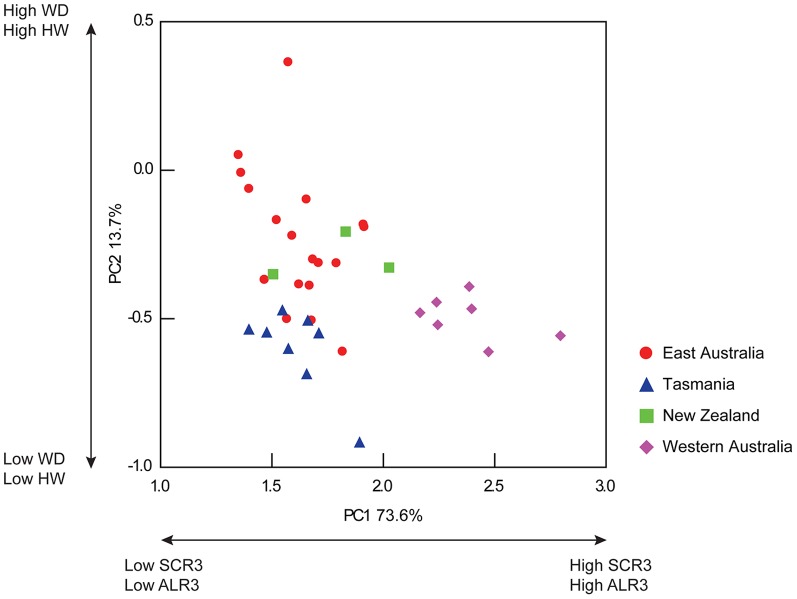
No strong interaction between the independent variable (coast) and the co-variate (MLd) was recorded (Pillai Trace = 1.937, F = 1.709, df = 48,45, p = 0.04), therefore the General Linear Model was run without the interaction. A significant difference was recorded among four coasts for the multivariate model based upon 16 morphological traits (and MLd as co-variate) measured from 36 mature male octopods (Pillai Trace = 2.070, F = 2.503, df = 48,54, p = 0.001; Table 2). Visualisation of the male PC biplot showed individuals from the east coast of Australia, Tasmania and New Zealand could not be distinguished from one another, and were characterised by relatively small SCR3 and ALR3 (Figure 4). Western Australian individuals formed a distinct group separate from east coast individuals. Individuals from Western Australia were characterised as having greater SCR3 and ALR3 (PC1) in comparison to individuals from east Australia, Tasmania and New Zealand. No distinctions based upon WD and HW among locations were detected (PC2).
Table 2. Summary of mature male morphological measurements taken from preserved museum specimens of Octopus tetricus (east Australia and Tasmania), O. cf. tetricus (Western Australia and O. gibbsi (New Zealand).
| Catalogue # | Institution | Location | MLd | MW | HW | AW | SDn | WD | ALL3 | ALR3 | SDeL2 | SDeL3 | SDeR2 | SDeR3 | SCL3 | SCR3 | LL | CL | TOL |
| C126244 | AM | East Australia | 69.5 | 67.2 | 46.9 | 23.9 | 12.3 | 93 | 172 | 193 | 17.5 | 16.1 | 16.6 | 15.8 | 162 | 135 | 3.6 | 1.7 | 11.7 |
| C171685 | AM | East Australia | 65 | 67.2 | 46.9 | 23.9 | 12.3 | 93 | 279 | 249 | 11 | 11.3 | 11.3 | 10.9 | 220 | 136 | 2.9 | 1.5 | 9.7 |
| F160334 | MV | East Australia | 54.3 | 67.2 | 46.9 | 23.9 | 12.3 | 93 | 147 | 163 | 17.5 | 8.2 | 7.8 | 7.4 | 187 | 143 | 1.5 | 0.6 | 7.5 |
| F77273 | MV | East Australia | 80.4 | 49.9 | 44.6 | 19.3 | 9.7 | 66 | 311 | 266 | 12.1 | 11.8 | 12.1 | 11.5 | 211 | 158 | 3.1 | 1.2 | 11.3 |
| F77274 | MV | East Australia | 86.1 | 59.6 | 31.4 | 15.9 | 10 | 86 | 340 | 284 | 13.6 | 11.9 | 12.4 | 13 | 197 | 133 | 3.9 | 1.7 | 14 |
| F78281b | AM | East Australia | 63.7 | 64.3 | 45.2 | 24.4 | 12.3 | 91 | 227 | 198 | 17.5 | 16.1 | 16.6 | 15.8 | 159 | 128 | 2.7 | 1.1 | 10.7 |
| F80438 | MV | East Australia | 89.6 | 58.7 | 43.1 | 24 | 9.8 | 99 | 307 | 285 | 17.5 | 16.1 | 16.6 | 15.8 | 242 | 153 | 3.7 | 1.7 | 12.9 |
| F80439 | MV | East Australia | 69.9 | 55.3 | 37.8 | 16.2 | 9.7 | 63 | 298 | 242 | 12.7 | 11.4 | 10.9 | 10.7 | 234 | 154 | 3 | 1.5 | 10.8 |
| F80440 | MV | East Australia | 110 | 66.6 | 40.9 | 21.3 | 12.1 | 101 | 435 | 370 | 14.7 | 14.9 | 14.6 | 15 | 190 | 144 | 4.6 | 2.3 | 15.9 |
| F80445 | MV | East Australia | 80.1 | 53.5 | 41.7 | 23.5 | 11.7 | 73 | 289 | 287 | 18.5 | 17.5 | 17.3 | 16.4 | 217 | 121 | 4.1 | 1.3 | 13.1 |
| F200319 | MV | East Australia | 93.6 | 55.2 | 48.7 | 30.1 | 13.6 | 78 | 365 | 333 | 17.6 | 16.5 | 17 | 16.1 | 217 | 153 | 3.5 | 1.5 | 15.1 |
| F200324 | MV | East Australia | 86.7 | 59.5 | 44.1 | 25.5 | 12.2 | 88 | 289 | 307 | 18.5 | 17.8 | 19.6 | 17.2 | 217 | 138 | 3 | 1.3 | 14.7 |
| F200323 | MV | East Australia | 85.6 | 51.8 | 39 | 20.5 | 12.1 | 74 | 289 | 260 | 14.9 | 14.5 | 16.2 | 14.2 | 217 | 139 | 2.6 | 0.7 | 12.8 |
| F200321 | MV | East Australia | 61.7 | 64.3 | 45.2 | 24.4 | 12.3 | 91 | 211 | 181 | 17.5 | 16.1 | 16.6 | 15.8 | 201 | 127 | 3 | 1.5 | 9.4 |
| F182058 | MV | East Australia | 121 | 88.1 | 65.5 | 40 | 17.3 | 114 | 289 | 426 | 25.7 | 24.7 | 24.7 | 25.7 | 217 | 143 | 4.2 | 2.1 | 19.7 |
| F182057 | MV | East Australia | 113.7 | 100.5 | 64.3 | 35.4 | 16.5 | 113 | 289 | 390 | 22.4 | 22.6 | 24.7 | 21.7 | 217 | 150 | 4.8 | 2.1 | 16 |
| F200317 | MV | East Australia | 122.4 | 80.6 | 52.9 | 28.6 | 16.4 | 142 | 399 | 456 | 31.9 | 26.8 | 30.2 | 29.1 | 142 | 142 | 5.2 | 2.3 | 14.6 |
| F200318b | MV | East Australia | 84.4 | 57.1 | 34.1 | 17.4 | 9.6 | 81 | 345 | 292 | 14.1 | 16.1 | 14.4 | 13.1 | 220 | 156 | 3.9 | 1.3 | 15.5 |
| F180706 | MV | Tasmania | 113.2 | 82.7 | 58.7 | 22.9 | 14 | 82 | 470 | 365 | 18.7 | 19.5 | 23 | 19.7 | 195 | 136 | 4.5 | 2 | 18.1 |
| F180698 | MV | Tasmania | 101 | 70.9 | 37.5 | 19.5 | 10.9 | 72 | 525 | 287 | 17.6 | 16.5 | 17 | 16.1 | 217 | 140 | 3.4 | 1.8 | 15 |
| F180707 | MV | Tasmania | 134.9 | 71 | 52.5 | 29.2 | 16.3 | 81 | 580 | 468 | 21.1 | 25.6 | 23 | 22.3 | 224 | 146 | 5.7 | 2.8 | 17.6 |
| F180699 | MV | Tasmania | 90.8 | 54 | 34.6 | 17.7 | 11.7 | 81 | 315 | 301 | 17.6 | 16.5 | 17 | 16.1 | 203 | 143 | 3.6 | 1.5 | 15.6 |
| F180700 | MV | Tasmania | 110.9 | 67.8 | 37.5 | 22 | 11.1 | 86 | 408 | 338 | 17.6 | 16.5 | 17 | 16.1 | 212 | 139 | 3.8 | 1.7 | 14.6 |
| F180697 | MV | Tasmania | 100.8 | 56.2 | 30.2 | 17.2 | 9.9 | 86 | 403 | 360 | 17.6 | 16.5 | 17 | 16.1 | 218 | 146 | 3.4 | 2.1 | 16.7 |
| F180696 | MV | Tasmania | 92.5 | 56.8 | 40.7 | 16.8 | 9.9 | 61 | 413 | 251 | 17.6 | 16.5 | 17 | 16.1 | 201 | 132 | 3.7 | 1.6 | 12.3 |
| F180702 | MV | Tasmania | 85 | 58.4 | 34.4 | 16.1 | 9.2 | 68 | 267 | 268 | 17.6 | 16.5 | 17 | 16.1 | 231 | 143 | 3.9 | 1.7 | 13.8 |
| NMNZM.118421 | O'Shea [16] | New Zealand | 135.5 | 97 | 62.5 | 23.9 | 12.3 | 142 | 532 | 418 | 17.6 | 16.5 | 21.3 | 21.3 | 217 | 150 | 4.1 | 2.2 | 14.6 |
| NMNZM.118305 | O'Shea [16] | New Zealand | 121.5 | 55.5 | 52 | 23.9 | 12.3 | 107 | 393 | 360 | 17.6 | 16.5 | 22.9 | 17 | 217 | 162 | 3 | 1.2 | 14.6 |
| NMNZM.118425 | O'Shea [16] | New Zealand | 89.5 | 59.5 | 37.2 | 23.9 | 12.3 | 62 | 365 | 192 | 17.6 | 16.5 | 11.5 | 11.6 | 217 | 142 | 3 | 1.5 | 14.6 |
| 310 6-83-1 | AM | Western Australia | 90 | 74.6 | 56.1 | 27.2 | 12.9 | 110 | 557 | 469 | 17.3 | 15.8 | 16 | 14.9 | 257 | 201 | 4.5 | 1.6 | 24.3 |
| F200330 | MV | Western Australia | 95.8 | 79.1 | 53.9 | 23.7 | 11.2 | 114 | 420 | 444 | 16.7 | 15.9 | 15.1 | 14.3 | 281 | 209 | 3.5 | 1.7 | 14.3 |
| F160306 | MV | Western Australia | 91.7 | 76.1 | 54.5 | 26.3 | 14.3 | 115 | 458 | 421 | 17.3 | 15.8 | 15.4 | 14.9 | 250 | 177 | 4.2 | 1.9 | 18.7 |
| F200327 | MV | Western Australia | 111.7 | 66.6 | 52.2 | 22.9 | 12.6 | 98 | 458 | 426 | 17.3 | 15.8 | 15.4 | 14.9 | 283 | 192 | 4.4 | 1.6 | 13.5 |
| F200329 | MV | Western Australia | 114.2 | 74.6 | 56.4 | 30.3 | 13.1 | 96 | 381 | 384 | 17.9 | 15.6 | 15 | 15.5 | 230 | 177 | 4.6 | 2.4 | 14.7 |
| F200328 | MV | Western Australia | 163.4 | 83.5 | 65.8 | 34.3 | 14.1 | 107 | 559 | 544 | 17.3 | 15.8 | 15.4 | 14.9 | 291 | 218 | 3.9 | 1.7 | 26.8 |
| F200326a | MV | Western Australia | 127 | 68 | 54 | 25.7 | 12.4 | 127 | 365 | 520 | 17.6 | 16.5 | 17 | 16.1 | 217 | 207 | 5 | 2.1 | 11.9 |
Missing data (shown in bold) has been replaced by the global mean of the respective trait.
Institutions – AM = Australian Museum, Sydney, MV = Museum Victoria.
Figure 4. Principal component biplot of male individuals of the tetricus complex.
X axis represents PC1 (explaining 73.6% of total variation) and is driven primarily by the SCR3 and ALR3. Y axis represents PC2 (explaining 13.7% of total variation) and is driven primarily by WD and HW.
DFA showed a significant difference among individuals from east Australia, Tasmania, New Zealand and Western Australia (Pillai Trace = 1.201, F = 16.020, df = 6, 64, p = <0.001). DFA assigned 100% (n = 7) of male individuals from Western Australia to a single group comprised solely of Western Australian individuals (Table 3). DFA assigned 83% (n = 15) of east Australian individuals to the east Australian group, with 17% (n = 3) allocated to the Tasmanian group. Furthermore, 88% (n = 7) of Tasmanian individuals were assigned to the Tasmanian group, whilst 12% (n = 1) were grouped with east Australian individuals. All individuals from New Zealand (n = 3) were allocated into the east Australian group.
Table 3. Male Discriminant Function Analysis: Jackknifed classification matrix.
| East Australia | Tasmania | New Zealand | Western Australia | % correct | |
| East Australia | 15 | 0 | 3 | 0 | 83 |
| Tasmania | 3 | 0 | 0 | 0 | 0 |
| New Zealand | 1 | 0 | 7 | 0 | 88 |
| Western Australia | 0 | 0 | 0 | 7 | 100 |
Ranking of CLs determined male SCR3 to be the most important variable in detecting variance among groups (Table S8 in File S1). Step-down analysis performed on male SCR3 showed a significant difference among coasts (F = 41.775, df = 3, p = <0.001). Tukey's post-hoc analysis showed no significant difference among east Australia, Tasmania and New Zealand (p = >0.6), however Western Australia differed significantly from all three of these locations (p = <0.001). Analysis of ALR3 (second highest ranked variable) showed a significant difference among coast once the co-variate and SCR3 were included in the model (F = 5.333, df = 3, p = 0.01). Tukey's post-hoc analysis showed no significant difference between individuals from east Australia, Tasmania and Western Australia (p = >0.1), whilst individuals from New Zealand differed significantly from both eastern and Western Australia (p = 0.02 and 0.01 respectively). Analysis of SCL3 (third highest ranked trait) showed no significant difference among coasts once the co-variate, SCR3 and ALR3 were included in the model (F = 0.410, df = 3, p = 0.7). Due to a non-significant result, stepdown analysis was discontinued.
Females
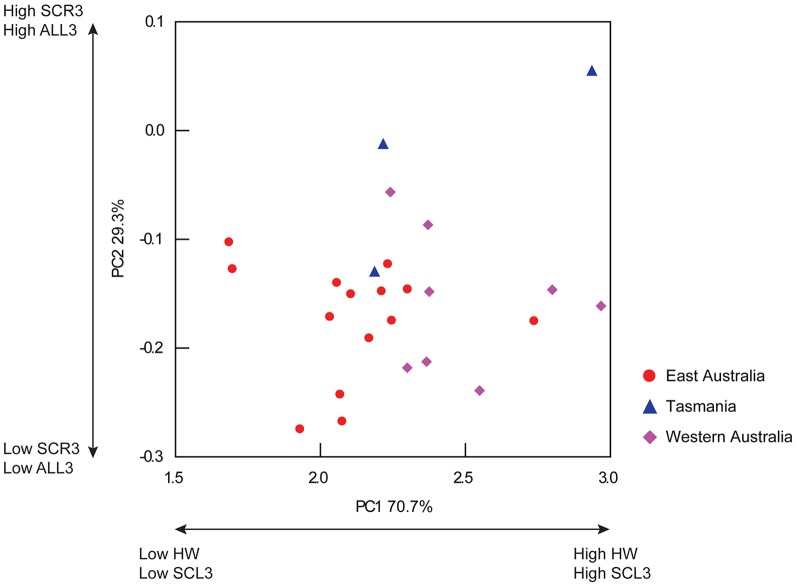
No interaction between the independent variable (coast) and the co-variate (MLd) was recorded (Pillai Trace = 1.083, F = 1.574, df = 18, 24, p = >0.1), therefore the model was run without the interaction. No significant difference was recorded among three locations for the multivariate model based upon nine morphological traits (and MLd as co-variate) measured from 25 mature female octopods (Pillai Trace = 0.122, F = 1.989, df = 18, 28, p = 0.05; Table 4). Visualisation of the female PC biplot showed overlap of individuals from east Australia, Tasmania and Western Australia along PC1 and PC2, which were primarily driven by HW/SCL3 and SCR3/ALL3 respectively (Figure 5). Although non-significant, female individuals from Western Australia generally possessed greater HW and SCL3 in relation to individuals from east Australia.
Table 4. Summary of mature female morphological measurements taken from preserved museum specimens of Octopus tetricus (east Australia and Tasmania), O. cf. tetricus (Western Australia) and O. gibbsi (New Zealand).
| Catalogue # | Institution | Location | MLd | MW | HW | AW | SDn | WD | ALL3 | ALR3 | SCL3 | SCR3 |
| C156208 | AM | East Australia | 67.6 | 65.4 | 44.6 | 20.7 | 12.2 | 84 | 212 | 232 | 197 | 194 |
| C171669 | AM | East Australia | 62.1 | 65.4 | 44.6 | 20.7 | 12.2 | 84 | 240 | 208 | 218 | 199 |
| F78082 | AM | East Australia | 117.8 | 65.4 | 31.6 | 19.5 | 12 | 96 | 315 | 335 | 218 | 229 |
| F78281a | AM | East Australia | 70.3 | 65.4 | 44.6 | 20.7 | 12.2 | 84 | 323 | 209 | 235 | 222 |
| F78283b | AM | East Australia | 53.2 | 65.4 | 44.6 | 20.7 | 12.2 | 84 | 158 | 327 | 163 | 222 |
| F80442a | MV | East Australia | 96 | 64.9 | 34.7 | 16.8 | 9.5 | 69 | 358 | 356 | 247 | 234 |
| F80442b | MV | East Australia | 126.3 | 74.8 | 38.1 | 16.9 | 11.8 | 88 | 452 | 418 | 238 | 254 |
| F80446 | MV | East Australia | 60.3 | 45.2 | 34.4 | 14.1 | 8.5 | 59 | 230 | 216 | 201 | 183 |
| F85370a | MV | East Australia | 100 | 65.4 | 44.6 | 20.7 | 12.2 | 84 | 287 | 400 | 216 | 221 |
| F200320 | MV | East Australia | 120.2 | 65.4 | 44.6 | 20.7 | 12.2 | 84 | 323 | 435 | 235 | 224 |
| F200319 | MV | East Australia | 104.5 | 80.1 | 52.2 | 28.8 | 15.2 | 100 | 437 | 409 | 265 | 290 |
| F200318a | MV | East Australia | 104.8 | 70.1 | 43.1 | 21 | 14.1 | 86 | 311 | 356 | 214 | 222 |
| F200318c | MV | East Australia | 84.4 | 47.7 | 31.5 | 13.4 | 9 | 52 | 259 | 243 | 204 | 198 |
| F78283c | AM | East Australia | 52.6 | 65.4 | 44.6 | 20.7 | 12.2 | 84 | 152 | 327 | 235 | 222 |
| F180704 | MV | Tasmania | 102.2 | 64.9 | 42.8 | 20 | 12.5 | 74 | 323 | 369 | 235 | 230 |
| F180701 | MV | Tasmania | 99.7 | 65.8 | 43.8 | 19.5 | 12.1 | 72 | 418 | 367 | 216 | 196 |
| F180705 | MV | Tasmania | 135.4 | 88.7 | 57.5 | 25.7 | 17.2 | 99 | 524 | 327 | 302 | 222 |
| NMNZM.90320 | O'Shea [16] | New Zealand | 48 | 35.7 | 30 | 20.7 | 6 | 45 | 141 | 313 | 206 | 194 |
| NMNZM.131569 | O'Shea [16] | New Zealand | 48.9 | 28 | 27 | 20.7 | 5.9 | 40 | 283 | 141 | 235 | 145 |
| NMNZM.118426 | O'Shea [16] | New Zealand | 137 | 90.5 | 55 | 20.7 | 16.8 | 112 | 424 | 485 | 235 | 243 |
| F200334 | MV | Western Australia | 114.1 | 97 | 61.5 | 31.4 | 15 | 139 | 474 | 327 | 235 | 222 |
| F160320 | MV | Western Australia | 84.2 | 59.2 | 44.6 | 20 | 20 | 75 | 304 | 327 | 264 | 264 |
| F160321 | MV | Western Australia | 90.9 | 64.6 | 53.9 | 19.5 | 11.2 | 101 | 396 | 438 | 264 | 264 |
| F160325 | MV | Western Australia | 97.3 | 59.3 | 48 | 17.5 | 9.7 | 82 | 350 | 355 | 257 | 266 |
| F80447 | MV | Western Australia | 87.4 | 69.3 | 45.2 | 19.6 | 11.5 | 79 | 356 | 322 | 241 | 186 |
| F200327 | MV | Western Australia | 88.3 | 64.3 | 51.9 | 20.4 | 12 | 82 | 240 | 277 | 290 | 253 |
| F200331 | MV | Western Australia | 119.6 | 69 | 57.4 | 25.7 | 13.2 | 119 | 457 | 327 | 276 | 222 |
| F160302 | MV | Western Australia | 71.4 | 68.9 | 51.8 | 22 | 13.2 | 97 | 302 | 305 | 236 | 200 |
Missing data (shown in bold) has been replaced by the global mean of the respective trait.
Institutions – AM = Australian Museum, Sydney, MV = Museum Victoria.
Figure 5. Principal component biplot of female individuals of the tetricus complex.
X axis represents PC1 (explaining 70.7% of total variation) and is driven primarily by HW and SCL3. Y axis represents PC2 (explaining 29.3% of total variation) and is driven primarily by SCR3 and ALL3.
DFA showed a significant difference among individuals from east Australia, Tasmania and Western Australia (Pillai Trace = 0.678, F = 5.637, df = 4, 44, p = <0.01). DFA assigned 93% (n = 13) of east Australian female individuals into the correct group, whilst 7% (n = 1) were placed into the Western Australian group (Table 5). 67% of individuals from Tasmania (n = 2) were placed into the correct group, whilst 33% (n = 1) were considered to belong to the east Australian group. 38% (n = 3) of female individuals from Western Australia were correctly assigned, whilst 50% (n = 4) and 12% (n = 1) were assigned to east Australian and Tasmanian groups respectively.
Table 5. Female Discriminant Function Analysis: Jackknifed classification matrix.
| East Australia | Tasmania | Western Australia | % correct | |
| East Australia | 13 | 0 | 1 | 93 |
| Tasmania | 1 | 2 | 0 | 67 |
| Western Australia | 4 | 1 | 3 | 38 |
Discussion
Species level relationships
The main focus of this study was to resolve the taxonomic status of the Australasian tetricus complex. Molecular and morphological results are consistent with the hypothesis that disjunct populations of Octopus tetricus from Australia's east coast (including Tasmania), and from Western Australia are separate species. In addition, findings of this study support the hypothesis that O. gibbsi of New Zealand is synonymous with east Australian O. tetricus [2]. Consequently, we propose that the species name O. gibbsi be considered a junior synonym of O. tetricus Gould, 1852, and will hereafter be included in reference to O. tetricus.
In the present study, interspecific variation of COI between eastern Octopus tetricus and western O. cf. tetricus was over one order of magnitude (∼18 times) greater than intraspecific variation within each of these populations; a marked ‘barcoding gap’ consistent with the ‘ten times rule’ of Hebert et al., [48]. This study estimated interspecific divergence of COI sequences between O. tetricus and O. cf. tetricus to be 3.4%, similar to congeneric differences previously reported for octopods [49], [50]. For example, within the family Octopodidae interspecific variation was found to be 1–2% and 2–3.3% for the octopod genera Pareledone [51] and Thaumeledone [49] respectively. The interspecific variation found between O. tetricus and O. cf. tetricus (3.4%) displayed higher species-level differentiation than the 1.3% divergence recommended by Undheim et al., [50] for O. vulgaris. Low nucleotide sequence divergence between octopod species in this and previous studies contrasts with higher levels recorded among moths, butterflies and birds, which range from 5.8–9.1% [48], [52], [53].
GMYC analysis suggested Western Australian Octopus cf. tetricus is a distinct species from O. tetricus, as well as supporting the synonymy of O. gibbsi with O. tetricus. However, GMYC analysis detected a second cryptic Western Australian species, which conflicts with the phylogenetic and morphological results of this study (which show no such cryptic speciation). This may be due to gaps in knowledge (i.e. more species exist than is currently known), although more likely reflects the tendency for GMYC analyses to ‘over-split’ taxa [35].
Talavera et al., [35] investigated the ability of GMYC analysis to delineate species using the well resolved European butterflies. Their analysis revealed 16 unexpected cryptic species, which (although the authors acknowledged that at least some of these cryptic species may represent real entities) was considered to be a failure of the model due to the high levels of intraspecific variability recorded within butterflies. As interspecific variability between Octopus cf. tetricus and O. tetricus was far greater relative to the low intraspecific variability within each individual group (see above), the discovery of a second cryptic Western Australian species is considered likely to be an artefact of ‘over-splitting’ by GMYC analysis.
Multivariate morphological analyses showed congruence in detecting significant differences between individuals from east Australia/New Zealand and Western Australia; although females appear to be a less reliable morphological discriminator of species identity. Male morphology was able to successfully discriminate between Octopus tetricus and O. cf. tetricus. Sucker numbers on the males third right arm explained the most variation between O. tetricus and O. cf. tetricus, with O. cf. tetricus having significantly greater sucker numbers. Males third right arms (left in some species) possess the hectocotylus, a copulatory organ used to pass sperm to the female during mating. The hectocotylus is comprised of the ligula and calamus, which provide a limit to the emergence of new suckers at a relatively early stage of ontogeny [54]. Toll [54] investigated sucker counts on the males hectocotylised arm (HASC) among 12 species of the sub-family Octopodinae, and demonstrated its value in identification and delimitation of otherwise morphologically similar octopods. Toll [54] showed sucker numbers on the hectocotylised arm to be relatively fixed, with different species appearing to be characterised by a narrow range of values for HASC, which he proposed were genetically defined. This assumption appears to be supported by congruence between molecular and HASC data obtained in this study. Consistency of sucker counts despite fixation, preservation [54] or environmental influence further reinforces the usefulness of male HASC in cryptic cephalopod taxonomy.
Biogeographic factors
Speciation between Octopus tetricus and O. cf. tetricus is likely the result of reproductive isolation due to allopatric eastern and western distributions. Divergence of O. tetricus (east Australian, Tasmania and New Zealand populations) and O. cf. tetricus (from Western Australia) were estimated to have occurred somewhere within the last 3.2–6.9 million years. This coincides with cooling of the previously tropical Miocene seas along the southern Australian coastline and the rising of the Bassian Isthmus (a historic land-bridge joining Tasmania and mainland Australia) during the Pliocene era, potentially dividing populations of a common tetricus complex ancestor in two. Glacial-interglacial epochs during the early Pleistocene resulted in northward progression of cooler waters, initiating the retreat of numerous wide-spread subtropical species along the eastern and western coasts, isolating populations which allowed for genetic differentiation to commence [55].
More recently oceanographic, climatic and ecological factors have likely maintained contemporary disjunction following the final inundation of the Bassian Isthmus 14,000 years ago. For example, the southern coast of Australia possesses extensive expanses with limited reef habitat in the Great Australian Bight and east of Wilson's Promontory in south-east Victoria. Limited reef habitat has been proposed as a factor in genetic divergence of populations and speciation events in other southern marine taxa such as decapods, echinoderms [56], [57], and gastropods [58], [59]. However studies conducted on O. gibbsi (treated as O. tetricus) among reefs in Northern New Zealand found reef habitat was not essential for successful settlement [17], and O. tetricus were often found in lairs within sandy bottomed estuaries along the southern coast of New South Wales (M. Amor, personal observation). The Great Australian Bight is also associated with sharp drops in sea surface temperature (SST), which is a likely explanation for maintenance of allopatric distributions between east and west taxa.
The absence of significant genetic differentiation between New Zealand and east Australian Octopus tetricus populations suggests ongoing gene flow across the Tasman Sea; a 2000 km wide marine body separating the two landmasses. Due to the benthic shallow-water habit of Octopus tetricus adults [15], connectivity between New Zealand and east Australian populations is likely attributable to trans-Tasman dispersal during the planktonic larval stage; although adults of the genus Octopus can raft on floating wood or drifting macroalgae [60], which may function as a rare mode of passive trans-Tasman migration.
A number of other southern Australasian marine taxa display similar trans-Tasman genetic homogeneity, including the southern rock lobster, Jasus edwardsii [61], [62], [63] and morwong (cheilodactylid) fishes [64], [65]. Planktonic larval durations (PLD) for the Octopodinae appear much shorter (35–60 days; reviewed in Villanueva, [66]) than those of the lobster J. edwardsii (2 years [67]) and cheilodactylid fishes (1 year [68]). Octopus paralarvae appear to be active and often constant swimmers [13], [69], potentially facilitating dispersal within surface currents. However, simulation based oceanographic modelling studies suggests that in the absence of rafting, a period of several months is required for even a low probability of successful trans-Tasman dispersal [70]. Octopod paralarvae have been observed rafting on macroalgal and other drift debris [71], which may function as habitat for post-settlement juveniles until arrival at suitable shallow-water habitat. Additionally, paralarvae of some octopods can delay settlement in the absence of suitable habitat [72]. These ‘super-paralarvae’ obtain larger sizes and more developed swimming capabilities, while retaining paralarval morphological characters (reviewed in Villanueva and Norman, [69]), and may facilitate trans-Tasman dispersal for Octopus tetricus. Further investigation into physiological, behavioural and ecological aspects of paralarval life histories would further our understanding of the dispersive capabilities of O. tetricus.
Evidence of range shifts and implications of climate change
This study is the first to verify the presence of Octopus tetricus in the temperate waters off Flinders Island, Tasmania. This suggests the southern distributional limit of O. tetricus along the Australian mainland (currently recognised as Eden, New South Wales) is underestimated and requires resurveying, in fact O. tetricus has been sighted as far south as Cape Conran, Victoria (M. Amor personal observation, 2013). Temperate coastal waters in eastern Tasmania appear to be warming at approximately four times the global ocean warming average due to climate change driven strengthening of the Eastern Australian Current [73]. This has been linked to recent range expansions of a number of sub-tropical and tropical marine species in Tasmanian waters, including 22 fish species, eastern rock lobster, leatherback turtle and two species of box jellyfish [74]. Coastal warming in Tasmania may have resulted in current temperatures exceeding the lower thermal limits of O. tetricus paralarvae, potentially allowing population establishment outside of their previously known range, as has been suggested for the sea urchin Centrostephanus rodgersii [75]. Investigation of the potential impacts of O.tetricus range expansion on native ecosystems and commercial fisheries should be given high priority.
Broader phylogenetic relationships
Mitochondrial DNA analyses placed the Australasian tetricus complex within a monophyletic clade along with Japanese and Chinese Octopus vulgaris, supporting previous speculations that these taxa are closely related [76]. The current study estimated that the tetricus complex and Japanese/Chinese O. vulgaris arose from a common ancestor following an ‘anti-tropical’ divergence event that took place between ∼5.4–11.7 ma. This estimated time of divergence is consistent with mid-Miocene climatic warming and the emergence of intervening tropical waters at lower latitudes [77]; suggesting vicariant isolation of a once common subtropical ancestor into Northern and Southern Hemisphere populations. Warming of equatorial waters during the mid-Miocene has also been implicated in trans-equatorial divergences for a number of marine taxa, especially reef fishes [78], [79], [80]. In addition, anti-tropical affinities between other subtropical Australasian-Japanese/Asian octopods have been noted. For example, Amphioctopus kagoshimensis Ortmann, 1888 from subtropical Japan and the morphologically indistinguishable taxon Amphioctopus cf. kagoshimensis recently discovered at similar latitudes in Australasian waters are predicted to represent closely related relicts of a wider distributed ancestry [76]. The ability of molecular analyses to detect cryptic species suggests that future molecular work would clarify the taxonomic, phylogenetic and palaeogeographical relationships between seemingly cryptic anti-tropical cephalopod species pairs.
Paraphyletic relationships within the vulgaris complex revealed in this study directly question the purported cosmopolitan distribution of Octopus vulgaris, and supports hypotheses regarding the existence of numerous cryptic vulgaris-like species [2], [76], [81]. Norman and Kubodera [76] previously suggested the possibility of an Asian vulgaris-like species ranging from Taiwan to Japan that was distinctly separate from genuine O. vulgaris, originally described from the Mediterranean Sea and Atlantic Ocean. Findings of this study support this theory of speciation between Atlantic and Pacific vulgaris-like species. However, the results of this study were based on samples from extremes in the distribution of O. vulgaris. Future work aimed at resolving the taxonomy of this species complex should include individuals from a representative range of the entire O. vulgaris distribution.
Conclusions and future directions
This study is the first attempt to resolve the taxonomy of the Australasian Octopus tetricus species complex. Molecular and morphological results support east Australian Octopus tetricus as a distinct species from Western Australian O. cf. tetricus, which requires future formal taxonomic description. Additionally, New Zealand's O. gibbsi was found to be synonymous with east Australian and Tasmanian O. tetricus. Paraphyletic relationships within the Octopus vulgaris complex revealed in this study adds support to hypotheses regarding the existence of numerous cryptic vulgaris-like species, warranting taxonomic revision of the O. vulgaris species complex to aid in the management of this significant global marine resource.
Supporting Information
Supporting tables. Table S1, Specimen information for individuals of which molecular sequencing was undertaken during the present study. Table S2, Specimen information for individuals accessed via GenBank for use in the present study. Table S3, Specimen information for individuals of which morphological traits were recorded during the present study. Table S4, Canonical correlation (CC) output for male octopod multivariate analysis. Table S5, Canonical loadings (CL) output for male octopod multivariate analysis. Table S6, Principal components (PC) calculated from canonical correlation and canonical loading outputs from male octopod multivariate analysis. Table S7, Eigenvalues and principal component (PC) variance contribution outputs from male octopod multivariate analysis. Table S8, Ranked canonical loadings (CL) from male octopod multivariate analysis; based upon contribution to principal components (PC). Table S9, Canonical correlation (CC) output for female octopod multivariate analysis. Table S10, Canonical loadings (CL) output for female octopod multivariate analysis. Table S11, Principal components (PC) calculated from canonical correlation and canonical loading outputs from female octopod multivariate analysis. Table S12, Eigenvalues and principal component (PC) variance contribution outputs from female octopod multivariate analysis. Table S13, Timing of divergence estimates (Tamura-Nei genetic distance) for Octopus tetricus (East Australia and New Zealand) and O. cf. tetricus (Western Australia). Table S14, Timing of divergence estimates (Tamura-Nei genetic distance) for the Australasian tetricus complex and Japanese/Chinese representatives of the Octopus vulgaris group.
(DOCX)
Acknowledgments
We thank Michelle Guzik (University of Adelaide, South Australia), Stephen Leporati (Fisheries and Marine Research Laboratories, Western Australia) and Alvaro Roura (Institute of Marine Research, Spain) for providing tissue samples. We thank Jorge Ramos Castillejos (University of Tasmania) for providing tissue samples and whole specimens. We thank Julian Finn (Museum Victoria) for historical collection of samples and project support, and Dave Staples and Chris Rowley (Museum Victoria) for their valued assistance in working with the museum's cephalopod collection. We thank Ira Cooke (La Trobe University, Victoria) and Keyne Monro (Monash University, Victoria) for their assistance with statistical analyses. The quality of this manuscript was greatly improved by the comments and suggestions of two anonymous reviewers.
Funding Statement
A La Trobe University Faculty of Science, Technology and Engineering grant supported this study. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Carlini DB, Young RE, Vecchione M (2001) A Molecular Phylogeny of the Octopoda (Mollusca: Cephalopoda) Evaluated in Light of Morphological Evidence. Mol Phylogen Evol 21: 388–397. [DOI] [PubMed] [Google Scholar]
- 2. Norman M, Hochberg G (2005) The current state of octopus taxonomy. Proceedings of the International Workshop and Symposium of Cephalopod International Advisory Council, Phuket, 2003 Phuket Marine Biological Center Special Publication 66: 127–154. [Google Scholar]
- 3. Guzik MT, Norman MD, Crozier RH (2005) Molecular phylogeny of the benthic shallow-water octopuses (Cephalopoda: Octopodinae). Mol Phylogen Evol 37: 235–248. [DOI] [PubMed] [Google Scholar]
- 4.FAO (2009) Global Production Statistics 2007. Available: http://www.fao.org/figis/servlet/TabSelector.
- 5. Acosta-Jofre MS, Sahade R, Laudien J, Chiappero MB (2012) A contribution to the understanding of phylogenetic relationships among species of the genus Octopus (Octopodidae: Cephalopoda). Scientia Marina 76: 311–318. [Google Scholar]
- 6.Edgar G (2000) Australian Marine Life: The Plants and Animals of Temperate Waters (Revised Edition). Sydney, Australia: Reed New Holland. 544 p. [Google Scholar]
- 7. Nottage JD, West RJ, Montgomery SS, Graham K (2007) Cephalopod diversity in commercial fisheries landings of New South Wales, Australia. Rev Fish Biol Fish 17: 271–281. [Google Scholar]
- 8.Norman MD, Finn JK, Hochberg FG (2014) Family Octopodidae. In P. Jereb, C.F.E. Roper, M.D. Norman & J.K. Finn eds. Cephalopods of the world. An annotated and illustrated catalogue of cephalopod species known to date. Volume 3. Octopods and Vampire Squids. FAO Species Catalogue for Fishery Purposes. No. 4, Vol. 3. Rome, FAO. pp. 36–215. [Google Scholar]
- 9.REDMAP (2011). Available: http://www.redmap.org.au/species/view/267/gloomy-octopus/.REDMAP. Accessed 2012 February 28.
- 10. Roper CFE, Sweeney MJ, Nauen CE (1984) FAO Species Catalogue. Volume 3. Cephalopods of the World: An Annotated and Illustrated Catalogue of Species of Interest to Fisheries. FAO Fisheries Synopsis. 3: 1–277. [Google Scholar]
- 11.Joll LM, editor (1983) Octopus tetricus. New York: Academic Press. 325–334 p. [Google Scholar]
- 12. Joll LM (1976) Mating, egg-laying and hatching of Octopus tetricus (Mollusca, Cephalopoda) in laboratory. Mar Biol 36: 327–333. [Google Scholar]
- 13. Joll LM (1977) Growth and food intake of Octopus tetricus (Mollusca, Cephalopoda in aquaria. Australian Journal of Marine and Freshwater Research 28: 45–56. [Google Scholar]
- 14. Joll LM (1978) Observations on embryonic development of Octopus tetricus (Mollusca, Cephalopoda). Australian Journal of Marine and Freshwater Research 29: 19–30. [Google Scholar]
- 15.Norman MD (2000) Cephalopods: A World Guide. Hackenheim, Germany: ConchBooks. 318 p. [Google Scholar]
- 16.O'Shea S (1999) The Marine Fauna of New Zealand: Octopoda (Mollusca: Cephalopoda): NIWA Biodiversity Memoir. 280 p. [Google Scholar]
- 17. Anderson TJ (1997) Habitat selection and shelter use by Octopus tetricus . Mar Ecol Prog Ser 150: 137–148. [Google Scholar]
- 18.Donnan Laboratories (2001) Extraction of DNA from tissue: High salt method. Version 1.0. School of Biological Sciences, University of Liverpool, L69 7ZD. UK. Available: http://www,genomics.liv.ac.uk/animal/research/isolation.pdf. Accessed 2011 January 25.
- 19. Simon C, Paabo S, Kocher TD, Wilson AC (1990) Evolution of mitochondrial ribosomal-RNA in insects as shown by the polymerase chain-reaction. Molecular Evolution 122: 235–244. [Google Scholar]
- 20. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial Cytochrome C Oxidase subunit from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3: 294–299. [PubMed] [Google Scholar]
- 21. Allcock AL, Strugnell J, Johnson M (2008) How useful are the recommended counts and indices in the systematics of the Octopodidae (Mollusca: Cephalopoda). Biol J Linn Soc 95: 205–218. [Google Scholar]
- 22. Kaneko N, Kubodera T, Iguchis A (2011) Taxonomic Study of Shallow-Water Octopuses (Cephalopoda: Octopodidae) in Japan and Adjacent Waters using Mitochondrial Genes with Perspectives on Octopus DNA Barcoding. Malacologia 54: 97–108. [Google Scholar]
- 23. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 24. Posada D (2008) jModelTest: Phylogenetic model averaging. Mol Biol Evol 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 25. Akaike H (1974) A new look at the statistical model identification. IEEE Transactions on Automatic Control 19: 716–723. [Google Scholar]
- 26. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate Maximum-Likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 27. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 28.Rambaut A, Drummond AJ (2003) Tracer 1.3 Oxford University. Available: http://tree.bio.ed.ac.uk/software/tracer.
- 29.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol: 512–526. [DOI] [PubMed]
- 31. Strugnell JM, Watts PC, Smith PJ, Allcock AL (2012) Persistent genetic signatures of historic climatic events in an Antarctic octopus. Mol Ecol 21: 2775–2787. [DOI] [PubMed] [Google Scholar]
- 32. Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, et al. (2006) Sequence-Based Species Delimitation for the DNA Taxonomy of Undescribed Insects. Syst Biol 55: 595–609. [DOI] [PubMed] [Google Scholar]
- 33. Drummond A, Suchard M, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monaghan MT, Wild R, Elliot M, Fujisawa T, Balke M, et al. (2009) Accelerated Species Inventory on Madagascar Using Coalescent-Based Models of Species Delineation. Syst Biol 58: 298–311. [DOI] [PubMed] [Google Scholar]
- 35.Talavera G, Dincă V, Vila R (2013) Factors affecting species delimitations with the GMYC model: insights from a butterfly survey. Methods in Ecology and Evolution: n/a-n/a.
- 36.Ezard T, Fujisawa T, Barraclough T (2009) Splits: Species' Limits by Threshold Statistics. R package version 1.0. Available: http://R-Forge.R-project.org/projects/splits/.
- 37.Team RDC (2008) R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria. ISBN 3-900051-07-0. Available: http://www.R-project.org.
- 38. Voight JR (1995) Sexual dimorphism and niche divergence in a mid-water octopod (Cephalopoda: Bolitaenidae). Biological Bulletin 189: 113–119. [DOI] [PubMed] [Google Scholar]
- 39. Voight JR (1991) Enlarged suckers as an indicator of male maturity in octopus. Bull Mar Sci 49: 98–106. [Google Scholar]
- 40. Iribarne OO (1991) Life history and distribution of the small south-western Atlantic octopus, Octopus tehuelchus . J Zool 223: 549–565. [Google Scholar]
- 41. Norman MD, Sweeney MJ (1997) The shallow-water octopuses (Cephalopoda: Octopodidae) of the Philippines. Invertebr Taxon 11: 89–140. [Google Scholar]
- 42.Systat Software, Inc. (2009) Systat, Version 13. Available: www.systat.com.
- 43. Berner D (2011) Size correction in biology: how reliable are approaches based on (common) principal component analysis? Oecologia 166: 961–971. [DOI] [PubMed] [Google Scholar]
- 44. Voight JR (1991) Morphological variation in octopod specimens - reassessing the assumption of preservation-induced deformation. Malacologia 33: 241–253. [Google Scholar]
- 45.Quinn G, Keough M (2002) Introduction to Multivariate Analyses. In: Experimental Design and Analysis for Biologists. New York: Cambridge University Press. [Google Scholar]
- 46.Roy SN, Bargmann RE (1958) Tests of multiple independence and the associated confidence bounds. The Annals of Mathematical Statistics: 491–503.
- 47.Quinn G, Keough M (2002) Experimental Design and Analysis for Biologists. New York: Cambridge University Press. 557 p. [Google Scholar]
- 48. Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM (2004) Identification of birds through DNA barcodes. PLoS Biol 2: e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Strugnell JM, Collins MA, Allcock AL (2008) Molecular evolutionary relationships of the octopodid genus Thaumeledone (Cephalopoda: Octopodidae) from the Southern Ocean. Antarct Sci 20: 245–251. [Google Scholar]
- 50. Undheim EAB, Norman JA, Thoen HH, Fry BG (2010) Genetic identification of Southern Ocean octopod samples using mtCOI. C R Biol 333: 395–404. [DOI] [PubMed] [Google Scholar]
- 51. Allcock AL, Strugnell JM, Prodohl P, Piatkowski U, Vecchione M (2007) A new species of Pareledone (Cephalopoda: Octopodidae) from Antarctic Peninsula Waters. Polar Biol 30: 883–893. [Google Scholar]
- 52. Hebert PDN, Cywinska A, Ball SL, DeWaard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London Series B-Biological Sciences 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moore WS (1995) Inferring phylogenies from mtDNA variation - mitochondrial-gene trees versus nuclear-gene trees. Evolution 49: 718–726. [DOI] [PubMed] [Google Scholar]
- 54. Toll R (1988) The use of arm sucker number in Octopodid systematics (Cephalopoda: Octopoda). Am Malacol Bull 6: 207–211. [Google Scholar]
- 55.Wilson B, Allen G (1987) Major components and distribution of marine fauna. pp 43–68 in: Dyne, G.R. & Walton, D.W. (eds.) Fauna of Australia: Volume 1A: General articles. Canberra: Australian Government Publishing Service. [Google Scholar]
- 56. O'Hara TD, Poore GCB (2000) Patterns of distribution of southern Australian marine echinoderms and decapods. J Biogeogr 27: 1321–1335. [Google Scholar]
- 57. O'Loughlin P, Waters JM, Roy MS (2003) A molecular and morphological of the asterinid, Patiriella gunnii (Gray) (Echinodermata: Asteroidea). Mem Mus Vic 60: 181–195. [Google Scholar]
- 58.Dartnall A (1974) Littoral biogeography: Biogeography and ecology in Tasmania (ed. by W.D. Williams). The Hague: Dr. Junk.
- 59. Waters JM, King TM, O'Loughlin PM, Spencer HG (2005) Phylogeographical disjunction in abundant high-dispersal littoral gastropods. Mol Ecol 14: 2789–2802. [DOI] [PubMed] [Google Scholar]
- 60. Thiel M, Gutow L (2005) The ecology of rafting in the marine environment. II. The rafting organisms and community. Oceanography and Marine Biology an Annual Review 43: 279–418. [Google Scholar]
- 61. Brasher DJ, Ovenden JR, Booth JD, White RWG (1992) Genetic subdivision of Australian and New Zealand populations of Jasus verreauxi (Decapoda, Palinuridae) - Preliminary evidence from the mitochondrial genome. N Z J Mar Freshwat Res 26: 53–58. [Google Scholar]
- 62. Ovenden JR, Brasher DJ, White RWG (1992) Mitochondrial DNA analyses of the red rock lobster Jasus edwardsii supports an apparent absence of population subdivision throughout Australasia. Mar Biol 112: 319–326. [Google Scholar]
- 63. Booth JD, Ovenden JR (2000) Distribution of Jasus spp. (Decapoda: Palinuridae) phyllosomas in southern waters: implications for larval recruitment. Mar Ecol Prog Ser 200: 241–255. [Google Scholar]
- 64. Grewe PM, Smolenski AJ, Ward RD (1994) Mitochondrial DNA diversity in jackass morwong (Nemadactylus macropterus: Teleostei) from Australian and New Zealand waters. Can J Fish Aquat Sci 51: 1101–1109. [Google Scholar]
- 65. Burridge CP, Smolenski AJ (2003) Lack of genetic divergence found with microsatellite DNA markers in the tarakihi Nemadactylus macropterus . N Z J Mar Freshwat Res 37: 223–230. [Google Scholar]
- 66. Villanueva R (1995) Experimental rearing and growth of planktonic Octopus vulgaris from hatching to settlement. Can J Fish Aquat Sci 52: 2639–2650. [Google Scholar]
- 67. Booth JD, Phillips BF (1994) Early life history of spiny lobster. Crustaceana 66: 271–294. [Google Scholar]
- 68. Burridge CP (1999) Molecular Phylogeny of Nemadactylus and Acantholatris (Perciformes: Cirrhitoidea: Cheilodactylidae), with Implications for Taxonomy and Biogeography. Mol Phylogen Evol 13: 93–109. [DOI] [PubMed] [Google Scholar]
- 69.Villanueva R, Norman MD (2008) Biology of the planktonic stages of benthic octopuses. Oceanography and Marine Biology: An Annual Review, Vol 46. Boca Raton: Crc Press-Taylor & Francis Group. pp. 105-+.
- 70. Chiswell SM, Wilkin J, Booth JD, Stanton B (2003) Trans-Tasman Sea larval transport: Is Australia a source for New Zealand rock lobsters? Mar Ecol Prog Ser 247: 173–182. [Google Scholar]
- 71. Smale MJ, Buchan PR (1981) Biology of Octopus vulgaris off the east coast of South Africa. Mar Biol 65: 1–12. [Google Scholar]
- 72. Strugnell J, Norman M, Drummond A, Cooper A (2004) Neotenous origins for pelagic octopuses. Curr Biol 14: R300–R301. [DOI] [PubMed] [Google Scholar]
- 73. Ridgway KR (2007) Long-term trend and decadal variability of the southward penetration of the East Australian Current. Geophys Res Lett 34: L13613. [Google Scholar]
- 74.REDMAP (2013). Available: http://www.redmap.org.au/region/tas/sightings/latest/.REDMAP. Accessed 2013 February 26.
- 75.Ling SD, Johnson CR, Frusher SD, Ridgway KR (2009) Overfishing reduces resilience of kelp beds to climate-driven catastrophic phase shift. Proceedings of the National Academy of Sciences. [DOI] [PMC free article] [PubMed]
- 76. Norman M, Kubodera T (2006) Taxonomy and biogeography of an Australian subtropical octopus with Japanese affinities. Proceedings of the 7th and 8th symposia on collection building and natural history studies in Asia and the Pacific Rim 1: 171–189. [Google Scholar]
- 77.Frakes I, McGowran B, Bowler J (1987) Evolution of Australian environments. pp 1–16 in: Dyne, G.R. & Walton, D.W. (eds.) Fauna of Australia: Volume 1A: General articles. Canberra: Australian Government Publishing Service. [Google Scholar]
- 78. Burridge CP, White RWG (2000) Molecular phylogeny of the antitropical subgenus Goniistius (Perciformes: Cheilodactylidae: Cheilodactylus): evidence for multiple transequatorial divergences and non-monophyly. Biol J Linn Soc 70: 435–458. [Google Scholar]
- 79. Valentine JW (1984) Neogene marine climate trends - Implications for biogeography and evolution of the shallow-sea biota. Geology 12: 647–650. [Google Scholar]
- 80. White BN (1986) The isthmian link, antitropicality and american biogeography - distributional history of the Atherinopsinae (Pisces, Atherinidae). Systematic Zoology 35: 176–194. [Google Scholar]
- 81. Teske P, Oosthuizen A, Papadopoulos I, Barker N (2007) Phylogeographic structure of Octopus vulgaris in South Africa revisited: identification of a second lineage near Durban harbour. Mar Biol 151: 2119–2122. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting tables. Table S1, Specimen information for individuals of which molecular sequencing was undertaken during the present study. Table S2, Specimen information for individuals accessed via GenBank for use in the present study. Table S3, Specimen information for individuals of which morphological traits were recorded during the present study. Table S4, Canonical correlation (CC) output for male octopod multivariate analysis. Table S5, Canonical loadings (CL) output for male octopod multivariate analysis. Table S6, Principal components (PC) calculated from canonical correlation and canonical loading outputs from male octopod multivariate analysis. Table S7, Eigenvalues and principal component (PC) variance contribution outputs from male octopod multivariate analysis. Table S8, Ranked canonical loadings (CL) from male octopod multivariate analysis; based upon contribution to principal components (PC). Table S9, Canonical correlation (CC) output for female octopod multivariate analysis. Table S10, Canonical loadings (CL) output for female octopod multivariate analysis. Table S11, Principal components (PC) calculated from canonical correlation and canonical loading outputs from female octopod multivariate analysis. Table S12, Eigenvalues and principal component (PC) variance contribution outputs from female octopod multivariate analysis. Table S13, Timing of divergence estimates (Tamura-Nei genetic distance) for Octopus tetricus (East Australia and New Zealand) and O. cf. tetricus (Western Australia). Table S14, Timing of divergence estimates (Tamura-Nei genetic distance) for the Australasian tetricus complex and Japanese/Chinese representatives of the Octopus vulgaris group.
(DOCX)