Abstract
Genetic deficits and loss of function for the triggering receptor expressed in myeloid cells 2 (TREM2; encoded at chr6p21.1), a transmembrane spanning stimulatory receptor of the immunoglobulin/lectin-like gene superfamily, have been associated with deficiencies in phagocytosis and the innate immune system in Alzheimer’s disease. In this study, we provide evidence that TREM2 is downregulated in samples of sporadic Alzheimer hippocampal CA1 compared with age-matched controls. A nuclear factor-κB (NF-κB)-sensitive miRNA-34a (encoded at chr1p36.22), upregulated in Alzheimer’s disease, was found to target the 299 nucleotide human TREM2 mRNA 3′-untranslated region (3′-UTR) and downregulate the expression of a TREM2-3′-UTR reporter vector. A stabilized anti-miRNA-34a (AM-34a) quenched this pathogenic response. The results suggest that an epigenetic mechanism involving an NF-κB-mediated, miRNA-34a-regulated downregulation of TREM2 expression may shape innate immune and phagocytic responses that contribute to inflammatory neurodegeneration.
Keywords: Alzheimer’s disease, amyloidosis, hippocampal CA1, innate immune response, microglial cells, miRNA-34a, NF-κB, phagocytosis, TREM2
Introduction
Alzheimer’s disease (AD) is a complex, progressive, and multifactorial brain dysfunction whose incidence is reaching epidemic proportions (http://www.alz.org/downloads/facts_figures 2012.pdf). Age-related deficits in the innate immune response and a compartmentalized proinflammatory degenerative neuropathology have long been associated with this progressive neurological disorder [1–4]. The mechanisms by which innate immune responses contribute to neuroinflammation and neurodegeneration remain incompletely understood. Microglial cells are a major resident myeloid-derived, TREM2-producing cell type in central nervous system (CNS) and are thought to fulfill important functions in immune surveillance, cell–cell interactions, tissue debris clearance, and the resolution of latent inflammatory reactions [3–7]. Absence of TREM2 expression on microglia not only impairs their capacity to phagocytose cellular debris but also increases their production of proinflammatory cytokines [4,6]. This idea is attractive because deficits in TREM2 could in part explain the loss of effective homeostatic phagocytic functions by microglial cells, the ensuing buildup of cellular debris such as amyloid-β (Aβ42) peptides, a progressive, ‘smoldering’, proinflammatory response associated with Aβ42 accumulation, and the chronic overproduction of proinflammatory cytokines.
In this study, to understand in more detail the molecular neurobiology and regulation of TREM2, we analyzed TREM2 expression in Alzheimer hippocampal CA1 samples and in cultured human primary brain cells. TREM2 protein levels in AD compared with age-matched control hippocampal CA1 were found to be downregulated. Inducible, upregulated proinflammatory microRNAs (miRNAs) have recently been associated with the downregulation of AD-relevant neuroimmune genes and immunological system deficits [8–14]. Using miRNA arrays, we next examined miRNA complexity and speciation to find a significant upregulation in miRNA-34a, a microglial-enriched miRNA previously shown to be under regulatory control by the proinflammatory transcription factor, nuclear factor-κB (NF-κB) [1,9,13]. Several independent bioinformatics algorithms next predicted that a 22 nucleotide region within the 299 nucleotide TREM2 mRNA 3′-untranslated region (3′-UTR) (chr6p21.1) is targeted by the human miRNA-34a (chr1p36.22). Transfection of a TREM2-3′-UTR luciferase reporter vector into microglial cells, when stressed, exhibited downregulation in the expression of the TREM2 reporter, and a stabilized anti-miRNA-34a (AM-34a) quenched these actions. Altogether, the results suggest that a miRNA-34a-mediated downregulation of TREM2 may be involved with impaired phagocytosis, innate immunological deficits, and inflammatory degeneration, which are characteristic features of the AD process.
Materials and methods
Reagents, AD and control tissues, and microglial cell cultures
Hydrogen peroxide solution (H2O2; 30 wt % in H2O; Sigma-Aldrich Chemical Company, St Louis, Missouri, USA) was used at a concentration of 60 µM for 1 h for the microglial cell cultures. Locked nucleic acids (LNA) and anti-miRNAs (AM-34a, AM-183) were purchased from Ambion (Invitrogen, Carlsbad, California, USA). Alzheimer’s and age-matched control tissues were analyzed for total miRNA using miRNA arrays, and TREM2 abundance was analyzed using Western analysis [9,11,14]. All Alzheimer samples were from adults and were obtained from the hippocampal CA1 region; the ApoE genotypes were not completely known. Murine CRL-2467 (C3H/HeJ) microglial cells were cultured according to the manufacturer’s protocols (ATCC, Manassas, Virginia, USA); transfections were performed using FuGENE6 transfection reagent as previously described [1,9,11]. Microglia-enriched cultures, used at 3 days and containing ~90% microglial cells and 10% astroglial and oligodendroglial cells, were prepared and assayed according to established methods [15,16].
Extraction of total RNA and protein and quality control
Total RNA and proteins were simultaneously isolated using TRIzol (Invitrogen) [1,9,11,12]. RNA quality was assessed using an Agilent Bioanalyzer 2100 (Lucent Technologies, Santa Clara, California, USA/Caliper Technologies, Hopkinton Massachusetts, USA) [12,14]. The RNA integrity number (RIN) values were typically between 8.0 and 9.0 [1,9,14]. The protein concentrations were determined using a dotMETRIC microassay (sensitivity 0.3 ng protein/ml; Millipore, Billerica, Massachusetts, USA) [11,12,14].
miRNA arrays and anti-miRNAs
miRNA labeling, hybridization, miRNA arrays, and RT-PCR analysis were performed as previously described [7,10–18]. LED-northern dot blot analysis was performed using a modified Bio-Dot microfiltration blotting device (detection limit=0.05 fM for a single miRNA species, apparatus #170-6545; BioRad Life Science Research, Hercules, California, USA) [9,11,12,19]. Ongoing studies suggest that LED-northern dot blots are a significant advancement over classical northern blotting techniques [18,19]. The miRNA and anti-miRNA (AM), as LNA oligonucleotides included a control miRNA-183 (5′-TATGGCACTGGTAGAATTCACT-3′), an anti-miRNA-183 (AM-183; 5′-AGTGAATTCTACCAGTGCCATA-3′), an anti-miRNA-34a (AM-34a; 5′-TCTTCCTGCTTTGTCTCTGCCT-3′), and a scrambled anti-miRNA-34a (AMsc-34a; 5′-CTTCTCGTTCTGTCTCTGTCTC-3′; Applied Biosystems/Ambion, Austin, Texas, USA or Exiqon Inc., Woburn, Massachusetts, USA [9,14] andwere used at an ambient concentration of 5–20 nM for a total treatment time of 12 or 36 h after H2O2-based treatment [9,11].
Western analysis of TREM2 and β-actin in brain tissues and microglial cells
Western immunoblotting was performed for quantification of TREM2 and β-actin protein using human-specific primary antibodies directed against the control protein marker β-actin (3598–100; Sigma-Aldrich Chemical Company) or human or murine TREM2 (B3; sc-373828, H160; sc-49764 or M227; sc-48765; Santa Cruz Biotechnologies, Santa Cruz, California, USA) [9,11].
Statistical analysis and data interpretation
All miRNA arrays were analyzed as previously described [1,11,12,14,18]. Statistical procedures for protein abundance were analyzed by a two-way factorial analysis of variance (P, ANOVA) using programs and procedures in the SAS language (Statistical Analysis Institute, Cary, North Carolina, USA) [11,14,18]. Only P-values less than 0.05 (ANOVA) were considered statistically significant. Figures were generated using Photoshop CS2 version 9.0.2 (Adobe, San Jose, California, USA).
Results
miRNA and LED-northern analysis of AD and age-matched control hippocampus samples
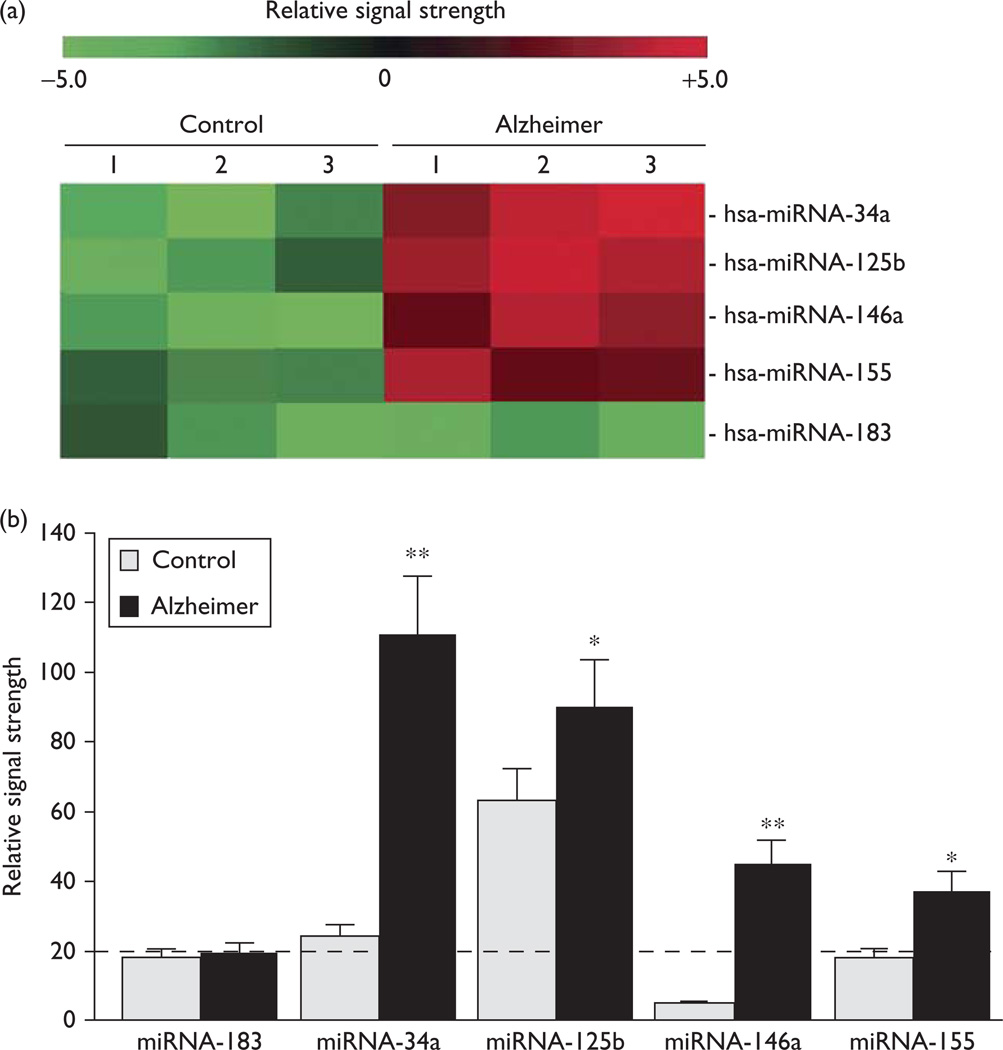
There were no significant differences in age, postmortem interval (PMI), total RNA yield, or quality between AD and control hippocampal CA1 samples used in this analysis, although drug history for this small sample set was not known. Of the five miRNAs found to be significantly upregulated (Fig. 1), miRNA-34a and miRNA-146a showed the greatest upregulation in AD samples, between 4.6- and nine-fold over controls (P < 0.01, ANOVA). Interestingly, all five miRNAs have previously been characterized as being under regulatory control by the proinflammatory transcription factor NF-κB and are inducible, according to the studies by Lukiw and colleagues [12,14,18]. However, as regards the upregulation of miRNA-34a in AD samples, it should be noted that these studies have limitations because of the small number of hippocampal CA1 (a total of N = 27 brains) samples, and that other miRNAs may participate in regulating the expression of TREM2.
Fig. 1.
Color-coded cluster analysis showing microRNA (miRNA) abundance in human hippocampal CA1. Fluorescent-based miRNA array data for the five most significantly upregulated miRNAs in control and Alzheimer hippocampal CA1 samples are shown in (a); all tissues were obtained from the hippocampal CA1 of the study group (controls N = 3; AD N = 3). There were no significant differences in the age (71.8±4.4 vs. 72.1±5.1 year, P~0.88), PMI (mean 2.0±1.8 vs. 2.0±1.5 h, P~0.94), RNA A260/280 indices (2.08±0.5 vs. 2.05±0.4, P~0.95), or RNA 28S/18S (1.48 vs. 1.44, P~0.95) between the age-matched controls and brain samples of Alzheimer patients, respectively. The RNA integrity numbers (RIN) ranged between 8.0 and 9.0. No significant differences in total RNA yield between the control and Alzheimer samples were noted; (b) similar results were obtained from an LED-northern dot blot assay [1,19]: the dashed horizontal line at 20.0 indicates control miRNA-183 signal in Alzheimer’s disease for the ease of comparison; miRNA-183 is an unchanging control miRNA that has been used as an internal control in previous studies [14,19]; miRNAs were analyzed by LC Sciences (Houston, Texas, USA) using miRNA array panels containing 1898 individual human miRNA targets. Interestingly, miRNA-34a, miRNA-125b, miRNA-146a, and miRNA-155 have each been previously characterized as being inducible, NF-κB-regulated miRNAs; *P<0.05; **P<0.01 (analysis of variance) [8–16].
miRNA-34a immune-relevant mRNA targets and downregulation of TREM2 protein
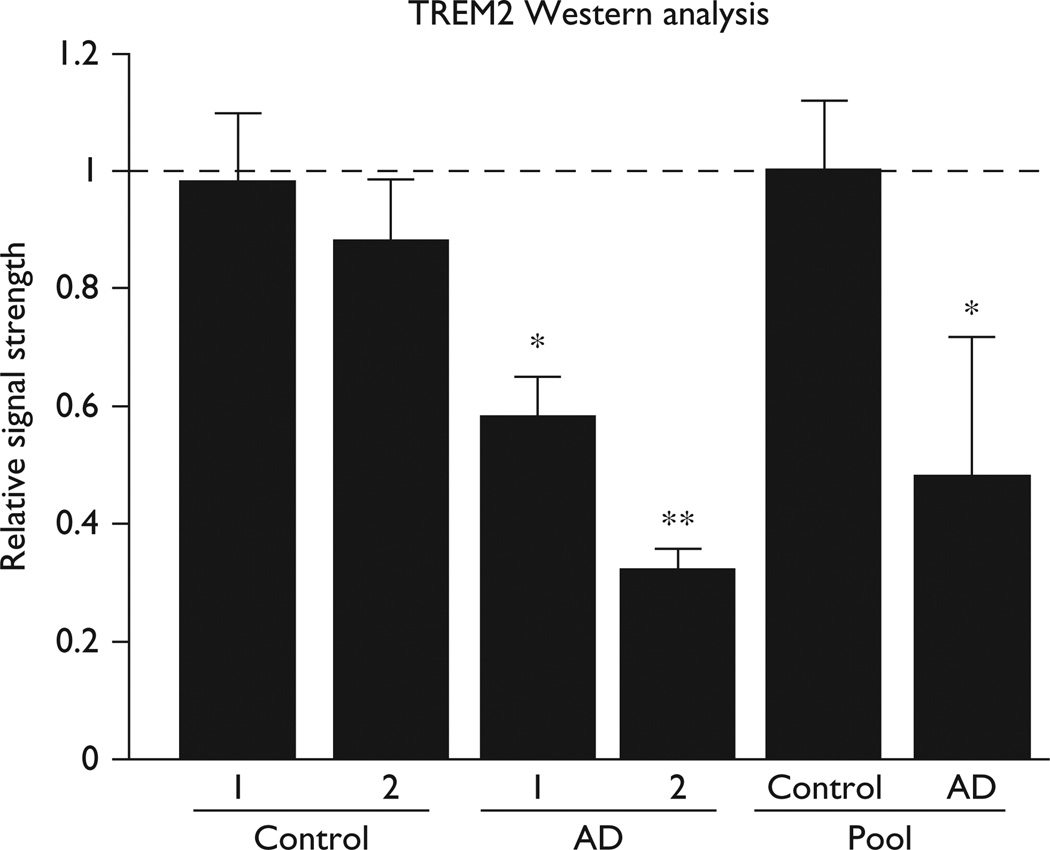
Because upregulated proinflammatory miRNAs have been associated with downregulation in the expression of Alzheimer-relevant neuroimmune genes and immunological system deficits, we systematically searched for potential miRNA-34a targets, and a strong candidate was the hsa-miRNA-34a-TREM2 mRNA 3′-UTR interaction (Fig. 2). Subsequently, Western analysis indicated that TREM2 protein levels were reduced in Alzheimer hippocampal CA1 samples and the mean of nine AD samples ranged between 0.32 and 0.58 of age-matched controls (Fig. 3) [20].
Fig. 2.

hsa-miRNA-34a-TREM2-mRNA-3′-UTR complementarity map. The free energy of association (EA) between hsa-miRNA-34a and the TREM2 mRNA-3′-UTR is ~16 kcal/mol; the miRNA-34a seed sequence 3′-UGUGACGG-5′ is highlighted in yellow; the complementary TREM2-3′-UTR recognition sequence 5′-ACACTGCT-3′ is highlighted in red; an ‘|’ indicates a full hydrogen bond between miRNA-34a and the TREM2-mRNA-3′-UTR and a ‘:’ indicates a partial hydrogen bond; the hsa-miRNA-34a recognition feature is located about midway in the 299 basepair TREM2-3′-UTR; several other brain miRNAs located within the TREM2-3′-UTR and may also affect TREM2 mRNA stability and regulate its expression (data not shown); notably, the TREM2 gene has no strong NF-κB binding site within at least 11 kb of its transcription start site; ribonucleotide sequences and alignment derived using miRBASE algorithms (European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton UK; http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/detail_view.pl?transcript_id=ENST00000373113); [3–6,8,20].
Fig. 3.
Western analysis in control and AD hippocampal CA1. Quantitation of levels of the 25.4 kDa TREM2 protein compared with the 42.2 kDa β-actin (ACTB; 374 amino acids) in the same sample; the highest abundance of TREM2 was found in the hippocampal CA1 at levels about 5 × more abundant compared with the neocortex; the two controls and two AD samples are obtained from short PMI tissues having a PMI of 2 h or less (further described in the text and legend to Fig. 1); results using TREM2 antibody [sc-373828 shown; the pools of control (N = 9)] and AD (N = 12) were derived from pooled whole brain extracts of short PMI tissues having a mean PMI of ~2 h; age range 66–74 for control and AD samples; TREM2 protein levels in AD samples and pools ranged between 0.32 and 0.58 of controls; *P<0.05; **P<0.01 (analysis of variance) [8–16]. AD, Alzheimer’s disease; PMI, postmortem interval.
Functional validation of hsa miRNA-34a-TREM2-3′-UTR interaction
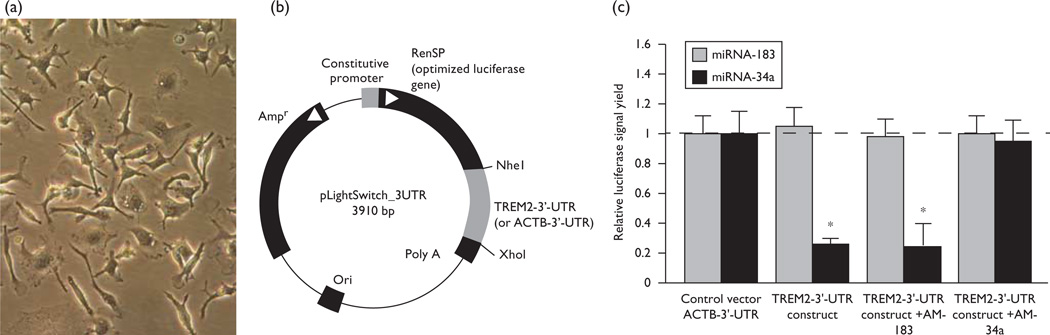
To establish that miRNA-34a is in fact involved with TREM2 expression, we used a TREM2-mRNA-3′-UTR expression vector luciferase reporter assay (pLightSwitch -3′-UTR; Cat #S801178; Switchgear Genomics, Menlo Park, California, USA). The results indicated that in stressed murine microglial cells, compared with several different controls, including a human β-actin (ACTB) 3′-UTR, a related brain-enriched miRNA-183, and an anti-miRNA-183 or scrambled anti-miRNA-34a, the miRNA-34a is involved in the downregulation of TREM2 expression.
Discussion
miRNAs are a recently discovered class of single stranded noncoding ribonucleotide regulators that, through base-pair complementarity, bind to the 3′-UTR of highly selective target mRNAs and direct the post-transcriptional repression of that mRNA’s genetic information (Figs 1 and 2) [8,9,21]. A family of CNS-enriched miRNAs, upregulated in brains of AD patients, are known to be under NF-κB control and are involved in the regulation of innate immune sensing and inflammatory responses, neurotrophism, synaptogenesis, and amyloidogenesis [9–14]. For example, an NF-κB-regulated miRNA subfamily including miRNA-9, miRNA-125b, miRNA-146a, and miRNA-155 targets the 3′-UTR of the key innate immune system-related and inflammation-related regulatory protein complement factor H (CFH), resulting in significant decreases in CFH expression, with subsequent stimulation of the innate immune response and chronic NF-κB activation [1,9,14]. Interestingly, the brain-abundant miRNA-34a has recently been implicated in endothelial cell senescence in the vasculature, in part through the NAD-dependent deacetylase sirtuin-1 (SIRT1) [21–23].
TREM2 is a glycosylated innate immune receptor expressed on the plasma membrane by a subset of myeloid cells including immature dendritic cells, tissue macrophages, and myeloid-derived microglia and is an integral part of the evolutionarily ancient innate immune and complement signaling system [2,3]. Signaling through TREM2 or its adaptor protein TYROBP (also called DAP12) is known to play neuroprotective roles through the clearance of noxious cellular debris from within the CNS, the resolution of damage-associated inflammation, and the phagocytosis of pathogens, which is accompanied by the release of reactive oxygen species [2–6]. Downregulated TREM2 may well interfere with the brain’s natural ability to deal with excessive levels of the Aβ42 peptide, resulting in their accumulation, self-aggregation, and ultimate formation into senile plaque [5,6]. Interestingly, extracellular biologically active TREM2 in the human CSF suggests a mobile phagocytotic and anti-inflammatory surveillance and signaling system; hence, soluble forms of TREM2 may extend the influence of TREM2 well outside of the cell types that express them [4–7,17]. Regional variation in TREM2 expression in the CNS may contribute to the varying sensitivities of different brain regions to similar pathological signals [5]. Rare mutations of TREM2 (or of its coupling protein, DAP12, also known as TYROBP) are currently associated with the progressive presenile dementing illnesses such as Nasu–Hakola syndrome, polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (POSL), and/or AD [4–7]. Given these anti-inflammatory and phagocytotic roles of TREM2 in the brain, loss of function or downregulation of TREM2 may have analogous proinflammatory and/or amyloidogenic effects. In other words, mutations in TREM2 leading to defective TREM2 or downregulation in the expression of a biologically active TREM2 may have the same end effects. Interestingly, the recently described deficiencies and involvement of CFH and TREM2 in AD underscores an emerging interwoven role of potential microbial pathogen involvement, Aβ42 peptide participation, and an altered innate immune response in driving chronic inflammatory aspects of the Alzheimer process [21–24]. These complex and progressive pathologies might be prevented by combined antibiotic, antiviral, and anti-inflammatory approaches, in part by using anti-miRNA (AM)-based strategies and/or stabilized miRNA derivatives.
Conclusion
This study is the first to show miRNA-based regulation of TREM2 and provides five new observations: (a) a proinflammatory NF-κB-regulated miRNA-34a is increased in abundance in AD hippocampal CA1 (Fig. 1); (b) TREM2 expression is downregulated in AD hippocampal CA1 (Fig. 2); (c) using complementarity algorithms, a miRNA-34a recognition feature is apparent in the human TREM2-3′UTR (Fig. 3); (d) a TREM2-3′UTR-luciferase reporter vector is significantly down-regulated in stressed microglial cells, and (e) this downregulation can be rescued to homeostatic levels by adding exogenous AM-34a (Fig. 4). These findings underscore the importance of miRNA as epigenetic regulators of sporadic AD-relevant gene expression. The potential of anti-miRNA (AM) for improving our understanding of the pathogenesis of inflammatory neurodegenerative diseases and for developing efficacious AM-based treatment strategies may be substantial, not only in AD but also in other progressive age-related degenerations of the human CNS.
Fig. 4.
Functional validation of hsa miRNA-34a-TREM2–3′UTR interaction. (a) CRL-2467 murine microglial cells, phase contrast 20×. (b) TREM2-mRNA-3′-UTR expression vector luciferase reporter assay (pLightSwitch-3′UTR; Cat #S801178; Switchgear Genomics). In this vector, the entire 299 nucleotide TREM2 3′-UTR was ligated into the unique Nhe1-Xho1 site. The transfected cells were treated exogenously with a control LNA-protected miRNA-183 or miRNA-34a and/or anti-miRNA-183 (AM-183), anti-miRNA-34a (AM-34a), or a scrambled anti-miRNA-34a (AMsc-34a) [18,19]. (c) Although the control vector ACTB-3′-UTR showed no significant effects on the relative luciferase signal yield after treatment with either miRNA-183 or miRNA-34a (dashed horizontal line set to 1.0), the TREM2-mRNA-3′-UTR vector exhibited reduced luciferase signal to a mean of 0.22-fold over controls. An exogenously added anti-miRNA-183 (AM-183) control or a scrambled anti-miRNA-34a (AMsc-34a; data not shown) showed no effect on the luciferase signal, whereas an anti-miRNA-34a (AM-34a) restored luciferase expression to near homeostatic levels; N = 3; *P<0.01 (analysis of variance). The results suggest an miRNA-34a-mediated downregulation of TREM2 expression in stressed microglial cells may be related to the downregulation of other immune system genes by proinflammatory miRNAs (such as complement factor H) [8,9,14] and/or an impairment in cellular phagocytosis or related signaling [5–7,20,21].
Acknowledgements
The authors thank Drs. L. Carver, E. Head, W. Poon, G. Tejada, H. LeBlanc, C. Eicken, and C. Hebel for human brain tissues or extracts, miRNA array work, and initial data interpretation and D. Guillot and A.I. Pogue for expert technical assistance. Additional hippocampal and other brain tissues were provided by the Memory Impairments and Neurological Disorders (MIND) Institute and the Alzheimer’s Disease Research Center of the University of California, Irvine (UCI-ADRC; NIA P50 AG16573). Research on miRNA in the Lukiw laboratory involving AD innate immune response and neuroinflammation was supported through Translational Research Initiative Grants from LSUHSC, Alzheimer Association Investigator-Initiated Research Grant IIRG-09–131729, and NIA Grants AG18031 and AG038834. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
These studies were presented in part at the 42nd Annual Society for Neuroscience Meeting, New Orleans Louisiana, 13–17 October 2012.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Lukiw WJ, Bhattacharjee S, Dua P, Alexandrov PN. Common micro RNAs (miRNAs) target complement factor H (CFH) regulation in Alzheimer’s disease (AD) and in age-related macular degeneration (AMD) Int J Biochem Mol Biol. 2012;3:105–116. [PMC free article] [PubMed] [Google Scholar]
- 2.Saijo K, Crotti A, Glass CK. Regulation of microglia activation and deactivation by nuclear receptors. Glia. 2013;61:104–111. doi: 10.1002/glia.22423. [DOI] [PubMed] [Google Scholar]
- 3.Aiyaz M, Lupton MK, Proitsi P, Powell JF, Lovestone S. Complement activation as a biomarker for Alzheimer’s disease. Immunobiology. 2012;217:204–215. doi: 10.1016/j.imbio.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 4.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 5.Schmid CD, Sautkulis LN, Danielson PE, Cooper J, Hasel KW, Hilbush BS, et al. Heterogeneous expression of the triggering receptor expressed on myeloid cells-2 on adult murine microglia. J Neurochem. 2002;83:1309–1320. doi: 10.1046/j.1471-4159.2002.01243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neumann H, Takahashi K. Essential role of the microglial triggering receptor expressed on myeloid cells-2 (TREM2) for central nervous tissue immune homeostasis. J Neuroimmunol. 2007;184:92–99. doi: 10.1016/j.jneuroim.2006.11.032. [DOI] [PubMed] [Google Scholar]
- 7.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, et al. TREM2 Variants in Alzheimer’s Disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Wang K, Chen X, Meng H, Song M, Wang Y, et al. Transcriptional activation of microRNA-34a by NF-kB in human cancer cells. BMC Mol Biol. 2012;13:4. doi: 10.1186/1471-2199-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukiw WJ, Zhao Y, Cui JG. An NF-kB-sensitive miRNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soreq H, Wolf Y. NeurimmiRs: microRNAs in the neuroimmune interface. Trends Mol Med. 2011;17:548–555. doi: 10.1016/j.molmed.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Cui JG, Li YY, Zhao Y, Bhattacharjee S, Lukiw WJ. Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-kB in stressed human astroglial cells and in Alzheimer disease. J Biol Chem. 2010;285:38951–38960. doi: 10.1074/jbc.M110.178848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukiw WJ. NF-κB-regulated micro RNAs (miRNAs) in primary human brain cells. Exp Neurol. 2012;235:484–490. doi: 10.1016/j.expneurol.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veremeyko T, Starossom SC, Weiner HL, Ponomarev ED. Detection of miRNAs in microglia by real-time PCR in normal CNS and during neuroinflammation. J Vis Exp. 2012 doi: 10.3791/4097. doi:pii: 4097. 10.3791/4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukiw WJ, Alexandrov PN. Regulation of complement factor H (CFH) by multiple miRNAs in Alzheimer’s disease (AD) brain. Mol Neurobiol. 2012;46:11–19. doi: 10.1007/s12035-012-8234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirrlinger J, Resch A, Gutterer JM, Dringen R. Oligodendroglial cells in culture effectively dispose of exogenous hydrogen peroxide: comparison with cultured neurons, astroglial and microglial cells. J Neurochem. 2002;82:635–644. doi: 10.1046/j.1471-4159.2002.00999.x. [DOI] [PubMed] [Google Scholar]
- 16.Giulian D, Baker TJ. Characterization of ameboid microglia isolated from developing mammalian brain. J Neurosci. 1986;6:2163–2178. doi: 10.1523/JNEUROSCI.06-08-02163.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. NeuroReport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 18.Li YY, Cui JG, Dua P, Pogue AI, Bhattacharjee S, Lukiw WJ. Differential expression of miRNA-146a-regulated inflammatory genes in human primary neural, astroglial and microglial cells. Neurosci Lett. 2011;499:109–113. doi: 10.1016/j.neulet.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukiw WJ, Alexandrov PN, Zhao Y, Hill JM, Bhattacharjee S. Spreading of Alzheimer’s disease inflammatory signaling through soluble micro-RNA. NeuroReport. 2012;23:621–626. doi: 10.1097/WNR.0b013e32835542b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.TREM2 gene expression data. Available at: http://www.genecards.org/cgi-bin/carddisp.pl?gene=TREM2. [Google Scholar]
- 21.Qin B, Yang H, Xiao B. Role of microRNAs in endothelial inflammation and senescence. Mol Biol Rep. 2012;39:4509–4518. doi: 10.1007/s11033-011-1241-0. [DOI] [PubMed] [Google Scholar]
- 22.Sonkoly E, Ståhle M, Pivarcsi A. MiRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–140. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, Lukiw WJ. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70:462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- 24.Schulte LN, Westermann AJ, Vogel J. Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res. 2013;41:542–553. doi: 10.1093/nar/gks1030. [DOI] [PMC free article] [PubMed] [Google Scholar]