Generally, a protein molecule should form its native structure to perform its biological function in the living cell. However, the failure of proper folding, i.e. misfolding, occurs occasionally and this causes aggregation and/or degradation of the protein, which is potentially linked to protein misfolding diseases, e.g. prion disease and Parkinson’s disease.1 Therefore, it is critical to understand the misfolding mechanism, as well as protein folding, in order to develop preventative procedures.
Among the numerous available proteins, horse heart cytochrome c (Cyt c) is one of the most intensively studied model system (Figure 1(a)).2 Cyt c has a fully unfolded structure under a certain concentration of denaturant, and external ligands such as carbon monoxide (CO) are easily bound to the iron in the heme group. Many previous experiments have been conducted in order to examine the folding mechanism of Cyt c using the unfolded Cyt c bound with CO. An optical pump pulse has been used to initiate folding by dissociating CO from the heme group and its time-dependent processes have been probed using various spectroscopic techniques.3–7 Although the existing studies have proposed the possible kinetic models for the folding of Cyt c, a unified kinetic model has not yet been established due to the complexities that are caused by the misligation between histidine 26/33 (His26/33) and iron. This abnormal interplay disturbs the binding of methionine 80 (Met80) to the iron and may lead to misfolding.
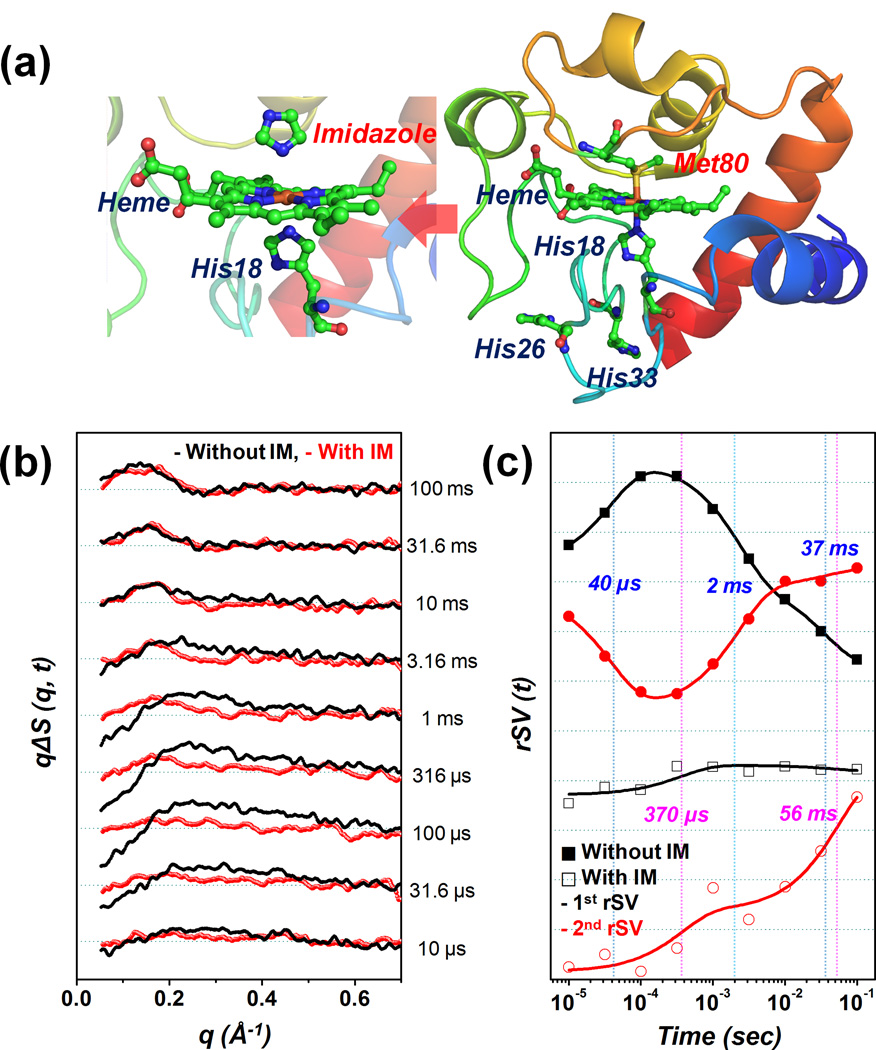
Figure 1.
(a) (Right) Native structure of horse heart cytochrome c (PDB: 1HRC) and (left) the transient binding between the imidazole and iron on the heme. (b) Time-resolved X-ray solution scattering data for the folding of cytochrome c after dissociating CO from the heme group. The red and black lines indicate the presence and absence of imidazole (IM), respectively. (c) The first and second right singular vectors scaled using singular values from the SVD. In order to determine the time constants for each case, the components were fitted simultaneously using the sum of the exponentials.
In this study, we explore the effect of misligation on the folding pathway of Cyt c using pump-probe (time-resolved) X-ray solution scattering.8–10 We employ imidazole as an inhibitor that blocks the misligation. Imidazole has a similar structure to that of histidine (Figure 1(a)); due to this structural similarity, imidazole can transiently bind to iron.11 This interaction can remove the complex ligand exchange process of misligated residues such as histidine 26 or 33. It is expected that the addition of imidazole at 50 times greater content than that of Cyt c will suppress the misligation between the His26/33 and heme through the transient binding of imidazole to the heme.
Figure 1(b) presents two sets of difference scattering curves (i.e. with and without imidazole), ΔS(q, t), which are obtained by subtracting the laser-off scattering curve (reference) measured at −5 µs from the laser-on scattering curve at various positive time delays. (Refer to the Supporting Information for further experimental details.) A quantitative kinetic analysis of the difference scattering curves was implemented using the singular value decomposition (SVD). (Refer to the Supporting Information for further details.) From the SVD analysis for each data set, the first two time-dependent components (right singular vectors; rSVs) weighted by their corresponding singular values were simultaneously fit according to the sum of exponentials in order to determine the apparent time constants. This yielded 40 (± 4) µs, 2.3 (± 0.2) ms, and 37 (± 9) ms for the sample without imidazole, and 370 (± 230) µs and 56 (± 41) ms for the sample with imidazole (Figure 1(c)).
Among the time constants determined for the case without imidazole, the 40 µs time constant is the candidate for the misligation between the His26/33 and heme. According to previous time-resolved circular dichroism (TRCD) and transient absorption (TA) experiments,5,6 the time constant of several tens of µs (50 µs) was determined through a kinetic analysis and was assigned to the misligation process between the His26/33 and heme. Hagen et al.3 also reported the misligation process with a time constant of approximately 100 µs. Even though the value of the time constant related to the misligation varies depending on the probing method and experimental conditions, the previous results have demonstrated the existence of misligation at several tens to hundreds of µs.3–6 The 40 µs kinetic component accompanies the appearance of a negative signal in the small angle region (< 0.15 Å−1). The negative signal that is already present at 10 µs becomes more pronounced as time progresses (Figure 1(b)). The negative signal that is associated with the time constant of 40 µs in this study can be linked to the transient binding between the His26/33 and heme.
In contrast, in the presence of imidazole, the difference scattering curve at the early µs is quite flat for the entire q range and forms a strong positive signal in the small angle region (< 0.25 Å−1) as time passes from 10 µs to 100 ms (Figure 1(b)). The absence of a strong negative signal in the presence of imidazole indicates that the associated time constant (370 µs) is unlikely due to misligation.
The positive signals at the small q values (< 0.25 Å−1) begin to increase from early ms without imidazole and hundreds of µs with imidazole. This positive feature, which was developed using time constants of 2.3 ms without imidazole and 370 µs with imidazole, can be assigned to the Cyt c folding dynamics that accompanies the global conformational change of the protein. Based on previous studies,3,4,7 the normal ligation between the Met80 and heme has already formed at the early ms, thereby the folded form (or native form) is the most populated in the late ms through the contraction of global protein structure. These considerations enable the suggestion that the formation of the native form is accelerated when imidazole is present. This finding is consistent with the rapid mixing experiment12 combined with the fluorescence measurement, the results of which suggested the faster formation of the folded structure in the presence of imidazole. Although the fluorescence quenching only reflects the local structural change between the tryptophan 59 (Trp 59) and heme, and the folding trigger differs to that used in this study (COphotolysis), the structural contraction of the unfolded form is clearly promoted through the suppression of misligation.
The two time constants (2.3 ms and 37 ms) observed in the absence of imidazole may correspond with the ligand replacement from His26/33 to Met80 (which causes folding) and the successive minor rearrangement to form the native structure, respectively. In the presence of imidazole, the complex kinetics including the ligand exchange process was replaced by a single component (56 ms). In the difference scattering curve, an oscillatory feature (peak-to-valley shape) appears around 10 ms in the middle angle region (0.1–0.4 Å−1) and becomes prominent at 100 ms in both cases. The build-up of this feature may be linked to the minor structural changes, e.g. the rearrangement of the secondary structure or some residues, rather than the major folding of global protein structure.
In summary, the time-resolved X-ray solution scattering data provide evidence that the misligation between the His26/33 and heme is suppressed with the addition of imidazole as a misligation inhibitor. The suppression of misligation accelerates the formation of the folded structure in the Cyt c folding. Compared with the previous experiment12 using the fluorescence quenching method, which is only sensitive to local structural change, the result in this study unravels the effect of misligation on Cyt c folding with respect to the global conformational change. Additional experiments with mutants of Cyt c that are designed to block the misligation and more detailed analyses should be conducted in order to establish the global kinetic framework of the Cyt c folding.
Supplementary Material
Acknowledgments
This work was supported by Institute for Basic Science (IBS) [CA1401-01]. Use of the BioCARS Sector 14 at the APS was supported by NIH National Institute of General Medical Sciences grant P41GM103543. The timeresolved set up at Sector 14 was funded in part through collaboration with P. Anfinrud (NIH/NIDDK) through the Intramural Research Program of the NIDDK. Use of the APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. DE-AC02-06CH11357.
Footnotes
This paper is to commemorate Professor Myung Soo Kim's honourable retirement.
Supporting Information. Experimental details and singular value decomposition (SVD) analysis.
References
- 1.Dobson CM. Nature. 2003;426:884. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Bushnell GW, Louie GV, Brayer GD. J. Mol. Biol. 1990;214:585. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 3.Hagen SJ, Latypov RF, Dolgikh DA, Roder H. Biochemistry. 2002;41:1372. doi: 10.1021/bi011371a. [DOI] [PubMed] [Google Scholar]
- 4.Jones CM, Henry ER, Hu Y, Chan CK, Luck SD, Bhuyan A, Roder H, Hofrichter J, Eaton WA. Proc. Natl. Acad. Sci. USA. 1993;90:11860. doi: 10.1073/pnas.90.24.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldbeck RA, Thomas YG, Chen EF, Esquerra RM, Kliger DS. Proc. Natl. Acad. Sci. USA. 1999;96:2782. doi: 10.1073/pnas.96.6.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen EF, Wood MJ, Fink AL, Kliger DS. Biochemistry. 1998;37:5589. doi: 10.1021/bi972369f. [DOI] [PubMed] [Google Scholar]
- 7.Choi J, Yang C, Kim J, Ihee H. J. Phys. Chem. B. 2011;115:3127. doi: 10.1021/jp106588d. [DOI] [PubMed] [Google Scholar]
- 8.Kim TW, Lee JH, Choi J, Kim KH, van Wilderen LJ, Guerin L, Kim Y, Jung YO, Yang C, Kim J, Wulff M, van Thor JJ, Ihee H. J. Am. Chem. Soc. 2012;134:3145. doi: 10.1021/ja210435n. [DOI] [PubMed] [Google Scholar]
- 9.Cammarata M, Levantino M, Schotte F, Anfinrud PA, Ewald F, Choi J, Cupane A, Wulff M, Ihee H. Nat. Methods. 2008;5:988. doi: 10.1038/nmeth.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ihee H, Lorenc M, Kim TK, Kong QY, Cammarata M, Lee JH, Bratos S, Wulff M. Science. 2005;309:1223. doi: 10.1126/science.1114782. [DOI] [PubMed] [Google Scholar]
- 11.Muthukrishnan K, Nall BT. Biochemistry. 1991;30:4706. doi: 10.1021/bi00233a010. [DOI] [PubMed] [Google Scholar]
- 12.Chan CK, Hu Y, Takahashi S, Rousseau DL, Eaton WA, Hofrichter J. Proc. Natl. Acad. Sci. USA. 1997;94:1779. doi: 10.1073/pnas.94.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.