This study provides novel insight into the formation and resolution of telomere associations, which have been observed during key cellular processes such as mitosis, meiosis, and carcinogenesis. TRF1, a core component of the telomere protein complex, is a mediator of telomere associations in mammalian cells.
Abstract
Telomere associations have been observed during key cellular processes such as mitosis, meiosis, and carcinogenesis and must be resolved before cell division to prevent genome instability. Here we establish that telomeric repeat-binding factor 1 (TRF1), a core component of the telomere protein complex, is a mediator of telomere associations in mammalian cells. Using live-cell imaging, we show that expression of TRF1 or yellow fluorescent protein (YFP)-TRF1 fusion protein above endogenous levels prevents proper telomere resolution during mitosis. TRF1 overexpression results in telomere anaphase bridges and aggregates containing TRF1 protein and telomeric DNA. Site-specific protein cleavage of YFP-TRF1 by tobacco etch virus protease resolves telomere aggregates, indicating that telomere associations are mediated by TRF1. This study provides novel insight into the formation and resolution of telomere associations.
INTRODUCTION
Telomeres are specialized protective structures at the ends of linear chromosomes composed of short tandem DNA repeats and associated proteins (Blackburn, 1991). Telomere integrity is maintained by shelterin, a six-subunit complex composed of TRF1, TRF2, TIN2, TPP1, POT1, and RAP1 (de Lange, 2005). TRF1 binds telomeres as a dimer and is present at telomeres throughout the cell cycle (Zhong et al., 1992; Chong et al., 1995; Bianchi et al., 1997). TRF1 contains a dimerization domain and a Myb DNA–binding domain, connected by a spatially flexible linker (Broccoli et al., 1997; Bianchi et al., 1999). TRF1 has been described as a negative regulator of telomere length because long-term overexpression of TRF1 leads to gradual telomere shortening, whereas expression of a dominant-negative TRF1 mutant leads to telomere elongation (van Steensel and de Lange, 1997). TRF1 deletion is lethal in the mouse embryo, but no evidence of impaired telomere length or deregulated telomerase activity was observed (Karlseder et al., 2003), suggesting that TRF1 has an essential function that is separate from its role in telomere length regulation.
TRF1 has been implicated in the formation of telomere associations during cell division and carcinogenesis. Associations between sister telomeres have been observed during mitosis upon depletion of tankyrase 1 (a negative regulator of TRF1), with cells arresting in early anaphase with unresolved sister telomeres (Dynek and Smith, 2004). Telomere aggregates between telomeres from multiple chromosomes have been observed under both physiological and pathological conditions. During meiotic prophase I of diverse eukaryotes, telomeres naturally cluster together on one side of the nucleus to form a telomere bouquet arrangement, which is believed to facilitate the alignment of chromosomes before homologous recombination and cell division. Taz1, the TRF1/TRF2 fission yeast orthologue, is required for stable association of telomeres during formation of the telomere bouquet (Cooper et al., 1998) and promotes semiconservative DNA replication through telomeres (Miller et al., 2006). Large telomere aggregates (Chuang et al., 2004; Goldberg-Bittman et al., 2008; Gadji et al., 2012), as well as TRF1 overexpression and depletion (Matsutani et al., 2001; Miyachi et al., 2002; Yamada et al., 2002a, b; Oh et al., 2005; Lin et al., 2006), have been observed in various human cancers; however, the link between telomere aggregation and elevated TRF1 levels is not clear. In vitro, TRF1 protein concentrations at near-saturating levels induce parallel pairing and clustering of telomeric DNA tracts (Griffith et al., 1998; Kim et al., 2003).
Recent studies using enhanced green fluorescent protein–TRF1 implicated TRF1 as a stem cell marker, in that elevated TRF1 levels were found in stem cells and correlated with the formation of teratomas and chimeras (Schneider et al., 2013). However, the role of TRF1 levels in telomere dynamics and telomere associations in actively dividing stem cells remains unclear. Elevated TRF1 levels have also been linked to cell cycle progression; TRF1 protein levels appear to increase in mitosis and decrease in G1 and S phase (Shen et al., 1997; Zhu et al., 2009). TRF1 association with telomeric chromatin was also observed to increase in mitosis and decrease as cells exit mitosis in Xenopus cell extracts (Nishiyama et al., 2006). However, it is not known how TRF1 levels affect telomere associations in cells or how important the regulation of TRF1 levels is for proper telomere maintenance.
Here we use high-resolution live-cell imaging to demonstrate that elevated TRF1 levels induce telomere associations in mouse embryonic stem cells. This study demonstrates that precise regulation of TRF1 levels is critical for resolution of telomeres before mitosis to allow proper cell cycle progression and provides novel insight into the role of TRF1 in telomere associations.
RESULTS
Generating cell populations expressing defined TRF1 protein levels
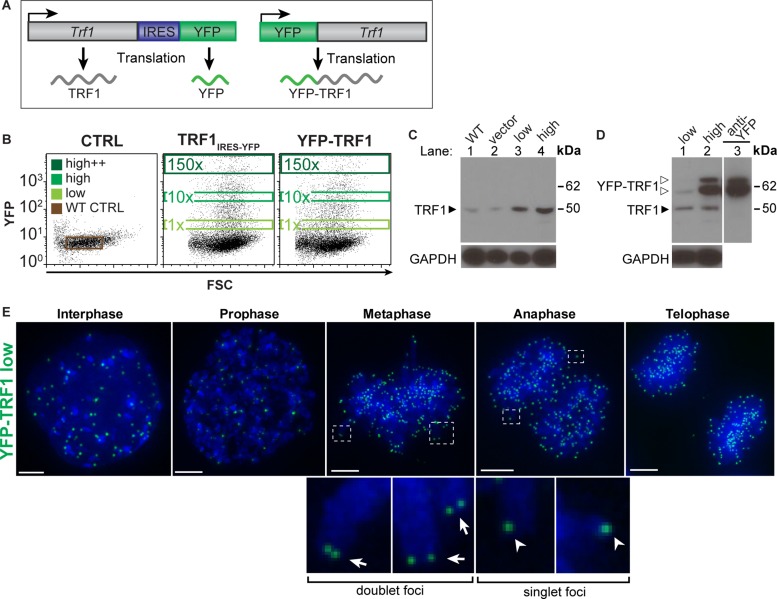
To investigate the role of TRF1 in telomere associations, we followed cells expressing defined levels of fluorescently labeled TRF1 by live-cell imaging. Constructs encoding either Trf1 fused to Venus yellow fluorescent protein (YFP-TRF1; Nagai et al., 2002) or Trf1 cotranscribed but translated separately from YFP by an internal ribosome entry site (IRES) domain (TRF1IRES-YFP) were transfected into mouse embryonic stem (ES) cells (Figure 1A). These cells were sorted at 24 h posttransfection by fluorescence-activated cell sorting (FACS) with gating for low (1×), high (10×), or high++ (150×) YFP fluorescence levels (Figure 1B). Western blot analysis shows that YFP can be used as an indicator of TRF1 protein levels in both the YFP-TRF1 and TRF1IRES-YFP strategies (Figure 1, B–D). Two bands around 62 kDa were observed in extracts from cells overexpressing YFP-TRF1. The nature of these two bands is unclear. Despite this uncertainty, we estimate the levels of transfected YFP-TRF1 protein in the low and high populations to be 0.5- to 1-fold and 5- to 10-fold endogenous TRF1 protein levels, respectively (Figure 1D).
FIGURE 1:
Generating cell populations expressing defined TRF1 protein levels. (A) Schematic of constructs encoding YFP-TRF1 (top) and TRF1 translated separately from YFP (bottom), driven by a CAG promoter. (B) FACS plots showing wild-type control, vector control (IRES-YFP), TRF1IRES-YFP, and YFP-TRF1 transfected cells at 24 h posttransfection and gates used to sort negative, low, high, and high++ YFP populations. Numbers in gates indicate fold difference in median YFP levels compared with the low population. (C) Western blot analysis of wild-type control, vector control (IRES-YFP), and TRF1IRES-YFP low and high sorted cell populations (lanes 1–4, respectively) and (D) YFP-TRF1 low (lane 1) and high (lane 2) sorted cell populations using anti-TRF1 antibody or anti-YFP antibody (D, lane 3). Bottom, GAPDH loading control. The molecular weights of TRF1 (solid arrowhead) and YFP-TRF1 (open arrowheads) are indicated. (E) Localization of low YFP-TRF1 (green) over the cell cycle with DAPI DNA stain (blue). Foci doublets in metaphase (insets, arrows) and singlets in anaphase (insets, arrowheads). Images are maximum-intensity projections.
Telomere dynamics can be visualized by fluorescently labeled TRF1
To determine whether the low–YFP-TRF1 population can be used to track telomere dynamics, we imaged cells stably expressing low YFP-TRF1 levels over the cell cycle (Figure 1E and Supplemental Movie S1). In interphase, YFP-TRF1 foci were distributed throughout the nuclear volume. In metaphase and anaphase, YFP-TRF1 foci localized to sister-chromatid ends, suggesting that low levels of YFP-TRF1 correctly localize to telomeres over the cell cycle. As expected, individual TRF1 foci nearly doubled from G1 to G2/M upon sister chromatid separation, with numbers corresponding closely to the expected number of telomeres (Supplemental Figure S1). Therefore cells expressing low levels of YFP-TRF1 can be used to study the dynamic behavior of telomeres in living cells, in line with previous reports (Smith and de Lange, 1997; Mattern et al., 2004).
High TRF1 levels induce TRF1 bridges and mitotic bypass
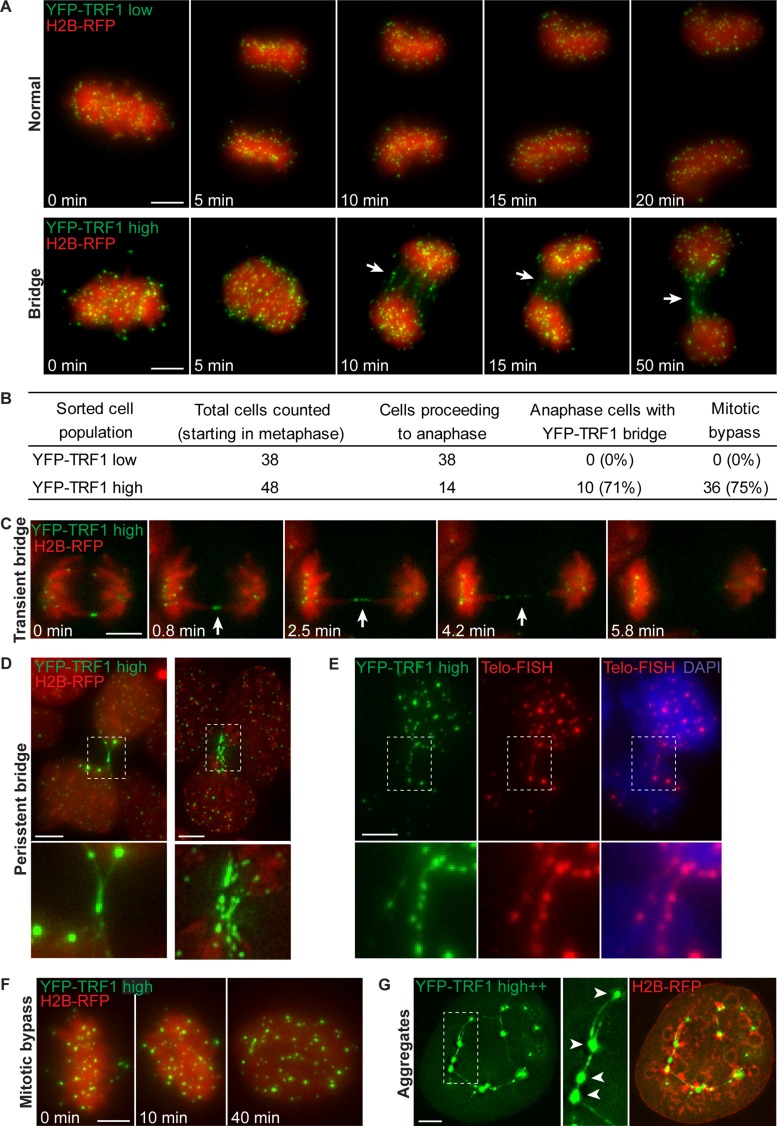
We used live-cell imaging to follow cells with high and high++ YFP-TRF1 levels through mitosis, starting in metaphase. To visualize chromosomes, the cells also stably expressed histone 2B fused to mCherry red fluorescent protein (H2B-RFP; Shaner et al., 2004). Of the cells that entered anaphase from the high YFP-TRF1 population, the majority (71%) exhibited chromatin bridges composed of several (∼1–15) thin fibers of YFP-TRF1 connecting the chromosomes of segregating daughter cells (Figure 2, A and B, and Supplemental Movie S2).
FIGURE 2:
TRF1 overexpression induces anaphase bridges containing TRF1, mitotic bypass, and TRF1 aggregates. (A) Time-lapse images of cells expressing low or high YFP-TRF1 (green) levels and H2B-RFP (chromosomes; red). Note the formation of YFP-TRF1 bridges (arrows) between segregating chromosomes, which persist into interphase (bottom). Images are maximum-intensity projections (Supplemental Movies S1 and S2). (B) Quantification of YFP-TRF1 bridges and mitotic bypass from two independent experiments. Percentages are given in brackets. (C) Time-lapse images showing transient YFP-TRF1 bridge (arrowheads). Images are of a single z-section (Supplemental Movie S3). (D) Examples of persistent TRF1 bridges between daughter cells in interphase. (E) YFP-TRF1 (green) bridges overlap with telomere FISH probe labeling telomeric DNA (red). Note that images of YFP-TRF1 in fixed cells were acquired before telomere FISH, and images were juxtaposed from previously recorded slide coordinates. (F) Time-lapse images showing mitotic bypass. Note that this cell appears to progress from metaphase directly to interphase without undergoing cell division. (G) Example of an interphase cell expressing high++ YFP-TRF1 exhibiting several large, intense aggregates of YFP-TRF1 foci (arrowheads) connected by thin stretches of YFP-TRF1. Scale bar, 5 μm.
We observed two types of bridges—transient and persistent. Transient bridges were composed of thin fibers of YFP-TRF1, which appeared to gradually lengthen and become thinner over time until the bridge was no longer visible, suggesting that bridges may be resolved during mitosis (Figure 2C and Supplemental Movie S3). Transient anaphase bridges almost always severed within the telomere region before nuclear membrane reformation and then rapidly retracted toward opposing poles. This suggests that physical tension rather than cytokinesis is the primary mechanism of transient anaphase bridge resolution. Persistent bridges did not resolve during mitosis and remained in interphase of the following cell cycle (characterized as a TRF1 bridge connecting two cells with decondensed interphase chromatin; Figure 2, A and D). Persistent bridges were often composed of multiple TRF1 fibers with high amounts of YFP-TRF1 within the bridge. These results suggest that the severity of the bridge determines the outcome of mitosis; minor bridges may be resolved during mitosis, whereas more-severe bridges persist for the duration of mitosis.
TRF1 bridges were also induced by TRF1 alone, translated separately from YFP (Supplemental Figure S2), suggesting that the bridges observed in YFP-TRF1–overexpressing cells are not a dominant-negative effect of the fluorescent protein fusion to TRF1. Because TRF1 proteins bind telomere repeats (Broccoli et al., 1997), we reasoned that the TRF1 bridges likely contain telomeric DNA. To confirm this, we performed fluorescence in situ hybridization (FISH) using a telomere probe to label telomeric DNA within the chromatin bridges. YFP-TRF1 bridges overlapped closely with signals from the telomere FISH probe (Figure 2E), showing that chromatin bridges are composed of YFP-TRF1 protein and tracts of telomeric DNA.
To determine whether telomere associations undergo DNA repair, we assayed for colocalization of TRF1 bridges with regulator of telomere length 1 (RTEL1). RTEL1 is required for telomere maintenance (Ding et al., 2004; Vannier et al., 2012) and suppresses homologous recombination during DNA repair (Barber et al., 2008; Uringa et al., 2011, 2012). We observed that RTEL1-YFP formed distinct foci that specifically colocalized to the extremities of persistent TRF1 bridges (Supplemental Figure S3). Of the cells exhibiting persistent RFP-TRF1 bridges, 5–30% contain RTEL1-YFP foci that colocalize with the persistent bridges (n = 30). Note that RTEL1-YFP photobleached rapidly during image acquisition. However, RTEL1 did not colocalize with the majority of transient telomere bridges or telomere aggregates. In addition, we did not observe a clear correlation between TRF1 aggregate size and RTEL1 colocalization. This is consistent with our previous study showing that, at any given time, the majority of RTEL1 and TRF1 foci do not colocalize (Uringa et al., 2012).
To determine how elevated TRF1 levels affects cell cycle progression, we followed cells expressing high YFP-TRF1 by live-cell imaging. We observed that 75% of cells that started in metaphase bypassed mitosis and failed to divide to form separate nuclei (mitotic bypass). These cells instead proceeded to form a single (tetraploid) nucleus in interphase (Figure 2, B and F). By contrast, none of the cells in the low–YFP-TRF1 population underwent mitotic bypass. Of note, daughter cells in interphase that remained linked by persistent YFP-TRF1 bridges (Figure 2, A and D) were not included as having bypassed mitosis; however, these cells were occasionally observed to fuse and then undergo mitosis, giving rise to tetraploid cells that often underwent apoptosis (unpublished data). This is consistent with studies showing that persistent telomere dysfunction in p53-deficient cells leads to tetraploidization and mitotic bypass (Davoli et al., 2010).
High TRF1 levels induce telomere associations
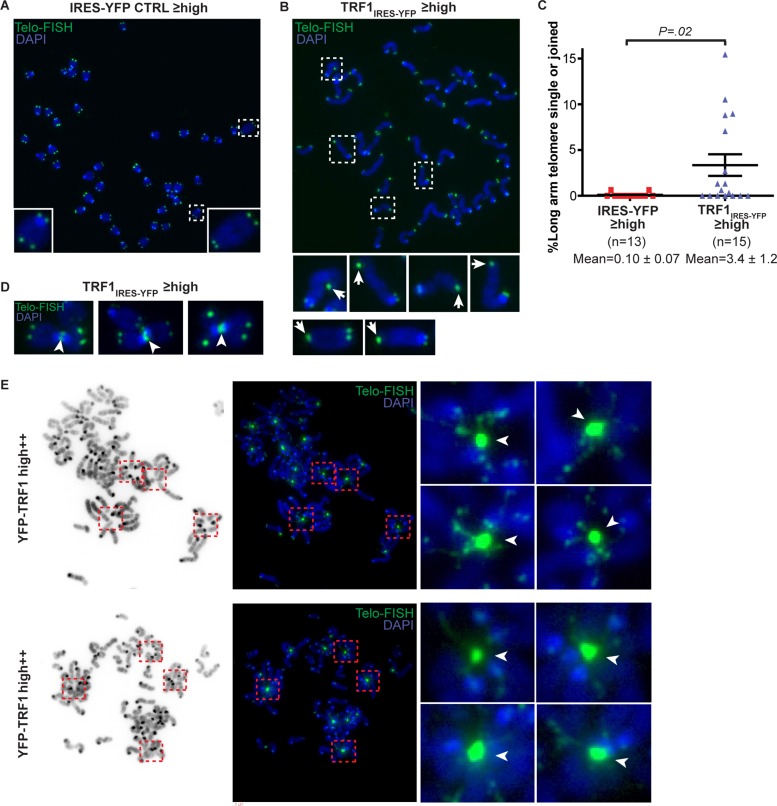
In cells expressing high++ levels of YFP-TRF1, we observed interphase cells with large aggregates of YFP-TRF1 connected by thin stretches of YFP-TRF1 (Figure 2G), suggesting that telomere associations may occur between multiple chromosomes. To test this hypothesis, we prepared metaphase chromosome spreads of cells overexpressing TRF1IRES-YFP, YFP-TRF1, and an IRES-YFP control. To increase the amount of cells used for metaphase spreads, we set FACS gates for sorting to include cells with high or greater YFP levels. We labeled telomeres using a telomere-specific FISH probe. Normally, sister telomeres are resolved into distinct foci. We observed single or joined sister telomeres at the long arm in up to 15% of telomeres in TRF1IRES-YFP-overexpressing cells (Figure 3, B and C). Of note, we observed these telomere associations despite using hypotonic treatment in our preparation of metaphase chromosome spreads, which may release some but not all proteins from chromosomes (Ohnuki, 1968; Dynek and Smith, 2004). In IRES-YFP control cells, single and joined telomeres were less frequent (<1%; Figure 3A). Occasionally, we observed telomere associations between telomeres from different chromosomes in TRF1IRES-YFP-overexpressing cells (Figure 3D). This was more pronounced in YFP-TRF1–overexpressing cells, in which large, intense aggregates of telomere foci were observed, indicating that telomeres from different chromosomes were joined (Figure 3E). Each telomere aggregate was surrounded by multiple (up to seven) radially distributed chromosomes. Multiple telomeric signals characteristic of fragile telomere phenotypes were also observed (Sfeir et al., 2009). The chromosomes appeared thinner immediately adjacent to telomere aggregates, suggesting that the chromosomes are physically stretched from the attachment point at the aggregate. These results suggest that TRF1 overexpression can induce two forms of telomere associations—single or joined sister telomeres—as well as telomere aggregates between multiple chromosomes.
FIGURE 3:
TRF1 overexpression induces telomere associations. Examples of metaphase chromosome spreads in mouse ES cells transiently expressing high or greater YFP levels for (A) vector control (IRES-YFP) or (B) TRF1IRES-YFP at ∼72 h posttransfection. Telomere DNA was labeled by a telomere FISH probe conjugated to Cy5 (green), and DNA was stained with DAPI (blue). (C) Column scatter plot showing percentage of single or joined telomeres at long arm. The p value was calculated using an unpaired two-tailed t test. Mean ± SEM is indicated. (D) Examples of telomere associations between metaphase chromosomes (arrowheads) in TRF1IRES-YFP overexpressing cells and (E) YFP-TRF1 overexpressing cells at ∼72 h posttransfection. Inverted DAPI images (E, left). Insets show telomere aggregates for which each telomere aggregate is surrounded by multiple (up to seven) radially distributed chromosomes. Gamma was set to 0.5 for images in E to visualize lower-intensity signals.
Development of TRF1 cleavage assay in mammalian cells
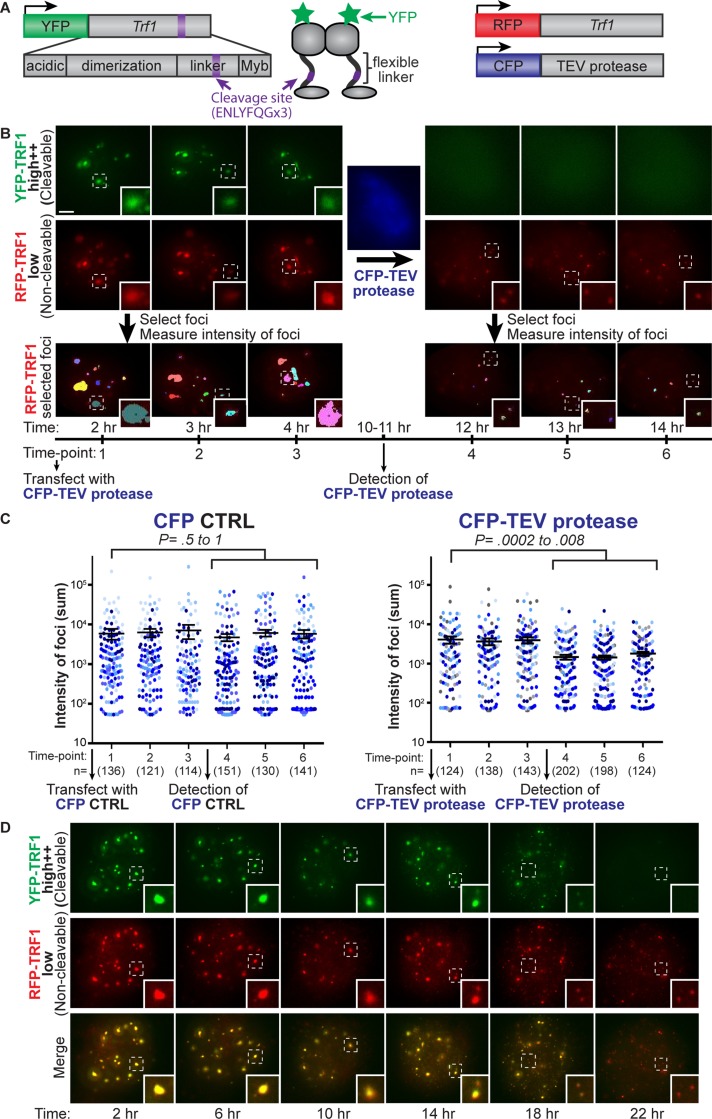
Because we observed that TRF1 protein levels must be tightly regulated for telomere resolution, we reasoned that telomere associations could result from TRF1 protein physically bridging telomeres together. To test this hypothesis, we used tobacco etch virus (TEV) protease to rapidly cleave TRF1 protein and followed the progression of telomere resolution by live-cell imaging. If telomere associations are mediated by TRF1, then cleavage of TRF1 protein should resolve telomere aggregates. To make TRF1 protein cleavable, we inserted a TEV protease recognition site in the flexible linker of YFP-TRF1 (YFP-TRF1TEV; Figure 4A). Because the flexible linker is poorly conserved, we reasoned that the TEV protease recognition site should not interfere with cleavable TRF1 binding to telomeres before induction of cleavage by TEV protease. To enable inducible expression of cleavable TRF1, we also fused YFP-TRF1TEV to an FK506-binding protein (FKBP) degradation domain, which can be stabilized by Shield-1 (Banaszynski et al., 2006). However, this system was not strong enough to induce the TRF1 levels required for telomere aggregation. Therefore we generated a stable clone expressing low levels of FKBP-YFP-TRF1TEV and transiently transfected these cells with FKBP-YFP-TRF1TEV in the presence of Shield-1 to overexpress and stabilize the cleavable FKBP-YFP-TRF1TEV protein. Note that a second transfection with FKBP-YFP-TRF1TEV was required, most likely because not enough of the overexpressed FKBP-YFP-TRF1TEV was stabilized by Shield-1 to induce telomere aggregates in the stable clone. Cells were then sorted for high++ levels of FKBP-YFP-TRF1TEV (FACS gating was the same as in Figure 1B, high++ gate) to be imaged at 24–48 h posttransfection and ∼3–10 h postsorting. TRF1 cleavage was induced by transient transfection with TEV protease fused to cerulean cyan fluorescent protein (CFP; Rizzo et al., 2004), driven by a cytomegalovirus early enhancer/chicken β-actin (CAG) promoter or a phosphoglycerate kinase (PGK) promoter as specified (Figure 4A). To facilitate nuclear entry of TEV protease, TEV protease was also flanked by nuclear localization signals as previously reported (Pauli et al., 2008). To visualize telomeres before and after detection of CFP-TEV protease by live-cell imaging, cells stably expressed low levels of noncleavable RFP-TRF1 (Figure 4A). Because RFP-TRF1 was expressed at levels comparable to endogenous TRF1 (unpublished data), we expected that RFP-TRF1 could be used as a faithful indicator of telomere distribution and would not induce telomere aggregates by itself.
FIGURE 4:
Resolution of telomere aggregates by TRF1 cleavage. (A) Schematic of constructs for TRF1 cleavage assay: cleavable FKBP-YFP-TRF1TEV showing TRF1 domains with cleavage site (purple) inserted in flexible linker, noncleavable RFP-TRF1 to visualize telomeres, and CFP-TEV protease driven by a CAG promoter (or a PGK promoter in D). (B) Projection images of cells expressing high levels of cleavable FKBP-YFP-TRF1TEV (green) and low levels of noncleavable RFP-TRF1 (red) for three time points before and after detection of CFP-TEV protease (blue). RFP-TRF1 foci selected using automatic thresholding of fluorescence intensity (bottom), with each color representing one selected focus. (C) Column scatter plot showing average intensity of foci (sum) on a log scale for three time points before and after detection of CFP-TEV protease or CFP control, where each dot represents an individual selected focus, and each color represents an individual cell. The number of selected foci (n) for each time point is indicated in brackets. Time-lapse movies of nine and seven cells are represented for CFP-TEV protease and a CFP control, respectively (legend). The p values for T = 1 compared with T = 4–6 was calculated using an unpaired two-tailed t test. Error bars, SEM. (D) Projection images from a time-lapse movie of cells expressing high levels of cleavable FKBP-YFP-TRF1TEV (green) and low levels of noncleavable RFP-TRF1 (red) upon transfection with PGK-CFP-TEV protease, showing time after transfection. Note that FKBP-YFP-TRF1TEV fluorescence becomes diffuse (indicating that cleavage has occurred), and noncleavable RFP-TRF1 aggregates become smaller and more uniform (insets; see Supplemental Movie S4). Scale bar, 5 μm.
To visualize TRF1 cleavage in real time, we transiently transfected cells stably expressing low levels of noncleavable RFP-TRF1 (red) and high++ levels of cleavable FKBP-YFP-TRF1TEV (green) with CFP-TEV protease (blue) or a CFP control, followed by live-cell fluorescence imaging. Before detection of CFP-TEV protease, large aggregates of TRF1 foci were observed as overlapping signals in both the YFP and RFP fluorescence channels, indicating that both the cleavable (green) and noncleavable (red) TRF1 fusion proteins colocalize (Figure 4B, time points 1–3). The detection of nuclear CFP (blue) fluorescence indicated successful expression and nuclear entry of CFP-TEV protease (Figure 4B). If TRF1 cleavage was successful, we expected that the cleaved N-terminal portion of FKBP-YFP-TRF1TEV would dissociate from telomeres (Figure 5B). Indeed, we observed a distinct change in YFP localization from distinct foci to diffuse fluorescence, which occurred simultaneously with detection of CFP-TEV protease (Figure 4B). This suggests that both the detection of CFP-TEV protease and diffuse YFP localization can serve as timing indicators of TRF1 cleavage. Therefore we developed a novel assay that combines site-specific protein cleavage of TRF1 by TEV protease with live-cell fluorescence imaging in a mammalian system.
FIGURE 5:
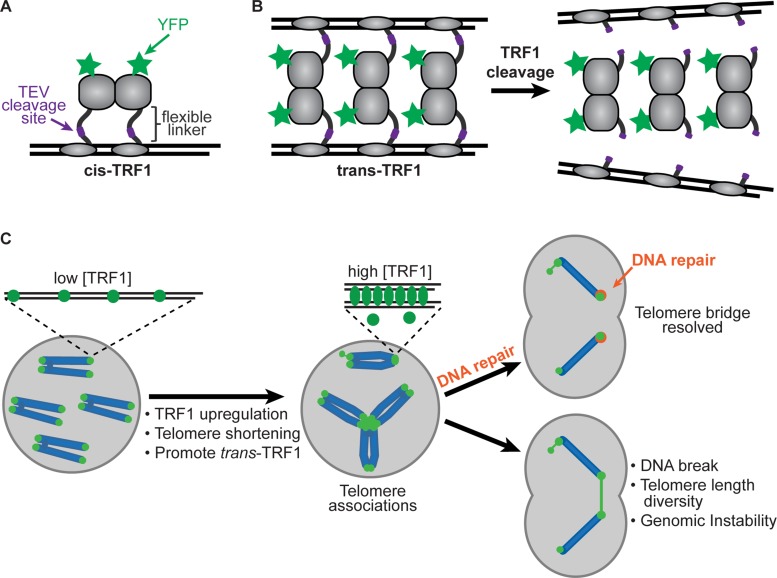
Model of TRF1-mediated telomere associations. (A) Cis-TRF1 conformation showing YFP on N-terminus and TEV cleavage site inserted in flexible linker domain. (B) Trans-TRF1 conformation may promote associations between telomeres under conditions in which telomere-binding sites are saturated with high levels of TRF1. (C) Model showing TRF1-mediated telomere associations in a cellular context and potential consequences. Telomere associations may occur by increase in TRF1 levels, decrease in telomere length (assuming TRF1 levels remain relatively constant), or under conditions that promote the trans-TRF1 conformation. Two main types of telomere associations may form: between sister telomeres and between telomeres from different chromosomes. Persistent telomere bridges may recruit DNA repair proteins, such as the helicase RTEL1, for resolution and repair. Telomere bridges may lead to DNA breakage within chromosomes or telomeres, and broken ends may continue to fuse and break in subsequent cell cycles, giving rise to genomic instability.
Resolution of telomere aggregates by TRF1 cleavage
Because RFP-TRF1 indicates telomere distribution within the cells, this enabled us to monitor telomere aggregates before and after detection of CFP-TEV protease. Images were acquired for three time points in individual cells before and after detection of CFP-TEV protease (Figure 4B). RFP-TRF1 foci were selected using automatic selection of threshold fluorescence intensity, and the intensity (sum) of each focus was measured (Figure 4B, bottom). After detection of CFP-TEV protease, the RFP-TRF1 foci were smaller and more uniform, indicating that telomere aggregates were resolved (Figure 4B, insets, and Figure 4C, right). We also observed an increase in the number of foci after TRF1 cleavage; however, this increase was difficult to measure above background fluorescence by automatic selection of threshold fluorescence (Figure 4B). In contrast, cells transfected with a CFP control did not show a significant change in the intensity (Figure 4C, left) or number of foci. In cells overexpressing YFP-TRF1 without the cleavage site, large telomere aggregates remained upon detection of CFP-TEV protease, suggesting that TEV protease cleavage is specific to TRF1 at the inserted cleavage site, as opposed to nonspecific sites. Of note, cells often underwent mitosis upon detection of CFP-TEV protease, suggesting that upon TRF1 cleavage, cells were released from a mitotic block and resumed cell cycle progression (unpublished data).
To determine how lower expression of TEV protease would affect telomere resolution, we generated cells in which CFP-TEV protease was driven by a lower-expressing PGK promoter. The change in fluorescence of FKBP-YFP-TRF1TEV from distinct foci to diffuse fluorescence served as the indicator of TRF1 cleavage. We observed that the FKBP-YFP-TRF1TEV fluorescence became gradually diffuse at a slower rate with the PGK-CFP-TEV protease than with the CAG-CFP-TEV protease. This corresponded closely with a dramatic increase in the number of noncleavable RFP-TRF1 foci and a decrease in focal intensity; however, foci numbers were difficult to measure above background fluorescence by automatic thresholding (Figure 4D and Supplemental Movie S4). Taken together, these results suggest that telomere aggregates are mediated by TRF1 and cleavage of TRF1 dimers resolves telomere associations and allows cell cycle progression.
Telomere aggregation before telomere resolution affects cell cycle progression and survival
Previous studies showed that truncated TRF1 containing the Myb domain but not the dimerization domain does not bind TTAGGG repeats (Bianchi et al., 1997). On the basis of this study, we expected that removal of TRF1 and associated shelterin components from the telomere upon TRF1 cleavage could lead to telomere dysfunction/deprotection and induction of apoptosis. After the induction of telomere aggregation by overexpressing FKBP-YFP-TRF1TEV for 24-48 h, TEV protease was expressed. We found that, on average, 14 h after CFP-TEV protease detection, cleavage of TRF1 leads to cell cycle arrest, followed by apoptosis in >80% of cells (n = 14). In these live-cell imaging experiments, cell cycle arrest was defined as absence of cell division over 48 h, and apoptosis was observed by brightfield imaging as membrane blebbing and cell breakdown. Of note, one cell underwent cell division and did not undergo apoptosis over the 48-h imaging duration.
We next determined whether TEV protease itself or the cleaved TRF1 products could negatively affect cell cycle progression and cell survival. Cells stably expressing noncleavable RFP-TRF1 (to visualize telomere foci) were cotransfected with PGK-CFP-TEV protease and cleavable FKBP-YFP-TRF1TEV and followed by live-cell imaging 24 h after transfection. We found that the telomere phenotype appeared normal in these cells, with no observed telomere bridges or large telomere aggregates. These cells actively divided, and telomere bridges were not observed in anaphase of mitotic cells (n = 10). However, two mitotic cells exhibited discrete lagging RFP-TRF1 foci between the segregating daughter cells in anaphase. Furthermore, we did not observe large telomere aggregates in interphase cells 24 h after transfection, and the number of noncleavable RFP-TRF1 foci was similar to that in an untransfected control (Supplemental Figure S4).
Taken together, our observations suggest that although the assay components themselves appear to be nontoxic, telomere clustering and cleavage can be toxic once telomere clusters are formed.
DISCUSSION
We developed a system for visualization of telomere and chromosome dynamics using multicolor, high-resolution live-cell imaging. Using this system, we examined cells expressing defined levels of TRF1 protein and demonstrated that elevated TRF1 levels (≥10 times endogenous TRF1 levels) induce telomere anaphase bridging, TRF1 aggregates, and mitotic bypass. Our findings provide insight into how high TRF1 levels influence telomere dynamics and telomere associations in rapidly dividing pluripotent stem cells and support the concept that precise regulation of cellular TRF1 levels is essential for telomere resolution and proper mitotic progression.
Analysis of telomeric DNA distribution in metaphase chromosomes revealed two forms of telomere associations—single or joined sister telomeres—as well as telomere aggregates between multiple chromosomes. This is consistent with previous in vitro studies showing that moderate TRF1 concentrations (two monomers per telomere repeat) lead to parallel pairing of DNA probes containing tracts of telomeric DNA, whereas high TRF1 concentrations (greater than five monomers per telomere repeat) lead to aggregates containing many DNA molecules (Griffith et al., 1998), suggesting that the phenotypic outcome of TRF1 overexpression is dependent on the ratio of TRF1 to telomere repeats.
We developed a novel assay that combines site-specific protein cleavage of TRF1 by TEV protease with live-cell fluorescence imaging in a mammalian system. We observed that the cellular localization of fluorescent cleavable TRF1 changed from distinct foci to diffuse fluorescence at the same time as detection of nuclear TEV protease, suggesting that TRF1 was successfully cleaved. We observed that telomere aggregates induced by TRF1 overexpression are resolved upon TEV protease–mediated TRF1 cleavage, suggesting that telomere associations result primarily from TRF1-mediated protein interactions. Our findings demonstrate for the first time that telomere associations are mediated by TRF1 in a mammalian system.
Elevated TRF1 levels have been observed under physiological conditions, such as in pluripotent stem cells (Schneider et al., 2013) and during mitosis (Shen et al., 1997; Nishiyama et al., 2006; Zhu et al., 2009). The phenotypes we observe at exogenous high TRF1 levels may be indicative of TRF1 function under physiological conditions, at which endogenous TRF1 levels may increase to a certain degree, but under precisely regulated conditions. For instance, TRF1-mediated telomere associations could function to hold telomeres in close proximity during meiotic telomere bouquet formation to facilitate alignment of chromosomes before homologous recombination (Cooper et al., 1998), as a cohesion mechanism between sister telomeres from replication until mitosis (Azzalin and Lingner, 2004; Dynek and Smith, 2004), to facilitate telomere recombination and repair, and to create localized areas for limiting telomere maintenance factors such as telomerase.
TRF1-mediated telomere associations could also play key roles under pathological conditions, such as carcinogenesis. It will be important to determine whether there is a link between telomere aggregation observed in human cancer cells (Chuang et al., 2004; Goldberg-Bittman et al., 2008; Gadji et al., 2012) and the elevated TRF1 levels observed in such cells (Matsutani et al., 2001; Oh et al., 2005).
We find that embryonic mouse ES cells, which lack a functional p53 checkpoint (Aladjem et al., 1998), progress through mitosis in the presence of unresolved telomeres, giving rise to anaphase bridges at telomeres and mitotic bypass. In contrast, 3T3 mouse fibroblasts undergo mitotic arrest upon TRF1 overexpression (unpublished data), consistent with previous studies showing mitotic arrest in the presence of unresolved telomeres in HeLa cells (Dynek and Smith, 2004). It will be important to determine how cell type, checkpoint control, and differentiation affect cell cycle progression in the presence of unresolved telomeres.
A potential mechanism for TRF1-mediated telomere associations could be that as telomeres shorten (by aging or disease), the relative concentration of TRF1 per telomere repeat increases (assuming that TRF1 levels remain relatively constant), leading to TRF1-mediated telomere associations (Figure 5C). Normal human cells display persistent telomere cohesion and anaphase delay before senescence, which can be overcome by overexpression of tankyrase, a major regulator of TRF1 levels (Kim and Smith, 2014). Imbalance between number of telomere repeats and TRF1 levels could result in relative overexpression of TRF1, resulting in the telomere associations that have been described. Flexible TRF1 dimers could mediate telomere associations by binding telomeres in a trans conformation, with the DNA-binding domain of each monomer binding opposing telomeres (Figure 5B), in contrast to a cis conformation, with binding to adjacent sites on the same telomere (Figure 5A). This is in line with previous in vitro studies showing that TRF1 protein alone induces parallel pairing and looping of telomeric DNA tracts when the ratio of telomere binding sites is limiting compared with TRF1 levels (Griffith et al., 1998; Bianchi et al., 1999). Our findings show that elevated levels of TRF1 induce telomere associations in cells, which manifest as telomere bridges and mitotic bypass (Figure 5C). In our studies, the telomere binding sites were likely limiting relative to TRF1 protein levels, supporting the hypothesis that competition for binding sites results in TRF1 dimers binding in a trans conformation.
Telomeres initially held in close proximity by a TRF1-mediated protein interaction may eventually progress to interactions between telomeric DNA. DNA-mediated interactions can arise via processes such as recombination, telomere fusion, stalled telomere replication, or DNA catenation. Although we occasionally observed metaphase chromosomes positioned end to end, the predominant phenotype in TRF1-overexpressing cells was sister telomere associations and telomere aggregates surrounded by multiple radially distributed chromosomes. Large telomere aggregates formed almost immediately after TRF1 overexpression, indicating that the primary outcome of elevated TRF1 levels is telomere associations as opposed to stalled telomere replication, which would be expected to lead primarily to sister telomere associations. Furthermore, since each telomere aggregate involves multiple (up to seven) chromosomes, it is unlikely that this is a result of end-to-end covalent fusions, which would be expected to lead to a linear positioning of chromosomes (van Steensel et al., 1998; Karlseder et al., 1999) in which each fusion point involved a maximum of two chromosomes linked end to end.
It is feasible that the close proximity of clustered telomeres facilitates recombination events such as telomere strand exchange by invasion of the 3′ single-stranded telomere overhang in-trans. RTEL1 may be recruited to these sites to reverse inappropriate strand invasion, in line with previous studies showing that RTEL1 dissociates displacement loops (D loops) formed at the site of strand invasion (Barber et al., 2008; Vannier et al., 2012). We observed that RTEL1 forms distinct foci that specifically localize to the extremities of persistent bridges. It is possible that TRF1 bridges that persisted into the following cell cycle are more likely to have undergone recombination events that require recruitment of DNA repair proteins such as RTEL1. We previously showed that RTEL1 is recruited to distinct foci at sites of DNA repair; RTEL1 foci colocalized with almost all 53BP1 and FANCD2 foci upon treatment with DNA-damaging agents (Uringa et al., 2012). Resolution of telomere aggregates may be important for telomere protection by allowing individual telomeres to fold back in a telomere loop (T-loop) by strand invasion in-cis. It is tempting to speculate that the persistent TRF1 bridges flanked by RTEL1 foci observed in this study (Supplemental Figure S3), may be similar to previously described ultrafine bridges flanked by Fanconi anemia complex proteins (Chan et al., 2009). Whereas ultrafine bridges were only rarely observed at telomeres, TRF1 bridges were observed almost exclusively at telomeres.
Telomere associations may lead to a combination of downstream events. Telomere bridges leading to chromatin breaks continue to fuse and break in subsequent cell cycles, giving rise to genomic instability (McClintock, 1941). Anaphase bridges have been reported to lead to chromosome breaks at variable positions (McClintock, 1941), with a preference for breakage at fragile sites—unstable regions of the genome that are prone to gaps, breaks, and genome rearrangements (Durkin and Glover, 2007; Chan et al., 2009). Of note, telomeres have been reported to resemble fragile sites (Sfeir et al., 2009). We speculate that if chromatin bridges predominantly break at or near telomeric DNA, telomere associations could contribute to the marked heterogeneity in telomere length at any given telomere (Lansdorp et al., 1996; Martens et al., 1998; Figure 5C).
Our studies show that telomere associations are mediated by TRF1 and provide insight into the resolution and repair of these associations. Future studies will likely further reveal how this mechanism is mediated, what other factors within and outside the shelterin complex play a role, and how telomere associations affect cellular processes under physiological and pathological conditions.
MATERIALS AND METHODS
Plasmid constructs
Fluorescent TRF1 and H2B constructs were fused to fluorescent proteins as specified in the foregoing text. CAG-IRES-YFP was also fused to the N-terminus of a nuclear membrane–localizing domain, importin α1 fragment, as previously described (Okita et al., 2004). RTEL1 was fused to YFP and an FKBP degradation domain (FKBP12-L106P) as previously described (Banaszynski et al., 2006; Uringa et al., 2012). These cells were grown in the presence of Shield-1 to stabilize RTEL1-YFP and transiently transfected with RFP-TRF1 for ∼72 h. An FKBP degradation domain was fused to the N-terminus of cleavable TRF1 (FKBP-YFP-TRF1TEV). Expression of all constructs was driven by a CAG promoter, except for RTEL1-YFP, driven by a PGK promoter, and CFP-TEV protease, driven by a CAG or a PGK promoter as specified.
Cell culture, transfection, and FACS analysis
Mouse ES cell lines (Mus musculus, 129 strain) were grown under standard culture conditions on 0.1% gelatin-coated dishes in the presence of leukemia inhibitory factor, as described previously (Gertsenstein et al., 2002). Transfection was performed using Effectene transfection reagent (Qiagen, Valencia, CA). Cell populations expressing defined TRF1 levels were obtained by transfecting cells with YFP-TRF1, TRF1IRES-YFP, or FKBP-YFP-TRF1TEV followed by FACS (BD/Cytopeia Influx; BD Biosciences, San Jose, CA). FACS data were analyzed using FlowJo software (Treestar, Ashland, OR). The generation of RTEL1-deficient cell lines was previously described (Ding et al., 2004).
Preparation of whole-cell extracts and Western blotting
Whole-cell extracts were prepared of cells transiently expressing TRF1IRES-YFP or YFP-TRF1 and sorted for low, high, or high++ YFP levels grown to ∼80% confluence. At 48 h posttransfection, cells were harvested by trypsinization, incubated in NuPAGE LDS-reducing sample buffer (Invitrogen, Carlsbad, CA), and stored at −20°C until use. Samples were heated to 70°C for 10 min before separation by SDS–PAGE (Invitrogen). After transfer to a nitrocellulose membrane (Bio-Rad, Hercules, CA), membrane strips were blocked (5% nonfat dry milk and 0.1% Tween 20 in phosphate-buffered saline [PBS-MT]) for 1 h and incubated overnight with rabbit anti-mouse TRF1 (1:500 in PBS-MT; a kind gift from Y. Shinkai), anti–green fluorescent protein (1:50,000 in PBS-MT; Ab290; Abcam, Cambridge, UK), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Research Diagnostics, Cleveland, OH) loading control. Proteins were detected by chemiluminescence (SuperSignal West Pico; Pierce, Thermo Scientific, Rockford, IL).
Immunostaining and TRF1 foci count
A cell clone stably expressing low levels of YFP-TRF1 was generated, and these cells were fixed in suspension with 4% formaldehyde in PBS for 20 min, permeabilized in 0.2% NP-40 in PBS, blocked with 1% bovine serum albumin, incubated with rabbit anti–phospho-histone H3 (Ser-10) (06-570; Upstate, Millipore, Billerica, MA) conjugated to goat anti-rabbit Cy5 (111-175-003; Jackson ImmunoResearch, West Grove, PA), and stained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were sorted into G1-, S-, and G2/M-based DAPI DNA content, and an anti–phospho-histone H3 (Ser-10; Supplemental Figure S1) using the BD/Cytopeia cell sorter. Image stacks of 20 whole cells were acquired from each sorted cell population (z-section spacing, 0.2 μm). Image analysis was performed using Volocity (Improvision, Perkin Elmer, Coventry, UK) to select TRF1 foci.
Live-cell imaging
Cells were grown in No. 1.5 LabTek II chambered coverglasses (Nalge NUNC, Rochester, NY). To adhere gelatin to the glass, the glass was coated with 0.1% gelatin, air dried, fixed with 4% paraformaldehyde for 2 h, and rinsed thoroughly with PBS. Cells grown using this method had normal morphology compared with standard gelatin-coated plastic dishes. Live-cell imaging was performed using a DeltaVision RT system (Applied Precision, Seattle, WA) with an Olympus IX inverted microscope and a CoolSnap HQ charge-coupled device camera (Roper Scientific, Tucson, AZ). Excitation and emission filters used were as follows: CFP, 436/10 and 465/30 nm; YFP, 492/18 and 535/30 nm; and RFP, 580/20 and 630/60 nm. Images were acquired using a 60× PlanApo 1.4 numerical aperture oil objective (Olympus). An environmental chamber was used at 37°C with 5% CO2 perfusion. Image analysis was performed using SoftWoRx Suite (Applied Precision) and Volocity.
TRF1 bridges and mitotic bypass
Cells were imaged starting in metaphase with image stacks (15 images; z-section spacing, 1 μm) acquired at 5-min intervals. Exposure was 0.2 s, and neutral density filters (transmission 10%) were used to minimize photobleaching. To obtain populations of cells with high and high++ TRF1 levels, cells stably expressing low YFP-TRF1 levels were transfected a second time with YFP-TRF1 and sorted by FACS, followed by imaging at ∼20–30 h posttransfection and ∼3–10 h postsorting.
Metaphase spread preparation and telomere FISH
Sorted cells overexpressing IRES-YFP, TRF1IRES-YFP, or YFP-TRF1 were treated with Colcemid (0.1 μg/ml) for 4 h before trypsin harvest. After washing and hypotonic swelling (75 mM KCl) for 3–5 min at 37°C, cells were fixed twice in methanol–acetic acid (3:1) at room temperature for 15 min, spread onto a precleaned slide, and air-dried overnight. Telomere FISH was previously described (Lansdorp et al., 1996) using a Cy5-conjugated (C3TA2)3 peptide nucleic acid probe (Boston Probes, Applied Biosystems, Grand Island, NY). Images were acquired on an Axioplan 2 microscope (Carl Zeiss, Jena, Germany) with Isis 5 software (Metasystems) using an AxioCam MRm camera (Carl Zeiss).
TRF1 cleavage assay
The same automatic thresholding of fluorescence intensity was used for all time points. Time points were selected at which the entire cell was in focus and ranged from 1 to 10 h before and after detection of CFP-TEV protease. TEV protease was fused to nuclear localization signals at the N- and C-termini (CFP-NLS-TEV protease-NLS-NLS), to facilitate nuclear entry of TEV protease (Pauli et al., 2008). The TEV protease recognition site ENLYFQGx3 was inserted in the flexible hinge region of mTrf1 using similar methods to Pauli et al. (2008). An HpaI site was introduced into TRF1 (amino acid position 338) by site-directed mutagenesis using the oligonucleotide 5′-GAAACAACGATGGAAGTTAACCGAAGAACC-3′. The TEV protease recognition site ENLYFQGx3 was then inserted at the HpaI site using the oligonucleotide 5′-PHOSPHATE-AACGCTCTAGAGAATTTGTATTTTCAGGGTGCTTCTGAAAACCTTTACTTCCAAGGAGAGCTCGAAAATCTTTATTTCCAGGGAGTT-3′.
Supplementary Material
Acknowledgments
We thank Ester Falconer and Geraldine Aubert for critical comments on the manuscript, Yoichi Shinkai for anti-mouse TRF1 antibody, Timm Schroeder for providing YFP and CAG-IRES-YFP plasmids, Roger Y. Tsien for providing the RFP plasmid, and Mike Schertzer for generating fluorescent TRF1 and H2B fusion plasmids. This work is funded by Terry Fox Foundation PPG Grant 18006.
Abbreviations used:
- CAG
chicken β-actin
- CFP
cyan fluorescent protein
- ES
embryonic stem
- FACS
fluorescence-activated cell sorting
- FKBP
FK506-binding protein
- IRES
internal ribosome entry site
- PGK
phosphoglycerate kinase
- RFP
red fluorescent protein
- RTEL1
regulator of telomere length 1
- TEV
tobacco etch virus
- TRF1
telomeric repeat-binding factor 1
- YFP
Venus yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-10-0564) on May 14, 2014.
REFERENCES
- Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J. Cell biology. Telomere wedding ends in divorce. Science. 2004;304:60–62. doi: 10.1126/science.1096809. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- Chuang TC, et al. The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol. 2004;2:12. doi: 10.1186/1741-7007-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Ding H, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- Gadji M, et al. Profiling three-dimensional nuclear telomeric architecture of myelodysplastic syndromes and acute myeloid leukemia defines patient subgroups. Clin Cancer Res. 2012;18:3293–3304. doi: 10.1158/1078-0432.CCR-12-0087. [DOI] [PubMed] [Google Scholar]
- Gertsenstein M, Lobe C, Nagy A. ES cell-mediated conditional transgenesis. Methods Mol Biol. 2002;185:285–307. doi: 10.1385/1-59259-241-4:285. [DOI] [PubMed] [Google Scholar]
- Goldberg-Bittman L, Kitay-Cohen Y, Quitt M, Hadary R, Fejgin MD, Yukla M, Amiel A. Telomere aggregates in non-Hodgkin lymphoma patients at different disease stages. Cancer Genet Cytogenet. 2008;184:105–108. doi: 10.1016/j.cancergencyto.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Griffith J, Bianchi A, de Lange T. TRF1 promotes parallel pairing of telomeric tracts in vitro. J Mol Biol. 1998;278:79–88. doi: 10.1006/jmbi.1998.1686. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Kachatrian L, Takai H, Mercer K, Hingorani S, Jacks T, de Lange T. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol Cell Biol. 2003;23:6533–6541. doi: 10.1128/MCB.23.18.6533-6541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Smith S. Persistent telomere cohesion triggers a prolonged anaphase. Mol Biol Cell. 2014;25:30–40. doi: 10.1091/mbc.E13-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Han S, You YH, Chen DJ, Campisi J. The human telomere-associated protein TIN2 stimulates interactions between telomeric DNA tracts in vitro. EMBO Rep. 2003;4:685–691. doi: 10.1038/sj.embor.embor872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, Raap AK, Tanke HJ. Heterogeneity in telomere length of human chromosomes. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- Lin X, Gu J, Lu C, Spitz MR, Wu X. Expression of telomere-associated genes as prognostic markers for overall survival in patients with non-small cell lung cancer. Clin Cancer Res. 2006;12:5720–5725. doi: 10.1158/1078-0432.CCR-05-2809. [DOI] [PubMed] [Google Scholar]
- Martens UM, Zijlmans JM, Poon SS, Dragowska W, Yui J, Chavez EA, Ward RK, Lansdorp PM. Short telomeres on human chromosome 17p. Nat Genet. 1998;18:76–80. doi: 10.1038/ng0198-018. [DOI] [PubMed] [Google Scholar]
- Matsutani N, Yokozaki H, Tahara E, Tahara H, Kuniyasu H, Haruma K, Chayama K, Yasui W. Expression of telomeric repeat binding factor 1 and 2 and TRF1-interacting nuclear protein 2 in human gastric carcinomas. Int J Oncol. 2001;19:507–512. [PubMed] [Google Scholar]
- Mattern KA, Swiggers SJ, Nigg AL, Lowenberg B, Houtsmuller AB, Zijlmans JM. Dynamics of protein binding to telomeres in living cells: implications for telomere structure and function. Mol Cell Biol. 2004;24:5587–5594. doi: 10.1128/MCB.24.12.5587-5594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- Miyachi K, Fujita M, Tanaka N, Sasaki K, Sunagawa M. Correlation between telomerase activity and telomeric-repeat binding factors in gastric cancer. J Exp Clin Cancer Res. 2002;21:269–275. [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Muraki K, Saito M, Ohsumi K, Kishimoto T, Ishikawa F. Cell-cycle-dependent Xenopus TRF1 recruitment to telomere chromatin regulated by Polo-like kinase. EMBO J. 2006;25:575–584. doi: 10.1038/sj.emboj.7600964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh BK, Kim YJ, Park C, Park YN. Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am J Pathol. 2005;166:73–80. doi: 10.1016/S0002-9440(10)62233-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuki Y. Structure of chromosomes. I. Morphological studies of the spiral structure of human somatic chromosomes. Chromosoma. 1968;25:402–428. doi: 10.1007/BF02327721. [DOI] [PubMed] [Google Scholar]
- Okita C, Sato M, Schroeder T. Generation of optimized yellow and red fluorescent proteins with distinct subcellular localization. BioTechniques. 2004;36:418–422. doi: 10.2144/04363ST01. 424. [DOI] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Schneider RP, Garrobo I, Foronda M, Palacios JA, Marion RM, Flores I, Ortega S, Blasco MA. TRF1 is a stem cell marker and is essential for the generation of induced pluripotent stem cells. Nat Commun. 2013;4:1946. doi: 10.1038/ncomms2946. [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shen M, Haggblom C, Vogt M, Hunter T, Lu KP. Characterization and cell cycle regulation of the related human telomeric proteins Pin2 and TRF1 suggest a role in mitosis. Proc Natl Acad Sci USA. 1997;94:13618–13623. doi: 10.1073/pnas.94.25.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, de Lange T. TRF1, a mammalian telomeric protein. Trends Genet. 1997;13:21–26. doi: 10.1016/s0168-9525(96)10052-4. [DOI] [PubMed] [Google Scholar]
- Uringa EJ, Lisaingo K, Pickett HA, Brind'amour J, Rohde JH, Zelensky A, Essers J, Lansdorp PM. RTEL1 contributes to DNA replication, repair and telomere maintenance. Mol Biol Cell. 2012;23:2782–2792. doi: 10.1091/mbc.E12-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uringa EJ, Youds JL, Lisaingo K, Lansdorp PM, Boulton SJ. RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 2011;39:1647–1655. doi: 10.1093/nar/gkq1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 Dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Yamada K, Yagihashi A, Yamada M, Asanuma K, Moriai R, Kobayashi D, Tsuji N, Watanabe N. Decreased gene expression for telomeric-repeat binding factors and TIN2 in malignant hematopoietic cells. Anticancer Res. 2002a;22:1315–1320. [PubMed] [Google Scholar]
- Yamada M, Tsuji N, Nakamura M, Moriai R, Kobayashi D, Yagihashi A, Watanabe N. Down-regulation of TRF1, TRF2 and TIN2 genes is important to maintain telomeric DNA for gastric cancers. Anticancer Res. 2002b;22:3303–3307. [PubMed] [Google Scholar]
- Zhong Z, Shiue L, Kaplan S, de Lange T. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol Cell Biol. 1992;12:4834–4843. doi: 10.1128/mcb.12.11.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Meng L, Hsu JK, Lin T, Teishima J, Tsai RY. GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J Cell Biol. 2009;185:827–839. doi: 10.1083/jcb.200812121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.